Abstract
Until recently, relationships between evidence of colonization or infection by specific microbial species and the development, persistence or exacerbation of pulmonary disease have informed our opinions of airway microbiology. However, recent applications of culture-independent tools for microbiome profiling have revealed a more diverse microbiota than previously recognized in the airways of patients with chronic pulmonary disease. New evidence indicates that the composition of airway microbiota differs in states of health and disease and with severity of symptoms and that the microbiota, as a collective entity, may contribute to pathophysiologic processes associated with chronic airway disease. Here, we review the evolution of airway microbiology studies of chronic pulmonary disease, focusing on asthma, chronic obstructive pulmonary disease and cystic fibrosis. Building on evidence derived from traditional microbiological approaches and more recent culture-independent microbiome studies, we discuss the implications of recent findings on potential microbial determinants of respiratory health or disease.
Keywords: 16S ribosomal RNA, asthma, COPD, cystic fibrosis, microbiota, next-generation sequencing, PhyloChip
An emerging body of evidence in the field of human microbial ecology is redefining our perception of the human superorganism and the delicate balance that exists between the host immune response and the microbial populations that inhabit multiple niches in the human host. From seminal studies of the gastrointestinal microbiome and obesity [1–3] to more recent findings linking airway microbiome composition to bronchial hyper-responsiveness in asthmatics on inhaled corticosteroids [4], it is clear that chronic inflammatory diseases represent a significantly more complex interaction between microbial communities and host inflammatory response than was previously appreciated. Here, we review the recent literature in the field of airway microbiome research, focusing first on the initial observations that spurred microbiota investigations in three major chronic airway diseases: asthma, chronic obstructive pulmonary disease (COPD) and cystic fibrosis (CF). Given the nascency of airway microbiome research, this article also includes recent literature on the upper airway microbiota in states of health and disease. While the findings are discussed independently by disease or anatomic distinction, as the field moves forward a unifying theory on the role of the airway microbiome in modulating chronic airway disease seems likely to emerge.
Specific microbial species & their association with chronic airway diseases
Given the large body of previous literature providing evidence of links between specific microbial species and chronic airway disease based on traditional microbiologic approaches, key findings in asthma, COPD and CF are discussed first.
Asthma
Acute respiratory infections are well-known triggers of asthma exacerbations, but evidence of colonization or infection by specific microbial species have also been linked with the development or presence of asthma. Amongst the bacterial species most studied in this context are the atypical organisms, Chlamydophila (previously known as Chlamydia) pneumoniae and Mycoplasma pneumoniae. Despite a relatively large body of epidemiologic literature investigating links between these organisms and chronic asthma, the evidence overall remains inconclusive (reviewed in [5]). Several studies have reported serologic evidence of atypical bacterial infection associated with the onset of asthma [6–8]. Others [9,10], including a larger study of 104 pediatric patients with newly diagnosed asthma and 120 matched healthy control patients [9], did not find differences in the serologic prevalence of C. pneumoniae-specific antibodies, regardless of the different detection methods used. However, among patients with severe asthma, IgG seropositivity to C. pneumoniae has been interestingly associated with a greater estimated annual decline in lung function, particularly in individuals with nonatopic adult-onset asthma [11].
A contributing factor to the controversy regarding an association between atypical bacteria and asthma has been the difficulty in detecting and diagnosing infection by these organisms. Laboratory culture-based identification, in general, represents a very insensitive diagnostic tool, particularly since these organisms are fastidious and difficult to grow under conventional conditions. Thus, serologic tests for antibodies directed against these species have commonly been employed in studies to determine evidence of infection. However, this approach is complicated by variability in available test methods and interpretation, as well as the relatively high seroprevalence of antibodies against these organisms in the general population [5]. More recently, several studies investigating links between atypical bacteria and asthma have applied targeted PCR-based amplification methods aimed at detecting these specific species using nucleic acids extracted from respiratory specimens [12–14]. Results from these studies have further supported a link between the presence of atypical bacteria in airway samples and chronic asthma, though detection of these species using DNA-based approaches neither indicates the viability of the organism in the respiratory tract nor distinguishes states of colonization versus infection. In a study of 95 patients with persistent asthma and 58 healthy controls, evidence of C. pneumoniae infection in induced sputum by positive PCR and/or immunoglobulin measurements, was more frequent in asthmatic subjects, particularly in those with poorly controlled or nonatopic asthma [14]. Lower airway specimens obtained from asthmatic and healthy subjects by bronchoscopy (bronchial biopsies or bronchoalveolar lavage fluid) have also been analyzed using species-specific PCR for M. pneumoniae or C. pneumoniae [13]. In this study, 31 out of 55 asthmatic patients (56%) were PCR-positive as compared with one out of 11 healthy controls (9%), suggesting that lower airway colonization or infection by atypical bacteria is more prevalent among adults with chronic stable asthma. However, whether these organisms represent the causative agent or are biomarkers of a distinct airway microbiota has not yet been established.
Evidence for a relationship between other bacterial species detected in the respiratory tract and asthma development has been recently described. In a study of 321 neonates from whom hypopharyngeal samples were obtained and cultured at 1 month of age, 21% of neonates had evidence of colonization by Streptococcus pneumoniae, Haemophilus influenzae, Moraxella catarrhalis or a combination of these organisms [15]. Colonization by one or more of these organisms was significantly associated with persistent wheeze during the first 5 years of life, and with the prevalence of asthma at age 5 years (33% in those with neonatal colonization versus 10% without neonatal colonization). Blood eosinophil counts and total IgE at 4 years of age were also significantly increased in children colonized neonatally with one or more of these species. Infection by these same bacteria has also been associated with acute wheezy episodes in children (H. influenzae, M. catarrhalis and S. pneumoniae; overall odds ratio [OR]: 2.9) independent of a similar association observed with viral infections (OR: 2.8) [16]. Although this does not confirm causality, these data collectively suggest that multiple bacterial species may either be prognostic of asthma risk or actively contribute to pathogenic processes associated with this disease. However, further RNA-based studies are necessary to delineate their role in this disease.
Infection by viruses, an established cause of acute wheezing, bronchiolitis and asthma exacerbation among infants and young children, has also been linked with subsequent development of asthma [17–19]. In the Childhood Origins of Asthma Study, among 259 children followed up to 6 years of age, viral respiratory illness with wheezing between birth and 3 years of age due to respiratory syncytial virus (RSV), rhinovirus (RV) or both, was associated with increased risks for asthma at 6 years of age (RSV OR: 2.6; RV OR: 9.8; both RSV and RV OR: 10) [17]. Similar observations were made in an Australian study of 198 children at high risk for atopy followed from birth to 5 years of age [19]. In this birth cohort, the detection of RV in the setting of a wheezing-associated lower respiratory tract illness in the first year of life was related to a diagnosis of asthma at age 5 years (OR: 2.9).
Positive associations have also been reported between RSV infection and the onset of asthma in children [17,20]. Although the greatest risk for asthma at age 6 years in the Childhood Origins of Asthma Study was observed with RV-related wheezing illness in the first 3 years of life, a smaller but statistically significant increase in asthma risk was also observed with RSV-related wheezing illnesses [17]. Longitudinal analyses of a smaller size birth cohort in Sweden followed since infancy after initial hospitalization for RSV bronchiolitis have also noted an increased risk for asthma and allergic sensitization when the children were sequentially evaluated at 7.5 [21], 13 [22] and, most recently, 18 years of age [20]. At each of these time points, asthma prevalence was significantly greater in the group who experienced RSV infection in infancy compared with the control group (e.g., 39 vs 9% at age 18). These findings suggest the potential for a prolonged effect of early RSV bronchiolitis on the development or persistence of asthma into early adulthood.
Finally, two more recently discovered viral species associated with acute respiratory illnesses, human metapneumovirus (hMPV) and bocavirus, have been considered in relation to asthma (reviewed in [23]). More research is needed to further define their contribution to asthma exacerbations, as well as to evaluate potential relationships between infection by these viruses and risk of developing asthma. To date, only one study has attempted to examine the latter research question in relation to hMPV [24]. The investigators found that hMPV-associated bronchiolitis in infancy conferred the greatest risk for a diagnosis of asthma in preschool years (OR: 15.9). Interestingly in this same study, the observed risk for asthma associated with hMPV infection was larger than that noted RSV-related bronchiolitis [24].
Chronic obstructive pulmonary disease
The role of specific microbial species in the pathogenesis of COPD has also been much studied over the past two decades. Despite some ongoing debate, the preponderance of current evidence suggests that bacterial infection plays a causative role in the pathogenesis of acute exacerbations [25–28], while the role of bacterial colonization or infection in the initial development of COPD is less clear. Exacerbations have been associated in particular with the acquisition of new strains of H. influenzae, M. catarrhalis, S. pneumoniae or Pseudomonas aeruginosa [25,26,28], which result in greater airway inflammation compared with exacerbations not associated with a new strain [29]. In an analysis of 177 exacerbations over a 2-year period, new strain-associated exacerbations demonstrated higher levels of IL-8, TNF-α and neutrophil elastase in sputum, as well as serum C-reactive protein, compared with exacerbations not associated with a new strain [29]. Moreover, clinical symptomology is related to the degree of airway inflammation, with sputum neutrophil elastase and serum C-reactive protein showing the strongest correlation with clinical symptom scores at exacerbation [29].
