Abstract
Global mapping of rRNA structure by traditional methods is prohibitive in terms of time, labor and expense. High throughput selective 2′ hydroxyl acylation analyzed by primer extension (hSHAPE) bypasses these problems by using fluorescently labeled primers to perform primer extension reactions, the products of which can be separated by capillary electrophoresis, thus enabling long read lengths in a cost effective manner. The data so generated is analyzed in a quantitative fashion using SHAPE Finder. This approach was used to map the flexibility of nearly the entire sequences of the 3 largest rRNAs from intact, empty yeast ribosomes. Mapping of these data onto near-atomic resolution yeast ribosome structures revealed the binding sites of known trans-acting factors, as well as previously unknown highly flexible regions of yeast rRNA. Refinement of this technology will enable nucleotide-specific mapping of changes in rRNA structure depending on the status of tRNA occupancy, the presence or absence of other transacting factors, due to mutations of intrinsic ribosome components or extrinsic factors affecting ribosome biogenesis or in the presence of translational inhibitors.
Key words: ribosome, hSHAPE, structure, rRNA
Introduction
Unlike mammalian cells, yeast is genetically malleable and large quantities of cells can be grown efficiently and inexpensively. These properties have made yeast the model organism for the study of eukaryotic ribosomes. While bacterial and archaeal ribosomes have been crystallized by a growing number of laboratories, similar structures of eukaryotic ribosomes have proven more elusive, thus making the study of this important branch of the tree of life more difficult. In addition to near-atomic level cryo-EM studies,1 structural mapping of the yeast ribosome has mostly been performed via chemical probing methods (reviewed in ref. 2). Classical chemical probing techniques employ a variety of base-specific (e.g., kethoxal, CMCT and DMS), and non-specific reagents (e.g., hydroxyl radical probing) combined with the use of radioactively labeled oligonucleotides in primer extension reactions which are resolved using large format urea-PAGE. These approaches are limited for two reasons. Specifically, the resolution of the primer extension reactions through gels is limited to less than 150 nt, and the chemical probes have low dynamic ranges, thus limiting the ability to discern the relative reactivity of individual bases. A newer technology, hSHAPE3,4 uses fluorescently labeled oligonucleotide primers instead of radioactivity; the extension reactions are resolved by capillary electrophoresis, thus allowing interrogation of up to 600 nucleotides using a single primer. In addition, a single chemical, 1-methyl-7-nitroisatoic acid (1M7) is used. 1M7 is not base specific like the traditional chemicals DMS, CMCT and kethoxal, instead its substrate is the flexible 2′-OH group of the ribose; it has the advantage of producing highly quantifiable signals over a wide dynamic range as well as simultaneously interrogating every nucleotide position. Thus, hSHAPE enables quantitative interrogation of the entire rRNA content of a ribosome in a cost effective and timely manner.
In the current study, application of hSHAPE to salt-washed, puromycin treated yeast ribosomes enabled interrogation of ∼95% of all of the bases in the three largest rRNA species: 25S, 18S and 5.8S rRNAs; in highly quantitative, rapid and cost effective manner, revealing the most flexible bases in these three rRNAs. While many of these mapped to rRNA loops identified as being on the surface of the yeast ribosome, others corresponded to bases in molecular hinges that are known or suspected of being involved in transmission of structural and functional information through the ribosome, or in expansion segments (ES) not found in the better characterized bacterial or archael ribosomes. Others corresponded to known sites of interaction with trans-acting factors, suggesting that this method can be used to map similar, but currently uncharacterized interactions. The quantitative nature of the data revealed that rRNA bases that participate in intersubunit bridges that form and deform during the translation elongation cycle showed intermediate levels of reactivity. This supports recent observations that the ribosome is intrinsically free to transit between a large number of conformational states at physiologic temperatures.5,6 In the future, coupling of the information gleaned by applying hSHAPE to ribosomes arrested at different translational states and/or complexed with various translational inhibitors combined with molecular dynamics simulations and atomic resolution yeast ribosome structures will enable the visualization of the myriad structural changes undergone by individual bases of the yeast ribosome during the process of protein synthesis.
Results
In order to overcome the limitations posed by traditional analyses of rRNA structural changes by use of denaturing urea-PAGE, hSHAPE, a capillary electrophoresis based fluorescent detection technology7 was applied to highly intact ribosomes purified using a chromatographic purification scheme.8 Previous work done by the Weeks lab has used hSHAPE to analyze secondary structure of deproteinized and separate 16S and 23S subunits of E. coli rRNA, but not on fully intact ribosomes.9 In the current study, salt washed, intact ribosomes were pre-treated with puromycin to ensure tRNA removal, enabling a clear view of the flexibility of the empty yeast ribosome, i.e., its “ground state”. As opposed to the traditional method in which resolution is limited to ∼150 nt, read lengths using hSHAPE were ∼300–600 nt. This enabled interrogation of the entirety of the yeast 18S, 25S and 5.8S rRNAs using only 20 primers (Table 1). The 5S rRNA was not probed, as its short length (121 nt) makes it a poor candidate for hSHAPE. Total coverage of the reactivity of the ribosome was 94.4% (5,054 out of 5,354 nt), with reactivity of 60.7% of the 5.8S rRNA (96 out of 158 nt), 94.7% of the 18S rRNA (1,705 out of 1,800 nt) and 95.7% of the 25S rRNA (3,253 out of 3,396 nt). In addition, the reactivity of each nucleotide was highly quantifiable, allowing us to distinguish not only which bases were flexible, but also the degree of flexibility at every position. An example of a sample read covering approximately 700 nt generated from primer 1,180 of the SSU is shown in Figure 1. With regard to variability, each primer set was used to probe ribosomes multiple times in order to optimize reaction conditions. During this process, conditions that maximized reproducibility were identified. While the final data reported here were generated from single reads, they were generated from ribosomes that were isolated and treated with 1M7 at different times. Importantly, regions of sequence overlap between reads from different primers were found to be highly reproducible.
