Abstract
Several Acinetobacter strains have utility for biotechnology applications, yet some are opportunistic pathogens. We compared strains of seven Acinetobacter species (baumannii, Ab; calcoaceticus, Ac; guillouiae, Ag; haemolyticus, Ah; lwoffii, Al; junii, Aj; and venetianus, Av-RAG-1) for their potential virulence attributes, including proliferation in mammalian cell conditions, haemolytic/cytolytic activity, ability to elicit inflammatory signals, and antibiotic susceptibility. Only Ah grew at 102 and 104 bacteria/well in mammalian cell culture medium at 37°C. However, co-culture with colonic epithelial cells (HT29) improved growth of all bacterial strains, except Av-RAG-1. Cytotoxicity of Ab and Ah toward HT29 was at least double that of other test bacteria. These effects included bacterial adherence, loss of metabolism, substrate detachment, and cytolysis. Only Ab and Ah exhibited resistance to killing by macrophage-like J774A.1 cells. Haemolytic activity of Ah and Av-RAG-1 was strong, but undetectable for other strains. When killed with an antibiotic, Ab, Ah, Aj and Av-RAG-1 induced 3 to 9-fold elevated HT29 interleukin (IL)-8 levels. However, none of the strains altered levels of J774A.1 pro-inflammatory cytokines (IL-1β, IL-6 and tumor necrosis factor-α). Antibiotic susceptibility profiling showed that Ab, Ag and Aj were viable at low concentrations of some antibiotics. All strains were positive for virulence factor genes ompA and epsA, and negative for mutations in gyrA and parC genes that convey fluoroquinolone resistance. The data demonstrate that Av-RAG-1, Ag and Al lack some potentially harmful characteristics compared to other Acinetobacter strains tested, but the biotechnology candidate Av-RAG-1 should be scrutinized further prior to widespread use.
Introduction
The genus Acinetobacter comprises 27 known species and several unnamed provisional species of gram-negative, ubiquitous, non-motile, non-fermentative, coccobacilli [1]–[2]. Many members share phenotypic features, but are not well defined with respect to beneficial and harmful characteristics [3].
In recent years, certain strains of different Acinetobacter species have been developed for bioremediation of recalcitrant and harmful organic chemicals, as well as bioengineering of enzymes and diagnostic materials [4]–[8]. One promising example is the RAG-1 strain of A. venetianus, which has been shown to produce several oil-modifying substances such as emulsans, esterases, lipases and surfactants [9]–[11]. This strain was previously assigned to other species groups (A.calcoaceticus and A.lwoffi) [12] which are recognized as opportunistic pathogens. Further advancement of the Acinetobacter genus for beneficial applications requires a rigorous clarification concerning threats to drinking water [13]–[14] and linkages to nosocomial infections [15]–[20].
Clinically, A.baumannii (Ab) is most often identified as the cause of infection, but others include A.calcoaceticus (Ac), A.haemolyticus (Ah), A.lwoffii (Al) and A.junii (Aj) [20]–[26]. The majority of reported clinical cases involved pneumonia/pulmonary infections and septicaemia, but others included endocarditis, meningitis, burn and surgical wound infections, and urinary tract infections. Many clinical isolates have acquired multiple antibiotic resistance [27] [17] and can be more pan-resistant than even methicillin-resistant Staphylococcus aureus (MRSA) [28]. Furthermore, infections caused by Acinetobacter are not restricted to the clinical setting, and reports have emerged describing cases involving otherwise healthy individuals of varying ages, occurring in community settings, during wars, and following natural disasters [29]–[36].
To date there has been no comparative testing for potential virulence and toxic effects of Acinetobacter species/strains isolated from clinical and environmental sources, which should be necessary prior to any intended biotechnology application. Our research objective is to expand the repertoire of useful endpoints required to predict and rank pathogenicity potential of environmental Acinetobacter strains with the aim of reducing the use of more costly and laborious in vivo test methods. In previous work, we developed a set of pathogenicity-toxicity parameters that could differentiate between potentially harmful and non-toxic strains of Bacillus species [37]–[39]. Here we describe a side-by-side comparison of strains from seven Acinetobacter strains which were selected from both clinical and environmental sources (See Table 1). We used our previous test parameters, as well as others developed to study clinical Acinetobacter strains. The latter include assays which assess capacity to bind or adhere to mammalian cells [40]–[43], disrupt mammalian cell interactions (attachments) [41]–[42] [44]–[49], produce haemolytic or cytolytic activities [41] [50]–[51], and/or infect by way of proliferation in mammalian cell culture medium. Also included is an assessment of exposure-induced immune system responses such as release of pro-inflammatory cytokines and chemoattractants (interleukin (IL)-1β, IL-6, IL-8, and tumour necrosis factor-α (TNF-α)), as observed in vitro with laryngeal epithelial cells [48] and cultured mouse splenocytes [52].
Table 1. Acinetobacter strains used in this study.
| Bacterium | ATCC # | Abbrev. | Previous Genomospecies Designation | Biohazard Level* | Isolation Source of Strain as reported by ATCC |
| A. baumannii | 9955 | Ab | 2 | 2 | Human spinal fluid |
| A. calcoaceticus | 23055 | Ac | 1 | 2 | Soil (Type Strain) |
| A. guillouiae | 11171 | Ag | 11 | 1 | Sewage containing gas-works effluent (Type Strain) |
| A. haemolyticus | 17906 | Ah | 4 | 2 | Human sputum (Type Strain) |
| A. junii | 17908 | Aj | 5 | 2 | Human urine (Type Strain) |
| A. lwoffii | 15309 | Al | 8/9 | 2 | NA (Type Strain) |
| A. venetianus | 31012 | Av-RAG-1 | NA | 1 | Tar on beach (Type Strain) |
Classified by ATCC according to U.S. Public Health Service guidelines.
NA – Not Available.
Materials and Methods
Bacterial Culture and Monitoring
Table 1 contains a list of the bacteria used in this study, along with their acquisition sources, species abbreviations, and biohazard ranking as designated by ATCC. Strains were selected based on availability from known repositories (ATCC), and quantity of literature information on the each strain. Almost all strains selected were Type Strains (except Ab ATCC# 9955), although this was not part of the selection criteria. In all experiments, level two biosafety facility and procedures were used. Bacterial stocks were adjusted to108 cfu/mL, stored at −80°C and routinely checked for viability, morphology and homogeneity by plating on Luria-Bertani (LB)-agar. For assessment of Ag growth, nutrient broth (NB) plates were used, which permitted more rapid results compared to LB plates. Growth of bacteria was assessed at 28°C and 37°C with turbidity measurements (optical density at 450 nm (OD450)) over a 24 h period using an automated scanning multiwell spectrophotometer (Spectramax Plus 386, Molecular Devices Co., Sunnyvale CA). Additional monitoring for growth and viability of cultures was done by measuring bioreduction activity after incubating cultures with 0.5 mg/mL XTT (sodium 3′-{1-[(phenylamino)-carbonyl]-3,4-tetrazolium}-bis{4-methoxy-6-nitro} benzene sulphonic acid hydrate), as done previously with Bacillus organisms [53]. Bacterial reduction of the tetrazolium salt XTT to XTT formazan was measured at OD480 every 15 min.
Cell-free bacterial culture filtrates were prepared by inoculating 10 mL of nutrient broth with 108 cfu of bacteria. Following 24 h at 28°C or 37°C, cultures were centrifuged at 12,000×G and the supernatants were filtered through a 0.22 µm syringe filter (Millipore Corp, Bedford, MA).
Bacterial Antibiotic Susceptibility and Haemolytic Activity
The Minimal Inhibitory Concentration (MIC) assay was conducted as described by Seligy and Rancourt [54]. Antibiotics were purchased from the Sigma Chemical Company (Oakville, ON) and Invitrogen (Carlsbad, CA). Using 96-multi-well plates, 2×104 cfu of bacteria were added to each microwell containing an antibiotic dilution series in trypticase soy broth (TSB) (200 µL final volume). The final concentration of each antibiotic in the series was 24, 12, 6, 3, 1.5, 0.75, 0.38 and zero µg/ml. Since growth in TSB varied between strains, plates were incubated at 28°C or 37°C for 24 h, 48 h or 96 h to generate enough bacterial growth to evaluate antibiotic susceptibility. The bacterial metabolic status of each sample was determined by adding MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) to a final concentration of one mg/ml/well. Plates were then incubated at 28°C or 37°C for two hours and examined for purple crystals. The MIC was defined as the minimum antibiotic concentration resulting in no detectable MTT bioreduction, which indicates that no metabolic activity and proliferation of bacterial cells has occurred.
Haemolytic activity produced by various Acinetobacter strains was compared to that produced by the positive control (Bacillus cereus ATCC# 14579). Sterile, defibrinated sheep blood (Cedarlane Laboratories, Hornby, Ontario) was washed three times with PBS by repeated centrifugation and resuspension in 0.9% (w/v) saline, before resuspension at a final concentration of 5% (v/v) with cooled (45°C), autoclaved agar (1.2% (w/v)) containing 1.4% (w/v) pancreatic digest of casein, 0.5% (w/v) NaCl, 0.45% (w/v) peptone and 0.45% (w/v) yeast extract. After agar solidification, each strain was deposited (103 cfu/10 µL spot) in a grid pattern onto the surface, and incubated at 37°C for up to 6 days. Photographs were taken daily to monitor haemolytic progression.