Bacteria are associated with approximately 50% of exacerbations, with the most frequently isolated species being H. influenzae (20–30%). P. aeruginosa is increasingly recognized as an important pathogen in COPD. It is identified more frequently in advanced COPD and associated with approximately 5–10% of exacerbations [30]. P. aeruginosa infection in COPD may demonstrate short-term colonization followed by rapid clearance, but in some cases, longer-term persistence also may occur, which is characterized by both frequent turnover of clones as well as intraclonal microevolution leading to increased mutation rates, antibiotic resistance and greater biofilm production [25,31,32]. Additional bacteria that have been isolated during acute exacerbations include Staphylococcus aureus, Haemophilus haemolyticus, Haemophilus parainfluenzae and the bacterial family Enterobacteriaceae [30]. Although the pathogenic role of these organisms in COPD is unclear, Enterobacteriaceae may be of particular interest as they comprise a large family of Gram-negative bacteria traditionally associated with the GI tract and include many known pathogenic species. Given the clinical significance of many of these organisms outside the respiratory tract, the detection of Enterobacteriaceae in patients with advanced COPD [30] suggests their potential to contribute to the pathogenesis of COPD or of acute exacerbations.
Viruses also are detected in 20–48% of acute COPD exacerbations [27,30,33], most commonly rhinovirus and influenza. Increasingly, detection of RSV and hMPV in acute exacerbations has also been reported [34,35]. In severe exacerbations requiring hospitalization, coinfection by viruses and bacteria resulted in significantly longer hospital stays (≥10 days) [27]. Greater decline in lung function and more severe clinical symptoms are also observed among patients experiencing exacerbations in which viruses are involved [27,36]. These clinical associations probably relate to the greater degree of airway inflammation seen in exacerbations involving bacterial and viral coinfection [27,36]. This suggests relevant interactions between viruses and bacteria in augmenting pathogenicity, such as increased adhesion of H. influenzae or S. pneumoniae to respiratory epithelial cells by viral-mediated upregulation of bacterial receptor expression [37], or conversely, enhanced binding of rhinovirus due to increased receptor expression induced by H. influenzae [38].
In contrast to acute exacerbations, whether microbial colonization or infection contributes to the development of COPD has not been established. However, early colonization of the airways is evident in smokers at risk for COPD, as well as in patients with mild COPD, based on the detection of bacteria such as S. pneumoniae, H. influenzae and M. catarrhalis in protected bronchial brushings or bronchoalveolar lavage fluid [39]. In established COPD, chronic colonization by bacteria can be detected in up to 50% of patients during stable disease [30,39–42], and is accompanied by elevation in airway inflammatory markers [41–43]. Chronic bacterial colonization is also associated with more frequent exacerbations and greater airflow obstruction, which contribute to further progression of lung disease [42]. In summary, current evidence indicates that the establishment or presence of chronic bacterial colonization in patients with COPD is not an innocuous state and probably contributes to the development or progression of COPD.
Although the role of specific bacterial species in COPD has been most studied, other microbial species may potentially colonize the airways in COPD. In particular, associations between Pneumocystis jiroveci colonization and COPD have been noted [44]. A eukaryotic fungal pathogen, and a familiar cause of pneumonia in immuno-compromised hosts, Pneumocystis has been observed to colonize the airways of patients with COPD more often than those of healthy controls. Moreover, independent of smoking history, the prevalence of colonization is positively correlated with the degree of airflow obstruction and therefore the severity of COPD, as staged by the Global Health Initiative on Obstructive Lung Disease [44]. These findings suggest that the airway ecosystem of patients with more severe airflow obstruction represents a suitable environment for Pneumocystis colonization, which may affect COPD progression. Finally, aside from exacerbations, little research has been conducted on the potential role of viruses in chronic stable COPD. Adenovirus has been suggested in one previous study [45] as a potential colonizer in patients with severe emphysema based on an analysis of lung tissue resected from COPD patients. The numbers of alveolar epithelial cells expressing adenovirus E1A protein were increased five- to 40-fold in mild and severe emphysema, respectively, compared with controls [45]. The authors concluded that this suggested the presence of latent adenoviral infection in these patients.
Cystic fibrosis
CF is, from a microbiological standpoint, arguably one of the most studied chronic airway diseases. Due to mutations in the CF transmembrane regulator gene, CF affects multiple organs, though the primary cause of mortality is chronic airway colonization and pulmonary failure. P. aeruginosa is the primary recognized bacterial pathogen in CF airway disease. Colonization and infection by P. aeruginosa is strongly associated with morbidity and mortality in CF, and antimicrobial therapies integrated into the chronic management of CF are largely targeted at P. aeruginosa (e.g., inhaled tobramycin). However, a number of other bacterial and fungal pathogens have been associated with chronic inflammatory airway disease in CF patients. Interestingly, many of these infectious agents, in particular bacterial, exhibit age-dependent relative abundance. For example, relatively high numbers of H. influenzae are detected in pediatric patients; however, the burden of this pathogen is reduced and remains low as patients progress towards adulthood. On the contrary, P. aeruginosa exhibits the opposite dynamics, being present in relatively low numbers in younger patients but becoming highly prevalent as the patient’s age and disease progresses.
Given the extent of microbiological studies focused on CF, it is unsurprising that a relatively large number of bacterial species are associated with pulmonary infection and chronic colonization in this patient cohort. Table 1 details many of these species and their clinical relevance to CF airway disease. In several cases, coinfection by a number of species has been observed using culture-based approaches [46]. More recently, a number of unusual pathogens have been described in the airways of CF patients including Ralstonia species [47], members of the Pandoraea genus [48,49], the Streptococcus milleri group [50,51] and species within the Enterobacteriaceae [52]. While specific species, such as Pandoraea apista, have been associated with transmissible severe airway disease [49], it should be noted that several of these studies were performed using sputum samples, which opens up the possibility of an oral origin for these species. Nonetheless, given the oral–airway continuum and that bacterial clearance is particularly compromised in CF patients, it is wholly possible that these species play a key role in CF pulmonary disease. In addition, since the frequency of recovery of these species from CF pulmonary samples appears to have increased over recent years, these organisms certainly merit further investigation.
Table 1.
Bacterial species most commonly associated with cystic fibrosis airway disease.
| Species | Clinical significance | Ref. |
|---|---|---|
| Pseudomonas aeruginosa | One of the most important CF infectious agents Initial nonmucoid strains give rise to highly antibiotic-resistant mucoid strains Forms biofilms that are refractory to mucocillary clearance, host responses and antimicrobials |
[98] |
| Haemophilus influenzae | Isolated most frequently from young patients Does not chronically colonize the airways but can persist for extended periods of time – associated antibiotic-resistant hypermutable strains |
[99,100] |
| Staphylococcus aureus | Frequently recovered from CF airways (52% of patients) Chronic infection dynamics exhibit a peak in adolescence |
[101] |
| Burkholderia cepacia complex | Group of at least 17 Burkholderia species Important opportunistic pathogens Cause a progressive, invasive and fatal pulmonary disease known as cepacia syndrome |
[102,103] |
| Stenotrophomonas maltophilia | Frequently isolated from mid-to-late adolescent CF airways Chronic infection is unusual (10%) of cases |
[104] |
| Achromobacter xylosoxidans | Emerging CF pathogen Detected with increasing frequency from CF sputum samples Can result in chronic infection with antimicrobial resistant isolates |
[105] |
| Nontuberculous Mycobacterium | Nontuberculous mycobacterium associated with coinfection with Stenotrophomonas or Aspergillus species Patients with chronic Mycobacterium abscessus infection exhibit greater rates of lung function decline than those with no nontuberculous mycobacterium infection |
[106] |
| Streptococcus pneumoniae | Isolated most frequently from pediatric CF patients Associated with pulmonary exacerbations |
[106–108] |
CF: Cystic fibrosis.
As with asthma, viral infection also plays a role in acute CF pulmonary exacerbation. In a study of 103 pediatric CF patients in Sao Paulo (49 female, 54 male), samples obtained from participants included nasopharyngeal aspirates or nasal mucus specimens for viral analysis, in addition to sputum or oropharyngeal samples for microbial culture. The study examined samples (both exacerbation and clinically stable state) for influenza viruses A and B, human parainfluenza viruses 1, 2 and 3, human coronavirus, hMPV, adenovirus, human bocavirus and picornavirus using molecular-based quantitative-PCR assays. A significant relationship between the presence of respiratory viruses and pulmonary exacerbation was found (OR: 1.195; p = 0.010). In addition, specific rhinovirus subtypes A2 or C were also significantly associated with respiratory exacerbations (OR: 1.213; p ≥ 0.025) [53]. Other studies corroborate these findings demonstrating a relationship between viral infection and pulmonary exacerbation in CF patients [54,55]. More recently, a study has demonstrated a mechanism by which viral infection may cause more severe exacerbation in CF patients. Performed using primary CF airway mucocillary differentiated epithelial cell lines infected with mucoid P. aeruginosa, Chattoraj and colleagues demonstrated release of planktonic P. aeruginosa cells from the apical and basolateral surfaces of biofilms attached to the epithelial cell cultures, upon superinfection with rhinovirus [56]. The presence of free-swimming cells coincided with increases in chemokine production, suggesting that the rhinovirus-driven release of immunogenic P. aeruginosa cells may increase inflammatory responses during viral exacerbation. Clearly, these findings have implications for the mixed species populations known to exist in CF airways, as will now be discussed.
