Abstract
Background and Aims
Gene flow is important in counteracting the divergence of populations but also in spreading genes among populations. However, contemporary gene flow is not well understood across alpine landscapes. The aim of this study was to estimate contemporary gene flow through pollen and to examine the realized mating system in the alpine perennial plant, Arabis alpina (Brassicaceae).
Methods
An entire sub-alpine to alpine landscape of 2 km2 was exhaustively sampled in the Swiss Alps. Eighteen nuclear microsatellite loci were used to genotype 595 individuals and 499 offspring from 49 maternal plants. Contemporary gene flow by pollen was estimated from paternity analysis, matching the genotypes of maternal plants and offspring to the pool of likely father plants. Realized mating patterns and genetic structure were also estimated.
Key Results
Paternity analysis revealed several long-distance gene flow events (≤1 km). However, most outcrossing pollen was dispersed close to the mother plants, and 84 % of all offspring were selfed. Individuals that were spatially close were more related than by chance and were also more likely to be connected by pollen dispersal.
Conclusions
In the alpine landscape studied, genetic structure occurred on small spatial scales as expected for alpine plants. However, gene flow also covered large distances. This makes it plausible for alpine plants to spread beneficial alleles at least via pollen across landscapes at a short time scale. Thus, gene flow potentially facilitates rapid adaptation in A. alpina likely to be required under ongoing climate change.
Keywords: Alpine, Arabis alpina, contemporary gene flow, genetic structure, mating system, paternity analysis, pollen dispersal, spatial autocorrelation
INTRODUCTION
Gene flow, the transfer of genes among populations, is a key factor in determining genetic differentiation of populations (Mayr, 1963) and in spreading genes across a landscape (Wright, 1984). In heterogeneous environments, populations of sessile organisms like plants may be naturally isolated. Fragmented populations are more likely to be subject to genetic erosion and loss of genetic diversity owing to genetic drift (Ellstrand, 1992; Frankham et al., 2002). Thus, gene flow is important to maintain a network of connected local populations, thereby increasing a species' chances for survival.
In alpine ecosystems, the heterogeneity of the landscape and the resulting spatio-temporal isolation of plant populations (Till-Bottraud and Gaudeul, 2002; Körner, 2003) have resulted in contrasting hypotheses on the relevance of gene flow in establishing or maintaining population connectivity beyond the local scale. On the one hand, gene flow is considered a rare event because of natural fragmentation of populations, which results in genetic divergence and substantial genetic structure of populations. At a large spatial scale, the genetic structure of populations of alpine species has mainly resulted from historical factors such as the location of glacial refugia and subsequent recolonization along post-glacial expansion routes (Körner, 2003; Schönswetter et al., 2005; Alvarez et al., 2009; Thiel-Egenter et al., 2011). These historical processes have left distinct imprints in the genetic structure of alpine species, which are still detectable today (Schönswetter et al., 2005; Thiel-Egenter et al., 2011). Even at a smaller spatial scale, the alpine landscape is characterized by topographic, edaphic and microclimatic heterogeneity, which potentially causes population divergence and local adaptation in alpine plants (Körner, 2003). These environmental gradients maintain population genetic structure (Till-Bottraud and Gaudeul, 2002; Körner, 2003) as a consequence of limited gene flow by pollen or seed and mating patterns favouring selfing and clonality. This assumption is supported by the low abundance and short flying distances of pollinators observed in the alpine landscape (Bingham and Orthner, 1998). In addition, clonality and selfing have been shown to increase at higher altitudes, impeding the dispersal of genetic variation but ensuring local population persistence (Schröter, 1926; Bliss, 1962; Bliss, 1971; García-Camacho and Totland, 2009).
In alpine plants, low gene flow and the resulting lack of functional connectivity have rarely been demonstrated by empirical data. Recent studies in alpine plants report restricted (Ohsawa and Ide, 2008; Gonzalo-Turpin and Hazard, 2009) to substantial long-distance gene flow events (Raffl et al., 2008). Therefore, it is possible that gene flow facilitates the dispersal of genes more frequently and at larger distances in alpine landscapes than expected. This latter assumption is in agreement with evidence from non-alpine study organisms suggesting that gene flow occurs over large distances and at evolutionarily significant rates (Ellstrand, 1992, 2003). Gene flow via pollen in particular has been shown to disperse genes across large distances in non-alpine studies (e.g. Watrud et al., 2004; Kamm et al., 2009). In studies on alpine plant species, gene flow is indirectly studied from pollinator foraging distances or directly estimated from pollen movement measured by pollen dye (Ellstrand, 1992). Alternatively, gene flow has been estimated from population structure based on genetic differentiation such as Wright's FST (Wright, 1984). The parameter FST gives a historical estimate of gene flow based on pollen and seed dispersal, which is insensitive to small changes in allele frequencies in the contemporary landscape (Sork et al., 1999). Therefore, this parameter is not suited to study contemporary patterns of gene flow (Whitlock and McCauley, 1999). In contrast, pollinator foraging distance and pollen movement give a contemporary estimate of gene flow by pollen, but severely underestimate the real amount of gene movement (Ellstrand, 1992).
To study contemporary gene flow by pollen, segregating molecular markers among parental and offspring populations are suitable to be analysed. The genotypes of the mother plant and offspring are compared with a pool of all potential father plants within a population to detect the most likely pollen parent through paternity analysis (Sork et al., 1999; Holderegger et al., 2010). While population structure, relating to historical processes, has also been analysed widely in alpine plants, studies on contemporary gene flow in herbs across natural landscapes are rare. Moreover, studies in alpine plants that have been conducted with paternity analysis or by pollinator observation are limited to small-scale studies in field sites (e.g. Schmitt, 1980; Hirao et al., 2006; Brunet and Holmquist, 2009; Stöcklin et al., 2009).
Investigating contemporary gene flow is especially relevant in alpine ecosystems because alpine plants are particularly vulnerable to climate change (Theurillat and Guisan, 2001; Till-Bottraud and Gaudeul, 2002; Byars et al., 2007). In alpine organisms, climate change has resulted in specific biological effects such as range contractions and local extinctions (Hughes, 2000; McCarthy, 2001; Parmesan, 2006). By measuring contemporary gene flow, the dynamics of gene movement in environmentally heterogeneous alpine landscapes can be estimated (Sork et al., 1999). This will help in estimating whether alpine plants can adapt locally or migrate to new habitats at short time scales, i.e. a few generations, given the rapid changes caused by factors such as global warming (Till-Bottraud and Gaudeul, 2002; Thuiller, 2007).
