Abstract
Toll–IL-1 receptor (TIR) domain-containing adaptor molecule-1 (TICAM1, also called TRIF) is an important adaptor protein in TLR3 and TLR4 signaling pathways that mediate proinflammatory cytokine and IFN responses. Negative regulation of TICAM1 by exogenous viral protease or by endogenous caspase and proteasome have been reported to shut down TICAM1-mediated signaling. In this study, we discovered that down-regulation of TICAM1, but not other components in this signaling pathway, occurred in a natural process of TLR3 activation induced by double-stranded RNA or human rhinovirus (RV) infection in airway epithelial cells and various other cell types. TICAM1 was essential for IFN expression, and the loss of TICAM1 significantly elevated RV production. The low level of TICAM1 protein expression, caused by the prior double-stranded RNA treatment, led to a lack of IFN production upon additional treatment, suggesting receptor desensitization. In follow-up studies, TICAM1 down-regulation was found to be dependent on TLR3 but not RIG1, MDA5, or PKR and appeared to be regulated post-translationally. Neither proteasome nor caspase inhibitors could prevent TICAM1 down-regulation. Instead, a lysosome-mediated process appeared to be involved, suggesting a novel mechanism that is different from previous reports. In conclusion, TICAM1 down-regulation is an essential step in TLR3 activation, and its function is to stop TLR3-mediated IFN production.
Keywords: TLR3, airway, desensitization, TICAM1, virus
Clinical Relevance
The present report describes a novel TLR3 desensitization step. It significantly advances our understanding of respiratory antiviral defense and facilitates the development of effective treatment of viral-induced respiratory diseases and their exacerbations.
Toll-like receptor (TLR) signaling is crucial for activation of innate and adaptive immunity. There are at least 10 expressed human TLRs, and they share similarities in extracellular and intracellular domains (1). When engaging their cognate ligands, TLRs dimerize, triggering recruitment of intracellular proteins and initiating signaling (2). The molecular basis of this recruitment and the subsequent signaling depends on the conserved part of the intracellular domain, called the Toll/IL-1 receptor (TIR) domain, on these proteins (TLR and other participating proteins) (2). Upon TLR dimerization, a TIR–TIR interaction immediately forms to further recruit other TIR-containing proteins (2). The initial TIR-interacting proteins are called adaptors. Five adaptors have been identified: myeloid differentiation primary response protein 88 (MYD88), TIR domain-containing adapter (TIRAP, also called Mal), TIR-containing adapter molecule-1 (TICAM1, also called TRIF), TRIF-related adapter molecule (TRAM, also called TICAM2), and sterile α and HEAT-Armadillo motifs (SARM) (2). Recruiting different adaptors, thus forming different downstream signaling proteins, is responsible for the diversified biological responses induced by TLR. Among these adaptors, MYD88 and TICAM1 are the most studied, and TLR pathways can be essentially categorized as MYD88 dependent or TICAM1 dependent (2). The downstream signaling pathways mediated by MYD88 include activations of NF-κB, p38, and JNK, which further lead to proinflammatory cytokine production (3). The downstream effect of TICAM1 is more complicated, and the activation of IFN production has been mainly attributed to this adaptor (3). TLR3 and TLR4 use TICAM1 as an adaptor. TLR4 activation recruits MYD88 and TICAM1, whereas TLR3 activation appears to recruit only TICAM1. The interaction between TLR4 and TICAM1 is indirect and is mediated through another adaptor-TRAM, but the interaction between TLR3 and TICAM1 is thought to be direct (2, 3).
Airway epithelium is the first line of defense against viral infection, and it is the initial infection site and the route of entry of viral pathogens. In the airway, viral infections directly cause illnesses and exacerbate existing chronic diseases, such as asthma (4), chronic obstructive pulmonary disease (5), and cystic fibrosis (6). Because TLR3 recognizes double-stranded (ds)RNA, a viral replicating mimicker, it has been implicated in antiviral defense in a variety of model systems (7). In the airway, TLR3 has been implicated in the infections of influenza (8, 9) and respiratory syncytial virus (8, 10). We and others have also reported TLR3 activation in human rhinovirus (RV) infection (11–13). Most recently, TLR3 signaling was demonstrated as the earliest activated pathway by RV infection to further regulate the downstream RIG1 and MDA5 pathways (14). Although TICAM1 was not explicitly examined in the report, its function as the sole adaptor to TLR3 suggests that TICAM1 may play important role in anti-RV defense.
The importance of antiviral systems (e.g., TLR3-TICAM1) is usually in parallel with the evasive mechanism used by the viruses. Serine protease-NS3/4A from hepatitis C virus was the first to be demonstrated to cleave and degrade TICAM1, thereby inhibiting IFN production (15). Recently, two endogenous mechanisms have been reported to terminate TLR3-initiated signaling by cleaving and/or degrading TICAM1: (1) the activation of caspases (16) and (2) the activation of 26S proteasome (17). Thus, regulation at the level of TICAM1 may be a new and important point to control TLR-induced signaling. In this report, we have discovered a novel negative regulatory mechanism that occurred in the natural process of TLR3 activation that differs from previously reported mechanisms.
Materials and Methods
Viruses, Chemicals, Inhibitors, and Antibodies
RV16 stock was amplified and purified based on the previous published protocol (18). Viral titers were determined by plaque assay (18). The brief protocols of viral purification and the sources of chemicals, inhibitors, and antibodies are described in the online supplement.
Cell Culture, dsRNA Treatment, and RV Infection
Human tissues were obtained from the National Disease Research Interchange or from the University Medical Center. The University of Arizona Institutional Review Board approved all procedures involved in tissue procurement. We have previously successfully established primary airway tracheobronchial epithelial (TBE) cell cultures (19, 20). The detailed conditions for the primary cells and cell lines are described in the online supplement. dsRNA or RV infection was performed based on our protocol as described previously (12, 21). Specific conditions and timing are described in the figure legends.
