Abstract
The functional contribution of CNV to human biology and disease pathophysiology has undergone limited exploration. Recent observations in humans indicate a tentative link between CNV and weight regulation. Smith-Magenis syndrome (SMS), manifesting obesity and hypercholesterolemia, results from a deletion CNV at 17p11.2, but is sometimes due to haploinsufficiency of a single gene, RAI1. The reciprocal duplication in 17p11.2 causes Potocki-Lupski syndrome (PTLS). We previously constructed mouse strains with a deletion, Df(11)17, or duplication, Dp(11)17, of the mouse genomic interval syntenic to the SMS/PTLS region. We demonstrate that Dp(11)17 is obesity-opposing; it conveys a highly penetrant, strain-independent phenotype of reduced weight, leaner body composition, lower TC/LDL, and increased insulin sensitivity that is not due to alteration in food intake or activity level. When fed with a high-fat diet, Dp(11)17/+ mice display much less weight gain and metabolic change than WT mice, demonstrating that the Dp(11)17 CNV protects against metabolic syndrome. Reciprocally, Df(11)17/+ mice with the deletion CNV have increased weight, higher fat content, decreased HDL, and reduced insulin sensitivity, manifesting a bona fide metabolic syndrome. These observations in the deficiency animal model are supported by human data from 76 SMS subjects. Further, studies on knockout/transgenic mice showed that the metabolic consequences of Dp(11)17 and Df(11)17 CNVs are not only due to dosage alterations of Rai1, the predominant dosage-sensitive gene for SMS and likely also PTLS. Our experiments in chromosome-engineered mouse CNV models for human genomic disorders demonstrate that a CNV can be causative for weight/metabolic phenotypes. Furthermore, we explored the biology underlying the contribution of CNV to the physiology of weight control and energy metabolism. The high penetrance, strain independence, and resistance to dietary influences associated with the CNVs in this study are features distinct from most SNP–associated metabolic traits and further highlight the potential importance of CNV in the etiology of both obesity and MetS as well as in the protection from these traits.
Author Summary
Genetic factors play a large role in obesity. However, despite recent technical progress in the search for genetic variants, the identities of causative and contributory genetic factors remain largely unknown. Whereas nucleotide sequence variation has been studied extensively with respect to its potential contribution to obesity, copy number variations (CNV), in which genes exist in abnormal numbers of copies mostly due to duplication or deletion, have only more recently been observed to be associated with human obesity. In this report, we utilize chromosome engineered mouse strains harboring a deletion or duplication CNV to address the potential functional impact of CNVs on weight control and metabolism. We show that the duplication CNV leads to lower body weight; it is also metabolically advantageous and protects from diet-induced obesity and metabolic syndrome (MetS). The deletion CNV causes a “mirror” phenotype with increased body weight and MetS–like phenotypes. Importantly, these effects manifest regardless of the genetic background and do not appear to be attributable to any single gene. These findings demonstrate experimentally that CNV can be causative for weight and metabolic phenotypes and highlight the potential relevance and importance of CNV in the etiology of obesity/MetS and the protection from these traits.
Introduction
The significance of copy number variation (CNV) in human genetic variation is now indisputable [1], [2]. However, in contrast to the revolutionary progress achieved in the discovery of CNVs and delineating the mechanisms for their formation, our current knowledge of the downstream functional mechanisms by which CNVs contribute to trait manifestations is limited. Functional contributions of CNV to human biology have only been examined in a few physiological systems including the neuropsychiatric/behavioral fields [2], [3].
About 400 million people worldwide are classified as obese [4] and are likely to suffer from premature mortality and obesity-associated morbidities, such as hyperglycemia, dyslipidemia, hypertension and metabolic syndrome (MetS) [5]. The etiologies for obesity include genetic contributions [4], but the identities of the specific genetic factors remain largely unknown. Single nucleotide polymorphisms (SNPs) identified through linkage and genome-wide association studies (GWAS) explain only 1–2% of the variation in obesity phenotypes as measured by BMI [6], [7], [8].
Recent observations in humans indicate a tentative link between CNV and weight regulation. Deletions at 16p11.2 were associated with a highly penetrant form of obesity often found with hyperphagia and intellectual disabilities, whereas the reciprocal duplication conveys a 8.3 fold increased risk for being clinically underweight [9], [10]. These comprehensive studies on patients added to the clinical observations of obesity associated with CNV that have been noted for several chromosomal syndromes and genomic disorders including Down [11] and Prader-Willi syndromes [12]. However, there is no experimental data that proves the causative role of the CNV in the abnormality in weight regulation, nor is there any study on the biology underlying this tentative link.
Potocki-Lupski syndrome (PTLS, MIM 610883) [13], [14] is an intellectual disability and multiple congenital anomalies (ID/MCA) syndrome due to a heterozygous interstitial duplication CNV in chromosome 17p11.2. Mildly lowered total cholesterol and LDL were noted for some PTLS patients [13]. The reciprocal deletion CNV of the same interval causes a distinct ID/MCA disorder known as Smith-Magenis syndrome (SMS, MIM 182290) [15], [16], [17]. Obesity and hypercholesterolemia are phenotypes of SMS [16], [18], [19]. By chromosomal engineering, we previously constructed mouse models of PTLS and SMS carrying the duplication [Dp(11)17] or deletion [Df(11)17] of a 2 Mb chromosomal segment that includes the majority of the mouse region syntenic to the PTLS/SMS common recurrent CNV interval (Figure S1) [20]. Both Dp(11)17/+ and Df(11)17/+ mice partially recapitulate the respective human phenotypes such as craniofacial abnormalities [21], [22], altered learning, memory and social interaction [20], [23], [24], [25], and display a transcriptome [25] that is distinct from their wild type (WT) littermates.
In the context of exploring the biological link between CNV and weight control, we now utilize these mouse models to investigate the detailed metabolic consequences of the PTLS duplication CNV and the reciprocal SMS deletion CNV. To simplify data analyses, all experiments were performed with male animals.
Results
Phenotypes reciprocal to metabolic syndrome in Dp(11)17 animals
First, we found that, similar to previous observations on several genetic backgrounds (C57BL/6J/129S5 and N7 or N12 congenic C57BL/6J) [20], [25], [26], the Dp(11)17/+ mice on an isogenic (N>17) C57BL/6J background also display significantly reduced body weight compared to their WT littermates (Figure 1A, 1B). In contrast, and again in accordance with earlier reports on different backgrounds (C57BL/6J/129S5 and N7 or N12 congenic C57BL/6J) [20], [25], the Df(11)17/+ mice with the reciprocal deletion CNV on a pure (N>10) 129S5 background are significantly heavier than their WT littermates after 15 weeks of age (Figure 1C, 1D). Thus, the reciprocal duplication and deletion CNVs not only change body weight in opposing directions, but also, strikingly, do so in a highly penetrant manner that is independent of the genetic background. Highly penetrant weight change phenotypes were recently also observed in humans for two obesity-associated deletion CNVs on 16p11.2 and the reciprocal duplication of one of them associated with leanness [9], [10], [27]. The high penetrance differentiates CNV-associated obesity from SNP associated obesity in which, except for some very rare mutations in a few genes of the leptin/melanocortin pathway (LEP, MC4R, etc.), almost all variants have low penetrance [28], [29].
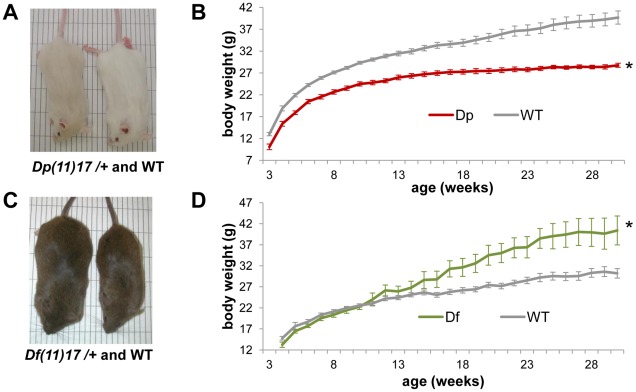
Figure 1. Dp(11)17/+ mice have reduced and Df(11)17/+ mice have increased body weight.
(A) A Dp(11)17/+ male (23 weeks old) and its WT littermate are shown, both on isogenic C57BL/6Tyr c-Brd background. Dp(11)17/+ mice appear gray because of a coat color marker in the construct used to chromosome engineer this strain [20]. (B) Growth curve of Dp(11)17/+ (red) and WT littermates (gray) reveal decreased weights for the duplication CNV mutants throughout their life span (*p<0.001 for by ANOVA with repeated measures). (C) A Df(11)17/+ male (32 weeks old) and its WT littermate on pure 129S5 background (D) Growth curve of Df(11)17/+ (green) and WT littermates (gray) reveal increased weights for the deletion CNV mutants (*p<0.05 by ANOVA). (B, D): n = 10–25 mice for each data point, results are expressed as mean ± s.e.m.
