Abstract
Background and Aims
Brassica rapa and B. oleracea are the progenitors of oilseed rape B. napus. The addition of each chromosome of B. oleracea to the chromosome complement of B. rapa results in a series of monosomic alien addition lines (MAALs). Analysis of MAALs determines which B. oleracea chromosomes carry genes controlling specific phenotypic traits, such as seed colour. Yellow-seeded oilseed rape is a desirable breeding goal both for food and livestock feed end-uses that relate to oil, protein and fibre contents. The aims of this study included developing a missing MAAL to complement an available series, for studies on seed colour control, chromosome homoeology and assignment of linkage groups to B. oleracea chromosomes.
Methods
A new batch of B. rapa–B. oleracea aneuploids was produced to generate the missing MAAL. Seed colour and other plant morphological features relevant to differentiation of MAALs were recorded. For chromosome characterization, Snow's carmine, fluorescence in situ hybridization (FISH) and genomic in situ hybridization (GISH) were used.
Key Results
The final MAAL was developed. Morphological traits that differentiated the MAALs comprised cotyledon number, leaf morphology, flower colour and seed colour. Seed colour was controlled by major genes on two B. oleracea chromosomes and minor genes on five other chromosomes of this species. Homoeologous pairing was largely between chromosomes with similar centromeric positions. FISH, GISH and a parallel microsatellite marker analysis defined the chromosomes in terms of their linkage groups.
Conclusions
A complete set of MAALs is now available for genetic, genomic, evolutionary and breeding perspectives. Defining chromosomes that carry specific genes, physical localization of DNA markers and access to established genetic linkage maps contribute to the integration of these approaches, manifested in the confirmed correspondence of linkage groups with specific chromosomes. Applications include marker-assisted selection and breeding for yellow seeds.
Keywords: Brassica rapa var. trilocularis, B. oleracea var. alboglabra, MAALs, characterization of C chromosomes, plant morphology, seed colour control, FISH, GISH, chromosome homoeology, chromosome structural changes, linkage groups, crop plant breeding
INTRODUCTION
The addition of single alien chromosomes or chromosome arms from a related species can add considerable value to the genetic stock of many common crops. This has often been achieved using defined crossing schemes where the dissected alien genome is represented by alien mono-, di- or telosomic additions to the chromosome complement of the host genome. Such interspecific or intergeneric addition lines are suitable materials for fundamental genetic and cytogenetic research, and for use in applied plant breeding (Chang and de Jong, 2005). Stocks of addition lines have been developed for a variety of crop plants. Wheat (Triticum aestivum) carrying barley (Hordeum vulgare) chromosomes has been useful for mapping genes to chromosome arms on the basis of transcript information (Bilgic et al., 2007), and for studies of the morphology and stability of these lines (Szakács and Molnár-Láng, 2010). The contribution of alien chromosomes to phenotype has also been documented in a complete set of oat (Avena sativa) lines carrying maize (Zea mays) chromosomes (Rines et al., 2009). Homoeology between alien and background chromosomes can be monitored by chromosome pairing and intergenomic recombination during meiosis, as documented in the Japanese bunching onion (Allium fistulosum) carrying chromosomes of A. cepa (Barthes and Ricroch, 2001), and in tomato (Solanum lycopersicum) carrying chromosomes of the wild relative Solanum lycopersicoides (Ji and Chetelat, 2003). Transfer of desirable genes of relevance to plant breeding has been the goal after addition of chromosomes of wild relatives to sugar beet (Gao et al., 2001), potato (Dong et al., 2005) and rice (Khush, 2010).
Genome dissection of crop brassicas and related species through the development of monosomic alien addition lines (MAALs) and disomic addition lines (Budahn et al., 2008) is summarized in reviews by Prakash et al. (2009) and Ziolkowski et al. (2011). Assignment of genes to specific chromosomes and transfer of desirable genes have been the main objectives in these studies. Background genomes that hosted individual Brassica oleracea chromosomes (2n = 18, genome CC) were those of B. rapa (2n = 20, genome AA) (Quiros et al., 1987; Chen et al., 1992) and Raphanus sativus (2n = 18) (Kaneko et al., 1987; Budahn et al., 2008). Up to seven out of the nine possible B. rapa–B. oleracea MAALs have been characterized by C genome-specific isozyme and restriction fragment length polymorphism (RFLP) markers, and by documenting gene duplication, intergenomic recombination, alien chromosome transmission and occurrence of deletions (Quiros et al., 1987; McGrath and Quiros, 1990; McGrath et al., 1990; Hu and Quiros. 1991). Using a pair of parent lines distinct from those used by the Quiros group, our work has previously led to the development of eight B. rapa var. trilocularis–B. oleracea var. alboglabra MAALs, with analysis focused on cytological chromosome differentiation and identification, and use of flower colour, seed colour, isozyme and random amplified polymorphic DNA (RAPD) markers for genetic differentiation between lines (Chen et al., 1992, 1997a, b; Cheng et al., 1994a, b, 1995; Jørgensen et al., 1996; Heneen and Brismar, 2001; Heneen and Jørgensen, 2001; Hasterok et al., 2005).
Here we report the development of the previously undeveloped MAAL, and making available all nine MAALs of B. rapa var. trilocularis–B. oleracea var. alboglabra. Although the C chromosome in one of the established lines is represented by only one chromosome arm, this does provide greater resolution for phenotypic traits determined by that arm. The use of C genome- and C chromosome-specific microsatellites (simple sequence repeats, SSRs) as molecular markers was decisive for the characterization of the available MAALs and development of the final MAAL (Geleta et al., 2012). Detailed documentation of seed coat colour for the different MAALs provides grounds for dissection of the genetic factors that control this trait, which is of agronomic interest (Shirzadegan and Röbbelen, 1985; Slominski et al., 1999). Behaviour of the C chromosome during diakinesis of pollen mother cells, and preference when pairing with background A chromosomes, was documented in all lines. The identification of the alien C chromosome was greatly facilitated by applying a new fluorescence in situ hybridization (FISH) technique that allows for integrating the cytogenetic and genetic maps (Xiong et al., 2010, 2011; Xiong and Pires, 2011). Genomic in situ hybridization (GISH) also contributed to the identification of the alien C chromosome. The combined FISH, GISH and SSR data provide a platform for determination of the correspondence between the cytological and linkage group (LG) designations of the C chromosomes.
MATERIALS AND METHODS
Plant material
Eight available Brassica rapa–B. oleracea MAALs (Chen et al., 1992, 1997a; Cheng et al., 1994a; Heneen and Jørgensen, 2001) were derived from a cross between a yellow-flowered and yellow-seeded B. rapa var. trilocularis (yellow sarson, K-151), and a white-flowered and black-seeded B. oleracea var. alboglabra (no. 4003) (Fig. 1). This original cross gave rise to the resynthesized B. napus (no. 7406, genomes AACC) which after backcrossing to B. rapa produced sesquidiploids (genomes AAC). These were selfed or backcrossed to the AA parent and produced progeny of expected aneuploids (AA + 1–9 C chromosomes) and parental AA plants. Aneuploids and their progeny were studied cytologically and with RAPD markers, for the detection and selection of monosomics that were carriers of the different C chromosomes (Fig. 1).
Fig. 1.
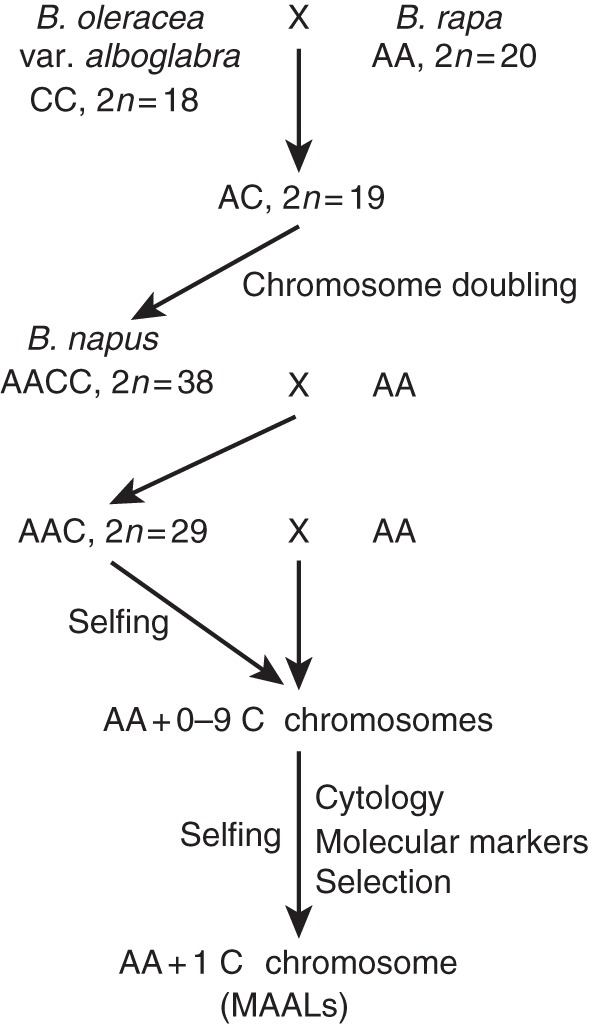
Scheme showing the origin of the developed Brassica MAALs.
Chronologically, and applying the numerical designation system of Cheng et al. (1995), the extra C chromosome was defined as C4 (Cheng et al., 1995), C8 and C9 (Chen et al., 1997a), C1 (Chen et al., 1997b), and C2, C3, C5 and C6/7 (referred to as chromosomes D, E, G and F, respectively; Heneen and Brismar, 2001; Heneen and Jørgensen, 2001). Chromosomes C5, C8 and C9 were later verified as rDNA carriers (Hasterok et al., 2005). During the propagation of this material, C3 lost one chromosome arm, and the maintained MAAL of C3 with this deletion is referred to as C3d. In addition to the maintained MAAL with a seemingly intact C4, a line was developed with a C4 that had a shorter short arm due to a deletion. This MAAL is referred to as C4d. Plants of the parental B. rapa var. trilocularis (K-151) and B. oleracea var. alboglabra (no. 4003) species and their resynthesized B. napus (no. 7406) were also raised. Backcrosses between B. napus and B. rapa were repeated in order to produce new batches of sesquidiploids and aneuploids in an effort to develop the missing MAAL.
