Abstract
Background
Alkali stress is an important agricultural contaminant and has complex effects on plant metabolism. The aim of this study was to investigate whether the alkali stress has different effects on the growth, ion balance, and nitrogen metabolism in old and young leaves of rice plants, and to compare functions of both organs in alkali tolerance.
Methodology/Principal Findings
The results showed that alkali stress only produced a small effect on the growth of young leaves, whereas strongly damaged old leaves. Rice protected young leaves from ion harm via the large accumulation of Na+ and Cl− in old leaves. The up-regulation of OsHKT1;1, OsAKT1, OsHAK1, OsHAK7, OsHAK10 and OsHAK16 may contribute to the larger accumulation of Na+ in old leaves under alkali stress. Alkali stress mightily reduced the NO3 − contents in both organs. As old leaf cells have larger vacuole, under alkali stress these scarce NO3 − was principally stored in old leaves. Accordingly, the expression of OsNRT1;1 and OsNRT1;2 in old leaves was up-regulated by alkali stress, revealing that the two genes might contribute to the accumulation of NO3 − in old leaves. NO3 − deficiency in young leaves under alkali stress might induce the reduction in OsNR1 expression and the subsequent lacking of NH4 +, which might be main reason for the larger down-regulation of OsFd-GOGAT and OsGS2 in young leaves.
Conclusions/Significance
Our results strongly indicated that, during adaptation of rice to alkali stress, young and old leaves have distinct mechanisms of ion balance and nitrogen metabolism regulation. We propose that the comparative studies of young and old tissues may be important for abiotic stress tolerance research.
Introduction
Alkali stress is an important agricultural contaminant and has complex effects on plant metabolism. There are 831 million hectares of soil in the world that are affected by salt stress. Of this area, alkalinized soils underlie 434 million hectares, while saline soils underlie 397 million hectares [1]. Soil alkalization causes severe problems in some areas. For example, more than 70% of land area in northeast China is alkaline grassland [2], sometimes with a soil pH>10 [3]. Plants on these soils endure concomitant salt and alkali (high-pH) stresses, making it difficult to grow, and only a few species of plants can survive on such soils. Some reports have clearly demonstrated that alkaline salt stress (NaHCO3 and/or Na2CO3) and neutral salt stress (NaCl and/or Na2SO4) are two distinct types of stress that affect plants and that they should be referred to as alkali stress and salt stress, respectively [4]. In fact, alkali stress has been shown to cause much stronger destructive effects on plants than salt stress [5], [6], [7]. However, to date, salt stress studies have generally emphasized NaCl [8], while little attention has been given to alkali stress.
It is well known that salt stress has different effects on old and young tissues [8]. For example, salt stress produces distinct effects on the growth, compatible solutes accumulation and ion metabolism of old and young leaves [9], [10], [11], [12], [13], [14]. Salt stress in a soil generally involves osmotic stress and ion-induced injury [15]. Comparison of alkali stress with salt stress reveals an added high-pH effect of alkali stress. The high-pH environment surrounding the roots can cause metal ions and phosphorus to precipitate, and the loss of the normal physiological functions of the roots and the destruction of root cell structure [5], [6], [7]. Alkali stress can inhibit the absorption of inorganic anions such as Cl−, NO3 − and H2PO4 −, greatly affects the selective absorption of K+–Na+, and breaks the ionic balance in tissue [6], [16]. Although several attentions have been given to alkali stress [1], [5], [6], [7], [16], [17], [18], the comparative study of old and young tissues was lacking. Old and young tissues may play different roles in alkali tolerance as in salt tolerance. Thus, the understanding of comparative effects of alkali stress on old and young tissues may be important for alkali tolerance research.
The maintenance of K+ and Na+ homeostasis is crucial for alkali tolerance [6], [16]. Many transporters of K+ and Na+ have been identified to date. In leaves of plants, salt overly sensitive (SOS) salt tolerance pathway may play important roles in Na+ eduction [8], [19]. In addition, some members of the high affinity K+ transporter (HKT) family, such as OsHKT1; 5 and AtHKT1;1, mediate Na+ exclusion from leaves via Na+ removal from the xylem sap [8]. Our previous studies showed that alkali stress may strongly affect assimilation and/or uptake of nitrate in rice [20] and other plants [6], [16]. Thus, nitrogen metabolism regulation may be important for rice alkali tolerance. NO3 − is reduced to nitrite by nitrate reductase (NR) and then to NH4 + by nitrite reductase (NiR). NH4 + is incorporated into organic molecules by glutamine synthetase (GS) and glutamate synthase (GOGAT) or alternative glutamate dehydrogenase (GDH) pathway [21]. Glutamine synthetase (GS) primarily exists as two isozymes with different subcellular localisations: GS1 in the cytosol and GS2 in chloroplasts/plastids. In rice, OsGS1;1, OsGS1;2 and OsGS1;3, encode GS1. OsGS1;1 and OsGS1;2 are especially abundant in the aerial parts and roots, respectively, whereas OsGS1;3 is present only in the spikelets [22].
In this study, we chose rice plants as the experimental material. The study was designed to investigate whether alkali stress has different effects on the growth, ion balance, and nitrogen metabolism in the old and young leaves of rice, and to compare functions of both organs in rice alkali tolerance.