This is the first study to investigate contemporary gene flow by pollen in an alpine plant on the basis of an exhaustive sampling at the landscape scale. We chose the alpine perennial plant Arabis alpina (Brassicaceae) to describe contemporary gene flow patterns with paternity analysis because of its recent development as an evolutionary and ecological model species for alpine plants (Ansell et al., 2008; Poncet et al., 2010; Tedder et al., 2011). Only recently, A. alpina was found to be largely selfing in populations of the Alps (Ansell et al., 2008; Buehler et al., 2011), presumably owing to the loss of a functional self-incompatibility system in the course of post-glacial re-colonization (Tedder et al., 2011). Accordingly, we expected to find limited gene flow through pollen because of high selfing and restricted outcrossing, which would probably result in strong spatial autocorrelation. In turn, information on effective seed dispersal is not available in A. alpina, which is potentially a main determinant of spatial genetic structure. The objectives of our study were (1) to investigate the extent to which individuals are functionally connected by contemporary pollen transfer at the landscape scale; (2) to estimate the realized mating pattern; and (3) to determine the fine-scale spatial genetic structure of this species in an alpine landscape of approx. 2 km2. For this, we sampled all individuals of A. alpina in a sub-alpine to alpine landscape and genotyped adult individuals and their offspring/seeds at 18 nuclear microsatellites.
MATERIALS AND METHODS
Study species
The alpine perennial Arabis alpina (Brassicaceae) is a pioneer plant species with a wide distribution in the northern hemisphere, ranging from eastern Canada to the Ural Mountains and tropical East Africa (Koch et al., 2006). In Central Europe, A. alpina occurs in the montane to alpine zones in calcareous open habitats (Schultze-Motel, 1986). It is found on scree fields, along small streams and wells, in moist ravines and on humus-rich rock floors. Arabis alpina is described as diploid (2n = 16) and reproduces both sexually through insect-pollinated seed and asexually by stoloniferous growth (Schultze-Motel, 1986). It has small white flowers frequently visited by insects from various groups, and its seeds are about 1 mm in size with wings extending around the edges, potentially facilitating wind dispersal (Schultze-Motel, 1986). Previous studies showed that A. alpina is self-compatible (Tedder et al., 2011) and highly selfing in populations studied from the European Alps (Ansell et al., 2008; Buehler et al., 2011).
Study area and sampling
Our study area was located on a sub-alpine to alpine landscape of approx. 2 km2 in Urnerboden, Switzerland (Fig. 1). This landscape encompasses a riverbank, a small ravine and a scree field. Along the riverbank at around 1350 m a.s.l., A. alpina occurs frequently over a stretch of about 700 m. The small ravine is located along a mountainside extending from 1350 to 1700 m a.s.l., over a length of about 1 km. In the rock/scree field at 1750 m a.s.l., A. alpina occurs on dry scree as well as on nutrient-rich alpine pastures surrounding the scree field. In this landscape, A. alpina grows as scattered individuals or in small groups. We exhaustively mapped and sampled all A. alpina plants in summer 2008, including all distinctly separated rosettes. We repeatedly searched the entire area to be confident that full sampling was achieved. Fresh leaf material was collected from 595 plants considered to have potentially flowered and was immediately stored in silica gel. Open-pollinated seeds were collected from all fruiting adult plants. The collected seeds were stored at room temperature until DNA extraction. We mapped the location of individuals and the relative distance to neighbouring plants by hand and recorded co-ordinates every 10 m as well as separately for every mother plant from which we sampled seeds using a hand-held GPS receiver. For paternity analysis, we randomly selected 49 mother plants, and we genotyped all adult plants and at least ten seeds per mother plant (n = 499 seeds).
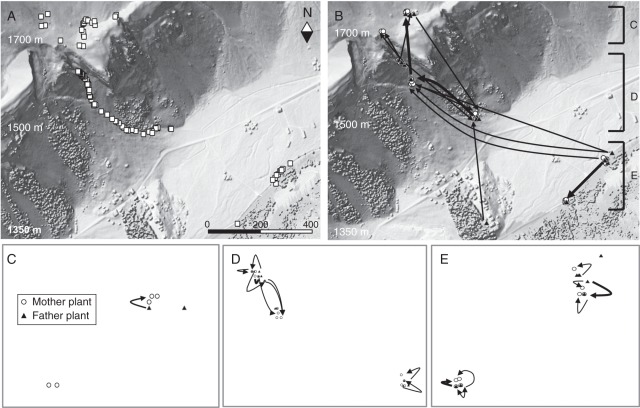
Fig. 1.
Distribution of Arabis alpina in Urnerboden Switzerland. (A) Spatial clusters of individual A. alpina plants are represented by squares. (B–E) Patterns of contemporary gene flow by pollen of A. alpina are indicated by lines connecting mating partners (mother plants shown as circles and father plants shown as triangles) as detected in genetic paternity analysis. Thin lines represent one mating event and thick lines represent two mating events. (B) Long-distance gene flow events (>100 m) across the landscape, and close-ups of the scree field (C), ravine (D) and river (E) are shown for short-distance gene flow events (<100 m). The positions of the close-ups along the North–South axis are marked in B.
DNA extraction and microsatellite genotyping
To genotype adult plants, DNA was isolated from 10 mg of lyophilized material ground using a disruptor mill (Retsch, Haan, Germany). Total genomic DNA was extracted following the DNeasy 96 plant kit protocol (Qiagen, Hombrechtikon, Switzerland). For paternity analysis, embryos and cotyledons were excised from seeds after placing them in H2O at room temperature for 12 h. Disruption and DNA extraction of the seeds were done as described for adult plants.
We used 18 of the 19 polymorphic nuclear simple sequence repeat (nSSR) markers (excluding locus 3X6R) described in Buehler et al. (2011). The nSSR loci were amplified in four multiplex PCRs (Table 1). Multiplex PCRs with fluorescent dye-labelled primers were performed in 10 µL volumes containing 1–2 ng of DNA, 4·6 µL of 2× Master Mix (Multiplex PCR Kit, Qiagen) and 0·1 µL of each primer (5 µm). PCRs were performed on Veriti thermocyclers (Applied Biosystems, Foster City, CA, USA). Initial denaturation was set at 94 °C for 15 min, and was followed by 27–29 cycles (multiplex 1 and 3, 28 cycles; multiplex 2, 29 cycles; multiplex 4, 27 cycles) at 94 °C for 30 s, 57 °C for 90 s, 72 °C for 30 s, ending with a final extension at 72 °C for 30 min. PCR products were run with ROX 400 HD as internal size standard (Applied Biosystems) on a 3130xl Genetic Analyzer (Applied Biosystems), and electropherograms were analysed using GENEMAPPER 3·7 (Applied Biosystems).
Table 1.
