Abstract
E Unus pluribum, or “Of One, Many”, may be at the root of decoding the RNA sequence-structure-function relationship. RNAs embody the large majority of genes in higher eukaryotes and fold in a sequence-directed fashion into three-dimensional structures that perform functions conserved across all cellular life forms, ranging from regulating to executing gene expression. While it is the most important determinant of RNA structure, the nucleotide sequence is generally not sufficient to specify a unique set of secondary and tertiary interactions due to the highly frustrated nature of RNA folding. This frustration results in folding heterogeneity, a common phenomenon wherein a chemically homogeneous population of RNA molecules folds into multiple stable structures. Often, these alternative conformations constitute misfolds, lacking the biological activity of the natively folded RNA. Intriguingly, a number of RNAs have recently been described as capable of adopting multiple distinct conformations that all perform, or contribute to, the same function. Characteristically, these conformations interconvert slowly on the experimental timescale, suggesting that they should be regarded as distinct native states. We discuss how rugged folding free energy landscapes give rise to multiple native states in the Tetrahymena Group I intron ribozyme, hairpin ribozyme, sarcin-ricin loop, ribosome, and an in vitro selected aptamer. We further describe the varying degrees to which folding heterogeneity impacts function in these RNAs, and compare and contrast this impact with that of heterogeneities found in protein folding. Embracing that one sequence can give rise to multiple native folds, we hypothesize that this phenomenon imparts adaptive advantages on any functionally evolving RNA quasispecies.
1. Introduction
The discovery three decades ago that certain RNA molecules, termed ribozymes, catalyze chemical reactions in a manner similar to protein enzymes demonstrated an unexpected level of functional versatility of RNA that may have spawned life in the form of an RNA world.1–3 Accordingly, over the last decade a large number of non-protein coding RNAs (ncRNAs) have been discovered that play essential roles in all aspects of modern life.4–7 These roles include regulation of gene expression,8,9 post-transcriptional RNA processing,2,3,10 protein biosynthesis,11 and essential genomic processing in pathogens.12–16 It was also discovered that some ribozyme motifs are broadly distributed among a wide set of organismal genomes.17–20 Moreover, in vitro selection has generated ribozymes with additional activities such as aminoacyl-RNA synthesis,21 self-replication,22 and organic synthesis,23 all functions postulated to have played a pivotal role in the RNA world. Finally, a rapidly increasing number of crystal structures has shed light onto the impressive complexity of the underlying RNA structures.24–26 Clearly, RNA has the capacity to assume a wide variety of functions based on the ability of its sequence to encode versatile three-dimensional structures, yet our understanding of the RNA sequence-structure-function relationship is still in its infancy.
Self-cleaving ribozymes are ideal model systems for the study of sequence-structure-function relationships in ncRNA, since their activity (and hence, proper folding) can be quickly and easily assayed.24,25,27–30 These ribozymes catalyze a site-specific transesterification of the phosphate-ribose backbone resulting in the formation of two cleavage products: a 5′ product bearing a 2′–3′ cyclic phosphate, and a 3′ product bearing a 5′-OH group. The activity of these ribozymes depends on the presence of metal cations – especially divalents such as Mg2+ – that stabilize the active tertiary structure of an RNA and may also confer a direct chemical rate enhancement.31 Additionally, the relatively small size of most self-cleaving ribozymes allows for convenient in vitro transcription of large amounts of sample for biochemical assays and chemical synthesis to incorporate site-specific modifications and labels for chemogenetic and biophysical studies.
Perhaps due to the ease of relating folding to activity, studies of ribozymes have revealed a peculiar propensity of RNA to adopt alternate conformations, a phenomenon often described as conformational heterogeneity. While the first reversible misfolding of RNA was reported in leucyl-tRNA (tRNALeu),32,33 the prevalence of alternate kinetically stable structures in RNA became more apparent with studies of several self-cleaving ribozymes. In most cases, such conformational heterogeneity was attributed to “misfolded” (inactive or less active) ribozymes or long-lived folding intermediates.34–37 In fact, virtually all ribozymes, as well as many other RNAs, are prone to this type of conformational heterogeneity, leading to a persistent view in the field that alternative folding is a nuisance to be avoided.38,39
A particularly intriguing example of folding heterogeneity was recently characterized in the Tetrahymena group I intron (TG1I) ribozyme (Fig. 1).40 Upon binding of substrate to its 5′-end, the TG1I ribozyme forms a helix termed P1 that subsequently swings by ~6 nm to dock the substrate into the preformed active site and form the active complex (Fig. 1A). Using single-molecule fluorescence resonance energy transfer (smFRET) measurements and cleavage activity assays, the authors provided evidence that subpopulations of the ribozyme exhibiting widely variable (>800-fold) docking equilibrium constants are, surprisingly, all catalytically active. In fact, 94% of all molecules within these different populations maintain the same rate constant of catalysis (Figs. 1E,F). While only a small fraction of molecules spontaneously switches between subpopulations on an experimentally accessible time scale (i.e., the heterogeneity is relatively static), molecules can be induced to redistribute among subpopulations by refolding through the removal and subsequent reintroduction of Mg2+ ions (Figs. 1B,C,D).40 This finding strongly suggests that several active, or “native”, states arise from conformational differences rather than changes in chemistry or local environment.
Fig. 1.
Single molecule observations of the TG1I ribozyme reveal folding heterogeneity that manifests in multiple native states.40 (A) Structural representation143 of TG1I ribozyme docking as monitored by smFRET experiments. The sequence of helix P1, composed of the 5′-end of the ribozyme and the substrate RNA strand, is shown with donor fluorophore (D) attached to the 3′-end of the substrate strand. The TG1I ribozyme contains a 3′-extension that is hybridized to a DNA oligonucleotide (grey) with a 3′-acceptor fluorophore and 5′-biotin for surface immobilization and 3′-acceptor. (B) Schematic representation of the deeply furrowed folding landscape of the TG1I ribozyme. Molecules (red, blue, and green) redistribute between three docking conformations after partial denaturation by removal (with EDTA) and addition of Mg2+. (C, D) The three energy wells manifest as single molecule distributions of vastly different docking free energy (ΔGdock), between which molecules (color-coded) redistribute from before (C) to after (D) denaturation. (E) The overall distribution of docking behaviors was binned into five color-coded categories. (F) Cleavage assays of molecules representing these five docking categories (color-coded) were all shown to display kinetics similar to the global average (solid line). In part adapted with permission from ref.40.
This work on the TG1I ribozyme provides a strong impetus to revisit questions about the origin and possible biological function of folding heterogeneity in RNA. In fact, evidence of very similar behavior has been accruing for a number of functional RNAs over the past decade. In the following we will discuss how the physical properties of RNA give rise to a propensity for heterogeneous folding. Providing further examples, we will show that such folding behavior is commonplace, and in some cases clearly contributes to RNA function. Finally, we will speculate as to the significance of this behavior in the context of molecular adaptability and evolution.
2. The free energy landscape of RNA folding is rugged and frustrated
Among biopolymers, RNA possesses a number of characteristics that make its folding behavior unique. First, the multitude of dihedral angles in the phosphate-ribose backbone of RNA results in an immense range of possible topologies (or folds) for even relatively short RNAs. Second, the relative dominance of only a few types of base pairing interactions (Watson-Crick A·U and G·C, as well as common G·U wobble pairs) results in a “frustrated” folding landscape with a large number of nearly degenerate secondary structures. Third, the ability of RNA to form highly stable duplexes, cooperatively reinforced by a large number of hydrogen bonds and base-stacking interactions, gives its folding landscape a deeply furrowed character, resulting in alternate secondary and tertiary structures that may take very long to interconvert once formed.41 Together, these factors give rise to a high propensity to form kinetically trapped alternate folds. The ribose-phosphate backbone contains six rotatable dihedral angles per nucleotide (conventionally labeled α-ζ, Fig. 2A), or even more when the constrained torsional angles of the sugar ring (ν0-ν4, Fig. 2A) and the glycosidic bond dihedral (χ, Fig. 2A) are considered. This number compares to only two such angles per amino acid in proteins, resulting in a much wider range of possible conformations in RNA than in peptides of comparable size. While Watson-Crick base pairing in standard A-form helical stems places strict restraints on the possible torsional angles, single-stranded regions of RNA molecules – formally junctions, loops, and bulges (Fig. 2B) – still have a multitude of possible conformations. As a result, the folding topology of an RNA can be rendered quite complex through the formation of multiple helical junctions and tertiary interactions such as pseudoknots, ribose zippers, kissing-loop interactions, and tetraloop-receptor interactions (Fig. 2C).42–47
Fig. 2.