Real-Time PCR
Real-time PCR was performed as described previously (22). The primers are listed in Table 1. The details are described in the online supplement. The results were usually calculated as fold induction over control as described previously (12), except for the RV-positive stranded RNA. Because of the lack of RV RNA in the control, the real-time result was presented in relative abundance as described above and as described in our previous report (21).
TABLE 1.
REAL-TIME PRIMERS
| Gene | Primers |
| IFN-β | Forward: ATTGCCTCAAGGACAGGATG |
| Reverse: GCTGCAGCTGCTTAATCTCC | |
| IFN-λ1 | Forward: GGACGCCTTGGAAGAGTCACT |
| Reverse: AGAAGCCTCAGGTCCCAATTC | |
| RV16 | Forward: GCTGTGCAGTTGGATGTGAT |
| Reverse: AAAGCCATGATGCAATCTCC | |
| TLR3 | Forward: ATTGGGCAAGAACTCACAGG |
| Reverse: AGCATCAGTCGTTGAAGGCT | |
| TRIF | Forward: ACTGAACGCAGCCTACTCAGC |
| Reverse: ATGACATGTGGCTCCCAAAAG | |
| GAPDH | Forward: CAATGACCCCTTCATTGACC |
| Reverse: GACAAGCTTCCCGTTCTCAG |
Flow Cytometry
A total of 1 × 105 cells were left alone (nonpermeable) or made permeable by treatment with Triton X-100. Then, the cells were stained with 1 mg/ml anti-TLR3 antibody (clone TLR3.7; eBioscience Inc.) or mouse IgG1 isotype control (eBioscience Inc., San Diego, CA) for 1 hour. A second antibody conjugated with Alexa488 (Invitrogen, Carlsbad, CA) was used to detect the positively stained cells. The cells were then fixed with 1% paraformaldehyde and analyzed on a BD LSR Cytometer (BD Biosciences, San Jose, CA) measuring 10,000 cells. The data were expressed as the mean fluorescence intensity of anti-TLR3–stained cells minus the mean fluorescence intensity of isotype control–stained cells.
Small Interference RNA and Transient Transfection
Control small interference RNA (siRNA) was purchased from Ambion (Austin, TX). siRNA against TLR3 (GGTATAGCCAGCTAACTAGAA) (12), TRIF (GACCAGACGCCACTCCAAC) (23), RIG1 (GGAAGAGGTGCAGTATATT) (15), and MDA5 (GGTGAAGGAGCAGATTCAG) (15) were synthesized by Ambion (Austin, TX). siRNA was transfected into cells using lipofectamine 2000 (Invitrogen) based on the manufacturer's instructions. Successful knockdown of the target was confirmed by real-time RT-PCR and Western blot.
Live Cell Imaging
Cells were grown on 35-mm glass-bottom dishes for live cell imaging and transfected with TICAM1-CFP. At 12 hours after transfection, cells were gently washed once with 1× PBS, and phenol red-free medium was added. Lysozyme was highlighted with lysoTracker (Invitrogen). All images were acquired using confocal microscope (LSM 510 meta; Carl Zeiss, Thornwood, NY).
Statistical Analysis
Experimental groups were compared using a two-sided Student's t test, with significance level set at P < 0.05. When data were not distributed normally, significance was assessed with the Wilcoxon matched-pairs signed-ranks test, and P < 0.05 was considered to be significant. When multiple groups were compared, ANOVA with Tukey–Kramer method was used. The analyses were assisted by biostatisticians in Integrative Health Sciences Facility Core of the Southwest Environmental Health Science Center (http://swehsc.pharmacy.arizona.edu/integrative/areas/biostatistics.php).
Results
dsRNA Treatment Dramatically Down-Regulated TICAM1 Levels in Airway Epithelial Cells and in Various Other Cell Types
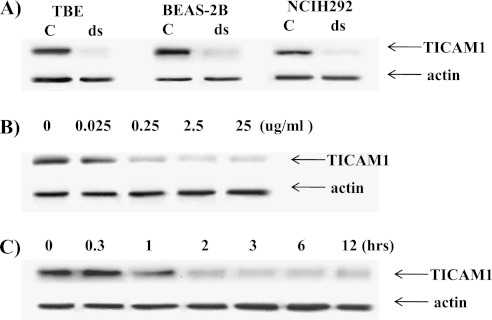
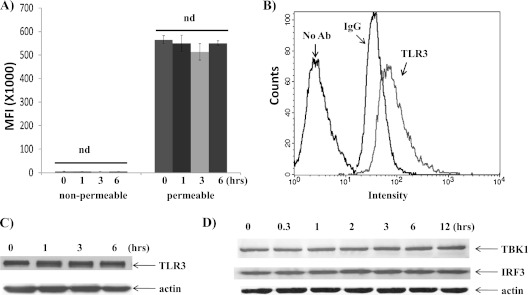
To characterize epithelial dsRNA response, we examined the expressions of various components of TLR signaling. The treatment of dsRNA dramatically down-regulated TICAM1 in two epithelial cell lines (BEAS-2B and NCIH292) and in primary cells (TBE) (Figure 1A). The reduction appeared to be time dependent (Figure 1B) and dose dependent (Figure 1C). The half-life of TICAM1 under 25 μg/ml dsRNA treatment in NCIH292 appeared to be approximately 1 hour. The other major components in this signaling pathway were not significantly changed. TLR3 expression was determined in permeable and nonpermeable conditions. The TLR3 staining signal under nonpermeable conditions mainly came from the cell surface, whereas the signal under permeable conditions came from the intracellular compartment. In epithelial cells, TLR3 expression was largely confined in the intracellular compartment and appeared not to be affected by dsRNA treatment (Figure 2A). An example histogram is included to show the antibody specificity (Figure 2B). To confirm the flow data, we analyzed total TLR3 protein using Western blot by a different antibody (see Materials and Methods). There was a slight, but insignificant, increase of TLR3 upon dsRNA treatment (Figure 2C). Nonetheless, dsRNA did not down-regulate TLR3 (Figures 2A and 2C). Similarly, the total protein level of TBK1 and IRF3 (two downstream signal components) were not affected by dsRNA treatment (Figure 2D). Therefore, this down-regulation at the protein level must not be caused by the outright cellular cytotoxicity and appeared to be limited to TICAM1. The specificity of TICAM1, TBK1, and IRF3 antibodies were confirmed by siRNA (see Figures E1 and E2 in the online supplement). To determine whether or not TICAM1 down-regulation existed only in airway epithelial cells, we examined two other major lung cell types: primary lung fibroblast and primary lung macrophages. dsRNA decreased TICAM1 level in these cells (Figures 3A and 3B). Thus, TICAM1 down-regulation by dsRNA appeared not to be an epithelial-specific, but rather a universal, phenomenon. To further understand the consequence and mechanism of this finding, we focused on the airway epithelial cells in a follow-up study.