In addition to having reduced weight, adult Dp(11)17/+ mice on an isogenic (N>17) C57BL/6J background are also leaner than WT males, as measured via ECHO-MRI whole body scans (Echo medical systems, Texas) and manual dissection. Dp(11)17/+ mice have a significantly lower percentage of both whole body fat mass and epididymal white adipose tissue (EWAT) (Figure 2A, 2B), as well as a significantly higher percentage of lean mass (Figure 2A). These findings are in accordance with our previous reports of reduced abdominal fat in Dp(11)17/+ mice on different strain backgrounds (N7 [26] and N12 C57BL/6J [25]), further demonstrating the strain-independent manifestation of the metabolic phenotypes caused by the duplication CNV. Moreover, adult Dp(11)17/+ mice display significantly reduced fasting total serum cholesterol (TC) and LDL levels (Figure 2C) as well as a cardioprotective decrease of TC/HDL ratio (Figure 2C). Interestingly, the change in TC and LDL resembles the clinical observations in PTLS patients [13], despite the mechanistic differences in lipid metabolism between human and mouse [30]. Consistent with their lower adiposity [31], the serum leptin concentration is decreased in Dp(11)17/+ mice (Figure 2D).
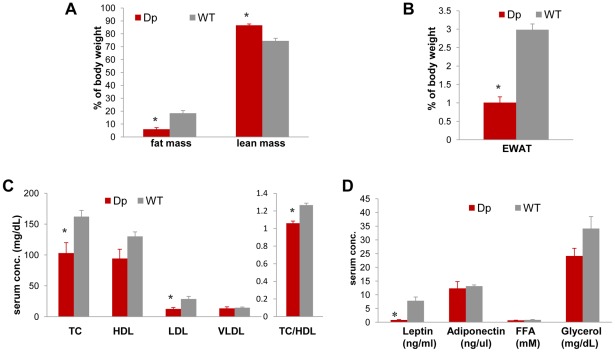
Figure 2. Dp(11)17/+ mice (red) are also leaner and have reduced serum TC, LDL, TC/HDL ratio, and leptin.
(A) Less relative total fat mass (*p = 0.000088) and more relative lean mass (*p = 0.00012) was identified in Dp(11)17/+ mice with ECHO-MRI system. (B) Dp(11)17/+ animals also possess smaller epididymal white adipose tissue pad (EWAT) (*p = 0.0020). Fasting serum profile revealed (C) reduced TC (*p = 0.021), LDL (*p = 0.01), TC/HDL ratio (*p = 0.0007) and (D) reduced leptin (*p = 0.021) in Dp(11)17/+ mice. All comparisons were made with two-tailed t-test; results are expressed as mean ± s.e.m. from measurements of (A) 6 Dp(11)17/+ and 10 WT at 21–22 wks (B) 6 Dp(11)17/+, 7 WT at 41 wks (C) 5 Dp(11)17/+ and 6 WT at 20–22 wks (D) 6 Dp(11)17/+ and 4 WT of 20–21 wks.
Furthermore, the intraperitoneal glucose tolerance test (IP-GTT) demonstrates an overall improved glucose clearance in Dp(11)17/+ mice compared to WT littermates; the difference in their serum glucose concentration becomes significant at 120 minutes (Figure 3A). The plasma insulin levels during the GTT were significantly lower in the Dp(11)17/+ animals throughout the test, suggesting that the improvement in glucose tolerance was not due to increased insulin production by the pancreas, but likely the result of improved insulin sensitivity (Figure 3B). Indeed, in the insulin tolerance test (ITT), insulin injection lowered blood glucose levels significantly faster in Dp(11)17/+ than in WT mice, further corroborating their increased insulin sensitivity (Figure 3C and 3D). Intriguingly, the circulating concentration of adiponectin is not changed in Dp(11)17/+ mice (Figure 2D), suggesting that adiponectin-independent pathways are involved in the alteration of their insulin sensitivity [32].
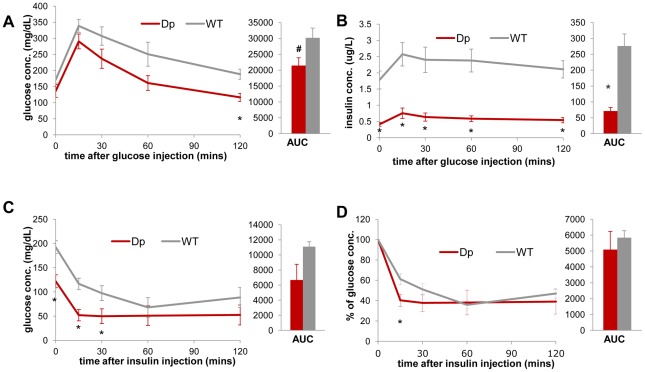
Figure 3. Dp(11)17/+ mice (red) display improved insulin sensitivity compared to WT mice (gray).
During IP-GTT (6 hr fasting, 1.5 mg glucose/g body weight), Dp(11)17/+ mice demonstrate (A) lower blood glucose (*p = 0.006 for 120 minutes post injection; # p = 0.052 for the area under curve (AUC)) and (B) lower blood insulin level (*p = 0.0037, 0.0026, 0.0051, 0.0031 and 0.0015 for the time points 0, 15, 30, 60 and 120 minutes; *p = 0.002 for AUC). During IP-ITT (4–6 hrs fasting, 1 mU insulin/g body weight), Dp(11)17/+ mice also demonstrate lower blood glucose concentration, shown as both actual concentration (C) (*p = 0.011, 0.004 and 0.037 for 0, 15 and 30 mins post insulin injection) and percentage of the initial glucose concentration (D) (*p = 0.038 for 15 mins post insulin injection). All comparisons were made with two-tailed t-test; results are expressed as mean ± s.e.m. from measurements of (A, B) n = 5 Dp(11)17/+ and 6 WT at 30 wks (C, D) 4 Dp(11)17/+ and 4 WT mice at 20–22 wks. All AUCs are computed until 120 minutes, for the entire length of the time curves.
The Dp(11)17 CNV results in higher intrinsic metabolic activity
We found that the reduction in body weight of Dp(11)17/+ mice is not simply due to an altered energy intake or increased activity since they consume identical amounts of food after four weeks of age (Figure 4A) and have similar activity levels to those of their WT littermates (Figure 4B). Thus, intrinsic changes in energy expenditure likely explain the observed phenotypes. Indeed, as assayed by indirect calorimetry, Dp(11)17/+ mice demonstrate an overall higher oxygen consumption (VO2) per lean mass and higher respiratory exchange ratio (RER) than WT mice, indicating higher energy expenditure than their WT littermates (Figure 4C–4F). Western blotting suggested an elevated expression level of protein uncoupled 1 (UCP1) in brown adipose tissue (BAT) of the Dp(11)17/+ mice (Figure 4G, 4H), although there is considerable variability in UCP1 expression among different samples. UCP1 is a key component of thermogenesis in BAT [33], this difference may partially explain the higher intrinsic energy expenditure of the Dp(11)17/+ mice. Interestingly, Dp(11)17/+ mice also appear to have a trend toward slightly higher body temperature (34.56±0.34°C) than WT littermates (33.9±0.90°C). We did not observe differences in the expression levels of UCP2 and UCP3 in BAT between Dp(11)17/+ and WT mice, nor did we observe differences in other signature metabolism genes (Glut4, AP2 in BAT, and Lpk, Fas, Acc1, Srebp1c, Tnxip in the liver) (Figure S4A, S4B).
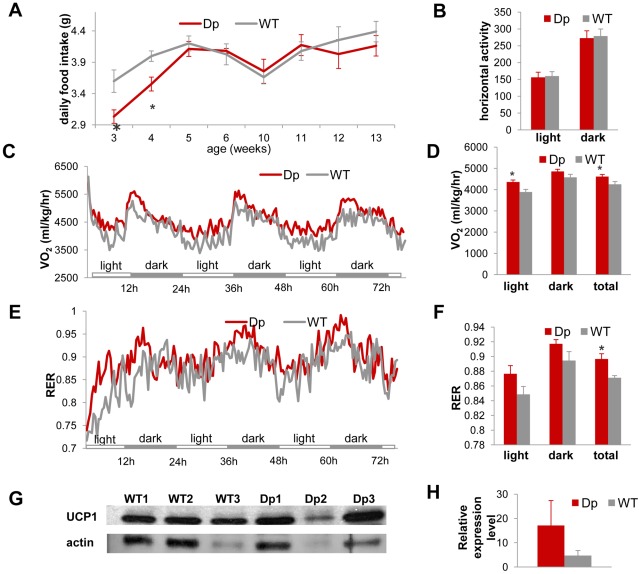
Figure 4. Dp(11)17/+ mice (red) have similar food intake and activity levels, but higher energy expenditure than WT mice (gray), which may be partially accounted for by the difference in expression levels of UCP1 in the BAT tissue.