Mammalian Cell Culture and Exposures
Human HT29 colonic epithelial cells and mouse J774A.1 macrophage were obtained from the American Type Culture Collection (Rockville, MD). Cell monolayers were maintained at less than 80% confluence in mammalian cell culture medium (MCCM; Dulbecco's Modified Eagles Medium containing 25 mM glucose, 10% fetal bovine serum and 2 mM glutamine with/without 50 µg/mL gentamicin). Exposures were conducted by exposing mammalian cells to 102 to 106 cfu for zero to 24 h using 96-well cell culture plates (100 µL final). Some experiments designed to study the effects of bacterial protein products included the addition of a protease inhibitor cocktail (Complete™ Tablets; Roche Diagnotics, Penszberg, Germany) during Acinetobacter exposure.
The effects of bacterial exposure on mammalian cell morphology were monitored by fluorescence and confocal microscopy. Cell monolayers were pre-grown on glass coverslips, fixed for 5 min with 4% (w/v) paraformaldehyde in PBS (pH 7.4), stained with 0.25% (v/v) SYTOX™ green for 10 min, and for 5 min each with 100 µg/mL Texas Red-conjugated wheat germ agglutinin and rhodamine-phalloidin (1∶40), according to the manufacturers = procedures (Molecular Probes Inc, Eugene, OR). Results were viewed and photographed with a Nikon TE-2000 Eclipse microscope equipped with epifluorescence optics and either a Nikon Coolpix 990 digital camera or a Nikon C1 confocal with 488, 543 and 633 lasers.
Quantitative changes in metabolism of mammalian cells during bacterial exposure was monitored by measuring MTT bioreduction activity as described previously [37]–[38]. Following exposures, the culture supernatants were removed and processed for cytokine content as described in the next section. Fresh mammalian culture media supplemented with MTT at a final concentration of one mg/mL was added to the wells. The plates were incubated for two hours at 37°C and the wells were rinsed twice with PBS to remove non-adherent cells and bacteria. Following addition of 100 µL/well DMSO (Sigma), solubilized-formazan color change was measured at OD505. Bioreduction activity of exposed cells was expressed as percentage activity compared to control PBS-treated cells.
Cytokine Measurements
Cytokine activity was measured using supernatants from individual exposures. Culture supernatants (100 µL) were transferred to membrane-containing 96-well (0.45 µm hydrophilic Multiscreen7 plates, Millipore Corp, Bedford, MA). The filtrates were collected through a vacuum manifold into empty 96-well plates. The bacteria and debris-free filtrates were frozen at −80°C until further analysis for cytokine content. After thawing, levels of IL-1β, IL-6, IL-8 and TNF-α were measured using a multiplex liquid bead array assay (BioRad, Laboratories Inc., Mississauga, ON), and validated using enzymatic immunosorbant assays [55] with the following types of antibodies: capture antibodies, human anti-IL-8 (4 µg/mL; R&D Systems, Minneapolis, MN), murine anti-IL-1β (4 µg/ml), anti-IL-6 (0.5 µg/ml), anti-TNF-α (0.5 µg/ml) (PeproTech, Rocky Hill, NJ); coating antibodies, biotinylated anti-human IL-8 (20 ng/mL; R&D Systems), biotinylated anti-murine IL-1β (100 ng/ml), anti-IL-6 (300 ng/ml), anti-TNF-α (300 ng/ml; PeproTech). Recombinant IL-8 (R&D Systems), IL-1β, IL-6, or TNF-α (PeproTech) were used as standards. Lipopolysaccharide (LPS; 0.1 to 1.0 ng/mL) from Escherichia coli, Salmonella typhimurium and Serratia marcescens (Sigma, Oakville, ON) was used as a positive control for cytokine expression studies.
Macrophage Bactericidal Assay
Killing of bacteria by macrophage was tested using ∼80% confluent J774A.1 monolayers (∼105 cells per 33 mm2 well) and 103 bacteria in 100 µL of mammalian cell culture medium without antibiotic. Several incubation times were tested and optimal conditions were determined to be four hours at 37°C. Following incubation, the entire well contents were scraped, serially diluted in PBS and then plated on LB-agar. Colonies were enumerated over a period of 18 to 72 h at 37°C. Control experiments included wells with only bacteria, wells containing only J774A.1 with no bacteria, and bacteria with HT29 epithelial cells.
PCR, Southern Hybridization and Sequencing
Primers for reported virulence factor genes (epsA and ompA; [40] [47] [56] and the quinolone resistance determining regions (QRDRs) of the constitutive genes parC and gyrA, where mutations may confer fluoroquinolone resistance [57]–[60], were designed from the Acinetobacter baumannii ATCC17978 genome. Primer sequences were cross referenced to other complete Acinetobacter genomes through Microbial Genomes Blast (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi) to ensure primers were not strain specific (Table 2). Fragments were amplified from 100 ng of genomic DNA template using standard reaction conditions (AmpliTaq core reagents, ABI, Carlesbad, CA) with 0.6 µM of each primer and the following reaction parameters: 30 cycles 94°C, 15 sec; 50°C, 30 sec; 72°C, 1∶30. PCR reactions were fractionated by gel electrophoresis (1% agarose) and amplicons were purified using a gel extraction spin kit (Qiagen, Germantown, MD).
Table 2. PCR Primers Used in this Study and Resulting Amplicon Sizes.
| Gene | Accession | Forward Primer | Reverse Primer | PCR Amplicon/ORF Size (bp) |
| ompA | AY485227 | CGCTTCTGCTGGTGCTGAAT | CGTGCAGTAGCGTTAGGGTA | 531/1317 |
| epsA | NC_011595 | AGCAAGTGGTTATCCAATCG | ACCAGACTCACCCATTACAT | 451/1101 |
| gyrB | CT025946.2 | ATGAGCGTATCGGAAATCCG | ACGTCAACAATACCTGATTTACC | 485/2713 |
| parC | CU459141.1 | GGCTGGTCTTCTTCACGAATA | CCTTGCGCATCATGCGACAG | 581/2220 |
Genomic DNA (two µg) from each Acinetobacter strain was digested with 10 units of EcoRI (New England BioLabs, Pickering, ON) at 37°C for three hours followed by electrophoresis on (1% agarose gels, 60 V, four hours). DNA contents of gels were pre-treated (15 min acid depurination in 0.25 M HCl, 30 min denaturation in 0.5 M NaOH, 1.5 M NaCl, 30 min neutralization in 0.5 M NaCl, 1.5 M Tris HCl pH 7.5) before overnight transfer onto Nytran supercharge nylon membranes (Whatmann, Piscataway, NJ) in 20× SSC (3 M NaCl, 0.3 M sodium citrate, pH 7.0). Purified Ab ompA and epsA PCR products were labelled with 32P-dCTP using the Bioprime random labelling kit (GE Healthcare, Baie d'Urfe, QC) and hybridized overnight to membranes at 42°C in UltraHyb hybridization buffer (50% formamide) (Ambion, Austin, TX). Blots were washed twice with 2× SSC, 0.1% sodium dodecyl sulphate (SDS) for 10 min at 42°C then twice with 0.5× SSC, 0.1% SDS for 15 min at 65°C. Membranes were exposed to phosphorimager screens for two hours then scanned with the Typhoon scanner (GEHealthcare, Baie d'Urfe, QC).
Amplicons of gyrA and parC were sequenced using BigDyeTerminator v3.1 sequencing kit (Applied Biosystems, Streetsville, ON) and gene specific forward and reverse primers. Reactions were set up with 1/8th reaction mix, sequencing buffer, 0.5 µM primer and 50 ng PCR template. Reactions were purified using Centri-Sep cycle sequencing clean up columns (Princeton Separations, Adelphia, NJ). Sequencing reactions were run on an Applied Biosystems 3130xl Genetic Analyzer on a 36 cm capillary array using POP-7™ polymer (Applied Biosystems, Streetsville, ON). Sequences were assembled and translated using VectorNTI v11 software (Invitrogen, Carlsbad, CA).
Statistical Analysis
The significance of the differences in cytokines produced by mammalian cells in response to different Acinetobacter strains was done by comparing their levels with an analysis of variance (ANOVA) followed by a post-hoc Tukey Multiple Comparison Test. Statistical analyses were done with SigmaPlot software (Systat Software Inc, San Jose, CA).