Figure 1.
Double-stranded RNA (dsRNA) down-regulated TICAM1 in airway epithelial cells. (A) The cells were treated with dsRNA at 25 μg/ml for 3 hours, and protein was collected for Western blot analysis of TICAM1. Antiactin antibody was used as a control for equal loading. TBE = primary tracheobronchial epithelial cells. BEAS-2B is a T-antigen immortalized human airway bronchial epithelial cell line; NCIH292 is a human mucoepidermoid carcinoma cell line. (B) NCIH292 cells were treated with different dosages of dsRNA for 3 hours, and protein was collected for Western blot analysis. (C) NCIH292 cells were treated with 25 μg/ml of dsRNA for different times as indicated, and protein was collected for Western blot analysis. Actin was used as a loading control. All images are representative of at least three independent experiments.
Figure 2.
(A) Flow cytometry analysis of TLR3 expression by dsRNA treatments. The nonpermeable condition was used to determine the cell surface expression, and the permeable condition was for intracellular expression. MFI = mean fluorescence intensity (n = 5); nd = no difference. (B) An example of histogram of flow cytometry under permeable condition. IgG = isotype-matching (IgG1) antibody control; No Ab = no antibody control; TLR3 = TLR3.7 antibody staining. (C) Western blot analysis of TLR3 protein levels. (D) Western blot analyses of TBK1 and IRF3 protein levels. The cells were treated with 25 μg/ml of dsRNA for different times as indicated, and protein was collected for Western blot analysis. Actin was used as a loading control. All the images are representative of at least three independent experiments.
Figure 3.
dsRNA down-regulated TICAM1 in other cell types. (A) Primary lung fibroblasts. (B) Primary lung macrophages. The cells were treated with dsRNA at 25 μg/ml for 3 hours, and protein was collected for Western blot analysis of TICAM1. Antiactin antibody was used as a control for equal loading. All images are representative of at least three independent experiments.
RV Could Down-Regulate TICAM1 level, and TICAM1 Was Required to Induce IFN Production and Anti-RV Response
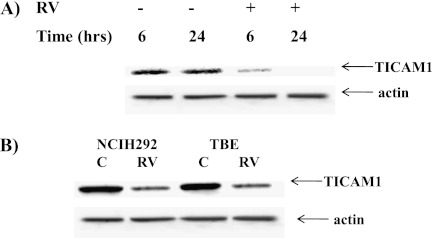
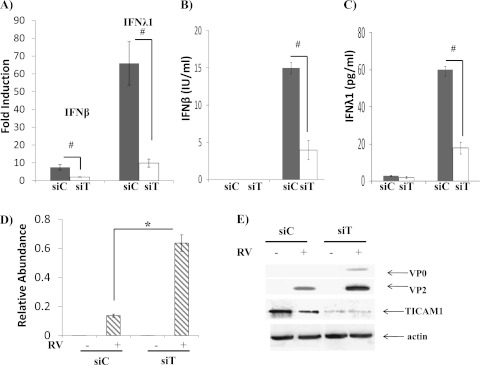
Because dsRNA was used as a viral replicating mimicker, the next question was if the down-regulation of TICAM1 could be triggered by real viral infection. We and others have reported before that human RV could activate the TLR3 pathway (11–13) in airway epithelial cells. Thus, we hypothesized that infecting cells with RV could down-regulate TICAM1. Because HeLa cells were primarily used to generate RV stock, we first tested the time course of RV infection in these cells and found that the TICAM1 down-regulation was started at 6 hours after RV infection and was completed at 24 hours after RV infection (Figure 4A). In addition, only replicating virus could down-regulate TICAM1 (Figure E3). We then tested NCIH292 cells and primary TBE cells at 24 hours after RV infection. TICAM1 was indeed significantly decreased in these cells (Figure 4B), but the magnitude was not as significant as in HeLa cells, which may be caused by the reduced infectivity of RV in these cells as compared with HeLa cells (24). Nonetheless, RV infection was also able to down-regulate TICAM1 in epithelial cells. For easy cell handling and gene manipulation, we used both BEAS-2B and NCIH292 epithelial lines in the subsequent studies to understand the consequence and mechanism of this down-regulation. All the data were identical in both cell lines. Thus, for clarity, only NCIH292 data are presented. To determine the consequence of this observation, we examined the role of TICAM1 in RV infection. RV significantly induced IFN gene expressions (i.e., IFN-β and -λ1) (Figure 5A) as well as the protein secretions of IFN-β (Figure 5B) and IFN-λ1 (Figure 5C). TICAM1 knockdown dramatically reduced IFN inductions (Figures 5A–5C), which highly elevated RV production as measured by the increase of RV genomic RNA (Figure 5D) and of the mature RV coat proteins VP0 and VP2 (Figure 5E) (21). These observations were the first to demonstrate the critical role of TICAM1 in anti-RV defense and were consistent with the previous reports regarding the pivotal role of its upstream receptor (i.e., TLR3) (14) or downstream effectors (i.e., IFN-β and IFN-λ) (21, 25, 26) in anti-RV defense.
