Abstract
Rationale: Based on surface brushings and bronchoalveolar lavage fluid, Hilty and coworkers demonstrated microbiomes in the human lung characteristic of asthma and chronic obstructive pulmonary disease (COPD), which have now been confirmed by others.
Objectives: To extend these findings to human lung tissue samples.
Methods: DNA from lung tissue samples was obtained from nonsmokers (n = 8); smokers without COPD (n = 8); patients with very severe COPD (Global Initiative for COPD [GOLD] 4) (n = 8); and patients with cystic fibrosis (CF) (n = 8). The latter served as a positive control, with sterile water as a negative control. All bacterial community analyses were based on polymerase chain reaction amplifying 16S rRNA gene fragments. Total bacterial populations were measured by quantitative polymerase chain reaction and bacterial community composition was assessed by terminal restriction fragment length polymorphism analysis and pyrotag sequencing.
Measurement and Main Results: Total bacterial populations within lung tissue were small (20–1,252 bacterial cells per 1,000 human cells) but greater in all four sample groups versus the negative control group (P < 0.001). Terminal restriction fragment length polymorphism analysis and sequencing distinguished three distinct bacterial community compositions: one common to the nonsmoker and smoker groups, a second to the GOLD 4 group, and the third to the CF-positive control group. Pyrotag sequencing identified greater than 1,400 unique bacterial sequences and showed an increase in the Firmicutes phylum in GOLD 4 patients versus all other groups (P < 0.003) attributable to an increase in the Lactobacillus genus (P < 0.0007).
Conclusions: There is a detectable bacterial community within human lung tissue that changes in patients with very severe COPD.
Keywords: COPD, bacteria, microbiome
At a Glance Commentary
Scientific Knowledge on the Subject
Using established and new molecular techniques this study provides a methodology for the quantification and identification of bacteria, culturable and nonculturable, within human lung tissue.
What This Study Adds to the Field
The finding that the bacteria in lung tissue from very severe chronic obstructive pulmonary disease is different than in nonsmokers, smokers, and patients with cystic fibrosis expands on previous research on the bacterial microbiome in healthy individuals and in moderate chronic obstructive pulmonary disease that primarily used bronchoalveolar lavage and bronchial brushings.
Chronic obstructive pulmonary disease (COPD) is a major public health problem projected to be the fourth leading cause of death worldwide by 2020 (1). Although persistent inhalation of toxic particles and gases are the major risk factors, with tobacco smoking being the best example of this type of risk, only a fraction (∼15%) of smokers actually develop COPD (2–4). Although smokers have a dysfunctional immune system with an increased risk of developing microbial infections, which can be alleviated on cessation (5), the development and increasing disease severity of COPD progressively worsens the inflammatory cell burden (6). Recent evidence suggests that narrowing and destruction of the smaller bronchioles accounts for the increased resistance to flow that occurs in the small conducting airways less than 2 mm in diameter (7). Moreover, this process begins before the onset of emphysematous destruction of the alveolar surface that reduces the elastic recoil force available to drive air out of the lung (7). Quantitative studies of the histology of these lesions show that they are associated with the accumulation of infiltrating CD4+ T cells and B cells that form tertiary lymphoid organs in increasing numbers in severe (Global Initiative for COPD [GOLD] 3) and very severe (GOLD 4) COPD (6). These results support the hypothesis that the innate and adaptive immune response contributes to the pathology in peripheral lung lesions in COPD, but the antigens that drive this response remain to be determined (8–13). Some have suggested that autoantigens drive the pathogenesis of COPD (14), whereas others propose that the emergence of new strains of foreign microbes (9–11, 15–18) initiates a cycle of infection, inflammation, and dysfunctional repair, which drives the progression of COPD (10, 15). The increase in frequency and severity of exacerbations that occurs with the progression of COPD might be a manifestation of this vicious cycle of infection and inflammation (19). The recent report of Hilty and coworkers (20) of bronchial brush samples showing that a microbiome exists in the normal smoker’s lung and that this microbiome changes in COPD has stimulated additional studies of the lung microbiome based on either bronchial brushings, bronchoalveolar lavage (BAL) specimens, endotracheal aspirates, or lung tissue (21, 22). This current study uses lung tissue samples and extends these previous findings by providing an in-depth analysis of the bacterial microbiome in lung tissue from patients with very severe COPD (GOLD 4), nonsmoking and smoking control subjects, and individuals with cystic fibrosis (CF). We characterized the bacterial community in these samples by amplification and analysis of the 16S rRNA gene by a combination of methods including quantitative polymerase chain reaction (QPCR), terminal restriction fragment length polymorphism (TRFLP) analysis, and pyrotag sequencing. Some of the results of these studies have been previously reported in the form of an abstract (23).
Methods
Subjects
Lungs from nonsmokers (n = 8) and smokers (n = 8) were obtained by surgical resection for lung cancer or from transplant donors whose lungs were released for research when no suitable recipient was located. Very severe COPD (GOLD 4) (n = 8) or CF (n = 8) lungs were obtained from patients treated by lung transplantation. Nonsmoker and smoker subjects were matched for age and sex to the GOLD 4 subjects. Informed consent was obtained from all patients and from the next of kin of those who donated their lungs for transplantation. This study was approved by appropriate committees of the institutions involved.