Results
Microbial Growth and Influence of Mammalian Cells
The proliferation capacity of the different Acinetobacter strains was compared in a bacterial medium (LB) and mammalian cell culture medium at 37°C. The results summarized in Table 3 are derived from experiments using bioreduction of XTT as a sensitive measure of bacterial growth and viability. In LB medium, the majority of strains reduced XTT at low to high inoculation concentrations. In contrast, almost all strains failed to reduce XTT in mammalian cell culture medium at low seeding densities, aside from Ah, which readily grew in all test conditions. Bioreduction was improved with high inoculation concentrations, and when HT29 cells were present. The exception was Av-RAG-1, which was highly bioreductive compared to all other strains in LB, but showed the lowest bioreduction in mammalian cell culture medium and no bioreduction in the presence of HT29 cells. A similar assessment using macrophage-like J774A.1 cells was not possible due to killing of most strains (see Section on Macrophage-like J774A.1 Exposures).
Table 3. Acinetobacter Growth-related Bioreduction Activity.
| Test Bacterium | Optical Density of XTT-Formazan Produced at 24 h* | ||||||||
| LB | DMEM | DMEM+HT29 | |||||||
| 102 cfu | 104 cfu | 106 cfu | 102 cfu | 104 cfu | 106 cfu | 102 cfu | 104 cfu | 106 cfu | |
| Ab | 0.40±0.25 | 2.3±0.1 | >3.2† | 0.00 | 0.00 | 1.14±0.02 | 0.00 | 1.13±0.1 | 2.42±0.19 |
| Ac | 0.01±0.02 | 0.01±0.01 | 0.67±0.13 | 0.00 | 0.00 | 0.4±0.04 | 0.11±0.01 | 0.26±0.01 | 2.28±0.27 |
| Ag | 0.00 | 0.03±0.02 | 0.89±0.05 | 0.00 | 0.00 | 0.12±0.03 | 0.18±0.03 | 0.23±0.02 | 0.33±0.14 |
| Ah | 1.50±0.11 | 1.73±0.16 | 2.18±0.20 | 0.12±0.08 | 1.28±0.14 | 1.63±0.14 | 0.64±0.08 | 0.92±0.11 | 1.43±0.15 |
| Aj | 0.04±0.06 | 0.03±0.05 | 2.95±0.14 | 0.00 | 0.00 | 1.47±0.17 | 0.08±0.01 | 0.42±0.07 | 1.57±0.16 |
| Al | 0.00 | 0.03±0.02 | 1.56±0.13 | 0.00 | 0.00 | 1.14±0.15 | 0.21±0.02 | 0.30±0.05 | 1.56±0.13 |
| RAG-1 | 1.85±0.08 | 2.39±0.06 | 2.92±0.06 | 0.00 | 0.00 | 0.53±0.04 | 0.00 | 0.00 | 0.00 |
Data represent means ± standard deviation from six replicates.
Upper Detection Limit of plate reader.
Haemolytic Activity
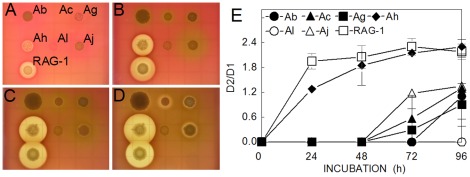
The haemolytic potential of Acinetobacter strains was tested by quantifying the lysis of sheep red blood cells embedded in agar plates. All strains except Ac formed colonies within 24 h (Figure 1A), but only Ah and Av-RAG-1 produced large clearing zones, indicative of β-haemolysis comparable to that produced by the B.cereus positive control. With longer incubations, Ac, Ag, and Aj produced a weak response (α-haemolysis), beginning as faint grey-green rings (Figure 1B, C and D), gradually forming a thin clearing zone. No haemolytic activity (γ-haemolysis) was produced by Al colonies. The quantification of the haemolytic activity summarized in Figure 1E was derived from four replicate plates by comparing the diameters of the lytic zones (D2) with those of the corresponding colonies (D1). Further tests of the filtrates derived from 24 h cultures of these bacteria grown in nutrient broth confirmed that only Ah and Av-RAG-1 produced appreciable erythrocyte cytolysis (data not shown).
Figure 1. Expression of haemolytic activity.
Sheep blood agar plates were inoculated with 103 cfu/spot of each Acinetobacter species, and photographed at 24 h (A), 48 h (B), 72 h (C) and 97 h (D). Graph (E) shows ratios of the diameters of the colonies with their lytic zones (D2) divided by the diameters of the colonies only (D1). Data points are the means ± standard deviation from 4 separate plate assays.
Mammalian Cell Toxicity
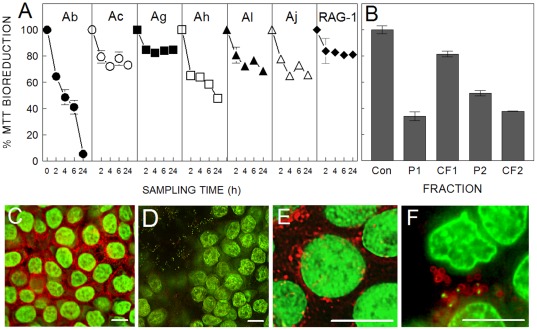
Loss of metabolism, measured by bioreduction of MTT, and abnormal cell morphology were used as toxicity indicators for assessing HT29 and J774A.1 exposures to bacteria cells and/or their extracellular products. The data summarized in Figure 2A show that all strains caused at least 15–20% loss in total HT29 bioreduction capacity within two hours of exposure. At this stage there were no significant morphological changes other than decreased intracellular formazan staining. With longer exposure durations (18 h to 24 h), the most damaging strains were Ab and Ah, causing 95% and 50% drops in bioreduction per well, respectively.
Figure 2. Toxicity of Acinetobacter towards human colonic epithelial cells.
(A) HT29 cell monolayers were exposed to 106 cfu/100 µL for up to 24 h and analyzed for bioreduction activity as described in Materials and Methods. (B) Bioreduction activity of HT29 cells following a 24-h exposure to Ab culture fractions in the presence of gentamicin. Bacterial cultures were centrifuged and separated into pellet (P1) and culture filtrate (CF1). The pellet was resuspended in fresh media, vigorously agitated and recentrifuged to yield a secondary pellet (P2) and filtrate (CF2). (C–F) Confocal micrographs of HT29 treated with PBS alone (C, E) or 106 cfu/100 µL of Ab (D) or Ah (E) for 18 h, and then fixed and permeabilized as described in the Materials and Methods. Cells in C and D were stained with SYTOX™ green and rhodamine-conjugated phalloidin. Panels E and F were stained with SYTOX™ green and Texas Red-conjugated wheat germ agglutinin. Error bars in A and B represent the mean of three exposures ± standard deviation. Bars in C–F represent 10 µm.
Bacterial culture fractions of Ab were prepared to determine if the toxicant was associated with the bacterial cell or was an extracellular component. Cultures were separated into a bacterial cell pellet fraction (P1), and cell-free, culture filtrate fraction (CF1) by centrifugation and filtration. Figure 2B shows that when exposures were done with P1 in the presence of the antibiotic, gentamicin, HT29 metabolic loss and detachment were observed, as reflected by the 65% drop in bioreduction. In contrast, CF1 exposure resulted in only ∼20% bioreduction loss, and no cell detachment. If the bacterial cultures were agitated prior to filtration, the resultant filtrate (CF2) caused both toxicity and cell detachment, suggesting that the effectors were loosely associated with bacterial cell itself. Also, the toxic component of Ab was resistant to heat (100°C, 30 min).
Compared to Ab and Ah, Av-RAG-1 was least toxic in exposures to HT29 (Figure 2A). The diminished effects were attributed to the inability of Av-RAG-1 to grow in the presence of HT29 cells (Table 3). However, culture filtrates generated from Av-RAG-1 cultures in LB, were haemolytic and also caused ∼60% loss in bioreduction of HT29 cells (data not shown). This toxic component was released into the medium during culture of Av-RAG-1 without cell agitation prior to filtration.
Microscopic examination during Ab and Ah exposures showed that the damage could be attributed to a combination of diminished bioreduction capacity and numbers of attached HT29 cells. To detail this observation further, exposed cells were fixed and stained with dyes for the nucleus and fibrous actin (Figures 2C and 2D), or membrane polysaccharides (Figures 2E and 2F). Figures 2D and 2F show HT29 monolayers that were disrupted by Ab and Ah exposures. These exposures were all done at 18 h and not 24 h because very few HT29 remained attached at the latter time point. Ab caused HT29 monolayers to detach in entire sheets, whereas Ah caused individual cells to detach. In either case, inclusion of a protease inhibitor cocktail during the exposure prevented cell detachment. These inhibitors were neither toxic to HT29 nor to the bacteria as measured by MTT and XTT viability assays. However, microscopic examination of the wells supplemented with protease inhibitors revealed that Ab and Ah exposures still resulted in almost complete loss of HT29 intracellular bioreduction activity (ie., the cells exhibited no internal formazan deposits). Cells incubated with bacteria showed loss of actin-staining in Ab-exposed cells (Figure 2D) and wheat germ agglutinin-binding in Ah-exposed cells (Figure 2F) compared to corresponding unexposed controls (Figure 2C and 2E, respectively). Also apparent during microscopy, was bacterial adherence to the monolayer cells and surrounding well surfaces (Figures 2D and 2F). This binding was also observed in MTT assays, which indicated that formazan contributed by bacteria was highest in these exposures. Bacterial adherence was also demonstrated with fluorescence from wheat germ agglutinin–bound Ah (red stain in Figure 2F). The binding of lectins to the capsular polysaccharide of one strain of A.venetianus has been documented previously [61], but not for Av-RAG-1.