Figure 4.
Rhinovirus (RV) down-regulated TICAM1 in HeLa, NCIH292, and primary TBE cells. (A) RV16 (MOI = 10) was used to infect HeLa cells for 6 and 24 hours. Cellular protein was collected for Western blot analysis of TICAM1. (B) RV16 was used to infect NCIH292 cells or TBE cells for 24 hours. Antiactin antibody was used as a control for equal loading. All the images are representative of at least three independent experiments.
Figure 5.
TICAM1 was required for IFN production and epithelial anti-RV response. RV-induced IFN expressions were dependent on TICAM1. Control siRNA (siC) and TICAM1 siRNA (siT) were transfected into NCI-H292 cells. Twenty-four hours later, NCIH292 cells were infected with RV16 at MOI = 10 for 24 hours. RV was then thoroughly washed out as described elsewhere (21). Total cellular RNA, protein, and media were collected 24 hours later. (A) IFN-β and IFN-λ1 expressions were determined by real-time PCR. The data are presented as the fold inductions comparing infected RV with mock (saline)-infected control cells. #P < 0.05 when comparing siC and siT (n = 5). (B) ELISA was used to determine the concentration of IFN-β in culture media. The data are presented as IU/ml. #P < 0.05 (n = 5). (C) ELISA was used to determine the concentration of IFN-λ1 in culture media. The data are presented as pg/ml. #P < 0.05 (n = 5). (D) RV-positive strand RNA was determined by real-time PCR. The data are presented as the relative abundance due to the lack of RV in mock-infected cells as described. #P < 0.05 when siC-transfected cells were compared with siT-transfected cells (n = 5). (E) Western blot analysis was used to determine TICAM1 and RV coat proteins (VP0 and VP2) as described elsewhere (21). All images are representative of at least three independent experiments.
TICAM1 Down-Regulation Desensitized TLR3
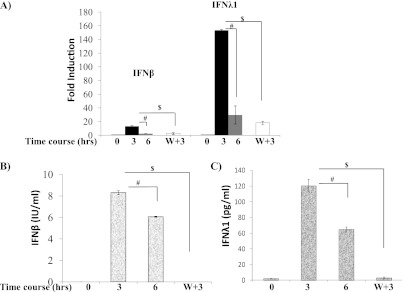
Because TICAM1 is required for IFN production by dsRNA (Figure E1) and by RV (Figures 5A–5C), we reasoned that the decrease of TICAM1 by dsRNA (see Figure 1) might lead to cellular nonresponsiveness to additional dsRNA treatment. IFN gene expressions (Figure 6A) and protein secretions (Figures 6B and 6C) were significantly increased at 3 hours and decreased at 6 hours, which suggests the existence of a negative regulatory mechanism. After 6 hours, retreating the cells with dsRNA could not reactivate IFN expression at the mRNA (Figure 6A) and protein (Figures 6B and 6C) levels. The lack of IFN production upon additional challenge was consistent with the down-regulation of TICAM1 (see Figure 1).
Figure 6.
Significant loss of TICAM1 desensitized TLR3. (A) NCIH292 cells were treated with 25 μg/ml of dsRNA for 0, 3, and 6 hours as indicated in the figure. W+3: The cells were thoroughly washed after dsRNA treatment for 6 hours. Then, the cells were rechallenged with dsRNA for an additional 3 hours. IFN-β and IFN-λ1 expressions were determined by real-time PCR. The data are presented as the fold inductions comparing dsRNA-treated with saline (dsRNA solvent)-treated control cells. #P < 0.05 when the samples treated with dsRNA for 3 hours were compared with samples treated for 6 hours (n = 5). $P < 0.05 when 3-hour treatments were compared with W+3-hour treatments (n = 5). (B) NCIH292 cells were treated based on the protocol as described in A. Culture medium was collected, and ELISA assay was used to determine the concentration of IFN-β. The data are presented as IU/ml. #P < 0.05 when the samples treated with dsRNA for 3 hours were compared with samples treated for 6 hours (n = 5). $P < 0.05 when 3-hour treatments were compared with W+3-hour treatments (n = 5). (C) NCIH292 cells were treated based on the protocol as described in A. Culture medium was collected, and ELISA assay was used to determine the concentration of IFN-λ1. The data are presented as pg/ml. #P < 0.05 when the samples treated with dsRNA for 3 hours were compared with samples treated for 6 hours (n = 5). $P < 0.05 when 3-hour treatments were compared with W+3-hour treatments (n = 5).
TICAM1 Down-Regulation Was Dependent on TLR3 but Not on Secreted IFNs
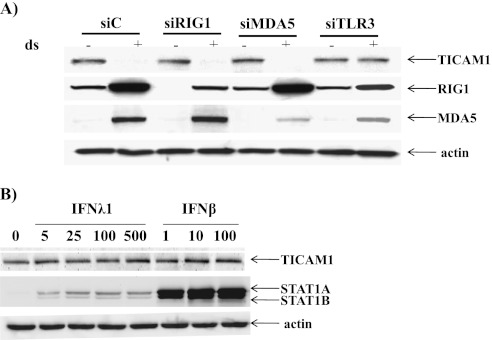
dsRNA primarily activates TLR3-mediated signaling (2); therefore, TICAM1 down-regulation might also depend on TLR3. Indeed, knockdown of TLR3 by siRNA could restore the TICAM1 level (Figure 7A). In contrast, the knockdown of RIG1 or MDA5 had no effect on TICAM1 (Figure 7A). The basal levels of RIG1 and MDA5 were very low, and dsRNA treatment significantly increased the levels of both proteins (Figure 7A). This finding was consistent with the recent report regarding the low level of RIG1 and MDA5 in noninfected airway epithelial cells (14). Because IFN autocrine/paracrine plays an important role in dsRNA/RV-induced gene expression (21), we tested if IFN alone could be responsible for TICAM1 down-regulation. Even at a very high concentration (100 IU/ml for IFN-β and 500 ng/ml for IFN-λ1), neither IFN-β nor -λ1 could down-regulate TICAM1 (Figure 7B). Based on the activation (phosphorylation) of STAT1, IFN-β, even at 1 IU/ml, could significantly activate STAT1 and appeared to be significantly stronger than IFN-λ1. However, this discrepancy may merely reflect the quality of these two IFN preparations. The commercially available IFN-β was sold by the functional unit (IU), whereas IFN-λ1 was sold by protein content (ng). The weight-based activity of IFN-λ1 was unclear. Thus, whether or not IFN-β is truly stronger than IFN-λ1 requires further investigation.