Diversity of RNA structures. (A) RNA has many dihedral angles (blue) that contribute to a large number of conformational degrees of freedom per nucleotide. These include angles of torsion about the bonds of the RNA backbone (α-ζ), the nucleosidic bond (χ), and pseudo-rotation angles within the ribose ring (ν0–ν4). (B) Hydrogen bonding (pairing) between complementary bases gives rise to several common secondary structure motifs, consisting of the RNA backbone (blue) held in various arrangements by base pairs (black line segments) and often resulting in short segments of unpaired nucleotides (short grey lines). (C) Common tertiary structure motifs build hierarchically onto secondary structure elements. (D) The small alphabet of RNA often results in a frustrated folding landscape with many possible conformations stabilized by alternative base-pairing. In this schematic, three segments of a linear RNA molecule are marked in blue, black, and red to illustrate the variety of possible secondary (2°) and tertiary (3°) structures arising from a single primary (1°) sequence of nucleotides.
Several advances have been made towards classifying combinations of dihedral angles into structural motifs based on mono- or dinucleotide units, reducing the number of empirically observed conformations considerably. In one approach, backbone dihedral angles were organized into so-called “suites”, where a conformation is defined between adjacent sugar residues as a group of two sets of angles: δ-ε-ζ and α-β-γ-δ.48 Along with careful quality-based filtering of crystallographic source data, this system enables the classification of empirically observed RNA backbone conformations into 42 suites, later expanded to 46 conformers in an effort that unified a handful of other approaches.49 Still, the number of conformations available to even a short oligonucleotide would be staggering in the absence of other information, posing a serious obstacle to decoding the relationship between sequence and structure.
Fortunately, the complexity of RNA folding is reduced considerably by its hierarchical nature wherein secondary structure typically folds before tertiary structure (Fig. 2D).39,50 Hybridization of complementary segments of an RNA sequence occurs in as little as microseconds.28,51 Once formed, an A-form RNA helix is stabilized by base pairing and base stacking interactions worth about −1 to −3 kcal/mol per base pair.41,50 As a result, even short RNA oligomers of ~10 base pairs (bp) may have half-lives of dissociation on the order of minutes or hours near room temperature.52,53 On the other hand, tertiary interactions (loop-loop hydrogen bonding, base-phosphate and base-sugar interactions) frequently form and interconvert on the timescale of milliseconds to seconds and are comparatively weak.54–56 Because of this difference in kinetics and stability, secondary structure places relatively rigid constraints on accessible tertiary structures; conversely, isolated stem-loops of a larger RNA structure often fold properly even in absence of tertiary interactions.41 While it simplifies the prediction of RNA structure, the stability of secondary structure also gives rise to a deeply furrowed free energy landscape of folding with large barriers separating different folded states.
Finally, the folding free energy landscape of RNA is described as “frustrated” by a large number of possible alternative folds (Fig. 2D). This description derives from the tendency of RNA to become trapped in local minima and require great energy to overcome these barriers and attain “native” structure. Due to the diminutive 4-nucleobase alphabet of RNA, there is a high probability that any two sequences of nucleotides will have coincidentally complementary regions. Even random RNA sequences containing tens of nucleotides are predicted to be approximately 50% base-paired,57 implying the existence of numerous possible alternative secondary structures. Modern software packages use partition function approaches to predict RNA secondary structure, yielding more reliable predictions of base pairing and thermodynamic parameters than considering only the minimum-energy structure.58–61 Clearly, the number and impact of possible alternate secondary structures are significant.
3. Multiple active species with slow interconversion are observed in a number of RNAs
Due to the complex and rugged folding free energy landscape of RNA, long-lived variations in structure, or so-called static heterogeneities, arise in RNAs as diverse as those found in plant virus satellites, the eukaryotic ribosome, and artificial selections of ligand-binding aptamers.40,54,62–64 Misfolding is perhaps a trivial example, but it is important to consider that one function’s trash may be another function’s treasure. An example is the adoption of different secondary structures for the purpose of switching between active and inactive forms of a ribozyme, such as at different stages in the replication cycle of a pathogen. For instance, the hepatitis delta virus (HDV) ribozyme must be active in order to cleave concatemeric linear transcripts of HDV RNA into unit-length fragments for ligation into circular copies of the genome. Once this task is accomplished, however, the ribozyme becomes inactive by adopting alternate base pairing patterns as part of a long, 70% self-complementary, rod-like structure of the RNA genome of HDV, presumably for the purpose of packaging and delivery of the intact genome to new host cells.16
Here, we focus on another type of static heterogeneity: the presence of distinct species that all contribute, in varying degrees, to the same nominal function. In investigating such phenomena, several questions must be addressed:
What impact does the structural heterogeneity have on function?
How robust is the heterogeneity to changes in experimental conditions?
Does the heterogeneity arise from purely conformational differences, i.e., can covalent modification or mutation be ruled out?
If the differences are conformational, do they represent alternate secondary structures or more subtle variations in tertiary structure? Studies of heterogeneity in model systems have begun to answer these questions.
Such studies often involve a combination of ensemble and single-molecule methods, utilizing the relative strengths of each approach to deduce the nature of the heterogeneity40,63–65 Ensemble assays of RNA are rapid, well established for many systems, and can employ a variety of detection methods, the two most common of which are fluorescence and autoradiography. While requiring relatively large amounts of sample material, ensemble assays often require minimal modification and processing of sample material. Single-molecule fluorescence based assays generally require precise fluorescent labeling, specialized equipment, and in-depth statistical analysis, but work with low amounts of sample. Additionally, single-molecule assays allow correlations to be drawn between conformational behavior and catalytic efficiency without physical separation. Only through utilizing these techniques in conjunction can the nature of these heterogeneities in RNA be addressed, as illustrated in the following with arguably the most prominent examples.
3.1 The hairpin ribozyme
One of the most thoroughly characterized examples of static heterogeneity in an RNA is found in the hairpin ribozyme, a self-cleaving and self-ligating small ribozyme first discovered in the negative strand of the tobacco ringspot virus (TRSV) satellite RNA.66,67 Central to catalytic activity of the ribozyme is the docking of its two helix-loop-helix domains (A and B) by tertiary interactions between the nucleotides in bulges present in each helix to form the active site. At room temperature, in the presence of Mg2+ ions, this docking is readily reversible (Fig. 3A). Despite its much smaller size, the docking of a helix (domain A) into the catalytic core (domain B) of the hairpin ribozyme, enabled by a flexible hinge region, superficially resembles docking of helix P1 into the TG1I ribozyme (Compare Figs. 1A and 2A).
Fig. 3.
Folding heterogeneity of the hairpin ribozyme.55,65,69 (A) smFRET assays detect docking heterogeneity of the hairpin ribozyme. Donor (D) and acceptor (A) fluorophores are attached to the 3′- and 5′-ends of the RzA strand, respectively, while the RzB strand carries a 5′-biotin for surface immobilization. Whereas all active HpRz molecules display a single docking rate constant (kdock), four distinct undocking rate constants (kundock,1–4) are observed. (B) Each docking rate constant leads to a distinct cleavage time course (numbered as in panel A), which taken together account for the overall cleavage observed in ensemble assays (1+2+3+4). (C) Reaction pathway of the hairpin ribozyme and resulting single-molecule multiple-turnover cleavage data. The docked, undocked and product released states are indicated. Three single molecule time trajectories demonstrate catalytic proficiency of each of the distinct subpopulations they represent (undocking rate constants and the fraction of molecules undocking with this rate constant are indicated). (D) Structural heterogeneity upon EMSA in which two structural forms of the hairpin ribozyme are resolved (Top and Bottom species). Separation of the two component strands RzA and RzB of each species by denaturing polyacrylamide gel electrophoresis (D-PAGE) yields four RNAs, as indicated, that were annealed in all possible combinations and analyzed by EMSA. The resulting fractions of Top and Bottom species are given. (E) High-resolution FT-ICR mass spectrometry of each of the four RNA strands isolated by the procedure described in panel D reveals identical isotope envelopes (insets) and average masses (consistent with the predicted masses) of the corresponding strands from the Top and Bottom species. In part adapted with permission from refs. 65,69.