Phagocytosis and Macrophage Bactericidal Activity
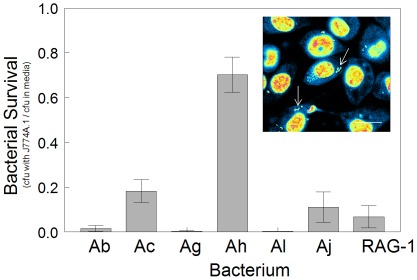
The capacity of J774A.1 to phagocytize bacteria and prevent infection in vitro, was examined using confocal microscopy, and coupled with tests for bacterial cell viability during exposure. Using the SYTOX™ stain, cells of each bacterial strain were initially detected on the apical surface of J774A.1 cells, but after ∼5 min they were seen to internalize and migrate towards the macrophage's basolateral surface. The photo inset of Figure 3 is typical of J774A.1 cells that have phagocytised Ah at the time of maximal uptake (60 min).
Figure 3. Phagocytosis and bactericidal activity of J774A.1 macrophage.
Survival of bacteria was monitored by enumerating cfu's following 4-h incubation to 103 cfu/100 µL with or without monolayers of J774A.1. Bacterial survival is expressed as the ratio cfu = s recovered from incubations with J774A.1 in mammalian culture medium (DMEM+supplements) and from mammalian culture medium alone. Inset: Phagocytosis of bacteria (Ah shown as example) was monitored by exposing J774A.1 monolayers to 106 cfu/100 µL for 60 min, and then fixing and staining with SYTOX nucleic acid stain. Internalization was visualized using a confocal microscope, and artificial graded colour intensity (SYTOX intensity from blue to red) was used to highlight bacteria (arrows) within J774A.1. Data points represent the means of five separate experiments ∀ standard deviation. Bar represents 10 µm.
Experiments were done to determine whether certain strains had the ability to resist being killed by J774A.1. As shown in Figure 3 with 4-h exposure assays, the viability of most Acinetobacter strains was reduced by ≥80% when bacteria and macrophage were co-incubated, compared to bacteria in mammalian cell culture medium alone (or with HT29). The viability for most strains was lowest whether their cell numbers were low (103 cfu/100 µL) or high (104–106 cfu/100 µL well). However, Ah consistently exhibited high viability, even at concentrations as low as 10–100 cfu/100 µL. At higher concentrations, recovered Ah cfu's were greater than the initial seeding concentration. Neither pre-activation of J774A.1 with IFN-γ, nor longer co-incubations of 7 to 24 h reduced Ah cfu numbers. Examination of time-course experiments by microscopy and colony enumerations revealed that the residual external bacteria were replicating, and not the internalized bacteria. This observation suggested that the surviving cfu = s measured in Figure 3 were primarily of external origin.
In other experiments bacteria were co-incubated with J774A.1 in wells with a 0.2 µm filter barrier between them. This exposure system resulted in no killing and demonstrated that phagocytosis or close interaction was needed for J774A.1-induced killing to occur. Parallel exposures conducted with HT29 epithelial cells did not result in lowered viability.
Colonic Epithelial Cell Cytokine Production
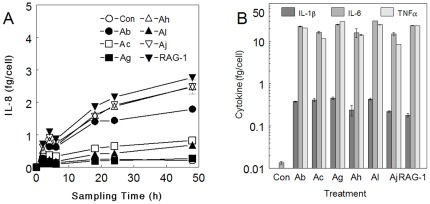
Production of cytokines was monitored during exposures to determine if Acinetobacter strains could initiate epithelial inflammatory responses. Cytokine levels were quantified using multiplex liquid bead arrays for GM-CSF, IL-1β, MIP-1β, IL-6, IL-8, IL-12 and TNF-α, and verified with double antibody sandwich immunoassays. HT29 cells most consistently produced IL-8 during Acinetobacter exposures. Experiments with Ab and Ah indicated that in the absence of antibiotic, the build-up of IL-8 peaked at 6–8 h and was markedly reduced thereafter. The observed drop in levels was likely related to HT29 death and detachment, but also IL-8 degradation during bacterial growth. Inclusion of antibiotic throughout the exposure regime resulted in sustained levels of ILB8 (Figure 4A). Unexposed HT29 produced a low (constitutive) level of IL-8 in the supernatant. Exposure to Acinetobacter strains in the presence of antibiotic resulted in increased extracellular IL-8 levels which persisted for at least 48 h. The induced IL-8 production, measured by both multi-bead array and ELISA, could be divided into two statistically divisible groups (p<0.001 at 48 h). Strains of Ab, Ah, Aj and Av-RAG-1 induced between 1.7 and 3 fg IL-8 per HT29 cell, whereas Ac, Ag and Al generated levels of ≤1 fg/cell.
Figure 4. Mammalian cell inflammatory cytokine levels during exposure to test bacteria.
HT29 (A) and J774A.1 (B) cells were exposed to 106 cfu/100 µL test bacteria for various time intervals. Exposure supernatants were filtered and used in liquid array bead assays for IL-8 (A) or IL-1β, IL-6 and TNF-α (B). Experiments were reproduced twice using both the multiplex liquid bead assay and ELISA, both yielding results within 15% variability of each other. Data points are means of three exposures ∀ standard deviation.
Macrophage Cell Cytokine Production
Macrophage-like J774A.1 cells were tested for cytokine production in exposures similar to those for HT29 cells. The J774A.1 did not produce significant levels of neutrophil chemoattractants, such as KC, but instead produced IL-1β, IL-6 and TNF-α. These three cytokines are involved in the initiation of the acute phase response (APR). In 24-h exposures with gentamicin, all bacteria were strong inducers of the three cytokines, resulting in extracellular expression levels of 0.3 fg/cell or 0.75 ng/mL for IL-1β, 15 fg/cell or 38 ng/mL for IL-6 and 15 fg/cell or 38 ng/mL for TNF-α (Figure 4B). The cytokine levels were comparable to those produced by J774A.1 in control experiments using commercial preparations of LPS (0.1 to 1.0 ng/mL) from Escherichia coli, Salmonella typhimurium and Serratia marcescens.
Presence of Virulence-related Genes
To determine if the strains differed in genes that have been reported to be overt toxins (espA (K1 capsular polysaccharide), ompA (outer membrane protein A)) [25] [47] [56], primers targeting those genes were made and used in PCR amplifications for amplicon size comparisons. In all cases, the appropriate sized amplicons were generated, suggesting that all the strains possessed similar gene segments.
QRDR of gyrA and parC genes
To determine if the strains differed in the QRDRs, PCR and nucleotide sequencing was carried out for the parC and gyrA genes. Sequences translated in silico were aligned with strain Ab AYE, known to have the amino acid substitution conferring resistance. Sequences from all strains lacked the leucine residues associated with fluoroquinoline resistance (Figures 5A for gyrA and Figure 5B for parC).
Figure 5. Alignment of Quinolone Resistance Determining Regions.
ClustalW alignment of coding regions of gyrA (A) and parC (B) genes derived from test Acinetobacter strains compared to Ab AYE strain with known fluoroquinolone resistance mutations. The alignment demonstrates that the test strains lack a Ser-Leu transition necessary for fluoroquinolone resistance.
Antibiotic Resistance
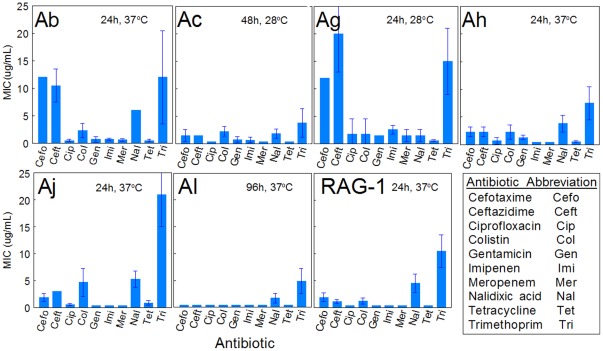
As a functional analysis of strain susceptibility towards a fluoroquinolone antibiotic and also several other antibiotic classes, bacterial MTT bioreduction activity towards a panel of antibiotics was tested. Figure 6 shows the resulting antibiograms, which clearly demonstrate that Ab was most resistant, albeit at low antibiotic concentration, to some test antibiotics. Ag and Aj also showed selective antibiotic resistance at low levels. Of these antibiotics, ciprofloxacin is a secondary fluoroquinolone. None of the strains showed high resistance to ciprofloxacin, which is consistent with the sequencing results in Figure 5 showing absence of mutations conveying fluoroquinolone resistance in genes, parC and gyrA.
Figure 6. Antibiotic Susceptibility Assays.
Antibiograms generated from growing each Acinetobacter strain in trypticase soy broth in the presence of different concentrations and classes of antibiotics for durations and temperatures indicated in the figure. The minimum inhibitory concentration (MIC) is the lowest concentration that is effective in preventing bacterial MTT bioreducton. Data are means of four experiments with vertical bars indicating standard deviation.