Table 1.
Oligonucleotide primers used in the current study
| Subunit | Primer name | Sequence | Location | ddNTP used (primary/secondary) |
| 18S | 224 | 5′ TCA AAG AGT CCG AAG ACA TTG 3′ | 224–244 | A/T |
| 18S | 448 | 5′ TCA CTA CCT CCC TGA ATT AGG 3′ | 448–468 | A/T |
| 18S | 597 | 5′ GCT TTT TAA CTG CAA CAA CTT TAA TAT AC 3′ | 597–625 | T/C |
| 18S | 969 | 5′ TCC CCT AAC TTT CGT TCT TG 3′ | 969–988 | A/T |
| 18S | 1180 | 5′ CCC CGT GTT GAG TCA AAT TAA G 3′ | 1180–1201 | T/C |
| 18S | 1572 | 5′ TTG CGC TTA CTA GGA ATT CCT C 3′ | 1572–1593 | T/G |
| 18S | 1780 | 5′ TAA TGA TCC TTC CGC AGG TTC 3′ | 1780–1800 | A/T |
| 25S | 356 | 5′ AGT TCT TTT CAT CTT TCC ATC ACT G 3′ | 356–380 | T/A |
| 25S | 644 | 5′ CTC CTT GGT CCG TGT TTC 3′ | 644–691 | C/A |
| 25S | 949 | 5′ GCT ATC CTG AGG GAA ACT TCG 3′ | 949–969 | C/A |
| 25S | 25-2 | 5′ GAC TTC CAT GGC CAC CG 3′ | 1232–1248 | A/C |
| 25S | 1466 | 5′ CCC ACT TCA GTC TTC AAA GTT C 3′ | 1466–1487 | T/C |
| 25S | 1700 | 5′ CAA TTC CGG GGT GAT AAG C 3′ | 1700–1718 | A/C |
| 25S | 2128 | 5′ CAA TGC TAT GTT TTA ATT AGA CAG TCA G 3′ | 2128–2155 | C/T |
| 25S | 2632 | 5′ GGA CAT CTG CGT TAT CGT TTA AC 3′ | 2632–2654 | C/A |
| 25S | 2836 | 5′ CAA TGT CGC TAT GAA CGC TTG 3′ | 2836–2856 | T/C |
| 25S | 25-7 | 5′ CCT GAT CAG ACA GCC GC 3′ | 3059–3075 | A/C |
| 25S | 3225 | 5′ GCC AAG TGC ACC GTT G 3′ | 3225–3240 | A/T |
| 25S | 3′ | 5′ AGA CAA CAA AGG CTT AAT CTC AGC 3′ | 3366–3389 | A/C |
| 5.8S | 5.8S | 5′ AAA TGA CGC TCA AAC AGG C 3′ | 1–19 | C/T |
The primer name corresponds to the 5′ base complementary to the indicated yeast rRNA species. Each primer spanned the indicated locations on the rRNAs. Each oligonucleotide was labeled with the following fluorescent molecules: 6FAM, VIC, NED and PET. NED (primary) and PET (secondary) labeled oligonucleotides were employed for sequencing reactions using the indicated dideoxynucleotides. VIC labeled oligonucleotides were used for primer extension reactions of DMSO (control) rRNAs. 6FAM labeled oligonucleotides were employed for primer extension reactions of 1M7 (experimental) treated rRNAs.
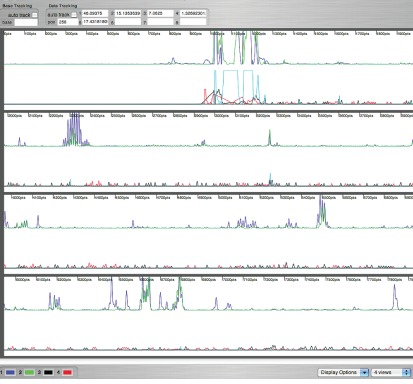
Figure 1.
Sample chromatogram of quantitative data analysis walkthrough. Figure shows a sample chromatogram representative of raw data after analysis using SH APE finder. This chromatogram shows an entire dataset covering approximately 700 nt generated from primer 1180 of the SSU. Data were subsequently quantified by removing negative values, normalization and assessment of final levels. 6FAM labeled primers (blue) were used for analysis of 1M7 treated (+) rRNAs, VIC labeled primers (green) were used for analysis of DMSO (−) treated rRNAs, NED labeled primers (black) were used for ddTTP sequencing reactions, and PET labeled primers (red) were used for ddCTP sequencing reactions.