Methods
Plant Growth Conditions
Tong-35, a major rice cultivar in north China, was chosen as the test organism. Seeds were germinated and grown in petri dishes for 6 d in a growth cabinet (30°C during the day and 25°C during the night, 16/8 h photoperiod at 250 µmol m−2 s−1). Seedlings were then transferred to buckets containing 2000 mL of sterile nutrient solution for solution culture. The nutrient solution was replaced daily. The buckets were placed in a growth chamber that was maintained at 27.0±1.5°C during the day and 22.0±1.5°C during the night, under a 16/8 h photoperiod at 250 µmol m−2 s−1. The nutrient solution used in this work accorded to the components described by the International Rice Research Institute [23], and contained 1.44 mM NH4NO3, 0.32 mM NaH2PO4, 0.6 mM K2SO4, 1.0 mM CaCl2, 1.6 mM MgSO4, 0.072 mM Fe-EDTA, 0.2 mM Na2SiO3, 9.1 µM MnCl2, 0.154 µM ZnSO4, 0.156 µM CuSO4, 18.5 µM H3BO3 and 0.526 µM H2MoO4 at pH 5.2.
Stress Treatment
Two alkaline salts (NaHCO3 and Na2CO3) were selected based on the salt components and pH in the majority of alkaline soils in northeast China. Two alkaline salts were mixed in a 9∶1 molar ratio (NaHCO3∶Na2CO3) as the alkali stress treatment. The total salt concentration was set at 50 mM (pH 9.10). After 22 days of growth in hydroponic medium, rice plants were subjected to alkali stress by transferring them to another bucket containing 2000 mL of the treatment solution amended with the above nutrients and 50 mM stress salts. A bucket including 20 seedlings represented one replicate, and there were four replicates per treatment. 8 buckets of seedlings were randomly divided into 2 sets, four buckets per set. Each bucket was considered as one replicate with four replicates per set, one set was used as control, and another set was treated with alkali stress. Namely, the experiment has four biological replicates. Treatment solutions were replaced daily. The nutrient solution without stress salts was used as a control. The 20 seedlings in each bucket were harvested after treatment for 6 d.
Measurements of Physiological Indices
Membrane permeability can be reflected by the electrolyte leakage rate, which was determined with the ameliorated method of Lutts et al. (1996) [24]. One fresh whole leaf from each bucket was washed three times with deionized water to remove surface adhered electrolytes, then was placed in a closed cuvette containing 20 mL of deionized water at 25°C for 5 h. The electrical conductivity of the solution (EC1) was determined with a conductivity gauge. After this the cuvette was autoclaved at 100°C for 20 min, and the electrical conductivity of the solution (EC2) was determined. Electrolyte leakage rate can be defined as follows: Electrolyte leakage rate (%) = (EC1/EC2)×100.
The young and old leaves of 10 seedlings in each bucket were separated and mixed, then immediately frozen in liquid nitrogen and then stored at −70°C for RNA isolation and the measurements of pigments. Carotenoids, chlorophyll a and chlorophyll b were determined with the method of Zhu (1993) [25], and expressed in mg g−1 FW. Another 10 seedlings in each bucket were washed with distilled water, after which the old (second leaf at bottom) and young leaves were separated and freeze-dried. Then the dry samples of plant material were levigated and mixed for physiological index measurements. Dry samples of plant material (50 mg) were treated with 10 mL deionized water at 100°C for 2 h, and the extract used to determine the contents of free inorganic ions and organic acids (OAs). The contents of NO3 −, Cl−, H2PO4 −, SO4 2− and oxalic acid were determined by ion chromatography (DX-300 ion chromatographic system; AS4A-SC ion-exchange column, CD M-II electrical conductivity detector, mobile phase: Na2CO3/NaHCO3 = 1.7/1.8 mM; DIONEX, Sunnyvale, USA). Other OAs were also determined by ion chromatography (DX-300 ion chromatographic system; ICE-AS6 ion-exclusion column, CDM-II electrical conductivity detector, AMMS-ICE II suppressor, mobile phase: 0.4 mM heptafluorobutyric acid; DIONEX, Sunnyvale, USA). A flame photometer was used to determine K+ and Na+ contents. Ammoniacal nitrogen and soluble sugars were measured, respectively, using ninhydrin and anthrone methods [26].
Quantitative Real Time PCR Analysis
We extracted the total RNA from the young and old leaves of seedlings grown under stress or non-stress conditions using TRIzol reagent (Invitrogen). The RNA was treated with DNaseI (Invitrogen), reverse-transcribed using SuperScriptTM RNase H-Reverse Transcriptase (Invitrogen), and then subjected to real time PCR analysis using gene-specific primers. The functions and sequence informations of the genes used in this study had been reported. The gene-specific primers and corresponding references are listed in Table S1 online. PCR amplification was conducted with an initial step at 95°C for 1 min followed by 40 cycles of 5 s at 95°C, 10 s at 60°C and 30 s at 72°C. Amplification of the target gene was monitored every cycle by SYBR Green. Amplification of the rice UBQ5 (GenBank Accession AK061988) mRNA was used as an internal quantitative control [27], [28], [29]. The relative expression of the target genes was calculated using the ΔCt method [30]. We optimized PCR reaction system, after which the amplification efficiencies of each target gene and reference gene were approximately equal.