Discussion
This paper summarizes several in vitro bacterial and mammalian cell-based assays that permit differentiation between potentially hazardous or virulent Acinetobacter strains from relatively safe strains. The assays that were useful in discriminating the virulence of these bacterial strains are summarized in Table 4. As we have cautioned for other genera [62], broad species-level conclusions should not be generalized from the test strains examined here. The analyses conducted would need to be repeated for each strain under evaluation.
Table 4. Summary of Assays for Comparing Potential Virulence Characteristics of Acinetobacter.
| Pathogenic Characteristic* | Ab | Ac | Ag | Ah | Aj | Al | Av-RAG-1 |
| Bacterial Growth with Mammalian Cells | + | + | (+) | + | + | + | − |
| Haemolytic Activity | (+) | (+) | (+) | + | (+) | − | + |
| Mammalian Cell Detachment/Lysis | + | (+) | (+) | + | (+) | (+) | (+) |
| Bacterial Survival with J774A.1 | − | (+) | − | + | (+) | − | (+) |
| HT29 Neutrophil Chemoattractant (IL-8) | (+) | − | − | + | + | − | + |
| Growth with Antibiotics** | + | − | + | (+) | + | (+) | (+) |
+ signifies substantial growth or activity.
(+) signifies low level and/or delayed activity.
− signifies negligible growth or activity
Note that tests for induction of J774A.1 pro-inflammatory cytokines (IL-1β, Il-6, TNF-α) and presence of known virulence gene segments (OmpA, EpsA) were excluded from the summary table since they failed to discriminate between bacterial strains.
Growth for this purpose is defined as MTT bioreduction in the presence of at least 5 µg/mL antibiotic.
In initial experiments, we screened strains for their capacity to multiply or grow in mammalian cell culture medium at 37°C as a simple determinant of potential virulence. In previous studies of the B.cereus-group bacteria, XTT bioreduction was used to measure growth of each strain in culture media alone or in the presence of mammalian cells [53]. In preliminary studies we found that the rates of XTT reduction by the different Acinetobacter strains were similar in LB medium, but lower than the previously observed rates of Bacillus spp by ∼4–5 times. However, similar to the Bacilli, most of the Acinetobacter strains were unable to grow or grew poorly in mammalian cell culture medium at the lowest inoculation concentrations, unless supplemented with either mammalian cells or mammalian cell-conditioned medium (data not shown). The exception was the Ah strain, which grew well, with or without HT29 cells. In contrast, Av-RAG-1 did not grow well in mammalian culture medium even in the presence of HT29 or conditioned medium.
Another indicator of virulence is the production of toxic or lytic by-products by various strains. In experiments using the sheep blood agar plate assay, both Ah and Av-RAG-1 exhibited strong haemolytic activity (referred to as β-haemolysis) whereas all other strains exhibited weak (α- haemolysis) or no (γ-haemolysis) capacity. However, Av-RAG-1 was not able to grow in mammalian medium in absence or presence of HT29 like other strains. Our data match that of several other reports in that only Ah and Av-RAG-1 were haemolytic [63]–[66]. However, none of these studies described the haemolysis in a semi-quantitative manner, as we have done here. In another study, 526 Acinetobacter strains were tested for haemolytic activity with human, sheep and bovine erythrocytes, and only 16% of them exhibited β-haemolysis while the majority were α-haemolytic [51]. This study included 24 Ah strains of which 17 (70%) were β-haemolytic. Some of the strains used by Gospodarek et al [51] may have been misclassified or variable in expression of haemolytic acivity since an absence of haemolytic activity was seen in 20 nosocomial Ab isolates tested with rabbit erythrocytes [41]. Furthermore, Antunes and colleagues [50] have recently demonstrated that there is significant inter-strain variation in the haemolytic capacity of four different Ab isolates, which is also dependent on the source (horse or sheep) of blood used in the assays. The genetic factor(s) contributing to this haemolytic activity remains to be identified.
As a further screen for toxic and lytic products, we used two mammalian cell models. In exposures using the colonic epithelial HT29 bioreduction assay [37], toxicity ensued rapidly within two hours of exposure most notably for Ab and Ah compared to the other strains. Further studies showed that loss of mammalian cell bioreductive capacity may be underestimated due to bacterial adherence, which would have contributed to total MTT formazan measured per assay well. With crude fractionation, it was shown that the Ab-mediated detachment of HT29 monolayers was cell wall -associated, likely proteinaceous and could be the same component causing the loss of HT29 bioreduction capacity. In contrast, the toxic agent(s) of Ah was not necessarily associated with the bacterial cell, since bacteria-free culture filtrates also caused hemolysis and cytolysis. Furthermore, while Av-RAG-1 did not grow in presence or absence of HT29 it did produce an Ah-like, filterable, extracellular-cytotoxic activity (as well as haemolytic activity) when grown in LB broth. This activity was heat sensitive, whereas the Ab toxicant was not. [63]–[66]
Several studies have identified cytotoxic factors from various Ab strains. In particular, Ab produces outer membrane vesicles that contain a number of virulence-related factors [67]. One of these factors, Ab outer membrane protein A (AbOmpA), is suggested to play an important role in pathogenesis as evidenced by its pleiotropic effects. Purified AbOmpA was localized to the mitochondria of HEp-2 human laryngeal epithelial cells, and resulted in the release of pro-apoptotic molecules such as cytochrome c and apoptosis-inducing factor into the cytosol, caspase-3 activation, and DNA fragmentation [56]. None of our strains demonstrated a difference in the presence of genes for ompA or another reported virulence factor gene, epsA. Our studies used the same Ab strain as in previous work of Choi and colleagues [40], and yet we did not observe Ab invasion of HT29 colonic epithelial cells, supporting earlier observations that other cell types were not as permissive for invasion as respiratory epithelial cells. Comparison of wild-type and AbOmpA-negative isogenic mutants of Ab by Choi et al [40] demonstrated that AbOmpA was also involved in epithelial cell attachment, which we have observed during our studies.
Since there are concerns about antibiotic resistance of some Acinetobacter strains [68]–[70], we also tested inhibitory effects (MIC assay) of several antibiotics, and in particular, if the test strains carry known mutations in the constitutive genes gyrA and parC conferring fluoroquinolone resistance [57]–[58]. The genes for gyrA and parC were sequenced, and then translated and aligned in silico. None of the tested strains had the serine to leucine transition (gyrA codon 83 and/or parC codon 80 or 84) implicated in the fluoroquinolone resistance phenotype (Figures 5A and 5B). Functional susceptibility to fluoroquinolone was confirmed for all strains with an MIC assay. Furthermore, this assay demonstrated that the Ab strain exhibited the most resistance to the antibiotics in general, and the least effective antibiotics towards all strains were cefotaxime, ceftazidime, and trimethoprim. The MIC assay also revealed that none of the strains were resistant to the fluoroquinolone, ciprofloxacin.
As a measure of immune response, we screened for bacterial-induced release of select cytokines known to be associated with our mammalian cell models [39]. For HT29, all of the Acinetobacter strains induced IL-8 levels that were consistent with those recorded for other non-invasive bacteria [71]. This neutrophil chemoattractant has been used previously to differentiate between pathogens and generally harmless bacteria [71]–[74]. The present study showed that Ab induced less IL-8 production in HT29 cells, compared to Aj, Ah, and RAG-1. Our results agree with a recent study by de Breij and colleagues [1] who reported that human epithelial cells produced less IL-8 in response to A. baumannii strains than to A. junii strains. They also suggested that A. baumannii appeared to be more virulent compared to the other strains, since infection caused by A. baumannii was associated with reduced capacity of bacterial elimination from the host. Furthermore, we agree with their comment that outgrowth may have caused increased elevation of the cytokines, as we see substantial growth in the presence of HT29 cells over a 24-h period (Table 3). However, our cytokine experiments were done in the presence of antibiotic to avoid the effect of differential growth on cytokine production, but also to limit any cytotoxicity associated with proliferating bacteria.
In contrast to the high levels of HT29 IL-8 release, J774A.1 released several pro-inflammatory cytokines, but none which were useful for differentiating between the strains tested. The similarity in the levels of J774A.1 cytokines, regardless of test bacterial strain, is consistent with known mechanisms of macrophage receptor-mediated recognition of bacterial pathogens. That is, cytokine induction is largely a result of pathogen-associated molecular patterns (PAMPs) made up of well-conserved microbial surface moieties, such as LPS or peptidoglycan (see reviews: [75]–[78].