Figure 7.
TICAM1 down-regulation was dependent on TLR3 but not on IFNs. (A) TLR3 was required for TICAM1 down-regulation. NCI-H292 cells were transfected with control siRNA (siC), TLR3 siRNA (siTLR3), RIG1 siRNA (siRIG1), and MDA5 siRNA (siMDA5). Twenty-four hours later, NCI-H292 cells were treated with 25 μg/ml of dsRNA for 3 hours, and cellular protein was collected for Western blot analysis of TICAM1, RIG1, and MDA5. Actin was used as the loading control. (B) IFN was not responsible for TICAM1 down-regulation. Cells were treated with increasing doses of IFN-λ1 (5, 25, 100, and 500 ng/ml) or IFN-β (1, 10, and 100 IU/ml) for 3 hours. Total cellular proteins were collected for Western analyses of TICAM1, phosphorylated (activated) STAT1A, and STAT1B. Actin was used as a loading control. All images are representative of at least three independent experiments.
TICAM1 Down-Regulation Was Likely to Be Caused by a Novel Post-Transcriptional Mechanism
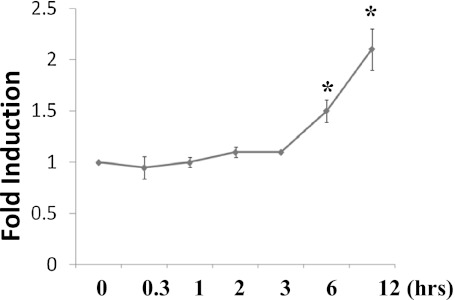
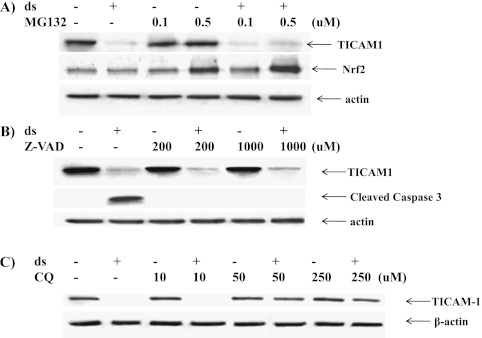
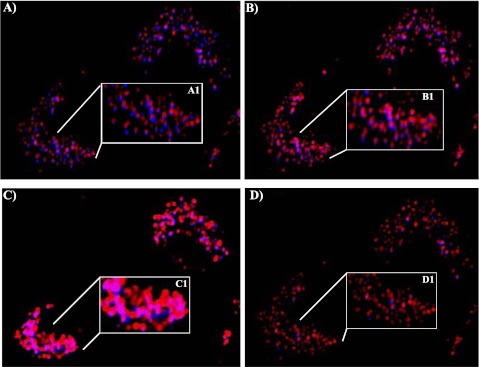
We tested whether TICAM1 down-regulation was caused by the decrease of gene transcription. dsRNA treatment increased TICAM1 mRNA in a significant but limited manner at 6 and 12 hours after treatment (Figure 8), suggesting that the decrease of TICAM1 was not controlled at a transcriptional level. Because caspase (16) and 26S proteasome (17) have been reported to cleave and degrade TICAM1, we tested MG132 (proteasome inhibitor) and zVAD (caspase inhibitor). Both inhibitors were proven functional in the previous studies (16, 17), but neither inhibitor could prevent the TICAM1 reduction by dsRNA treatment (Figures 9A and 9B). To verify that both inhibitors were functional in our system, we tested their documented targets. MG132 has been shown to increase cellular Nrf2 level by preventing its proteasome-mediated degradation (27). zVAD is a classical inhibitor for caspase activation (16). As expected, MG132 successfully prevented Nrf2 degradation (Figure 9A), and zVAD blocked caspase activation (Figure 9B), suggesting that both inhibitors worked well in our system. Thus, the down-regulation of TICAM1 appeared not to be mediated by the previously reported mechanisms. After screening various inhibitors, chloroquine, a lysozyme blocker, was the only one able to prevent TICAM1 down-regulation in a dose-dependent manner (Figure 9C). Consistently, we found that TICAM1 was originally located adjacent to lysozyme under the nontreated condition (Figure 10A) but started to translocate into lysozyme after 20 minutes of dsRNA treatment (Figure 10B). The lysozymal compartment appeared to be significantly expanded by dsRNA treatment (Figures 10B and 10C). At 1 hour, most TICAM1 proteins were in lysozymes (Figure 10C). At 3 hours, TICAM1 was mostly degraded (Figure 10D). Thus, lysozymal degradation appeared to be responsible for TICAM down-regulation by dsRNA treatment.
Figure 8.
TICAM1 mRNA was increased by dsRNA treatment. Cells were treated with dsRNA, and TICAM1 mRNA was collected at various time points as indicated and determined by the real-time PCR. *P < 0.05 (n = 5).
Figure 9.