Characteristics of 18 nuclear microsatellite loci for Arabis alpina in a sub-alpine to alpine landscape
| Adults |
Offspring |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Locus | NA | HE | HO | FIS | NA | HE | HO | FIS | Labels | Multiplex |
| 3JUY | 6 | 0·74 | 0·22 | 0·70 | 6 | 0·68 | 0·23 | 0·67 | NED | 1 |
| 4MDH | 5 | 0·48 | 0·13 | 0·73 | 4 | 0·41 | 0·07 | 0·83 | FAM | 1 |
| 5GTC | 10 | 0·70 | 0·26 | 0·63 | 6 | 0·54 | 0·15 | 0·72 | HEX | 1 |
| 9VSH | 12 | 0·45 | 0·18 | 0·61 | 8 | 0·44 | 0·13 | 0·71 | FAM | 1 |
| A1T8T | 4 | 0·64 | 0·24 | 0·62 | 3 | 0·65 | 0·15 | 0·77 | NED | 1 |
| A4JW7 | 5 | 0·50 | 0·15 | 0·71 | 5 | 0·58 | 0·13 | 0·77 | HEX | 1 |
| DJSE | 4 | 0·55 | 0·20 | 0·64 | 4 | 0·57 | 0·09 | 0·84 | FAM | 1 |
| 3Q19 | 6 | 0·40 | 0·16 | 0·61 | 5 | 0·47 | 0·14 | 0·69 | NED | 2 |
| 6U3A | 5 | 0·41 | 0·14 | 0·66 | 5 | 0·53 | 0·06 | 0·88 | NED | 2 |
| A25GM | 6 | 0·71 | 0·22 | 0·69 | 5 | 0·67 | 0·23 | 0·65 | HEX | 2 |
| DEET | 7 | 0·45 | 0·18 | 0·59 | 6 | 0·59 | 0·08 | 0·86 | FAM | 2 |
| 46HI | 5 | 0·49 | 0·19 | 0·61 | 5 | 0·38 | 0·09 | 0·76 | HEX | 3 |
| 7PJQ | 7 | 0·64 | 0·22 | 0·66 | 4 | 0·63 | 0·12 | 0·81 | HEX | 3 |
| BWF1 | 7 | 0·45 | 0·12 | 0·74 | 7 | 0·34 | 0·10 | 0·72 | FAM | 3 |
| 79PO | 10 | 0·73 | 0·25 | 0·66 | 9 | 0·76 | 0·17 | 0·77 | HEX | 4 |
| A8E90 | 4 | 0·67 | 0·17 | 0·75 | 5 | 0·69 | 0·13 | 0·81 | FAM | 4 |
| A93Q | 3 | 0·18 | 0·06 | 0·65 | 2 | 0·30 | 0·06 | 0·79 | FAM | 4 |
| EB54 | 5 | 0·61 | 0·20 | 0·66 | 6 | 0·54 | 0·17 | 0·69 | HEX | 4 |
| Mean | 6·17 (2·38) | 0·54 (0·15) | 0·18 (0·05) | 0·66 (0·05) | 5·28 (1·67) | 0·54 (0·13) | 0·13 (0·05) | 0·76 (0·07) | ||
Shown are the number of alleles (NA), expected heterozygosity (HE), observed heterozygosity (HO) and inbreeding coefficient (FIS) for each locus in the adult plants and offspring, with standard errors in parentheses. Furthermore, for each locus, the fluorescent labelking (Labels) and the multiplex PCR in which the locus was amplified (Multiplex) are given.
Genotypic diversity and clonal structure
The average number of alleles per locus, expected (HE) and observed (HO) heterozygosity, and the global inbreeding coefficient (FIS) were calculated separately for adult plants and offspring using GENEPOP 4.0.10 (Raymond and Rousset, 1995; Rousset, 2008) and FSTAT 2.9.3 (Goudet, 1995). To characterize clonal structure, we searched for identical multilocus genotypes (ignoring missing data). We calculated the cumulative probability of finding two matching multilocus genotypes by chance and the estimated expected number of individuals considering the sample size. These calculations, considering both unrelated and sibling individuals, were performed with GENALEX 6.41 (Peakall and Smouse, 2006). The probability of identity, i.e. finding the same multilocus genotype by chance and not as ramets of the same genet, was 1·5 × 10−11. In our sample, it was thus expected to find 9·4 × 10−9 individuals showing the same multilocus genotype. Respective values considering siblings were 1·4 × 10−5 and 8·7 × 10−3. Given these estimates, we henceforth assume that any individuals showing the same allele combinations across all 18 loci resulted from clonal propagation.
Paternity analysis
Paternity analysis was carried out with 49 mother plants and ten open-pollinated offspring per mother plant using CERVUS 3.0.3 (Marshall et al., 1998; Kalinowski et al., 2007). CERVUS uses a likelihood-based approach and assigns paternity according to the highest logarithm of the likelihood (LOD score). LOD scores are calculated by determining the likelihood of assignment of a parent relative to the likelihood of arbitrary parents. We applied the following simulation parameters to find the confidence level of paternity analysis assignment: 10 000 simulated mating events; 1000 candidate father plants; eight as the minimum number of loci; 0·8 as the proportion of candidate parents sampled; genotyping error rate of zero (Slate et al., 2000; Slavov et al., 2005); 0·66 as the rate of inbreeding (FIS value given by the GENEPOP analysis above); all adult plants treated as candidate father plants. While setting the genotyping error rate to 0·01 (as calculated from replicates) did not change the outcome (D. Buehler, unpubl. res.), we used the population-wide inbreeding coefficient to avoid generating a bias when paternity analysis relied on the maternal plants sampled. However, mating system analysis (see below) gave a very similar inbreeding estimate (0·67), which also did not affect the outcome of our study. As in Rathmacher et al. (2010), allele frequencies used in the simulation step were based on the A. alpina adult plants. We subsequently added alleles private to the offspring as P = 0·0001 to the frequency data file. In the paternity analysis, we used 95 % as strict and 80 % as relaxed confidence levels as recommended by Marshall et al. (1998). Effective pollen dispersal distances were calculated as the straight line distance from the mother plant to the most likely pollen parent, reflecting minimum flight distances of pollen-carrying insects. Curve fitting estimation for the distribution of pollen dispersal distances was done using TABLECURVE 2D (Systat, Erkrath, Germany).