Statistical Analysis
Statistical analysis of the data was performed using the statistical program SPSS 13.0 (SPSS, Chicago, USA). All data were represented by an average of the four biological replicates and the standard errors (S.E.). Statistically significant between old and young leaves at same stress condition was determined by t test.
Results
Growth and Ion Accumulation
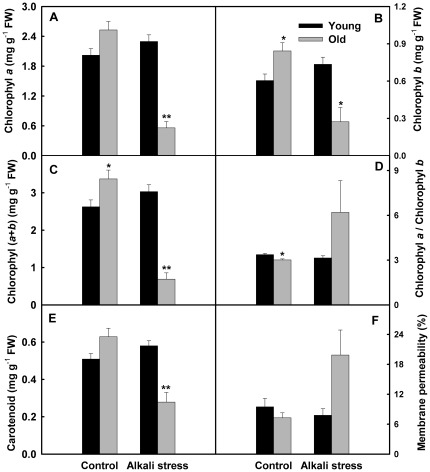
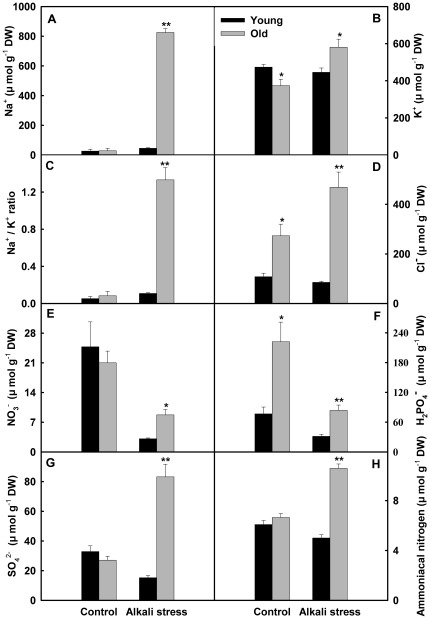
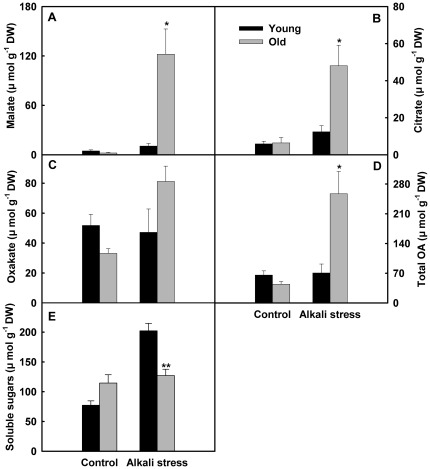
Alkali stress showed a stronger inhibition effect on the growth of old leaves than that of young leaves (Fig. 1). Alkali stress only has small effect on young leaves, whereas mightily increased electrolyte leakage rate and decreased the contents of carotenoids, chlorophyll a and chlorophyll b (Fig. 1). In addition, alkali stress clearly changed chlorophyll a/chlorophyll b ratio in old leaves. The effects of alkali stress on Na+, K+, Na+/K+ ratio, Cl−, SO4 2−, ammoniacal nitrogen, and organic acids in young leaves were no significant, but alkali stress strongly stimulated their accumulation in old leaves (Figs. 2 and 3). Moreover, malate, citrate and oxalate were the dominant components of both young and old leaves under alkali stress, while only trace amounts of succinate, acetate, formate and lactate were detected. Thus, we only listed the results of malate, citrate and oxalate in Fig. 3. Alkali stress reduced the NO3 − contents in both old and young leaves, with reduction in young leaves greater than in old leaves. Under alkali stress, the NO3 − content in old leaves was much higher than that in young leaves (Fig. 2E). Alkali stress decreased the H2PO4 − contents in both old and young leaves (Fig. 2F). Alkali stress elevated the soluble sugars content in young leaves but did not influence its content in old leaves (Fig. 3E).
Figure 1. Effects of alkali stress on the growth in young and old leaves of rice seedlings.
The values are means (± SE) of four biological replicates. Statistically significant between organs at same stress condition was determined by t-test, and marked as * (P<0.05) and ** (P<0.01). The seedlings were subjected to 50 mM alkali stress (NaHCO3∶Na2CO3 = 9∶1; pH 9.10) stresses for 6d.
Figure 2. Effects of alkali stress on the contents of inorganic ions in young and old leaves of rice seedlings.
The values are means (± SE) of four biological replicates, and each replicate consisted of a pool of 10 plants. Statistically significant between organs at same stress condition was determined by t-test, and marked as * (P<0.05) and ** (P<0.01). The seedlings were subjected to 50 mM alkali stress (NaHCO3∶Na2CO3 = 9∶1; pH 9.10) stresses for 6d.
Figure 3. Effects of alkali stress on the contents of organic acids (OA) and soluble sugars in young and old leaves of rice seedlings.
The values are means (± SE) of four biological replicates, and each replicate consisted of a pool of 10 plants. Statistically significant between organs at same stress condition was determined by t-test, and marked as * (P<0.05) and ** (P<0.01). The seedlings were subjected to 50 mM alkali stress (NaHCO3∶Na2CO3 = 9∶1; pH 9.10) stresses for 6d.