Microbiota community composition in airway health & disease
Previous research has charted evidence of a role for specific organisms in the pathogenesis of asthma, COPD or CF. This has been achieved primarily through the use of culture-based approaches or targeted molecular approaches to identify specific microbial species. The sensitivity of such methods for detecting other microbiota is inherently limited, resulting in lost opportunities to investigate potential relationships between the broader microbial community and airway disease. Nonetheless, these findings have provided the foundation for applying broader culture-independent tools to interrogate the microbiome associated with chronic inflammatory airway disease in a variety of patient populations.
The application of newer culture-independent tools that enable interrogation of the microbial community without prior expectation of the organisms present, including fastidious or nonculturable species, has dramatically impacted on our knowledge of human microbial diversity in a variety of organ systems [57–60]. The most widely applied approach to study bacterial communities has been analysis based on the 16S ribosomal RNA (rRNA) gene, which is ubiquitous to all bacteria. This gene is comprised of conserved stretches of sequence that can be used to design universal primers to amplify the gene from the majority of known bacterial species. These regions are interspersed with variable sequence regions, the sequences of which can be used to assign identity and phylogeny of the organisms in a mixed community. While several molecular approaches for 16S rRNA-based analysis exist, high-throughput tools such as 16S rRNA microarray platforms or next-generation sequencing applications have been more recently applied to provide profiles of microbial community composition at high resolution. In particular, these two complementary approaches have the capacity to provide the greatest resolution in profiling bacterial communities [61–65]. In contrast to the study of bacterial communities, fewer inroads have been made in molecular approaches for global analysis of viral or fungal microbiota, although there have been some recent efforts to advance these areas [66–70].
From recent studies of the human microbiome, a wealth of information has emerged about the diversity of microbiota in specific organ niches, with increasing efforts to gain insight into functional aspects of the microbiome and to better understand the role of microbiota in disease pathogenesis [71]. Most of this knowledge has come from the large body of literature on the role of gut bacterial communities in health and disease [72]. By contrast, our knowledge and understanding of the airway microbiome in health and disease is lagging, particularly with respect to the lower respiratory tract. However, the rate of research efforts focused on the airway microbiome has increased and recent studies of the upper and lower airway microbiota in the context of airway health or disease are now discussed.
Upper airway microbiota in health & disease
Sampling of the upper respiratory tract can be performed with greater ease compared with the lower respiratory tract. In particular, relationships between the oral microbiota and periodontal disease have been examined in several studies by 16S rRNA analysis approaches, results of which have demonstrated multiple types of subgingival/periodontal microbiota to be important in periodontitis [73]. It is worth noting that the abundance of information on the oral microbiota generated over the past several years has been catalogued in the Human Oral Microbiome Database [58].
Recent efforts have been made to characterize the oral and nasopharyngeal microbiota in healthy individuals, including bacteria and fungal communities [66,74,75]. Zaura et al. sampled several intra-oral sites (given potential microbial community variation in different oral niches) in three healthy subjects, including the buccal and palatal surfaces, tongue and saliva [75]. Using 454-pyrose-quencing analysis of 16S rRNA amplicon libraries, they observed that 66% of all the sequence reads, translating into 72% of higher taxa (i.e., classified at genus level or above), were shared across the three individuals, suggestive of a ‘core’ oral bacterial community in healthy individuals. The most predominant of these taxa were Streptococcus, Corynebacterium, Neisseria, Rothia and Veillonella species; all genera that have previously been identified in the oral cavity [58]. However, as the authors acknowledge, the diversity and richness of the oral microbiota are probably underestimated in this study because of the relatively limited sequence read depth performed for each sample and their criteria for selection of sequences for analysis (minimum of five reads). Typically, while this approach provides good information on the dominant phylotypes in a given community, it frequently neglects the less abundant or rarer bacterial phylotypes, which have recently been shown to contribute the greatest functional capacity to a given community [76]. This underscores the need for high-resolution profiling approaches to fully understand community structure and its relationship with clinical variables.
Nonbacterial microbiota in the respiratory tract have not been well characterized and is an area of unmet research need. While there is evidence of the involvement of specific fungal species in COPD [77,78] and CF [79–81], little is known of the airway fungal microbiota in the pathogenesis of asthma, COPD or CF. Moreover, even less is understood regarding potential interkingdom microbial interactions that may impact functional expression of the collective microbiome and in turn, host responses and manifestations of airway disease. The oral fungal microbiome was recently studied in 20 healthy individuals from whom oral rinse samples were analyzed using pan-fungal primers followed by pyrosequencing [66]. In total, 101 fungal species were identified and of these, in a given individual, only between nine and 23 were known culturable species. Candida spp. were the most frequently isolated, in 75% of subjects, followed by the genera Cladosporium, Aureobasidium, Saccharomycetales, Aspergillus, Fusarium and Cryptococcus. While four genera (Candida, Cladosporium, Aureobasidium and Saccharomycetales) were detected in at least ten subjects (50% of participants), the authors also suggest the existence of a ‘basal’ state oral mycobiome, consisting of 15 genera present in at least 20% of study subjects. However, a large number of diverse fungi were putatively identified in this study, with 53% of the identified genera observed only once across samples. These findings provide an initial baseline for studying fungal microbiota along the respiratory tract and demonstrate that within the oral cavity which houses a diversity of bacterial species, a diverse fungal microbiota can coexist. Future investigations of the fungal mycobiome in other respiratory niches such as the lower airways, and potentially in relation to chronic inflammatory airway disease, would be of great interest.
In healthy individuals the nasopharyngeal niche appears to harbor a more varied, yet distinct, community of microbiota compared with the oropharynx [74,82,83]. The predominant phyla represented in the nostril are the Firmicutes and Actinobacteria [74], including the bacterial families Staphylococcaeae, Propionibacteriaceae, and Corynebacteriaceae [82]. In the oropharynx, the predominant phyla are Firmicutes, Proteobacteria and Bacteroidetes. Thus, the nostril and oropharynx exhibit distinct microbiota distributions at the phylum level, with the former more similar to that found on the skin and the latter more reminiscent of the distribution of gastrointestinal-associated microbiota [74]. However, the specific region sampled in the nasopharynx (i.e., a more ‘outer’ area such as the nostril vs a more ‘inner’ part of the nasal tract) may influence the distribution observed, as oropharyngeal-associated communities have also been noted in samples obtained from a more posterior aspect of the nasal cavity [82].
The oral and nasopharyngeal microbiota in a ‘nonhealthy’ state was recently explored by Charlson et al., examining the impact of cigarette smoke exposure on the microbial community found in these two niches [84]. As the major risk factor for COPD, cigarette smoke may promote microbial colonization and respiratory infection by a number of mechanisms, including disruption of mucociliary clearance, enhanced binding of bacteria to epithelial cells and impairment of host immune responses [84]. 16S rRNA pyrosequencing analysis of samples from 29 asymptomatic current smokers and 33 healthy nonsmokers, demonstrated that bacterial communities from smokers clustered separately from nonsmokers [84]. Smokers exhibited greater variation in the types and relative abundance of bacteria inhabiting their oropharynx or nasopharynx, indicative of a more heterogeneous or potentially diverse bacterial community. The investigators identified specific microbiota that appeared to distinguish a smoking-associated bacterial community from that of nonsmokers. At the general level in oropharyngeal samples, the presence of Capnocytophaga, Megasphaera, Veillonella, Haemophius and Neisseria spp. best characterized a smoking-associated community. In the nasopharynx, smokers also demonstrated greater relative abundance of several bacterial families, primarily belonging to the Firmicutes phylum, including Aerococcaceae, Eubacteriaceae, Lachnospiraceae, Peptostreptococcaceae and Erysipelotrichaceae. The main conclusion of this study is that smoke exposure alters normal microbiota community structure, which may facilitate enhanced pathogen colonization of the upper respiratory tract [84]. Normal commensal microbiota have been shown to modulate host immune responses in respiratory mucosa in the setting of infections by Mycoplasma pulmonis or influenza A virus [85,86].