Structure mapping.
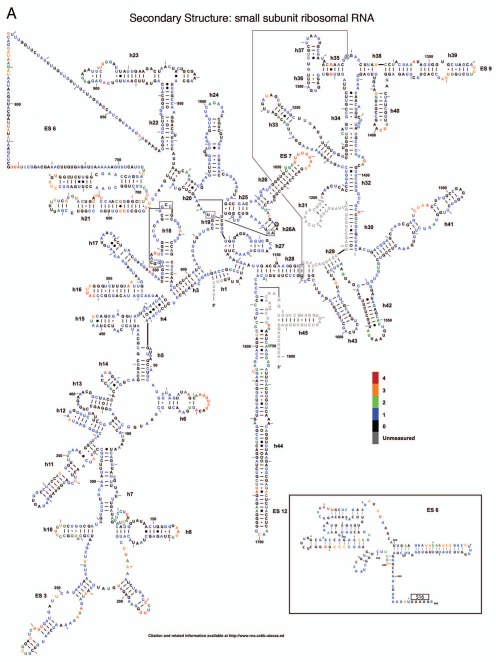
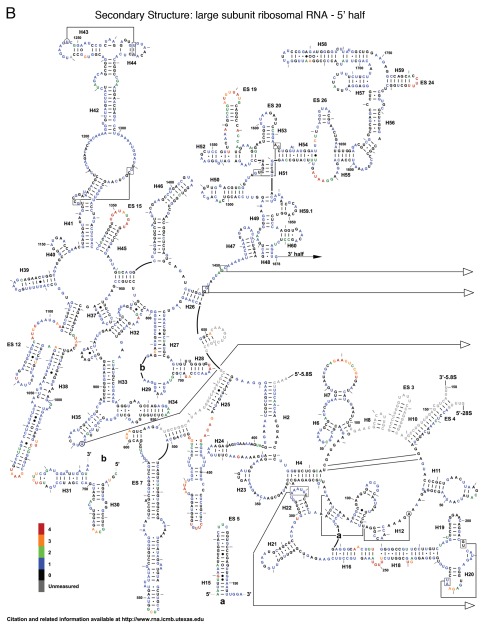
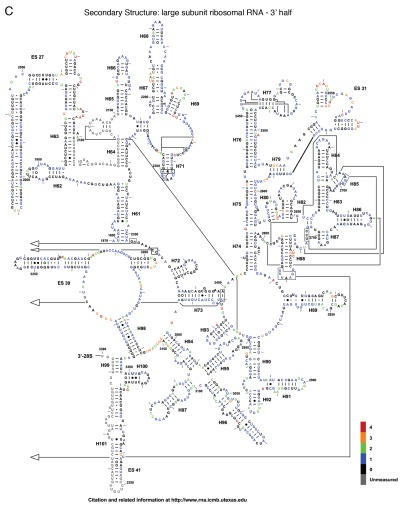
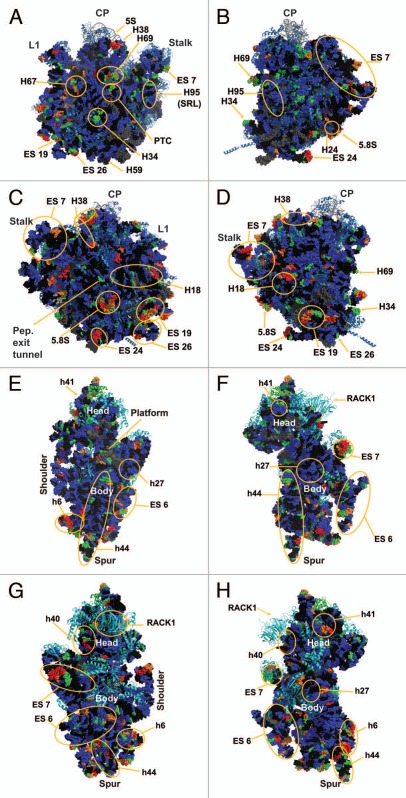
The reactivity of all 5,354 nucleotides of the three yeast rRNAs are depicted as flat, two-dimensional maps in Figures 2A–C. Included as an insert in Figure 2A is a two-dimensional map of ES6 based on the cryo-EM structure of the yeast ribosome,1 which postulates an alternative structure to the standard non-ordered region and from the variant previously proposed in reference 10. Figure 3A–H shows these data mapped onto a series of snapshots of the 3D structure of the 60S and 40S ribosomal subunits. These 3D structural maps allow us to examine how the various flexible regions interact with known trans-acting factors, e.g., ribosomal proteins and the influence of the intersubunit bridges on rRNA flexibility. The ribosomal subunit maps are presented in Figure 3. The large subunit is depicted in Figures 3A–D, beginning with the “crown view” (Fig. 3A), which shows the intersubunit face of the LSU. Figures 3B–D are serially rotated 90° clockwise, showing the elongation factor binding region (3B), the solvent face containing the nascent peptide exit tunnel (3C), and the deacylated-tRNA exit side (3D). Figures 3E and F show the small subunit. Figure 3E depicts the intersubunit face of the SSU including the mRNA platform and decoding center. Similar to described above, Figures 3F–H are serially serially rotated 90° clockwise, showing the elongation factor binding face (3F), the solvent accessible rear of the LSU (3G), and the deacylated tRNA exit side (3H). Labels depict regions containing highly flexible bases mapped from Figure 2. The pymol 3D structures can be downloaded from dinmanlab.umd.edu/hshape/. This pymol structure allows fine grain manipulation of the image to allow a clear depiction of any region of the ribosome that is discussed further in the paper. The maps are color coded to indicate the degree of reactivity of each base with 1M7. Bases colored red were the most reactive, followed by orange, green and blue. Bases colored black were non-reactive. Grey was used to indicate bases that could not be resolved, either due to their proximity to the 3′ ends of the 3 rRNAs, their proximity to highly modified bases, which masked signals of nearby bases due to their ability to promote strong stops to reverse transcriptase reactions, or their proximity to the 3′ ends of a primer. Inspection of these Figures reveals that 82% of bases were non-reactive or only weakly reactive. This is due to the highly structured nature of the ribosome, in which most of the rRNA bases participate in base pairing interactions, in tertiary interactions among bases or interact with ribosomal proteins. The most flexible bases tended to be found in regions of the rRNAs that are unstructured or in terminal loops, and in internal loops, which may function as hinges.
Figure 2.
Two dimensional flexibility maps of yeast 18S, 25S and 5.8S rRNAs. A 2D structure map of yeast rRNAs was downloaded from http://www.rna.ccbb.utexas.edu. (A) 18S rRNA. Inset is 2D structure of ES6 based on 3D structure.1 Bases interacting with ribosomal protein S16 (i.e., U861-A865) are indicated. A 2D structure map of yeast rRNAs was downloaded from www.rna.ccbb.utexas.edu. (B) the 5′ half of 25S + 5.8S rRNA. Bases are colored coded based on their relative reactivities with 1M7. (C) the 3′ half of 25S rRNA. Grey, unmeasured base; Black, 0-unreactive; Blue, 1-weakly reactive; Green, 2-moderate reactivity; Orange, 3-strong reactivity; Red, 4-highly reactive.