Mating system
To characterize the effective mating system in our study population of A. alpina, the mother plants and the open-pollinated offspring were analysed with MLTR v3.3 (Ritland, 2002). On the basis of maximum likelihood procedures, the program estimates the multi- and single-locus outcrossing rates (tm and ts, respectively), biparental inbreeding (tm – ts), maternal inbreeding (F), correlated selfing rate (rs) and the multilocus paternity correlation (rp). The correlated selfing rate indicates among-family variation in mating system; rs = 1 signifies that siblings are all either selfed or outcrossed and rs = 0 indicates that selfing rates do not vary among families. The multilocus paternity correlation is the proportion of full-sibs among outcrossed sibs and indicates whether offspring are the result of single or multiple paternities. The reciprocal of rp (Nep = 1/rp) gives the effective number of pollen donors (Smouse et al., 2001; Fernández-Manjarres and Sork, 2005). All seeds belonging to a mother plant were grouped as a family, and default settings for the estimated parameters were used in MLTR for the analysis of families (i.e. t = 0·9, rt = 0·1, rp = 0·1, F = 0·1). We used 1000 bootstraps to derive confidence intervals with resampling done over families.
Spatial genetic structure
To estimate the genetic structure of the adult plants in our study landscape, we used spatial autocorrelation analysis (Smouse and Peakall, 1999) as implemented in GENALEX 6.41 (Peakall and Smouse, 2006). Pairwise spatial autocorrelation coefficients (r) were computed as a multilocus average for diploid individuals and co-dominant markers. Correlograms were generated with eight size classes of even sample size: 0–30, 30–60, 60–120, 120–240, 240–480, 480–960 and 960–1920 m for a data set with all individuals and a data set excluding clonal individuals. A second analysis was performed, also with and without clonal individuals, to test for spatial genetic structure for the data set with all individuals at distances <30 m, with size classes of 5 m. We used 1000 bootstraps to estimate 95 % confidence intervals for the significance of r against no spatial structuring, i.e. random distribution.
RESULTS
Genotypic diversity and clonal structure
Over the entire landscape, adult plants and offspring of A. alpina showed high inbreeding coefficients (FIS = 0·66 and FIS = 0·76, respectively), while mean genetic diversity was identical for both categories (HE = 0·54; Table 1). The number of alleles per locus ranged from three to 12, with a total of 111 and 95 alleles (genotyped for adults and offspring, respectively) over the 18 loci (Table 1). Five alleles were private to the offspring. Five cases of triplicate samples taken from rosettes assumed to be ramets of the same plant gave identical multilocus genotypes, providing high confidence in the reliability of our markers. Only 28 clonal groups of adult plants were determined. There were 16 clonal groups with individuals located within 5 m, and 12 clonal groups with individuals located at distances of 40–330 m between them. Nineteen clonal groups consisted of only two plants, while nine clonal groups consisted of 3–7 plants.
Paternity analysis and pollen flow distance
Ten offspring were typed at fewer than eight loci and were excluded from the analysis. CERVUS analysis resulted in an exclusion probability of 0·969 and a clear assignment of 249 (51 %) of the 499 offspring analysed to one most likely father plant. The remaining 240 offspring (49 %) were unassigned to a potential father plant. Of these, 122 (51 %) offspring had several potential father plants, whereas 118 (49 %) offspring had no potential father plants assigned in the study area. Of the assigned offspring, 86 (35 %) were assigned with a confidence level of 95 %, and 163 (65 %) were assigned at an 80 % confidence level. Overall, 210 (84 %) offspring resulting from selfing were detected. Outcrossing events were rare, as only 39 of the assigned offspring (16 %) were outcrossed (23 at a confidence level of 95 % and 16 more at a confidence level of 80 %). Eighteen of the 49 mother plants produced only selfed offspring and mother plants had 1–3 outcrossed offspring.
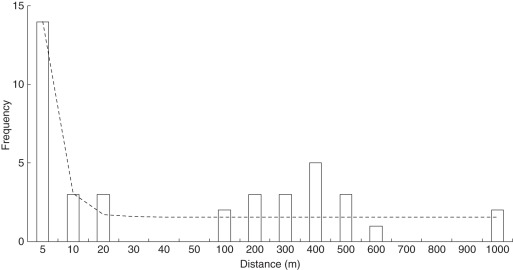
The distance of effective pollen dispersal, i.e if not selfed, ranged from ≤5 m to about 1000 m (Figs 1 and 2). Altogether, 36 % (14 counts) of the 39 effective outcrossed pollen travelled ≤5 m and about 51 % (20) were dispersed to ≤20 m. Two pollen (6 % of outcrossing gene flow) were dispersed over a distance of 1000 m. Pollen dispersal occurred over an altitudinal difference of up to 350 m (Fig. 1). The curve best fitting the empirical data on pollen flow was a general non-linear power equation; pollen dispersal distance = 1·548 + 1672·60x−3·045 (R2 = 0·804; P = 0·001, Fig. 2).
Fig. 2.
Frequency distribution of distances of contemporary gene flow by pollen in Arabis alpina as detected in genetic paternity analysis. The dashed line represents the best-fitting curve (y = 1·548 + 1672·60x−3·045; R2 = 0·804, P = 0·001).
Mating system
Outcrossing rates were considerably low (tm = 0·315; ts = 0·118) indicating a high degree of selfing in A. alpina (Table 2). Biparental inbreeding was substantial (tm – ts = 0·197) and suggests frequent mating among relatives. The estimate for inbreeding in the mother plants was also high (F = 0·674). The correlated selfing rate rs was 0·305, and the multilocus paternity correlation rp was 0·712. Hence, a large number of offspring were the result of single paternity. The number of effective pollen donors (Nep = 1/rp) was 1·4 individuals.
Table 2.
Realized mating pattern analysis of Arabis alpina on a sub-alpine to alpine landscape
| Parental inbreeding, F | 0·674 (0·050) |
| Multilocus outcrossing rate, tm | 0·315 (0·035) |
| Single-locus outcrossing rate, ts | 0·118 (0·019) |
| Biparental inbreeding, tm – ts | 0·197 (0·025) |
| Correlated selfing rate, rs | 0·305 (0·062) |
| Multilocus paternity correlation, rp | 0·712 (0·059) |
| Number of effective pollen donors, Nep | 1·400 |
Errors of estimates based on 1000 bootstraps are shown in parentheses.
Spatial genetic structure
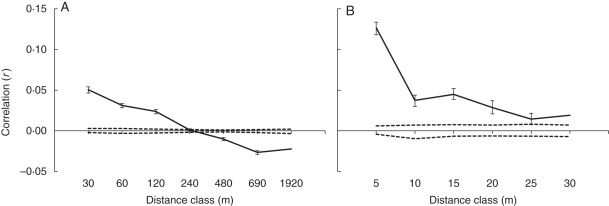
In a correlogram showing autocorrelation coefficients as a function of distance classes and a corresponding 95 % confidence interval, the distance class at which the estimate of r is no longer significant provides an approximation of the extent of detectable positive spatial genetic structure (Peakall et al., 2003). In the data set considering all samples, the intercept occurred at 261 m, indicating that in distance classes 0–240 m, individuals were genetically more related than expected from random distribution at a significance level of P = 0·05 (Fig. 3A). The correlogram for the spatial distance class for <30 m showed that individuals located at 5 m distance were substantially genetically related (Fig. 3B). The correlograms excluding clonal individuals gave similar results, supporting that clones did not affect spatial structuring (results not shown).