Morphology
Morphological features of relevance for the differentiation of monosomics, representing the different C chromosomes were recorded. These primarily included the colour of sown seeds, number and size of cotyledons, even or puckered leaf surface, green or faint anthocyanin coloration of the stem, flower colour, plant and silique size as well as the seed coat colour of harvested seeds.
Chromosome number determination and chromosome nomenclature
Chromosome numbers were determined from meiotic analysis of pollen mother cells. Flower buds were fixed in ethanol–acetic acid (3:1) and stained in Snow's carmine as previously applied on this material (Chen et al., 1992; Cheng et al., 1994b; Heneen and Jørgensen, 2001). Emphasis was placed on studying early diakinesis since differences in chromatin condensation patterns between the A and C chromosomes at this stage were distinctive, permitting the differentiation between these chromosomes (Cheng et al., 1994b; Heneen et al., 1995; Heneen and Jørgensen, 2001).
The cytological numerical designation of the chromosomes of B. rapa (AA) and B. oleracea (CC) is according to our earlier reports (Cheng et al., 1995; Heneen et al., 1995). Identification of the C chromosomes in the developed MAALs was based on size, defined or inferred centromeric position and the heterochromatic nature of the NOR (nucleolar organizer region) and its proximal chromatin. For the sake of simplicity in the following text, AA and CC will be used when referring to the parental genomes and the A-genome and C-genome chromosomes will be referred to as A1–A10 and C1–C9. Another commonly used, and internationally agreed designation system of the A and C chromosomes is based on molecular genetic LGs (Parkin et al., 2005; http://www.brassica.info/resource/maps/lg-assignments.php; J. Wang et al. 2011). This nomenclature system has been integrated with, and consolidated by FISH-based physical localization of DNA probes (Howell et al., 2002; Xiong and Pires, 2011).
The nomenclature system of the C genome (Cheng et al., 1995) is based on arranging the chromosomes in groups depending on centromeric position, and within groups according to size, and so differs from the system based solely on chromosome size applied by Armstrong et al. (1998) and Howell et al. (2002). Similarly, it was not possible to ascertain the correspondence between all chromosomes when comparing the systems of Cheng et al. (1995) and Xiong and Pires (2011). Accordingly, we were not previously able to designate all the C chromosomes of the eight established MAALs in terms of LGs. However, the current FISH and GISH physical mapping of the MAALs, and the molecular SSR characterization of these lines (Geleta et al., 2012) made it possible to determine the correspondence between the hitherto used cytological designations (A1–A10 and C1–C9) according to Cheng et al. (1995) and the LG designations (sensu Parkin et al., 2005) hereafter referred to as LG-A1–LG-A10 and LG-C1–LG-C9. Both cytological and LG designations will be used in the following text.
FISH: multiple target
The use of immature buds as sources of mitotic cells as well as pollen mother cells, the choice of DNA probes and the application of FISH were according to a new technique developed for the characterization and differentiation of Brassica A and C chromosomes (Xiong et al., 2010, 2011; Xiong and Pires, 2011). In this method, genetically mapped bacterial artificial chromosome (BAC) probes of B. rapa were used to identify the locations of repetitive elements in both B. rapa and B. oleracea. A second hybridization was done on the same chromosome spreads using a C genome repeat, giving a result similar to a GISH experiment. The probes used in the first round comprised those specific to 5S and 45S rDNA, repeated DNA sequences in eight chromosome pairs of B. rapa by using BAC KBrB072L17, and repeated DNA sequences specific to two chromosome pairs of B. rapa by using BAC KBrH092N24. The probes used in the second round were those for the repetitive centromeric DNA sequences CentBr1 and CentBr2 and for repetitive DNA sequences that are C genome specific by using BAC BNIH 123L05 which gives a GISH-like labelling. In addition, the BAC KBrH117M18 probe specific to LG-A3 and LG-C3, followed by the C genome-specific probe BAC BNIH 123L05 were used.
GISH: labelled C-genome DNA
GISH alone, and GISH following FISH using the BAC BoB004H11 probe specific to the LG-C4, was carried out by S. Armstrong and E. Howell at the University of Birmingham according to Howell et al. (2008).
RESULTS
Development of the missing MAAL
The backcross between the resynthesized B. napus (no. 7406) and the parental B. rapa (K-151), that led to the development of the eight available MAALs, was repeated to provide new batches of sesquidiploids and aneuploids, in an effort to develop the missing MAAL (Fig. 1). Pollination of 34 flowers of the AACC parent with pollen from the AA parent resulted in 35 seemingly viable well-developed seeds expected to have the AAC constitution. Plants raised from these seeds were either selfed or backcrossed to the AA parent. Selfing of 160 flowers yielded 14 seeds, while backcrossing of 66 flowers resulted in 80 seeds. As expected, more viable zygotes and seeds were produced after pollination with balanced B. rapa pollen in the case of backcrossing than after fusion of frequently produced unbalanced gametes when the AAC plants were selfed. Plants raised from seeds obtained after backcrossing/selfing are largely aneuploids (AA + 1–9 C chromosomes) and to a minor extent parental euploids (AA).
Of the seeds obtained after selfing and backcrossing, and their progeny seeds, a total of 52 were sown, giving rise to 47 viable plants. Of these, 23 comparatively less vigorous plants, possibly representing aneuploids, were chosen for cytological characterization and SSR marker analysis (Geleta et al., 2012). Emphasis was placed, in the first instance, on finding plants labelled by C genome-specific SSR primer pairs that did not label monosomics of the eight available MAALs. Such SSRs, inferred to be possibly specific for the undefined C chromosome, labelled six presumed aneuploid plants. Of these plants, two were sterile and did not yield any seeds. Meiotic chromosome number was estimated on three of the four remaining plants. Two plants had 2n = 23 and one monosomic plant had 2n = 21. The monosomic plant, monosomic progeny of this plant and certain monosomic plants among the progeny of one of the plants with 2n = 23 were shown by SSR analysis to be carriers of the undefined C chromosome (Geleta et al., 2012). This confirmed that we had successfully generated the starting material for the previously unavailable MAAL. The C chromosome in this line has been designated C7, and the previously available line C6/7 is now designated as C6 (see below).
Morphology
A general characteristic of plants carrying additional C chromosomes in all the MAALs studied is decreased vigour, reduced stature and decreased fertility compared with sibling euploid AA plants. This is also reflected in silique size being smaller in monosomics than in the euploid parent B. rapa (Fig. 2A). Other morphological features that discriminate between the parental species and between monosomics carrying different C chromosomes are listed in Table 1. These characters comprise the number of cotyledons, anthocyanin coloration of the stem, the appearance of the leaf surface, flower colour and colour of the seed coat (Fig. 2B–H). Seed coat colour (henceforth referred to as seed colour) is the most valuable character for differentiation between MAALs and between carriers of C chromosomes and euploids in the progeny of the MAALs. Therefore, data on seed colour are presented in more detail (Fig. 3, Table 2).
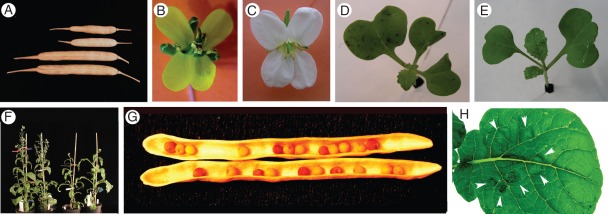
Fig. 2.
Morphological features of relevance for the differentiation of plants that carry the alien C chromosome. (A) General small size of siliques exemplified by siliques of a C2-carrier plant and large siliques of the parental Brassica rapa species. (B) Yellow flower of B. rapa, the colour characteristic of all MAALs except the MAAL for C4. (C) White flower colour characteristic of B. oleracea var. alboglabra and of C4 carriers. (D, E) C1 carriers: (D) three cotyledons; (E) two cotyledons, one almost double the size of the other. (F) Two white-flowered C4 carriers to the left and two smaller yellow-flowered C4d carriers to the right. (G) Eight brown seeds and 14 yellow seeds in a silique of a C4 carrier. (H) Puckered surface of a leaf of a C5 carrier.
Table 1.
Morphological features characteristic of the parental AA and CC species and of monosomics carrying different types of C chromosomes
| Material | No. of cotyledons | Stem anthocyanin | Leaf surface | Flower colour | Colour of seeds produced |
|---|---|---|---|---|---|
| AA | 2 | − | Even | Yellow | Yellow |
| CC | 2 | + | Even | White | Black |
| C1 | 3 | + | Even | Yellow | Dark brown |
| C2 | 2 | − | Even | Yellow | Yellow + yellow with brown spots/patches |
| C3d | 2 | + | Even | Yellow | Yellow + yellow with light brown dots |
| C4 | 2 | + | Even | White | Yellow + brown with dark spots |
| C4d | 2 | + | Even | Yellow | Yellow + faint brown with dark spots |
| C5 | 2 | − | Puckered | Yellow | Yellow + yellow with brown spots/patches |
| C6 | 2 | − | Even | Yellow | Yellow + yellow with brown spots/patches |
| C7 | 2 | − | Even | Yellow | Yellow + yellow with brown spots/patches |
| C8 | 2 | − | Even | Yellow | Yellow |
| C9 | 2 | + | Even | Yellow | Yellow |
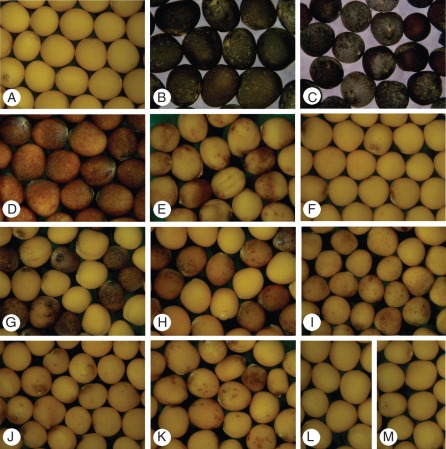
Fig. 3.