Figure 3.
Three dimensional flexibility map of the yeast rRNA. The data presented in Figure 2 were mapped onto a cryo-EM reconstruction of the yeast ribosome.1 Bases are colored based on their relative reactivity with 1M7. Grey, unmeasured; black, unreactive (0); blue, weak (1); green, moderate (2); orange, strong (3); red, highly reactive (4). Helixes and regions of interest are marked. (A–D) 60S subunit: (A) crown view, (B) tRNA entry face, (C) solvent exposed “rear” of the LSU including the peptide exit tunnel, (D) tRNA exit face. CP denotes the Central Protuberance. Stalk is the P1/P2 stalk (L7/L11 in bacteria). L1 shows the L1 stalk. (E–H): 40s subunit: (E) intersubunit face, (F) tRNA entry face, (G) rear solvent face, (H) tRNA exit face. Labeled landmarks include the Head, Body, mRNA Platform, Shoulder and H44 Spur.
Small subunit (SSU) analysis.
The SSU of the ribosome may be divided into four domains:11 the most significant changes in these domains are discussed. In the 5′ domain of the SSU, helix 6, also known as the “spur”, contains a loop with numerous flexible bases. This flexibility is consistent with previous studies in E. coli which indicate this is among the most conformationally dynamic regions of the ribosome.12 In the core domain, A780 to U782, located in expansion segment 6 which has no E. coli equivalent, were previously shown to be reactive with CMCT;10 these were also highly reactive with 1M7. In the core domain a long series of highly flexible bases were observed in expansion segment 7 at the head of helix 26. Conversely, helix 27 contained largely unreactive or weakly reactive bases, consistent with its position buried within the SSU. U912 (A702 in E. coli) was highly reactive; given that this base has been shown to be protected when the ribosome is in the “classical state” but deprotected in the “hybrid state”,13 its reactivity in the this study supports current models in which “ground state” ribosomes can freely transition through a large variety of conformational variations at physiological temperatures without significant thermodynamic penalty.5,6 In the 3′ major domain, the distal loops of helices 40 and 41 were both highly reactive; helix 40 has been shown in E. coli to interact with rpS1,14 and the 3D structure shows RACK1 in close proximity to this helix. This is consistent with these ribosomes having been salt-washed, i.e., RACK1 was not present in these samples. Similarly, while helix 41 in E. coli interacts with rpS7,15 the three-dimensional model in Figure 3 shows rpS19e to be in close proximity to this helix. In the 3′ minor domain of the SSU, the decoding center flexible bases were found in the bulge region at the base of helix 44. In the three-dimensional structure of the 80S ribosome, these bases are located near Helix 69 of the LSU and participate in the B2a and B3 bridges; these bases are involved in information exchange across the two ribosomal subunits.11,16 Additionally, bases in extension segment E6 were recently found to form the eukaryote-specific eB12 intersubunit bridge with the eukaryote-specific C-terminus of L19e.16 Such flexibility is important for information exchange within the ribosome, allowing the ribosome to transit between different structural/informational states.