Fig. 3.
Analysis of spatial genetic structure of Arabis alpina in a sub-alpine to alpine landscape using spatial autocorrelation analysis. The correlograms show average correlation coefficients between genetic distance among pairs of individuals plotted against distance classes: (A) for eight distance classes across the entire range of the study area, and (B) for a section of the correlogram with 5 m distance classes up to 30 m. Dashed lines delimit 95 % confidence intervals under the hypothesis of randomly distributed genotypes.
DISCUSSION
A considerable number of studies have been published describing historical gene flow in alpine plants as inferred from population genetic structure at large spatial scales or by direct observation of pollinators at small spatial scales (e.g. Hirao et al., 2006; Ohsawa and Ide, 2008; Paun et al., 2008; Gonzalo-Turpin and Hazard, 2009; Stöcklin et al., 2009; Meirmans et al., 2011). However, to our knowledge, studies describing contemporary gene flow at the landscape scale are lacking. The aim of this study was to estimate contemporary gene flow by pollen using paternity analysis and to examine the mating system in the alpine plant A. alpina. We detected maximum gene flow distances up to 1 km. However, A. alpina had a high selfing rate (FIS = 0·66) and a low number of outcrossing events (16 %) within the study population. Spatial genetic structure, indicative of gene flow within a given area, showed that individuals growing in close proximity were genetically more related than expected from random distribution. Below we will discuss these findings in the context of alpine plants.
Long-distance gene flow
Approximately 5 % of outcrossing pollen found in this study was dispersed at much larger maximum distances (≤1 km) than detected in other studies on alpine plants (e.g. Schmitt, 1980; Hirao et al., 2006; Brunet and Holmquist, 2009; Stöcklin et al., 2009). For instance, in the alpine plants Epilobium fleischeri, Geum reptans and Campanula thyrsoides, pollen dispersal was inferred from pollinator movement at a maximum distance of only 40 m (Stöcklin et al., 2009). In another study, paternity analysis was used to measure contemporary gene flow in the alpine plant Aquilegia coerulea across population patches located up to 150 m apart (Brunet and Holmquist, 2009). The results of this study showed that pollinators frequently dispersed pollen among patches. However, gene flow detection was limited to the maximum distance between population patches. Therefore, studies using pollen flow measurements on small spatial scales or pollen flow inferred from pollinator observation bias information about the potential of gene flow in alpine plants, ultimately rendering gene flow a rare event (Ellstrand, 1992). The long-distance dispersal detected in our study is, thus, reasonable, even though our results also in part reflect both the size of our study landscape (2 km2) and the method used to quantify pollen flow. It remains to be studied the ways in which attractive flowers enhance – by attracting pollinators over large distances – or limit long-distance gene flow by pollen – owing to the local supply of sufficient (but see Wirth et al., 2011). Moreover, it is unknown how much of the selfing results from autonomous selfing or from insect-induced pollen deposition (within or among flowers of the same genet). Only field experiments including insect exclusion will be informative in this regard.
Outcrossing pollen in A. alpina was mostly dispersed within a 5-m radius of the mother plant. This is reflected in the general findings of other gene flow studies, which describe pollen flow as a numerically frequent event close to the source plant and decreasing with distance (Ellstrand, 1992; Kamm et al., 2009). In alpine plants, gene flow inferred from pollinator movement has shown that pollinators frequently visit flowers which are located close together (Schmitt et al., 1980; Stöcklin et al., 2009). Pollinator abundance, pollinator flight distances and differences in plant phenology directly limit gene flow in alpine areas (Hirao and Kudo, 2004; García-Camacho and Totland, 2009). In alpine environments, low pollinator abundance has been detected and pollinator activity depends on the frequency of adverse weather conditions and the seasonal phenology of alpine plants (Hirao et al., 2006). However, long-distance pollen flow was not restricted by differences in elevation in our study (Fig. 1). Therefore, asynchronous flowering patterns may have been only partially responsible for restricting gene flow in A. alpina. Instead, a combination of the above-described factors may explain the low number of outcrossing events and high frequency of pollen dispersal over short distances, while the dominance of selfing remains important in describing gene flow in A. alpina.
We detected a high percentage of offspring (49 %) with unresolved paternity. These offspring resulted either from gene flow of unsampled individuals within the landscape (cryptic gene flow) or from pollen immigration via gene flow from outside the study landscape (pollen inflow). This same result was also detected when a 1 % genotyping error was assumed in paternity analysis. In our landscape, cryptic gene flow is plausible as a result of working with a small herb over a large area. Some individuals might have been left unsampled within the landscape despite an exhaustive search. However, pollen inflow may have occurred originating from scree fields at higher elevations or from ravines located at approx. 2 km distance. Again, genotyping errors could account for undetected paternity to a certain degree, but this effect cannot be adequately quantified.
Selfing, realized mating patterns and spatial genetic structure
In this study, a substantial number of offspring (84 %) analysed in the paternity analysis resulted from selfing. This corresponds to the generally held view that alpine plants often self, especially with increasing altitude, even though much evidence for such a presumed trend comes from observational or correlative inference (Bliss, 1971; but see Gugerli et al., 1998; Bingham and Ranker, 2000). As a result of pollen limitation, selfing, but also clonal proliferation, may be seen as a reproductive assurance mechanism (García-Camacho and Totland, 2009).
The investigation of the realized mating system in A. alpina also confirmed that sexual reproduction occurred mostly by selfing. Mother plants had low outcrossing rates, which often resulted in offspring from single paternity. Due to few effective pollen donors, inferred pollen availability was low. In a recent study, Tedder et al. (2011) showed that the self-incompatibility system, as is found in other members of the Brassicaceae (Bateman, 1954), was largely non-functional in populations of A. alpina from the Central and Western Alps, while it still functions in populations from the Appenines in Italy. Accordingly, A. alpina populations in the Central and Western Alps, which comprised our study landscape, are characterized by high levels of inbreeding, suggesting a regional change in breeding system possibly during the course of post-glacial re-colonization (Ansell et al., 2008). This corresponds to the distinct phylogeographic patterns found in the Central Alps, indicating independent origins and/or local glacial refugia (Ehrich et al., 2007; Ansell et al., 2008; Alvarez et al., 2009). Tedder et al. (2011) concluded that populations of A. alpina in the Central Alps lack an inbreeding avoidance mechanism. This suggests that in our landscape, self-compatible plants colonized the area, or, less likely, a loss of self-incompatibility has occurred locally.