DNA Extraction from Lung Samples
Tissue samples were collected from a transverse slice of either right or left lung at the level of the hilum. Preliminary experiments showed variation in bacterial cells dependent on lung height with the level of the hilum showing the highest total bacteria (data not shown). The procedure for DNA extraction is described in the online supplement. Lung samples from subjects with CF (n = 8) served as positive control samples because it is well established that their lungs contain bacteria (24), and sterile water served as the negative control.
QPCR Analysis
QPCR amplified a 293-bp target spanning the hypervariable region V2 of the 16S rRNA gene (25, 26) using primers, and cycling conditions (20, 27, 28) described in the online supplement. Bacterial numbers were normalized to the single copy gene Rpp40 (29) after a correction factor for bacterial numbers in the negative control samples. Further details are in the online supplement.
TRFLP
TRFLP used the restriction enzyme HHaI (30) to digest FAM-labeled 881-bp PCR products spanning the hypervariable regions V1-V3 (see online supplement) (25).
Pyrotag Sequencing
Nested PCR for pyrosequencing used first-round primers and cycling conditions for the 881-bp TRFLP target and second-round primers to amplify a 550-bp sequence spanning the V1-V3 hypervariable regions (see online supplement). All CF samples were also analyzed by a nonnested PCR using second-round primers to compare the two methods. PCR products were sequenced using the GS-FLX 454 platform (Genome Quebec Innovation Centre, McGill University, Montreal, Canada).
Data Analysis
QPCR and phyla analysis of the pyrotag sequencing data were analyzed using a Kruskal-Wallis nonparametric test, with Bonferroni correction for multiple comparisons. TRFLP fingerprints were analyzed for presence-absence and relative abundance of peaks by PC-ORD (MJM Software Design, Gleneden Beach, OR) plus nonmetric multidimensional scaling (31) and a multiresponse permutation procedure (32) measuring T, A, and P values. Pyrotag sequence qvality filtering, binning into operational taxonomic units (OTUs), and taxonomic assignment were conducted (see online supplement). Relative OTU abundances were calculated for each sample before any statistical analysis was performed. Principal coordinates analysis and pairwise comparisons using PERMANOVA with Bonferroni corrections were used to compare sample groups. Bacterial diversity was assessed using Simpson inverse index (33). Ginkgo was used to identify whether the presence or abundance of a species could be attributed to a particular sample group (34, 35). Q values to account for multiple comparisons (i.e., the number of OTUs) were obtained using qvality (36) with a false-discovery rate cutoff of 0.05.
Results
Subjects
GOLD 4 and CF groups have reduced FEV1 percent predicted and FVC percent predicted, compared with the nonsmoker and smoker control groups. The age (32.6 ± 8.7, mean ± SD) of the subjects with CF was younger than the other groups as expected for patients with CF undergoing lung transplantation (Table 1). Computed tomography images of the lungs and inspection of the cut surfaces of the slices of the frozen lung sections showed that no bronchiectasis was present in any of the patients with COPD (GOLD 4). It was not possible to document the presence of gastroesophageal reflux within this group of patients with COPD (GOLD 4) from the information available. More information on the patient population can be found in the online supplement.
TABLE 1.
DEMOGRAPHIC AND CLINICAL CHARACTERISTICS OF STUDY PARTICIPANTS
| Nonsmoker Control | Smoker Control | COPD (GOLD 4) | Cystic Fibrosis | |
| N | 8 | 8 | 8 | 8 |
| Age | 56.3 | 56.9 | 58.8 | 32.6* |
| Sex | 5 F/3 M | 5 F/3 M | 3 F/5 M | 2 F/5 M/1 unknown |
| FEV1pp | 88.8 ± 13.4 | 94.3 ± 15.3 | 15.4 ± 2.4† | 35.0 ± 22.9‡ |
| FEV1 | 2.55 ± 0.79 | 2.83 ± 0.84 | 0.50 ± 0.14† | 1.27 ± 0.88‡ |
| FVCpp | 87.2 ± 10.5 | 98.5 ± 18.1 | 47.7 ± 11.3† | 51.4 ± 22.4‡ |
| FVC | 3.17 ± 1.07 | 3.70 ± 1.12 | 1.97 ± 0.69† | 2.29 ± 1.10‡ |
| FEV1/FVC | 80.80 ± 4.82 | 76.65 ± 5.07 | 26.83 ± 7.85† | 53.76 ± 20.05‡ |
| Pack-years | 0.00 ± 0.00 | 46.00 ± 12.24§ | 38.83 ± 14.97║ | 0.00 ± 0.00 |
| Sustained smokers | 0 | 8 | 4 | 0 |
Definition of abbreviations: COPD = chronic obstructive pulmonary disease; GOLD = Global Initiative for COPD; pp = percent predicted.
P < 0.015 lower than all other groups.
P < 0.0001 lower than nonsmoker and smoker control groups.
P < 0.03 lower than nonsmoker and smoker control groups.
P < 0.0001 higher than nonsmoker and cystic fibrosis group.
P < 0.01 higher than nonsmoker and cystic fibrosis group.