(A) Dp(11)17/+ mice have similar amount daily food intake to WT mice after 4 wks of age, although they consume less food at 3 wks (*p = 0.001) and 4 wks (*p = 0.048). (B) VersaMax system (Accuscan Inc., Ohio) using the beam block technique implemented in home cages revealed no difference in horizontal activity level between Dp(11)17/+ and WT animals. (C, D) Oxygen consumption measured using the CLAMS system (Columbus Ins., Ohio) for over three days documented higher energy expenditure of Dp(11)17/+ mice in the light phases alone (*p = 0.0095) and during the entire day (*p = 0.044). (E, F) Respiratory exchange ratio (RER) measured using the CLAMS system for over three days again confirmed higher metabolic activity of Dp(11)17/+ mice (*p = 0.00151). (G) Western blot for UCP1 expression in BAT tissue of three Dp(11)17/+ and three WT mice with antibody AB3036 (Millipore). The same blot was normalized to actin blotting using MAB1501 (Millipore). (H) Normalized intensity of UCP1 signals in Dp(11)17/+ vs. WT mice (17.13±10.21 vs. 4.74±2.05, p = 0.35). The measurements are from (A) 5–13 Dp(11)17/+ and 5–11 WT at different time points (B) to (F) 12 Dp(11)17/+ and 7–10 WT at 25–32 wks (G) 3 Dp(11)17/+ and 3 WT at 30 wks.
Thus, while on a regular chow diet, Dp(11)17/+ mice are leaner than their WT littermates, and they have lower serum TC/LDL levels and reduced leptin concentration. Dp(11)17/+ mice also appear to be more insulin sensitive and have higher energy expenditure but display no difference in activity level in comparison to WT mice. These traits are reciprocal or antithetical to those of metabolic syndrome and most appear to manifest independent of the genetic background of the mouse strain.
Importantly, except for the parameters related to energy metabolism, a comprehensive serum analysis did not observe any difference in other serum chemistry parameters between Dp(11)17/+ and WT animals (Table S1). Also, under daily evaluation by veterinarian staff in our mouse facility, no overt illness was observed in Dp(11)17/+ mice. Combined with the fact that Dp(11)17/+ animals also have identical activity level and food intake to WT mice, the metabolic traits we observed in Dp(11)17/+ mice are unlikely due to any illness related to the duplication CNV, but rather a direct effect from the CNV.
The Dp(11)17 CNV protects against diet-induced obesity (DIO)
Next, we investigated potential influences of the Dp(11)17 CNV on the genetic susceptibility to diet induced obesity (DIO). First, we placed Dp(11)17/+ and WT mice on a HF (60% calories from fat) diet for three weeks (19 to 22) after 19 weeks of a normal diet. After these three weeks, the WT mice had dramatically increased body weight compared to control WT mice of the same age that have been kept on a regular chow (RC) diet. The Dp(11)17/+ mice, however, did not display significant weight gain compared to other Dp(11)17/+ animals on RC (Figure 5A). More specifically, the weight gain in WT mice is predominantly due to an increase in the amount of fat mass that is only observed in WT and not Dp(11)17/+ mice (Figure 5A). Indeed, while BAT and liver remain of similar sizes (relative to body weight) between the two genotypes after HF diet, the WAT tissues (from four body locations: epididymal, mesenteric, retroperitoneal and inguinal WAT) are much smaller in Dp(11)17/+ than WT mice (Figure 5B, 5C), accompanied by smaller-sized adipocytes (Figure 5D). Furthermore, for WT mice, HF diet resulted in a marked decrease in glucose clearance during GTT but no change in blood insulin level; whereas the glucose clearance in Dp(11)17/+ mice is much less affected by the HF diet (Figure 6A, 6B). This difference in the extent of HF diet mediated insulin resistance between WT and Dp(11)17/+ mice was further confirmed in ITT experiments, which demonstrate even more significant differences between the two genotypes after HF diet (Figure 6C, 6D).
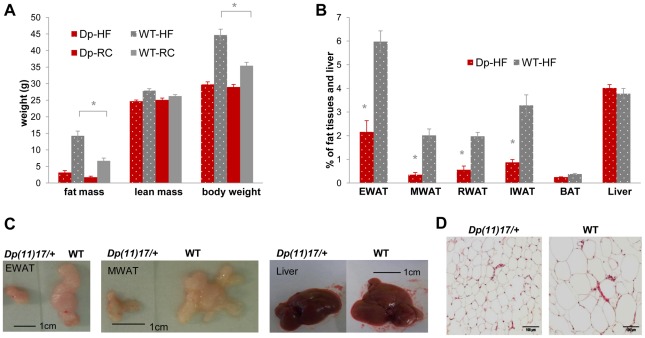
Figure 5. Dp(11)17/+ mice (red) display resistance to diet-induced obesity compared to WT littermates (gray) after a high-fat diet (HF) feeding from 19 to 22 weeks.
(A) Only WT, but not Dp(11)17/+ mice have significant (*p = 0.00028) weight gain after three weeks of HF (19–22 wks) that is mainly due to fat mass increase (*p = 0.00030). (B) Body weight percentage of epididymal (EWAT), mesenteric (MWAT), retroperitoneal (RWAT) and inguinal (IWAT) white adipose tissues are all higher in WT mice post HF than Dp(11)17/+ mice (*p = 0.000033 for EWAT, p = 0.00021 for MWAT, p = 0.000033 for RWAT, p = 0.000048 for IWAT). Liver and brown adipose tissues (BAT) remain similar. (C) Dissected EWAT, MWAT and liver are compared between WT and Dp(11)17/+ mice post HF diet. EWAT and MWAT, but not the liver, are much smaller in Dp(11)17/+ mice. (D) Histology of the EWAT adipocytes from Dp(11)17/+ and WT mice, demonstrating smaller adipocytes in Dp(11)17/+ mice after HF feeding. The measurements are from (A): 11 Dp(11)17/+ and 12 WT at 22 wks post HF feeding compared to 6 Dp(11)17/+ and 10 WT on RC at 21–22 wks (B) 8 Dp(11)17/+ and 9 WT post HF feeding. Dp/WT mice after HF diet: red/gray bars with dotted pattern; Dp/WT mice with RC: red/gray bars without pattern.
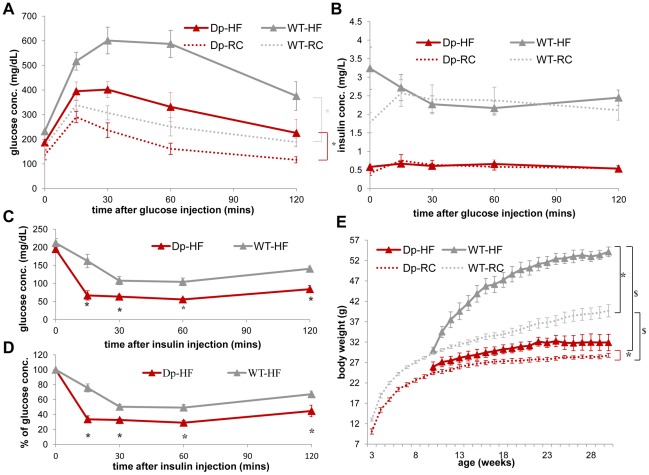
Figure 6. WT mice (gray) display a greater increase in insulin resistance than Dp(11)17/+ mice (red) after HF diet from 19 to 22 weeks as well as an increased weight gain after a long-term HF diet.
(A) During IP-GTT, WT mice display more dramatically decreased glucose clearance rate (*p = 0.00011) after HF diet than Dp(11)17/+ mice (*p = 0.03). (B) The insulin level of both genotypes are not impacted by HF diet. (C, D) During IP-ITT, Dp(11)17/+ mice after HF diet demonstrate lower blood glucose concentration than WT mice, shown as both actual concentration in (C) (*p = 0.002, 0.007, 0.003 and 0.002) and percentage of the initial glucose concentration in (D) (*p = 0.00015, 0.00063, 0.0026, 0.041); whereas the differences are only partially significant under RC as shown in Figure 3 (C, D). (E) Body weight of both genotypes after HF diet from 10 to 30 weeks. *: comparison for each genotype between HF and RC; $: comparison between the genotypes under the same diet condition. After HF feeding, both genotypes gain weight (WT: *p<0.001; Dp(11)17/+: *p = 0.048). Dp(11)17/+ mice are still lighter than WT littermates after HF ($p<0.0005), similar to those fed with RC ($p<0.001). The curves for normal diet (data points without triangles) are the same as shown in Figure 1B. All comparisons were made with ANOVA with repeated measures (A, B, E) or two-tailed t-test (C, D); results are expressed as mean ± s.e.m. and obtained from measurements of n = 5 Dp(11)17/+ and 7 WT after HF diet. Dp/WT mice after HF diet: red/gray solid line with triangle markers; Dp/WT mice with RC: red/gray dashed line without marker.
Next, to explore the long-term impact of the Dp(11)17 CNV in DIO, we examined Dp(11)17/+ males along with their WT littermates on a 42% fat HF diet starting from week 3 for 20 weeks. While the high-fat diet causes massive weight gain in WT mice and literally “supersizes” these animals, it produces only a minimal increase in body weight in the Dp(11)17/+ mice, confirming the salient resistance of the Dp(11)17/+ genotype to diet-induced weight gain (Figure 6E). In aggregate, these findings demonstrate the salubrious effect of this duplication CNV in that it provides protection against diet-induced obesity and insulin resistance.