Lower airway microbiota & relationships to asthma, COPD & CF
Airway microbiome in asthma
Recent culture-independent studies indicate that a resident microbiota community exists in the lower respiratory tract [4,83,87], in contrast to the traditional view that the lower airways are a completely sterile environment in healthy individuals. However, it remains to be proven definitively whether the detected presence of lower airway microbiota in apparently healthy individuals represents true colonization of the lower airways, the presence of transient inhaled species with regular turnover of the microbial community, oropharyngeal contamination in the collection of lower airway samples or even linked potentially to individuals incorrectly categorized as truly healthy. Given the practical challenges in sampling the lower airways in human subjects, distinguishing among the first three of these possibilities is difficult. While it is perhaps less surprising that a rich microbial community is present in COPD airways by culture-independent analyses, recent findings have also demonstrated that a diverse community of microbiota in the bronchi is associated with chronic stable asthma [4,83]. Using a traditional 16S rRNA clone library and sequencing analysis, Hilty et al. analyzed respiratory samples from 24 adults (11 with asthma, five with COPD and eight healthy controls) and 20 children (13 with ‘difficult’ asthma, seven controls) [83]. Although the clone library approach for 16S rRNA-based community analysis provides significantly less microbial community resolution compared with next-generation sequencing or PhyloChip (Affymetrix Corporation®, Santa Clara, CA, USA) microarray methods [62], pathogenic members of the Proteobacteria phylum, in particular Haemophilus spp., were more commonly found in bronchial brushings from adult patients with airway disease (asthma or COPD) compared with controls. This was similarly observed in analysis of bronchoalveolar lavage fluid from pediatric asthma patients. By contrast, members of the Bacteroidetes phylum, especially Prevotella spp., were more often identified in healthy adult controls. Notably, all asthmatic patients in the study were prescribed corticosteroid therapies (inhaled or oral), which have unknown effects on the microbiome. This study also investigated potential geographical differences in the microbial community between the upper and lower respiratory tract by comparing samples from the nasopharynx (NP), oropharynx and left upper lung lobe (LUL). NP microbiota appeared to be distinct from that found in the oropharynx or LUL. Among patients with airway disease (asthma or COPD), LUL microbial communities differed from that found in the LUL or oropharynx of healthy controls, suggesting distinct differences in community composition in diseased bronchi compared with healthy states. Interestingly, smokers were observed to have lower 16S rRNA copy numbers. While 16S rRNA copy number cannot be directly compared with measures of bacterial community diversity or richness (i.e., the number of different phylotypes), since some bacteria can possess multiple copies of the 16S rRNA gene, it is interesting to note that others have observed smokers to exhibit greater variation in the types and relative abundance of bacteria inhabiting the nasal or oropharynx, compared with nonsmokers [84]. Despite limitations of a small sample size, limited analysis for potential differences in airway microbiota between asthmatic and COPD subjects and the unknown potential impact of corticosteroid use on the findings, the study suggests that characteristic bacterial microbiota may colonize the lower airways of patients with stable asthma.
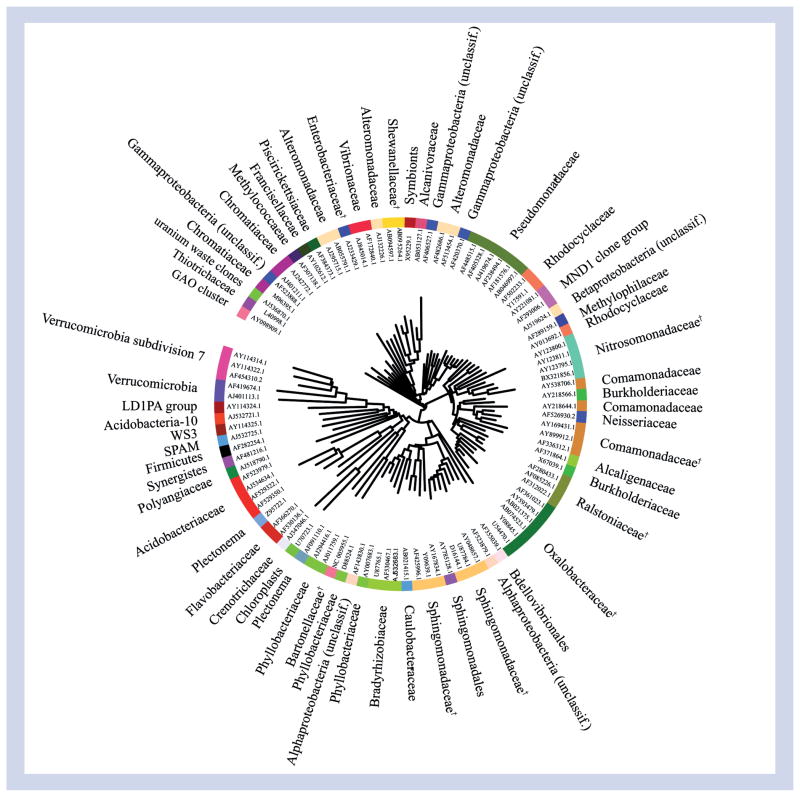
In a larger study of 75 asthmatic and nonasthmatic healthy adults, chronic stable asthma was also found to be associated with differences in the composition and structure of the bronchial microbiota community compared with healthy subjects [4]. Huang et al. examined the microbial community sampled by triplicate protected specimen bronchial brushings, using a high-density bacterial microarray (16S rRNA PhyloChip), as well as quantitative PCR analysis [4]. The G2 PhyloChip platform is capable of detecting approximately 8500 bacterial taxa in parallel. Owing to the nature of this assay (hybridization based), low- and high-abundance community members are detected with equal efficiency, permitting a high-resolution profile of the community to be generated. Asthmatic subjects exhibited greater bacterial burden and significantly higher bacterial diversity in their bronchial microbiota community. All asthmatics in this study had stable mild-to-moderate disease and were taking standardized inhaled corticosteroid therapy. This study further investigated potential relationships between characteristics of the microbial community and clinical features of asthma. In independent analyses, both bacterial diversity and microbiota community composition were strongly correlated with bronchial hyper-responsiveness. In particular, greater bronchial hyper-responsiveness was highly correlated with the relative abundance of approximately 100 specific bacterial phylotypes, including bacterial families within Proteobacteria such as Oxalobacteraceae, Pseudomonadaceae, Sphingomonadaceae and Comamonadaceae (Figure 1). Many representative species in these families possess either pathogenic potential or chemical functions that conceivably could contribute to known pathophysiologic processes in asthma. These findings thus may be of particular interest as they suggest the possibility of functional interactions between specific components of the airway microbiome and the host that may impact pathophysiologic features of asthma.
Figure 1. Phylogenetic tree based on representative 16S rRNA gene sequences of the approximately 100 bacterial taxa highly correlated with more severe bronchial hyper-responsiveness (p < 0.01; q < 0.015) in asthmatics on inhaled corticosteroids.
Colors represent different bacterial families.
†Taxa with member species previously associated with clinical disease or possessing notable functional features.
Unclassif.: Unclassified.
Reproduced with permission from [4].
Airway microbiome in COPD
In COPD, bacterial colonization or infection of the lower airways has long been noted by culture methods, but recent culture-independent investigations by 16S rRNA analysis approaches have revealed much greater community richness in COPD airways than previously appreciated [87,88]. In a PhyloChip-based study of COPD patients hospitalized for respiratory failure owing to severe exacerbations, more than 1200 bacterial taxa were identified in lower airway samples [88]. In addition to bacterial families containing the typical pathogens associated with COPD exacerbations (e.g., Pasteurelleaceae/H. influenzae, Pseudomonadaceae/P. aeruginosa, Streptococcaceae/S. pneumoniae, Moraxellaceae/M. catarrhalis), a multitude of other families with potentially pathogenic member species were identified. These included members of the Enterobacteriaceae, Burkholderiaceae, Mycoplasmataceae, Alteromondaceae and many others. COPD patient samples harbored an average of 411 bacterial taxa despite concurrently receiving broad-spectrum antimicrobial therapies. In addition, a core microbiota comprised of 75 taxa present in all subjects was identified, representing 27 distinct families (e.g., Pseudomonadaceae, Enterobacteriaceae, Campylobacteraceae, Helicobacteraceae and others) and again including many potentially pathogenic species.
Although this highly select cohort of severely exacerbating COPD patients all harbored P. aeruginosa, the patients clustered into two separate groups distinguished by significant differences in community richness. Patients with a richer bacterial community had received ventilatory support for a significantly shorter duration, and this microbiota community was more commonly populated by members of Clostridiaceae, Lachnospiraceae, Bacillaceae and Peptostreptococcaceae. Many of these represent normal commensal microbiota in the upper respiratory tract that may help maintain community stability and functional homeostasis in the microbiome, as has been observed in the gut [89]. These findings suggest that members of the airway microbiota community, other than typical respiratory pathogens, may have important roles in determining states of clinical instability or stability in COPD.
A comparison of the lower airway microbiota among patients with COPD, ‘healthy’ smokers without evidence of COPD and nonsmoking controls was performed by analysis of bronchoalveolar lavage fluid collected from 14 subjects (four with COPD, seven smokers with normal spirometry, and three healthy nonsmokers) [87]. Out of the four COPD subjects, three had mild obstructive disease and one had severe disease by Global Health Initiative on Obstructive Lung Disease stage criteria. Although the subject numbers were small, nonsmoking controls demonstrated heterogeneity in their lower airway bacterial communities similar to that observed in healthy nonsmokers and two of the COPD patients with mild disease. By contrast, very limited bacterial community diversity was observed in the two other COPD patients, one of whom had severe disease. Across all subjects, the predominant phyla found in the lungs were the Proteobacteria, Firmicutes and Bacteroidetes, as also observed in other studies [4,83]. The authors suggest that the existence of a ‘core’ pulmonary microbiome in healthy subjects comprised of bacterial genera with the greatest number of sequence reads in their study. These included Pseudomonas, Streptococcus, Prevotella and Fusobacterium spp. Finally, microanatomical geographic differences in microbiota composition may exist in the diseased lung. In analysis of COPD lung explants, differences in bacterial community composition were noted, depending on which bronchus was sampled within the same lung. Species highly associated with microanatomical variation in community composition included Haemophilus, Stenotrophomonas or Pseudomonas spp., suggesting that these organisms may represent keystone species that drive ecosystem functioning and, as a result, define both community composition and host responses in these discrete niches. Moreover the data suggest that, similar to the GI tract, niche-specific microbiota also exist in the airway.