(A) TICAM1 down-regulation was not affected by proteasome inhibition. NCI-H292 cells were treated with MG132 at the indicated dose or with DMSO (solvent control) for 1 hour, and then dsRNA or saline (in control) was added. Cellular protein was collected 3 hours later for Western blot analysis of TICAM1 and Nrf2, respectively. (B) TICAM1 down-regulation was not affected by caspase inhibition. NCIH292 cells were treated with zVAD at the indicated dose or DMSO (solvent control) for 1 hour, and then dsRNA or saline (in control) was added. Cellular protein was collected 3 hours later for Western blot analysis of TICAM1 and cleaved caspase 3 (indication of caspase 3 activation). (C) TICAM1 down-regulation was prevented by chloroquine (CQ). NCIH292 cells were treated with CQ at the indicated dose or DMSO (solvent control) for 1 hour, and then dsRNA or saline (in control) was added. Cellular protein was collected 3 hours later for Western blot analysis of TICAM1. Antiactin antibody was used as a control for loading. All images are representative of at least three independent experiments.
Figure 10.
Analysis of TICAM1 translocation using live cell imaging. TICAM-CFP was transfected into the cells. After 12 hours, dsRNA was treated and recorded at various time points. (A) Nontreated cells, time 0. (B) Twenty minutes after the addition of dsRNA. (C) One hour after the addition of dsRNA. (D) Three hours after the addition of dsRNA. Original magnification: ×20. Two cells were shown in the image (lower left and upper right corners). A1, B1, C1, and D1 were the magnified image from the indicated area in A, B, C, and D, respectively. Blue: TICAM1-CFP. Red: lysozymes labeled with lysoTracker. Purple: the overlap between TICAM1-CFP and lysosome. All images are representative of at least three independent experiments.
Discussion
The importance of the TLR system cannot be overemphasized in innate and adaptive immunity. The adaptor proteins are not only responsible for determining which pathway will be activated in response to various TLR ligands; they also play an essential role in the regulation of the TLR signaling cascade. Abundant information about the regulation of MYD88-depedent signaling pathway, particularly the one to activate NF-κB, has become available in recent years (2, 28). In contrast, the study on the regulation of TICAM1-dependent TLR signaling, particularly at the level of TICAM1, is relatively limited. The PIAS (Protein Inhibitors of Activated STAT) family was first found to interact with TICAM1, IRF3, and IRF7 to inhibit the activation of the IFN-sensitive response element (29). Exogenous vaccinia viral product-A46R has been shown to interact with TICAM1 and to inhibit IRF3 activation (30). Another layer of control is to directly decrease the protein level of TICAM1, thereby inhibiting the downstream signaling (mainly IFN production). Hepatitis C virus serine protease NS3/4A was found to shut off TLR3-mediated antiviral defense by directly cleaving and degrading TICAM1 (15). In addition, cellular caspases were found to cleave and degrade TICAM1 during dsRNA-induced apoptosis (16), suggesting that targeting TICAM1 may be a common mechanism to shut down TLR3 signaling. To reinforce this notion, integrin αM has recently been shown to negatively regulate TLR3 and TLR4 signaling by directly degrading TICAM1 through the 26S proteasome-mediated mechanism (17). Therefore, down-regulating the adaptor (i.e., TICAM1) may be an emerging regulatory mechanism of TLR signaling.
In the present study, we have shown that TICAM1 down-regulation is a natural step in the process of TLR3 activation. In the previous report using polio viral infection, TICAM1 (or TRIF) was not found to be cleaved by polioviral-induced caspase activation; rather, these caspases cleaved Cardif, another adaptor protein downstream of RIG1 and MDA5 (16). In this report, we found that RV, belonging to the same Piconavirus family as polio virus, could significantly down-regulate TICAM1. Although RV genome encodes several proteases, their activations are regulated autonomously through autosplicing of the polyprotein that is translated from RV genomic RNA (31). Considering the requirement of cellular TLR3 in TICAM1 down-regulation, these RV-derived proteases were less likely to be involved. In addition, we have demonstrated that the lack of TICAM1 could substantially reduce dsRNA/RV-induced IFN expression and enhance RV production. Although the importance of TICAM1 was implied in the previous report (14), we are the first to provide the experimental evidence that TICAM1 was indeed indispensable in anti-RV defense. Thus, RV, a TLR3-TICAM1–activating virus, appeared to take advantage of this step, in which TICAM1 was severely down-regulated, to escape the cellular antiviral immunity.
TLR3 appeared to be desensitized by the lack of IFN production after the first round of activation, which was clearly correlated with TICAM1 down-regulation, suggesting that it may play a potentially significant role in this process. Although we have shown that the protein levels of other key components (i.e., TLR3, TBK1, and IRF3) were not significantly changed, our test was not exhaustive. It is also possible that other alterations, which were not at protein expression level (e.g., TBK1 activity, protein–protein interaction), may be responsible for this desensitization. Thus, further study is needed to determine the precise mechanism of this desensitization. Desensitization is a ubiquitous biological process to shut off the signal. It has been well characterized in G-protein–coupled receptor signaling (32) and growth factor receptor signaling (33). However, the studies on TLR desensitization are limited. Desensitization of TLR2, -4, and -6 has been reported (34, 35), but the underlying mechanisms are unknown. Except for some speculations (36), there has been no study on the desensitization of nucleotide-sensing TLRs (TLR3, -7, -8, and -9). Thus, this is the first report to describe the desensitization of this type of TLR. Our results suggest that the biological function of TLR3 desensitization was perhaps to limit IFN production, which could induce cell growth inhibition and cell death if being excessively produced. However, the loss of IFN response might also open a window for secondary infection. Further study using in vivo models is ongoing to elucidate this process.