Rai1 gain alone is insufficient to account for CNV–associated metabolic derangements
We next examined whether the metabolic traits conveyed by the duplication CNV are due to the copy number gain of a single gene. The typical CNV interval of PTLS/SMS encompasses over 40 human genes; one of them, retinoic acid induced 1 (RAI1), is considered the “predominant” causative gene in the deletion CNV interval mediating the majority of SMS clinical findings through haploinsufficiency [15], [16], [17]. Also, for PTLS, RAI1 is a major dosage sensitive gene contributing to the phenotype, as suggested by duplication mapping in humans [34] and the rescue of selected phenotypes after normalizing the gene dosage of Rai1 to n = 2 in Dp(11)17/Rai1− animals [26]. To examine the contribution of the RAI1/Rai1 gene to the metabolic phenotypes of PTLS, we compared the metabolic profile of TgRai1 animals [35] that overexpress Rai1 but do not have copy number change of most of the surrounding genomic regions to that of Dp(11)17/+ mice. Although the regulation of Rai1 expression in TgRai1 mice is mechanistically different from that in Dp(11)17/+ mice, in which Rai1 is localized in a large genomic segment that has a well-defined duplication of the genome, expression studies demonstrated that the Rai1 “steady state” expression level is similar in TgRai1 [35] and Dp(11)17/+ mice [25] (1.5 fold that of WT). Interestingly, TgRai1 animals display an initial growth retardation; however, they eventually normalize their body weight by 20 weeks of age [35]. This is distinct from the Dp(11)17/+ mice, whose difference in body weight when compared to their WT littermates remains and even exacerbates as they age (Figure 1). Further, TgRai1 animals do not demonstrate the dramatically altered body composition and serum chemistry displayed by Dp(11)17/+ mice (Figure S2A, S2B). Finally, again in striking contrast to the remarkably improved insulin sensitivity and glucose clearance of Dp(11)17/+ mice, TgRai1 animals demonstrate no significant differences in their blood glucose or plasma insulin during GTT when compared with WTs (Figure S2C, S2D). We conclude that the dosage or steady state expression level of RAI1/Rai1 is unlikely the sole or major contributor to the obesity opposing and protective metabolic phenotypes observed in the PTLS mice.
The Df(11)17 deletion CNV conveys metabolic syndrome like phenotypes in both mouse and human
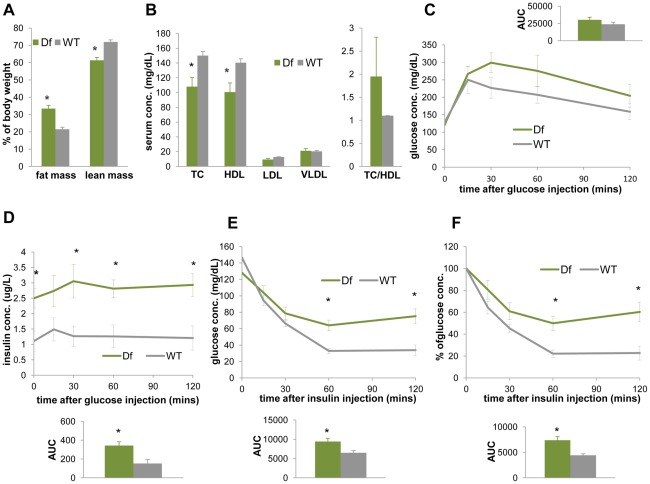
After studying the Dp(11)17/+ mice, we sought to characterize the metabolic profile of the Df(11)17/+ deletion mice on a fully congenic (N>10) 129S5 background. In mirror image contrast to the observations in Dp(11)17/+ mice, Df(11)17/+ mice have not only increased body weight, but also significantly increased percentage of body fat and decreased percentage of lean mass (Figure 7A). These findings are again in accordance with our previous reports of increased size of abdominal fat pad in Df(11)17/+ mice on mixed [20] and congenic (N>12) C57BL/6J [25] strain backgrounds. Intriguingly, Df(11)17/+ mice also have reduced TC, similar to Dp(11)17/+ mice (Figure 7B). However, in contrast to the reduction of the atherogenic LDL in the case of Dp(11)17/+ animals, the reduced TC of Df(11)17/+ mice is the result of a reduced HDL, a cardioprotective species of plasma lipoprotein (Figure 7B). The TC/HDL ratio appears higher in Df(11)17/+ mice although the difference is not significant (Figure 7B). During the GTT, Df(11)17/+ mice display an impaired glucose tolerance phenotype (Figure 7C) accompanied by significantly higher plasma insulin levels during the test as compared with WT mice (Figure 7D), suggesting that Df(11)17/+ mice indeed have increased insulin resistance. This interpretation is bolstered by a blunted blood glucose decrement in response to insulin injection during an insulin tolerance test (ITT) in Df(11)17/+ mice as compared to WT mice (Figure 7E and 7F). The increased insulin resistance and impaired glucose tolerance in Df(11)17/+ mice, along with the increased body weight, relative adiposity and reduced HDL further document that, metabolically, Df(11)17/+ mice display endophenotypes that resemble a bona fide metabolic syndrome. We did not find significant differences in the level of UCP1 protein between Df(11)17/+ and WT mice (Figure S4C, S4D).
Figure 7. Df(11)17/+ mice (green) are obese, have reduced TC, HDL, and display reduced insulin sensitivity in comparison to WT mice (gray).
(A) ECHO-MRI identified elevated fat mass (*p = 0.0041) and reduced lean mass (*p = 0.0034) in Df(11)17/+ animals. (B) Df(11)17/+ animals have lower serum TC (*p = 0.033) and lower HDL (*p = 0.039), but no significantly change in TC/HDL ratio. IP-GTT documented (C) similar blood glucose levels but (D) significantly higher insulin levels (*p = 0.015, 0.028, 0.012 and 0.013 at 0, 30, 60, 120 mins post injection and *p = 0.011 for AUC) in Df(11)17/+ animals. During IP-ITT, Df(11)17/+ mice retain higher blood glucose concentration, shown as both (E) actual concentration (*p = 0.026 and 0.008 for 60 and 120 mins post insulin injection and *p = 0.018 for AUC) and (F) percentage of the initial glucose concentration (*p = 0.01 for both 60 and 120 min after injection and *p = 0.0086 for AUC). All comparisons were made with two-tailed t-test; results are expressed as mean ± s.e.m. from measurements of (A) 5 Df(11)17/+ and 7 WT mice at 32–36 wks (B) 6 Df(11)17/+ and 6 WT mice at 34–37 wks (C, D) 5 Df(11)17/+ and 5 WT mice at 37–41 wks (E, F) 5 Df(11)17/+ and 6 WT mice at 33–37 wks. All AUCs are computed until 120 minutes, for the entire length of the time curves.
From studies of human patients, a meta-analysis of 105 cases [16] including both children and adults concluded that 33.3% of the SMS patients are overweight (BMI>24). In a study of 49 SMS children (0.6 to 17.6 years), Smith et al. [19] observed that SMS boys had a significantly higher BMI than the published age-matched standards. To systematically address the potential obesity directly caused by the SMS deletion CNV in the context of the population norm, we compared 179 height and 216 weight measurements from 76 subjects with SMS aged between newborn and 46 years (Figure S3) to the population mean of the same age/gender from the center for disease control and prevention (http://www.cdc.gov/growthcharts/). We found that both male and female SMS individuals of all age categories in this cohort are shorter than the general population (Figure S3A, S3D, S3G). Male SMS subjects do not have significant weight abnormalities (Figure S3B, S3H). Females below 11 years have weights below the population mean, whereas those older than 12 years appear heavier than the population mean, although the difference is not significant (Figure S3E, S3H). Most importantly, male SMS subjects older than 2 years and female subjects older than 20 years have significantly higher BMI values than the general population (Figure S3C, S3F, S3I). These observations are consistent with the interpretation that the SMS deletion CNV indeed causes a higher BMI in humans and thus conveys an increased risk for obesity. Smith et al [19] also found that the mean fasting TC of SMS patients in their cohort was significantly higher than published pediatric age-matched norms. The aggregate of weight gain and elevated TC in human SMS patients together with the weight gain and insulin resistance in the Df(11)17/+ mice is consistent with MetS-like traits as part of the SMS endophenotypes.
