Abstract
A full understanding of the mechanism of post-transcriptional regulation requires more than simple two-state prediction (binding or not binding) for RNA binding proteins. Here we report a sequence-based technique dedicated for predicting complex structures of protein and RNA by combining fold recognition with binding affinity prediction. The method not only provides a highly accurate complex structure prediction (77% of residues are within 4 Å RMSD from native in average for the independent test set) but also achieves the best performing two-state binding or non-binding prediction with an accuracy of 98%, precision of 84% and Mathews correlation coefficient (MCC) of 0.62. Moreover, it predicts binding residues with an accuracy of 84%, precision of 66% and MCC value of 0.51. In addition, it has a success rate of 77% in predicting RNA binding types (mRNA, tRNA or rRNA). We further demonstrate that it makes more than 10% improvement either in precision or sensitivity than PSI-BLAST, HHPRED and our previously developed structure-based technique. This method expects to be useful for highly accurate genome-scale, high-resolution prediction of RNA-binding proteins and their complex structures. A web server (SPOT) is freely available for academic users at sparks.informatics.iupui.edu.
Key words: protein-RNA complexes, structure prediction, RNA binding sites, fold recognition, binding RNA types
Introduction
Significant new interest in RNA-binding proteins (RBPs) are resulted from the discovery and characterization of microRNAs in post-transcriptional regulation and the implication of RBPs in many human diseases including HIV/AIDS, cancer and neurodegenerative disorders.1 RBPs are encoded in large number (thousands) because their diversity appears to increase during evolution of post-transcriptional machinery and the increase in number of introns. Despite of their importance, many of these RBPs are yet to be uncovered and/or characterized. Computational prediction methods are therefore essential as initial steps for function annotation and characterization.
Function prediction for RBPs can be roughly classified into four levels of resolutions with different levels of details (low, medium, high and the highest). The first low level of prediction is a simple two-state classification of binding or non-binding to RNA. The next medium level is the location of RNA binding residues of RBPs. A high resolution prediction is to predict the RNA type that the RBP would bind. This prediction would provide further deeper understanding of the RBP function. The highest resolution will involve the prediction of the actual binding RNA sequence and its binding complex structure with the predicted RBP.
Most computational methods developed so far attempted to detect the sequence homologous and/or evolutionary relationship between un-characterized and characterized proteins.2,3 The assumption made in these methods is that homologous sequences have the same biological function. However, this is true only for orthologs. Even with help of sequence homology, there is less than half of identified proteins are annotated.4 Moreover, many proteins have hidden function of RNA binding.5,6 Thus, it is necessary to develop sequence-based techniques that can detect function similarity in the absence of high sequence homology to known RBPs.
Several sequence-based classifiers for RBP prediction are based on support-vector machines (SVM) and limited to the low resolution prediction of binding or non-binding proteins.7–12 Early studies7,8 did not exclude homologous sequences from training or testing. Moreover, all these techniques were trained and tested in a balanced set with equal number of positive (RBP) and negative (Non-RBP) data sets.9–12 The reported Mathews correlation coefficient value for RBP classification is 0.53 for a sequence-based SVM classifier (5-fold cross validation on 134 RNA binding and 134 non-binding proteins)11 and 0.72 for a structure-based SVM classifier for a dataset of 76 RNA binding proteins and 246 nonnucleic-acid binding proteins (leave-one-out test).13 The performance of these techniques for such a balance set likely becomes worse when applied to a real world situation where RBP is about 15% of all proteins.14
Other methods make medium resolution prediction of RNA binding residues (or binding sites) directly based on either sequence-based11,15–22 or structure-based13,23–27 information. The best reported values for Mathews correlation coefficient are between 0.47–0.51.11,25,26 One issue associated with these techniques is that they will predict RNA binding sites even for the proteins that do not bind RNA.
This work is inspired by our structure-based prediction of DNA and RNA binding proteins [SPOT-Stru (DNA),28 SPOT-Stru (RNA) 29]. We found that structural alignment to known protein-RNA complexes coupled with binding assessment with a statistical energy function based on distance-scaled finite ideal gas reference (DFIRE) state yields a highly accurate (98%) prediction of RBPs with a reasonable sensitivity of 36% and Mathews correlation coefficient (MCC) of 0.57 for a large benchmark of 212 RNA binding and 6,761 non-RNA binding domains. Its applications on additional APO and HOLO benchmarks and structural genomics targets yielded consistent accuracy and/or sensitivity.
This structure-based technique, however, has a limited application because the structures for the majority of proteins are unknown. The success of this structure-based technique motivates us to develop a sequence-based technique by coupling structure prediction with binding prediction, an approach proven successful for protein-DNA binding prediction.30 Here we perform structure prediction by using the latest version of our fold recognition technique called SPARKS X31 that is among the best performing single automatic servers in several critical assessment of structure prediction (CASP) meetings (CASP 6,32 CASP 7,33 and CASP 931). While many template-based structure prediction methods exist, the coupling between fold recognition and binding affinity prediction provides the first dedicated high-resolution function prediction for RBPs.