Utilizing smFRET assays to quantify the kinetics of these docking and undocking transitions, the Walter and Chu groups first uncovered kinetic heterogeneities in the hairpin ribozyme in 2002.55 Only by utilizing smFRET could the behaviors of individual molecules be observed without the ensemble averaging inherent to bulk assays. For smFRET detection, the opposite ends of one strand of the RNA were labeled with the fluorophores Cy3 (smFRET donor) and Cy5 (acceptor), which exhibit high FRET efficiency when proximal to each other in the docked state and low FRET efficiency in the undocked state (Fig. 3A). Samples were immobilized on a derivatized quartz slide, and Cy3 and Cy5 emission intensities of single molecules were observed by total internal reflection fluorescence (TIRF) microscopy.55,68 Although the rate constant of docking was invariant across all molecules, subpopulations of ribozyme molecules were observed to undock with four distinct rate constants spanning three orders of magnitude. Furthermore, individual molecules rarely (<5% of observed molecules) switched between these kinetic regimes even over 3 hours incubation – each molecule retained a “memory” of its undocking rate constant. Strikingly, the multiple undocking populations were observed to all convert to product from the docked state; i.e., the faster the undocking relative to the cleavage/ligation rate constants, the more docking/undocking cycles become necessary on average for a successful substrate turnover (Figs. 3B and 3C). Thus, the existence of distinctly undocking, yet catalytically active, native states quantitatively explains the multi-exponential cleavage kinetics noted in ensemble assays of the hairpin ribozyme (Figs. 3A and 3B).55,69
Importantly, it was shown that this heterogeneity is not caused by the surface immobilization used in smFRET measurements. First, bulk cleavage assays of ribozymes with the same modifications (Cy3, Cy5, and biotin) yield similar kinetic parameters to those obtained by smFRET techniques.55,69,70 Second, the same kinetic heterogeneity was observed by smFRET when the Lilley and Ha groups captured ribozymes on the slide surface by encapsulation within phospholipid vesicles, rather than through direct biotin-streptavidin interaction.71
Surprisingly, the level of undocking heterogeneity proved not to be affected by either changes in Mg2+ concentration72 or site-specific mutations or modifications with impact on the undocking rate constants; the rate constant of each subpopulation simply shifted by about the same factor.69 In subsequent work, evidence was presented that the molecular heterogeneities of the hairpin ribozyme, while extremely long-lived, are not due to any detectable covalent modification.65 More specifically, through the use of electrophoretic mobility shift assays (EMSAs) on polyacrylamide gels, two distinct, either slow- or fast-migrating species of the ribozyme were resolved (as top and bottom bands, respectively). Upon elution from the gel and further analysis of the fluorophore distance distributions by time-resolved FRET (trFRET), the two species showed marked differences; while less than 40% of the slow-migrating species adopts the docked conformation, greater than 80% of the fast-migrating species does. Analysis by smFRET showed that the top band is enriched in the fastest undocking subpopulation, which is expected to be less docked and compact, consistent with its lower mobility, whereas the bottom band is enriched in the most slowly undocking subpopulation, expected to reside largely in the compact docked conformation.65
This differential enrichment within two separable species opened up an opportunity to determine whether the hairpin ribozyme subpopulations can be induced to redistribute. To this end, the individual 5′- and 3′-segment strands (termed RzA and RzB) of each species were further separated by denaturing gel electrophoresis, removing all base pairing between them. When the RzA and RzB strands of the slow-migrating species were re-annealed and re-analyzed by EMSA, the RNA redistributed into fast- and slow-migrating species as before, but not the RzA and RzB strands originating from the fast-migrating species, which continued to preferentially form the fast-migrating species (Fig. 3D). Accordingly, mixing RzA and RzB strands originating from different species yields intermediate levels of the two EMSA bands (Fig. 3D). This asymmetry suggests that the molecular heterogeneity leading to formation of the catalytically more active fast-migrating species is maintained even upon full denaturation of all interstrand base pairs.65
In the same work, we were also able to show by high-resolution (<1 amu) mass spectrometry that the RzA and RzB strands from the fast- and slow-migrating strands are identical in mass (Fig. 3E).65 In addition, both chemically synthesized and in vitro transcribed ribozyme behave similarly, and footprinting revealed only minor differences in secondary structure between the EMSA separated species. While these observations do not rule out mass-neutral covalent modifications as the source of conformational heterogeneity, such as certain UV-induced crosslinks, they do strongly support the notion that more subtle conformational73 or topological differences40 are at play.
In summary, the hairpin ribozyme folds into multiple active populations with disparate global dynamics but only subtle differences in secondary structure. Compared to the multiple native states of the TG1I ribozyme many parallels are observed, although the native states of the hairpin ribozyme are separated by larger free energy barriers and, consequently, do not interconvert quite as freely upon refolding as those of the TG1I ribozyme (Fig. 4).
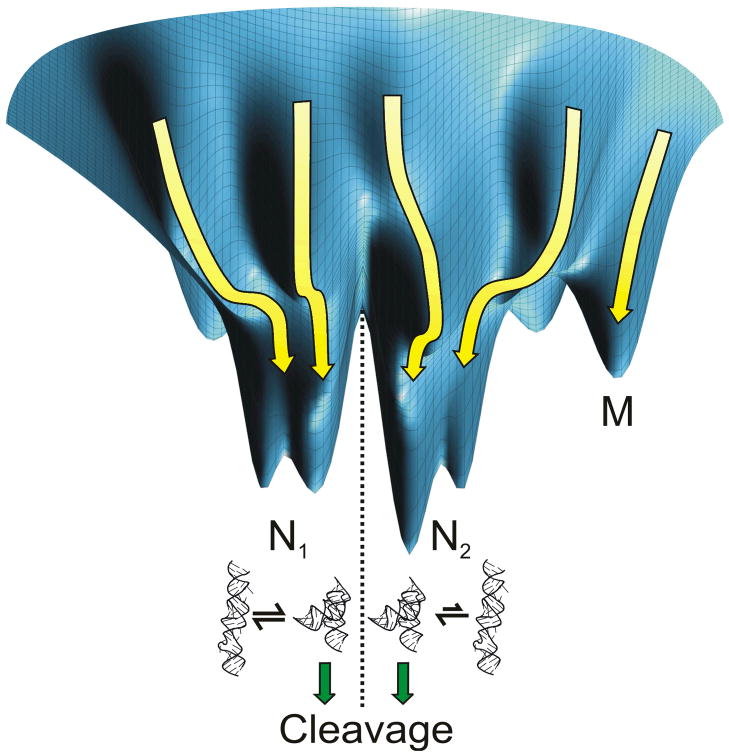
Fig. 4.
Schematic representation of the rugged conformational free energy landscape (blue surface) of the hairpin ribozyme. An individual molecule folds along one of many possible pathways (yellow arrows) to one of multiple native states (N1, N2) separated by relatively large energy barriers. These native states sample similar conformations, albeit with different kinetics, and thus possess similar cleavage activity. Alternatively, the molecule may enter a trapped misfolded state (M) that is non-functional.
3.2 The sarcin-ricin loop
Domain B of the hairpin ribozyme shares sequence homology and a version of an RNA structural motif termed an S-turn with the toxin-sensitive sarcin-ricin loop (SRL) of the large subunit ribosomal RNA (rRNA, Fig. 5A).65 Intriguingly, conformational heterogeneity has also been observed in this stem-loop motif that is highly conserved across all kingdoms.62 Several 27-nt versions of the sarcin-ricin loop (SRL) from rat were transcribed in vitro and analyzed by EMSA, revealing two species of identical length but with different electrophoretic mobilities (Fig. 5B), similar to our observations on the hairpin ribozyme. No heat-induced interconversion of the two species was observed, suggesting that the heterogeneity is thermodynamically quite stable. Intriguingly, the slow-migrating species (constituting 30–50% of the total SRL) is resistant to cleavage by the endoribonuclease restrictocin (a sarcin analog) when compared to the native substrate (70–50% of total SRL, Fig. 5B).62 As with the hairpin ribozyme, chemically synthesized SRL shows the same heterogeneity. While it is unknown whether this phenomenon persists in vivo, or how it might affect translation by the ribosome, i.e., which of the species represents a native state, it is tempting to speculate that the presence of multiple folds could confer an adaptive advantage through partial resistance to different toxins. Conformational heterogeneity may thus be akin to sequence variation of rRNA; in E. coli alone there are seven different rRNA sequences, each with minor sequence discrepancies that may confer increased adaptability to the cell.
Fig. 5.
Folding heterogeneity of the sarcin-ricin loop (SRL). (A) Comparative cartoon representations of crystal structures of loop B of the docked hairpin ribozyme and the SRL reveal a common, conserved S-turn motif (blue).65 (B) In vitro transcribed SRL was either purified by denaturing (DPAGE) or non-denaturing polyacrylamide gel electrophoresis (NPAGE or EMSA), subjected to restrictocin cleavage, and the products over time analyzed by gel electrophoresis, as indicated.62 EMSA in particular reveals a non-interconvertable, slow-migrating S* species that is relatively resistant to restrictocin cleavage. Adapted with permission from refs. 62,65.
