Abstract
Epithelial-mesenchymal transition (EMT) and the underlying mechanisms and signaling pathways regulating such transitions have generated a lot of interest among cancer researchers. Much of this can be attributed to the apparent similarities in the molecular processes regulating embryonic EMT that can be recapitulated during tumor progression and metastasis. It appears that both embryonic and oncogenic EMT are regulated by an intricate interplay of transcriptional and post-transcriptional programs, and the recent discovery of a transcript-selective translational regulatory pathway controlling expression of EMT-associated mRNAs demonstrates the high fidelity and tight regulation associated with the process of EMT and metastatic progression. Heterogeneous nuclear ribonucleoprotein E1 (hnRNP E1) is emerging as a critical and integral modulator of TGFβ-induced EMT and subsequent tumor metastasis. Through its RNA-binding ability, hnRNP E1 binds distinct 3′-UTR structural elements present in mRNA transcripts required for EMT and translationally silences their expression. Translational silencing, mediated by hnRNP E1, occurs specifically at the translation elongation step through effects on the eukaryotic elongation factor-1 A1 (eEF1A1), and is relieved by Akt2-mediated phosphorylation. Interestingly, modulation of either the steady-state expression or the posttranscriptional modification of hnRNP E1 has a temporo-spatial effect on translational repression, tumorigenesis and cancer metastasis.
Key words: EMT, TGFβ, metastasis, hnRNP E1, eEF1A1, post-transcriptional regulation
Introduction
Epithelial-mesenchymal transition (EMT) is a process in which cells undergo a developmental switch from a polarized, epithelial phenotype to a highly motile mesenchymal phenotype. EMT is not only a fundamental process during normal embryonic development and in adult tissue homeostasis, but is also a critical step in metastatic progression of tumors.1–3 Changes in cell-cell adhesion, remodeling of extracellular matrix and enhanced migratory activity, are typical characteristics associated with EMT, which cumulatively enable tumor cells to metastasize.1–3 Numerous cytokines and autocrine growth factors, including TGFβ, have been implicated in EMT.4,5
A carcinoma cell activates EMT programs through different regulatory pathways, many of which remain to be identified. It is known, however, that differentiation to the mesenchymal phenotype arms the cancer cell with all the ammunition needed to initiate its journey through the various hurdles of metastatic progression, including dissociation from the primary tumor, invasion of surrounding tissue, intravasation into lymphatic or vascular vessels, extravasation and finally micrometastatic colonization at the secondary site.5
Several transcriptional programs have been identified which act in concert to induce EMT. Of note are the Snail,6 Slug,7 Goosecoid,8 FOXC2,9 ZEB1,6 ZEB2/Sip16 and Twist6 transcription factors. Twist1 was initially shown to be a limiting transcriptional upregulator of programs required for EMT and metastatic progression.10 Twsit2, along with Twist1, has a proven role in embryonic development.10 Additionally, both the Twist isoforms are normally overexpressed in different human cancers.11 Interestingly, Twist1 and Twist2 not only potentiate metastatic progression, but also override, together with oncogenic Ras, oncogene-induced premature senescence.12
Post-transcriptional regulatory events have also been implicated in key molecular pathways in cancer development and metastatic progression.13 Y-box binding protein-1 (YB-1), a DNA/RNA-binding protein, has been shown to translationally regulate, through a cap-independent mechanism, mRNA transcripts encoding EMT-promoting factors including the transcription factor Snail.14 We have previously demonstrated that the EMT-promoting mRNAs disabled-2 (Dab2) and interleukin-like EMT inducer (ILEI) are translationally regulated by TGFβ.15 It is clear that the regulatory processes controlling EMT are complex and involve many pleiotropic transcriptional and posttranscriptional regulators. Perhaps, the multiplicity of these factors and pathways is a means by which a cell reactivates context-dependent signaling to activate EMT.