The new technique, called SPOT-Seq, is initially trained and validated on a dataset of 174 RNA-binding and 5,765 nonbinding protein chains, so that it can compare to other methods. SPOT-Seq achieves the highest MCC value of 0.61, when compared to PSI-BLAST, the commonly used sequence-to-profile homology search technique34 (MCC = 0.48), HHPRED, a profile-profile fold-recognition technique based on the hidden Markov model35 (MCC = 0.50), SPARKS X fold recognition method31 (MCC = 0.57), and the structure-based prediction technique (SPOT-Stru, MCC = 0.56). More than 10% improvement in either sensitivity or precision or both are observed. Further expansion of test and training sets (431 RBPs) and template library (1,164 binding domains and chains) confirms the MCC of 0.62, accuracy of 98%, precision of 84% and sensitivity of 47%.
Result
Low resolution two-state prediction.
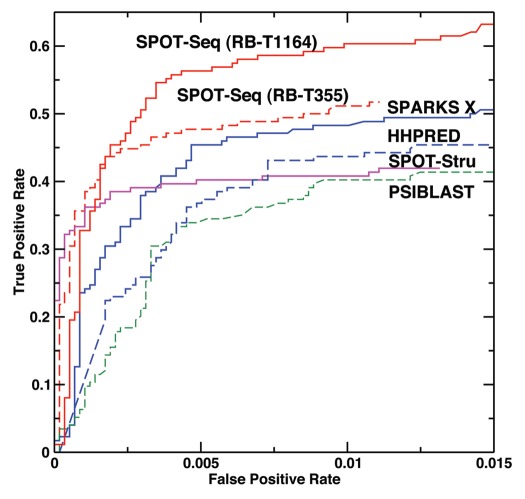
Cross validation. Figure 1 compares the performance of PSI-BLAST,34 fold recognition method HHPRED,35 SPARKS X,31 structure-based method SPOT-Stru (RNA)29 and SPOT-Seq from this work by cross validation after removing all templates with >30% sequence identity to the target. The results are also quantitatively summarized in Table 1 based on thresholds optimized for the highest Mathews correlation coefficient. These results are obtained by taking one chain sequence from either RB-C174 or NB-C5765 and predicting whether it binds or does not bind to RNA. This large unbalanced dataset with 3% binding sequences is employed to mimic real situation where binding proteins are a small portion of all proteins. Table 1 indicates that SPARKS X improves 12% over PSI-BLAST in sensitivity and 5% in precision with similar level of accuracy. On the other hand, SPARKS X improves over HHPRED35 mostly in precision (11%) at similar level of sensitivity and accuracy. The structure-based technique (SPOT-Stru), although has a much higher precision than the fold-recognition technique (SPARKS X) (94% versus 75%), but with a significantly lower sensitivity (35% versus 45%). Introduction of binding affinity prediction further improves the precision from 75% in SPARKS X to 85% in SPOT-Seq without much change in sensitivity or accuracy.
Figure 1.
True positive rate versus false positive rate as given by PSI-BLAST (Green, dashed line), SPOT-Stru (Magenta), HHPRED (Blue, dashed line), SPARKS X (Blue, Solid line) and SPOT-Seq (Red, dashed line for the RB-T355 template library and solid line for the RB-T1164 template library) for the low-resolution two-state prediction (binding vs. no binding).
Table 1.
Method comparison for predicting RNA-binding proteins
| Method | Sensitivity | Accuracy | Precision | MCC |
| PSI-BLASTa | 33% | 98% | 70% | 0.48 |
| HHPREDa | 43% | 98% | 64% | 0.51 |
| SPARKS Xa | 45% | 98% | 75% | 0.57 |
| SPOT-Stru (RNA)b | 35% | 98% | 94% | 0.56 |
| SPOT-Seq (this work)a | 45% | 98% | 85% | 0.61 |
Sequence-based method. bStructure-based method.
Discriminating binding from non-binding within the same fold. According to the SCOP annotation,38 there are 44 folds shared by both RNA-binding and non-RNA-binding proteins in RB-C174 and NB-C5765. As shown in Table 2, the majority (849/861) non-RNA binding proteins are filtered by SPARKS X while SPOT-Seq further reduces the number of false positives from 12 to 8 and leads to a very low false positive rate of 0.9%. At the mean time, SPOT-Seq increases the true positive rate to 37% (50/134) from 28% (37/134) given by SPARKS X. The result confirms that both fold recognition technique and energy calculation contributes to the power of distinguishing the RNA-binding proteins from nonbinding ones even within the same fold.
Table 2.