Airway microbiome in CF
Culture-independent efforts in the field of CF airway microbiota have a relatively longer history. The accumulating body of literature and anecdotal evidence from the clinical laboratory for the coexistence of multiple species in the airways of CF patients prompted Rogers and colleagues, in 2003, to perform the first culture-independent analysis of adult CF patient bronchoscopic and sputum samples [90]. They demonstrated that multiple distinct 16S rRNA PCR products were present in each patient sample, indicating the presence of distinct bacterial phylotypes in this niche [90]. Moreover, subsequent sequencing efforts identified, in addition to P. aeruginosa and Stenotrophomona smaltophilia, multiple other species typically associated with the oral or gastrointestinal cavity, including Prevotellaoris, Fusobacterium gonidoformans and Bacteroides fragilis, amongst others; species not previously associated with CF airways [90]. The investigators provided a seminal follow-up study, based on RNA rather than DNA, to profile active members of the CF airway microbiota in adult patients [91]. Total RNA extracted from 71 sputum samples was reverse transcribed and used as a template for 16S rRNA PCR amplification. Amplicons generated by this approach represent viable community members, actively transcribing their 16S rRNA gene. Profiling of the amplicons identified 248 distinct bands, each originating from a viable bacterial member of the CF airway, thus providing the first evidence for an active, diverse airway microbiota in a chronic pulmonary disease. Across patient samples, bacterial community richness (number of distinct phylotypes detected) was as high as 37 viable phylotypes in one patient, substantially greater than the typical one or two pulmonary pathogens reported through culture-based clinical laboratory testing to colonize CF airways. Follow-up cloning efforts from three patients in the study and sequencing 53 of these clones identified the presence of additional species not previously identified in CF airways including two Abiotrophia species, Mycoplasma salivarium, Ralstoniataiwanensis, Rothiamucilaginosa, Treponemavincentii and Veillonellaatypicai [92]. These seminal studies demonstrated that a diversity of viable bacterial species not typically associated with the CF airways were present in these patients that could putatively contribute to airway disease. This ground-breaking work laid the foundation for a number of subsequent studies that have dramatically expanded our understanding of the airway microbiota.
Subsequent culture-independent studies using progressively higher-resolution tools have revealed an even more complex and diverse microbiota in the airways of CF patients [93–95]. These studies have further expanded the diversity of organisms detected in the airways of CF patients. However, it is necessary to determine relationships between the composition of these assemblages and host health status as a means to identify key features of the microbiome that contribute to patient health. In a cross-sectional study of respiratory samples collected from 45 clinically stable CF patients (aged between 9 months and 72 years), who had not received antibiotics for acute pulmonary exacerbation within 2 months of sample collection, Cox and colleagues examined changes in airway microbiota composition within this age-stratified, cross-sectional study [95]. They demonstrated a significant negative correlation (−0.48; p < 0.0003) existed between patient age and pulmonary function, confirming that, compared with the younger population, older CF patients in the cohort exhibited poorer airway health. The authors also found that compared with the younger patients, older individuals exhibited significant losses in bacterial community richness (number of types of bacteria present), evenness (relative distribution of community members) and diversity (index calculated based on richness and evenness metrics). In addition, the communities present in the airways of older CF patients were comprised of phylogenetically related organisms primarily belonging to the family Pseudomonadaceae. This study demonstrated that, as in chronic inflammatory disease of the GI tract, which is associated with loss of bacterial diversity and domination of the community by a handful of species, the airway microbiota exhibits the same hallmarks in CF patients with the most severe inflammatory status. Limitations of the study include its cross-sectional nature and the specimen types examined: expectorated sputum and deep throat swabs from adult and pediatric patients, respectively. Although such specimens are always subject to contamination by oral microbiota, these are the standard types of samples obtained clinically to infer airway microbiology in treating CF patients. Nonetheless, this study provided the first evidence that airway microbiota composition is associated with airway function status, suggesting that microbiota composition could provide a robust diagnostic or prognostic marker for airway health.
The potentially important contributions and role of oropharyngeal-associated microbiota in lower airways disease should not be overlooked. The finding in lower respiratory tract specimens of organisms typically associated with the oropharynx always raises the question of oral contamination. However, this may not necessarily be the case, since recent evidence suggests that the presence of these species in the airways may not be wholly innocuous. Given the complexity of airway microbiota that has been revealed in association with chronic airway disease, particularly CF, interactions among microbial communities undoubtedly occur and are likely to affect the functional or phenotypic expression of one or more organisms, including the microbiota community as a collective whole. Two studies have elegantly demonstrated the pathogenic contributions of oropharyngeal flora in the presence of a primary pathogen, namely P. aeruginosa. Using a rat lung infection model, Duan and colleagues showed that pulmonary damage was significantly enhanced when animals were coinfected by P. aeruginosa and oropharyngeal flora isolated from CF patients, compared with infection by P. aeruginosa alone [96]. This was mediated by auto-inducer-2 quorum sensing and led to upregulation of a number of P. aeruginosa virulence factors. Similar findings were observed in a more recent study by the same investigative group in which a polymicrobial infection Drosophila melanogaster model was used [97]. Oropharyngeal-derived strains that themselves were not pathogenic to the host significantly worsened fly survival when coinfection with P. aeruginosa (PA01 strain) was performed. Thus, the finding of oropharyngeal-associated bacterial microbiota in the lower airways should not automatically be construed as innocuous bystanders in airway disease.
As we move into the age of high-throughput ‘omics’, new tools developed to provide comprehensive profiles of the species present in a given community, as well as both the functional capacity of, and expression profiles from, their collective pan-genome, will provide unparalleled insights into the relationship between the human host and its microbial inhabitants. Though niche-specificity exists in discrete niches, it is comforting that, already, we can begin to observe consistent trends in data generated across mucosal surfaces in distinct anatomical niches in the human host. For example, chronic inflammation in either the GI tract or airway results in reduced microbiota diversity at both of these sites, though the composition of these depleted microbiota is distinct, niche specific and probably driven by local selective pressures. Clearly, there is a great need for highly integrated translational research efforts across large cohorts of patients. However, with the recent development of molecular tools and platforms to provide the comprehensive microbial profiles necessary to interrogate the complex communities that inhabit human mucosal surfaces, the field of airway microbiota research is poised for expansion and studies that address key gaps in our understanding of airway health.
Expert commentary
The dogma that only a handful of bacterial species were associated with chronic airway disease has been supplanted by recent culture-independent studies that have revealed that the airway microbiome of obstructive pulmonary disease patients represents a far more diverse consortium of bacterial species than previously believed. It is important to note that previous dogma was primarily based on data generated in an era predating the application of more sensitive culture-independent tools for microbial community profiling. This is particularly pertinent given that the majority of known bacterial species are currently unculturable under conventional laboratory conditions. Emerging culture-independent data suggest that respiratory microbiota differ in states of pulmonary disease and health, and with disease severity. This strongly suggests that, as has been demonstrated for the gastrointestinal microbiota, shifts in airway microbiome composition may drive specific immunophenotypes associated with distinct chronic airway diseases. Thus this emerging field opens up the possibility of microbiota manipulation as a novel therapeutic strategy for treatment or management of chronic airway disease.
Five-year view
Initial investigations in CF airway microbiota suggest a relationship between airway microbiome composition and severity of disease. Building upon recent studies in chronic airway disease, future investigations of the airway microbiome in differing disease phenotypes or across gradients of disease severity, particularly in asthmatic, COPD or CF populations, will likely identify microbial drivers of these disease states, and potentially lead to improved patient stratification and patient-tailored therapy. Although phylogenetic profiling has provided us with a wealth of information on the diversity of species present in the airway microbiota, functional studies to better establish links between microbiome behavior and host immune responses will provide a better understanding of pathophysiologic processes in chronic inflammatory diseases of the airways. The development and application of tools for airway microbiome profiling at both the structural and functional level has been rapid. Studies integrating these high-resolution tools appropriately in well-designed clinical studies that include larger subject numbers, encompass health and disease states and ascribe particular attention to airway sample source, will confirm or disprove current evidence.
Key issues.
Much of our knowledge of airway microbiology is based upon culture-based or species-specific molecular detection
Specific bacterial species have been associated with the risk for, and established, asthma, though these organisms may biomarkers for a specific airway microbiome composition.
Bacterial species linked with chronic obstructive pulmonary disease include Haemophilus influenzae, Moraxella catarrhalis and Streptococcus pneumoniae; organisms that have also been associated with risk for childhood asthma development.
Many more organisms are associated with chronic pulmonary disease in cystic fibrosis (CF) patients. These include emerging such as Ralstonia, Pandoraea and Enterococcus species, as well as the Streptoccusmilleri group.
Culture-independent studies of asthmatic bronchoscopic samples have demonstrated relationships between airway microbiota composition and the severity of bronchial hyper-responsiveness.
Chronic obstructive pulmonary disease airway microbiota remain rich in bacterial species (mean number of phylotypes: 411), despite administration of broad-spectrum antimicrobials for acute exacerbation.
CF airway microbiota have been examined by a number of culture-independent approaches. As the resolution of the these airway samples has increased, so too has the number of types of bacteria identified in this patient population.