Large subunit (LSU) analysis.
The large subunit can be divided into 6 domains. Beginning with domain I, Helix 18 contains a highly flexible loop centered around nucleotide 250. Examination of the three-dimensional structure in Figure 3 shows this loop to be exposed to solvent and not masked by any intrinsic ribosomal proteins. One possible interpretation is that this evolutionarily conserved bulge may serve as a binding site for other ligands or trans-acting factors. Similarly, the region of Helix 25 that leads into expansion segment 7 was observed to be highly flexible. This may allow the entire expansion segment to flex around this hinge as a mobile rRNA domain. Domain II of the LSU contains Helix 38, which harbors numerous hinge bases, while its distal loop (the A-site finger) is involved in formation of the B1a intersubunit bridge. This structure has been shown to be highly flexible, enabling it to participate in information exchange among the functional centers of the ribosome.17 The high degree of flexibility observed in the current study, which would allow interaction with both S15 and S18 (S13 and S19 E. coli), especially at joint regions, also seem to indicate that the ribosome is empty and thus not locked into any single conformation.18 The flexible tip of Helix 34, also in domain II, lies in close proximity to rpS13 in the three-dimensional ribosome structure. This participates in the B4 bridge, another component of the information exchange network between rRNA and protein.11,16,19 In domain III, expansion segment 19 of Helix 52, centered around U1570, and a bulge on expansion segment 26, centered around A1814 showed solvent exposure profiles and flexibility profiles similar to those seen with Helix 18. The distal loop of Helix 69 in domain IV, A2255–A2262 (A1912–A1919 in E. coli), which is involved in the dynamic rearrangement of the B2a bridge during the elongation cycle,11,16 also contained the highly flexible bases required for such interactions, again indicating that the ribosomes probed here are not preferentially distributed toward any single conformational state. The reactive bases in Helix 67 in the three-dimensional structure lie very close to reactive bases in helix 23 of the SSU, indicating a possible intersubunit interaction between these two structural elements. In the core of the peptidyltransferase center in domain V, the universally conserved A2820 (E. coli A2451) was highly flexible, consistent with prior observations in E. coli.20 Also in the peptidyltransferase center are two reactive bases, U2954 (E. coli U2585) and U2955 (E. coli U2586), which lie along the aa-tRNA accommodation corridor through which aa-tRNAs transit from eEF1A into the peptidyl-transferase center.21 The hSHAPE analysis confirms the strong reactivity of specific bases in this structural element, serving as quality control indicators and revealing the conformational plasticity of the “ground state” ribosome. Additionally, A2971 (E. coli A2602) located in Helix 93 in domain V of the large subunit rRNA was previously shown to be a highly reactive in both E. coli,20 and yeast ribosomes22 in the absence of aa-tRNA bound to the ribosomal A-site. In the three-dimensional structure, G2620 (E. coli G2252) lies opposite A2971 (E. coli A2602), i.e., at the tip the “P-loop”. This base was observed to be moderately reactive, although much of the rest of Helix 80 was largely unreactive, consistent with its being buried inside the core of the LSU. A3027 (A2660 E. coli), located in the distal tip of Helix 95 (the Sarcin- Ricin loop) in domain VI, showed a moderate level of reactivity. However the remainder of this loop, while solvent exposed, was nonreactive; consistent with the highly structured nature of this of this helix which contains numerous noncanonical basepairs.23 Additionally, this loop is known to be involved in interactions with EF-G in T. thermophiles ribosomes, which is surrounded by domain III, V and the G domain.24 The 5.8S rRNA acts as an extension of the LSU at the 5′ end. It was largely unreactive, though one solvent exposed loop was found to be highly flexible.
Discussion
The traditional method of rRNA structural analysis suffers from inherent limitations, including run length, quantifiability of results and reliance on radioactive materials. The hSHAPE method overcomes these issues by increasing read lengths (up to 600 nt), enabling simple signal quantitation and using fluorescent reporters. Application of hSHAPE allowed interrogation of 95% of the bases in the three largest rRNA species of the yeast ribosome in a fraction of the time and effort that would otherwise have been required using traditional methods employing large format urea-PAGE and autoradiography. Quantization of the results using SHAPEfinder3 was the rate limiting step in data analysis due to the high level of manual manipulation required to gain maximal benefit from the program. While hSHAPE was applied to ribosomes probed with the ribose 2′-OH specific reagent 1M7 in the current study, this general method should also be applicable to ribosomes probed using other methods, e.g., in-line probing, hydroxyl radical analysis and base specific methods, e.g., DMS, CMCT and kethoxal. Indeed, 1M7 and the base specific chemicals would be an excellent complement to each other, allowing the differential probing of the backbone and the base at each nucleotide.