Examination of 44 SCOP folds shared by both RNA-binding (RB-C174) and non-binding (NB-C5765) proteins
| SCOP Fold ID | Dataset (RB/NB) | SPARKS X (RB/NB) | SPOT-Seq (RB/NB) |
| d.58 | 14/70 | 4/1 | 11/1 |
| b.40 | 11/39 | 2/0 | 1/0 |
| c.26 | 9/18 | 8/0 | 7/0 |
| a.4 | 9/96 | 1/2 | 2/4 |
| b.34 | 8/21 | 2/0 | 2/0 |
| g.41 | 6/5 | 1/0 | 1/0 |
| d.104 | 6/1 | 3/0 | 6/0 |
| c.55 | 6/62 | 3/3 | 4/0 |
| e.8 | 5/3 | 2/0 | 2/0 |
| d.79 | 5/10 | 1/0 | 2/0 |
| d.50 | 4/3 | 2/0 | 3/0 |
| b.121 | 4/26 | 0/0 | 0/0 |
| d.52 | 3/5 | 0/0 | 0/0 |
| d.41 | 3/5 | 2/0 | 3/0 |
| d.14 | 3/10 | 0/0 | 0/0 |
| b.43 | 3/10 | 2/0 | 2/0 |
| a.2 | 3/13 | 1/0 | 1/0 |
| g.39 | 2/10 | 0/0 | 0/0 |
| d.67 | 2/1 | 0/0 | 0/0 |
| d.218 | 2/7 | 0/0 | 0/0 |
| c.51 | 2/6 | 0/0 | 0/0 |
| b.122 | 2/3 | 2/0 | 2/0 |
| a.118 | 2/40 | 0/0 | 0/0 |
| d.157 | 1/8 | 0/0 | 0/0 |
| d.1 | 1/2 | 0/1 | 0/0 |
| c.97 | 1/6 | 0/2 | 0/0 |
| c.9 | 1/1 | 0/0 | 0/0 |
| c.66 | 1/27 | 1/1 | 1/1 |
| c.62 | 1/5 | 0/0 | 0/0 |
| c.52 | 1/17 | 0/0 | 0/0 |
| c.37 | 1/70 | 0/1 | 0/2 |
| c.23 | 1/41 | 0/0 | 0/0 |
| c.1 | 1/136 | 0/0 | 0/0 |
| b.82 | 1/28 | 0/0 | 0/0 |
| b.46 | 1/1 | 0/0 | 0/0 |
| b.44 | 1/1 | 0/0 | 0/0 |
| b.38 | 1/4 | 0/0 | 0/0 |
| b.2 | 1/23 | 0/1 | 0/0 |
| a.7 | 1/20 | 0/0 | 0/0 |
| a.30 | 1/3 | 0/0 | 0/0 |
| a.160 | 1/1 | 0/0 | 0/0 |
| a.156 | 1/2 | 0/0 | 0/0 |
| a.144 | 1/1 | 0/0 | 0/0 |
| a.137 | 1/7 | 0/0 | 0/0 |
| Total | 134/861 | 37/12 | 50/8 |
Medium resolution binding-residue prediction.
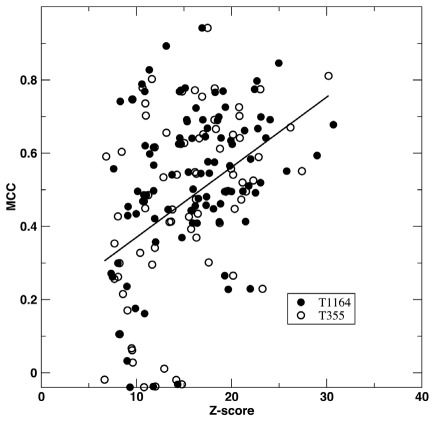
The predicted binding complex between a target and a template RNA allows us to infer RNA binding residues for the target. We define an amino-acid residue as RNA-binding if any heavy atoms of the residue are less than 4.5 Å away from any heavy atoms of a RNA base. For a few proteins, we found that it is necessary to perform crystal symmetry operation to yield correct information on binding residues. We examine the accuracy of binding-residue prediction by focusing on true positive prediction of 78 proteins from the cross validation test on RB-C174/NB-C5765. Compared to native binding residues, we achieved 53% in sensitivity, 85% in accuracy and 63% in precision. The MCC value is 0.47. This value is significantly lower than 0.72, the MCC value given by SPOT-Stru. This suggests that structural alignment allows a better detection of RNA binding regions than model complex structures, predicted by SPARKS X due to inaccuracy of models predicted. In other words, SPARKS X improves over SPOT-Stru in sensitivity of detecting RNA-binding proteins (low resolution prediction) while reducing the accuracy of predicting binding regions (medium resolution prediction). Figure 2 displays 78 MCC values (open circles) for predicted binding residues as a function of Z-score. Clearly, there is a trend that higher Z-scores (high confidence in the accuracy for the model structure) leads to higher MCC values. However, there exist a few proteins with poorly predicted binding regions when Z-score <15.
Figure 2.
Medium resolution prediction of RNA-binding sites. MCC values for predicted RNA-binding residues are shown as a function of fold recognition Z-scores. Results of RB-C174 tested on small and expanded template libraries of RB-T355 (open circles) and RB-T1164 (closed circles) are shown. The line from linear regression is employed to illustrate the trend.
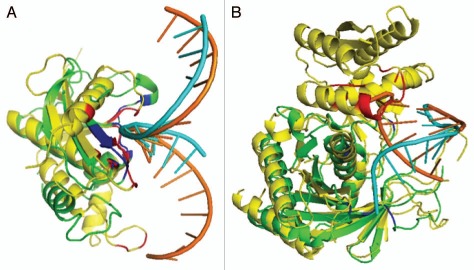
Figure 3 shows two examples: one with a reasonable prediction of binding residues but the other with a poor prediction. For the human Rnase H1 (target 2qk9A, Fig. 3A), predicted (orange) and actual (magenta) RNA structures are located in a similar location, the predicted binding region (in Blue) is also close to the native binding region (in Red). The MCC value for predicted binding residues is 0.65 with a sensitivity of 97% and an accuracy of 93%. However, the predicted and actual RNA structures for the target A. fulgidus Piwi protein (PDB ID# 1ytuB, Fig. 3B) are different. The native structure binds with double helix RNA and the binding residues are represented as red, but the predicted structure based on the template (3f73A) binds with a single strand RNA that only partially overlaps with native RNA structure. This leads to wrongly predicted binding residues (in blue). This error is likely caused by the fact that predicted protein structure (green) for 1ytuB is only a part of the actual native structure.