QPCR
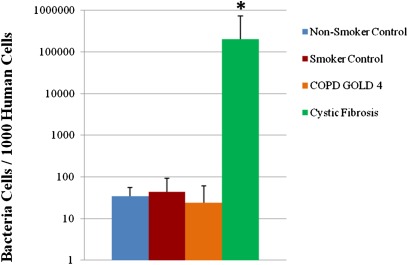
Total abundance of bacteria measured in lung tissue was extremely low and near the detection limit of the QPCR assay. The average bacterial population range detected in the negative control group (n = 24) was 0–588 cells per sample, which was significantly lower than that of the other experimental groups (P < 0.001). The CF group had a higher bacterial population than any of the other four experimental groups (P < 0.001), which ranged from 2,712–1,075,846 cells per sample. There was no significant difference in bacterial populations among the nonsmoker, smoker, and GOLD 4 groups (P > 0.05), which ranged from 20–1252 cells per sample. With the application of the correction factor to account for the contaminating bacteria and number of host cells in the specimen there still were no significant differences among the nonsmoking, smoking, and GOLD 4 groups (Figure 1), which showed a range of between 10 and 100 bacterial cells per 1,000 human cells. In contrast, the number of bacteria per 1,000 human cells in lung tissue from the CF group was three to four orders of magnitude greater.
Figure 1.
Quantitative polymerase chain reaction results of the average number of bacterial cells in 1,000 human cells in the four groups of lung samples (n = 8). *P = 0.0002 versus all other groups. COPD = chronic obstructive pulmonary disease; GOLD = Global Initiative for COPD.
TRFLP
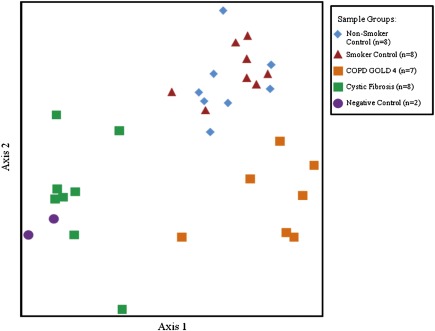
TRFLP analysis determined that two of four negative control samples, seven of eight GOLD 4 samples, and all other samples yielded sufficient amplicons for further analysis. The TRFLP results using nonmetric multidimensional scaling (Figure 2) indicated that the composition of the lung bacterial communities in the samples clustered into three distinct groups, corresponding to the nonsmoker plus smoker groups, the GOLD 4 group, and the CF group, with the two negative control samples clustering with the CF group.
Figure 2.
Nonmetric multidimensional scaling analysis of the terminal restriction fragment length polymorphism profiles that provided adequate number of amplicons. Three distinct groups can be discerned, with the negative control subjects clustering close to the cystic fibrosis sample group. COPD = chronic obstructive pulmonary disease; GOLD = Global Initiative for COPD.
T values from the multiresponse permutation procedure analysis, where negative numbers indicate less similarity among groups and positive values indicate greater similarity (37), demonstrate that these three clusters of communities were all significantly different from one another (Table 2). The number of negative control samples yielding TRFLP results was insufficient to conclude that they should be grouped separately. The T value for the nonsmoker versus smoker groups indicates that the bacterial communities are similar to each other. The A value, where positive numbers indicate better within-group agreement and negative value less (38), is weakly negative suggesting that the two groups were not distinct enough to be separated.
TABLE 2.
MULTIRESPONSE PERMUTATION PROCEDURE ANALYSIS OF TRFLP PRODUCTS
| T Value | A Value | P Value | |
| Smoker control vs. cystic fibrosis | −7.92 | 0.24 | 1.26 × 10−5 |
| Smoker control vs. COPD (GOLD 4) | −7.80 | 0.35 | 0.0001 |
| Smoker control vs. nonsmoker control | 1.04 | −0.03 | 0.86 |
| Cystic fibrosis vs. COPD (GOLD 4) | −5.68 | 0.19 | 0.0002 |
| Cystic fibrosis vs. nonsmoker control | −7.37 | 0.21 | 8.86 × 10−6 |
| COPD (GOLD 4) vs. nonsmoker control | −7.80 | 0.34 | 0.0001 |
Definition of abbreviations: COPD = chronic obstructive pulmonary disease; GOLD = Global Initiative for COPD; TRFLP = terminal restriction fragment length polymorphism.
Pyrotag Sequencing
Nested PCR was necessary to obtain sufficient amplicons from the samples with low bacterial densities. Pyrotag analysis of the CF lung samples with and without nested PCR yielded indistinguishable community compositions (P > 0.05) (see Figure E1 in the online supplement). Thus, the nested PCR was concluded not to bias pyrotag sequencing and was used for all further analysis.
A principal coordinates analysis of the total pyrotag OTUs revealed four distinct clusters of bacterial communities (see Figure E2A), which was consistent with the TRFLP data. All six negative controls samples yielded sufficient amplicons for pyrotag analysis with their bacterial communities clustered as a distinct group, except for one sample clustering with the CF group for which only six samples were analyzed because the other two spots in the sequencing run were used to check within-sample repeatability (data not shown). The communities from the nonsmoker and smoker groups clustered together, whereas those in the GOLD 4 group and CF group were each distinct. On subtraction of OTUs that shared 3% or greater similarity to reads from the negative control samples, the ordination trends persisted, with greater distinction among the three remaining clusters (see Figure E2B).
Pairwise comparisons of the sequencing results among the four patient groups (Table 3) showed nonsmoker and smoker control groups were not different. The GOLD 4 and CF groups were different from nonsmoker and smoker groups and from each other (P < 0.05). After subtraction of OTUs that shared 3% or greater similarity to reads from the negative controls, the results of the comparisons did not change (Table 3).
TABLE 3.