Ion Metabolism
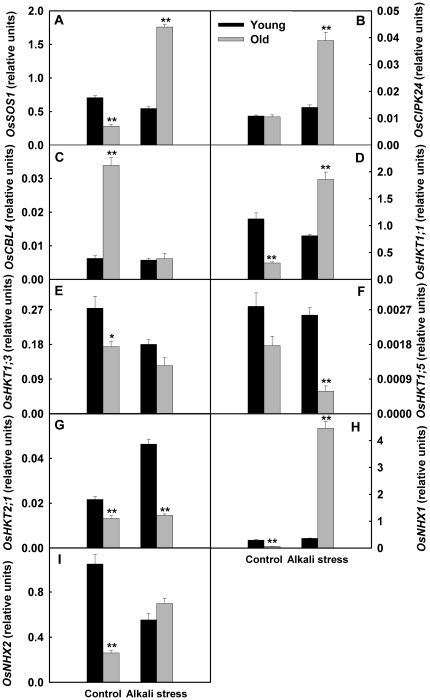
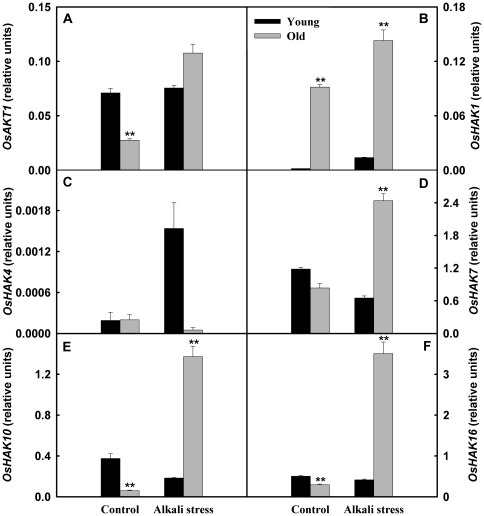
The responses of the genes related to K+/Na+ metabolism to alkali stress were diverse. The expression levels of several genes, such as OsSOS1, calcineurin B-like interacting protein kinase 24 (OsCIPK24), Na+/H+ exchanger 1 (OsNHX1), OsNHX2, OsHKT1;1, low affinity K+ transporter 1 (OsAKT1), KUP/HAK/KT K+ transporter 1 (OsHAK1), OsHAK7, OsHAK10, and OsHAK16, in old leaves were clearly up-regulated by alkali stress, but their expression levels in young leaves were not affected (Figs. 4 and 5). Alkali stress only produced small effects on expression of calcineurin B-like 4 (OsCBL4) and OsHKT1;5 in young leaves, but reduced their expression levels in old leaves. Interestingly, alkali stress did not influence OsHAK4 expression in old leaves, whereas strongly stimulated its expression in young leaves (Fig. 5C). The expression levels of OsHKT1;3 in both organs were reduced by alkali stress (Fig. 4E).
Figure 4. Effects of alkali stress on the expression of OsSOS pathway genes and OsHKT gene family in young and old leaves of rice seedlings.
The values are means (± SE) of four biological replicates, and each replicate consisted of a pool of 10 plants. Statistically significant between organs at same stress condition was determined by t-test, and marked as * (P<0.05) and ** (P<0.01). The seedlings were subjected to 50 mM alkali stress (NaHCO3∶Na2CO3 = 9∶1; pH 9.10) stresses for 6d.
Figure 5. Effects of alkali stress on the expression of OsAKT1 and OsHAK gene family in young and old leaves of rice seedlings.
The values are means (± SE) of four biological replicates, and each replicate consisted of a pool of 10 plants. Statistically significant between organs at same stress condition was determined by t-test, and marked as * (P<0.05) and ** (P<0.01). The seedlings were subjected to 50 mM alkali stress (NaHCO3∶Na2CO3 = 9∶1; pH 9.10) stresses for 6d.
NH4 + Assimilation
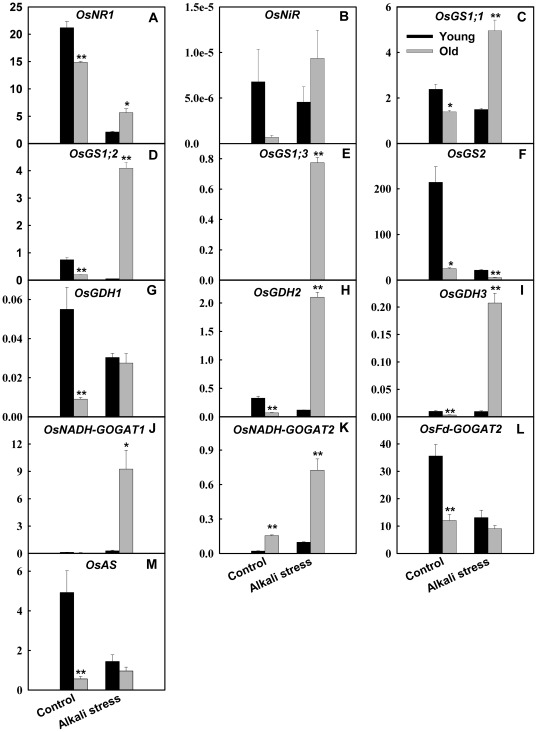
OsGS2 was dominant OsGs family member in leaves, and its expression level in leaves was much higher than other members (Fig. 6). Similarity, OsFd-GOGAT was dominant OsGOGAT family member in leaves. Alkali stress only has small effects on the expression of OsFd-GOGAT and OsGS2 in old leaves, and mightily reduced their expression in young leaves. Alkali stress only produced small effects on expression of OsGS1 family, OsNADH-GOGAT1, OsNADH-GOGAT2, OsGDH2, and OsGDH3 in young leaves, and strongly stimulated their expression in old leaves (Fig. 6). Alkali stress increased the expression level of OsGDH1 in old leaves, whereas decreased its expression in young leaves. Alkali stress down-regulated the expression of asparagine synthetase (OsAS) in young leaves (Fig. 6M), did not influence its expression in old leaves. Alkali stress down-regulated the OsNR1 expression in both tissues, with reduction in young leaves greater than in old leaves (Fig. 6A). Alkali stress reduced the expression of OsNiR in young leaves, and increased its expression in old leaves.