Figure 3.
Comparison between the predicted (green) and actual (yellow) complex structures for two targets with RNA structures colored in cyan for predicted and orange for native RNA structure and binding regions colored in Red for native structure and Blue for predicted structure. (A) Target 2qk9A predicted with template 1zbiB (sequence identity between them is 13%). (B) Target 1ytuB predicted with template 3f73A3 (sequence identity between them is 2.0%).
High resolution prediction of binding RNA types.
The next resolution level of function prediction is to predict the types of RNA that bind to the target protein. We manually classified the types of RNA included in our template library, according to the annotation of DAVID.41 In the template library (RBT-355), 272 are annotated into 5 types of RNA-binding proteins. There are 189 proteins binding with tRNA, 148 binding with rRNA, 47 binding with mRNA, 25 binding with synthetic RNA and 7 binding with SRP RNA. Because some RNAs have more than one function, the total number of proteins with annotated RNA binding types is less than the number of RNAs grouped according to function.
The ability of our method to predict the type of binding RNA is examined by analyzing 78 true positives (RNA-binding domains). These 78 RNA-binding domains contain 48 tRNA-binding proteins, 34 rRNA-binding proteins, 10 mRNA-binding proteins, 3 synthetic RNA-binding protein, 3 SRP RNA-binding proteins. If we use the template RNA in the predicted complex structure to predict the binding RNA type for the target protein, we achieve success rates of 90% (43/48) for tRNA, 91% (31/34) for rRNA and 70% (7/10) for mRNA. Table 3 listed all mis-predicted RNA types. They are between tRNA, rRNA and mRNA.
Table 3.
Mis-predicted binding types for tRNA, mRNA and rRNA-binding proteins
| Native tRNA | Pred. Type | Native mRNA | Pred. Type | Native rRNA | Pred. Type |
| 1jj2U | rRNA | 1yz9A | - | 1mzpA | - |
| 1mzpA | -a | 2gxbB | - | 1yz9A | - |
| 1ytyA | mRNA | 2ozbA | tRNA | 2bh2A | tRNA |
| 2i82A | rRNA | 2rfkA | tRNA | ||
| 3bt7A | rRNA |
The RNA type in the template is not annotated.
The highest resolution: protein-RNA complex structure.
To examine the quality of predicted structures, we used TM-Score from TM-Align42 to compare the native and predicted structures which is 1 for perfect agreement and about 0.2 between two random structures. For 78 correctly predicted targets, the average TM-Score is 0.73. One can also measure the structure similarity by the fraction of residues in model structure has an root-mean-squared distance (RMSD) of 4 Å or less. We found that the medium value is 72%. We found that one structure for the target 2j035 (50S ribosomal protein L31) was predicted poorly (TM-Score < 0.4 and only 22% residues has RMSD < 4 Å). This large error in predicted structure is caused by the non-globular shape of the native structure (a small 59 residue protein with a radius of gyration of 23.4 Å). We further found that the structural accuracy of binding regions is higher than that of whole proteins. For example, the binding regions of 15 targets have more than 95% residues with RMSD < 5 Å. By comparison, only 8 targets satisfy the same criterion for the whole protein.
As an illustrative example, Figure 3A showed the predicted and actual complex structures with RNA for target 2qk9A (human Rnase H1). The template 1zbiB (Bacillus halodurans RNase H catalytic domain) was located with a Z-score of 18.0 and the binding energy of −1.62. In this example, 50% aligned residues of native structure and predicted structure has RMSD < 4 Å, much lower than the medium value of 72%. This is largely due to a helix near binding region in the template, but only a coil in the native structure. Yet, the binding region is reasonably accurately modeled based on the proximity of blue and red colors (a MCC value of 0.65, a sensitivity of 97% and an accuracy of 93%). This remote homologous template is identified despite of a low sequence identity of 13.3%. In this example, the conformation of RNA is also modeled correctly. For Figure 3B, only a portion of the target A. fulgidus Piwi protein (PDB ID# 1ytuB) is predicted. This portion was predicted with a TM-Score of 0.75. The sequence identity between the target and template (3f73A3) is 2.0%.
Discrimination against DNA binding proteins.
We tested the ability of SPOT-Seq for separating RNA and DNA binding proteins by applying the method to the dataset of 250 non-redundant DNA binding proteins (DBPs) collected by us previously in reference 28. We employed the thresholds for Z-score and binding affinity obtained by optimizing the MCC value for RB-C174/NB-C5765. Only 5 out of 250 DBPs are predicted as RBPs. Among these 5 predicted RBPs, four have high sequence identity (>77%) with the templates and are known for binding with both RNA and DNA. The remaining target 1sfuA (the viral Zalpha domain43) is a remote homolog of the template 2gxbB with a sequence identity of 27.1%. 1sfuA was also predicted as RNA-binding proteins in previous structure-based study.29 This poxvirus protein is E3L protein that has a Z-alpha motif similar with ADAR1 (double-stranded RNA adenosine deaminase) which is known to bind with Z-RNA.44,45 Thus, there is no false positive from DNA-binding proteins.
Effect of the expanded template library.