Pediatric CF patient airways, which exhibit good pulmonary function and mild disease, possess a diverse community of contrast, older CF patients with more severe disease exhibit communities with reduced richness (number of types of skewed communities (highly dominated consortia) of low diversity.
Footnotes
For reprint orders, please contact reprints@expert-reviews.com
Financial & competing interests disclosure
YJ Huang is supported by NIH award 1K23HL105572 and a Cystic Fibrosis Foundation PACE award. SV Lynch is supported by NIH awards AI075410, A113916 and a Cystic Fibrosis Foundation award. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 2.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444(7122):1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 3.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102(31):11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang YJ, Nelson CE, Brodie EL, et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J Allergy Clin Immunol. 2011;127:372–381. e1–e3. doi: 10.1016/j.jaci.2010.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnston SL, Martin RJ. Chlamydophila pneumoniae and Mycoplasma pneumoniae: a role in asthma pathogenesis? Am J Respir Crit Care Med. 2005;172(9):1078–1089. doi: 10.1164/rccm.200412-1743PP. [DOI] [PubMed] [Google Scholar]
- 6.Hahn DL, Dodge RW, Golubjatnikov R. Association of Chlamydia pneumoniae (strain TWAR) infection with wheezing, asthmatic bronchitis, and adult-onset asthma. JAMA. 1991;266(2):225–230. [PubMed] [Google Scholar]
- 7.Sirmatel F, Ustunsoy H, Sirmatel O, Akdemir I, Dikensoy O. The relationship between Chlamydia pneumoniae seropositivity and peripheral vascular diseases, acute myocardial infarction and late-onset asthma. Infection. 2003;31(5):367–368. doi: 10.1007/s15010-003-2130-9. [DOI] [PubMed] [Google Scholar]
- 8.Yano T, Ichikawa Y, Komatu S, Arai S, Oizumi K. Association of Mycoplasma pneumoniae antigen with initial onset of bronchial asthma. Am J Respir Crit Care Med. 1994;149(5):1348–1353. doi: 10.1164/ajrccm.149.5.8173777. [DOI] [PubMed] [Google Scholar]
- 9.Korppi M, Paldanius M, Hyvarinen A, Nevalainen A, Husman T. Chlamydia pneumoniae and newly diagnosed asthma: a case–control study in 1 to 6-year-old children. Respirology. 2004;9(2):255–259. doi: 10.1111/j.1440-1843.2004.00558.x. [DOI] [PubMed] [Google Scholar]
- 10.Routes JM, Nelson HS, Noda JA, Simon FT. Lack of correlation between Chlamydia pneumoniae antibody titers and adult-onset asthma. J Allergy Clin Immunol. 2000;105(2 Pt 1):391–392. doi: 10.1016/s0091-6749(00)90093-9. [DOI] [PubMed] [Google Scholar]
- 11.Ten Brinke A, Van Dissel JT, Sterk PJ, Zwinderman AH, Rabe KF, Bel EH. Persistent airflow limitation in adult-onset nonatopic asthma is associated with serologic evidence of Chlamydia pneumoniae infection. J Allergy Clin Immunol. 2001;107(3):449–454. doi: 10.1067/mai.2001.113047. [DOI] [PubMed] [Google Scholar]
- 12.Kocabas A, Avsar M, Hanta I, Koksal F, Kuleci S. Chlamydophila pneumoniae infection in adult asthmatics patients. J Asthma. 2008;45(1):39–43. doi: 10.1080/02770900701815735. [DOI] [PubMed] [Google Scholar]
- 13.Martin RJ, Kraft M, Chu HW, Berns EA, Cassell GH. A link between chronic asthma and chronic infection. J Allergy Clin Immunol. 2001;107(4):595–601. doi: 10.1067/mai.2001.113563. [DOI] [PubMed] [Google Scholar]
- 14.Specjalski K, Jassem E. Chlamydophila pneumoniae, Mycoplasma pneumoniae infections, and asthma control. Allergy Asthma Proc. 2011;32(2):9–17. doi: 10.2500/aap.2011.32.3431. [DOI] [PubMed] [Google Scholar]
- 15.Bisgaard H, Hermansen MN, Buchvald F, et al. Childhood asthma after bacterial colonization of the airway in neonates. N Engl J Med. 2007;357(15):1487–1495. doi: 10.1056/NEJMoa052632. [DOI] [PubMed] [Google Scholar]
- 16.Bisgaard H, Hermansen MN, Bonnelykke K, et al. Association of bacteria and viruses with wheezy episodes in young children: prospective birth cohort study. BMJ. 2010;341:c4978. doi: 10.1136/bmj.c4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson DJ, Gangnon RE, Evans MD, et al. Wheezing rhinovirus illnesses in early life predict asthma development in high-risk children. Am J Respir Crit Care Med. 2008;178(7):667–672. doi: 10.1164/rccm.200802-309OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kotaniemi-Syrjanen A, Vainionpaa R, Reijonen TM, Waris M, Korhonen K, Korppi M. Rhinovirus-induced wheezing in infancy – the first sign of childhood asthma? J Allergy Clin Immunol. 2003;111(1):66–71. doi: 10.1067/mai.2003.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kusel MM, De Klerk NH, Kebadze T, et al. Early-life respiratory viral infections, atopic sensitization, and risk of subsequent development of persistent asthma. J Allergy Clin Immunol. 2007;119(5):1105–1110. doi: 10.1016/j.jaci.2006.12.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sigurs N, Aljassim F, Kjellman B, et al. Asthma and allergy patterns over 18 years after severe RSV bronchiolitis in the first year of life. Thorax. 2010;65(12):1045–1052. doi: 10.1136/thx.2009.121582. [DOI] [PubMed] [Google Scholar]
- 21.Sigurs N. A cohort of children hospitalised with acute RSV bronchiolitis: impact on later respiratory disease. Paediatr Respir Rev. 2002;3(3):177–183. doi: 10.1016/s1526-0542(02)00191-4. [DOI] [PubMed] [Google Scholar]
- 22.Sigurs N, Gustafsson PM, Bjarnason R, et al. Severe respiratory syncytial virus bronchiolitis in infancy and asthma and allergy at age 13. Am J Respir Crit Care Med. 2005;171(2):137–141. doi: 10.1164/rccm.200406-730OC. [DOI] [PubMed] [Google Scholar]
- 23.Dulek DE, Peebles RS., Jr Viruses and asthma. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbagen.2011.01.012. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Garcia ML, Calvo C, Casas I, et al. Human metapneumovirus bronchiolitis in infancy is an important risk factor for asthma at age 5. Pediatr Pulmonol. 2007;42(5):458–464. doi: 10.1002/ppul.20597. [DOI] [PubMed] [Google Scholar]
- 25.Murphy TF, Brauer AL, Eschberger K, et al. Pseudomonas aeruginosa in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2008;177(8):853–860. doi: 10.1164/rccm.200709-1413OC. [DOI] [PubMed] [Google Scholar]
- 26.Murphy TF, Brauer AL, Grant BJ, Sethi S. Moraxella catarrhalis in chronic obstructive pulmonary disease: burden of disease and immune response. Am J Respir Crit Care Med. 2005;172(2):195–199. doi: 10.1164/rccm.200412-1747OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papi A, Bellettato CM, Braccioni F, et al. Infections and airway inflammation in chronic obstructive pulmonary disease severe exacerbations. Am J Respir Crit Care Med. 2006;173(10):1114–1121. doi: 10.1164/rccm.200506-859OC. [DOI] [PubMed] [Google Scholar]
- 28.Sethi S, Evans N, Grant BJ, Murphy TF. New strains of bacteria and exacerbations of chronic obstructive pulmonary disease. N Engl J Med. 2002;347(7):465–471. doi: 10.1056/NEJMoa012561. [DOI] [PubMed] [Google Scholar]
- 29.Sethi S, Wrona C, Eschberger K, Lobbins P, Cai X, Murphy TF. Inflammatory profile of new bacterial strain exacerbations of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2008;177(5):491–497. doi: 10.1164/rccm.200708-1234OC. [DOI] [PubMed] [Google Scholar]
- 30.Sethi S, Murphy TF. Infection in the pathogenesis and course of chronic obstructive pulmonary disease. N Engl J Med. 2008;359(22):2355–2365. doi: 10.1056/NEJMra0800353. [DOI] [PubMed] [Google Scholar]
- 31.Martinez-Solano L, Macia MD, Fajardo A, Oliver A, Martinez JL. Chronic Pseudomonas aeruginosa infection in chronic obstructive pulmonary disease. Clin Infect Dis. 2008;47(12):1526–1533. doi: 10.1086/593186. [DOI] [PubMed] [Google Scholar]
- 32.Rakhimova E, Wiehlmann L, Brauer AL, Sethi S, Murphy TF, Tummler B. Pseudomonas aeruginosa population biology in chronic obstructive pulmonary disease. J Infect Dis. 2009;200(12):1928–1935. doi: 10.1086/648404. [DOI] [PubMed] [Google Scholar]
- 33.Bafadhel M, Mckenna S, Terry S, et al. Acute exacerbations of COPD: identification of biological clusters and their biomarkers. Am J Respir Crit Care Med. 2011 doi: 10.1164/rccm.201104-0597OC. 201104-0597OCv1. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 34.De Serres G, Lampron N, La Forge J, et al. Importance of viral and bacterial infections in chronic obstructive pulmonary disease exacerbations. J Clin Virol. 2009;46(2):129–133. doi: 10.1016/j.jcv.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kherad O, Kaiser L, Bridevaux PO, et al. Upper-respiratory viral infection, biomarkers, and COPD exacerbations. Chest. 2010;138(4):896–904. doi: 10.1378/chest.09-2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilkinson TM, Hurst JR, Perera WR, Wilks M, Donaldson GC, Wedzicha JA. Effect of interactions between lower airway bacterial and rhinoviral infection in exacerbations of COPD. Chest. 2006;129(2):317–324. doi: 10.1378/chest.129.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Avadhanula V, Rodriguez CA, Devincenzo JP, et al. Respiratory viruses augment the adhesion of bacterial pathogens to respiratory epithelium in a viral species- and cell type-dependent manner. J Virol. 2006;80(4):1629–1636. doi: 10.1128/JVI.80.4.1629-1636.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sajjan US, Jia Y, Newcomb DC, et al. H. influenzae potentiates airway epithelial cell responses to rhinovirus by increasing ICAM-1 and TLR3 expression. FASEB J. 2006;20(12):2121–2123. doi: 10.1096/fj.06-5806fje. [DOI] [PubMed] [Google Scholar]