Seed colour of the Brassica material studied. (A) Yellow in B. rapa var. trilocularis (K-151); (B) black in B. oleracea var. alboglabra (no. 4003); (C) black, dark grey and brown in resynthesized B. napus (no. 7406); (D) brown seeds harvested from a C1 carrier; (E) selected seeds with brown spots/patches from a C2 carrier; (F) bulk seeds harvested from a C3d carrier; seeds with faint brown dots are difficult to detect by the naked eye; (G, H) mixtures of easily distinguishable yellow and brown seeds originating from C4 and C4d carriers, respectively; the brown seeds are slightly lighter in colour in the case of C4d; (I–K) selected seeds with brown spots/patches from plants carrying C5, C6 and C7, respectively; (L, M) yellow seeds harvested from plants with C8 and C9, respectively.
Table 2.
Frequency of plants producing pigmented seeds and of plants with alien chromosomes in relation to colour of seed of origin, and frequency of pigmented seeds in seed mixtures, in seven MAALs harbouring genes that affect seed colour
| MAAL* |
No. of sown seeds† |
No. of raised plants producing seeds† |
% of plants with pigmented seeds | No. of plants with chromosome numbers |
% of mono- and disomics | No. of harvested seeds† |
% of pigmented seeds | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cyt. | LG | B | S/P | S/P + Y | Y | B | B + Y | S/P + Y | Y | 20 | 21 | 22 | Total | B | S/P | Y | |||
| C1 | C9 | 122 | 37 | 85 | 30·3 | ||||||||||||||
| 9 | 9 | 100·0 | |||||||||||||||||
| 37 | 37 | 0·0 | |||||||||||||||||
| C2 | C1 | 37 | 37 | 100·0 | 1899 | 639 | 1260 | 33·6 | |||||||||||
| 20 | 17 | 3 | 100·0 | ||||||||||||||||
| 15 | 15 | 0·0 | |||||||||||||||||
| 24 | 23 | 1 | 4·2 | ||||||||||||||||
| C3d | C5 | 65 | 22 | 43 | 33·8 | ||||||||||||||
| 10 | 10 | 100·0 | |||||||||||||||||
| 7 | 1 | 5 | 1 | 85·7 | |||||||||||||||
| 10 | 10 | 0·0 | |||||||||||||||||
| 9 | 8 | 1 | 11·1 | ||||||||||||||||
| C4 | C3 | 58 | 56 | 2 | 96·6 | 1832 | 636 | 1196 | 34·7 | ||||||||||
| 51 | 45 | 6 | 100·0 | ||||||||||||||||
| 30 | 1 | 29 | 3·3 | ||||||||||||||||
| 41 | 41 | 0·0 | |||||||||||||||||
| C4d | C3 | 28 | 26 | 2 | 92·9 | 2164 | 795 | 1369 | 36·7 | ||||||||||
| 5 | 5 | 100·0 | |||||||||||||||||
| C5 | C4 | 22 | 22 | 100·0 | 1764 | 573 | 1191 | 32·5 | |||||||||||
| 16 | 1 | 15 | 93·8 | ||||||||||||||||
| 10 | 10 | 0·0 | |||||||||||||||||
| 12 | 10 | 2 | 16·7 | ||||||||||||||||
| C6 | C6 | 29 | 20 | 9 | 69·0 | 2297 | 738 | 1559 | 32·1 | ||||||||||
| 16 | 4 | 12 | 75·0 | ||||||||||||||||
| C7 | C2 | 30 | 19 | 11 | 63·3 | 3298 | 1055 | 2243 | 32·0 | ||||||||||
| 8 | 8 | 100·0 | |||||||||||||||||
| 4 | 4 | 0·0 | |||||||||||||||||
* Cytological (Cyt.) and corresponding linkage group (LG) designations according to Table 4.
† Seeds are brown (B), brown spotted or patchy (S/P), or yellow (Y).
Differences between the parental species regarding the characters mentioned above relate to the yellow flower colour (Fig. 2B) and yellow seed colour (Fig. 3A) specific to the AA parent, and anthocyanin coloration of stem, white flower colour (Fig. 2C) and black seed colour (Fig. 3B) specific to the CC parent. White flower colour and black seed colour are dominant characters. In accordance, resynthesized B. napus from these parental species produces a mixture of black, dark brown and dark grey seeds (Fig. 3C).
MAAL for C1
Dark brown seeds (Fig. 3D) were grown. The occurrence of three cotyledons in some seedlings (Fig. 2D) raised from these seeds has been established to be correlated with the presence of C1. These plants produce dark brown seeds. The chromosome number 2n = 21 and the presence of C1 prevailed in 15 tested plants originating from tri-cotyledonous seedlings. Seedlings with two heteromorphic cotyledons, one almost double the size of the other (Fig. 2E), were commonly also carriers of C1. Plants from this line, regardless of the number or shape of cotyledons, produce either yellow seeds, similar to those of the B. rapa parent, or dark brown seeds (Fig. 3D). Cytological analysis has shown that all plants that produce brown seeds are carriers of C1, while plants producing yellow seeds are euploids with 2n = 20 (Table 2). The fact that a C1-carrier plant produces only brown seeds irrespective of the euploid or monosomic nature of these seeds indicates that the maternal inheritance of the seed colour character is controlled by C1 (Heneen and Brismar, 2001). The frequency of progeny plants producing brown seeds reflects the transmission rate of C1, amounting to 30·3 % (Table 2). The MAAL for C1 is the only line where plants produce either one of two homogenous types of seeds based on seed colour.
MAAL for C2
Seeds expected to be C2 carriers are yellow with brown spots/patches (Fig. 3E). Plants raised from these seeds produce either only yellow seeds or a mixture of yellow seeds and brown-spotted/patchy yellow seeds. It was found that the brown-spotted/patchy seeds (Fig. 3E) give rise to C2-carrier plants which in turn produce a mixture of seed types, while plants raised from pure yellow seeds are largely of the parental B. rapa type and produce just yellow seeds (Table 2). The frequency of the brown-spotted/patchy seeds indicated the transmission rate of this chromosome (33·6 %, Table 2).
MAAL for C3d
Selected yellow seeds with faint brown dots were grown. Plants raised from these seeds produce yellow seeds (Fig. 3F). It is difficult to detect differences between these yellow seeds by the naked eye, but with the aid of a stereo microscope some plants can be discerned as producers of a mixture of pure yellow and faint brown-spotted seeds (33·8 %, Table 2). The faint brown-spotted seeds are largely producers of C3d-carrier plants that generate seed mixtures (Table 2).
MAALs for C4 and C4d
For the MAAL for C4, brown seeds with dark spots (Fig. 3G) were sown. Plants were observed to produce either yellow or white flowers (Fig. 2B, C). The white-flowered plants turned out to be carriers of C4, which harbours the dominant gene for white flower colour. The yellow-flowered plants were euploids. One C4-carrier offspring plant (2n = 21) was found to have yellow flowers. Closer cytological examination of this plant showed that it had a deletion in the short arm of its C4 chromosome (see below). Apparently, the deletion carrying the gene for white flower colour allows the background gene for yellow colour to be expressed. This plant was the starting point of a daughter MAAL designated C4d (with a deleted segment in the short arm of C4). Differences in plant size also characterized the yellow-flowered C4d carriers, being slightly smaller than the white-flowered C4 carriers (Fig. 2F). Seeds selected for sowing in the case of MAAL for C4d were faint brown with dark spots (Fig. 3H). In MAALs for C4 and C4d, plants produced either yellow seeds or a mixture of easily differentiable yellow seeds and brown seeds with dark spots (Figs 2G and 3G, H). The brown seeds in both lines gave rise to C chromosome carrier plants which in turn produced a mixture of seeds (Table 2). From seed counts, the transmission frequencies of C4 and C4d amounted to 34·7 and 36·7 %, respectively (Table 2).
MAALs for C5, C6 and C7
In plants that are carriers of C5, the leaves express a distinctive character by having a puckered appearance (Fig. 2H) from the seedling stage onwards and throughout plant growth and development. The puckered leaf surface is visually easily detectable, and chromosome analysis of 15 plants with such leaves revealed that they had 2n = 21 and were carriers of C5. Seeds selected for sowing from MAALs for C5–C7 are yellow with brown spots/patches (Fig. 3I–K). Plants raised from these seeds produced either only yellow seeds or a mixture of yellow seeds and brown-spotted/patchy yellow seeds (Fig. 3I–K). In these lines, the brown spots/patches were less pronounced than in the MAAL for C2. Seeds with brown spots/patches largely gave rise to C-carrier plants that produced a mixture of seeds (Table 2). Accordingly, the frequencies of pigmented seeds in these lines (32·0–32·5 %, Table 2) are approximate values of the transmission frequencies of these chromosomes.
MAALs for C8 and C9
Yellow seeds were sown and the resulting plants produced only yellow seeds (Fig. 3L, M), thus implying that these two chromosomes do not carry genes controlling or affecting seed colour.
Maintenance and availability of the MAALs
For the C1-carrier MAAL, plants solely producing brown seeds were propagated for the maintenance of the line. Seedlings with three cotyledons or two cotyledons that markedly differ in size are usually carriers of C1. In the case of MAALs for C2, C4, C4d and C5–C7, brown or brown-spotted/patchy seeds, expected to be C carriers, were separated from pure yellow seeds produced by C-carrier plants and propagated. For MAAL C3d, either bulk seeds, or selected seeds with faint brown dots, produced by C3d carriers were propagated. At the plant level, puckered leaves are a reliable indicator of a C5-carrier state. For MAALs for C8 and C9 that produce only yellow seeds, bulk seeds produced by C carriers were propagated. In the case of these two lines in particular, but also relevant for all other lines, cytological and molecular SSR analyses are required for verification of the C-carrier plants.
Limited amounts of bulk seeds of MAALs for C1, C3d, C8 and C9, as well as selected pigmented seeds of MAALs for C2, C4, C4d and C5–C7, together with the parental lines B. rapa var. trilocularis (K-151) and B. oleracea var. alboglabra (no. 4003) and their resynthesized B. napus (no. 7406) have been delivered to the gene bank NordGen (www.nordgen.org) in Alnarp, Sweden. Accession numbers of these materials and correspondence of the numerals of the C chromosomes in the MAAL to the LG designations (see below) are presented in Supplementary Data Table S1. A description of the material and recommended method for propagation of the different MAALs will be supplied by the gene bank on request.