Emerging Mechanism of Post-Transcriptional Regulation of TGFβ-Mediated EMT and Metastatic Progression
Protein expression levels depend on the rate of transcription, as well as other defined control mechanisms, such as mRNA stability,16 nuclear export and mRNA localization,17 translational regulation,18 and finally protein degradation.19 Post-transcriptional regulation is mainly controlled by the binding of RNA Binding Proteins (RBPs) to regulatory regions (cis elements) in the untranslated regions (UTRs) of mRNAs. The Human Genome Project reported the mean lengths of 5′-untranslated regions (UTRs) and 3′-UTRs of human mRNAs as 300 nt and 770 nt, respectively, compared with the mean coding length of 1,340 nt (International Human Genome Sequencing Consortium 2001).20 More importantly the 3′-UTRs shows high evolutionary conservation (in comparison to the less conserved 5′-UTRs), suggesting critical roles for the non-coding region at the 3′-end of mRNAs.
Previous work in our lab15 and others21,22 has shown that regulation of gene expression at the post-transcriptional level plays an indispensable role in TGFβ-induced EMT and metastasis. We initially identified a transcript-selective translational regulatory pathway in which a ribonucleoprotein (mRNP) complex binds to a 33-nucleotides long 3′-UTR regulatory BAT (TGFβ activated translation) element and silences translation of Dab2 and ILEI mRNAs, which are involved in mediating EMT.15 mRNPs are diverse macromolecular assemblies consisting of both protein and RNA components and modulate critical roles in the maturation of most RNAs and in the translation of mRNAs.23,24 hnRNP E1 was initially identified as a critical component of the BAT binding mRNP complex.15 We found that TGFβ activates a kinase cascade terminating in the phosphorylation of hnRNP E1, by isoform-specific stimulation of protein kinase Bβ/Akt2, inducing the release of the mRNP complex from the 3′-UTR element, in turn resulting in the reversal of translational silencing and increased expression of Dab2 and ILEI transcripts that mediates EMT (Fig. 1A).15
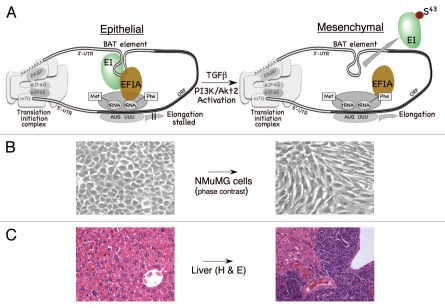
Figure 1.
TGFβ-activated translational (BAT) silencing mechanism mediating EMT and metastasis. (A) eEF 1A1 forms a complex with hnRNP E1 and the BAT element and silences translational elongation. Given the necessity for cognate-codon interaction with the ribosomal A site, it is likely that the formation of the BAT mRNP complex occurs post-delivery of the aminoacyl-tRNA to the ribosome. The ability of the BAT mRNP complex to inhibit eEF 1A1-dependent elongation suggests that the 3′-UT R is interacting with the 5′-UT R in a circularized model to facilitate its proximity to the 80S ribosome (left part). Following TGFβ-stimulation, activated Akt2 phosphorylates hnRNP E1 ar Ser43, which disrupts the mRNP complex and allows translation of Dab2 and ILEI mRNAs (right part). (B) Phase contrast image of control (left part) and NMuMG cells treated with TGFβ for 24 h (right part). TGFβ treatment causes distinct morphological changes associated with EMT, as discussed in the text. (C) Silencing of hnRNP E1 renders noninvasive NMuMG cells to form metastatic tumors in allograftic studies. H&E staining of liver tissue from mice injected with either NMuMG cells (left part) or SH14 cells (NMuMG cells harboring a stable shRNA against hnRNP E1) (right part).