Figure 6. Effects of alkali stress on the expression (relative units) of genes involved in NH4 + assimilation in young and old leaves of rice seedlings.
The values are means (± SE) of four biological replicates, and each replicate consisted of a pool of 10 plants. Statistically significant between organs at same stress condition was determined by t-test, and marked as * (P<0.05) and ** (P<0.01). The seedlings were subjected to 50 mM alkali stress (NaHCO3∶Na2CO3 = 9∶1; pH 9.10) stresses for 6d.
Nitrogen Uptake
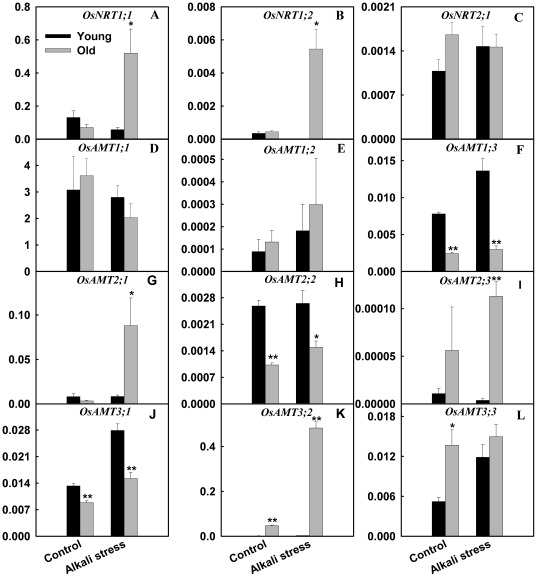
The expression levels of nitrate transporter 1;1 (OsNRT1;1) and OsNRT1;2 in old leaves were up-regulated by alkali stress, while their expression in young leaves decreased under alkali stress (Fig. 7). However, alkali stress did not influence the expression of OsNRT2;1 in both tissues. Responses of ammonium transporter (OsAMT) family members to alkali stress were diverse. For example, alkali stress strongly stimulated the expression of OsAMT1;3 in young leaves, while increased the expression of OsAMT2;3 and OsAMT3;2 in old leaves (Fig. 7).
Figure 7. Effects of alkali stress on the expression (relative units) of OsAMT and OsNRT gene families in young and old leaves of rice seedlings.
The values are means (± SE) of four biological replicates, and each replicate consisted of a pool of 10 plants. Statistically significant between organs at same stress condition was determined by t-test, and marked as * (P<0.05) and ** (P<0.01). The seedlings were subjected to 50 mM alkali stress (NaHCO3∶Na2CO3 = 9∶1; pH 9.10) stresses for 6d.
Discussion
Growth
It is well-known that salt stress has stronger inhibition effect on the growth of old leaves than that of young leaves. This may be an adaptive strategy of plants to salt stress, and protects young organs via sacrificing old organs. We also had found this phenomenon in many extreme halophytes and glycophytes (unpublished data). In present study, we have observed that alkali stress has a stronger injury effect on old leaves than that of young leaves. Alkali stress strongly damaged the membrane system and photosynthetic pigment of old leaves even changed the ratio between different pigments (Fig. 1D).
Ion Balance
Alkali stress showed a stronger effect on the ion balance in rice old leaves than young leaves. The results indicated that the effects of alkali stress on Na+, K+, Na+/K+ ratio, Cl−, SO4 2− and ammoniacal nitrogen in young leaves were no significant, but alkali stress strongly stimulated their accumulation in old leaves (Fig. 2). This may be an adaptive response of rice plants to alkali stress. Rice plants may protect young leaves from ion injury via a large accumulation of toxicant ions in old leaves. We also observed that, under alkali stress, both first and second leaves at bottom (old leaves) accumulated larger Na+ but other five leaves (one young leaf and four maturate leaves) all accumulated the low concentration of Na+ (unpublished data). Old leaf cells have larger vacuole, but young leaf cells only have dispersed miniature vacuoles. Rice plants can compartmentalize toxicant ions like Na+ into the larger vacuoles of old leaves to avoid the ion toxicity of whole green part (Fig. 2). Rice may have a specific regulatory mechanism of Na+ transmission into old leaves.