- 39.Soler N, Ewig S, Torres A, Filella X, Gonzalez J, Zaubet A. Airway inflammation and bronchial microbial patterns in patients with stable chronic obstructive pulmonary disease. Eur Respir J. 1999;14(5):1015–1022. doi: 10.1183/09031936.99.14510159. [DOI] [PubMed] [Google Scholar]
- 40.Rosell A, Monso E, Soler N, et al. Microbiologic determinants of exacerbation in chronic obstructive pulmonary disease. Arch Intern Med. 2005;165(8):891–897. doi: 10.1001/archinte.165.8.891. [DOI] [PubMed] [Google Scholar]
- 41.Sethi S, Maloney J, Grove L, Wrona C, Berenson CS. Airway inflammation and bronchial bacterial colonization in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2006;173(9):991–998. doi: 10.1164/rccm.200509-1525OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang M, Li Q, Zhang XY, Ding X, Zhu D, Zhou X. Relevance of lower airway bacterial colonization, airway inflammation, and pulmonary function in the stable stage of chronic obstructive pulmonary disease. Eur J Clin Microbiol Infect Dis. 2010;29(12):1487–1493. doi: 10.1007/s10096-010-1027-7. [DOI] [PubMed] [Google Scholar]
- 43.Wen Y, Reid DW, Zhang D, Ward C, Wood-Baker R, Walters EH. Assessment of airway inflammation using sputum, BAL, and endobronchial biopsies in current and ex-smokers with established COPD. Int J Chron Obstruct Pulmon Dis. 2010;5:327–334. doi: 10.2147/COPD.S11343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morris A, Sciurba FC, Norris KA. Pneumocystis: a novel pathogen in chronic obstructive pulmonary disease? COPD. 2008;5(1):43–51. doi: 10.1080/1541255070181756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Retamales I, Elliott WM, Meshi B, et al. Amplification of inflammation in emphysema and its association with latent adenoviral infection. Am J Respir Crit Care Med. 2001;164(3):469–473. doi: 10.1164/ajrccm.164.3.2007149. [DOI] [PubMed] [Google Scholar]
- 46.Mc Manus TE, Moore JE, Crowe M, Dunbar K, Elborn JS. A comparison of pulmonary exacerbations with single and multiple organisms in patients with cystic fibrosis and chronic Burkholderia cepacia infection. J Infect. 2003;46(1):56–59. doi: 10.1053/jinf.2001.1077. [DOI] [PubMed] [Google Scholar]
- 47.Coenye T, Vandamme P, Lipuma JJ. Infection by Ralstonia species in cystic fibrosis patients: identification of R. pickettii and R. mannitolilytica by polymerase chain reaction. Emerg Infect Dis. 2002;8(7):692–696. doi: 10.3201/eid0807.010472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Atkinson RM, Lipuma JJ, Rosenbluth DB, Dunne WM., Jr Chronic colonization with Pandoraea apista in cystic fibrosis patients determined by repetitive-element-sequence PCR. J Clin Microbiol. 2006;44(3):833–836. doi: 10.1128/JCM.44.3.833-836.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jorgensen IM, Johansen HK, Frederiksen B, et al. Epidemic spread of Pandoraea apista, a new pathogen causing severe lung disease in cystic fibrosis patients. Pediatr Pulmonol. 2003;36(5):439–446. doi: 10.1002/ppul.10383. [DOI] [PubMed] [Google Scholar]
- 50.Grinwis ME, Sibley CD, Parkins MD, Eshaghurshan CS, Rabin HR, Surette MG. Characterization of Streptococcus milleri group isolates from expectorated sputum of adult patients with cystic fibrosis. J Clin Microbiol. 2009;48(2):395–401. doi: 10.1128/JCM.01807-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parkins MD, Sibley CD, Surette MG, Rabin HR. The Streptococcus milleri group – an unrecognized cause of disease in cystic fibrosis: a case series and literature review. Pediatr Pulmonol. 2008;43(5):490–497. doi: 10.1002/ppul.20809. [DOI] [PubMed] [Google Scholar]
- 52.Canton R, Morosini MI, Ballestero S, et al. Lung colonization with Enterobacteriaceae producing extended-spectrum β-lactamases in cystic fibrosis patients. Pediatr Pulmonol. 1997;24(3):213–217. doi: 10.1002/(sici)1099-0496(199709)24:3<213::aid-ppul7>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 53.De Almeida MB, Zerbinati RM, Tateno AF, et al. Rhinovirus C and respiratory exacerbations in children with cystic fibrosis. Emerg Infect Dis. 2010;16(6):996–999. doi: 10.3201/eid1606.100063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smyth AR, Smyth RL, Tong CY, Hart CA, Heaf DP. Effect of respiratory virus infections including rhinovirus on clinical status in cystic fibrosis. Arch Dis Child. 1995;73(2):117–120. doi: 10.1136/adc.73.2.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wat D, Gelder C, Hibbitts S, et al. Is there a role for influenza vaccination in cystic fibrosis? J Cyst Fibros. 2008;7(1):85–88. doi: 10.1016/j.jcf.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 56.Chattoraj SS, Ganesan S, Faris A, Comstock A, Lee WM, Sajjan US. Pseudomonas aeruginosa suppresses interferon response to rhinovirus infection in cystic fibrosis, but not in normal bronchial epithelial cells. Infect Immun. 2011;79(10):4131–4145. doi: 10.1128/IAI.05120-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bik EM, Eckburg PB, Gill SR, et al. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci USA. 2006;103(3):732–737. doi: 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dewhirst FE, Chen T, Izard J, et al. The human oral microbiome. J Bacteriol. 2010;192(19):5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308(5728):1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011;9(4):244–253. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brodie EL, Desantis TZ, Parker JP, Zubietta IX, Piceno YM, Andersen GL. Urban aerosols harbor diverse and dynamic bacterial populations. Proc Natl Acad Sci USA. 2007;104(1):299–304. doi: 10.1073/pnas.0608255104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Desantis TZ, Brodie EL, Moberg JP, Zubieta IX, Piceno YM, Andersen GL. High-density universal 16S rRNA microarray analysis reveals broader diversity than typical clone library when sampling the environment. Microb Ecol. 2007;53(3):371–383. doi: 10.1007/s00248-006-9134-9. [DOI] [PubMed] [Google Scholar]
- 63.Lemos LN, Fulthorpe RR, Triplett EW, Roesch LF. Rethinking microbial diversity analysis in the high throughput sequencing era. J Microbiol Methods. 2011;86(1):42–51. doi: 10.1016/j.mimet.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 64.Petrosino JF, Highlander S, Luna RA, Gibbs RA, Versalovic J. Metagenomic pyrosequencing and microbial identification. Clin Chem. 2009;55(5):856–866. doi: 10.1373/clinchem.2008.107565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Van Den Bogert B, De Vos WM, Zoetendal EG, Kleerebezem M. Microarray analysis and barcoded pyrosequencing provide consistent microbial profiles depending on the source of human intestinal samples. Appl Environ Microbiol. 2011;77(6):2071–2080. doi: 10.1128/AEM.02477-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ghannoum MA, Jurevic RJ, Mukherjee PK, et al. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 2010;6(1):e1000713. doi: 10.1371/journal.ppat.1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kistler A, Avila PC, Rouskin S, et al. Pan-viral screening of respiratory tract infections in adults with and without asthma reveals unexpected human coronavirus and human rhinovirus diversity. J Infect Dis. 2007;196(6):817–825. doi: 10.1086/520816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Paulino LC, Tseng CH, Strober BE, Blaser MJ. Molecular analysis of fungal microbiota in samples from healthy human skin and psoriatic lesions. J Clin Microbiol. 2006;44(8):2933–2941. doi: 10.1128/JCM.00785-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang D, Coscoy L, Zylberberg M, et al. Microarray-based detection and genotyping of viral pathogens. Proc Natl Acad Sci USA. 2002;99(24):15687–15692. doi: 10.1073/pnas.242579699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reyes A, Haynes M, Hanson N, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466(7304):334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dethlefsen L, Mcfall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449(7164):811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fujimura KE, Slusher NA, Cabana MD, Lynch SV. Role of the gut microbiota in defining human health. Expert Rev Anti Infect Ther. 2010;8(4):435–454. doi: 10.1586/eri.10.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wade WG. Has the use of molecular methods for the characterization of the human oral microbiome changed our understanding of the role of bacteria in the pathogenesis of periodontal disease? J Clin Periodontol. 2011;38(Suppl 11):7–16. doi: 10.1111/j.1600-051X.2010.01679.x. [DOI] [PubMed] [Google Scholar]