Cytology
Differentiation of A and C chromosomes
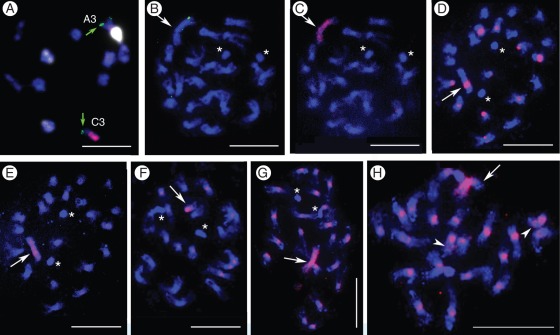
Differences in chromatin condensation patterns of the two parental AA and CC species observed during diakinesis in pollen mother cells often made it possible to differentiate between the chromosomes when present in the same cell. The chromosomes of B. rapa usually exhibit highly condensed heavily stained pericentric regions, and less condensed and less stained regions towards the chromosome ends (Fig. 4A, C). This is especially evident in chromosomes with median and sub-median centromeres (Fig. 4C). The ten bivalents of B. rapa are identifiable as members of two groups depending on centromeric position, or as the nucleolar bivalent (Fig. 4C). The chromosomes of B. oleracea, on the other hand, at a corresponding stage of chromatin condensation exhibit no or slight differentiation of chromatin condensation (Fig. 4B). These species differences led to the distinction between the A and C chromosomes, as exemplified in diakinesis configurations of a sesquidiploid (Supplementary Data Fig. S1A), an aneuploid (2n = 22) with C6 and C8 (Fig. S1B) and MAALs with C5, C6 or C7 appearing as univalents or components of multivalents (Fig. S1C–F).
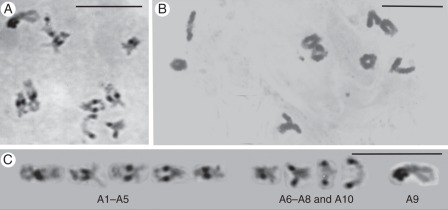
Fig. 4.
Diakinesis chromosomes. (A, C) Brassica rapa (AA); differential condensation and staining marking the heavily stained heterochromatic pericentric regions, numeric designations of chromosomes with median/sub-median centromeres (A1–A5), sub-median/sub-terminal centromeres (A6–A8 and A10) and the nucleolar pair (A9) are according to Cheng et al. (1995). (B) B. oleracea var. alboglabra (CC); chromosomes are generally homogenously stained. Scale bars = 10 µm.
Chromosome structural changes and substitution
Eight of the nine developed MAALs contain seemingly intact C chromosomes, judged by their size. C3 is represented by only one chromosome arm in the C3d line (see below). Another line with a deletion was the MAAL for C4d, which has a deletion in the short arm of C4 (see below). Further examples of structural chromosome changes in progeny plants from the MAAL for C4 are presented in Supplementary Data Fig. S2A–E. In a B. rapa offspring plant (2n = 20), one chromosome of the nucleolar pair had a deletion in the heterochromatic short arm (Fig. S2A) and, in a monosomic plant (2n = 21), the C4 chromosome was exceptionally large, probably resulting from a duplication or translocation (Fig. S2B). Of interest is the finding of an additional mini-chromosome in a C4-carrier progeny plant (Fig. S2C–E). The mini-chromosome divided in a normal way (Fig. S2D, E) and was found in all analysable meiotic cells in this plant, indicating that it has a functional centromere. In addition to finding AA euploids as well as mono- and disomics with alien chromosomes (2n = 21 and 22) among the progeny of monosomics, plants with a substitution (2n = 20, 19 A chromosomes + 1 C chromosome) were also found (Supplementary Data Fig. S2F).
Behaviour of the C chromosome during meiosis
By comparing the centromere position of the C chromosome in the newly developed MAAL with that of C6/7 (Cheng et al., 1995), the latter was designated C6 and the former C7. A detailed chart showing examples of the most frequently encountered patterns of behaviour for the C chromosomes in all developed MAALs is presented in Fig. 5. The C chromosomes appear as univalents, or components of bivalents, trivalents or pentavalents. The less condensed C univalents exhibit differential chromatin condensation generally portraying a more condensed state of the short arm. When the C chromosome pairs with a homoeologous A chromosome, they form a heteromorphic bivalent, while the unpaired A chromosome appears as a univalent. Thus, it is necessary to be careful not always to infer univalents as representing the alien chromosome, without confirming their identity.
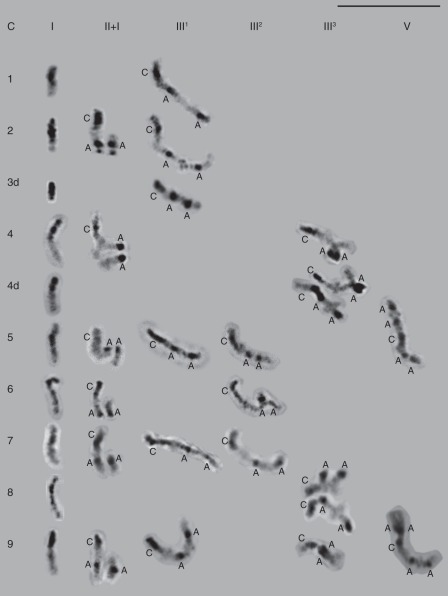
Fig. 5.
The behaviour of the C chromosome during diakinesis in the MAALs. It either remains unpaired as a univalent (I), or pairs with one A chromosome forming a heteromorphic bivalent, while the other A chromosome remains as a univalent (II + I), or appears as part of a trivalent when pairing with a pair of A chromosomes with median/sub-median centromeres (III1), sub-median/sub-terminal centromeres (III2), or the nucleolar organizers (III3), or appears as part of a pentavalent (V) when pairing with two pairs of A chromosomes. In the case of C4d and C8, two trivalents representing short and long arm associations of the C chromosome with the nucleolar A pair are depicted. Scale bar = 10 µm.
The frequencies with which the C chromosome remains as a univalent and when it pairs in different combinations with A chromosomes are given in Table 3. The frequency of univalent C chromosomes is highest (90·2 %) in the MAAL for C3d. This is apparently due to the fact that C3d is a short chromosome, representing only one arm. A seemingly intact C chromosome that generally remains unpaired as a univalent is C8 (82·5 %), whereas the other C chromosomes appear as univalents in the range 38·9–68·1 %. Chromosome pairing provides information on homoeological relationships between the extra C chromosome and the chromosomes of the AA background (Fig. 5). C1 and C2 with median centromeres and C3d which originates from a chromosome with a median/sub-median centromere (C3) revealed their preferential pairing with A chromosomes with a median/sub-median centromere (Fig. 5, Table 3). Other C chromosomes with an affinity to pair with A chromosomes having a median/sub-median centromere are C5, C7 and C9 (Fig. 5, Table 3). Homoeologous pairing with A chromosomes having a sub-median/sub-terminal centromere occurs with C chromosomes (C5–C7) having sub-median centromeres. Homoeologies to the nucleolar A9 chromosomes are expressed mainly by C4, C4d, C8 and C9, and to a minor degree by C6. Pentavalents resulting from pairing of the extra C chromosome with two types of A pairs are recorded in the MAALs for C5 and C9 (Fig. 5, Table 3).
Table 3.
The frequency of diakinesis cells in which the C chromosome stays as a univalent (I), and when it pairs with an A chromosome forming a heteromorphic bivalent (II), and when it is part of a trivalent after pairing with a pair of A chromosomes with median/submedian centromeres (III1), or submedian/subterminal centromeres (III2), or when it pairs with the nucleolar chromosomes (III3), and when it pairs with two pairs of A chromosomes forming a pentavalent (V).
| MAAL* |
% of cells with |
|||||||
|---|---|---|---|---|---|---|---|---|
| Cyt. | LG | No. of cells | I | II | III1 | III2 | III3 | V |
| C1 | C9 | 100 | 39·0 | 61·0 | ||||
| C2 | C1 | 193 | 59·1 | 8·8 | 32·1 | |||
| C3d | C5 | 441 | 90·2 | 0·7 | 9·1 | |||
| C4 | C3 | 573 | 51·3 | 0·4 | 48·3 | |||
| C4d | C3 | 537 | 38·9 | 0·2 | 60·9 | |||
| C5 | C4 | 124 | 44·3 | 6·5 | 12·9 | 25,0 | 11·3 | |
| C6 | C6 | 348 | 68·1 | 1·2 | 29·0 | 1·7 | ||
| C7 | C2 | 80 | 62·5 | 5·0 | 32·5 | |||
| C8 | C7 | 343 | 82·5 | 17·5 | ||||
| C9 | C8 | 350 | 39·7 | 0·3 | 9·4 | 48·0 | 2·6 | |
| Average† | 55·8 | 2·8 | 39·7 | 1·7 | ||||
*Cytological (Cyt.) and corresponding linkage groups (LG) designations according to Table 4.
†Averages excluding the data for the structurally changed chromosomes C3d and C4d.