In our data set, we detected that individuals were genetically more similar than expected under random distribution up to a distance of about 260 m (Fig 3). Spatial genetic structure is expected to be prominent in alpine plant populations as a result of habitat fragmentation and spatial isolation (Till-Bottraud and Gaudeul, 2002). However, there are several life history traits, such as mating system and means of dispersal, which determine the degree of spatial autocorrelation. For example, the outcrossing Brassicaceae species Biscutella laevigata shows spatial autocorrelation over much shorter distances than A. alpina (Parisod and Bonvin 2008), which may be explained by mating system. However, even within this species, Parisod and Joost (2010) found significant differences between two populations classified as leading vs. trailing edge. Clonally propagated individuals were rare in our landscape and did not visibly affect the correlogram, but clonal reproduction may become more important at even higher elevations where A. alpina approaches its upper limit of occurrence. However, a specific, hypothesis-driven sampling with appropriate replication would be required for rigorous testing of increased clonality at higher elevations. A closer look at the genetic structure found for A. alpina in the study population showed that individuals located within 5 m were highly related, indicating that a large fraction of gene dispersal frequently occurs over such short distances. This assumption was congruent with the result of the paternity analysis, in which the highest frequency of pollen dispersal occurred at <5 m distance from the source (Fig. 2), and the mating system, which indicated high selfing. Therefore, the genetic structure in the landscape mirrors the contemporary pollen dispersal and mating patterns in A. alpina.
Conclusions
Alpine landscapes are especially prone to be affected by climate change (IPCC, 2007), and alpine organisms are showing the first signs of range contractions or distribution changes (Hughes, 2000; McCarthy, 2001; Parmesan, 2006; Frei et al., 2010). Various studies have shown that alpine plants usually possess ample variation at neutral genetic loci (Stöcklin et al., 2009), and genetic variation at putatively adaptive loci has also been described (e.g. Manel et al., 2010; Poncet et al., 2010). Therefore, alpine plants potentially harbour genetic variation suitable for adaptation to changing environmental and climatic conditions. However, the survival of alpine species under climate change does not only depend on suitable standing genetic variation, but also on an organism's ability to spread beneficial genes to those areas where they match future environmental conditions (Byars et al., 2009). The question of whether alpine organisms are able to adapt at short time scales remains open (Till-Bottraud and Gaudeul, 2002; Thuiller, 2007) since it is strongly influenced by outcrossing gene flow and, thus, the mating system.
The present work challenges the common finding that gene flow only occurs across short distances in alpine areas (Pluess and Stöcklin 2004). To make general conclusions about gene flow in A. alpina and in alpine plants, additional landscapes or alpine species, representing a variety of mating and dispersal strategies, should be analysed. Unfortunately, there is a tremendous gap in empirical data on contemporary gene flow in alpine plants to date, particularly so at a large spatial scale. In predominantly outcrossing plants, gene flow is expected to occur more frequently than shown here for the highly selfing plant A. alpina. However, in this study, our landscape-scale analysis, using molecular markers, showed that contemporary gene flow can reach large distances in A. alpina. Even a few long-distance gene flow events can spread genes, such as potentially advantageous alleles, among populations (Morjan and Rieseberg, 2004). The findings of this study suggest that gene flow has the potential to spread (beneficial) alleles in A. alpina at a short time scale to larger distances than previously expected, which might help this species to cope rapidly with climate change.
ACKNOWLEDGEMENTS
We thank Simone Prospero and Sarah Bryner for comments on the manuscript, and Thomas Wuest for software assistance. Two anonymous reviewers helped improve an earlier version of the article. Microsatellite analysis was carried out at the Genetic Diversity Centre, ETH Zurich. This work was supported by the Competence Center Environment and Sustainability BIOCHANGE project of the ETH-domain.
LITERATURE CITED
- Alvarez N, Thiel-Egenter C, Tribsch A, et al. History or ecology? Substrate type as a major driver of spatial genetic structure in Alpine plants. Ecology Letters. 2009;12:632–640. doi: 10.1111/j.1461-0248.2009.01312.x. [DOI] [PubMed] [Google Scholar]
- Ansell SW, Grundmann M, Russell SJ, Schneider H, Vogel JC. Genetic discontinuity, breeding-system change and population history of Arabis alpina in the Italian Peninsula and adjacent Alps. Molecular Ecology. 2008;17:2245–2257. doi: 10.1111/j.1365-294X.2008.03739.x. [DOI] [PubMed] [Google Scholar]
- Bateman AJ. Self-incompatibility systems in angiosperms. 2. Iberis-Amara. Heredity. 1954;8:305–332. [Google Scholar]
- Bingham RA, Orthner AR. Efficient pollination of alpine plants. Nature. 1998;391:238–239. [Google Scholar]
- Bingham RA, Ranker TA. Genetic diversity in alpine and foothill populations of Campanula rotundifolia (Campanulaceae) International Journal of Plant Sciences. 2000;161:403–411. doi: 10.1086/314272. [DOI] [PubMed] [Google Scholar]
- Bliss LC. Adaptations of arctic and alpine plants to environmental conditions. Arctic. 1962;15:117–144. [Google Scholar]
- Bliss LC. Arctic and alpine plant life cycles. Annual Review of Ecology and Systematics. 1971;2:405–438. [Google Scholar]
- Brunet J, Holmquist KGA. The influence of distinct pollinators on female and male reproductive success in the Rocky Mountain columbine. Molecular Ecology. 2009;18:3745–3758. doi: 10.1111/j.1365-294X.2009.04304.x. [DOI] [PubMed] [Google Scholar]
- Buehler D, Graf R, Holderegger R, Gugerli F. Using the 454 pyrosequencing-based technique in the development of nuclear microsatellite loci in the alpine plant Arabis alpina (Brassicaceae) American Journal of Botany. 2011;98:e103–e105. doi: 10.3732/ajb.1000488. [DOI] [PubMed] [Google Scholar]