Na+ enters plant cells through the K+ transporter pathways and non-selective cation channels [31]. Under salt stress the in vivo Na+ metabolism of plants has least three processes: compartmentalization (at cellular and/or tissue levels), exclusion (from shoots into roots) and transportation (in vasculatures) of the ions. In Arabidopsis, the salt overly sensitive protein 1 (SOS1) functions in Na+ exclusion from root epidermal cells into the rhizosphere, which also may play a role in retrieving Na+ from leaf under severe salt stress [8]. The Ca2+-responsive AtSOS3–AtSOS2 (AtCIPK24-AtCBL4) protein kinase pathway mediates regulation of the expression and activities of Na+ transporters such as AtSOS1 and AtNHX, a Na+/H+ exchanger that mediates Na+ compartmentalization into vacuoles [31]. The rice SOS salt tolerance pathway has been identified and its functions have been shown as similar to that of the SOS pathway in Arabidopsis [32]. In Arabidopsis and some other plant species, the Na+/H+ exchanger (NHX) family has been shown to function in Na+ compartmentalization into vacuoles [8]. In rice leaves, SOS salt tolerance pathway and NHX family may play a role in retrieving Na+ from leaf cells to the vascular tissue and Na+ compartmentalization, separately [8]. Our results revealed that, under alkali stress, Na+ homeostasis of rice leaves might have a sophisticated regulating network. Under alkali stress, rice was able to change the ion distribution at whole plant level via altered expression of critical genes involved in ion balance. For example, the expression of several genes, such as OsSOS1, Os CIPK24, OsNHX1, OsNHX2, OsHKT1;1, OsAKT1, OsHAK1, OsHAK7, OsHAK10, OsHAK16, in old leaves was clearly stimulated by alkali stress, but their expression levels in young leaves were not affected (Figs. 4 and 5). Under alkali stress, the increased expression of OsSOS1, OsCIPK24 and OsNHX family might be helpful for Na+ compartmentalization and retrieving Na+ from old leaf cells to vascular tissue. Up-regulated expression of OsHKT1;1, OsAKT1, OsHAK1, OsHAK7, OsHAK10 and OsHAK16 might contribute to the larger accumulation of Na+ in old leaves of alkali stressed-rice. However, alkali stress did not increase the expression of OsHKT1;3, OsHKT1;5, and OsHKT2;1 in rice old leaves (Fig. 4), suggesting that it was unlikely that the three genes contributed to the accumulation of Na+ in old leaves.
Ionic imbalance in plants is mainly caused by the influx of superfluous Na+ [6], [16]. Plants usually accumulate inorganic anions, such as Cl−, NO3 − and SO4 2−, or synthesized organic anions to maintain ionic balance and pH homeostasis [16]. Our results revealed that, under alkali stress, young leaves did not accumulate Cl−, SO4 2−, and organic acids, but old leaves accumulated the high concentrations of organic acids, Cl− and SO4 2−. This may be an adaptative response to Na+ excess in old leaves (Figs. 2 and 3), and plays important roles in the maintaining ionic balance and pH homeostasis in old leaves. In addition, we found that alkali stress stimulated the accumulation of soluble sugars in young leaves but did not increase its content in old leaves (Fig. 3E). This may be an adaptive response of rice to alkali stress, and soluble sugar possibly plays an important osmotic role in the young leaves of alkali-stressed rice.
Nitrogen Nutrition
Plant survival and growth in saline environments is a result of adaptive processes such as ion transport and compartmentation, compatible solutes synthesis and accumulation. Many of these compatible solutes are N-containing compounds, such as amino acids and betaines, hence the nitrogen metabolism is of central importance for salt tolerance [33]. However, interference between salinity and nitrogen nutrition is a very complex network affecting almost all processes in plant metabolism and development. Our results indicated that alkali stress strongly influenced the nitrogen metabolism of rice leaves. Alkali stress reduced the NO3 − contents in both old and young leaves, but the reduction in young leaves was greater that in old leaves (Fig. 2E). Under alkali stress, the NO3 − content in old leaves was much higher than that in young leaves. The lacking of NO3 − in roots may be main reason for the deficiency of NO3 − in rice leaves under alkali stress. It has been reported that alkali stress limited NO3 − uptake of rice roots, and reduced NO3 − content of the roots [20], which may lead to the persistent lacking of NO3 − in stems and leaves. As the old leaf cells have larger vacuole (only miniature vacuoles for young leaf), these scarce NO3 − may be principally stored in rice old leaves to keep the NO3 − supply in stems and leaves. Rice may limit the transmission of NO3 − into young leaves. Accordingly, the two genes coding nitrate transporter, OsNRT1;1 and OsNRT1;2, in old leaves were up-regulated by alkali stress, while their expression levels in young leaves were down-regulated (Fig. 7). This revealed that the two genes might contribute to the accumulation of NO3 − in the old leaves of alkali stressed-rice.