3.3 The ribosome
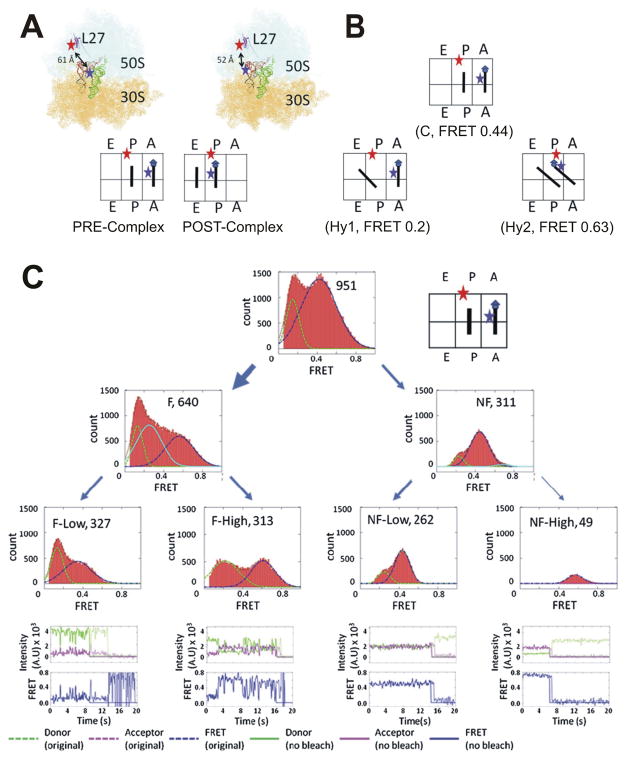
Given that a small stem-loop of the ribosome exhibits profound folding heterogeneity, it comes as no surprise that the bacterial ribosome, at ~2.4 MDa and with three RNA and >50 protein components the largest of all ribozymes, has recently been shown to display heterogenous intersubunit rotational dynamics in its pre-translocation complex (Fig. 6). Pre-translocation complexes (pre-complexes) occur when the acetyl(A)-site and peptidyl(P)-sites of the ribosome are occupied by tRNAs that have undergone a peptidyl transfer but have yet to translocate to the P- and E-sites, respectively. As shown in previous ensemble and single-molecule FRET based studies,74–77 the small (30S) and large (50S) subunits of the ribosome spontaneously ratchet among the classic pre-complex (with the two tRNAs occupying the A/A-P/P sites in the 30S/50S subunits) and two hybrid conformations (Hy1: A/A-P/E, Hy2: A/P-P/E). Subsequently, elongation factor-G catalyzes translocation to the post-translocation complex (P/P-E/E). Heterogeneity arises in that only ~2/3 of all ribosomal complexes observed by smFRET display dynamic fluctuation between the classic and hybrid states (Fig. 6).64,76 Of these, a roughly equal distribution of complexes exhibit transitions either between the Hy1and the classic state or the Hy1 and the Hy2 state. The remaining ~1/3 of static complexes are distributed among the low FRET classic and hybrid states with a high prevalence for the classic state, along with a small number of high FRET molecules occupying the POST state (Fig. 6C).64 The authors propose a qualitative folding free energy landscape of translocation with high energy barriers preventing reverse translocation and smaller minima/maxima for the fluctuating and nonfluctuating FRET states, downwardly trending towards the Hy2 state that is adopted just before translocation.64
Fig. 6.
Folding heterogeneity of the bacterial ribosome.64 (A) Structural and schematic representation of the fluorophore labeled E. coli ribosome in the pretranslocation (PRE) and posttranslocation (POST) complexes. Fluorophores attached to the L27 protein of the large subunit and A-site tRNA are used to characterize by smFRET their relative motions as the ribosome samples multiple PRE-complex conformations. (B) Ribosomes are observed in conformations including the classic state, C, along with two hybrid states, Hy1 and Hy2, with tRNA occupying various sites in the large and small subunit, each with a distinct FRET value, as indicated. (C) Ribosomes can be categorized by their dynamic (or fluctuating, F) or static (nonfluctuating, NF) occupancy of these conformations. Among the F molecules, roughly half exhibit low (0.2) to mid (0.44) FRET transitions while the other half exhibit low to high (0.63) transitions. Among the NF molecules, three categories emerge, where two categories with the low and mid FRET values of the C and Hy1 states, respectively, were described as a joint NF-Low category, whereas a high FRET category was assigned to POST complexes. Representative smFRET time trajectories accompany each of these four categories. Adapted with permission from ref. 64.
3.4 The AN58 aptamer
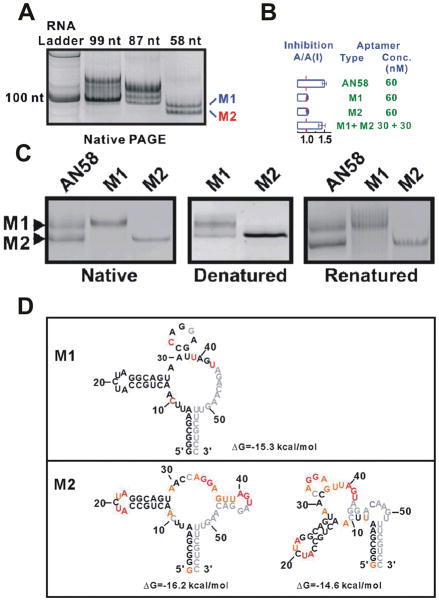
So far, we have discussed the static heterogeneities observed in naturally occurring RNAs, yet there exists at least one artificially selected RNA aptamer that shows similar behavior. The AN58 RNA is a truncation construct of the larger aptGluR2-99 aptamer, in vitro selected to block the GluR2 AMPA receptor by binding to regions thought to be essential to function.78 When AN58 was transcribed in vitro and purified by gel electrophoresis, two different, non-interconvertable populations (M1 and M2) were observed by EMSA (Fig. 7). Neither of these isoforms can individually inhibit the GluR2 AMPA receptor, yet when recombined, they show inhibition comparable to that of unseparated AN58 and very similar to that of the parent aptGluR2-99 (Fig. 7B). Sequencing by primer extension and glyoxal treatment followed by gel electrophoretic analysis showed that M1 and M2 have the same length and sequence. To elucidate any differences in secondary structure between M1 and M2, a combination of in-line probing and selective 2′-hydroxyl acetylation analyzed by primer extension (SHAPE) was utilized. Results from these footprinting assays suggested that M1 and M2 form short stem-loop structures in alternate regions of the unstructured 3′-region of the RNA (Fig. 7D).63 Thus, in contrast to the hairpin ribozyme, in this case alternate, very stable secondary structures appear to be responsible for the heterogeneous behavior. Yet AN58 maintains its static heterogeneity in a manner similar to both hairpin ribozyme and SRL (Fig. 7C).63
Fig. 7.
Folding heterogeneity of the AN58 aptamer.63 (A) AN58, a truncation of a larger AN99 aptamer, resolves as two discrete bands during EMSA (Native PAGE), termed M1 and M2, suggesting two different structures of chemically homogenous RNA. (B) While AN58 is capable of blocking the action of the GluR2 AMPA receptor channel, neither purified form (M1 or M2) can inhibit activity on its own, yet a mix of both M1 and M2 results in restoration of AN58 function. (C) M1 and M2 were separated from one another by EMSA and analyzed by EMSA (left panel), denatured by urea-containing polyacrylamide gel electrophoresis and visualized (middle panel, Denatured), then refolded and further analyzed by EMSA (right panel, Renatured), revealing a lack of interconversion between species. (D) Proposed secondary structures of M1 and M2. Adapted with permission from ref. 63.
In summary, while likely only the tip of the proverbial iceberg, the above five examples (TG1I and hairpin ribozymes, SRL, bacterial ribosome, and AN58 aptamer) arguably represent the best characterized occurrences of heterogeneous RNA behaviors. In each case, different copies of what is ostensibly a single chemical species with a defined nucleotide sequence are capable of adopting different conformations of similar or distinguishable native functionality.
4. Parallels with protein folding
Often, insights into RNA structure-function relationships trail those of proteins; it is therefore helpful to take a look at our current understanding of heterogeneities in protein folding. There are many similarities between the folding behavior of RNA and that of proteins. Like RNA, proteins fold hierarchically, with local interactions forming first and largely determining the overall structure of the folded polypeptide.79,80 Both RNA and proteins can adopt a given fold with very few specific sequence requirements.19,81 However, there are some important differences. Protein folding is mainly directed by fairly nonspecific hydrophobic collapse80,82 rather than by the formation of specific hydrogen bonds such as in tightly aligned (stacked) RNA base pairs. Furthermore, the native states of proteins are typically only marginally stabilized by 5–10 kcal/mol relative to their denatured states, which is comparable to the stability of an RNA duplex containing a mere 8–10 base pairs.83,84 In other words, a short RNA stem-loop folds with similar thermodynamic stability as an entire protein. This distinction is reflected by the fact that, whereas RNA secondary structure elements fold very stably in isolation,41 the secondary structure of proteins is often strongly influenced by context (such as tertiary interactions).85 Thus, one might expect to observe less profoundly heterogeneous folding in proteins than in RNA.