To search for the potential mechanism of dsRNA-induced TICAM1 down-regulation, we have first excluded the possibility of transcriptional control by demonstrating that the level of TICAM1 mRNA was not decreased but actually increased. Although we cannot exclude the possibility that the protein translation of TICAM1 might be decreased, the chances are that TICAM1 down-regulation may be controlled at a posttranslational level through protein turnover similar to the previously reported mechanisms (16, 17). However, two inhibitors from the previous reports (16, 17) had no effect. The treatment of dsRNA was able to induce caspase activation and apoptosis, but the inhibition using zVAD failed to prevent TICAM1 down-regulation. Similarly, MG132, a proteasome inhibitor, could not restore the TICAM level. Further studies using lysozyme blocker and immunofluorescence have demonstrated that TICAM1 appeared to enter and be degraded in the lysozymal compartment after dsRNA treatment. Thus, this is a novel mechanism that negatively regulates TLR3–TICAM1 signaling. TLR3 is primarily located in the intracellular organelles (e.g., endosome) (37). This is in contrast to RIG1 and MDA5, which are present in the cytosol. Consistently, we have shown that TICAM1 down-regulation was dependent on TLR3 but not on RIG1 or MDA5. In addition, although autocrined/paracrined IFN has been shown to affect dsRNA/RV-induced gene expression (21), it did not affect TICAM1 down-regulation. The endolysosomal compartment is one of the major cellular protein degradation factors (38). Thus, due to the proximity of TICAM1 to this compartment, it is not surprising that the endolysosomal proteases were responsible for its protein turnover. Lysozymal proteases consist of serine, cysteine, and aspartic proteases (38). For the two previously reported TICAM1 proteases, NS3/4A is a serine protease, and caspase is a cysteine protease. Thus, similar endolysosomal proteases may cleave and degrade TICAM1, which needs further study.
In summary, we have discovered a novel negative control of TLR3 signaling through TICAM1 down-regulation. Complete elucidation of the underlying mechanism and the biological consequence of this regulation will advance our understanding of TLR3 in innate and adaptive immunity against pathogen infections.
Supplementary Material
Acknowledgments
The authors thank Ms. Erika C. Barrett for editing the manuscript.
Footnotes
Supported by National Institutes of Health grant RO1AI061695 and by an Arizona Research Biomedical Commission pilot grant.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2011-0340OC on December 30, 2011
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Roach JC, Glusman G, Rowen L, Kaur A, Purcell MK, Smith KD, Hood LE, Aderem A. The evolution of vertebrate toll-like receptors. Proc Natl Acad Sci USA 2005;102:9577–9582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brikos C, O'Neill LAJ. Signalling of toll-like receptors. In: S. Bauer GH, editor. Toll-like receptors (TLRs) and innate immunity. Berlin: Springer; 2008. pp. 21–50.
- 3.Watters TM, Kenny EF, O'Neill LA. Structure, function and regulation of the Toll/IL-1 receptor adaptor proteins. Immunol Cell Biol 2007;85:411–419 [DOI] [PubMed] [Google Scholar]
- 4.Busse WW, Lemanske RF, Jr, Gern JE. Role of viral respiratory infections in asthma and asthma exacerbations. Lancet 2010;376:826–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Varkey JB, Varkey B. Viral infections in patients with chronic obstructive pulmonary disease. Curr Opin Pulm Med 2008;14:89–94 [DOI] [PubMed] [Google Scholar]
- 6.Lipuma JJ. The changing microbial epidemiology in cystic fibrosis. Clin Microbiol Rev 2010;23:299–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vercammen E, Staal J, Beyaert R. Sensing of viral infection and activation of innate immunity by toll-like receptor 3. Clin Microbiol Rev 2008;21:13–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boukhvalova MS, Sotomayor TB, Point RC, Pletneva LM, Prince GA, Blanco JC. Activation of interferon response through toll-like receptor 3 impacts viral pathogenesis and pulmonary toll-like receptor expression during respiratory syncytial virus and influenza infections in the cotton rat sigmodon hispidus model. J Interferon Cytokine Res 2010;30:229–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guillot L, Le Goffic R, Bloch S, Escriou N, Akira S, Chignard M, Si-Tahar M. Involvement of toll-like receptor 3 in the immune response of lung epithelial cells to double-stranded rna and influenza a virus. J Biol Chem 2005;280:5571–5580 [DOI] [PubMed] [Google Scholar]
- 10.Rudd BD, Smit JJ, Flavell RA, Alexopoulou L, Schaller MA, Gruber A, Berlin AA, Lukacs NW. Deletion of tlr3 alters the pulmonary immune environment and mucus production during respiratory syncytial virus infection. J Immunol 2006;176:1937–1942 [DOI] [PubMed] [Google Scholar]
- 11.Hewson CA, Jardine A, Edwards MR, Laza-Stanca V, Johnston SL. Toll-like receptor 3 is induced by and mediates antiviral activity against rhinovirus infection of human bronchial epithelial cells. J Virol 2005;79:12273–12279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu L, Lee PK, Lee WM, Zhao Y, Yu D, Chen Y. Rhinovirus-induced major airway mucin production involves a novel tlr3-EGFR-dependent pathway. Am J Respir Cell Mol Biol 2009;40:610–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Q, Nagarkar DR, Bowman ER, Schneider D, Gosangi B, Lei J, Zhao Y, McHenry CL, Burgens RV, Miller DJ, et al. Role of double-stranded RNA pattern recognition receptors in rhinovirus-induced airway epithelial cell responses. J Immunol 2009;183:6989–6997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Slater L, Bartlett NW, Haas JJ, Zhu J, Message SD, Walton RP, Sykes A, Dahdaleh S, Clarke DL, Belvisi MG, et al. Co-ordinated role of tlr3, rig-i and mda5 in the innate response to rhinovirus in bronchial epithelium. PLoS Pathog 2010;6:e1001178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M, Ray SC, Gale M, Jr, Lemon SM. Immune evasion by hepatitis C virus ns3/4a protease-mediated cleavage of the toll-like receptor 3 adaptor protein trif. Proc Natl Acad Sci USA 2005;102:2992–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rebsamen M, Meylan E, Curran J, Tschopp J. The antiviral adaptor proteins cardif and trif are processed and inactivated by caspases. Cell Death Differ 2008;15:1804–1811 [DOI] [PubMed] [Google Scholar]