- Byars SG, Papst W, Hoffmann AA. Local adaptation and cogradient selection in the alpine plant, Poa hiemata, along a narrow altitudinal gradient. Evolution. 2007;61:2925–2941. doi: 10.1111/j.1558-5646.2007.00248.x. [DOI] [PubMed] [Google Scholar]
- Byars SG, Parsons Y, Hoffmann AA. Effect of altitude on the genetic structure of an Alpine grass. Poa hiemata. Annals of Botany. 2009;103:885–899. doi: 10.1093/aob/mcp018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrich D, Gaudeul M, Assefa A, et al. Genetic consequences of Pleistocene range shifts: contrast between the Arctic, the Alps and the East African mountains. Molecular Ecology. 2007;16:2542–2559. doi: 10.1111/j.1365-294X.2007.03299.x. [DOI] [PubMed] [Google Scholar]
- Ellstrand NC. Gene flow by pollen: implications for plant conservation genetics. Oikos. 1992;63:77–86. [Google Scholar]
- Ellstrand NC. Current knowledge of gene flow in plants: implications for transgene flow. Philosophical Transactions of the Royal Society B: Biological Sciences. 2003;358:1163–1170. doi: 10.1098/rstb.2003.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Manjarres JF, Sork VL. Mating patterns of a subdivided population of the Andean oak (Quercus humboldtii Bonpl., Fagaceae) Journal of Heredity. 2005;96:635–643. doi: 10.1093/jhered/esi104. [DOI] [PubMed] [Google Scholar]
- Frankham R, Ballou JD, Bricoe DA. Introduction to conservation genetics. Cambridge: Cambridge University Press; 2002. [Google Scholar]
- Frei E, Bodin J, Walther G-R. Plant species' range shifts in mountainous areas – all uphill from here? Botanica Helvetica. 2010;120:117–128. [Google Scholar]
- García-Camacho R, Totland Ø. Pollen limitation in the Alpine: a meta-analysis. Arctic, Antarctic, and Alpine Research. 2009;41:103–111. [Google Scholar]
- Gonzalo-Turpin H, Hazard L. Local adaptation occurs along altitudinal gradient despite the existence of gene flow in the alpine plant species Festuca eskia. Journal of Ecology. 2009;97:742–751. [Google Scholar]
- Goudet J. FSTAT (version 1·2): a computer program to calculate F-statistics. Journal of Heredity. 1995;86:485–486. [Google Scholar]
- Gugerli F. Effect of elevation on sexual reproduction in alpine populations of Saxifraga oppositifolia (Saxifragaceae) Oecologia. 1998;114:60–66. doi: 10.1007/s004420050420. [DOI] [PubMed] [Google Scholar]
- Hirao AS, Kudo G. Landscape genetics of alpine-snowbed plants: comparisons along geographic and snowmelt gradients. Heredity. 2004;93:290–298. doi: 10.1038/sj.hdy.6800503. [DOI] [PubMed] [Google Scholar]
- Hirao AS, Kameyama Y, Ohara M, Isagi Y, Kudo G. Seasonal changes in pollinator activity influence pollen dispersal and seed production of the alpine shrub Rhododendron aureum (Ericaceae) Molecular Ecology. 2006;15:1165–1173. doi: 10.1111/j.1365-294X.2006.02853.x. [DOI] [PubMed] [Google Scholar]
- Holderegger R, Buehler D, Gugerli F, Manel S. Landscape genetics of plants. Trends in Plant Science. 2010;15:675–683. doi: 10.1016/j.tplants.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Hughes L. Biological consequences of global warming: is the signal already apparent? Trends in Ecology and Evolution. 2000;15:56–61. doi: 10.1016/s0169-5347(99)01764-4. [DOI] [PubMed] [Google Scholar]
- Solomon S, Qin D, Manning M, et al., editors. IPCC. Climate change 2007: the physical science basis, contribution of working group I to the fourth assessment report of the intergovernmental panel on climate change. Cambridge: Cambridge University Press; 2007. [Google Scholar]
- Kalinowski ST, Taper ML, Marshall TC. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Molecular Ecology. 2007;16:1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x. [DOI] [PubMed] [Google Scholar]
- Kamm U, Rotach P, Gugerli F, Siroky M, Edwards P, Holderegger R. Frequent long-distance gene flow in a rare temperate forest tree (Sorbus domestica) at the landscape scale. Heredity. 2009;103:476–482. doi: 10.1038/hdy.2009.70. [DOI] [PubMed] [Google Scholar]
- Koch MA, Kiefer C, Ehrich D, Vogel J, Brochmann C, Mummenhoff K. Three times out of Asia Minor: the phylogeography of Arabis alpina L. (Brassicaceae) Molecular Ecology. 2006;15:825–839. doi: 10.1111/j.1365-294X.2005.02848.x. [DOI] [PubMed] [Google Scholar]
- Körner C. Alpine plant life: functional plant ecology of high mountain ecosystems. Berlin: Springer; 2003. [Google Scholar]
- Manel S, Poncet BN, Legendre P, Gugerli F, Holderegger R. Common factors drive adaptive genetic variation at different spatial scales in Arabis alpina. Molecular Ecology. 2010;19:3824–3835. doi: 10.1111/j.1365-294X.2010.04716.x. [DOI] [PubMed] [Google Scholar]
- Marshall TC, Slate L, Kruuk LEB, Pemberton JM. Statistical confidence for likelihood-based paternity inference in natural populations. Molecular Ecology. 1998;7:639–655. doi: 10.1046/j.1365-294x.1998.00374.x. [DOI] [PubMed] [Google Scholar]
- Mayr E. Animal species and evolution. Cambridge, MA: Harvard University Press; 1963. [Google Scholar]
- McCarthy JP. Ecological consequences of recent climate change. Conservation Biology. 2001;15:320–331. [Google Scholar]
- Meirmans PG, Goudet J Intrabiodiv Consortium, Gagiotti OE. Ecology and life history affect different aspects of the population structure of 27 high-alpine plants. Molecular Ecology. 2011;20:3144–3155. doi: 10.1111/j.1365-294X.2011.05164.x. [DOI] [PubMed] [Google Scholar]
- Morjan CL, Rieseberg LH. How species evolve collectively: implications of gene flow and selection for the spread of advantageous alleles. Molecular Ecology. 2004;13:1341–1356. doi: 10.1111/j.1365-294X.2004.02164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsawa T, Ide Y. Global patterns of genetic variation in plant species along vertical and horizontal gradients on mountains. Global Ecology and Biogeography. 2008;17:152–163. [Google Scholar]
- Parisod C, Bonvin G. Fine-scale genetic structure and marginal processes in an expanding population of Biscutella laevigata L. (Brassicaceae) Heredity. 2008;101:536–542. doi: 10.1038/hdy.2008.95. [DOI] [PubMed] [Google Scholar]