Table 4 examines the effect of the expanded template library at all four levels of prediction resolutions. It is clear that expanding template library from 355 to 1,164 protein domains and chains improves sensitivity from 45% to 56% at the expense of reducing precision from 85% to 81%. The effect on the ROC curve can be found in Figure 2. SPOT-Seq with RB-T355 has a higher sensitivity (or true positive rate) only at a very low false positive rate (<0.2%) while SPOT-Seq with RB-T1164 has a higher sensitivity at low to moderate false positive rates. The overall MCC value increases from 0.61 to 0.67 due to the expanded library.
Table 4.
SPOT-Seq performance for an expanded template library from T355 to T1164, an independent test C257 and two equally divided training and test sets (C216 and C215)
| Resolution | T355a | T1164a | |||
| Level | C174b | C174b | IC257b | C216b | C215b |
| Two-statec | |||||
| MCC | 0.61 | 0.67 | 0.60 | 0.62 | 0.62 |
| Accuracy | 98% | 98% | 97% | 98% | 98% |
| Precision | 85% | 81% | 82% | 84% | 84% |
| Sensitivity | 45% | 56% | 45% | 48% | 47% |
| Binding Residued | |||||
| MCC | 0.47 | 0.53 | 0.48 | 0.50 | 0.51 |
| Accuracy | 85% | 85% | 83% | 84% | 84% |
| Precision | 63% | 69% | 63% | 66% | 66% |
| Sensitivity | 53% | 60% | 58% | 59% | 60% |
| RNA-typee | |||||
| tRNA | 90% | 67% | 67% | 62% | 69% |
| (43/48) | (46/69) | (24/36) | (33/53) | (33/48) | |
| mRNA | 70% | 82% | 62% | 73% | 56% |
| (7/10) | (9/11) | (24/39) | (16/22) | (15/27) | |
| rRNA | 91% | 92% | 91% | 91% | 96% |
| (31/34) | (48/52) | (61/67) | (52/57) | (54/56) | |
| Complex Structuref | |||||
| TM-Score | 0.73 | 0.69 | 0.66 | 0.66 | 0.66 |
| RMSD(<4Å) | 72% | 78% | 76% | 76% | 77% |
| # (Whole) | 10% | 16% | 17% | 15% | 17% |
| #(Binding) | 19% | 33% | 25% | 29% | 25% |
The template sets of 355 and 1164 RBPs, respectively.
The target sets of C174 for training and cross validation, C257 for independent test. C174 and C257 are further randomly separated into C216 for training and cross validation and C215 for independent test.
Performance on low-resolution two-state prediction based on Mathews correlation coefficient and others.
Performance on medium-resolution prediction of RNA binding residues based on Mathews correlation coefficient and others.
Success rate of the high resolution prediction of bound RNA types (tRNA, mRNA and rRNA): the fraction of correctly predicted RNA binding types in actual number of proteins in that type.
The highest resolution of complex structure prediction based on the average structural similarity score (TM-Score), medium value for the percentage of aligned residues in the model structure with RMSD <4Å from the native structure, percentage of targets with 95% predicted residues within RMSD <5 Å from the native residues for the whole protein and binding regions only.
For binding residue prediction, expanding templates improve both precision (from 63% to 69%) and sensitivity (from 53% to 60%) without change to accuracy. This leads to an improved MCC value from 0.47 to 0.53. Figure 2 compared the MCC values as a function of Z-Score given by different template libraries. Expanding templates reduces the number of poorly predicted binding regions (MCC < 0.2) from 10 to 7.
The effect of the enlarged template library on prediction of RNA types is mixed. There is a reduction of success rate from 90% (43/48) to 67% (46/69) for tRNA, improved success rate from 70% (7/10) to 82% (9/11) for mRNA and unchanged success rate [91% (31/34) versus 92% (48/52)] for rRNA. This large fluctuation suggests that the dataset may be too small to assess the accuracy of RNA type prediction. The overall success rates are essentially the same (77% for RB-T355 and 78% for RB-T1164). We further examined the prediction ability on the highest resolution of protein-RNA complexes. We found that the average TM-score is reduced from 0.73 to 0.69 while the medium value for the fraction of residues with RMSD < 4 Å increases from 72% to 78%. This somewhat conflict result reveals the difficulty to consistently assess the quality of predicted structures.
Independent test.
Table 4 also displays the results of independent test on RB-IC257 based on the thresholds generated by the cross validation set of RB-C174/NB-C5765 with the template library of RB-T1164. Overall speaking, there is a somewhat reduction of performance in the two-state prediction (the MCC value reduced from 0.67 to 0.60). The most reduction is in the sensitivity from 56% to 45%. This reduction of sensitivity is somewhat expected because the RB-IC257 set contains low resolution X-ray structures and NMR structures. The performance of binding residue prediction for the independent test set is also reduced in accuracy (1%), precision (6%) and sensitivity (2%). The accuracy of predicted complex structures also decreases somewhat (TM-Score from 0.69 to 0.66 and the fraction of residues with RMSD < 4 Å from 78% to 76%). We hypothesize that the poorer performance for RB-IC257 may be because it was compiled by including low resolution X-ray structures, EM structures and NMR structures and recently solved structures. These structures lead to less accurate model structures.