PAIRWISE COMPARISONS OF PYROTAG SEQUENCING RESULTS OF THE 16S RRNA GENE AMONG THE EXPERIMENTAL GROUPS BEFORE AND AFTER SUBTRACTION OF OTU THAT SHARED 3% OR GREATER SIMILARITY TO READS FROM THE NEGATIVE CONTROL GROUPS
| P Value* | P Value (with negative subtracted)* | |
| Smoker control vs. cystic fibrosis | 0.0005 | 0.0003 |
| Smoker control vs. COPD (GOLD 4) | 0.0007 | 0.0008 |
| Smoker control vs. nonsmoker control | 0.8891 | 0.8597 |
| Cystic fibrosis vs. COPD (GOLD 4) | 0.0003 | 0.0009 |
| Cystic fibrosis vs. nonsmoker control | 0.0005 | 0.0005 |
| COPD (GOLD 4) vs. nonsmoker control | 0.0027 | 0.0022 |
| Negative control vs. nonsmoker control | 0.0011 | N/A |
| Negative control vs. Smoker control | 0.0006 | N/A |
| Negative control vs. COPD (GOLD 4) | 0.0019 | N/A |
| Negative control vs. cystic fibrosis | 0.0044 | N/A |
Definition of abbreviations: COPD = chronic obstructive pulmonary disease; GOLD = Global Initiative for COPD; OTU = operational taxonomic units.
All P values except for the smoker control versus the nonsmoker control remain significant after Bonferroni correction for multiple comparisons.
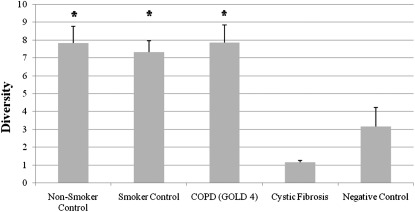
Pyrotag analysis identified greater than 1,400 OTUs (∼97% similarity level) in the lung samples that were distinct from the negative control samples. A list of the top 150 OTUs can be found in the online supplement (see Table E6). The experimental groups with low bacterial density in lung tissue (nonsmoker, smoker, and GOLD 4 groups) (Figure 1) had the highest diversity as assessed by Simpson inverse index (Figure 3). Among these three groups, the diversity did not differ significantly (P > 0.05). By comparison, diversity was much lower in the CF lung tissue and in the negative control groups indicating that a small number of organisms are dominant within the CF (24) and negative control groups.
Figure 3.
Simpson inverse index of bacterial diversity in the four groups of lung samples and the negative control subjects where higher values correspond to higher diversity. Histograms represent average ± SEM (n = 8 for each lung sample group, n = 6 for the negative control group). *P < 0.0001 versus the cystic fibrosis and negative control groups. COPD = chronic obstructive pulmonary disease; GOLD = Global Initiative for COPD.
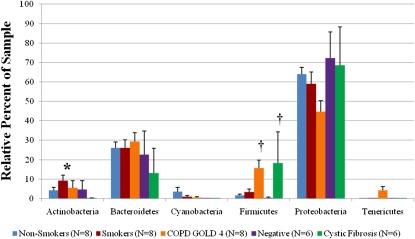
At the phylum level, community composition was generally similar among the experimental groups, with only three exceptions (Figure 4). The smoker group had a significantly greater abundance of Actinobacteria than all other groups (P < 0.007). The GOLD 4 and CF groups had a significantly greater abundance of Firmicutes than the remaining groups (P < 0.003). For the GOLD 4 group the primary increase in Firmicutes came from the Lactobacillus genus (see Figure E3A) and was consistently observed in seven of the eight samples (data not shown). By contrast, in the CF group, the increase in Firmicutes was mainly caused by the high abundance of the genus Streptococcus (see Figure E3A) but was only observed in one of the eight samples (data not shown). The CF and the GOLD 4 groups contained greater than 5% Burkholderia genus (see Figure E3B), whereas the other sample groups did not.
Figure 4.
Comparison among the nonsmoker control, smoker control, COPD (GOLD 4), cystic fibrosis, and negative control groups of the percent abundance of the major identified phyla within a sample. Histogram represents average ± SEM. *P < 0.007 versus all other groups. †P < 0.003 versus nonsmoking, smoking, and negative control groups. COPD = chronic obstructive pulmonary disease; GOLD = Global Initiative for COPD.
Indicator species analysis identified 52 OTUs correlating to one of the lung sample groups with P values of less than 0.05 (see Table E7) and 28 of these OTUs were specific to the GOLD 4 group. Of the listed GOLD 4 indicators (see Table E7), Lactobacillus and Bacteroidales species each accounted for 25%, whereas Burkholderia species represented 11% of these 28 OTUs. After false-discovery rate correction, 4 of these 28, remained significantly associated with the GOLD 4 group (Table 4), whereas none of those associating with other sample groups were significant (see Table E7). Of these four, three were classified as unknown Lactobacillus species and the other OTU classified to an unknown Burkholderia species. The Lactobacillus relative abundance in the GOLD 4 group was confirmed by a QPCR assay, which showed that it was not significantly different from that obtained from the pyrotag sequence analysis (see Figure E4).
TABLE 4.