Rai1 haploinsufficiency partially contributes to the Df(11)17-mediated phenotype
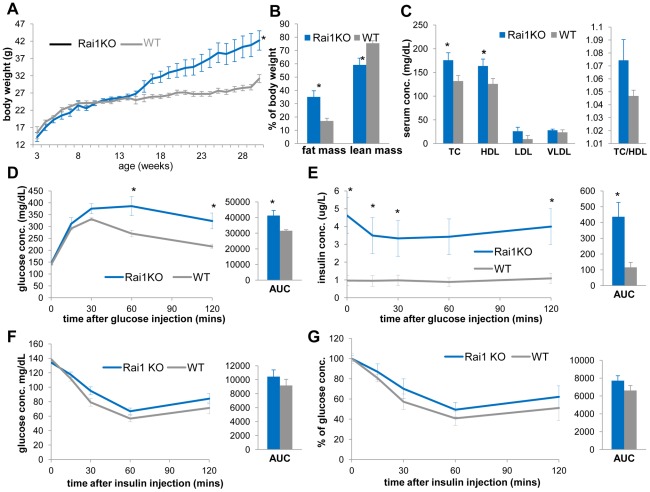
To explore the contribution of RAI1 copy number loss to the metabolic phenotypes of SMS, we also studied Rai1 +/− mice [36], [37] on the same 129S5 (N>10) strain background as the Df(11)17/+ mice. Similar to what we observed for Df(11)17/+ mice, and in accordance with a previous study [18] conducted on a different genetic background, Rai1 +/− males have both significantly increased body weight in adulthood (Figure 8A) and elevated overall proportion of body fat (Figure 8B). Also similar to the Df(11)17/+ mice, Rai1 +/− animals display an unchanged TC/HDL ratio, although they have both elevated TC and HDL levels, opposite to the reduced TC and HDL levels in Df(11)17/+ mice (Figure 8C). Elevated cholesterol was also observed in Rai1 +/− mice on C57BL/6J background in Burns et al [18], although the difference was not statistically significant, which may reflect the mixing of male and female mice and a potential dilution of the difference in male animals. Similarly, Burns et al also noted the increased proportion of body fat in both males and females, although the difference in their assay is only significant for the females. The differences in our findings for the male mice may result from different experimental approaches; ECHO-MRI is less subject to variations introduced by the individual experimentalist and potentially more objectively detects subtle differences in body composition. In addition, the impairment of Rai1 +/− in glucose clearance becomes significant at later time points during the GTT assay (Figure 8D); these animals also show higher plasma insulin at fasting and during the GTT (Figure 8E), although there is no significant difference in blood glucose during ITT (Figure 8F and 8G).
Figure 8. Rai1+/− mice (blue) are obese, have increased TC, HDL, and display reduced insulin sensitivity compared to WT mice (gray).
(A) The growth curve of Rai1+/− and WT littermates reveals increased body weights of Rai1+/− mice (*p = 0.028 by ANOVA with repeated measures). (B) Rai1+/− mice have increased body fat mass (*p = 0.014) and reduced lean mass (*p = 0.015). (C) Rai1+/− mice demonstrate higher TC (*p = 0.036) and HDL (*p = 0.048), but unchanged TC/HDL ratio. In GTT experiments, Rai1+/− mice have (D) higher blood glucose at 60 mins (*p = 0.042) and 120 mins (*p = 0.030 after glucose injection, and *p = 0.038 for total AUC) and (E) higher insulin levels throughout (*p = 0.0056, 0.0069, 0.029, 0.008 at 0, 15, 30 and 120 mins and #p = 0.058 at 60 mins after injection. For AUC, *p = 0.021). (F, G) IP-ITT resulted in similar glucose level change between Rai1+/− and WT mice, as shown by both actual concentration (F) and percentage of the initial glucose concentration (G). All comparisons were made with two-tailed t-test except (A) that used ANOVA; results are expressed as mean ± s.e.m. from measurements of (A) 10–25 Df(11)17/+ and 10–25 WT mice (B, C) 6 Rai1+/− and 8 WT at 30–31 wks (D, E) 5 Rai1+/− and 5 WT mice at 41–43 wks (F, G) n = 7 Rai1+/− and 6 WT mice at 33–36 wks. All AUCs are computed until 120 minutes, for the entire length of the time curves.
Overall, Rai1 +/− mice are similar to Df(11)17/+ mice in their increased body weight and total body fat percentage as well as hyperinsulinemia, impaired GTT and an unchanged TC/HDL ratio. Intriguingly, Edelman et al [16] observed a higher percentage of obesity in SMS patients with RAI1 point mutations (66.7%) than those with 17p11.2 deletions (12.9%). Although the number of SMS patients due to RAI1 point mutation in that report is small (n = 9), these data nevertheless support a significant role for RAI1 copy number loss in the overall metabolic phenotype of SMS, and also suggest possible contributions from other genes/genetic elements in the SMS deletion interval or the deletion per se [25].
Discussion
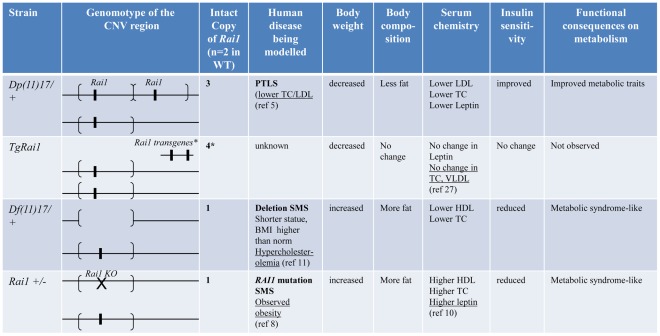
In summary (Figure 9), our detailed analyses of mouse models and human patients demonstrate that the duplication CNV of PTLS conveys highly penetrant metabolic consequences that are antithetical to or “mirror” [9] those observed in MetS. At the same time, it confers protection against the development of diet induced obesity and insulin resistance. These phenotypes are not manifest in the transgenic TgRai1 mice with a similarly increased level of Rai1 expression but without the duplication CNV. In contrast, the reciprocal deletion CNV causes phenotypes that are opposite to those observed with the duplication CNV and that resemble a bona fide metabolic syndrome (summarized in Figure 9).
Figure 9. Experimental findings for specific genetic/genomic variations in this report.
Genomotypes are shown similar to [57]. The segments flanked by brackets that encompass the Rai1 gene represent the CNV region, duplicated in Dp(11)17/+ or deleted in Df(11)17/+. *: TgRai1 strain has the insertion of Rai1 outside of chromosome 11; it gives similar Rai1 expression levels to those of the Dp(11)17/+ strain although the copy number has been determined as four [35]. Underlined results are from published reports.
The reciprocal/mirror phenotypes caused by the reciprocal CNV of Dp(11)17 and Df(11)17 is interesting. Reciprocal phenotypes associated with opposing gene/genome dosage alterations (i.e. copy number loss versus copy number gain) have been described for the complex neuropsychiatric traits of schizophrenia and autism, as well as microcephaly and macrocephaly, associated with, respectively, duplication/deletion CNV at 16p11.2 [38], [39], [40] and deletion/duplication of 1q21.1 [41], [42], [43], [44]. Indeed, another pair of weight regulation associated duplication/deletion CNVs at 16p11.2 was also related to reciprocal changes in BMI and manifest leanness/obesity [9], [10], [27]. These reciprocal traits with opposing dosage alterations are consistent with the model of diametrically opposing phenotypes for genomic sister disorders postulated by Crespi et al [40], [45], [46].
Importantly, neither the duplication nor the deletion CNV associated phenotypes we describe herein in both human patients and mouse models can be attributed solely to the RAI1/Rai1 gene, although RAI1/Rai1 dosage loss does appear to partially contribute to the deletion phenotypes.
Besides Rai1, another gene Srebf1 (coding for sterol regulatory element binding protein 1, Srebp1), that maps directly adjacent to RAI1/Rai1 in both human/mouse genome and functions as a key regulator in the biosynthesis of fatty acid and cholesterol, is the only gene in the SMS/PTLS interval known to be involved in energy metabolism. Overexpression of the active N-terminal portion of Srebp1 protein does not change the plasma lipid profile [47] and results in mild insulin resistance [48]. Both phenotypes are distinctly different from the metabolism phenotype we observed in Dp(11)17/+ mice, rendering the copy number gain of Srebf1 unlikely the reason for the Dp(11)17/+ metabolic phenotypes. Heterozygous Srebf1 knockout mice Srebf1+/− were described as “phenotypically normal” [49]. Srebf1−/− animals are 50–85% embryonic lethal, but the surviving mice display unchanged body weight and slightly reduced total cholesterol and triglycerides in plasma [49]. The copy number loss of Srebf1 in Df(11)17/+ mice is thus also unlikely a major contributor to the observed metabolic phenotypes.
Recently, a microRNA miR33b was found to be embedded in an intron of human SREBF1 [50], [51]. Together with its paralogue miR33a (embedded in the paralogue of SREBF1, SREBF2), miR33b targets the adenosine triphosphate-binding cassette transporter (ABCA1), decreases plasma HDL and boosts intracellular cholesterol levels in cooperation with SREBP proteins [50], [51], [52]. However, mouse Srebf1 does not contain mir33b [50], [51]; it is thus not a candidate accounting for the metabolic phenotype observed in Dp(11)17/+ and Df(11)17/+ mice. The potential role of miR33b in human PTLS/SMS metabolic manifestation will have to be studied with different models, such as those that introduce a human miR33b into the mouse genome.