The current study lays out the reactivity and flexibility of yeast ribosomes that have been salt-washed and puromycin treated, i.e., in the “ground, empty” state. The results of this study show that the most flexible regions of the ribosome tend to be localized in solvent accessible loops, in known flexible hinges, and in bases participating in intersubunit bridges (Figs. 2 and 3). Bases previously identified as reactive in yeast ribosomes using the old method, such as A2971, were found to be similarly reactive by hSHAPE. Consistency across species was also observed, as bases known to be flexible in E. coli, such as A2451, were found to be flexible in yeast (A2820). This indicates that hSHAPE is capable of picking up known differences, providing confidence in the results so obtained.
Importantly, bases that normally interact with trans-acting factors, e.g., RACK1, SRP and the elongation factors (eEF1A-tRNA and eEF2), but which are not present in salt-washed ribosomes, were found to be reactive. For example, the expansion segment 24 head of Helix 59 (in Domain III) is involved in formation of the C2 connection between the ribosome and signal recognition particle (SRP), while Helix 24 (Domain I) participates in the ribosome/SRP C3 connection.25 The high levels of reactivity with 1M7 of C1762-U1765 in H59 (U1534-G1537 in E. coli), and of G376 (G481 in E. coli) in H24 are consistent with the surface-exposed nature of these structures in the absence of SRP in salt-washed ribosomes. In the small subunit, the reactivity of helix 40 is an indicator of the absence of trans-acting factor RACK1. Thus, this analysis may enable identification of potential sites of interactions between the ribosome and other trans-acting factors.
While the classical two-dimensional map of the SSU expansion segment 6 does not predict any structure, cryo-EM three-dimensional reconstructions revealed this region to well-ordered.1 Superimposition of the hSHAPE onto this base-pairing scheme (see inset, Fig. 2A) is in good agreement with some, but not all of the cryo-EM generated structure. For example, the reactivity patterns of the bases from nt 700–807 in expansion segment E6 are in generally good agreement with the tertiary structure. The exception is the loop from nt 725–739, which is non-reactive by hSHAPE, consistent with its participation in the eukaryote-specific bridge eB12.16 In contrast, the high reactivities of nt 808–815 indicate that these are not base-paired, while the low reactivity of their counterparts (nt 828–834) suggests that these are protected. This also applies to nt 848–866. Only the terminal end of this segment is known to interact with a ribosomal protein (S16), while the remainder of this segment extends into the solvent at the foot of the SSU, suggesting that this may either interact with SX4,16 or as a binding site for soluble transacting factors. Similarly, the observed reactivity of other solvent exposed structures, e.g., h6 the SSU or H18 of the LSU, suggest that these too may be able to interact with soluble trans-acting factors. Furthermore, positions of previously suspected flexibility, such as those involved in the various intersubunit bridges, were shown to in fact be flexible and open to conformational changes. This further supports the open ratchet model of the ribosome whereby empty ribosomes can freely transition between various conformational states.
Having established the baseline reactivity of yeast rRNA bases in the current study, a more comprehensive set of examinations of the ribosome can begin. Initial probes of the ribosome in various functional states, e.g., through the various steps of the translation elongation cycle, will identify changes in base flexibility as mRNAs and tRNAs move through the ribosome. In essence, application of hSHAPE can be applied to illuminate all of the allosteric changes occurring in the entire ribosome as it transits all phases of the ribosome life-cycle including but not limited to: biogenesis, the multiple steps of initiation, elongation, termination and post-termination. Thus, application of hSHAPE to the ribosome will greatly facilitate characterization of the inner workings of this complex nanomachine at the single nucleotide level. Additional applications may also include probing the ribosome as it interacts with small molecule translational inhibitors, identifying changes in ribosome structure distal to their sites of interaction. In addition, this method provides means for assessing the effects of specific protein and rRNA mutations on the global structure of the ribosome.
Materials and Methods
Yeast strains and reagents.
Ribosomes were purified from yeast strain JD1370 (MATa trp1 ura3 leu2 PEP4::HIS3 NUC1::LEU2). Sulfolink was purchased from Pierce (20402). Chemicals were purchased from Sigma-Aldrich, and GTP was purchased from Fermentas (R0461). 1M7 was graciously provided by Dr. Kevin Weeks. RNase free water was purchased from Ambion (AM9938). Primers are listed in Table 1 and were purchased from Applied Biosystems.
Ribosome purification.
Ribosome purification was performed as previously described in reference 8, followed by puromycin treatment to remove endogenous tRNAs.26 In brief, yeast was grown to an OD595 of 1.0 in 500 ml of YPAD. Cells were broken using glass beads at 4°C with a Biospec Mini bead beater (Bartlesville, OK). Cellular debris was cleared by centrifugation at 30,000x g for 30 minutes in a Beckman-Coulter Optima Max E ultracentrifuge (Fullerton, CA). The supernatant was chromatographically purified.8 After elution from the column, ribosomes were treated on ice with 1 mM GTP and 1 mM puromycin (pH neutralized) for 10 minutes and at 30°C for 20 minutes, after which the chromatographic protocol was continued. After final purification step, ribosomes were spectrophotometrically quantified and stored at −80°C.