TOP FIVE BACTERIAL SPECIES ASSOCIATED WITH EACH SAMPLE GROUP
| Species Name | Biserial Correlation Coefficient | P Value | Q Value |
| Cystic fibrosis | |||
| Alcaligenaceae unclassified | 0.55 | 0.0186 | 0.291883 |
| Pseudomonas unclassified | 0.55 | 0.0064 | 0.164345 |
| Pseudomonas unclassified | 0.53 | 0.0083 | 0.203868 |
| Pseudomonadaceae unclassified | 0.48 | 0.0382 | 0.405884 |
| Alcaligenaceae unclassified | 0.47 | 0.0314 | 0.405884 |
| COPD (GOLD 4) | |||
| Lactobacillus unclassified | 0.69 | 0.0003 | 0.011299 |
| Burkholderia unclassified | 0.69 | 0.0004 | 0.014123 |
| Lactobacillus unclassified | 0.65 | 0.0007 | 0.023262 |
| Lactobacillus unclassified | 0.62 | 0.0008 | 0.025108 |
| Burkholderiales unclassified | 0.61 | 0.0024 | 0.07136 |
| Smoker control* | |||
| Aquabacterium unclassified | 0.51 | 0.0115 | 0.234045 |
| Rhodocyclaceae unclassified | 0.43 | 0.0424 | 0.405884 |
| Acidovorax caeni | 0.52 | 0.0496 | 0.424557 |
| Nonsmoker control | |||
| Comamonadaceae unclassified | 0.61 | 0.0032 | 0.09039 |
| Comamonadaceae unclassified | 0.55 | 0.0038 | 0.102226 |
| Brevundimonas diminuta | 0.53 | 0.0103 | 0.234045 |
| Diaphorobacter unclassified | 0.52 | 0.0114 | 0.234045 |
| Comamonadaceae unclassified | 0.52 | 0.0172 | 0.291883 |
Definition of abbreviations: COPD = chronic obstructive pulmonary disease; GOLD = Global Initiative for COPD.
Bold species represent those that passed the false-discovery rate q value of <0.05.
Smoker control group only had three species whose P value was significant.
Discussion
The bacterial densities detected in lung tissue samples examined in this study are lower than those reported in previous studies based on either BAL or protected bronchial brush samples (20, 21, 39). However, these previous reports used different methods to quantify total bacteria (20, 21, 39). Lower densities found in this study are attributable to the small epithelial surface area within the lung tissue samples compared with the epithelial surface areas sampled by washing or brushing the lung. Additionally, the low bacterial densities we detected may be partly caused by decreased contamination of tissue samples removed from frozen lung specimens compared with samples that are either brushed (20, 21, 39) or washed (20, 21) from the lung surface. Despite this distinction from other studies our results confirm those of Hilty and coworkers (20) showing no difference in total bacterial number between nonsmoker and smoker control groups compared with smokers with COPD. Moreover, TRFLP and pyrotag analyses in this study confirm that there are differences in the bacterial community in lung tissue from GOLD 4 patients compared with nonsmoker and smoker control subjects. Thus, a unique bacterial community is associated with lung tissue from patients with very severe COPD.
It is noteworthy that according to the Simpson inverse index the bacterial diversity in lung tissue from the GOLD 4 group was the same as in the nonsmoker and smoker control groups. By contrast, the CF lung had much lower diversity and much higher bacterial density, consistent with the expansion of a small number of bacterial species during the course of CF, and is consistent with previous studies of CF (40). Although the richness component of diversity measurements can be biased by PCR and sequencing errors, our conclusions about the relative diversity of these communities are valid because the groups have similar sample numbers and are compared using identical methods.
In the case of very severe COPD, there seems to be a shift in the relative abundance of a few populations without any one becoming dominant. This observation does not support the previous suggestion that the diversity of organisms is reduced in disease compared with that in healthy smokers and nonsmokers by the emergence of a few dominant genera in cases of COPD (20) and in more severe COPD (21). Our contrasting results from the lung tissue of GOLD 4 patients in our study and that of the other two may be explained in part by differences in study design. Hilty and coworkers (20) analyzed bronchial brushings from patients with moderate COPD, whereas Erb-Downward and coworkers (21) analyzed BAL samples and tissue from moderate and very severe patients with COPD. Also, unequal and inadequate sample size may be an issue. In the former, samples from five moderate patients with COPD were compared with eight control samples. In the latter, BAL samples from seven healthy smokers and three nonsmokers were compared with four patients with COPD, two with mild, one with moderate, and one with severe COPD, where the first two had patterns of diversity similar to most of the nonsmoker and smoker control subjects, whereas the other two had a much limited diversity.
The present results differ from those based on bronchial brushings and BAL in that they probably reflect the mircobiome in parenchymal tissue (i.e., gas exchanging alveolated tissue) rather than the airways because 90% of the lung volume is taken up by parenchyma and only 10% by nonparenchyma (i.e., airways, vessels, and interstitial tissue) (41). Although it was not possible to histologically examine the 30 mg of tissue used, to measure the microbiome, we were able to examine nearby samples to confirm that parenchyma was the dominate histologic feature of lung close to the sampled regions. Therefore, samples obtained by brushing and BAL likely have a better chance of sampling the airway content.
Our bacterial community composition results also differ from both of these studies. Huang and coworkers (22) provide a possible explanation from their study on the changes in diversity of microorganisms in the lung microbiome associated with length of intubation in patients with acute exacerbations of COPD. They found that individuals with higher bacterial community diversity also supported a higher number of Firmicutes and those with less bacterial diversity had a higher number of Proteobacteria (22). This is consistent with our findings, and in those with severe COPD (21), that a low bacterial diversity could lead to an increase in the Proteobacteria phylum and a higher bacterial diversity could lead to an increase in the Firmicutes phylum.