Huttunen and colleagues [79] have used IL-1β, IL-6, and TNF-α expression from RAW264.7 macrophage as toxicological indicators of hazard from indoor microbial sources. In their study, doses up to 107 cfu/mL (equivalent to our 106 cfu/100 µL well), caused different levels of acute phase response (APR) cytokines depending on the bacterial strain tested. Bacillus cereus was least inducing (eg. 3.1 ng/mL TNFα), Streptomyces californicus caused an intermediate level (6.1 ng/mL TNF-α) and Pseudomonas fluorescens was marginally better (6.9 ng/mL TNF-α). In preliminary comparative tests (data not shown), we found that when the macrophage-like cell line RAW264.7 was exposed to Acinetobacter strains, it expressed APR cytokines at similar levels as J774A.1. However, those levels were at least 5-fold higher than those observed by Huttunen and colleagues with the RAW264.7 cells. The difference is most probably due to macrophage densities during exposure, underlining the importance of expressing cytokine expression data on a per-cell basis. In addition, bacterial species or strain differences likely play a role in the differential APR cytokine expression levels observed here. The lack of a difference we observed with our test system suggests that APR cytokine expression is not discriminatory at the species level for Acinetobacter, but may be a more relevant indicator when comparing between genera.
Our data with J774A.1 cells clearly demonstrate that Ah is the most resistant to macrophage-induced inactivation. Unlike other claims that addition of 0.004–0.1% Triton X-100 facilitates release of internalized bacteria and improves colony recovery [80], we found that the detergent was ineffective between 0.004–0.05% in lysing J774A.1 cells, and inhibited bacterial growth above 0.05%.
In conclusion, this comparative study shows the utility of in vitro assays and molecular probes for assessing for growth, toxicity, infectivity and potential immune responses to aid in screening virulence potential of select Acinetobacter species or strains. Collectively, these analyses have demonstrated that the Ab and Ah strains used here are likely to be the most hazardous if used in a biotech process involving large scale release. In contrast, the environmental strain Ag was relatively benign for growth in mammalian environments, toxicity and intracellular infectivity. Further, the Av-RAG-1 bioremediation strain was relatively non-infectious, but it had strong haemolytic capacity and induced production of inflammatory chemokines. These observations suggest that Av-RAG-1 should be investigated in more detail for potential clinical effects prior to environmental use as an industrial-scale bioremediation agent. Furthermore, our results suggest that ompA and epsA are not the primary virulence factors for these test stains. The next phase of our research will focus on confirming these in vitro observations using analogous in vivo murine exposure scenarios as recently reported for screening Bacillus organisms [39].
Acknowledgments
The authors would like to thank Drs. Sabina Halappanavar and Dalibor Breznan for reviewing the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported by the Canadian Regulatory System for Biotechnology (CRSB). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.de Breij A, Dijkshoorn L, Lagendijk E, van der Meer J, Koster A, et al. Do Biofilm Formation and Interactions with Human Cells Explain the Clinical Success of Acinetobacter baumannii? PLoS ONE. 2010;5:e10732. doi: 10.1371/journal.pone.0010732. doi: 10.1371/journal.pone.0010732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Euzéby JP. List of Bacterial Names with Standing in Nomenclature: a folder available on the Internet. Int J Syst Bacteriol. 1997;47:590–592. doi: 10.1099/00207713-47-2-590. [DOI] [PubMed] [Google Scholar]
- 3.Dijkshoorn L, Nemec A. The Diversity of the Genus Acinetobacter. In: Gerischer U, editor. Acinetobacter molecular microbiology. Horizon Scientific Press; 2008. [Google Scholar]
- 4.Luckarift HR, Sizemore SR, Farrington KE, Fulmer PA, Biffinger JC, et al. 2011. Biodegradation of medium chain hydrocarbons by Acinetobacter venetianus 2AW immobilized to hair-based adsorbent mats. Biotechnology Progress. Available: http://www.ncbi.nlm.nih.gov/pubmed/21948333. Accessed 28 October 2011. [DOI] [PubMed]
- 5.Abdel-El-Haleem D. Acinetobacter: environmental and biotechnological applications. African Journal of Biotechnology. 2003;2:71–74. [Google Scholar]
- 6.Singh GB, Gupta S, Srivastava S, Gupta N. Biodegradation of Carbazole by Newly Isolated Acinetobacter spp. Bull Environ Contam Toxicol. 2011;87:522–526. doi: 10.1007/s00128-011-0382-0. doi: 10.1007/s00128-011-0382-0. [DOI] [PubMed] [Google Scholar]
- 7.Jung J, Noh J, Park W. Physiological and metabolic responses for hexadecane degradation in Acinetobacter oleivorans DR1. J Microbiol. 2011;49:208–215. doi: 10.1007/s12275-011-0395-8. doi: 10.1007/s12275-011-0395-8. [DOI] [PubMed] [Google Scholar]
- 8.Zhao Z, Selvam A, Wong JW-C. Synergistic effect of thermophilic temperature and biosurfactant produced by Acinetobacter calcoaceticus BU03 on the biodegradation of phenanthrene in bioslurry system. J Hazard Mater. 2011;190:345–350. doi: 10.1016/j.jhazmat.2011.03.042. doi: 10.1016/j.jhazmat.2011.03.042. [DOI] [PubMed] [Google Scholar]
- 9.Dams-Kozlowska H, Mercaldi MP, Panilaitis BJ, Kaplan DL. Modifications and applications of the Acinetobacter venetianus RAG-1 exopolysaccharide, the emulsan complex and its components. Appl Microbiol Biotechnol. 2008;81:201–210. doi: 10.1007/s00253-008-1664-2. doi: 10.1007/s00253-008-1664-2. [DOI] [PubMed] [Google Scholar]
- 10.Snellman EA, Sullivan ER, Colwell RR. Purification and properties of the extracellular lipase, LipA, of Acinetobacter sp. RAG-1. Eur J Biochem. 2002;269:5771–5779. doi: 10.1046/j.1432-1033.2002.03235.x. [DOI] [PubMed] [Google Scholar]
- 11.Su W-T, Chen W-J, Lin Y-F. Optimizing emulsan production of A. venetianus RAG-1 using response surface methodology. Appl Microbiol Biotechnol. 2009;84:271–279. doi: 10.1007/s00253-009-1957-0. doi: 10.1007/s00253-009-1957-0. [DOI] [PubMed] [Google Scholar]
- 12.Vaneechoutte M, Tjernberg I, Baldi F, Pepi M, Fani R, et al. Oil-degrading Acinetobacter strain RAG-1 and strains described as “Acinetobacter venetianus sp. nov.” belong to the same genomic species. Res Microbiol. 1999;150:69–73. doi: 10.1016/s0923-2508(99)80047-3. [DOI] [PubMed] [Google Scholar]
- 13.Bifulco JM, Shirey JJ, Bissonnette GK. Detection of Acinetobacter spp. in rural drinking water supplies. Appl Environ Microbiol. 1989;55:2214–2219. doi: 10.1128/aem.55.9.2214-2219.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simões LC, Simões M, Vieira MJ. Intergeneric coaggregation among drinking water bacteria: evidence of a role for Acinetobacter calcoaceticus as a bridging bacterium. Appl Environ Microbiol. 2008;74:1259–1263. doi: 10.1128/AEM.01747-07. doi: 10.1128/AEM.01747-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giamarellou H, Antoniadou A, Kanellakopoulou K. Acinetobacter baumannii: a universal threat to public health? Int J Antimicrob Agents. 2008;32:106–119. doi: 10.1016/j.ijantimicag.2008.02.013. doi: 10.1016/j.ijantimicag.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 16.Gootz TD, Marra A. Acinetobacter baumannii: an emerging multidrug-resistant threat. Expert Rev Anti Infect Ther. 2008;6:309–325. doi: 10.1586/14787210.6.3.309. doi: 10.1586/14787210.6.3.309. [DOI] [PubMed] [Google Scholar]
- 17.Keen EF, Robinson BJ, Hospenthal DR, Aldous WK, Wolf SE, et al. Prevalence of multidrug-resistant organisms recovered at a military burn center. Burns. 2010;36:819–825. doi: 10.1016/j.burns.2009.10.013. doi: 10.1016/j.burns.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 18.Luna CM, Aruj PK. Nosocomial Acinetobacter pneumonia. Respirology. 2007;12:787–791. doi: 10.1111/j.1440-1843.2007.01147.x. doi: 10.1111/j.1440-1843.2007.01147.x. [DOI] [PubMed] [Google Scholar]