It was recognized that NO3 − is reduced to nitrite by nitrate reductase (NR) and then to NH4 + by nitrite reductase (NiR). NH4 + from both nitrate reduction and soil are incorporated into organic molecules by glutamine synthetase (GS) and glutamate synthase (Fd-GOGAT and NADH-GOGAT) or alternative glutamate dehydrogenase (GDH) pathway (reviewed by Shi et al. 2010) [21]. The expression of OsFd-GOGAT in rice leaves was more abundant than other OsGOGAT gene family members, and the expression of OsGS2 also was more abundant than other OsGS gene family members (Fig. 6). Namely, OsFd-GOGAT and OsGS2 are principally expressed in leaves and played crucial roles in the assimilation of NH4 + from photorespiration and other metabolic process. Compared both tissues, we found that the effect of alkali stress on the nitrogen metabolism of young leaves was stronger than that of old leaves. Alkali stress did not influence the expression of OsFd-GOGAT and OsGS2 in old leaves, whereas mightily reduced their expression in young leaves. The decreased expression of OsFd-GOGAT and OsGS2 in young leaves might be a response to NO3 − deficiency. The NO3 − deficiency in young leaves might cause the large reduction in OsNR1 expression (Fig. 6A) and the subsequent lacking of free NH4 +, which might be main reason why alkali stress sharply down-regulated the expression of OsFd-GOGAT and OsGS2 in young leaves (Fig. 6). Moreover, the responses of the OsAS, OsGDH and OsAMT gene families in both tissues to alkali stress also showed that alkali stress has different effect on their nitrogen metabolism.
In summary, alkali stress only produced a small effect on the growth of young leaves, whereas strongly damaged the membrane system and photosynthetic pigment in old leaves. Old leaf cells have larger vacuole, but young leaf cells only have dispersed miniature vacuoles. Rice plants can compartmentalize toxicant ions, such as Na+ and Cl−, into the larger vacuoles of old leaves to avoid the ion toxicity of whole plant. Up-regulated expression of OsHKT1;1, OsAKT1, OsHAK1, OsHAK7, OsHAK10 and OsHAK16 might contribute to the larger accumulation of Na+ in old leaves of alkali stressed-rice. The effect of alkali stress on the nitrogen metabolism of young leaves was stronger than that of old leaves. Alkali stress reduced the NO3 − contents in both old and young leaves, but the reduction in young leaves was greater than that in old leaves. As old leaf cells have larger vacuole, these scarce NO3 − may be principally stored in rice old leaves. OsNRT1;1 and OsNRT1;2 might contribute to the accumulation of NO3 − in old leaves. NO3 − deficiency in young leaves might induce the large reduction in OsNR1 expression and the subsequent lacking of free NH4 +, which might be main reason why alkali stress sharply down-regulated the expression of OsFd-GOGAT and OsGS2 in young leaves. Our results strongly indicated that, during adaptation of rice to alkali stress, young and old leaves have distinct mechanisms of ion balance and nitrogen metabolism regulation. However, most of research of salt stress and alkali stress still use the mixed sample of young and old tissues, which only provides the limited insights into stress tolerance mechanism. We propose that the comparative studies of young and old tissues may be important for abiotic stress tolerance research.
Supporting Information
Gene-specific primers used in real time PCR analysis.
(DOC)
Acknowledgments
We thank two anonymous reviewers for the critical and constructive comments for further improving of the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported by the State Key Basic Research and Development Plan of China (2011CB100205), the Programme for Introducing Talents to Universities (No. B07017), the National Natural Science Foundation of China Project (Nos. 31072078 and 30970232), and the Fundamental Research Funds for the Central Universities (Nos. 11QNJJ020 and 10SSXT143. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wang Y, Ma H, Liu G, Xu C, Zhang D, et al. Analysis of gene expression profile of Limonium bicolor under NaHCO3 stress using cDNA microarray. Plant Mol Bio Rep. 2008;26:241–254. [Google Scholar]
- 2.Kawanabe S, Zhu T. Degeneration and conservational trial of Aneurolepidium chinense grassland in Northern China. J Japan Grassl Sci. 1991;37:91–99. [Google Scholar]
- 3.Zheng H, Li J. Form and dynamic trait of halophyte community. Beijing: Science Press; 1999. pp. 137–142. [Google Scholar]
- 4.Shi DC, Yin LJ. Difference between salt (NaCl) and alkaline (Na2CO3) stresses on Pucinellia tenuiflora (Griseb.) Scribn. et Merr. plants. Acta Bot Sin. 1993;35:144–149. [Google Scholar]
- 5.Yang C, Jianaer A, Li C, Shi D, Wang D. Comparison of the effects of salt-stress and alkali-stress on photosynthesis and energy storage of an alkali-resistant halophyte Chloris virgata. Photosynthetica. 2008;46:273–278. [Google Scholar]
- 6.Yang C, Shi D, Wang D. Comparative effects of salt and alkali stresses on growth, osmotic adjustment and ionic balance of an alkali-resistant halophyte Suaeda glauca (Bge.). Plant Growth Regul. 2008;56:179–190. [Google Scholar]