Because the three methods used in this study to analyze bacteria in lung samples were based on PCR amplification of the bacterial 16S rRNA, which is prone to contamination, negative control samples were included. QPCR results show that bacterial abundance in the negative control samples was significantly lower than in the lung samples. Also, TRFLP analysis indicates that the very low populations in the negative control groups compared with the nonsmoker, smoker, and GOLD 4 groups are not caused by contamination. Further, clustering in the pyrotag analysis confirms that the lung samples with low bacterial densities contain bacterial communities, which are distinct from the negative control samples. These two distinct methods to analyze bacterial diversity provide reasonable evidence that bacteria detected in the lung samples are not caused by contaminating bacteria. In this regard, it is interesting to note that neither Hilty and coworkers (20) nor Erb-Downward and coworkers (21) report the inclusion of negative control samples in their studies.
Although specific OTUs were significantly associated with very severe COPD, these sequences have not been closely affiliated with any described species and could not be identified with the taxonomic database that was used in this study. Statistical analyses show that the Firmicutes phylum is significantly associated with these COPD lungs and that Lactobacillus is the main genus associated with the increase in the Firmicutes. Our study also showed that OTUs belonging to individual species of the genus Lactobacillus were also significantly associated with very severe COPD, but again these OTUs were not affiliated to known sequences within our taxonomic database.
The Lactobacillus genus is commonly found in the gastrointestinal and reproductive tracts (42–44). Most Lactobacillus identified in humans are postulated to come from food (45) and have been identified in the stomach of healthy, noncancerous control subjects and are increased in patients with stomach cancer (46). Gastroesophageal reflux is considered an important risk factor for increased exacerbations and is one of the self-reported comorbidities in frequent exacerbators in COPD (19, 47–49). It is possible that Lactobacillus bacteria enter the lungs by gastroesophageal reflux or by endotracheal intubation during lung resection surgery. Recently, it has been shown that Lactobacillus can modulate the immune response to influenza A virus in mice and loss of this genus dampens this immune response (50). The increased presence of the Lactobacillus within the lung tissue could be a result of an inflammatory modulation toward a stimulus, such as influenza, and results in the formation of tertiary lymphoid follicles associated with the small airways. This could be attributed to either Lactobacillus being the target or acting as an immune modulator and aiding in the inflammatory response. Whether these bacteria remain in the lung and modulate inflammatory responses or become antigenic targets that drive the innate and adaptive immune responses within human lungs remains to be determined.
Indicator species analysis also showed that a Burkholderia genus OTU was significantly associated with the GOLD 4 group. Although the Burkholderia genus and in particular Burkholderia cepacia are implicated in CF disease (51), it is not commonly associated with COPD. Because the OTU did not compare well with any known species within the database that was used, a definitive conclusion cannot be made toward the possible role this unidentified bacterium might play in disease. A limitation of this study is the absence of a moderate COPD (GOLD 2) and severe COPD (GOLD 3) group. Thus, extrapolation of our findings to these two groups may not be possible. The emergence of Lactobacillus and Burkholderia within the GOLD 4 group could be either a sudden occurrence or gradually progressive, with a higher frequency of the two genera seen as disease severity intensifies.
The results of this study confirm that low numbers of bacteria can be detected in lung tissue using the molecular techniques that are currently available. Although the total numbers detected were not different among the lung specimens from nonsmoking, smoking, or GOLD 4 subjects, specimens from the nonsmoking and smoking subjects clearly harbor a different bacterial composition than specimens from very severe COPD subjects. Whether these organisms are targeted by the host immune system or they help modulate the response needs to be determined before proceeding to the question of whether the immune response they stimulate or incite drives the tissue response that leads to the lesions causing airflow limitation in COPD. However, recent studies (50, 52) on the bacterial microbiome and immune response strongly suggest that bacteria can play an active role in directing the immune responses and inflammatory processes in wide ranging diseases from viral infections to arthritis.
Supplementary Material
Acknowledgments
We would like to thank Martin Hartmann for developing the original pipeline for pyrotag analysis.
Footnotes
Supported by Merck external studies agreement IIS 38,978 (UBC F1003533), NIH HL084948, CIHR CIF-97687, and the Lavin family foundation to J.C. P.A.D. was supported by a grant from the Centre for Microbial Diversity and Evolution from the Tula Foundation. None of the agencies had any input into the study design; in the collection, analysis, and interpretation of data; in the writing of the report; and in the decision to submit the report for publication.