Our subsequent experiments have helped us identify eukaryotic elongation factor-1 A1 (eEF1A1) as the other functional component of the mRNP complex.25 The BAT element, hnRNP E1 and eEF1A1 form a ternary complex that mediates translational silencing at the translational elongation step. In non-stimulated cells, which exhibit epithelial characteristics, hnRNP E1 binds to eEF1A1 and blocks progression of the 80S ribosome by preventing the release of eEF1A1 from the ribosomal A-site post-GTP hydrolysis.25 In TGFβ-stimulated cells, which exhibit distinct mesenchymal properties, activated Akt2 phosphorylates hnRNP E1 at serine-43 residue and disrupts the mRNP complex, allowing eEF1A1-mediated translational elongation to proceed (Fig. 1A).15,25
The interaction between eEF1A1 and hnRNP E1 inhibits eEF1A1 release from the ribosomal A-site following hydrolysis of GTP; hence, eEF1A1-GDP remains locked on the ribosome, thereby preventing the subsequent translocation of the aminoacyl-tRNA to the peptidyl moiety of the growing peptide chain (Fig. 1).25 The ribosome is therefore stalled at the eEF1A1-dependent elongation stage, resulting in translational silencing.25 This translational regulatory mechanism is based on discrete RNA-protein and protein-protein interactions. Interaction between eEF1A1 and hnRNP E1 is driven by their inherent and independent binding capacity to the ‘BAT’ RNA element.25 Cumulatively, this allows for transcript-specific translational silencing activity in mRNAs that harbor the 33-nucleotide BAT element in their 3′-UTR.25 Structural fidelity (identical 2° structure), rather than a conserved sequence homology (different nucleotide sequences of Dab2 and ILEI BAT elements), of the BAT element is responsible for mediating this post-transcriptional regulation,15 and mutations that alter the 2° structure of the BAT element impair the ability of both hnRNP E1 and eEF1A1 to bind and mediate translational silencing.15,25
Implications of hnRNP E1-Mediated Regulation of Metastatic Progression
Our findings are of significance in that they highlight a novel regulatory mechanism for cancer progression and one that can ultimately provide a framework for the design of strategies and therapeutics selectively targeting the metastatic stage of tumorigenesis. Indeed, we demonstrate that knockdown of hnRNP E1 expression, and thus loss of translational silencing of EMT transcripts, in noninvasive breast epithelial cells not only induced EMT (Fig. 1B) but also enabled cells to form metastatic lesions (Fig. 1C). In allograftic tumor studies, hnRNP E1-knockdown cells readily formed large, slowly growing tumors. In contrast, normal mammary epithelial cells formed only tiny, regressing nodules. Moreover, we observed a downregulation of E-cadherin expression in hnRNP E1-knockdown-induced tumors, as well as upregulation of the EMT marker vimentin and strong expression of ILEI. Further, we showed that shRNA-mediated silencing of the EMT mRNA transcripts, Dab2 and ILEI, is sufficient to inhibit TGFβ-mediated EMT as analyzed morphologically and by loss of the mesenchymal markers N-cadherin and vimentin; whereas, overexpression of both Dab2 and ILEI is insufficient to induce constitutive EMT, independent of TGFβ signaling.15,26 Thus, while Dab2 and ILEI are required for TGFβ-mediated EMT, they are not sufficient, suggesting that hnRNP E1 might be regulating a cohort of mRNAs, including Dab2 and ILEI, that together contribute to TGFβ-induced EMT. In addition, knockdown of hnRNP E1 in the human breast cancer line MCF7 caused these cells to acquire EMT markers and to form lung metastases when injected intravenously.25 MCF cells normally form tumors, but no metastatic nodules at secondary sites. Collectively, our cellular and animal studies support a direct role for hnRNP E1, and the BAT mRNP complex, in mediating EMT and metastasis.
hnRNP E1 has a predominant nuclear localization, specifically in nuclear speckles.27–29 hnRNP E1 has a nonameric nuclear localization signal (NLS I), putatively located between its KH2 and KH3 domain; of note, it lacks the NLS II present in hnRNP E2.28 But the posttranscriptional regulatory pathway we have elucidated is cytoplasmic. This is evident by the fact that the Dab2 and ILEI mRNAs are already loaded onto the ribosomes. hnRNP E1 has also been reportedly observed in the cytoplasm.30 It will be interesting to elucidate at what step during its synthesis in the nucleus or subsequent export into the cytoplasm, hnRNP E1 binds with the 3′-UTR of Dab2 and ILEI mRNAs. It is also imperative to determine the outcome of the RNAs whose translation has been stalled. The stalled mRNAs are not degraded, as we do not see any mRNA degradation over time.15 Hence, there is a possibility that the stalled transcripts are localized to distinct cytoplasmic foci and that transcript-selective translation occurs in these same cellular compartments. This model, however, would need to take into account that not only the mRNAs be sequestered into specific cellular locales but that the associated ribosomes and translational machinery also be similarly colocalized.