Our current knowledge thus does not support a “single gene” contribution of the dosage change of Rai1, Srebf1 or any other known genetic element to the metabolic phenotypes of SMS/PTLS. However, it is distinctly possible that the copy number change of RAI1 and SREBF1 in cis, with one of them exerting an epistatic effect on the other or functioning as a modifier, is required for manifestation of the complete metabolic phenotype of PTLS/SMS. Further, the potential “cis” effect could also possibly involve other genetic elements in addition to RAI1 and SREBF1. These metabolic manifestations would then belong to the category of “contiguous gene syndromes” [53] or genomic disorders [54], [55] that require multiple genes/genetic/genomic factors to work in concert, a concept referred to as cis-genetics and in contrast to the trans interactions of alleles at one locus formalized by Mendelism [56]. Similar mechanisms have been proposed for the craniofacial phenotypes of the SMS/Df(11)17/+ mice, wherein the phenotypic penetrance is clearly modified by other genetic elements in the deletion interval although the copy loss of Rai1 appears to be responsible for most of the traits [21], [22].
Further, a number of other mechanisms, including gene interruption and gene fusion due to CNV breakpoints, position effect, the unmasking of a recessive allele by a deletion, as well as potential effects of transvection can contribute to the functional consequence of a CNV [57]. None of them can be displayed by single nucleotide variations (SNV). Recently, it has been demonstrated experimentally that a genomic structural change per se, as in a large CNV, can cause altered expression and functional perturbation of other loci/genes localized to the same chromosome, but outside of the CNV [25]. All these mechanisms can potentially contribute to the salient effect of the Dp(11)17 CNV on weight regulation and energy metabolism that does not appear to be attributed to the dosage change of any single gene or genetic element(s).
Overall, we show that a duplication CNV can result in a lean body phenotype, metabolic phenotypes in mirror image contrast to those observed in metabolic syndrome, and protect from diet induced obesity. Moreover, we demonstrate that these phenotypes are fully penetrant, independent of genetic background and resistant to environmental influences. Furthermore, we provide evidence that the CNV effects are due to more than dosage alteration of a single gene, a finding that highlights distinct functional significance of CNV as compared to SNVs. These findings confirm that CNVs can be causative for weight regulation and energy metabolism phenotypes and suggest that CNVs could play a major role in the common complex diseases of human obesity.
Materials and Methods
Animals
All animal studies were approved by Baylor College of Medicine IRB and carried out in accordance with Baylor IACUC. Mice were housed 2–5 per cage in a 12-hour light/12-hour dark cycle with access to food and water ad libitum.
Body composition and Serum analyses
Body composition was analyzed with the ECHO-MRI system (Echo medical systems, Texas). Mouse serum was prepared from blood obtained through cardiac puncture and analyzed with the COBAS Integra 400 plus analyzer (Roche). Plasma leptin, FFA, adiponectin and glycerol levels were measured by using a Mouse Leptin ELISA Kit (Millipore), NEFA C Test Kit (Wako), Mouse Adiponectin ELISA Kit (Millipore) and Serum/plasma Glycerol detection kit (Sigma), respectively.
Histology
Epididymal (EWAT), mesenteric (MWAT), retroperitoneal (RWAT) and inguinal (IWAT) white adipose tissues, as well as brown adipose tissues (BAT) and liver were dissected from mice deeply anesthetized with Isoflorane (Butler). Tissues were weighed and fixed in 4% neutral buffered formaldehyde (Fisher). Paraffin-embedded sections were stained with hematoxylin and eosin. Photomicrographs were captured by optic microscopy (Zeiss Axiostar Plus).
Activity and metabolic rate measurements
The locomotion activity assay was performed in home cages by using the VersaMax Animal Activity Monitoring System (AccuScan Instruments). Mice were acclimated in the monitoring environments for at least 24 hours before the experiment.
Energy expenditure was measured using the CLAMS System (Columbus Instruments). Animals were allowed to acclimatize in the chambers for 72 hours, and measurements were taken subsequently for 72 hr during the light cycle and dark cycle while mice were freely allowed to access food and water. Oxygen consumption was normalized to lean tissue mass.
Glucose Tolerance Test (GTT) and Insulin Tolerance Test (ITT)
For intraperitoneal GTT, 1.5 g of glucose/kg of body weight was injected after a 6-h fasting period. For ITT, an intraperitoneal injection of regular insulin (Humulin R; 1 unit/kg of body weight) was administered after a 4–6 h fasting. Blood glucose levels were measured using a glucometer (Life Scan).
Protein extraction, immunoblotting, and quantitative RT–PCR
Tissues were lysed in RIPA buffer with Complete Protease Inhibitor Cocktail (Roche). Protein concentration was determined with BCA protein assay kit (Pierce); each sample was separated by SDS-PAGE and electro-transferred to nitrocellulose membrane for immunoblot analyses. Western blots for UCP1 protein were performed with antibody AB3036 (Millipore), after which the same blot was normalized to actin using MAB1501 (Millipore). The ImmunoCruz Western Blotting Luminol reagent (SantaCruz) was used as the substrate.
RNA was isolated with Trizol (Invitrogen), cDNA synthesized with SuperScript III System (Invitrogen), and RT-PCR was performed on the Strategene MX3000 real time detection system using iQ SYBR Green PCR reagent kit (Biorad).
Statistical methods
Results are expressed as mean ± s.e.m. Comparisons between two groups were made using either two-tailed Student's t-test (EXCEL) or ANOVA repeated measures (SPSS), as appropriate. AUC analysis was performed using SigmaBlot. P<0.05 was considered to be statistically significant.
Supporting Information
Mouse models of SMS and PTLS. The region on mouse chromosome 11 syntenic to the human the SMS/PTLS region on human chromosome 17 (synteny is indicated by gray shaded regions). Key genes that demarcate the SMS/PTLS region are shown. The thick black horizontal line above denotes the region of the SMS/PTLS common deletion/duplication. Shown below is the region deleted/duplicated in Df(11)17/Dp(11)17 mice (bold horizontal bar with vertical bars on the end); asterisk * represents the knock-out mouse model of Rai1.
(PDF)
TgRai1 mice (pink) have similar body composition (A), similar serum leptin level (B), and display similar glucose and insulin levels during IP-GTT (C, D) compared to WT mice (gray). All comparisons were made with two-tailed t-test; results are expressed as mean ± s.e.m. from measurements of (A) 9 TgRai1 and 8 WT at 32–35 wks, via dissection of intra-abdominal fat pads (gonadal, retroperitoneal and mesenteric) and subcutaneous fat pads (dorsal, inguinal and groin) as a measure of total fat [18]. (B) 11 TgRai1 and 10 WT at 10–20 wks by leptin ELISA assay at the University of Cincinnati Mouse Metabolic Phenotyping Core per standard protocols and (C, D) 6 TgRai1 and 4 WT animals at 30–32 wks.
(PDF)
Z-scores for height_for_age (A, D, G), weight_for_age (B, E, H) and BMI_for_age (C, F, I) of SMS subjects are plotted as two way scatter plots (A to F) for male (black) (A, B, C) and female (gray) (D, E, F) separately and summarized as mean ± s.e.m. (G to I) with both genders. Subjects are grouped into group 1: 0–23 months; group 2: 2–5 years; group 3: 6–11 years; group 4: 12–19 years; group 5: ≥20years; asterisk (*): significant differences with comparison to the population normative values as calculated with 2-tailed one-sample t-test. All age groups from both genders differ from population norm in their height: for males, p<0.001 (group 1, 2), p = 0.002, 0.059 (#) and 0.006 for group 3 to 5; for females, p = 0.034 and 0.019 for group 1 and 5, p<0.001 for group 2 to 4. The weight of female group 1–3 differs from population norm: p<0.001 for group 1 and 2; p = 0.004 for group 3. Significantly higher BMI was found for male age group 2 to 4 (p = 0.04, 0.022, 0.007, 0.001) and female group 4 (p = 0.002).
(PDF)
Comparative expression analyses of (A, B) some signature metabolic genes in Dp(11)17/+ mice (red) and (C, D) UCP1 in Df(11)17/+ mice (green). (A, B) Relative mRNA abundance for a group of signature genes for energy metabolism: (A) AP2, Ucp1, Ucp2, Ucp3 and Glut4 (B) Lpk, Fas, Acc1, Srebp1c, Tnxip was determined in BAT (A) and liver (B) of Dp(11)17/+ and WT mice. None of the genes displayed significant expression difference between two genotypes: (A) Ap2 (p = 0.79), Ucp1 (p = 0.27), Ucp2 (p = 0.12), Ucp3 (p = 0.08) Glut4 (p = 0.83). (B) Lpk (p = 0.65), Fas (p = 0.15), Acc1 (p = 0.94), Srebp1c (p = 0.75), Tnxip (p = 0.62). (C) Western blot for UCP1 expression in BAT tissue of four Df(11)17/+ and four WT mice with antibody AB3036 (Millipore) and normalized to actin blotting using MAB1501 (Millipore). (D) Normalized intensity of UCP1 signals in Df(11)17/+ vs. WT mice (1.776±0.154 vs. 1.186±0.059, p = 0.154). The measurements are from (A, B) 3 Dp(11)17/+ and 3 WT at 30 wks. (C, D) 4 Df(11)17/+ and 4 WT at 30 wks.