Author Contributions: M.A.S., performed and designed experiments and wrote first draft; P.A.D., data analysis and intellectual contributions; S.H., intellectual contributions; W.M.E., tissue procurement; J.E.M., tissue procurement; J.V.G., intellectual contributions; J.C., tissue procurement; D.D.S., intellectual contributions; W.W.M., polymerase chain reaction materials and intellectual contributions; and J.C.H., conceived and designed experiments and intellectual contributions.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201111-2075OC on March 15, 2012
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Mathers C, Loncar D. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med 2006;3:2011–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lowell FC, Franklin W, Michelson AL, Schiller IW. Chronic obstructive pulmonary emphysema: a disease of smokers. Ann Intern Med 1956;45:268–274 [DOI] [PubMed] [Google Scholar]
- 3.Williams MH, Jr, Seriff NS. Chronic obstructive pulmonary disease: an analysis of clinical, physiologic, and roentgenologic features. Am J Med 1963;35:20–30 [DOI] [PubMed] [Google Scholar]
- 4.Fletcher C, Peto R. The natural history of chronic airflow obstruction. BMJ 1977;1:1645–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arcavi L, Benowitz NL. Cigarette smoking and infection. Arch Intern Med 2004;164:2206–2216 [DOI] [PubMed] [Google Scholar]
- 6.Hogg JC, Chu F, Utokaparch S, Woods R, Elliott WM, Buzatu L, Cherniack RM, Rogers RM, Sciurba FC, Coxson HO, et al. The nature of small-airway obstruction in chronic obstructive pulmonary disease. N Engl J Med 2004;350:2645–2653 [DOI] [PubMed] [Google Scholar]
- 7.McDonough JE, Yuan R, Suzuki M, Seyednejad N, Elliott WM, Sanchez PG, Wright AC, Gefter WB, Litzky L, Coxson HO, et al. The relationship between small airway obstruction and emphysema in chronic obstructive pulmonary disease. N Engl J Med 2011;365:1567–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greene CM, Low TB, O'Neill SJ, McElvaney NG. Anti-proline-glycine-proline or antielastin autoantibodies are not evident in chronic inflammatory lung disease. Am J Respir Crit Care Med 2010;181:31–35 [DOI] [PubMed] [Google Scholar]
- 9.Serres GD, Lampronb N, Forgeb JL, Rouleauc I, Bourbeaud J, Weiss K, Barret B, Boiving G. Importance of viral and bacterial infections in chronic obstructive pulmonary disease exacerbations. J Clin Virol 2009;46:129–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sethi S. Bacterial infection and the pathogenesis of COPD. Chest 2000;117:286–291 [DOI] [PubMed] [Google Scholar]
- 11.Curran T, Coyle PV, McManus TE, Kidney J, Coulter WA. Evaluation of real-time PCR for the detection and quantification of bacteria in chronic obstructive pulmonary disease. FEMS Immunol Med Microbiol 2007;1:112–118 [DOI] [PubMed] [Google Scholar]
- 12.Sikkel MB, Quint JK, Mallia P, Wedzicha JA, Johnston SL. Respiratory syncytial virus persistence in chronic obstructive pulmonary disease. Pediatr Infect Dis J 2008;27:63–70 [DOI] [PubMed] [Google Scholar]
- 13.Morimoto K, Gosselink JV, Kartono A, Hogg JC, Hayashi S, Ogawa E. Adenovirus E1a regulates lung epithelial ICAM-1 expression by interacting with transcriptional regulators at its promoter. Am J Physiol Lung Cell Mol Physiol 2008;296:361–371 [DOI] [PubMed] [Google Scholar]
- 14.Wood AM, de Pablo P, Buckley CD, Ahmad A, Stockley RA. Smoke exposure as a determinant of auto-antibody titre in α1-antitrypsin and COPD. Eur Respir J 2011;37:32–38 [DOI] [PubMed] [Google Scholar]
- 15.Sethi S, Evans N, Grant BJ, Murphy TF. New strains of bacteria and exacerbations of chronic obstructive pulmonary disease. N Engl J Med 2002;347:465–471 [DOI] [PubMed] [Google Scholar]
- 16.Banerjee D, Khair OA, Honeybourne D. Impact of sputum bacteria on airway inflammation and health status in clinical stable COPD. Eur Respir J 2004;23:685–691 [DOI] [PubMed] [Google Scholar]
- 17.Wilkinson TM, Patel IS, Wilks M, Donaldson GC, Wedzicha JA. Airway bacterial load and FEV1 decline in patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2003;167:1090–1095 [DOI] [PubMed] [Google Scholar]
- 18.Hill AT, Campbell EJ, Hill SL, Bayley DL, Stockley RA. Association between airway bacterial load and markers of airway inflammation in patients with stable chronic bronchitis. Am J Med 1999;109:288–295 [DOI] [PubMed] [Google Scholar]
- 19.Hurst JR, Vestbo J, Anzueto A, Locantore N, Mullerova H, Ral-Singer R, Miller B, Lomas DA, Agusti A, MacNee W, et al. Susceptibility to exacerbation in chronic obstructive pulmonary disease. N Engl J Med 2010;363:1128–1138 [DOI] [PubMed] [Google Scholar]
- 20.Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, Davies J, Ervine A, Poulter L, Pachter L, et al. Disordered microbial communities in asthmatic airways. PLoS ONE 2010;5:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erb-Downward JR, Thompson DL, Han MK, Freeman CM, McCloskey L, Schmidt LA, Young VB, Toews GB, Curtis JL, Sundaram B, et al. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS ONE 2011;6:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang YJ, Kim E, Cox MJ, Brodie EL, Brown R, Wiener-Kronish JP, Lynch SV. A persistent and diverse airway microbiota present during chronic obstructive pulmonary disease exacerbations. OMICS 2010;14:9–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sze MA, Gosselink JV, McDonough JE, Elliott WM, Adams S, Friedman J, Zhao Y, Varhol R, Miller D, He A, et al. The lung microbiome in COPD [abstract]. Am J Respir Crit Care Med 2011;183:A1017 [Google Scholar]