Role of hnRNP E1 as a Critical Mediator of Metastatic Progression
While EMT and metastatic progression has been shown to encompass a wide continuum of alterations in epithelial plasticity in response to different stimulus by cytokines, an important finding of our study is that a single factor, hnRNP E1, is responsible for silencing a TGFβ-mediated EMT program. In fact, our observations also suggest that hnRNP E1 might have a role in tumorigenesis per se. Previous reports suggest that this in fact may be the case, revealing that hnRNP E1 plays a critical and well-defined role in carcinogenesis. Recently, it has been shown that hnRNP E1 translationally suppresses PRL-3 phosphatase and prevents metastatic progression and that its overexpression causes Akt inactivation.22 hnRNP E1, along with hnRNP A1 and hnRNP D, has also been shown to form a heterotrimeric complex on telomeric repeats,31 indicative of its involvement in the maintenance of telomeric length that has been shown to play a role in oncogenic transformation.32 Downregulation of hnRNP E1 expression is also required for human papillomavirus (HPV) proliferation and subsequent incidence of cervical carcinoma from cervical dysplasia.33 hnRNP E1, hnRNP K and the Sm family protein FUS/TLS are intricate constituents of spreading initiation centers (SICs), macromolecular complexes of RNPs required for initiation of cell spreading during metastatic progression.34 In these studies, attenuation of hnRNP E1 by neutralizing antibodies was shown to stimulate cell spreading,34 corroborating our cellular analyses demonstrating that hnRNP E1 knockdown mediates increased cellular migration and invasiveness.15,25 Cellular reprogramming has also been associated with a decrease in the expression of hnRNP E1 in carcinoma CNE-2 cells35 and in mouse embryonic stem cells.36
Concluding Remarks
It is becoming increasingly clear that EMT and tumorigenesis is mediated by an interplay of transcriptional and post-transcriptional programs. As during development, when translational regulatory mechanisms activate maternally inherited mRNAs to mediate embryogenesis, it appears that EMT and metastasis may be similarly regulated. Whether these regulatory pathways are distinct or whether they are similar and simply aberrantly reactivated during metastasis remains to be determined.
Our data suggests that the hnRNP E1 translational pathway may in fact regulate a cohort of EMT and metastasis mRNA transcript of which Dab2 and ILEI represent just two. This postulate is based on the fact that while their functions are required, the combined expression of Dab2 and ILEI is not sufficient for mediating TGFβ-mediated EMT.15 In addition, while our findings suggest that the hnRNP E1-dependent translational regulatory mechanism is functionally significant in the metastatic progression of tumors, further studies are needed to determine whether this pathway is deregulated in cancer cells and tissues. Given the necessity of hnRNP E1 in the BAT mRNP complex, we speculate that expression levels and/or phosphorylation status of p-Akt2 and hnRNP E1, as well as the expression of Dab2 and ILEI, may be directly correlative with metastatic progression of tumors. Our work also suggests that in addition to, or super-imposed upon, gene expression analyses, post-transcriptional mapping will be extremely useful for future interrogation and correlation of genetic variance with predisposition to disease onset and progression.
Other interesting questions relating to the hnRNP E1-mediated translational mechanism are whether phosphorylation effects the subcellular localization or half-life of hnRNP E1. Resolution of the crystal structure of the phosphorylated form of hnRNP E1 will also lead to a clearer understanding of how post-translational modification attenuates hnRNP E1's RNA binding potential. Further investigation is essential to unravel the signaling pathways regulating hnRNP E1 expression, turnover and function. It is highly probable that hnRNP E1's different functions are specified and dictated by the topology of hnRNP E1 and/or its binding partners to a specific mRNA. Deciphering the various hnRNP E1 complexes in vivo through a comprehensive integration of genetic, biochemical, molecular biology and structural analyses will shed further light and will perhaps give us a well-defined tool for therapeutic targeting.