FISH and GISH
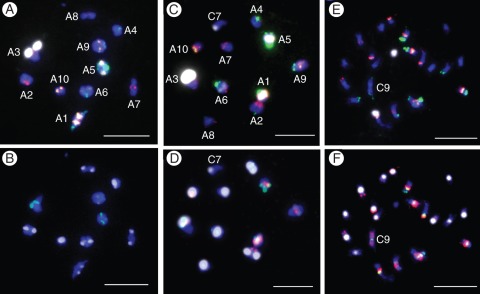
The use of FISH with 5S and 25S rDNA probes in B. oleracea var. alboglabra has been useful for characterizing C5 (LG-C4) carrying 5S rDNA, as well as C8 (LG-C7) and C9 (LG-C8) carrying 25S rDNA, as reported earlier (Howell et al., 2002; Hasterok et al., 2005). In addition, other FISH and GISH approaches have been applied to differentiate the C chromosomes and determine their LGs. The FISH approaches comprised the use of genome-, chromosome- and LG-specific probes, and, when applying GISH, labelled C genome DNA was used. Application of the multiple target FISH technique (Xiong and Pires, 2011) led to the differentiation of all the A chromosomes in B. rapa (Fig. 6A, B) and identification of the C chromosome in the five MAALs for C1, C3d, C4, C6 and C8 (Fig. 6C–F; Supplementary Data Fig. S3A–F). In Fig. 6 and Supplementary Data Fig. S3, the designations of the A and C chromosomes correspond to the LG numbers. For correspondence between LG and cytological numerical designations, see below (Table 4). Labelling with chromosome-specific probes is exemplified by the use of the LG-C3-specific BAC KBrH117M18 probe and the LG-C4-specific BAC BoB004H11 probe shown to be specific to chromosomes C4 and C5, respectively (Fig. 7A–C). Applying the 5S rDNA probe, followed by GISH, led to the detection and characterization of C5 (Fig. 7D, E). The use of GISH alone was enough for the detection of C chromosomes (Fig. 7F–H). In spite of labelling the pericentric regions of the A chromosomes by GISH, the C chromosomes are detectable as being labelled along their whole length. Thus unlabelled segments of the C chromosomes, and labelled sites other than the pericentric regions of the A chromosomes, may denote intergenomic introgression (Fig. 7H).
Fig. 6.
Identification of the A and C chromosomes after applying two rounds of multiple target FISH according to Xiong and Pires (2011). (A and B) Diakinesis chromosomes of B. rapa (K-151), using in the first round (A) probes for 5S rDNA (yellow), 45S rDNA (white), repeated DNA sequences in eight chromosome pairs of B. rapa by using BAC KBrB072L17 (green), and repeated DNA sequences specific to two pairs of B. rapa by using BAC KBrH092N24 (red), and applying in the second round (B) probes for the repetitive centromeric DNA sequences CentBr1 (white) and CentBr2 (green) and for repetitive DNA sequences that are C genome specific by using the BAC BNIH 123L05 (red). (C–F) Identification of the C chromosome in MAALs; (C, D) diakinesis chromosomes with C8 (LG-C7); (E, F) mitotic chromosomes with C1 (LG-C9). Scale bars = 10 µm.
Table 4.
Correspondence between cytological (Cyt.) and linkage group (LG) designations of the Brassica oleracea var. alboglabra chromosomes
| Evidence |
||||
|---|---|---|---|---|
| Cyt. | LG | SSR* | FISH and GISH | Figures |
| C1 | C9 | BnGMS185 | FISH: multiple target | Fig. 6E and F |
| BnGMS634 | ||||
| C2 | C1 | CB10277 | – | – |
| C3d | C5 | Na10B08 | FISH: multiple target | Fig. S3C and D |
| C4 | C3 | BRAS068 | FISH: multiple target | Fig. S3A and B |
| FITO-306 | FISH: specific BAC | Fig. 7A | ||
| FITO-505 | ||||
| C5 | C4 | BRAS003 | FISH: specific BAC | Fig. 7B and C |
| BnGMS408 | FISH: 5S rDNA and GISH | Fig. 7D and E | ||
| BnGMS490 | FISH: 5S and 25S rDNA† | |||
| C6 | C6 | CB10010 | FISH: multiple target | Fig. S3E and F |
| FITO-067 | ||||
| C7 | C2 | BnGMS280 | – | – |
| BnGMS454 | ||||
| CB10026 | ||||
| C8 | C7 | BRAS019 | FISH: multiple target | Fig. 6C and D |
| Fito-472 | FISH: 5S and 25S rDNA† | |||
| FITO-497 | ||||
| Na12-F03 | ||||
| Ol10-H04 | ||||
| C9 | C8 | BnGMS336 | FISH, 5S and 25S rDNA† | |
| BNGMS439 | ||||
| BNGMS509 | ||||
| CB10139 | ||||
| CB10179 | ||||
*From Table 1 in Geleta et al. (2012), with kind permission from Springer Science+Business Media.
† C5, C8 and C9 were characterized in the MAALs by Hasterok et al. (2005) and correspond to LG-C4, LG-C7 and LG-C8, respectively (Howell et al., 2002).
Fig. 7.
Identification of the C chromosome in MAALs after applying chromosome-specific BAC probes (A–C), the 5S rDNA probe followed by GISH with labelled C genome DNA hybridizing to pericentric regions of A chromosomes and to C chromatin (D, E), or only GISH (F–H), and detection of possible intergenomic introgression (H). (A) Diakinesis chromosomes with C4 (LG-C3) labelled with BAC KBrH117M18 specific for LG-C3 and LG-A3 (green) and BAC BNIH 123L05 specific for C genome chromosomes (red), and 45S rDNA probe (white); (B, C) mitotic chromosomes with C5 (LG-C4) labelled with BAC BoB004H11 probe (green) followed by GISH (red), arrows; (D, E) C5 labelled by 5S rDNA (red) and GISH (red) marked by arrows. (F–H) Mitotic chromosomes; (F) C3d mainly composed of one arm (arrow); (G) C5 (arrow); (H) C4 (arrow) whose distal region of the short arm is unlabelled possibly denoting introgressed A chromatin, also labelling of intercalary sites in two A chromosomes (arrowheads) possibly reflecting introgressed C chromatin. Asterisks denote satellites of the nucleolar A pair. Scale bars = 10 µm.
DISCUSSION
The findings in the present work touch upon four topics that will be discussed in some detail. The first topic relates to the development and characterization of the MAALs. Of relevance here was the access to the newly produced aneuploid material and the use of C genome- and C chromosome-specific SSRs for distinguishing between lines and for developing the missing line. The second topic deals with the black/brown seed colour control by genes on the C chromosomes in view of the interest in developing yellow-seeded oilseed rape. Genes with major effects on seed colour occur on two chromosomes and genes with minor effect on five other chromosomes. The third topic covers the cytological identification of the C chromosomes and their homoeology to A chromosomes. Chromosome pairing at diakinesis and the use of FISH and GISH were elucidative for this purpose. The fourth topic is on integrating cytological and genetic linkage data by defining the correspondence between these designation systems.
Development of the missing MAAL
Two approaches were crucial for the development of the missing MAAL. One was the development of a new batch of B. rapa–B. oleracea aneuploids as a source for the missing line. The second approach was the use of SSR markers for molecular characterization of the parental species, the eight available MAALs and the newly developed aneuploid material. SSR markers have been widely used for characterization of the C genome in B. oleracea and both the A and C genomes in B. napus (e.g. Lowe et al., 2004; Piquemal et al., 2005; Gao et al., 2007; Iniguez-Luy et al., 2008; Basunanda et al., 2010; J. Wang et al., 2011). Defining SSR markers that are specific to the currently used C genome and to each of the eight available MAALs led to the identification of SSR markers specific to the hitherto uncharacterized chromosome and discovering monosomics that carry this chromosome among the newly developed aneuploids (Geleta et al., 2012). In this way, for the first time, access to all nine MAALs is now possible through the gene bank NordGen (www.nordgen.org) where limited amounts of seeds of the MAALs, the AA and CC parents and the resynthesized B. napus are deposited.
The fact that the MAAL for C3d carries only one chromosome arm might be restrictive for certain purposes, although resolution of phenotypic traits residing on this arm will be more precise. In the MAAL for C4d, plants that carry C4d are yellow flowered, while plants carrying the intact C4 are white flowered, indicating the localization of the flower gene in the deleted segment. The location of the white flower gene on C4 has been shown in earlier works (Chen et al., 1992; Cheng et al., 1995), and has been mapped to the corresponding LG-C3 (Ramsay et al., 1996). Comparative studies on MAALs for C4 and C4d would be informative as to gene localization and eventual position effects of the deletion. Access to MAALs for C3d and C4d emphasizes the value of establishing MAALs with structurally altered C chromosomes.
Morphological features other than the flower colour that discriminate plants carrying different C chromosomes have been defined. These comprise the occurrence of three cotyledons indicating the presence of C1 and a puckered leaf surface in plants carrying C5. A puckered leaf morphology has been observed in one of the seven B. rapa–B. oleracea MAALs developed by McGrath et al. (1990). Seed colour was also a reliable and valuable character for discrimination of plants or seeds that are carriers or non-carriers of C chromosome(s) in progeny of up to seven of the nine MAALs.
Seed colour control by major and minor genes in B. oleracea
Development of the current MAALs has been valuable for defining the chromosomes that control the black seed colour of B. oleracea var. alboglabra and to determine the mode of this control. This knowledge is a prerequisite if attempts are being made to develop yellow-seeded oilseed rape from its progenitor species. Yellow-seeded B. rapa are common, whereas yellow-seeded B. oleracea are scarce. The interest in developing yellow-seeded oilseed rape (Chen et al., 1988; Meng et al., 1998; Rahman, 2001) relates to the fact that this character is associated with higher oil and protein contents and less fibre content, which are desirable food and livestock feed agronomic goals (Shirzadegan and Röbbelen, 1985; Slominski et al., 1999). Findings in the present work indicate that seven out of the nine C chromosomes carry genes that affect seed colour. C1 and C4 carry major genes for dark seed colour that lead to pigmentation of the entire seed coat. These genes are controlled maternally and through the embryo, respectively (Heneen and Brismar, 2001). In addition to these two major genes, biparental control through the embryo was further expressed as less pronounced pigmented spots/patches on a fraction of the yellow seeds produced by C2, C3, C5, C6 and C7 carriers, apparently due to the presence of at least five minor quantitative genes on these chromosomes. This is in accordance with the indirect finding that the currently used B. oleracea var. alboglabra possibly contains two independently dominant genes with major and additive effect for black seed colour and that other genes with minor effects may also be present (Chen and Heneen, 1992). It remains to be determined if pigmentation expressed as spots or patches is due to incomplete penetrance, epigenetic factors, DNA transposon insertion or other factors. In most studies of seed colour inheritance in B. napus, it has been concluded that three or four genes control this character both maternally and through the embryo (Shirzadegan, 1986; Van Deynze and Pauls, 1994; Rahman et al., 2001, 2010).
Of interest is the finding that the extent of seed pigmentation and plant vigour are less pronounced in C4d carriers compared with C4 carriers. This could be due to a position effect or due to the loss of minor quantitative genes for seed pigmentation and vigour in the deleted chromosome segment.