hSHAPE extension.
hSHAPE was performed following protocols previously described in reference 27 and 28. Ribosomes were reactivated using 50 µl 2x SHAPE ribosome buffer (160 mM HEPES pH 7.5, 100 mM NaCl, 5 mM MgOAc, 6 mM β-mercaptoethanol) to a final volume of 100 µl in RNase free water and incubated for 10 minutes at 30°C. The ribosomes were then divided into two 50 µl aliquots. 75 µl of 1x SHAPE ribosome buffer was added to each tube. To one tube, 12.5 µl of DMSO was added, while 12.5 µl of 60 mM 1M7 was added to the other tube. Reactions were allowed to proceed for 10 minutes at 30°C. Reactions were then precipitated overnight using 100% ethanol and 300 mM sodium acetate. After overnight ethanol precipitation, rRNAs were and purified using a RNaqueous micro kit (Ambion, AM1931) and rRNA concentrations were determined using a Nanodrop 1000 (Wilmington, DE).
Primer extension reactions were performed as follows. For each primer sequence (Table 1), four primers were constructed with the following fluorescent molecules: 6FAM, VIC, NED, and PET. Each primer was diluted to 2.5 pmols/µl. To 1 µg of 1M7 treated RNA (experimental sample), 2.5 pmols of 6FAM-labeled primer was added and brought to 6.5 µl with water. To 1 µg of DMSO treated RNA (control sample), 2.5 pmols of VIC-labeled primer was added and brought to 6.5 µl with water. For sequencing, 1 µg of untreated rRNA was added to 2.5 pmols of NED or PET-labeled primer brought to 5.5 µl with water. The samples were incubated at 65°C for five minutes and at 50°C for five minutes and then placed on ice. A master mix was made of 100 U Superscript 3 Reverse Transcriptase (Invitrogen 18080044), 10 mM dNTP mixture (Fermentas, R0182), 5X Superscript Buffer (Invitrogen 18080044) and 10 mM DTT and added to the samples. In addition, 10 mM of appropriate ddNTP (Fermentas) (See Table 1) was added to the sequencing samples. Samples were incubated at 50°C for one minute, 52°C for 45 minutes and 65°C for 5 minutes and placed on ice. The four extension reactions were combined and precipitated overnight in ethanol. Samples were washed twice with 70% ethanol and resuspended in Hi-Di Formamide (Applied Biosystems, 4311320). Fragment analyses using capillary electrophoresis were performed by Genewiz (South Plainfield, NJ).
SHAPEfinder analysis.
Data were analyzed using SHAPEFinder.3 All traces were passed through a processing pipeline using the following SHAPEfinder tools: baseline adjust, mobility shift, signal decay correction and align and integrate. A sample of this type of data is shown in Figure 1. After the data integration was complete, further data analyses were performed using Excel. Visual inspection of the data was performed and a value of 0 was assigned to any peak in which both the sequencing lanes showed a large peak to denote the presence of some non-1M7 mediated reverse transcription stoppage or in which the data showed a negative value. Data were assigned four levels of reactivity: 0 (for less than a trace's median value), 1 (for a value between the median and the mean), 2 (for a value between the mean and the 1st standard deviation), 3 (for a value between the 1st and the 2nd standard deviation) and 4 (for values above the 2nd standard deviation). Mean, median and standard deviation were calculated on a per trace basis. A level of 5 was used to indicate that there were no data available for that nucleotide. Levels were aligned to the known sequence for each rRNA. Where sequences overlapped, the values from the sequences whose primers were closest to the primer were chosen, unless the values were 0, in which the higher values were chosen. The data were plotted onto a Postscript file of the 2D map of the ribosome. Pymol was used to map this data onto the most recent cryo-EM structure of the yeast ribosome, PDB entries 3JYV, 3JYW, 3JYX.1 The raw data used to color pdb entries 3JYV, 3JYW, 3JYX, the completed pymol session, and python code used to color the pdb and utexas postscript files may be downloaded from: dinmanlab.umd.edu/hshape/. A: small subunit. B: large subunit.
Acknowledgements
We would like to thank Kevin Weeks and the members of his laboratory for the kind gift of 1M7, and for providing support and assistance with SHAPEFinder. Thanks also to Drs. Arturas Meskauskas and Karen Jacks for providing assistance and expertise in ribosome purification. This work was supported by a grant to J.D.D. from the National Institutes of Health (5R01 GM058859). J.A.L. was supported by a supplement to the parent grant through the American Recovery and Reinvestment Act (3R01GM058859-10A1S1). A.T.B. was partially supported by an NIH Virology Training Grant (T32 AI51967) and a University of Maryland College of Chemical and Life Sciences Drs. Wayne T. and Mary T. Hockmeyer Doctoral Fellowship Award.