(PDF)
Serum chemistry comparison of Dp(11)17/+ and WT mice. Results are expressed as mean ± s.e.m. and are calculated from the measurements of 5 Dp(11)17/+ and 6 WT males at 21–22 wks. For clarity, the measurements, but not the p-values are shown in bold.
(PDF)
Acknowledgments
We thank Dr. Vasumathi Theegala Reddy for her technical assistance and Dr. Chester Brown, Philip Boone, and Claudia Gonzaga-Jauregui for helpful discussion and critical reading of the manuscript.
Footnotes
JRL is a consultant for Athena Diagnostics, holds stock ownership in 23andMe and Ion Torrent Systems, and is a co-inventor on multiple United States and European patents for DNA diagnostics.
We acknowledge the Baylor Mouse Metabolism Core at Diabetes and Endocrinology Research Center, supported by P30DK079638, and Dr. Roger Price at the Baylor Comparative Pathology lab. This work was supported in part by the National Institute of Neurological Disorders and Stroke (National Institutes of Health) grant R01NS058529 to JRL; National Heart, Lung, and Blood Institute grant R01-HL51586 to LC; Texas Children's Hospital General Clinical Research Center grant M01RR00188; and Intellectual and Developmental Disabilities Research Centers grant P30HD024064. WG is a recipient of the Feodore-Lynen Fellowship from Alexander-von-Humboldt Foundation. ML is supported by the Houston Laboratory and Population Sciences Training Program of the Burroughs Wellcome Foundation.
References
- 1.Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu Rev Med. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- 2.Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horev G, Ellegood J, Lerch JP, Son YE, Muthuswamy L, et al. Dosage-dependent phenotypes in models of 16p11.2 lesions found in autism. Proc Natl Acad Sci U S A. 2011;108:17076–17081. doi: 10.1073/pnas.1114042108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee YS. The role of genes in the current obesity epidemic. Ann Acad Med Singapore. 2009;38:45–43. [PubMed] [Google Scholar]
- 5.Cornier MA, Dabelea D, Hernandez TL, Lindstrom RC, Steig AJ, et al. The metabolic syndrome. Endocr Rev. 2008;29:777–822. doi: 10.1210/er.2008-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hebebrand J, Volckmar AL, Knoll N, Hinney A. Chipping away the ‘missing heritability’: GIANT steps forward in the molecular elucidation of obesity - but still lots to go. Obes Facts. 2010;3:294–303. doi: 10.1159/000321537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacquemont S, Reymond A, Zufferey F, Harewood L, Walters RG, et al. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature. 2011;478:97–102. doi: 10.1038/nature10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walters RG, Jacquemont S, Valsesia A, de Smith AJ, Martinet D, et al. A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature. 2010;463:671–675. doi: 10.1038/nature08727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hawn J, Rice C, Nichols H, McDermott S. Overweight and obesity among children with Down syndrome: a descriptive study of children attending a Down syndrome clinic in South Carolina. J S C Med Assoc. 2009;105:64–68. [PubMed] [Google Scholar]
- 12.Butler MG. Prader-Willi Syndrome: Obesity due to Genomic Imprinting. Curr Genomics. 2011;12:204–215. doi: 10.2174/138920211795677877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Potocki L, Bi W, Treadwell-Deering D, Carvalho CM, Eifert A, et al. Characterization of Potocki-Lupski syndrome (dup(17)(p11.2p11.2)) and delineation of a dosage-sensitive critical interval that can convey an autism phenotype. Am J Hum Genet. 2007;80:633–649. doi: 10.1086/512864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Potocki L, Chen KS, Park SS, Osterholm DE, Withers MA, et al. Molecular mechanism for duplication 17p11.2- the homologous recombination reciprocal of the Smith-Magenis microdeletion. Nat Genet. 2000;24:84–87. doi: 10.1038/71743. [DOI] [PubMed] [Google Scholar]
- 15.Bi W, Lupski JR. Epstein CJ, Erickson RP, Wynshaw-Boris A, editors. RAI1, the Smith–Magenis, and Potocki–Lupski Syndromes. Inborn Errors of Development. 2008. pp. 1078–1088.
- 16.Edelman EA, Girirajan S, Finucane B, Patel PI, Lupski JR, et al. Gender, genotype, and phenotype differences in Smith-Magenis syndrome: a meta-analysis of 105 cases. Clin Genet. 2007;71:540–550. doi: 10.1111/j.1399-0004.2007.00815.x. [DOI] [PubMed] [Google Scholar]
- 17.Elsea SH, Girirajan S. Smith-Magenis syndrome. Eur J Hum Genet. 2008;16:412–421. doi: 10.1038/sj.ejhg.5202009. [DOI] [PubMed] [Google Scholar]
- 18.Burns B, Schmidt K, Williams SR, Kim S, Girirajan S, et al. Rai1 haploinsufficiency causes reduced Bdnf expression resulting in hyperphagia, obesity and altered fat distribution in mice and humans with no evidence of metabolic syndrome. Hum Mol Genet. 2010;19:4026–4042. doi: 10.1093/hmg/ddq317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith AC, Gropman AL, Bailey-Wilson JE, Goker-Alpan O, Elsea SH, et al. Hypercholesterolemia in children with Smith-Magenis syndrome: del (17) (p11.2p11.2). Genet Med. 2002;4:118–125. doi: 10.1097/00125817-200205000-00004. [DOI] [PubMed] [Google Scholar]
- 20.Walz K, Caratini-Rivera S, Bi W, Fonseca P, Mansouri DL, et al. Modeling del(17)(p11.2p11.2) and dup(17)(p11.2p11.2) contiguous gene syndromes by chromosome engineering in mice: phenotypic consequences of gene dosage imbalance. Mol Cell Biol. 2003;23:3646–3655. doi: 10.1128/MCB.23.10.3646-3655.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan J, Bi W, Lupski JR. Penetrance of craniofacial anomalies in mouse models of Smith-Magenis syndrome is modified by genomic sequence surrounding Rai1: not all null alleles are alike. Am J Hum Genet. 2007;80:518–525. doi: 10.1086/512043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan J, Keener VW, Bi W, Walz K, Bradley A, et al. Reduced penetrance of craniofacial anomalies as a function of deletion size and genetic background in a chromosome engineered partial mouse model for Smith-Magenis syndrome. Hum Mol Genet. 2004;13:2613–2624. doi: 10.1093/hmg/ddh288. [DOI] [PubMed] [Google Scholar]
- 23.Walz K, Spencer C, Kaasik K, Lee CC, Lupski JR, et al. Behavioral characterization of mouse models for Smith-Magenis syndrome and dup(17)(p11.2p11.2). Hum Mol Genet. 2004;13:367–378. doi: 10.1093/hmg/ddh044. [DOI] [PubMed] [Google Scholar]
- 24.Molina J, Carmona-Mora P, Chrast J, Krall PM, Canales CP, et al. Abnormal social behaviors and altered gene expression rates in a mouse model for Potocki-Lupski syndrome. Hum Mol Genet. 2008;17:2486–2495. doi: 10.1093/hmg/ddn148. [DOI] [PubMed] [Google Scholar]
- 25.Ricard G, Molina J, Chrast J, Gu W, Gheldof N, et al. Phenotypic consequences of copy number variation: insights from Smith-Magenis and Potocki-Lupski syndrome mouse models. PLoS Biol. 2010;8:e1000543. doi: 10.1371/journal.pbio.1000543. doi: 10.1371/journal.pbio.1000543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walz K, Paylor R, Yan J, Bi W, Lupski JR. Rai1 duplication causes physical and behavioral phenotypes in a mouse model of dup(17)(p11.2p11.2). J Clin Invest. 2006;116:3035–3041. doi: 10.1172/JCI28953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bochukova EG, Huang N, Keogh J, Henning E, Purmann C, et al. Large, rare chromosomal deletions associated with severe early-onset obesity. Nature. 2010;463:666–670. doi: 10.1038/nature08689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choquet H, Meyre D. Molecular basis of obesity: current status and future prospects. Curr Genomics. 2011;12:154–168. doi: 10.2174/138920211795677921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blakemore AI, Froguel P. Investigation of Mendelian forms of obesity holds out the prospect of personalized medicine. Ann N Y Acad Sci. 2010;1214:180–189. doi: 10.1111/j.1749-6632.2010.05880.x. [DOI] [PubMed] [Google Scholar]
- 30.Savage DB. Mouse models of inherited lipodystrophy. Dis Model Mech. 2009;2:554–562. doi: 10.1242/dmm.002907. [DOI] [PubMed] [Google Scholar]
- 31.Klempel MC, Varady KA. Reliability of leptin, but not adiponectin, as a biomarker for diet-induced weight loss in humans. Nutr Rev. 2011;69:145–154. doi: 10.1111/j.1753-4887.2011.00373.x. [DOI] [PubMed] [Google Scholar]