The interest in the seed colour character in oilseed rape has driven efforts to identify molecular markers that are closely linked to genes controlling this trait for use in marker-assisted selection (Van Deynze et al., 1995; Somers et al., 2001; Badani et al., 2006; Liu et al., 2006; Fu et al., 2007; Xiao et al., 2007; Rahman et al., 2010; Zhang et al., 2011). In our work on the MAALs for C1, a RAPD marker relatively close to the major gene for dark seed colour on this chromosome was defined, and absence of this marker in a C1-carrier progeny plant was accompanied by production of yellow seeds (Chen et al., 1997a). Progeny plants that lack the major genes for dark seed colour on C1 and C4 are of interest in breeding for yellow-seeded B. oleracea and B. napus. The manifestation of the seed colour character is also associated with seed coat thickness (Heneen and Brismar, 2007) and the accumulation of seed coat pigments. Transcriptome analysis and chemistry of seed coat pigmentation reveal a multitude of genetic factors that control pigmentation of the seed coat (Marles and Gruber, 2004; Akhov et al., 2009; Jiang and Deyholos, 2010).
Identification of alien C chromosomes and their homoeology to A chromosomes
Cytological characterization of the alien C chromosomes and the background A genome was based on comparisons with the previously described karyotypes of the parental species (Cheng et al., 1995) and diakinesis chromosomes of B. rapa (Cheng et al., 1994a; Heneen et al., 1995). When applying FISH on the MAALs, rDNA probes were used first, thus defining C5, C8 and C9 as carriers of 5S and 45S rDNA (Hasterok et al., 2005). This was expanded in the present work by using additional probes of other repetitive DNA sequences in multiple target FISH (Xiong and Pires, 2011), and using chromosome- and LG-specific probes, as well as GISH. This recent approach enabled the characterization of seven MAALs. A significant advantage of these different approaches was the identification of the C chromosomes in terms of their respective LGs. The use of FISH and GISH has enabled the distinction between the A and C genomes and identification of their individual chromosomes (Howell et al., 2008; Xiong and Pires, 2011). These approaches also provided evidence of what could be intergenomic introgression, as has been inferred earlier following RAPD characterization of the MAALs (Chen et al., 1992, 1997a, b; Jørgensen et al., 1996; Heneen and Jørgensen, 2001).
The pairing patterns at diakinesis marked the homoeologous relationships between the A and C chromosomes. These detailed data were restricted to the carmine-stained preparations, thus defining only whether the A chromosomes belonged to groups with specific centromeric positions, or if they were the easily differentiable nucleolar chromosomes. Intergenomic pairing in the present material indicated that chromosome homoeology is maintained between C and A chromosomes with similar centromeric and karyotypic positions, in spite of the extensive duplications and structural chromosome changes shown or inferred to have occurred during Brassica evolution (Parkin et al., 2003; Lysak et al., 2007). The lowest frequency of pairing of a seemingly intact alien chromosome was manifested by C8. This might be a consequence of rearrangements of homoeologous regions in C8 relative to their counterparts in the A genome, or vice versa.
The fact that many C chromosomes had more than one type of homoeological pairing partner among the A chromosomes most probably reflects the frequent presence of similar and homoeologous duplicate and triplicate sequences within and between the A and C genomes, as evidenced by molecular mapping and physical painting (McGrath et al., 1990; Parkin et al., 2003; Lysak et al., 2007), and more recently by whole-genome analysis (J. Wang et al., 2011; X. Wang et al., 2011). An example relating to the frequent pairing between C4 and the nucleolar chromosomes of the A genome (Table 3) is the prevailing homoeology shown by FISH between the long arm of LG-C3 (cytological C4) and the long arm of the nucleolar A chromosome (LG-A3) (Xiong and Pires, 2011). The relatively high frequency of homoeologous pairing in the MAALs, and the occurrence of chromosome structural changes, certainly contribute to prevalence of variations in the make-up of plants carrying alien chromosomes within a MAAL. Changes are likely to have happened in the background AA genome and in the alien chromosome during the development and propagation of MAALs. Such changes have been documented in resynthesized AACC oilseed rape (Song et al., 1995; Pires et al., 2004; Gaeta and Pires, 2010; Szadkowski et al., 2010; Xiong et al., 2011), in the sesquidiploid AAC (Nozaki et al., 2000; Leflon et al., 2006) and in the aneuploids and MAALs (Heneen and Jørgensen, 2001). Cytological differentiation of all B. napus chromosomes using FISH (Xiong and Pires, 2011) also made it possible to monitor the incidence of homoeologous chromosome compensation (replacement) following resynthesis and selfing. The highest rates of homoeologous pairing, reciprocal exchange, non-reciprocal transposition and chromosome compensation occur between A and C chromosomes that are largely syntenic along their entire length (Parkin et al., 1995, 2005; Udall et al., 2005; Gaeta et al., 2007; Xiong et al., 2011). The observed relatively low frequency of intergenomic multivalents involving C2 (LG-C1) and C7 (LG-C2) known to share a high degree of homology with their counterparts in the A genome may relate to genetic changes that have occurred in these chromosomes.
Correspondence of the cytological and molecular linkage group nomenclatures
The identification of the alien C chromosomes of the MAALs in terms of LGs by different FISH and GISH approaches together with the SSR markers that are linked to specific LGs (Geleta et al., 2012) were confirmative and complementary to each other as to the correspondence between seven of the cytological and LG designations (Table 4). The correspondence of C2 and C7 to LG-C1 and LG-C2, respectively is based on SSR evidence only. It is valuable for future users of the nine MAALs to be able to refer to the C chromosomes in terms of LG designations commonly agreed upon in gene and molecular linkage mapping (Parkin et al., 2005; see also http://www.brassica.info/resource/maps/lg-assignments.php; J. Wang et al., 2011). Knowledge of the LGs represented in the MAALs adds to the benefits of using these cytological stocks for detailed studies on gene mapping, physical DNA probing, intergenomic gene transfer and integration of these approaches (Armstrong et al., 1998; Howell et al., 2002; Snowdon, 2007). Of interest is to compare the cytological karyotype (Cheng et al., 1995) with the idiogram of the LGs based on the FISH work by Xiong and Pires (2011). Putting these results together (Supplementary Data Fig. S4) permitted direct comparisons between these two modes. In general, chromosome size and centromeric positions in the karyotype coincided well with those in the idiogram. Only in the idiogram of LG-C2 was the position of the centromere more median than in its corresponding C7.
Future prospects
The available stocks of B. rapa var. trilocularis–B. oleracea var. alboglabra MAALs are suitable materials for cytological and genetic studies on the alien C chromosomes and the AA background. For refinement of physical mapping of the B. oleracea var. alboglabra chromosomes, painting of chromosome arms or whole chromosomes would be desirable. This can be achieved by multicolour FISH of BAC contigs specific to the entire length of a C chromosome arm at a time, preferably at the pachytene stage, as applied on LG-A7 of B. rapa (Xiong et al., 2010). The advantage of applying this on the MAALs would be the presence of only one C chromosome in an AA background. Painting of the C chromosome will also be accompanied by partial painting of the homoeologous segments on the A chromosomes. This is valuable not only when the C chromosome is a univalent, but also when it pairs with A chromosomes, thus defining the exact identity of homoeologous chromosomes and the order of pairing preferences, portraying the extent of homoeology between the C chromosome and different A chromosomes. Thus, a closer analysis of heteromorphic bivalents and multivalents is highly desirable.
To complement the array of seemingly intact and defined structurally changed alien C chromosomes represented in the available MAALs, it is desirable to put in some effort towards developing a new MAAL with an intact C3. Having observed the advantages of maintaining MAALs with structurally changed C chromosomes, represented by MAALs for C3d and C4d, it is advisable to retain additional partial MAALs for detailed studies on gene and marker mapping, position effects and gene expression. Development of lines with stable mini-chromosomes could also be useful for gene mapping and gene transfer. Other materials that became available when working with the MAALs, and have not been discussed in the present work, are the euploid B. rapa plants which are siblings of the monosomics. They would be expected to carry introgressed C chromatin and are worth closer examination. Introgressed genetic material from the C genome might contain desirable genes for breeding improvement of B. rapa. Monosomics and their sibling B. rapa euploids, cytologically and molecularly monitored, could be valuable genetic resources.
Since a large number of A and C genome- and chromosome-specific SSRs are defined in the reported literature, it would be highly desirable to determine their physical localization on chromosomes. Physical detection of SSRs and their mapping on chromosomes applying non-denaturing FISH (Cuadrado and Jouve, 2010), in comparison with SSR positions on linkage maps, would be an appropriate approach to map meiotic recombination incidence along the chromosomes as well as approximate sites of SSR-linked genes of interest. This would also contribute to the integration of cytological, physical and genetic maps.
The MAALs are suitable for defining and mapping genes of agronomic interest on specific chromosomes. The application of next-generation sequencing on the MAALs by targeting the alien C chromosome or the background genome would enable the development of molecular markers for linkage mapping and marker-assisted selection, the profiling of the transcriptome relating to the alien C chromosome and monitoring of intergenomic introgression (Varshney et al., 2009). The knowledge acquired on the maternal and embryonal control of seed colour character in B. oleracea var. alboglabra and the expression of this character sets the stage for a better understanding of seed colour inheritance, mapping of major and minor controlling genes and definition of linked molecular markers, as well as transcriptome analysis of pigmentation patterns and chemistry. MAALs proved useful for studies on other agronomic characters, such as control of seed size (A.I. Stoute et al., unpubl. res.).
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We are indebted to Professor Baoyuan Chen (Bayer Crop Science Inc., Saskatoon, Canada) who initiated this work in 1988 in Sweden, and who provided seeds of the first developed partially characterized six monosomic addition lines before leaving for Canada in 1996. We appreciate the efforts of Göran Olsson to optimize plant growth conditions in the greenhouse. We also thank two anonymous reviewers for valuable suggestions to improve the manuscript. This research was supported by the Royal Physiographic Society in Lund, Sweden. Z.X. and J.C.P. were supported by the American National Science Foundation (DBI 0501712 and DBI 0638536). A.I.S., R.J.S., G.J.K. and S.K. were funded by the Biotechnology and Biological Sciences Research Council, UK (BB/F009721/1).