- 19.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008;21:538–582. doi: 10.1128/CMR.00058-07. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Apisarnthanarak A, Kiratisin P, Thongphubeth K, Yuakyen C, Mundy LM. Pseudo-outbreak of Acinetobacter lwoffii infection in a tertiary care center in Thailand. Infect Control Hosp Epidemiol. 2007;28:637–639. doi: 10.1086/513620. doi: 10.1086/513620. [DOI] [PubMed] [Google Scholar]
- 22.Cayô R, Yañez San Segundo L, Pérez del Molino Bernal IC, García de la Fuente C, Bermúdez Rodríguez MA, et al. Bloodstream infection caused by Acinetobacter junii in a patient with acute lymphoblastic leukaemia after allogenic haematopoietic cell transplantation. J Med Microbiol. 2011;60:375–377. doi: 10.1099/jmm.0.024596-0. doi: 10.1099/jmm.0.024596-0. [DOI] [PubMed] [Google Scholar]
- 23.Hung Y-T, Lee Y-T, Huang L-J, Chen T-L, Yu K-W, et al. Clinical characteristics of patients with Acinetobacter junii infection. J Microbiol Immunol Infect. 2009;42:47–53. [PubMed] [Google Scholar]
- 24.Smith AW, Alpar KE. Immune response to Acinetobacter calcoaceticus infection in man. J Med Microbiol. 1991;34:83–88. doi: 10.1099/00222615-34-2-83. [DOI] [PubMed] [Google Scholar]
- 25.Chen T-L, Siu L-K, Lee Y-T, Chen C-P, Huang L-Y, et al. Acinetobacter Baylyi as a Pathogen for Opportunistic Infection. J Clin Microbiol. 2008;46:2938–2944. doi: 10.1128/JCM.00232-08. doi: 10.1128/JCM.00232-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pellegrino FLPC, Vieira VV, Baio PVP, dos Santos RMR, dos Santos ALA, et al. Acinetobacter soli as a cause of bloodstream infection in a neonatal intensive care unit. J Clin Microbiol. 2011;49:2283–2285. doi: 10.1128/JCM.00326-11. doi: 10.1128/JCM.00326-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gordon NC, Wareham DW. Multidrug-resistant Acinetobacter baumannii: mechanisms of virulence and resistance. Int J Antimicrob Agents. 2010;35:219–226. doi: 10.1016/j.ijantimicag.2009.10.024. doi: 10.1016/j.ijantimicag.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 28.Livermore DM. The threat from the pink corner. Ann Med. 2003;35:226–234. doi: 10.1080/07853890310001609. [DOI] [PubMed] [Google Scholar]
- 29.Joly-Guillou M-L. Clinical impact and pathogenicity of Acinetobacter. Clinical microbiology and infection the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2005;11:868–873. doi: 10.1111/j.1469-0691.2005.01227.x. [DOI] [PubMed] [Google Scholar]
- 30.Ong CWM, Lye DCB, Khoo KL, Chua GSW, Yeoh SF, et al. Severe community-acquired Acinetobacter baumannii pneumonia: an emerging highly lethal infectious disease in the Asia-Pacific. Respirology. 2009;14:1200–1205. doi: 10.1111/j.1440-1843.2009.01630.x. doi: 10.1111/j.1440-1843.2009.01630.x. [DOI] [PubMed] [Google Scholar]
- 31.Regalado NG, Martin G, Antony SJ. Acinetobacter lwoffii: bacteremia associated with acute gastroenteritis. Travel Med Infect Dis. 2009;7:316–317. doi: 10.1016/j.tmaid.2009.06.001. doi: 10.1016/j.tmaid.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 32.Falagas ME, Karveli EA, Kelesidis I, Kelesidis T. Community-acquired Acinetobacter infections. Eur J Clin Microbiol Infect Dis. 2007;26:857–868. doi: 10.1007/s10096-007-0365-6. doi: 10.1007/s10096-007-0365-6. [DOI] [PubMed] [Google Scholar]
- 33.Hu J, Robinson JL. Systematic review of invasive Acinetobacter infections in children. Can J Infect Dis Med Microbiol. 2010;21:83–88. doi: 10.1155/2010/690715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sengstock DM, Thyagarajan R, Apalara J, Mira A, Chopra T, et al. Multidrug-resistant Acinetobacter baumannii: an emerging pathogen among older adults in community hospitals and nursing homes. Clin Infect Dis. 2010;50:1611–1616. doi: 10.1086/652759. doi: 10.1086/652759. [DOI] [PubMed] [Google Scholar]
- 35.Moreira Silva G, Morais L, Marques L, Senra V. Acinetobacter community-acquired pneumonia in a healthy child. Revista Portuguesa De Pneumologia. 2012;18:96–98. doi: 10.1016/j.rppneu.2011.07.006. doi: 10.1016/j.rppneu.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 36.Ozaki T, Nishimura N, Arakawa Y, Suzuki M, Narita A, et al. Community-acquired Acinetobacter baumannii meningitis in a previously healthy 14-month-old boy. J Infect Chemother. 2009;15:322–324. doi: 10.1007/s10156-009-0704-x. doi: 10.1007/s10156-009-0704-x. [DOI] [PubMed] [Google Scholar]
- 37.Tayabali AF, Seligy VL. Human cell exposure assays of Bacillus thuringiensis commercial insecticides: production of Bacillus cereus-like cytolytic effects from outgrowth of spores. Environ Health Perspect. 2000;108:919–930. doi: 10.1289/ehp.00108919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tayabali AF, Seligy VL. Cell integrity markers for in vitro evaluation of cytotoxic responses to bacteria-containing commercial insecticides. Ecotoxicol Environ Saf. 1997;37:152–162. doi: 10.1006/eesa.1997.1525. doi: 10.1006/eesa.1997.1525. [DOI] [PubMed] [Google Scholar]
- 39.Tayabali AF, Nguyen KC, Seligy VL. Early murine immune responses from endotracheal exposures to biotechnology-related Bacillus strains. Toxicological & Environmental Chemistry. 2011;93:314–331. doi: 10.1080/02772248.2010.526784. doi: 10.1080/02772248.2010.526784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi CH, Lee JS, Lee YC, Park TI, Lee JC. Acinetobacter baumannii invades epithelial cells and outer membrane protein A mediates interactions with epithelial cells. BMC Microbiology. 2008;8:216. doi: 10.1186/1471-2180-8-216. doi: 10.1186/1471-2180-8-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sechi LA, Karadenizli A, Deriu A, Zanetti S, Kolayli F, et al. PER-1 type beta-lactamase production in Acinetobacter baumannii is related to cell adhesion. Med Sci Monit. 2004;10:BR180–184. [PubMed] [Google Scholar]
- 42.Gohl O, Friedrich A, Hoppert M, Averhoff B. The thin pili of Acinetobacter sp. strain BD413 mediate adhesion to biotic and abiotic surfaces. Appl Environ Microbiol. 2006;72:1394–1401. doi: 10.1128/AEM.72.2.1394-1401.2006. doi: 10.1128/AEM.72.2.1394-1401.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee JC, Koerten H, van den Broek P, Beekhuizen H, Wolterbeek R, et al. Adherence of Acinetobacter baumannii strains to human bronchial epithelial cells. Res Microbiol. 2006;157:360–366. doi: 10.1016/j.resmic.2005.09.011. doi: 10.1016/j.resmic.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 44.Choi CH, Hyun SH, Kim J, Lee YC, Seol SY, et al. Nuclear translocation and DNAse I-like enzymatic activity of Acinetobacter baumannii outer membrane protein A. FEMS Microbiol Lett. 2008;288:62–67. doi: 10.1111/j.1574-6968.2008.01323.x. doi: 10.1111/j.1574-6968.2008.01323.x. [DOI] [PubMed] [Google Scholar]
- 45.Choi CH, Hyun SH, Lee JY, Lee JS, Lee YS, et al. Acinetobacter baumannii outer membrane protein A targets the nucleus and induces cytotoxicity. Cellular Microbiology. 2008;10:309–319. doi: 10.1111/j.1462-5822.2007.01041.x. doi: 10.1111/j.1462-5822.2007.01041.x. [DOI] [PubMed] [Google Scholar]
- 46.Lee JC, Oh JY, Kim KS, Jeong YW, Park JC, et al. Apoptotic cell death induced by Acinetobacter baumannii in epithelial cells through caspase-3 activation. APMIS. 2001;109:679–684. doi: 10.1034/j.1600-0463.2001.d01-132.x. [DOI] [PubMed] [Google Scholar]
- 47.Russo TA, Luke NR, Beanan JM, Olson R, Sauberan SL, et al. The K1 capsular polysaccharide of Acinetobacter baumannii strain 307-0294 is a major virulence factor. Infect Immun. 2010;78:3993–4000. doi: 10.1128/IAI.00366-10. doi: 10.1128/IAI.00366-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim SA, Yoo SM, Hyun SH, Choi CH, Yang SY, et al. Global gene expression patterns and induction of innate immune response in human laryngeal epithelial cells in response to Acinetobacter baumannii outer membrane protein A. FEMS Immunol Med Microbiol. 2008;54:45–52. doi: 10.1111/j.1574-695X.2008.00446.x. doi: 10.1111/j.1574-695X.2008.00446.x. [DOI] [PubMed] [Google Scholar]
- 49.Taylor D, Whatling C, Kearney JN, Matthews B, Holland KT. The effect of bacterial products on human fibroblast and keratinocyte detachment and viability. Br J Dermatol. 1990;122:23–28. doi: 10.1111/j.1365-2133.1990.tb08235.x. [DOI] [PubMed] [Google Scholar]