Ensemble kinetic experiments on protein folding have frequently observed trapped conformations. Many of these are non-native folding intermediates that generally persist for only seconds or less, as for lysozyme86 and staphylococcal nuclease.87 In other cases, states with native-like activity have been observed. For instance, upon refolding from 8 M urea, the majority of dihydrofolate reductase folds transiently into an intermediate that efficiently binds a substrate analog.88 Similar behavior was observed for RNase A, which has a long-lived folding intermediate possessing enzymatic activity similar to that of the native state in spite of some structural differences.89 In both of these proteins, the native state ultimately forms, even though RNase A takes from one to several minutes to fold to completion.
The advent of single-molecule fluorescence spectroscopy overcame the drawbacks of ensemble averaging and revealed numerous examples of heterogeneous activity and folding of single protein enzymes, showing that this property is the rule rather than the exception. Individual lipase molecules were shown to exhibit fluctuating substrate turnover kinetics, likely explained by conformational changes on the order of tens of milliseconds.90 Single-molecule studies of flavoenzymes, monitored by changes in the intrinsic fluorescence of the flavin cofactor in the course of its redox chemistry or electron transfer to a nearby tyrosine, found fluctuations as slow as 1 s−1 in both the substrate turnover kinetics and conformations of individual enzymes (Figs 8A–C),91,92 as did similar studies of horseradish peroxidase.93 Longer-lived fluctuations in activity, with lifetimes on the order of minutes, were observed for single molecules of bacteriophage λ exonuclease.94 The activity of electrophoretically purified lactate dehydrogenase molecules was found to vary by a factor of four in a manner that remained constant for a given single molecule over two hours, which the authors suggested could be due to different stable arrangements of monomers in the homotetrameric enzyme.95,96 Similar findings of static heterogeneity were made for alkaline phosphatase,97 β-galactosidase,98 and the DNA helicase RecBCD.99,100 Perhaps most intriguingly, recent folding studies of a GFP triple mutant with enhanced fluorescence emission termed GFPmut2 provide evidence of multiple native states, characterized by distinct chromophore switching kinetics, that do not interconvert over several hours unless refolded from the denatured state,101–103 providing strong evidence of long-lived conformational heterogeneity in native proteins (Figs. 8D and 8E).
Fig. 8.
Examples of folding heterogeneity in the proteins cholesterol oxidase91 and green fluorescent protein (GFP).103 (A) Turnover of substrates by an individual molecule of cholesterol oxidase gives rise to stochastic transitions between fluorescent and non-fluorescent states. (B) Although transitions between these two states occur stochastically, dwell times in the fluorescent state for two adjacent turnovers n and n+1 are slightly correlated, as shown by the diagonal feature (encircled by a white ellipse) of a conditional probability distribution. (C) In contrast, dwell times separated by 10 turnovers are not correlated. (D) Single GFPmut2 molecules exhibit spontaneous switching between anionic (fluorescence intensity in cyan) and neutral (grey) states. (E) The frequency K0 of switching events is not uniformly distributed, but occurs in clusters. When unfolding in guanidine hydrochloride, the switching rate KFIN of each molecule in the ~20 ms prior to the unfolding event is highly accelerated (KFIN ~ 440 Hz in red, ~ 720 Hz in green, and ~930 Hz in red) and is well predicted by the pre-unfolding K0 value, as shown by the clustering of similar colors. Although values of K0 are stable over 24 h, individual molecules may enter different parts of the distribution upon cycles of denaturation (inset). Adapted with permission from ref. 91,103.
In summary, like the RNA examples highlighted above, many native proteins exhibit conformational heterogeneity that generally lasts for milliseconds to seconds, but can persist for hours in some proteins. The commonly shorter timescale of most of these protein fluctuations may reflect the less rugged conformational landscape of proteins as compared with RNA, although there are clearly exceptions. As with RNA, the microscopic origin of this heterogeneity in proteins is generally unclear. As an exception, Polakowski et al. showed that at least some long-lived heterogeneity of enzyme activity can be attributed to covalent differences such as partial degradation of the peptide or post-translational modifications such as glycosylation, which persist in crudely purified samples.104 However, at least in the case of the GFP mutant GFPmut2, the differences between native states appear conformational in origin, as the states redistribute upon denaturation as is observed also for the TG1I ribozyme. The discovery of such behavior in both proteins and RNAs, in spite of their distinct biophysical properties, suggests that multiple similar native states may be a general feature of biopolymers of complex structure. The fact that wild-type GFP shows less conformational heterogeneity than GFPmut2 invokes the notion that natural evolution may in some cases select against it.
5. RNA conformational quasispecies may be natural facilitators of molecular evolution
In the context of evolution, genetic diversity within a population of organisms correlates with the fitness of that population.105,106 Such diversity confers upon the population resistance to parasites, toxins, and other environmental insults. Numerous examples for such effects have been observed, ranging from genetic resistance to certain human diseases,107–109 resistance of insects towards pesticides,110 and the appearance of antibiotic-resistant strains of bacteria.111 While a large amount of phenotypic diversity arises from genetic mutations, many other molecular sources of phenotypic variation have been elucidated over the last several decades, including the action of transcription factors and repressors,112 covalent modification of histones and DNA,113, RNA interference,114–116 riboswitches, 117–121 and alternative splicing.122,123 These mechanisms make a great degree of phenotypic diversity possible even in populations of genetically identical cells. In some cases, phenotypic variation can arise stochastically in a population, such as through the translation of low-copy-number mRNAs in cells124,125 or the presence of very low concentrations of transcriptional regulators.112 Thus, stochastic single-molecule events have an important impact on the fate of an entire organism and perhaps the fitness of the entire population of organisms.126
In the case of rapidly replicating systems with relatively high mutation rates, such as viruses or bacteria, molecular evolution is described by the quasispecies model, in which variation and selection occur not on the level of well-defined molecular species with nearly identical genetic makeup, but rather on the level of so-called quasispecies comprising clusters of related sequences replicating according to their aggregate fitness level.127–129 Due to high replication and mutation rates, the fitness of a single genotype, which will not likely be faithfully preserved in the offspring, becomes less important than the overall fitness of the cluster or quasispecies. In fact, the functional diversity of the quasispecies confers enhanced adaptability to dynamic environments, allowing for example viruses to rapidly evolve resistance to vaccines and antiviral drugs.130 To maximally exploit this evolutionary advantage, most viruses and organisms are found to maintain an inverse relationship between their mutational error rate and the length of their genome, i.e., they live close to an error threshold imposed by their genome length.127–129 Consequently, mutation-inducing drugs can cause this error threshold to be crossed, resulting in lethality.131,132
In light of the evidence presented here, the quasispecies model, formulated in the context of genotypic variation, may now need to be extended to conformational variability of RNA with a single sequence. Specifically, given the capability of RNA to form alternative active folds that are stable in vitro relative to the lifetime of RNA in vivo, we suggest that RNA might function and evolve as conformationally distinct, but functionally related conformational quasispecies. In this view, the source of functional variation is not solely provided by sequence, but by the inherent ruggedness and high degree of energetic degeneracy in the RNA folding landscape. In fit RNA quasispecies, then, alternate folding may constitute another level of adaptive phenotypic variation.
What advantages might such heterogeneity confer on quasispecies of RNA? First, it would enable molecules to achieve their function even if one particular conformer becomes a target for a toxin or nuclease. The observation of a conformational species of the sarcin-ricin loop resistant to cleavage by restrictocin62 provides a salient example of how such heterogeneity could confer an immediate advantage, provided that the resistant species is still biologically functional. Such conformational heterogeneity could constitute an attractive mode of adaptation, complementary to sequence variation, when it is necessary to respond to variable environmental challenges on short time scales because larger fractions of an RNA are immediately available with an altered folding behavior than are typically found to carry a specific (set of) mutation(s) leading to such behavior. Conversely, long-term exposure to a toxin or other insult would likely drive preferential selection of sequence variants that thermodynamically or kinetically prefer the formation of the resistant conformer(s).