LITERATURE CITED
- Akhov L, Ashe P, Tan YF, Datla R, Selvaraj G. Proanthocyanidin biosynthesis in the seed coat of yellow-seeded, canola quality Brassica napus YN01-429 is constrained at the committed step catalyzed by dihydroflavonol 4-reductase. Botany. 2009;87:616–625. [Google Scholar]
- Armstrong SJ, Fransz P, Marshall DF, Jones GH. Physical mapping of DNA repetitive sequences to mitotic and meiotic chromosomes of Brassica oleracea var. alboglabra by fluorescence in situ hybridization. Heredity. 1998;81:666–673. [Google Scholar]
- Badani AG, Snowdon RJ, Wittkop B, et al. Colocalization of a partially dominant gene for yellow seed colour with a major QTL influencing acid detergent fibre (ADF) content in different crosses of oilseed rape (Brassica napus) Genome. 2006;49:1499–1509. doi: 10.1139/g06-091. [DOI] [PubMed] [Google Scholar]
- Barthes L, Ricroch A. Interspecific chromosomal arrangements in monosomic addition lines of Allium. Genome. 2001;44:929–935. [PubMed] [Google Scholar]
- Basunanda P, Radoev M, Ecke W, Friedt W, Becker HC, Snowdon RJ. Comparative mapping of quantitative trait loci involved in heterosis for seedling and yield traits in oilseed rape (Brassica napus L.) Theoretical and Applied Genetics. 2010;120:271–281. doi: 10.1007/s00122-009-1133-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilgic H, Cho S, Garvin DF, Muehlbauer GJ. Mapping barley genes to chromosome arms by transcript profiling of wheat–barley ditelosomic chromosome addition lines. Genome. 2007;50:898–906. doi: 10.1139/g07-059. [DOI] [PubMed] [Google Scholar]
- Budahn H, Schrader O, Peterka H. Development of a complete set of disomic rape–radish chromosome-addition lines. Euphytica. 2008;162:117–128. [Google Scholar]
- Chang SB, de Jong H. Production of alien chromosome additions and their utility in plant genetics. Cytogenetic and Genome Research. 2005;109:335–343. doi: 10.1159/000082417. [DOI] [PubMed] [Google Scholar]
- Chen BY, Heneen WH. Inheritance of seed colour in Brassica campestris L. and breeding for yellow-seeded B. napus L. Euphytica. 1992;59:157–163. [Google Scholar]
- Chen BY, Heneen WK, Jonsson R. Resynthesis of Brassica napus L. through interspecific hybridization between B. alboglabra Bailey and B. campestris L. with special emphasis on seed colour. Plant Breeding. 1988;101:52–59. [Google Scholar]
- Chen BY, Simonsson V, Lannér-Harrera C, Heneen WK. A Brassica alboglabra addition line and its use for gene mapping, intergenomic gene transfer and generation of trisomics. Theoretical and Applied Genetics. 1992;84:592–599. doi: 10.1007/BF00224157. [DOI] [PubMed] [Google Scholar]
- Chen BY, Cheng BF, Jørgensen RB, Heneen WK. Production and cytogenetics of Brassica campestris–alboglabra chromosome addition lines. Theoretical and Applied Genetics. 1997a;94:633–640. [Google Scholar]
- Chen BY, Jørgensen RB, Cheng BF, Heneen WK. Identification and chromosomal assignment of RAPD markers linked with a gene for seed colour in a Brassica campestris–alboglabra addition line. Hereditas. 1997b;126:133–138. [Google Scholar]
- Cheng BF, Chen BY, Heneen WK. Addition of Brassica alboglabra Bailey chromosomes to B. campestris L. with special emphasis on seed colour. Heredity. 1994a;73:185–189. [Google Scholar]
- Cheng BF, Heneen WK, Chen BY. Meiotic studies on a Brassica campestris–alboglabra monosomic addition line and derived B. campestris primary trisomics. Genome. 1994b;37:584–589. doi: 10.1139/g94-083. [DOI] [PubMed] [Google Scholar]
- Cheng BF, Heneen WK, Chen BY. Mitotic karyotypes of Brassica campestris and Brassica alboglabra and identification of the B. alboglabra chromosome in an addition line. Genome. 1995;38:313–319. doi: 10.1139/g95-039. [DOI] [PubMed] [Google Scholar]
- Cuadrado Á, Jouve N. Chromosomal detection of simple sequence repeats (SSRs) using nondenaturing FISH (ND-FISH) Chromosoma. 2010;119:495–503. doi: 10.1007/s00412-010-0273-x. [DOI] [PubMed] [Google Scholar]
- Dong F, Tek AL, Frasca ABL, et al. Development and characterization of potato–Solanum brevidens chromosomal addition/substitution lines. Genetics and Genome Research. 2005;109:368–372. doi: 10.1159/000082421. [DOI] [PubMed] [Google Scholar]
- Fu FY, Liu LZ, Chai YR, et al. Localization of QTLs for seed color using recombinant inbred lines of Brassica napus in different environments. Genome. 2007;50:840–854. doi: 10.1139/g07-068. [DOI] [PubMed] [Google Scholar]
- Gaeta RT, Pires JC. Homoeologous recombination in allopolyploids: the polyploidy ratchet. New Phytologist. 2010;186:18–28. doi: 10.1111/j.1469-8137.2009.03089.x. [DOI] [PubMed] [Google Scholar]
- Gaeta RT, Pires JC, Iniguez-Luy F, Leon E, Osborn TC. Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. The Plant Cell. 2007;19:3403–3417. doi: 10.1105/tpc.107.054346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Guo D, Jung C. Monosomic addition lines of Beta corolliflora Zoss in sugar beet: cytological and molecular-marker analysis. Theoretical and Applied Genetics. 2001;103:240–247. [Google Scholar]
- Gao MQ, Li GN, Yang B, Qiu D, Farnham M, Quiros C. High-density Brassica oleracea linkage map: identification of useful new linkages. Theoretical and Applied Genetics. 2007;115:277–287. doi: 10.1007/s00122-007-0568-3. [DOI] [PubMed] [Google Scholar]
- Geleta M, Heneen WK, Stoute AI, et al. Assigning Brassica microsatellite markers to the nine C-genome chromosomes using Brassica rapa var. trilocularis–B. oleracea var. alboglabra monosomic alien addition lines. Theoretical and Applied Genetics. 2012 doi: 10.1007/s00122-012-1845-3. in press. doi:10.1007/s00122-012-1845-3. [DOI] [PubMed] [Google Scholar]
- Hasterok R, Wolny E, Kulak S, Zdziechiewicz A, Maluszynska J, Heneen WK. Molecular cytogenetic analysis of Brassica rapa–Brassica oleracea var. alboglabra monosomic addition lines. Theoretical and Applied Genetics. 2005;111:196–205. doi: 10.1007/s00122-005-1942-7. [DOI] [PubMed] [Google Scholar]
- Heneen WK, Brismar K. Maternal and embryonal control of seed colour by different Brassica alboglabra chromosomes. Plant Breeding. 2001;120:325–329. [Google Scholar]
- Heneen WK, Brismar K. Seed coat structure and colour in Brassica. Scanning. 2007;29:71–72. [Google Scholar]
- Heneen WK, Jørgensen RB. Cytology, RAPD, and seed colour of progeny plants from Brassica rapa–alboglabra aneuploids and development of monosomic addition lines. Genome. 2001;44:1007–1021. doi: 10.1139/g01-095. [DOI] [PubMed] [Google Scholar]
- Heneen WK, Chen BY, Cheng BF, et al. Characterization of the A and C genomes of Brassica campestris and B. alboglabra. Hereditas. 1995;123:251–267. [Google Scholar]
- Howell EC, Barker GC, Jones GH, et al. Integration of the cytogenetic and genetic linkage maps of Brassica oleracea. Genetics. 2002;161:1225–1234. doi: 10.1093/genetics/161.3.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell EC, Kearsey MJ, Jones GH, King GJ, Armstrong SJ. A and C genome distinction and chromosome identification in Brassica napus by sequential fluorescence in situ hybridization and genomic in situ hybridization. Genetics. 2008;180:1849–1857. doi: 10.1534/genetics.108.095893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Quiros CF. Molecular and cytological evidence of deletions in alien chromosomes for two monosomic addition lines of Brassica campestris–oleracea. Theoretical and Applied Genetics. 1991;81:221–226. doi: 10.1007/BF00215726. [DOI] [PubMed] [Google Scholar]
- Iniguez-Luy FL, Voort AV, Osborn TC. Development of a set of public SSR markers derived from genomic sequence of a rapid cycling Brassica oleracea L. genotype. Theoretical and Applied Genetics. 2008;117:977–985. doi: 10.1007/s00122-008-0837-9. [DOI] [PubMed] [Google Scholar]
- Ji Y, Chetelat RT. Homoeologous pairing and recombination in Solanum lycopersicoides monosomic addition and substitution lines in tomato. Theoretical and Applied Genetics. 2003;106:979–989. doi: 10.1007/s00122-002-1090-2. [DOI] [PubMed] [Google Scholar]
- Jiang YQ, Deyholos MK. Transcriptome analysis of secondary-wall-enriched seed coat tissues of canola (Brassica napus L.) Plant Cell Reports. 2010;29:327–342. doi: 10.1007/s00299-010-0824-x. [DOI] [PubMed] [Google Scholar]
- Jørgensen RB, Chen BY, Cheng BF, Heneen WK, Simonsen V. Random amplified polymorphic DNA markers of the Brassica alboglabra chromosome of a B. campestris–alboglabra addition line. Chromosome Research. 1996;4:111–114. doi: 10.1007/BF02259703. [DOI] [PubMed] [Google Scholar]
- Kaneko Y, Matsuzawa Y, Sarashima M. Breeding of the chromosome addition lines of radish with single kale chromosome. Japanese Journal of Breeding. 1987;37:438–452. [Google Scholar]
- Khush GS. Trisomics and alien addition lines in rice. Breeding Science. 2010;60:469–474. [Google Scholar]
- Leflon M, Eber F, Letanneur JC, et al. Pairing and recombination at meiosis of Brassica rapa (AA)×Brassica napus (AACC) hybrids. Theoretical and Applied Genetics. 2006;113:1467–1480. doi: 10.1007/s00122-006-0393-0. [DOI] [PubMed] [Google Scholar]