- 50.Antunes LCS, Imperi F, Carattoli A, Visca P. Deciphering the Multifactorial Nature of Acinetobacter baumannii Pathogenicity. PLoS One. n.d.;6. doi: 10.1371/journal.pone.0022674. doi: 10.1371/journal.pone.0022674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gospodarek E. [Hemolytic properties of bacteria belonging to the Acinetobacter genus]. Med Dosw Mikrobiol. 1993;45:317–322. [PubMed] [Google Scholar]
- 52.García A, Salgado F, Solar H, González CL, Zemelman R, et al. Some immunological properties of lipopolysaccharide from Acinetobacter baumannii. J Med Microbiol. 1999;48:479–483. doi: 10.1099/00222615-48-5-479. [DOI] [PubMed] [Google Scholar]
- 53.Seligy VL, Beggs RW, Rancourt JM, Tayabali AF. Quantitative bioreduction assays for calibrating spore content and viability of commercial Bacillus thuringiensis insecticides. Journal of Industrial Microbiology and Biotechnology. 1997;18:370–378. doi: 10.1038/sj.jim.2900404. [Google Scholar]
- 54.Seligy, Rancourt Antibiotic MIC/MBC analysis of Bacillus-based commercial insecticides: use of bioreduction and DNA-based assays. J Ind Microbiol Biotechnol. 1999;22:565–574. doi: 10.1038/sj.jim.2900625. [DOI] [PubMed] [Google Scholar]
- 55.Reen, DJ . Enzyme-linked Immunosorbant Assay (ELISA). In: Walker JM, editor. Basic protein and peptide protocols. Humana Press, Vol; 1994. 32 [Google Scholar]
- 56.Choi CH, Lee EY, Lee YC, Park TI, Kim HJ, et al. Outer membrane protein 38 of Acinetobacter baumannii localizes to the mitochondria and induces apoptosis of epithelial cells. Cell Microbiol. 2005;7:1127–1138. doi: 10.1111/j.1462-5822.2005.00538.x. doi: 10.1111/j.1462-5822.2005.00538.x. [DOI] [PubMed] [Google Scholar]
- 57.Vila J, Ruiz J, Goñi P, Jimenez de Anta T. Quinolone-resistance mutations in the topoisomerase IV parC gene of Acinetobacter baumannii. J Antimicrob Chemother. 1997;39:757–762. doi: 10.1093/jac/39.6.757. [DOI] [PubMed] [Google Scholar]
- 58.Vila J, Ruiz J, Goñi P, Marcos A, Jimenez de Anta T. Mutation in the gyrA gene of quinolone-resistant clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother. 1995;39:1201–1203. doi: 10.1128/aac.39.5.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fournier P-E, Vallenet D, Barbe V, Audic S, Ogata H, et al. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet. 2006;2:e7. doi: 10.1371/journal.pgen.0020007. doi: 10.1371/journal.pgen.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vallenet D, Nordmann P, Barbe V, Poirel L, Mangenot S, et al. Comparative analysis of Acinetobacters: three genomes for three lifestyles. PLoS ONE. 2008;3:e1805. doi: 10.1371/journal.pone.0001805. doi: 10.1371/journal.pone.0001805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baldi F, Pepi M, Capone A, della Giovampaola C, Milanesi C, et al. Envelope glycosylation determined by lectins in microscopy sections of Acinetobacter venetianus induced by diesel fuel. Research in Microbiology. 154:417–424. doi: 10.1016/S0923-2508(03)00128-1. doi: 10.1016/S0923-2508(03)00128-1. [DOI] [PubMed] [Google Scholar]
- 62.Tayabali AF, Nguyen KC, Seligy VL. Early murine immune responses from endotracheal exposures to biotechnology-related Bacillus strains. Toxicological & Environmental Chemistry. 2011;93:314–331. doi: 10.1080/02772248.2010.526784. doi: 10.1080/02772248.2010.526784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bouvet PJM, Grimont PAD. Taxonomy of the Genus Acinetobacter with the Recognition of Acinetobacter Baumannii Sp. Nov., Acinetobacter Haemolyticus Sp. Nov., Acinetobacter Johnsonii Sp. Nov., and Acinetobacter Junii Sp. Nov. and Emended Descriptions of Acinetobacter Calcoaceticus and Acinetobacter Lwoffii. Int J Syst Bacteriol. 1986;36:228–240. doi: 10.1099/00207713-36-2-228. [Google Scholar]
- 64.Lehmann V. HAEMOLYTIC ACTIVITY OF VARIOUS STRAINS OF ACINETOBACTER. Acta Pathologica Microbiologica Scandinavica Section B Microbiology and Immunology. 1973;81B:427–432. doi: 10.1111/j.1699-0463.1973.tb02226.x. doi: 10.1111/j.1699-0463.1973.tb02226.x. [DOI] [PubMed] [Google Scholar]
- 65.Lehmann V. HAEMOLYTIC ACTIVITY OF ACINETOBACTER CALCOACETICUS. Acta Pathologica Microbiologica Scandinavica Section B Microbiology and Immunology. 1971;79B:61–66. doi: 10.1111/j.1699-0463.1971.tb00033.x. doi: 10.1111/j.1699-0463.1971.tb00033.x. [DOI] [PubMed] [Google Scholar]
- 66.Vaneechoutte M, Nemec A, Musílek M, van der Reijden TJK, van den Barselaar M, et al. Description of Acinetobacter venetianus ex Di Cello et al. 1997 sp. nov. Int J Syst Evol Microbiol. 2009;59:1376–1381. doi: 10.1099/ijs.0.003541-0. doi: 10.1099/ijs.0.003541-0. [DOI] [PubMed] [Google Scholar]
- 67.Kwon S-O, Gho YS, Lee JC, Kim SI. Proteome analysis of outer membrane vesicles from a clinical Acinetobacter baumannii isolate. FEMS Microbiol Lett. 2009;297:150–156. doi: 10.1111/j.1574-6968.2009.01669.x. doi: 10.1111/j.1574-6968.2009.01669.x. [DOI] [PubMed] [Google Scholar]
- 68.Coyne S, Courvalin P, Périchon B. Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob Agents Chemother. 2011;55:947–953. doi: 10.1128/AAC.01388-10. doi: 10.1128/AAC.01388-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Neonakis IK, Spandidos DA, Petinaki E. Confronting multidrug-resistant Acinetobacter baumannii: a review. Int J Antimicrob Agents. 2011;37:102–109. doi: 10.1016/j.ijantimicag.2010.10.014. doi: 10.1016/j.ijantimicag.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 70.Garnacho-Montero J, Amaya-Villar R. Multiresistant Acinetobacter baumannii infections: epidemiology and management. Curr Opin Infect Dis. 2010;23:332–339. doi: 10.1097/QCO.0b013e32833ae38b. doi: 10.1097/QCO.0b013e32833ae38b. [DOI] [PubMed] [Google Scholar]
- 71.Jung HC, Eckmann L, Yang SK, Panja A, Fierer J, et al. A distinct array of proinflammatory cytokines is expressed in human colon epithelial cells in response to bacterial invasion. J Clin Invest. 1995;95:55–65. doi: 10.1172/JCI117676. doi: 10.1172/JCI117676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eckmann L, Kagnoff MF, Fierer J. Epithelial cells secrete the chemokine interleukin-8 in response to bacterial entry. Infect Immun. 1993;61:4569–4574. doi: 10.1128/iai.61.11.4569-4574.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Palfreyman RW, Watson ML, Eden C, Smith AW. Induction of biologically active interleukin-8 from lung epithelial cells by Burkholderia (Pseudomonas) cepacia products. Infect Immun. 1997;65:617–622. doi: 10.1128/iai.65.2.617-622.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schuerer-Maly CC, Eckmann L, Kagnoff MF, Falco MT, Maly FE. Colonic epithelial cell lines as a source of interleukin-8: stimulation by inflammatory cytokines and bacterial lipopolysaccharide. Immunology. 1994;81:85–91. [PMC free article] [PubMed] [Google Scholar]
- 75.Bianchi ME. DAMPs, PAMPs and alarmins: all we need to know about danger. J Leukoc Biol. 2007;81:1–5. doi: 10.1189/jlb.0306164. doi: 10.1189/jlb.0306164. [DOI] [PubMed] [Google Scholar]
- 76.Dalpke A, Heeg K. Signal integration following Toll-like receptor triggering. Crit Rev Immunol. 2002;22:217–250. [PubMed] [Google Scholar]
- 77.Vogel SN, Fitzgerald KA, Fenton MJ. TLRs: differential adapter utilization by toll-like receptors mediates TLR-specific patterns of gene expression. Mol Interv. 2003;3:466–477. doi: 10.1124/mi.3.8.466. doi: 10.1124/mi.3.8.466. [DOI] [PubMed] [Google Scholar]
- 78.Werling D, Jungi TW. TOLL-like receptors linking innate and adaptive immune response. Vet Immunol Immunopathol. 2003;91:1–12. doi: 10.1016/s0165-2427(02)00228-3. [DOI] [PubMed] [Google Scholar]
- 79.Huttunen K, Hyvärinen A, Nevalainen A, Komulainen H, Hirvonen M-R. Production of proinflammatory mediators by indoor air bacteria and fungal spores in mouse and human cell lines. Environ Health Perspect. 2003;111:85–92. doi: 10.1289/ehp.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Utaisincharoen P, Kespichayawattana W, Anuntagool N, Chaisuriya P, Pichyangkul S, et al. CpG ODN enhances uptake of bacteria by mouse macrophages. Clin Exp Immunol. 2003;132:70–75. doi: 10.1046/j.1365-2249.2003.02107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]