To verify this hypothesis, we randomly divided to RB-IC257 and RB-C174 into two independent sets of RB-C216 and RB-C215. We first employed RB-C216/T1164 to train the thresholds and found that these thresholds are nearly identical to those trained by RB-C174/T1164 [(8.04, −0.565) vs. (7.47, −0.561) for Z-score and energy thresholds]. Then, we applied these thresholds to RB-C215. The results are shown in Table 4. Indeed, we found that the results on RB-C216 and RB-C215 are essentially the same at every resolution level of function prediction except expected fluctuation for RNA-type prediction due to the small number of annotated RNA binding types. For example, the MCC value for the two-state prediction is 0.62 for training or test while the MCC values for the binding residue prediction are 0.50 and 0.51 for training and test, respectively.
Discussion
In this paper, we described the first technique that provides prediction of RNA binding proteins at all four levels of resolution. At the low resolution level of two-state prediction, its MCC value based on a large dataset of 216 binding proteins (or independent 215 binding proteins) and 5,765 non-binding proteins is 0.62 (0.62). This value is higher than 0.53, the best reported, sequencebased SVM classifier method (5-fold cross validation on 134 RNA binding and 134 non-binding proteins only).11 Its MCC values for the medium resolution prediction of RNA-binding residues [0.50 (0.51)] for RB-C216 (RB-C215) sets are comparable to 0.47 given by the same SVM classifier.11 More importantly, the high-resolution prediction of binding RNA types and binding complex structures are highly reliable. The overall success rates are 77% and 78% for training and test, respectively. The average TM-score for predicted structures is 0.66 (0.66 also for RB-C215). One important feature of SPOT-Seq is its ability to separate RNA from DNA binding proteins. It yields zero false positives when applied to 250 DNA binding proteins.
We would like to emphasize that we have purposely trained and tested SPOT-Seq in entire chains of proteins, rather than protein domains. This is to mimic the real-world situation that in most cases, protein domain boundaries are unknown. SPOT-Seq will allow direct identification of RNA-binding domains from the target chain as it searches for the best matching domain and/or chain from the template library.
SPOT-Seq has one obvious limitation. It relies on the availability of protein-RNA complexes as templates. It will not be able to predict RNA-binding proteins whose structures do not have a template in the template library or when its template in the library is difficult to recognize. We have used the RB-T355 library that includes both domains and chains with 95% sequence-identity cutoff for the purpose of maximizing available templates. The low sensitivity (45%) is in part due to lack of structurally matching templates. Although expanding the number of templates from T355 to T1164 improves sensitivity, it reduces precision at the same time because a low resolution RBP structure will more likely make a false match to a non-binding structure. More importantly, tripling the number of templates from 355 to 1,164 does not expand the structural space as much. For example, in the RB-IC257 set, there are 141 false negatives that have 52 targets with TM-score > 0.5 to the structures in T355. The number of structurally similar templates only increases by 24 to 76 targets when the number of templates expands to 1,164. It is clear that significantly more high-quality, non-redundant protein-RNA complex structures are needed with the current method in order to further advance the sensitivity and precision at the same time.
Knowing the fraction of correct predictions in all predictions (precision) is important. The final precision of 84% based on optimized MCC value on 216 positives and 5,765 negatives is likely a low estimation for the actual precision when applying to a genome because our non-binding proteins are significantly more than the number of our binding proteins. In fact, when including all 431 positives with 5,765 negatives (7% positives, still smaller than estimated 15% positives in a genome) as a single set, one would have a precision of 91% because the number of false positives20 does not change. Moreover, a close examination of those 20 false positives indicates that at least two are likely true positives. One is mitochondrial aconitate hydratase in Bos taurus (Bovine) (chain A in PDB ID #1c96). Although this protein is not annotated as RNA binding, aconitate hydratase 1 of E. Coli was found to bind with mRNA 3′-UTR binding.46 The second one is human Cytotoxic T-lymphocyte protein 4 (CTLA4) where RNA aptamer inhibitors were found recently in reference 47. More importantly, one half of 20 false positives are also identified as positive by our structure-based SPOT-Stru method. Thus, we expect that our SPOT-Seq has a precision close to 90% and the application of our method for genome-wide prediction will lead to highly accurate prediction results.
Finally, one important advantage of this SPOT technique is its reasonable speed. For example, it only takes 1,107 CPU hours (46 days) on a single processor PC to scan about 7,380 genes in yeast genomes. We will report these results in a separate paper. A freely available, easy-to-use webserver (SPOT) is available for academic users at sparks.informatics.iupui.edu.
Methods
Function prediction protocol.
The method proposed here is similar to the structure-based technique called SPOT-Stru (RNA) developed by us29 except that the structure is predicted by fold recognition technique called SPARKS X.31 The flow diagram is shown in Figure 4.
Figure 4.