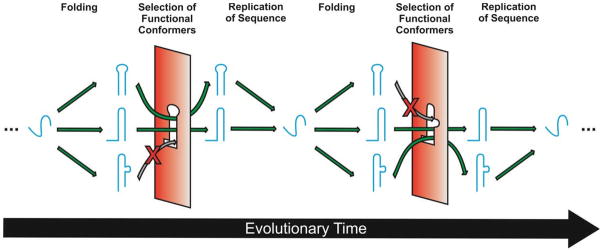
Second, the capability of an RNA of a single sequence to adopt multiple conformations that directly or indirectly act in concert may enable short RNA oligomers to adopt more sophisticated functions, such as found in the AN58 aptamer. While this aptamer was artificially selected in vitro, this type of behavior could be useful in nature due to its sequence economy. Furthermore, Huang et al. suggest that this type of dual-use sequence could provide a precursor to gene duplication and phenotype divergence for functional nucleic acids.63 Previous work, in which a single RNA sequence was designed to encode the folds and activities of both the HDV ribozyme and an RNA ligase ribozyme,133 similarly suggests that intersections in sequence space between neutral networks of distinct functional RNAs may be common, and could give rise to new folds and functions during evolution. In fact, the simplistic single RNA-single function paradigm does not do justice to the complexity of nature, where an RNA will always have to exert multiple functions in parallel. An example is the hairpin ribozyme that, like the HDV ribozyme, needs to cleave concatemeric replication intermediates of its satellite RNA into monomers, then ligate these into circles that function as rolling-circle replication substrates and are devoid of exonuclease-sensitive 5′- and 3′-ends so as to maintain their integrity as substrates.56 That is, catalytic activity is essential (and defines the “native” state) for one part of the replication cycle, but catalytic inactivity is critical (“native”) for another part. The existence of conformational isomers of the hairpin ribozyme with different docking-undocking equilibria may then ensure that some RNA molecules are always optimally performing one function while others optimally perform another function without losing all capacity for the former. We hypothesize that such conformational adaptability endows an RNA quasispecies with enhanced functionality in the face of dynamic evolutionary selection criteria (Fig. 9).
Fig. 9.
Schematic representation of a possible adaptive role for conformational quasispecies of RNA under evolutionary pressure. A single RNA sequence (blue) may fold into several stable conformers, or native states, with varying functionality. Changing environmental conditions may impose certain restrictions (red) on the fitness of conformers, but the success of a subset of these conformers will enable the replication of the sequence and the evolutionary survival of all stable (and kinetically accessible) conformers. If conditions are sufficiently variable, there is a clear survival advantage to maintaining a broad quasispecies of RNA folds and functions.
Of course, essentially all studies demonstrating multiple functional folded states of RNA have been conducted in vitro, and it remains to be seen whether such behaviors will be recapitulated in vivo. The one example of obligate folding heterogeneity was observed for a truncated sequence of an artificially selected aptamer, and observations of multiple native states in the hairpin and TG1I ribozymes were made using in vitro transcribed or chemically synthesized RNA that had been purified at least once by denaturing polyacrylamide gel electrophoresis. In nature, by contrast, RNA folds as it is transcribed from 5′- to 3′-end, which influences folding in important ways. For example, the segmental co-transcriptional folding of circularly permuted variants of the Tetrahymena group I intron was found to yield a higher percentage of natively folded RNA than refolding the entire sequence at once.134 Transcriptional speed and site-specific pausing were found to be important factors in the folding and function of the FMN riboswitch.135 The Varkud satellite ribozyme, shown to exhibit folding heterogeneity by smFRET56 and EMSA, folds into a much narrower range of conformations when purified without denaturation or refolding after transcription.136 A bioinformatic study found evidence that sequences of natural transcripts are selected for features that promote co-transcriptional folding into the correct native secondary structure.137 Interestingly, while the hairpin ribozyme was found to fold sequentially under kinetic control during in vitro transcription, the relative thermodynamic stability of competing helices was a larger determinant of folding in yeast cells,138 though kinetic traps can persist in vivo if they are sufficiently stable.139 The greater preference for thermodynamically stable structures in vivo could be due to RNA chaperones and other RNA-binding proteins in the cell39 that may serve to re-equilibrate kinetically trapped species via ATP-driven helicase activity or nonspecific stabilization of unfolded intermediates. In the case of CYT-19, an ATP-dependent DEAD-box helicase, there even appears to be some preference for unwinding duplexes within misfolded TG1IRz molecules, perhaps based on compactness of tertiary structure alone.140 Another DEAD-box helicase, Mss116, has been shown to stimulate the folding of a group II intron into its near-native state by promoting the formation of unstable intermediates and dynamic sampling of structures along the folding pathway of the intron.141,142 While still in their infancy, these studies of co-transcriptional RNA folding and RNA chaperone action suggest that RNA folding behavior should also be studied under conditions as similar as possible to those found in the native cellular environment.
Given the profound kinetic barriers found in some RNAs it seems likely that multiple native states of certain RNAs, either naturally evolved or engineered by humans, will persist in vivo even when folded co-transcriptionally in the presence of nucleic acid binding proteins. For natural RNAs, such heterogeneity may depend on the balance between energy requirements to redistribute kinetically trapped species and any (dis)advantages of maintaining a homogeneous over a heterogeneous population of native RNAs. Only in vivo testing will determine what roles conformational heterogeneity of RNA may have in living organisms. At least in theory, a shape-shifting RNA quasispecies, as observed in vitro, can be expected to impart evolutionary advantages.
Acknowledgments
The authors acknowledge funding from NIH grant GM062357.
References
- 1.Cech TR, Zaug AJ, Grabowski PJ. Cell. 1981;27:487–496. doi: 10.1016/0092-8674(81)90390-1. [DOI] [PubMed] [Google Scholar]
- 2.Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 3.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 4.Amaral PP, Dinger ME, Mercer TR, Mattick JS. Science. 2008;319:1787–1789. doi: 10.1126/science.1155472. [DOI] [PubMed] [Google Scholar]
- 5.Walter NG, Woodson SA, Batey RT. Non-protein coding RNAs. Springer; Heidelberg: 2009. [Google Scholar]
- 6.Moazed D. Nature. 2009;457:413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mercer TR, Dinger ME, Mattick JS. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 8.Mello CC, Conte D. Nature. 2004;431:338–342. doi: 10.1038/nature02872. [DOI] [PubMed] [Google Scholar]
- 9.Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- 10.Toor N, Rajashankar K, Keating KS, Pyle AM. Nat Struct Mol Biol. 2008;15:1221–1222. doi: 10.1038/nsmb.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 12.Prody GA, Bakos JT, Buzayan JM, Schneider IR, Bruening G. Science. 1986;231:1577–1580. doi: 10.1126/science.231.4745.1577. [DOI] [PubMed] [Google Scholar]
- 13.Forster AC, Symons RH. Cell. 1987;50:9–16. doi: 10.1016/0092-8674(87)90657-x. [DOI] [PubMed] [Google Scholar]
- 14.Hampel A, Tritz R. Biochemistry. 1989;28:4929–4933. doi: 10.1021/bi00438a002. [DOI] [PubMed] [Google Scholar]
- 15.Saville BJ, Collins RA. Cell. 1990;61:685–696. doi: 10.1016/0092-8674(90)90480-3. [DOI] [PubMed] [Google Scholar]
- 16.Lai MMC. Annu Rev Biochem. 1995;64:259–286. doi: 10.1146/annurev.bi.64.070195.001355. [DOI] [PubMed] [Google Scholar]
- 17.Salehi-Ashtiani K, Luptak A, Litovchick A, Szostak JW. Science. 2006;313:1788–1792. doi: 10.1126/science.1129308. [DOI] [PubMed] [Google Scholar]