- 7.Yang C, Wang P, Li C, Shi D, Wang D. Comparison of effects of salt and alkali stresses on the growth and photosynthesis of wheat. Photosynthetica. 2008;46:107–114. [Google Scholar]
- 8.Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- 9.Akram MS, Ali Q, Athar HUR, Bhatti AS. Ion uptake and distribution in Panicum antidotale Retz. under salt stress. Pak J Bot. 2006;38:1661–1669. [Google Scholar]
- 10.Ashraf M, O'Leary J. Ion distribution in leaves of salt-tolerant and salt-sensitive lines of spring wheat under salt stress. Acta Bot Neer. 1997;46:207–218. [Google Scholar]
- 11.Hajlaoui H, Ayeb NE, Garrec JP, Denden M. Differential effects of salt stress on osmotic adjustment and solutes allocation on the basis of root and leaf tissue senescence of two silage maize (Zea mays L.) varieties. Ind Crops Pro. 2010;31:122–130. [Google Scholar]
- 12.Nakamura T, Ishitani M, Harinasut P, Nomura M, Takabe T. Distribution of glycinebetaine in old and young leaf blades of salt-stressed barley plants. Plant Cell Physiol. 1996;37:873–877. [Google Scholar]
- 13.Vera-Estrella R, Barkla BJ, Garcia-Ramirez L, Pantoja O. Salt stress in Thellungiella halophila activates Na+ transport mechanisms required for salinity tolerance. Plant Physiol. 2005;139:1507–1517. doi: 10.1104/pp.105.067850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yasar F, Uzal O, Tufenkci S, Yildiz K. Ion accumulation in different organs of green bean genotypes grown under salt stress. Plant Soil Environ. 2006;52:476. [Google Scholar]
- 15.Munns R. Comparative physiology of salt and water stress. Plant Cell Environ. 2002;25:239–250. doi: 10.1046/j.0016-8025.2001.00808.x. [DOI] [PubMed] [Google Scholar]
- 16.Yang C, Chong J, Li C, Kim C, Shi D, et al. Osmotic adjustment and ion balance traits of an alkali resistant halophyte Kochia sieversiana during adaptation to salt and alkali conditions. Plant Soil. 2007;294:263–276. [Google Scholar]
- 17.Gao C, Wang Y, Liu G, Yang C, Jiang J, et al. Expression profiling of salinity-alkali stress responses by large-scale expressed sequence tag analysis in Tamarix hispid. Plant Mol Biol. 2008;66:245–258. doi: 10.1007/s11103-007-9266-4. [DOI] [PubMed] [Google Scholar]
- 18.Shi D, Wang D. Effects of various salt-alkaline mixed stresses on Aneurolepidium chinense (Trin.) Kitag. Plant Soil. 2005;271:15–26. [Google Scholar]
- 19.Shi H, Quintero FJ, Pardo JM, Zhu JK. The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell. 2002;14:465–477. doi: 10.1105/tpc.010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, Wu Z, Chen Y, Yang C, Shi D. Effects of salt and alkali stresses on growth and ion balance in rice (Oryza sativa L.). Plant Soil Environ. 2011;57:286–294. [Google Scholar]
- 21.Shi WM, Xu WF, Li SM, Zhao XQ, Dong GQ. Responses of two rice cultivars differing in seedling-stage nitrogen use efficiency to growth under low-nitrogen conditions. Plant Soil. 2010;326:291–302. [Google Scholar]
- 22.Kusano M, Tabuchi M, Fukushima A, Funayama K, Diaz C, et al. Metabolomics data reveal a crucial role of cytosolic glutamine synthetase 1; 1 in coordinating metabolic balance in rice. Plant J. 2011;66:456–466. doi: 10.1111/j.1365-313X.2011.04506.x. [DOI] [PubMed] [Google Scholar]
- 23.Yoshida S, Forno DA, Cock J. Laboratory Manual for Physiological Studies of Rice. Los Banos: International Rice Research Institute; 1976. 62 [Google Scholar]
- 24.Lutts S, Kinet J, Bouharmont J. NaCl-induced senescence in leaves of rice (Oryza sativa L.) cultivars differing in salinity resistance. Ann Bot. 1996;78:389–398. [Google Scholar]
- 25.Zhu GL. Laboratory Manual of Plant Physiology. Beijing: Beijing University Press; 1993. 45 [Google Scholar]
- 26.Zhang Z. Laboratory Manual of Plant Physiology. Beijing: Higher education press; 2004. pp. 160–172. [Google Scholar]
- 27.Jain M, Nijhawan A, Tyagi AK, Khurana JP. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biop Res Co. 2006;345:646–651. doi: 10.1016/j.bbrc.2006.04.140. [DOI] [PubMed] [Google Scholar]
- 28.Quinet M, Ndayiragije A, Lefèvre I, Lambillotte B, Dupont-Gillain CC, et al. Putrescine differently influences the effect of salt stress on polyamine metabolism and ethylene synthesis in rice cultivars differing in salt resistance. J Exp Bot. 2010;61:2719–2733. doi: 10.1093/jxb/erq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zang A, Xu X, Neill S, Cai W. Overexpression of OsRAN2 in rice and Arabidopsis renders transgenic plants hypersensitive to salinity and osmotic stress. J Exp Bot. 2010;61:777–789. doi: 10.1093/jxb/erp341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-[Delta][Delta] CT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Zhu JK. Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol. 2003;6:441–445. doi: 10.1016/s1369-5266(03)00085-2. [DOI] [PubMed] [Google Scholar]
- 32.Martinez-Atienza, Jiang X, Garciadeblas B, Mendoza I, Zhu JK, et al. Conservation of the salt overly sensitive pathway in rice. Plant Physiol. 2007;143:1001–1012. doi: 10.1104/pp.106.092635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Läuchli A, Lüttge U. Salinity: Environment-plants-molecules. Boston: Boston Kluwer Academic Publishers; 2002. pp. 229–248. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene-specific primers used in real time PCR analysis.
(DOC)