Acknowledgments
This work was supported by Grants CA55536 and CA154663 from the National Cancer Institute to P.H.H. G.S.H. and A.C. were supported by American Heart Association (Ohio Valley Affiliate) Pre-doctoral Fellowship awards 10PRE3870024 and 075080B, respectively.
References
- 1.Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 2.Zavadil J, Bottinger EP. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24:5764–5774. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- 3.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 4.Bierie B, Moses HL. TGF-beta and cancer. Cytokine Growth Factor Rev. 2006;17:29–40. doi: 10.1016/j.cytogfr.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 5.Massagué J. TGFbeta in Cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 7.Savagner P, Yamada KM, Thiery JP. The zinc-finger protein slug causes desmosome dissociation, an initial and necessary step for growth factor-induced epithelial-mesenchymal transition. J Cell Biol. 1997;137:1403–1419. doi: 10.1083/jcb.137.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J, et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci USA. 2010;107:15449–15454. doi: 10.1073/pnas.1004900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mani SA, Yang J, Brooks M, Schwaninger G, Zhou A, Miura N, et al. Mesenchyme Forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers. Proc Natl Acad Sci USA. 2007;104:10069–10074. doi: 10.1073/pnas.0703900104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 11.Yang J. Exploring the molecular basis of tumor metastasis by microarray analysis. Assay Drug Dev Technol. 2006;4:483–488. doi: 10.1089/adt.2006.4.483. [DOI] [PubMed] [Google Scholar]
- 12.Weinberg RA. Leaving home early: reexamination of the canonical models of tumor progression. Cancer Cell. 2008;14:283–284. doi: 10.1016/j.ccr.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 13.Van Der Kelen K, Beyaert R, Inze D, De Veylder L. Translational control of eukaryotic gene expression. Crit Rev Biochem Mol Biol. 2009;44:143–168. doi: 10.1080/10409230902882090. [DOI] [PubMed] [Google Scholar]
- 14.Evdokimova V, Tognon C, Ng T, Ruzanov P, Melynk N, Fink D, et al. Translational activation of Snail1 and other developmentally regulated transcription factors by YB-1 promotes an epithelial-mesenchymal transition. Cancer Cell. 2009;15:402–415. doi: 10.1016/j.ccr.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 15.Chaudhury A, Hussey GS, Ray PS, Jin G, Fox PL, Howe PH. TGF-beta-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nat Cell Biol. 2010;12:286–293. doi: 10.1038/ncb2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.García-Martínez J, Aranda A, Perez-Ortin JE. Genomic run-on evaluates transcription rates for all yeast genes and identifies gene regulatory mechanisms. Mol Cell. 2004;15:303–313. doi: 10.1016/j.molcel.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 17.Hieronymus H, Silver PA. A systems view of mRNP biology. Genes Dev. 2004;18:2845–2860. doi: 10.1101/gad.1256904. [DOI] [PubMed] [Google Scholar]
- 18.Beilharz TH, Preiss T. Translational profiling: the genome-wide measure of the nascent proteome. Brief Funct Genomics Proteomic. 2004;3:103–111. doi: 10.1093/bfgp/3.2.103. [DOI] [PubMed] [Google Scholar]
- 19.Beyer A, Hollunder J, Nasheuer HP, Wilhelm T. Post-transcriptional expression regulation in the yeast Saccharomyces cerevisiae on a genomic scale. Mol Cell Proteomics. 2004;3:1083–92. doi: 10.1074/mcp.M400099-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Reimann I, Huth A, Thiele H, Thiele BJ. Suppression of 15-lipoxygenase synthesis by hnRNP E1 is dependent on repetitive nature of LOX mRNA 3′-UTR control element DICE. J Mol Biol. 2002;315:965–974. doi: 10.1006/jmbi.2001.5315. [DOI] [PubMed] [Google Scholar]