- Liu ZW, Fu TD, Wang Y, et al. Development of SCAR and CAPS markers for a partially dominant yellow seed coat gene in Brassica napus L. Euphytica. 2006;149:381–385. [Google Scholar]
- Lowe AJ, Moule C, Trick M, Edwards KJ. Efficient large-scale development of microsatellites for marker and mapping applications in Brassica crop species. Theoretical and Applied Genetics. 2004;108:1103–1112. doi: 10.1007/s00122-003-1522-7. [DOI] [PubMed] [Google Scholar]
- Lysak MA, Cheung K, Kitschke M, Bureš P. Ancestral chromosomal blocks are triplicated in Brassicaceae species with varying chromosome number and gene size. Plant Physiology. 2007;145:402–410. doi: 10.1104/pp.107.104380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marles MS, Gruber MY. Histochemical characterisation of unextractable seed coat pigments and quantification of extractable lignin in the Brassicaceae. Journal of the Science of Food and Agriculture. 2004;84:251–262. [Google Scholar]
- McGrath JM, Quiros CF. Generation of alien chromosome addition lines from synthetic Brassica napus: morphology, cytology, fertility, and chromosome transmission. Genome. 1990;33:374–383. [Google Scholar]
- McGrath JM, Quiros CF, Harada JJ, Landry BS. Identification of Brassica oleracea monosomic alien chromosome addition lines with molecular markers reveals extensive gene duplication. Molecular and General Genetics. 1990;223:198–204. doi: 10.1007/BF00265054. [DOI] [PubMed] [Google Scholar]
- Meng JL, Shi SW, Gan L, Li ZY, Qu XS. The production of yellow-seeded Brassica napus (AACC) through crossing interspecific hybrids of B. campestris (AA) and B. carinata (BBCC) with B. napus. Euphytica. 1998;103:329–333. [Google Scholar]
- Nozaki T, Mishiba K, Mii M, Koba T. Construction of synteny groups of Brassica alboglabra by RAPD markers and detection of chromosome aberrations and distorted transmission under the genetic background of B. campestris. Theoretical and Applied Genetics. 2000;101:538–546. [Google Scholar]
- Parkin IAP, Sharpe AG, Keith DJ, Lydiate DJ. Identification of the A and C genomes of amphidiploid Brassica napus (oilseed rape) Genome. 1995;38:1122–1131. doi: 10.1139/g95-149. [DOI] [PubMed] [Google Scholar]
- Parkin IAP, Sharpe AG, Lydiate DJ. Patterns of genome duplication within the Brassica napus genome. Genome. 2003;46:291–303. doi: 10.1139/g03-006. [DOI] [PubMed] [Google Scholar]
- Parkin IAP, Gulden SM, Sharpe AG, et al. Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics. 2005;171:765–781. doi: 10.1534/genetics.105.042093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piquemal J, Cinquin E, Couton F, et al. Construction of an oilseed rape (Brassica napus L.) genetic map with SSR markers. Theoretical and Applied Genetics. 2005;111:1514–1523. doi: 10.1007/s00122-005-0080-6. [DOI] [PubMed] [Google Scholar]
- Pires JC, Zhao JW, Schranz ME, et al. Flowering time divergence and genomic rearrangements in resynthesized Brassica polyploids (Brassicaceae) Biological Journal of the Linnean Society. 2004;82:675–688. [Google Scholar]
- Prakash S, Bhat SR, Quiros CF, Kirti PB, Chopra VL. Brassica and its close allies: cytogenetics and evolution. Plant Breeding Reviews. 2009;31:21–187. [Google Scholar]
- Quiros CF, Ochoa O, Kianian SF, Douches D. Analysis of the Brassica oleracea genome by the generation of B. campestris–oleracea chromosome addition lines; characterization by isozymes and rDNA genes. Theoretical and Applied Genetics. 1987;74:758–766. doi: 10.1007/BF00247554. [DOI] [PubMed] [Google Scholar]
- Rahman MH. Production of yellow-seeded Brassica napus through interspecific crosses. Plant Breeding. 2001;120:463–472. [Google Scholar]
- Rahman MH, Joersbo M, Poulsen MH. Development of yellow-seeded Brassica napus of double low quality. Plant Breeding. 2001;120:473–478. [Google Scholar]
- Rahman M, Li G, Schroeder D, McVetty PBE. Inheritance of seed coat color genes in Brassica napus (L.) and tagging the genes using SRAP, SCAR and SNP molecular markers. Molecular Breeding. 2010;26:439–453. [Google Scholar]
- Ramsay LD, Jennings DE, Bohuon EJR, et al. The construction of a substitution library of recombinant backcross lines in Brassica oleracea for the precision mapping of quantitative trait loci. Genome. 1996;39:558–567. doi: 10.1139/g96-071. [DOI] [PubMed] [Google Scholar]
- Rines HW, Phillips RL, Kynast RG, et al. Addition of individual chromosomes of maize inbreds B73 and Mo17 to oat cultivars Starter and Sun II: maize chromosome retention, transmission, and plant phenotype. Theoretical and Applied Genetics. 2009;119:1255–1264. doi: 10.1007/s00122-009-1130-2. [DOI] [PubMed] [Google Scholar]
- Shirzadegan M. Inheritance of seed colour in Brassica napus L. Zeitschrift für Pflanzenzüchtung. 1986;96:140–146. [Google Scholar]
- Shirzadegan M, Röbbelen G. Influence of seed color and hull proportion on quality properties of seeds in Brassica napus L. Fette Seifen Anstrichmittel. 1985;87:235–237. [Google Scholar]
- Slominski BA, Simbaya J, Campbell LD, Rakow G, Guenter W. Nutritive value for broilers of meals derived from newly developed varieties of yellow-seeded canola. Animal Feed Science and Technology. 1999;78:249–262. [Google Scholar]
- Snowdon RJ. Cytogenetics and genome analysis in Brassica crops. Chromosome Research. 2007;15:85–95. doi: 10.1007/s10577-006-1105-y. [DOI] [PubMed] [Google Scholar]
- Somers DJ, Rakow G, Prabhu VK, Friesen KRD. Identification of a major gene and RAPD markers for yellow seed coat colour in Brassica napus. Genome. 2001;44:1077–1082. [PubMed] [Google Scholar]
- Song K, Lu P, Tang K, Osborn TC. Rapid genome change in synthetic polyploids of Brassica and its implications for polyploidy evolution. Proceedings of the National Academy of Sciences, USA. 1995;92:7719–7723. doi: 10.1073/pnas.92.17.7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szadkowski E, Eber F, Huteau V, et al. The first meiosis of resynthesized Brassica napus, a genome blender. New Phytology. 2010;186:102–112. doi: 10.1111/j.1469-8137.2010.03182.x. [DOI] [PubMed] [Google Scholar]
- Szakács É, Molnár-Láng M. Identification of new winter wheat–barley addition lines (6HS and 7H) using fluorescence in situ hybridization and the stability of the whole ‘Martonvásári 9 kr1’–Igri' addition set. Genome. 2010;53:35–44. doi: 10.1139/g09-085. [DOI] [PubMed] [Google Scholar]
- Udall JA, Quijada PA, Osborn TC. Chromosomal rearrangements derived from homeologous recombination in four mapping populations of Brassica napus L. Genetics. 2005;169:967–979. doi: 10.1534/genetics.104.033209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Deynze A, Pauls KP. The inheritance of seed colour and vernalization requirement in Brassica napus using doubled haploid populations. Euphytica. 1994;74:77–83. [Google Scholar]
- Van Deynze AE, Landry BS, Pauls KP. The identification of restriction fragment polymorphisms linked to seed colour genes in Brassica napus. Genome. 1995;38:534–542. doi: 10.1139/g95-069. [DOI] [PubMed] [Google Scholar]
- Varshney RK, Nayak SN, May GD, Jackson SA. Next-generation sequencing technologies for crop genetics and breeding. Trends in Biotechnology. 2009;27:522–530. doi: 10.1016/j.tibtech.2009.05.006. [DOI] [PubMed] [Google Scholar]
- Wang J, Lydiate DJ, Parkin IAP, et al. Integration of linkage maps for the amphidiploid Brassica napus and comparative mapping with Arabidopsis and Brassica rapa. BMC Genomics. 2011;12:101. doi: 10.1186/1471-2164-12-101. http://dx.doi.org/10.1186/1471-2164-12-101 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang H, Wang J, et al. The Brassica rapa Genome Sequencing Project Consortium. The genome of the mesopolyploid crop species Brassica rapa. Nature Genetics. 2011;43:1035–1039. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- Xiao SS, Xu JS, Li Y, et al. Generation and mapping of SCAR and CAPS markers linked to the seed coat color gene in Brassica napus using a genome-walking technique. Genome. 2007;50:611–618. doi: 10.1139/g07-044. [DOI] [PubMed] [Google Scholar]
- Xiong Z, Pires JC. Karyotype and identification of all homoeologous chromosomes of allopolyploid Brassica napus and its diploid progenitors. Genetics. 2011;187:37–49. doi: 10.1534/genetics.110.122473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z, Kim JS, Pires JC. Integration of genetic, physical, and cytogenetic maps for Brassica rapa chromosome A7. Cytogenetic and Genome Research. 2010;129:190–198. doi: 10.1159/000314640. [DOI] [PubMed] [Google Scholar]
- Xiong Z, Gaeta RT, Pires JC. Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proceedings of the National Academy of Sciences, USA. 2011;108:7908–7913. doi: 10.1073/pnas.1014138108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li X, Chen W, et al. Identification of two major QTL for yellow seed color in two crosses of resynthesized Brassica napus line 2127-17. Molecular Breeding. 2011;28:335–342. [Google Scholar]
- Ziolkowski PA, Lysak MA, Heneen WK. Cytogenetic studies in vegetable Brassicas. In: Sadowski J, Kole C, editors. Genetics, genomics and breeding of vegetable Brassicas. Boca Raton, FL: CRC Press; 2011. pp. 257–303. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.