- 18.Martick M, Horan LH, Noller HF, Scott WG. Nature. 2008;454:899–U857. doi: 10.1038/nature07117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Webb CH, Riccitelli NJ, Ruminski DJ, Luptak A. Science. 2009;326:953. doi: 10.1126/science.1178084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de la Pena M, Garcia-Robles I. RNA. 2010;16:1943–1950. doi: 10.1261/rna.2130310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Illangasekare M, Yarus M. Proc Natl Acad Sci USA. 1999;96:5470–5475. doi: 10.1073/pnas.96.10.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lincoln TA, Joyce GF. Science. 2009;323:1229–1232. doi: 10.1126/science.1167856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agresti JJ, Kelly BT, Jaschke A, Griffiths AD. Proc Natl Acad Sci USA. 2005;102:16170–16175. doi: 10.1073/pnas.0503733102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scott WG. Curr Opin Struct Biol. 2007;17:280–286. doi: 10.1016/j.sbi.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 25.Serganov A, Patel DJ. Nat Rev Genet. 2007;8:776–790. doi: 10.1038/nrg2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korostelev A, Noller HF. Trends Biochem Sci. 2007;32:434–441. doi: 10.1016/j.tibs.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 27.Walter NG. Mol Cell. 2007;28:923–929. doi: 10.1016/j.molcel.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Al-Hashimi HM, Walter NG. Curr Opin Struct Biol. 2008;18:321–329. doi: 10.1016/j.sbi.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cochrane JC, Strobel SA. Acc Chem Res. 2008;41:1027–1035. doi: 10.1021/ar800050c. [DOI] [PubMed] [Google Scholar]
- 30.Walter NG, Perumal S. In: Non-Protein Coding RNAs. Walter NG, Woodson SA, Batey RT, editors. Vol. 13. Springer; Berlin: 2009. pp. 103–127. Editon edn. [Google Scholar]
- 31.Johnson-Buck AE, McDowell SE, Walter NG. Met Ions Life Sci. 2011;9 doi: 10.1039/9781849732512-00175. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gartland WJ, Sueoka N. Proc Natl Acad Sci USA. 1966;55:948. doi: 10.1073/pnas.55.4.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindahl T, Adams A, Fresco JR. Proc Natl Acad Sci USA. 1966;55:941. doi: 10.1073/pnas.55.4.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walstrum SA, Uhlenbeck OC. Biochemistry. 1990;29:10573–10576. doi: 10.1021/bi00498a022. [DOI] [PubMed] [Google Scholar]
- 35.Esteban JA, Banerjee AR, Burke JM. J Biol Chem. 1997;272:13629–13639. doi: 10.1074/jbc.272.21.13629. [DOI] [PubMed] [Google Scholar]
- 36.Pan T, Sosnick TR. Nat Struct Biol. 1997;4:931–938. doi: 10.1038/nsb1197-931. [DOI] [PubMed] [Google Scholar]
- 37.Chadalavada DM, Knudsen SM, Nakano S, Bevilacqua PC. J Mol Biol. 2000;301:349–367. doi: 10.1006/jmbi.2000.3953. [DOI] [PubMed] [Google Scholar]
- 38.Treiber DK, Williamson JR. Curr Opin Struct Biol. 1999;9:339–345. doi: 10.1016/S0959-440X(99)80045-1. [DOI] [PubMed] [Google Scholar]
- 39.Russell R. Frontiers Biosci. 2008;13:1–20. doi: 10.2741/2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Solomatin SV, Greenfeld M, Chu S, Herschlag D. Nature. 2010;463:681–U117. doi: 10.1038/nature08717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sigler PB. Annu Rev Biophys Bioengin. 1975;4:477–527. doi: 10.1146/annurev.bb.04.060175.002401. [DOI] [PubMed] [Google Scholar]
- 42.Rastogi T, Beattie TL, Olive JE, Collins RA. EMBO J. 1996;15:2820–2825. [PMC free article] [PubMed] [Google Scholar]
- 43.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 44.Ferre-D’Amare AR, Zhou KH, Doudna JA. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- 45.Rupert PB, Ferre-D’Amare AR. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- 46.Klein DJ, Ferre-D’Amare AR. Science. 2006;313:1752–1756. doi: 10.1126/science.1129666. [DOI] [PubMed] [Google Scholar]
- 47.Lipfert J, Ouellet J, Norman DG, Doniach S, Lilley DM. Structure. 2008;16:1357–1367. doi: 10.1016/j.str.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murray LJW, Arendall WB, Richardson DC, Richardson JS. Proc Natl Acad Sci USA. 2003;100:13904–13909. doi: 10.1073/pnas.1835769100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Richardson JS, Schneider B, Murray LW, Kapral GJ, Immormino RM, Headd JJ, Richardson DC, Ham D, Hershkovits E, Williams LD, Keating KS, Pyle AM, Micallef D, Westbrook J, Berman HM. RNA. 2008;14:465–481. doi: 10.1261/rna.657708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brion P, Westhof E. Annu Rev Biophys Biomol Struct. 1997;26:113–137. doi: 10.1146/annurev.biophys.26.1.113. [DOI] [PubMed] [Google Scholar]
- 51.Porschke D, Eigen M. J Mol Biol. 1971;62:361–381. doi: 10.1016/0022-2836(71)90433-5. [DOI] [PubMed] [Google Scholar]
- 52.Turner DH, Sugimoto N, Freier SM. In: Nucleic Acids. Saenger W, editor. Springer; Berlin: 1990. pp. 201–227. Editon edn. [Google Scholar]
- 53.Herschlag D. J Biol Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 54.Zhuang XW, Bartley LE, Babcock HP, Russell R, Ha TJ, Herschlag D, Chu S. Science. 2000;288:2048. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- 55.Zhuang XW, Kim H, Pereira MJB, Babcock HP, Walter NG, Chu S. Science. 2002;296:1473–1476. doi: 10.1126/science.1069013. [DOI] [PubMed] [Google Scholar]
- 56.Pereira MJB, Nikolova EN, Hiley SL, Jaikaran D, Collins RA, Walter NG. J Mol Biol. 2008;382:496–509. doi: 10.1016/j.jmb.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gralla J, Delisi C. Nature. 1974;248:330–332. doi: 10.1038/248330a0. [DOI] [PubMed] [Google Scholar]
- 58.Zuker M. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]
- 59.Mccaskill JS. Biopolymers. 1990;29:1105–1119. doi: 10.1002/bip.360290621. [DOI] [PubMed] [Google Scholar]
- 60.Mathews DH. RNA. 2004;10:1178–1190. doi: 10.1261/rna.7650904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Markham NR, Zuker M. Nucleic Acids Res. 2005;33:W577–W581. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Korennykh AV, Plantinga MJ, Correll CC, Piccirilli JA. Biochemistry. 2007;46:12744–12756. doi: 10.1021/bi700931y. [DOI] [PubMed] [Google Scholar]
- 63.Huang Z, Pei WM, Han Y, Jayaseelan S, Shekhtman A, Shi H, Niu L. Nucleic Acids Res. 2009;37:4022–4032. doi: 10.1093/nar/gkp284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Altuntop ME, Ly CT, Wang YH. Biophys J. 2010;99:3002–3009. doi: 10.1016/j.bpj.2010.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ditzler MA, Rueda D, Mo JJ, Hakansson K, Walter NG. Nucleic Acids Res. 2008;36:7088–7099. doi: 10.1093/nar/gkn871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buzayan JM, Gerlach WL, Bruening G. Proc Natl Acad Sci USA. 1986;83:8859–8862. doi: 10.1073/pnas.83.23.8859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Buzayan JM, Gerlach WL, Bruening G, Keese P, Gould AR. Virology. 1986;151:186–199. doi: 10.1016/0042-6822(86)90041-3. [DOI] [PubMed] [Google Scholar]
- 68.Walter NG, Huang CY, Manzo AJ, Sobhy MA. Nat Methods. 2008;5:475–489. doi: 10.1038/nmeth.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rueda D, Bokinsky G, Rhodes MM, Rust MJ, Zhuang X, Walter NG. Proc Natl Acad Sci USA. 2004;101:10066–10071. doi: 10.1073/pnas.0403575101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu S, Bokinsky G, Walter NG, Zhuang X. Proc Natl Acad Sci USA. 2007;104:12634–12639. doi: 10.1073/pnas.0610597104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Okumus B, Wilson TJ, Lilley DMJ, Ha T. Biophys J. 2004;87:2798–2806. doi: 10.1529/biophysj.104.045971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bokinsky G, Rueda D, Misra VK, Rhodes MM, Gordus A, Babcock HP, Walter NG, Zhuang X. Proc Natl Acad Sci USA. 2003;100:9302–9307. doi: 10.1073/pnas.1133280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mortimer SA, Weeks KM. Proc Natl Acad Sci USA. 2009;106:15622–15627. doi: 10.1073/pnas.0901319106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ermolenko DN, Majumdar ZK, Hickerson RP, Spiegel PC, Clegg RM, Noller HF. J Mol Biol. 2007;370:530–540. doi: 10.1016/j.jmb.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 75.Ermolenko DN, Spiegel PC, Majumdar ZK, Hickerson RP, Clegg RM, Noller HF. Nat Struct Mol Biol. 2007;14:493–497. doi: 10.1038/nsmb1243. [DOI] [PubMed] [Google Scholar]
- 76.Munro JB, Altman RB, O’Connor N, Blanchard SC. Mol Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cornish PV, Ermolenko DN, Noller HF, Ha T. Mol Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huang Z, Pei W, Jayaseelan S, Shi H, Niu L. Biochemistry. 2007;46:12648–12655. doi: 10.1021/bi701036p. [DOI] [PubMed] [Google Scholar]