References
- 1.Taylor DJ, Devkota B, Huang AD, et al. Comprehensive molecular structure of the eukaryotic ribosome. Structure. 2009;17:1591–1604. doi: 10.1016/j.str.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meskauskas A, Dinman JD. Ribosomal protein L3: Gatekeeper to the A-site. Mol Cell. 2007;25:877–888. doi: 10.1016/j.molcel.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vasa SM, Guex N, Wilkinson KA, Weeks KM, Giddings MC. ShapeFinder: a software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. RNA. 2008;14:1979–1990. doi: 10.1261/rna.1166808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilkinson KA, Vasa SM, Deigan KE, Mortimer SA, Giddings MC, Weeks KM. Influence of nucleotide identity on ribose 2′-hydroxyl reactivity in RNA. RNA. 2009;15:1314–1321. doi: 10.1261/rna.1536209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aitken CE, Petrov A, Puglisi JD. Single ribosome dynamics and the mechanism of translation. Annu Rev Biophys. 2010;39:491–513. doi: 10.1146/annurev.biophys.093008.131427. [DOI] [PubMed] [Google Scholar]
- 6.Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature. 2010;466:329–333. doi: 10.1038/nature09206. [DOI] [PubMed] [Google Scholar]
- 7.Watts JM, Dang KK, Gorelick RJ, et al. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leshin JA, Rakauskaite R, Dinman JD, Meskauskas A. Enhanced purity, activity and structural integrity of yeast ribosomes purified using a general chromatographic method. RNA Biol. 2010;7:354–360. doi: 10.4161/rna.7.3.11648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure determination. Proc Natl Acad Sci USA. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alkemar G, Nygard O. Probing the secondary structure of expansion segment ES6 in 18S ribosomal RNA. Biochemistry. 2006;45:8067–8078. doi: 10.1021/bi052149z. [DOI] [PubMed] [Google Scholar]
- 11.Yusupov MM, Yusupova GZ, Baucom A, et al. Crystal structure of the ribosome at 5.5 a resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 12.Gao H, Sengupta J, Valle M, et al. Study of the structural dynamics of the E. coli 70S ribosome using real-space refinement. Cell. 2003;113:789–801. doi: 10.1016/s0092-8674(03)00427-6. [DOI] [PubMed] [Google Scholar]
- 13.Zhang W, Dunkle JA, Cate JH. Structures of the ribosome in intermediate states of ratcheting. Science. 2009;325:1014–1017. doi: 10.1126/science.1175275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sengupta J, Agrawal RK, Frank J. Visualization of protein S1 within the 30S ribosomal subunit and its interaction with messenger RNA. Proc Natl Acad Sci USA. 2001;98:11991–11996. doi: 10.1073/pnas.211266898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka I, Nakagawa A, Hosaka H, Wakatsuki S, Mueller F, Brimacombe R. Matching the crystallographic structure of ribosomal protein S7 to a three-dimensional model of the 16S ribosomal RNA. RNA. 1998;4:542–550. doi: 10.1017/s1355838298972004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ben Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 17.Rakauskaite R, Dinman JD. An arc of unpaired “hinge bases” facilitates information exchange among functional centers of the ribosome. Mol Cell Biol. 2006;26:8992–9002. doi: 10.1128/MCB.01311-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agirrezabala X, Frank J. Elongation in translation as a dynamic interaction among the ribosome, tRNA and elongation factors EF-G and EF-Tu Q. Rev Biophys. 2009;42:159–200. doi: 10.1017/S0033583509990060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Culver GM, Cate JH, Yusupova GZ, Yusupov MM, Noller HF. Identification of an RNA-protein bridge spanning the ribosomal subunit interface. Science. 1999;285:2133–2135. doi: 10.1126/science.285.5436.2133. [DOI] [PubMed] [Google Scholar]
- 20.Moazed D, Noller HF. Sites of interaction of the CCA end of peptidyl-tRNA with 23S rRNA. Proc Natl Acad Sci USA. 1991;88:3725–3728. doi: 10.1073/pnas.88.9.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanbonmatsu KY, Joseph S, Tung CS. Simulating movement of tRNA into the ribosome during decoding. Proc Natl Acad Sci USA. 2005;102:15854–15859. doi: 10.1073/pnas.0503456102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meskauskas A, Dinman JD. A molecular clamp ensures allosteric coordination of peptidyltransfer and ligand binding to the ribosomal A-site. Nucleic Acids Res. 2010;38:7800–7813. doi: 10.1093/nar/gkq641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Correll CC, Munishkin A, Chan YL, Ren Z, Wool IG, Steitz TA. Crystal structure of the ribosomal RNA domain essential for binding elongation factors. Proc Natl Acad Sci USA. 1998;95:13436–13441. doi: 10.1073/pnas.95.23.13436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. The structure of the ribosome with elongation factor G trapped in the post-translocational state. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halic M, Becker T, Pool MR, Spahn CM, Grassucci RA, Frank J, et al. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature. 2004;427:808–814. doi: 10.1038/nature02342. [DOI] [PubMed] [Google Scholar]
- 26.Triana F, Nierhaus KH, Chakraburtty K. Transfer RNA binding to 80S ribosomes from yeast: evidence for three sites. Biochem Mol Biol Int. 1994;33:909–915. [PubMed] [Google Scholar]
- 27.Mortimer SA, Weeks KM. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J Am Chem Soc. 2007;129:4144–4145. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- 28.Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]