- 24.Cox MJ, Allgaier M, Taylor B, Baek MS, Huang YJ, Daly RA, Karaoz U, Anderson GL, Brown R, Fujimura KE, et al. Airway microbiota and pathogen abundance in age-stratified cystic fibrosis patients. PLoS ONE 2010;5:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neefs J-M, Van de Peer Y, Hendriks L, De Watcher R. Compiliation of small ribosomal subunit RNA sequences. Nucleic Acids Res 1990;18:2237–2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brosius J, Palmer ML, Kennedy PP, Noller HF. Complete nucleotide sequence of a 16S ribsomal RNA gene from Escherichia coli. Proc Natl Acad Sci USA 1978;75:4801–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castillo M, Martin-Orue S, Manzanilla EG, Badiola I, Martin M, Gasa J. Quantification of total bacteria, enterobacteria and lactobacilli populations in pig digesta by real-time PCR. Vet Microbiol 2006;114:165–170 [DOI] [PubMed] [Google Scholar]
- 28.Grice EA, Kong HH, Renaud G, Young AC. NICS Comparative Sequencing Program, Bouffard GG, Blakesley RW, Wolfsberg TG, Turner ML, Segre JA. A diversity profile of the human skin microbiota. Genome Res 2008;18:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McNees AL, White ZS, Zanwar P, Vilchez RA, Butel JS. Specific and quantitative detection of human polyomaviruses BKV, JCV, and SV40 by real time PCR. J Clin Virol 2005;34:52–62 [DOI] [PubMed] [Google Scholar]
- 30.Liu W-T, Marsh TL, Cheng H, Forney LJ. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol 1997;63:4516–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferreira RBR, Gill N, Willing BP, Antunes LCM, Russell SL, Croxen MA, Finlay BB. The intestinal microbiota plays a role in Salmonella-induced colitis independent of pathogen colonization. PLoS ONE 2011;6:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biondini M, Bonham C, Redente E. Secondary successional patterns in a sagebrush (Artemisia tridentata) community as the relate to soil disturbance and soil biological activity. Vegetatio 1985;60:25–36 [Google Scholar]
- 33.Simpson EH. Measurement of diversity. Nature 1949;163:688 [Google Scholar]
- 34.De Cáceres M, Oliva F, Font X, Vives S. Ginkgo, a program for non-standard multivariate fuzzy analysis. Adv in Fuzzy Sets & Systems 2 2007;1:41–56 [Google Scholar]
- 35.Mouillot D, Culioli J-M, Chi TD. Indicator species analysis as a test of non-random distribution of species in the context of marine protected areas. Environ Conserv 2002;29:385–390 [Google Scholar]
- 36.Kall L, Storey JD, Noble WS. Qvality: non-parametric estimation of q-values and posterior error probabilities. Bioinformatics 2009;25:964–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Faultz JE. Effects of shelterwood management on flower-visiting insects and their floral resources. Entomology. Bozeman: Montana State; Master’s Thesis: 2005. p. 161. Available from: http://etd.lib.montana.edu/etd/2005/fultz/FultzJ0805.pdf.
- 38.McCune B, Rosentreter R, Ponzetti J, Shaw D. Epiphyte habitats in an old conifer forest in western Washington, USA. Bryologist 2000;103:417–427 [Google Scholar]
- 39.Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, Bushman FD, Collman RG. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med 2011;184:957–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Döring G, Parameswaran IG, Murphy TF. Differential adaptation of microbial pathogens to airways of patients with cystic fibrosis and chronic obstructive pulmonary disease. FEMS Microbial Rev 2011;35:124–146 [DOI] [PubMed] [Google Scholar]
- 41.Weibel ER. Morphometry of the human lung. New York: Academic Press; 1963. pp. 51–54 [Google Scholar]
- 42.Ling Z, Kong J, Liu F, Zhu H, Chen X, Wang Y, Li L, Nelson KE, Xia Y, Xiang C. Molecular analysis of the diversity of vaginal microbiota associated with bacterial vaginosis. BMC Genomics 2010;11:1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walter J, Britton RA, Roos S. Host-microbial symbiosis in the vertebrate gastrointestinal tract and the Lactobacillus reuteri paradigm. Proc Natl Acad Sci USA 2011;108:4645–4652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heavens D, Tailford LE, Crossman L, Jeffers F, MacKenzie DA, Caccamo M, Juge N. Genome sequence of a vertebrate gut symbiont Lactobacillus reuteri ATC 53608. J Bacteriol 2011;193:4015–4016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walter J. Mini-reviews: ecological role of lactobacilli in the gastrointestinal tract: implications for fundamental and biomedical research. Appl Environ Microbiol 2008;74:4985–4996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dicksved J, Lindberg M, Rosenquist M, Enroth H, Jansson JK, Engstrand L. Molecular characterization of the stomach microbiota in patients with gastric cancer and in controls. J Med Microbiol 2009;58:509–516 [DOI] [PubMed] [Google Scholar]
- 47.Rascon-Aguilar IE, Pamer M, Wludyka P, Cury J, Coultas D, Lambiase LR, Nahman NS, Vega KJ. Role of gastroesophageal reflux symptoms in exacerbations of COPD. Chest 2006;130:1096–1101 [DOI] [PubMed] [Google Scholar]
- 48.Liang BM, Feng YL. Association of gastroesophageal reflux disease symptoms with stable chronic obstructive pulmonary disease. Lung (In press) [DOI] [PubMed] [Google Scholar]
- 49.Lindberg A, Larsson LG, Ronmark E, Lundback B. Co-morbidity in mild-to-moderate COPD: comparison to normal and restrictive lung function. COPD 2011;8:421–428 [DOI] [PubMed] [Google Scholar]
- 50.Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho J, Murray TS, Iwasaki A. Microbiota regulates immune defense against respiratory tract influenza a virus infection. Proc Natl Acad Sci USA 2011;108:5354–5359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Vrankrijker A, Wolfs TF, van der Ent CK. Challenging and emerging pathogens in cystic fibrosis. Paediatr Respir Rev 2010;4:246–254 [DOI] [PubMed] [Google Scholar]
- 52.Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 2010;32:815–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.