- 79.Baldwin RL, Rose GD. Trends Biochem Sci. 1999;24:26–33. doi: 10.1016/s0968-0004(98)01346-2. [DOI] [PubMed] [Google Scholar]
- 80.Dill KA, Ozkan SB, Shell MS, Weikl TR. Annu Rev Biophys. 2008;37:289–316. doi: 10.1146/annurev.biophys.37.092707.153558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dalal S, Regan L. Protein Sci. 2000;9:1651–1659. doi: 10.1110/ps.9.9.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Matthews BW. Adv Protein Chem. 1995;46:249–278. doi: 10.1016/s0065-3233(08)60337-x. [DOI] [PubMed] [Google Scholar]
- 83.Freier SM, Kierzek R, Jaeger JA, Sugimoto N, Caruthers MH, Neilson T, Turner DH. Proc Natl Acad Sci USA. 1986;83:9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Markham NR, Zuker M. Methods Mol Biol. 2008;453:3–31. doi: 10.1007/978-1-60327-429-6_1. [DOI] [PubMed] [Google Scholar]
- 85.Minor DL, Jr, Kim PS. Nature. 1994;371:264–267. doi: 10.1038/371264a0. [DOI] [PubMed] [Google Scholar]
- 86.Kiefhaber T. Proc Natl Acad Sci USA. 1995;92:9029–9033. doi: 10.1073/pnas.92.20.9029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kamagata K, Sawano Y, Tanokura M, Kuwajima K. J Mol Biol. 2003;332:1143–1153. doi: 10.1016/j.jmb.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 88.Jennings PA, Finn BE, Jones BE, Matthews CR. Biochemistry. 1993;32:3783–3789. doi: 10.1021/bi00065a034. [DOI] [PubMed] [Google Scholar]
- 89.Schmid FX, Blaschek H. Eur J Biochem. 1981;114:111–117. doi: 10.1111/j.1432-1033.1981.tb06180.x. [DOI] [PubMed] [Google Scholar]
- 90.Flomenbom O, Velonia K, Loos D, Masuo S, Cotlet M, Engelborghs Y, Hofkens J, Rowan AE, Nolte RJ, Van der Auweraer M, de Schryver FC, Klafter J. Proc Natl Acad Sci USA. 2005;102:2368–2372. doi: 10.1073/pnas.0409039102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lu HP, Xun LY, Xie XS. Science. 1998;282:1877–1882. doi: 10.1126/science.282.5395.1877. [DOI] [PubMed] [Google Scholar]
- 92.Yang H, Luo G, Karnchanaphanurach P, Louie TM, Rech I, Cova S, Xun L, Xie XS. Science. 2003;302:262–266. doi: 10.1126/science.1086911. [DOI] [PubMed] [Google Scholar]
- 93.Edman L, Foldes-Papp Z, Wennmalm S, Rigler R. Chem Phys. 1999;247:11–22. [Google Scholar]
- 94.van Oijen AM, Blainey PC, Crampton DJ, Richardson CC, Ellenberger T, Xie XS. Science. 2003;301:1235–1238. doi: 10.1126/science.1084387. [DOI] [PubMed] [Google Scholar]
- 95.Xue Q, Yeung ES. Nature. 1995;373:681–683. doi: 10.1038/373681a0. [DOI] [PubMed] [Google Scholar]
- 96.Tan WH, Yeung ES. Anal Chem. 1997;69:4242–4248. [Google Scholar]
- 97.Craig DB, Arriaga EA, Wong JCY, Lu H, Dovichi NJ. J Am Chem Soc. 1996;118:5245–5253. [Google Scholar]
- 98.Craig DB, Dovichi NJ. Can J Chem. 1998;76:623–626. [Google Scholar]
- 99.Handa N, Bianco PR, Baskin RJ, Kowalczykowski SC. Mol Cell. 2005;17:745–750. doi: 10.1016/j.molcel.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 100.Bianco PR, Kowalczykowski SC. Nature. 2000;405:368–372. doi: 10.1038/35012652. [DOI] [PubMed] [Google Scholar]
- 101.Baldini G, Cannone F, Chirico G. Science. 2005;309:1096–1100. doi: 10.1126/science.1115001. [DOI] [PubMed] [Google Scholar]
- 102.Cannone F, Bologna S, Campanini B, Diaspro A, Bettati S, Mozzarelli A, Chirico G. Biophys J. 2005;89:2033–2045. doi: 10.1529/biophysj.105.064584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Baldini G, Cannone F, Chirico G, Collini M, Campanini B, Bettati S, Mozzarelli A. Biophys J. 2007;92:1724–1731. doi: 10.1529/biophysj.106.093567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Polakowski R, Craig DB, Skelley A, Dovichi NJ. J Am Chem Soc. 2000;122:4853–4855. [Google Scholar]
- 105.Franklin I. In: Sinauer Associates. Soulé ME, Wilcox BA, editors. Sunderland, Massachusetts: 1980. pp. 135–150. Editon edn. [Google Scholar]
- 106.Reed DH, Frankham R. Conserv Biol. 2003;17:230–237. [Google Scholar]
- 107.Allison AC. Exp Parasitol. 1957;6:418–447. doi: 10.1016/0014-4894(57)90032-2. [DOI] [PubMed] [Google Scholar]
- 108.Robertson DL, Hahn BH, Sharp PM. J Mol Evol. 1995;40:249–259. doi: 10.1007/BF00163230. [DOI] [PubMed] [Google Scholar]
- 109.Dean M, Carrington M, Winkler C, Huttley GA, Smith MW, Allikmets R, Goedert JJ, Buchbinder SP, Vittinghoff E, Gomperts E, Donfield S, Vlahov D, Kaslow R, Saah A, Rinaldo C, Detels R, O’Brien SJ. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 110.Uyenoyama M. Pesticide Resistance: Strategies and Tactics for Management. National Academy Press; Washington, D.C: 1986. [Google Scholar]
- 111.Lyon BR, Skurray R. Microbiol Rev. 1987;51:88–134. doi: 10.1128/mr.51.1.88-134.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Choi PJ, Cai L, Frieda K, Xie XS. Science. 2008;322:442–446. doi: 10.1126/science.1161427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wolffe AP, Matzke MA. Science. 1999;286:481–486. doi: 10.1126/science.286.5439.481. [DOI] [PubMed] [Google Scholar]
- 114.Hannon GJ. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 115.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 116.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR. Chem Biol. 2002;9:1043. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- 118.Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- 119.Winkler W, Nahvi A, Breaker RR. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 120.Winkler WC, Cohen-Chalamish S, Breaker RR. Proc Natl Acad Sci USA. 2002;99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mandal M, Breaker RR. Nat Rev Mol Cell Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- 122.Graveley BR. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 123.Blencowe BJ. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 124.Cai L, Friedman N, Xie XS. Nature. 2006;440:358–362. doi: 10.1038/nature04599. [DOI] [PubMed] [Google Scholar]
- 125.Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, Emili A, Xie XS. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dekel E, Alon U. Nature. 2005;436:588–592. doi: 10.1038/nature03842. [DOI] [PubMed] [Google Scholar]
- 127.Eigen M, Mccaskill J, Schuster P. J Phys Chem. 1988;92:6881–6891. [Google Scholar]
- 128.Schuster P. Orig Life Evol Biosph. 1993;23:373–391. doi: 10.1007/BF01582087. [DOI] [PubMed] [Google Scholar]
- 129.Biebricher CK, Eigen M. Curr Top Microbiol Immunol. 2006;299:1–31. doi: 10.1007/3-540-26397-7_1. [DOI] [PubMed] [Google Scholar]
- 130.Lauring AS, Andino R. PLoS Pathog. 2010;6:e1001005. doi: 10.1371/journal.ppat.1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Anderson JP, Daifuku R, Loeb LA. Annu Rev Microbiol. 2004;58:183–205. doi: 10.1146/annurev.micro.58.030603.123649. [DOI] [PubMed] [Google Scholar]
- 132.Bull JJ, Meyers LA, Lachmann M. PLoS Comput Biol. 2005;1:e61. doi: 10.1371/journal.pcbi.0010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Schultes EA, Bartel DP. Science. 2000;289:448–452. doi: 10.1126/science.289.5478.448. [DOI] [PubMed] [Google Scholar]
- 134.Heilman-Miller SL, Woodson SA. RNA. 2003;9:722–733. doi: 10.1261/rna.5200903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wickiser JK, Winkler WC, Breaker RR, Crothers DM. Mol Cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 136.Pereira MJ, Behera V, Walter NG. PLoS One. 2010;5:e12953. doi: 10.1371/journal.pone.0012953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Meyer IM, Miklos I. BMC Mol Biol. 2004;5:10. doi: 10.1186/1471-2199-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mahen EM, Harger JW, Calderon EM, Fedor MJ. Mol Cell. 2005;19:27–37. doi: 10.1016/j.molcel.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 139.Mahen EM, Watson PY, Cottrell JW, Fedor MJ. PLoS Biol. 2010;8:e1000307. doi: 10.1371/journal.pbio.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bhaskaran H, Russell R. Nature. 2007;449:1014–1018. doi: 10.1038/nature06235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Fedorova O, Solem A, Pyle AM. J Mol Biol. 2010;397:799–813. doi: 10.1016/j.jmb.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Karunatilaka KS, Solem A, Pyle AM, Rueda D. Nature. 2010;467:935–939. doi: 10.1038/nature09422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Guo F, Gooding AR, Cech TR. Molecular Cell. 2004;16:351–362. doi: 10.1016/j.molcel.2004.10.003. [DOI] [PubMed] [Google Scholar]