- Parisod C, Joost S. Divergent selection in trailing- versus leading-edge populations of Biscutella laevigata. Annals of Botany. 2010;105:655–660. doi: 10.1093/aob/mcq014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmesan C. Ecological and evolutionary responses to recent climate change. Annual Review of Ecology, Evolution and Systematics. 2006;37:637–669. [Google Scholar]
- Paun O, Schönswetter P, Winkler M, Consortium I, Tribsch A. Historical divergence vs. contemporary gene flow: evolutionary history of the calcicole Ranunculus alpestris group (Ranunculaceae) in the European Alps and the Carpathians. Molecular Ecology. 2008;17:4263–4275. doi: 10.1111/j.1365-294x.2008.03908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall R, Ruibal M, Lindenmayer DB. Spatial autocorrelation analysis offers new insights into gene flow in the Australian bush rat, Rattus fuscipes. Evolution. 2003;57:1182–1195. doi: 10.1111/j.0014-3820.2003.tb00327.x. [DOI] [PubMed] [Google Scholar]
- Pluess AR, Stöcklin J. Spatial population genetic structure in the clonal alpine plant Geum reptans (Rosaceae) American Journal of Botany. 2004;91:2013–2021. doi: 10.3732/ajb.91.12.2013. [DOI] [PubMed] [Google Scholar]
- Poncet BN, Herrmann D, Gugerli F, et al. Tracking genes of ecological relevance using a genome scan in two independent regional population samples of Arabis alpina. Molecular Ecology. 2010;19:2896–2907. doi: 10.1111/j.1365-294X.2010.04696.x. [DOI] [PubMed] [Google Scholar]
- Raffl C, Holderegger R, Parson W, Erschbamer B. Patterns in genetic diversity of Trifolium pallescens populations do not reflect chronosequence on alpine glacier forelands. Heredity. 2008;100:526–532. doi: 10.1038/hdy.2008.8. [DOI] [PubMed] [Google Scholar]
- Rathmacher G, Niggemann M, Köhnen M, Ziegenhagen B, Bialozyt R. Short-distance gene flow in Populus nigra L. accounts for small-scale spatial genetic structures: implications for in situ conservation measures. Conservation Genetics. 2010;11:1327–1338. [Google Scholar]
- Raymond M, Rousset F. GENEPOP (version 1·2): population genetics software for exact tests and ecumenicism. Journal of Heredity. 1995;86:248–249. [Google Scholar]
- Ritland K. Extensions of models for the estimation of mating systems using n independent loci. Heredity. 2002;88:221–228. doi: 10.1038/sj.hdy.6800029. [DOI] [PubMed] [Google Scholar]
- Rousset F. GENEPOP'007: a complete re-implementation of the GENEPOP software for Windows and Linux. Molecular Ecology Resources. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- Schmitt J. Pollinator foraging behavior and gene dispersal in Senecio (Compositae) Evolution. 1980;34:934–943. doi: 10.1111/j.1558-5646.1980.tb04031.x. [DOI] [PubMed] [Google Scholar]
- Schönswetter P, Stehlik I, Holderegger R, Tribsch A. Molecular evidence for glacial refugia of mountain plants in the European Alps. Molecular Ecology. 2005;14:3547–3555. doi: 10.1111/j.1365-294X.2005.02683.x. [DOI] [PubMed] [Google Scholar]
- Schröter C. Das Pflanzenleben der Alpen. Zürich, Switzerland: Raustein; 1926. [Google Scholar]
- Schultze-Motel W. In: Arabis alpina. In Gustav Hegi, Ilustrierte Flora von Mitteleuropa. Conert HJ, Hamann U, Schultze-Motel W, Wagenitz G, editors. Vol 4. Berlin: Parey; 1986. pp. 248–251. [Google Scholar]
- Slate J, Marshall T, Pemberton J. A retrospective assessment of the accuracy of the paternity inference program CERVUS. Molecular Ecology. 2000;9:801–808. doi: 10.1046/j.1365-294x.2000.00930.x. [DOI] [PubMed] [Google Scholar]
- Slavov GT, Howe GT, Gyaourova AV, Birkes DS, Adams WT. Estimating pollen flow using SSR markers and paternity exclusion: accounting for mistyping. Molecular Ecology. 2005;14:3109–3121. doi: 10.1111/j.1365-294x.2005.02620.x. [DOI] [PubMed] [Google Scholar]
- Smouse PE, Peakall R. Spatial autocorrelation analysis of individual multiallele and multilocus genetic structure. Heredity. 1999;82:561–573. doi: 10.1038/sj.hdy.6885180. [DOI] [PubMed] [Google Scholar]
- Smouse PE, Dyer RJ, Westfall RD, Sork VL. Two-generation analysis of pollen flow across a landscape. I. Male gamete heterogeneity among females. Evolution. 2001;55:260–271. doi: 10.1111/j.0014-3820.2001.tb01291.x. [DOI] [PubMed] [Google Scholar]
- Sork VL, Nason J, Campbell DR, Fernandez JF. Landscape approaches to historical and contemporary gene flow in plants. Trends in Ecology and Evolution. 1999;14:219–224. doi: 10.1016/s0169-5347(98)01585-7. [DOI] [PubMed] [Google Scholar]
- Stöcklin J, Kuss P, Pluess AR. Genetic diversity, phenotypic variation and local adaptation in the alpine landscape: case studies with alpine plant species. Botanica Helvetica. 2009;119:125–133. [Google Scholar]
- Tedder A, Ansell SW, Lao X, Vogel JC, Mable BK. Sporophytic self-incompatibility genes and mating system variation in Arabis alpina. Annals of Botany. 2011;108:699–713. doi: 10.1093/aob/mcr157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theurillat JP, Guisan A. Potential impact of climate change on vegetation in the European Alps: a review. Climate Change. 2001;20:77–109. [Google Scholar]
- Thiel-Egenter C, Alvarez N, Holderegger R, et al. Break zones in the distributions of alleles and species in alpine plants. Journal of Biogeography. 2011;38:772–782. [Google Scholar]
- Thuiller W. Biodiversity: climate change and the ecologist. Nature. 2007;448:550–552. doi: 10.1038/448550a. [DOI] [PubMed] [Google Scholar]
- Till-Bottraud I, Gaudeul M. Intraspecific genetic diversity in alpine plants. In: Körner C, Spehn EM, editors. Mountain biodiversity: a global assessment. London: Parthenon Publishing Group; 2002. pp. 23–34. [Google Scholar]
- Watrud LS, Lee HE, Fairbrother A, et al. Evidence for landscape-level, pollen-mediated gene flow from genetically modified creeping bentgrass with CP4 EPSPS as a marker. Proceedings of the National Academy of Sciences, USA. 2004;101:14533–14538. doi: 10.1073/pnas.0405154101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock M, Mccauley DE. Indirect measures of gene flow and migration: Fst ≠ 1/(4Nm+1) Heredity. 1999;82:117–125. doi: 10.1038/sj.hdy.6884960. [DOI] [PubMed] [Google Scholar]
- Wirth LR, Waser NM, Graf R, et al. Effects of floral neighborhood and genetic diversity of seeds in high-alpine cushion plants. Oecologia. 2011;167:427–434. doi: 10.1007/s00442-011-1985-1. [DOI] [PubMed] [Google Scholar]
- Wright S. Evolution and the genetics of populations. Chicago: University of Chicago Press; 1984. [Google Scholar]