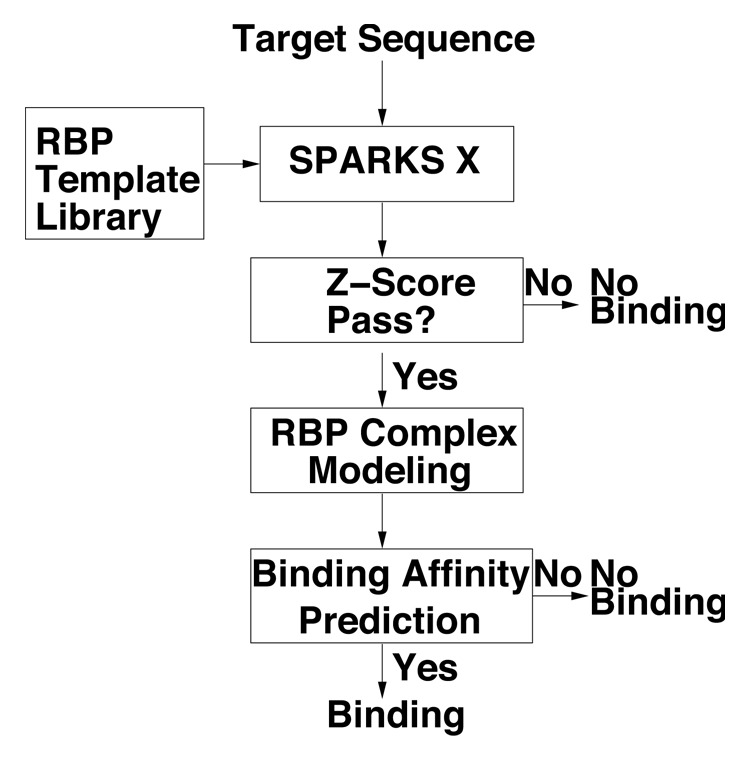
The flow diagram of the sequence-based prediction of RNA binding proteins (RBPs).
First, we perform fold recognition between the target sequence and templates in the template library of RBPs by SPARKS X.31 Our template library is built on a collection of RNA binding and non-binding proteins (see the section on Test/cross-validation datasets). SPARKS X31 attempts to match the sequence profile of the target sequence (generated from PSI-BLAST34) and predicted one-dimensional structural profiles (secondary structure, solvent accessibile surface area and backbone torsion angles from SPINE X36) to the corresponding profiles of the template structures in the library. The sequence-structure matching score is measured by Z-Score where
| (1) |
where Si is the alignment raw score between target and template i and Smean and σ are the average raw score and the standard deviation for all templates. Typically, a Z-score of higher than 6.0 is considered as a significant template hit.
If the Z-score for any of RBP templates (after removing all templates >30% sequence identity) is higher than a threshold to be determined, a complex structure of the target protein and template RNA is built by replacing template protein sequence with target protein sequence based on the sequence-to-structure alignment from SPARKS X. For this study, the gap region is not modeled for simplicity.
Using the complex structure of model target and template RNA we can estimate the binding affinity according to a statistical energy function based on the distance-scaled finite ideal-gas reference state37 that was extended to protein-RNA interaction (DRNA).29 In this work, we made no changes to the DRNA energy function. However, the binding affinity is evaluated with mainchain atoms and Cβ atoms only to avoid the need to build sidechains in this initial development of the technique. If the binding affinity is higher than a to-be-determined threshold, the target protein is predicted as RNA binding and its complex structure model serves as the basis for the high-resolution prediction of RNA binding function. For convenience, we shall label our method as SPOT-Seq.
Template library.
For comparison, we initially employ both binding and non-binding chains from the structure-based method SPOT-Stru (RNA).29 These 225 high-resolution RNA-binding protein chains are protein-RNA complex structures (the July 2009 release). They are divided into domains according to SCOP annotations38 or by automatic technique called DDOMAIN39 if SCOP annotations are not available. A domain is RNA-binding if it has at least five amino acid residues whose heavy atoms are within 4.5 Å from any heavy atoms of nucleotide functional groups. Redundancy in resulting domains is removed by using BLASTClust with 95% sequence identity cutoff.34 This leads to 250 domains as binding templates. To increase sensitivity, both original chains and domains are included in our template library and lead to a final template library of 355 RNA-binding protein structural templates (RB-T355). Non-binding templates are from the non-binding protein-domain sets of 6,761 domains obtained previously based on 25% sequence identity cutoff.29 We only include the original chains into the template library with a 25% sequence identity cutoff. The final number of templates after a 25% sequence identity cutoff is 5,765 (NB-C5765).
Test/cross validation datasets.
RB-C174 and NB-C5765. We built a cross-validation data set of RNA-binding sequences by removing redundant sequences of all sequences contained in RB-T355 with BLASTClust34 at a sequence identity 25% cutoff. A total of 174 sequences (RB-C174) remained. Only full chains (not domains) (RB-C174 for positive and NB-C5765 for negative sets) are employed for cross validation.
Expanded template library and independent test set.
The above template library was based on high-resolution X-ray structure (3 Å or less) on July 2009. To examine the effect for an expanded template library and provide an independent test set, we downloaded all pdb structures that contains RNA and protein on April 1, 2011. After removing the structures contained in the template library, we obtained 1,027 complex structures that are separated into chains and domains according to SCOP38 or DDOMAIN39 classifications. After removing domains with less than 60 residues or having less than 5 binding residues and redundant domains with more than 95% sequence identity by BLASTClust, we obtained 612 domains in addition to 250 domains previously obtained. Both domains and their respective chains are included in our new expanded library with a total of 1,164 templates including RB-T355. We shall label this library as RB-T1164.
There are a total of 566 chains contained in the new template library. These sequences are clustered with BLASTClust at a sequence identity of 25% cutoff among themselves and the sequences contained in RB-C174. This leads to an independent test dataset of 257 chains (RB-IC257). However, this independent test set cannot be considered as representative because it contains both high and low resolution structures. Thus, we randomly divide RB-C174 and RB-IC257 into two equal sets of 216 and 215 chains, respectively (RB-C216 and RB-C215). One will be used for final training and one for final testing.