- 21.Waerner T, Alacakaptan M, Tamir I, Oberauer R, Gal A, Brabletz T, et al. ILEI: a cytokine essential for EMT, tumor formation and late events in metastasis in epithelial cells. Cancer Cell. 2006;10:227–239. doi: 10.1016/j.ccr.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 22.Wang H, Vardy LA, Tan CP, Loo JM, Guo K, Li J, et al. PCBP1 suppresses the translation of metastasis-associated PRL-3 phosphatase. Cancer Cell. 2010;18:52–62. doi: 10.1016/j.ccr.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 23.Varani G, Nagai K. RNA recognition by RNP proteins during RNA processing. Annu Rev Biophys Biomol Struct. 1998;27:407–4045. doi: 10.1146/annurev.biophys.27.1.407. [DOI] [PubMed] [Google Scholar]
- 24.Cusack S. RNA-protein complexes. Curr Opin Struct Biol. 1999;9:66–73. doi: 10.1016/S0959-440X(99)80009-8. [DOI] [PubMed] [Google Scholar]
- 25.Hussey GS, Chaudhury A, Dawson AE, Lindner DJ, Knudsen CR, Wilce MC, et al. Identification of an mRNP complex regulating tumorigenesis at the translational elongation step. Mol Cell. 2011;41:419–431. doi: 10.1016/j.molcel.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prunier C, Howe PH. Disabled-2 (Dab2) is required for transforming growth factor beta-induced epithelial to mesenchymal transition (EMT) J Biol Chem. 2005;280:17540–17548. doi: 10.1074/jbc.M500974200. [DOI] [PubMed] [Google Scholar]
- 27.Mintz PJ, Patterson SD, Neuwald AF, Spahr CS, Spector DL. Purification and biochemical characterization of interchromatin granule clusters. EMBO J. 1999;18:4308–4320. doi: 10.1093/emboj/18.15.4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chkheidze AN, Liebhaber SA. A novel set of nuclear localization signals determine distributions of the alphaCP RNA-binding proteins. Mol Cell Biol. 2003;23:8405–8415. doi: 10.1128/MCB.23.23.8405-15.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chaudhury A, Chander P, Howe PH. Heterogeneous nuclear ribonucleoproteins (hnRNPs) in cellular processes: Focus on hnRNP E1's multifunctional regulatory roles. RNA. 2010;16:1449–1462. doi: 10.1261/rna.2254110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gamarnik AV, Andino R. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA. 1997;3:882–892. [PMC free article] [PubMed] [Google Scholar]
- 31.Ishikawa F, Matunis MJ, Dreyfuss G, Cech TR. Nuclear proteins that bind the pre-mRNA 3′ splice site sequence r(UUAG/G) and the human telomeric DNA sequence d(TTAGGG)n. Mol Cell Biol. 1993;13:4301–4310. doi: 10.1128/mcb.13.7.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.LaBranche H, Dupuis S, Ben-David Y, Bani MR, Wellinger RJ, Chabot B. Telomere elongation by hnRNP A1 and a derivative that interacts with telomeric repeats and telomerase. Nat Genet. 1998;19:199–202. doi: 10.1038/575. [DOI] [PubMed] [Google Scholar]
- 33.Pillai MR, Chacko P, Kesari LA, Jayaprakash PG, Jayaram HN, Antony AC. Expression of folate receptors and heterogeneous nuclear ribonucleoprotein E1 in women with human papillomavirus mediated transformation of cervical tissue to cancer. J Clin Pathol. 2003;56:569–574. doi: 10.1136/jcp.56.8.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Hoog CL, Foster LJ, Mann M. RNA and RNA binding proteins participate in early stages of cell spreading through spreading initiation centers. Cell. 2004;117:649–662. doi: 10.1016/S0092-8674(04)00456-8. [DOI] [PubMed] [Google Scholar]
- 35.Cao JX, Cui YX, Long ZJ, Dai ZM, Lin JY, Liang Y, et al. Pluripotency-associated genes in human nasopharyngeal carcinoma CNE-2 cells are reactivated by a unique epigenetic sub-microenvironment. BMC Cancer. 2010;10:68. doi: 10.1186/14712407-10-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baharvand H, Fathi A, Gourabi H, Mollamohammadi S, Salekdeh GH. Identification of mouse embryonic stem cell-associated proteins. J Proteome Res. 2008;7:412–423. doi: 10.1021/pr700560t. [DOI] [PubMed] [Google Scholar]