- 32.Kadowaki T, Yamauchi T, Kubota N, Hara K, Ueki K, et al. Adiponectin and adiponectin receptors in insulin resistance, diabetes, and the metabolic syndrome. J Clin Invest. 2006;116:1784–1792. doi: 10.1172/JCI29126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zingaretti MC, Crosta F, Vitali A, Guerrieri M, Frontini A, et al. The presence of UCP1 demonstrates that metabolically active adipose tissue in the neck of adult humans truly represents brown adipose tissue. FASEB J. 2009;23:3113–3120. doi: 10.1096/fj.09-133546. [DOI] [PubMed] [Google Scholar]
- 34.Zhang F, Potocki L, Sampson JB, Liu P, Sanchez-Valle A, et al. Identification of uncommon recurrent Potocki-Lupski syndrome-associated duplications and the distribution of rearrangement types and mechanisms in PTLS. Am J Hum Genet. 2010;86:462–470. doi: 10.1016/j.ajhg.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Girirajan S, Patel N, Slager RE, Tokarz ME, Bucan M, et al. How much is too much? Phenotypic consequences of Rai1 overexpression in mice. Eur J Hum Genet. 2008;16:941–954. doi: 10.1038/ejhg.2008.21. [DOI] [PubMed] [Google Scholar]
- 36.Bi W, Ohyama T, Nakamura H, Yan J, Visvanathan J, et al. Inactivation of Rai1 in mice recapitulates phenotypes observed in chromosome engineered mouse models for Smith-Magenis syndrome. Hum Mol Genet. 2005;14:983–995. doi: 10.1093/hmg/ddi085. [DOI] [PubMed] [Google Scholar]
- 37.Bi W, Yan J, Shi X, Yuva-Paylor LA, Antalffy BA, et al. Rai1 deficiency in mice causes learning impairment and motor dysfunction, whereas Rai1 heterozygous mice display minimal behavioral phenotypes. Hum Mol Genet. 2007;16:1802–1813. doi: 10.1093/hmg/ddm128. [DOI] [PubMed] [Google Scholar]
- 38.McCarthy SE, Makarov V, Kirov G, Addington AM, McClellan J, et al. Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet. 2009;41:1223–1227. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shinawi M, Liu P, Kang SH, Shen J, Belmont JW, et al. Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J Med Genet. 2010;47:332–341. doi: 10.1136/jmg.2009.073015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 41.Brunetti-Pierri N, Berg JS, Scaglia F, Belmont J, Bacino CA, et al. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet. 2008;40:1466–1471. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Consortium IS. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lupski JR. Schizophrenia: Incriminating genomic evidence. Nature. 2008;455:178–179. doi: 10.1038/455178a. [DOI] [PubMed] [Google Scholar]
- 44.Stefansson H, Rujescu D, Cichon S, Pietiläinen OP, Ingason A, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crespi BJ. Revisiting Bleuler: relationship between autism and schizophrenia. Br J Psychiatry. 2010;196:495; author reply 495-496. doi: 10.1192/bjp.196.6.495. [DOI] [PubMed] [Google Scholar]
- 46.Crespi B, Summers K, Dorus S. Genomic sister-disorders of neurodevelopment: an evolutionary approach. Evolutionary Applications. 2009;2:81–100. doi: 10.1111/j.1752-4571.2008.00056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shimano H, Horton JD, Hammer RE, Shimomura I, Brown MS, et al. Overproduction of cholesterol and fatty acids causes massive liver enlargement in transgenic mice expressing truncated SREBP-1a. J Clin Invest. 1996;98:1575–1584. doi: 10.1172/JCI118951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takahashi A, Shimano H, Nakagawa Y, Yamamoto T, Motomura K, et al. Transgenic mice overexpressing SREBP-1a under the control of the PEPCK promoter exhibit insulin resistance, but not diabetes. Biochim Biophys Acta. 2005;1740:427–433. doi: 10.1016/j.bbadis.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 49.Shimano H, Shimomura I, Hammer RE, Herz J, Goldstein JL, et al. Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J Clin Invest. 1997;100:2115–2124. doi: 10.1172/JCI119746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, et al. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rayner KJ, Suárez Y, Dávalos A, Parathath S, Fitzgerald ML, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brown MS, Ye J, Goldstein JL. Medicine. HDL miR-ed down by SREBP introns. Science. 2010;328:1495–1496. doi: 10.1126/science.1192409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmickel RD. Clinical genetics conference: progress in understanding muscle disease. J Pediatr. 1986;109:1071–1073. doi: 10.1016/s0022-3476(86)80303-1. [DOI] [PubMed] [Google Scholar]
- 54.Lupski JR. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–422. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- 55.Lupski JR. Genomic disorders ten years on. Genome Med. 2009;1:42. doi: 10.1186/gm42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lupski JR, Belmont JW, Boerwinkle E, Gibbs RA. Clan genomics and the complex architecture of human disease. Cell. 2011;147:32–43. doi: 10.1016/j.cell.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lupski JR, Stankiewicz P. Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet. 2005;1:e49. doi: 10.1371/journal.pgen.0010049. doi: 10.1371/journal.pgen.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mouse models of SMS and PTLS. The region on mouse chromosome 11 syntenic to the human the SMS/PTLS region on human chromosome 17 (synteny is indicated by gray shaded regions). Key genes that demarcate the SMS/PTLS region are shown. The thick black horizontal line above denotes the region of the SMS/PTLS common deletion/duplication. Shown below is the region deleted/duplicated in Df(11)17/Dp(11)17 mice (bold horizontal bar with vertical bars on the end); asterisk * represents the knock-out mouse model of Rai1.
(PDF)
TgRai1 mice (pink) have similar body composition (A), similar serum leptin level (B), and display similar glucose and insulin levels during IP-GTT (C, D) compared to WT mice (gray). All comparisons were made with two-tailed t-test; results are expressed as mean ± s.e.m. from measurements of (A) 9 TgRai1 and 8 WT at 32–35 wks, via dissection of intra-abdominal fat pads (gonadal, retroperitoneal and mesenteric) and subcutaneous fat pads (dorsal, inguinal and groin) as a measure of total fat [18]. (B) 11 TgRai1 and 10 WT at 10–20 wks by leptin ELISA assay at the University of Cincinnati Mouse Metabolic Phenotyping Core per standard protocols and (C, D) 6 TgRai1 and 4 WT animals at 30–32 wks.
(PDF)
Z-scores for height_for_age (A, D, G), weight_for_age (B, E, H) and BMI_for_age (C, F, I) of SMS subjects are plotted as two way scatter plots (A to F) for male (black) (A, B, C) and female (gray) (D, E, F) separately and summarized as mean ± s.e.m. (G to I) with both genders. Subjects are grouped into group 1: 0–23 months; group 2: 2–5 years; group 3: 6–11 years; group 4: 12–19 years; group 5: ≥20years; asterisk (*): significant differences with comparison to the population normative values as calculated with 2-tailed one-sample t-test. All age groups from both genders differ from population norm in their height: for males, p<0.001 (group 1, 2), p = 0.002, 0.059 (#) and 0.006 for group 3 to 5; for females, p = 0.034 and 0.019 for group 1 and 5, p<0.001 for group 2 to 4. The weight of female group 1–3 differs from population norm: p<0.001 for group 1 and 2; p = 0.004 for group 3. Significantly higher BMI was found for male age group 2 to 4 (p = 0.04, 0.022, 0.007, 0.001) and female group 4 (p = 0.002).
(PDF)
Comparative expression analyses of (A, B) some signature metabolic genes in Dp(11)17/+ mice (red) and (C, D) UCP1 in Df(11)17/+ mice (green). (A, B) Relative mRNA abundance for a group of signature genes for energy metabolism: (A) AP2, Ucp1, Ucp2, Ucp3 and Glut4 (B) Lpk, Fas, Acc1, Srebp1c, Tnxip was determined in BAT (A) and liver (B) of Dp(11)17/+ and WT mice. None of the genes displayed significant expression difference between two genotypes: (A) Ap2 (p = 0.79), Ucp1 (p = 0.27), Ucp2 (p = 0.12), Ucp3 (p = 0.08) Glut4 (p = 0.83). (B) Lpk (p = 0.65), Fas (p = 0.15), Acc1 (p = 0.94), Srebp1c (p = 0.75), Tnxip (p = 0.62). (C) Western blot for UCP1 expression in BAT tissue of four Df(11)17/+ and four WT mice with antibody AB3036 (Millipore) and normalized to actin blotting using MAB1501 (Millipore). (D) Normalized intensity of UCP1 signals in Df(11)17/+ vs. WT mice (1.776±0.154 vs. 1.186±0.059, p = 0.154). The measurements are from (A, B) 3 Dp(11)17/+ and 3 WT at 30 wks. (C, D) 4 Df(11)17/+ and 4 WT at 30 wks.
(PDF)
Serum chemistry comparison of Dp(11)17/+ and WT mice. Results are expressed as mean ± s.e.m. and are calculated from the measurements of 5 Dp(11)17/+ and 6 WT males at 21–22 wks. For clarity, the measurements, but not the p-values are shown in bold.
(PDF)