- 17.Han C, Jin J, Xu S, Liu H, Li N, Cao X. Integrin cd11b negatively regulates tlr-triggered inflammatory responses by activating syk and promoting degradation of myd88 and trif via cbl-b. Nat Immunol 2010;11:734–742 [DOI] [PubMed] [Google Scholar]
- 18.Wang W, Lee WM, Mosser AG, Rueckert RR. Win 52035-dependent human rhinovirus 16: assembly deficiency caused by mutations near the canyon surface. J Virol 1998;72:1210–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, Zhao YH, Di YP, Wu R. Characterization of human mucin 5b gene expression in airway epithelium and the genomic clone of the amino-terminal and 5′-flanking region. Am J Respir Cell Mol Biol 2001;25:542–553 [DOI] [PubMed] [Google Scholar]
- 20.Wu R, Zhao YH, Chang MM. Growth and differentiation of conducting airway epithelial cells in culture. Eur Respir J 1997;10:2398–2403 [DOI] [PubMed] [Google Scholar]
- 21.Chen Y, Hamati E, Lee PK, Lee WM, Wachi S, Schnurr D, Yagi S, Dolganov G, Boushey H, Avila P, et al. Rhinovirus induces airway epithelial gene expression through double-stranded rna and ifn-dependent pathways. Am J Respir Cell Mol Biol 2006;34:192–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kao CY, Chen Y, Thai P, Wachi S, Huang F, Kim C, Harper RW, Wu R. Il-17 markedly up-regulates beta-defensin-2 expression in human airway epithelium via jak and NF-kappaB signaling pathways. J Immunol 2004;173:3482–3491 [DOI] [PubMed] [Google Scholar]
- 23.Oshiumi H, Matsumoto M, Funami K, Akazawa T, Seya T. Ticam-1, an adaptor molecule that participates in toll-like receptor 3-mediated interferon-beta induction. Nat Immunol 2003;4:161–167 [DOI] [PubMed] [Google Scholar]
- 24.Mosser AG, Brockman-Schneider R, Amineva S, Burchell L, Sedgwick JB, Busse WW, Gern JE. Similar frequency of rhinovirus-infectible cells in upper and lower airway epithelium. J Infect Dis 2002;185:734–743 [DOI] [PubMed] [Google Scholar]
- 25.Contoli M, Message SD, Laza-Stanca V, Edwards MR, Wark PA, Bartlett NW, Kebadze T, Mallia P, Stanciu LA, Parker HL, et al. Role of deficient type III interferon-lambda production in asthma exacerbations. Nat Med 2006;12:1023–1026 [DOI] [PubMed] [Google Scholar]
- 26.Wark PA, Johnston SL, Bucchieri F, Powell R, Puddicombe S, Laza-Stanca V, Holgate ST, Davies DE. Asthmatic bronchial epithelial cells have a deficient innate immune response to infection with rhinovirus. J Exp Med 2005;201:937–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobayashi M, Yamamoto M. Molecular mechanisms activating the nrf2-keap1 pathway of antioxidant gene regulation. Antioxid Redox Signal 2005;7:385–394 [DOI] [PubMed] [Google Scholar]
- 28.Liew FY, Xu D, Brint EK, O'Neill LA. Negative regulation of toll-like receptor-mediated immune responses. Nat Rev Immunol 2005;5:446–458 [DOI] [PubMed] [Google Scholar]
- 29.Zhang J, Xu LG, Han KJ, Wei X, Shu HB. Piasy represses trif-induced isre and NF-kappaB activation but not apoptosis. FEBS Lett 2004;570:97–101 [DOI] [PubMed] [Google Scholar]
- 30.Stack J, Haga IR, Schroder M, Bartlett NW, Maloney G, Reading PC, Fitzgerald KA, Smith GL, Bowie AG. Vaccinia virus protein a46r targets multiple toll-like-interleukin-1 receptor adaptors and contributes to virulence. J Exp Med 2005;201:1007–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Savolainen C, Blomqvist S, Hovi T. Human rhinoviruses. Paediatr Respir Rev 2003;4:91–98 [DOI] [PubMed] [Google Scholar]
- 32.Luttrell LM. Transmembrane signaling by G protein-coupled receptors. Methods Mol Biol 2006;332:3–49 [DOI] [PubMed] [Google Scholar]
- 33.Goldkorn T, Ravid T, Khan EM. Life and death decisions: ceramide generation and egf receptor trafficking are modulated by oxidative stress. Antioxid Redox Signal 2005;7:119–128 [DOI] [PubMed] [Google Scholar]
- 34.Rostron AJ, Cork DM, Avlonitis VS, Fisher AJ, Dark JH, Kirby JA. Contribution of toll-like receptor activation to lung damage after donor brain death. Transplantation 2010;90:732–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Didierlaurent A, Goulding J, Patel S, Snelgrove R, Low L, Bebien M, Lawrence T, van Rijt LS, Lambrecht BN, Sirard JC, et al. Sustained desensitization to bacterial toll-like receptor ligands after resolution of respiratory influenza infection. J Exp Med 2008;205:323–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwok SK, Lee JY, Park SH, Cho ML, Min SY, Kim HY, Cho YG. Dysfunctional interferon-alpha production by peripheral plasmacytoid dendritic cells upon toll-like receptor-9 stimulation in patients with systemic lupus erythematosus. Arthritis Res Ther 2008;10:R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Bouteiller O, Merck E, Hasan UA, Hubac S, Benguigui B, Trinchieri G, Bates EE, Caux C. Recognition of double-stranded rna by human toll-like receptor 3 and downstream receptor signaling requires multimerization and an acidic ph. J Biol Chem 2005;280:38133–38145 [DOI] [PubMed] [Google Scholar]
- 38.Brix K. Lysosomal proteases: revival of the sleeping beauty. : Saftig P, editor Lysosomes. Georgetown, TX, New York, NY: Landes; Bioscience/Eurekah.com; 2005. pp. 259–264 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.