- 74.Lemon KP, Klepac-Ceraj V, Schiffer HK, Brodie EL, Lynch SV, Kolter R. Comparative analyses of the bacterial microbiota of the human nostril and oropharynx. MBio. 2010;1(3):e00129-10. doi: 10.1128/mBio.00129-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zaura E, Keijser BJ, Huse SM, Crielaard W. Defining the healthy ‘core microbiome’ of oral microbial communities. BMC Microbiol. 2009;9:259. doi: 10.1186/1471-2180-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Norris KA, Morris A. Pneumocystis infection and the pathogenesis of chronic obstructive pulmonary disease. Immunol Res. 2011;50(2–3):175–180. doi: 10.1007/s12026-011-8218-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Morris A, Alexander T, Radhi S, et al. Airway obstruction is increased in pneumocystis-colonized human immunodeficiency virus-infected outpatients. J Clin Microbiol. 2009;47(11):3773–3776. doi: 10.1128/JCM.01712-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Reihill JA, Moore JE, Elborn JS, Ennis M. Effect of Aspergillus fumigatus and Candida albicans on pro-inflammatory response in cystic fibrosis epithelium. J Cyst Fibros. 2011 doi: 10.1016/j.jcf.2011.06.006. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 80.Muller FM, Seidler M. Characteristics of pathogenic fungi and antifungal therapy in cystic fibrosis. Expert Rev Anti Infect Ther. 2010;8(8):957–964. doi: 10.1586/eri.10.72. [DOI] [PubMed] [Google Scholar]
- 81.Horre R, Marklein G, Siekmeier R, Reiffert SM. Detection of hyphomycetes in the upper respiratory tract of patients with cystic fibrosis. Mycoses. 2010 doi: 10.1111/j.1439-0507.2010.01897.x. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 82.Charlson ES, Bittinger K, Haas AR, et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. 2011 doi: 10.1164/rccm.201104-0655OC. 201104-0655OCv2. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hilty M, Burke C, Pedro H, et al. Disordered microbial communities in asthmatic airways. PLoS One. 2010;5(1):e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Charlson ES, Chen J, Custers-Allen R, et al. Disordered microbial communities in the upper respiratory tract of cigarette smokers. PLoS One. 2010;5(12):e15216. doi: 10.1371/journal.pone.0015216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Henriksson G, Helgeland L, Midtvedt T, Stierna P, Brandtzaeg P. Immune response to Mycoplasma pulmonis in nasal mucosa is modulated by the normal microbiota. Am J Respir Cell Mol Biol. 2004;31(6):657–662. doi: 10.1165/rcmb.2004-0207OC. [DOI] [PubMed] [Google Scholar]
- 86.Ichinohe T, Pang IK, Kumamoto Y, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci USA. 2011;108(13):5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Erb-Downward JR, Thompson DL, Han MK, et al. Analysis of the lung microbiome in the ‘healthy’ smoker and in COPD. PLoS One. 2011;6(2):e16384. doi: 10.1371/journal.pone.0016384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Huang YJ, Kim E, Cox MJ, et al. A persistent and diverse airway microbiota present during chronic obstructive pulmonary disease exacerbations. OMICS. 2010;14(1):9–59. doi: 10.1089/omi.2009.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Artis D. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat Rev Immunol. 2008;8(6):411–420. doi: 10.1038/nri2316. [DOI] [PubMed] [Google Scholar]
- 90.Rogers GB, Hart CA, Mason JR, Hughes M, Walshaw MJ, Bruce KD. Bacterial diversity in cases of lung infection in cystic fibrosis patients: 16S ribosomal DNA (rDNA) length heterogeneity PCR and 16S rDNA terminal restriction fragment length polymorphism profiling. J Clin Microbiol. 2003;41(8):3548–3558. doi: 10.1128/JCM.41.8.3548-3558.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rogers GB, Carroll MP, Serisier DJ, et al. Bacterial activity in cystic fibrosis lung infections. Respir Res. 2005;6:49. doi: 10.1186/1465-9921-6-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rogers GB, Carroll MP, Serisier DJ, Hockey PM, Jones G, Bruce KD. Characterization of bacterial community diversity in cystic fibrosis lung infections by use of 16s ribosomal DNA terminal restriction fragment length polymorphism profiling. J Clin Microbiol. 2004;42(11):5176–5183. doi: 10.1128/JCM.42.11.5176-5183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Harris JK, De Groote MA, Sagel SD, et al. Molecular identification of bacteria in bronchoalveolar lavage fluid from children with cystic fibrosis. Proc Natl Acad Sci USA. 2007;104(51):20529–20533. doi: 10.1073/pnas.0709804104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Klepac-Ceraj V, Lemon KP, Martin TR, et al. Relationship between cystic fibrosis respiratory tract bacterial communities and age, genotype, antibiotics and Pseudomonas aeruginosa. Environ Microbiol. 2010;12(5):1293–1303. doi: 10.1111/j.1462-2920.2010.02173.x. [DOI] [PubMed] [Google Scholar]
- 95.Cox MJ, Allgaier M, Taylor B, et al. Airway microbiota and pathogen abundance in age-stratified cystic fibrosis patients. PLoS One. 2010;5(6):e11044. doi: 10.1371/journal.pone.0011044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Duan K, Dammel C, Stein J, Rabin H, Surette MG. Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol Microbiol. 2003;50(5):1477–1491. doi: 10.1046/j.1365-2958.2003.03803.x. [DOI] [PubMed] [Google Scholar]
- 97.Sibley CD, Duan K, Fischer C, et al. Discerning the complexity of community interactions using a Drosophila model of polymicrobial infections. PLoS Pathog. 2008;4(10):e1000184. doi: 10.1371/journal.ppat.1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Høiby N, Ciofu O, Johansen HK, et al. The clinical impact of bacterial biofilms. Int J Oral Sci. 2011;3(2):55–65. doi: 10.4248/IJOS11026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Campos J, Roman F, Georgiou M, et al. Long-term persistence of ciprofloxacin-resistant Haemophilus influenzae in patients with cystic fibrosis. J Infect Dis. 1996;174(6):1345–1347. doi: 10.1093/infdis/174.6.1345. [DOI] [PubMed] [Google Scholar]
- 100.Hoiby N. Epidemiological investigations of the respiratory tract bacteriology in patients with cystic fibrosis. Acta Pathol Microbiol Scand B Microbiol Immunol. 1974;82(4):541–550. [PubMed] [Google Scholar]
- 101.Clode FE, Metherell LA, Pitt TL. Nosocomial acquisition of Burkholderia gladioli in patients with cystic fibrosis. Am J Respir Crit Care Med. 1999;160(1):374–375. doi: 10.1164/ajrccm.160.1.16011. [DOI] [PubMed] [Google Scholar]
- 102.Yang JH, Spilker T, LiPuma JJ. Simultaneous coinfection by multiple strains during Burkholderia cepacia complex infection in cystic fibrosis. Diagn Microbiol Infect Dis. 2006;54(2):95–98. doi: 10.1016/j.diagmicrobio.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 103.Drevinek P, Mahenthiralingam E. Burkholderia cenocepacia in cystic fibrosis: epidemiology and molecular mechanisms of virulence. Clin Microbiol Infect. 2010;16(7):821–830. doi: 10.1111/j.1469-0691.2010.03237.x. [DOI] [PubMed] [Google Scholar]
- 104.Gibson RL, Burns JL, Ramsey BW. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am J Respir Crit Care Med. 2003;168(8):918–951. doi: 10.1164/rccm.200304-505SO. [DOI] [PubMed] [Google Scholar]
- 105.Lambiase A, Catania MR, Del Pezzo M, et al. Achromobacter xylosoxidans respiratory tract infection in cystic fibrosis patients. Eur J Clin Microbiol Infect Dis. 2011;30(8):973–980. doi: 10.1007/s10096-011-1182-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Esther CR, Jr, Esserman DA, Gilligan P, Kerr A, Noone PG. Chronic Mycobacterium abscessus infection and lung function decline in cystic fibrosis. J Cyst Fibros. 2010;9(2):117–123. doi: 10.1016/j.jcf.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bauernfeind A, Bertele RM, Harms K, et al. Qualitative and quantitative microbiological analysis of sputa of 102 patients with cystic fibrosis. Infection. 1987;15(4):270–277. doi: 10.1007/BF01644137. [DOI] [PubMed] [Google Scholar]
- 108.Del Campo R, Morosini MI, De La Pedrosa EG, et al. Population structure, antimicrobial resistance, and mutation frequencies of Streptococcus pneumoniae isolates from cystic fibrosis patients. J Clin Microbiol. 2005;43(5):2207–2214. doi: 10.1128/JCM.43.5.2207-2214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]