Performance evaluation.
The performance of the method is evaluated by sensitivity [SN = TP/(TP + FN)], specificity [Sp = TN/(TN + FP)], accuracy [AC = (TP + TN)/(TP + FN + TN + FP)], precision [PR = TP/(TP + FP)], and Matthews correlation coefficient (MCC)
| (2) |
Here, TP, TN, FP, FN refer to true positives, true negatives, false positives and false negatives, respectively. A MCC value provides an overall assessment of the method performance with 1 for perfect agreement. One should note that sensitivity can also be called as coverage of true positive prediction while precision is fraction of corrected predictions in all positive predictions.
Other methods and threshold optimizations.
PSI-BLAST is employed for searching homologous sequences by searching against the NCBI non-redundant sequence library for four iterations. If a target has at least one template from RB-T355 with an E-value lower than a to-be-determined threshold, the target is considered as a RNA-binding protein. Any templates having >30% sequence identity with the target sequence is removed. The threshold is optimized by maximizing the MCC value.
SPARKS X is a method without the steps for building the complex structure and prediction of binding affinity in Figure 1. Z-Score threshold, optimized by maximizing the MCC value, is 7.
To assess the ability to detect RNA-binding proteins of SPARKS X, relative to other fold-recognition methods, we employed HHPRED as an example because it is one of the best fold-recognition techniques in CASP.40 HHPRED version 1.5.1 was downloaded from toolkit.tuebingen.mpg.de/hhpred/. Default parameters, options and scripts were used to generate HMM profiles for both targets and template proteins. We also tested the option “-mact” and results are essentially the same. Probability was used as a significant score in the prediction.
Two thresholds of Z-score and binding affinity for SPOT-Seq (i.e., SPARKS X + DRNA) are optimized by a grid-based search for the highest MCC value. The binding affinity threshold is obtained by considering the lowest energy value at different Z-scores of a given target. For the prediction of RNA-binding proteins, the Z-score threshold is 6.6 and the energy threshold is −0.28. For the expanded template library (RB-T1164), the Z-score threshold is 7.47 and the energy threshold is −0.561, respectively. This was optimized based on the dataset of RB-C174 and NB-C5765. A larger template library leads to stricter Z-score and energy thresholds to prevent false positives, as expected. The same thresholds are applied to independent test set of RB-IC257.
Acknowledgment
This work was supported by the National Institutes of of Health [grant R01 GM 085003].
References
- 1.Lukong KE, Chang KW, Khandjian EW, Richard S. RNA-binding proteins in human genetic disease. Trends Genet. 2008;24:416–425. doi: 10.1016/j.tig.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Bork P, Koonin EV. Predicting functions from protein sequences—where are the bottlenecks? Nat Genet. 1998;18:313–318. doi: 10.1038/ng0498-313. [DOI] [PubMed] [Google Scholar]
- 3.Friedberg I. Automated protein function prediction—the genomic challenge. Brief Bioinform. 2006;7:225–242. doi: 10.1093/bib/bbl004. [DOI] [PubMed] [Google Scholar]
- 4.Perez-Iratxeta C, Palidwor G, Andrade-Navarro MA. Towards completion of the earth's proteome. EMBO Rep. 2007;8:1135–1141. doi: 10.1038/sj.embor.7401117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsvetanova NG, Klass DM, Salzman J, Brown PO. Proteome-wide search reveals unexpected RNA-binding proteins in saccharomyces cerevisiae. PLoS ONE. 2010;5:12671. doi: 10.1371/journal.pone.0012671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scherrer T, Mittal N, Janga SC, Gerber AP. A screen for RNA-binding proteins in yeast indicates dual functions for many enzymes. PLoS ONE. 2010;5:15499. doi: 10.1371/journal.pone.0015499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai YD, Lin SL. Support vector machines for predicting rRNA-, RNA- and DNA-binding proteins from amino acid sequence. Biochim Biophys Acta. 2003;1648:127–133. doi: 10.1016/s1570-9639(03)00112-2. [DOI] [PubMed] [Google Scholar]
- 8.Han LY, Cai CZ, Lo SL, Chung MCM, Chen YZ. Prediction of RNA-binding proteins from primary sequence by a support vector machine approach. RNA. 2004;10:355–368. doi: 10.1261/rna.5890304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu X, Cao J, Cai Y, Shi T, Li Y. Predicting rRNA-, RNA- and DNA-binding proteins from primary structure with support vector machines. J Theor Biol. 2006;240:175–184. doi: 10.1016/j.jtbi.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 10.Shao X, Tian Y, Wu L, Wang Y, Jing L, Deng N. Predicting DNA- and RNA-binding proteins from sequences with kernel methods. J Theor Biol. 2009;258:289–293. doi: 10.1016/j.jtbi.2009.01.024. [DOI] [PubMed] [Google Scholar]
- 11.Spriggs RV, Murakami Y, Nakamura H, Jones S. Protein function annotation from sequence: prediction of residues interacting with RNA. Bioinformatics. 2009;25:1492–1497. doi: 10.1093/bioinformatics/btp257. [DOI] [PubMed] [Google Scholar]
- 12.Kumar M, Gromiha MM, Raghava GPS. SVM based prediction of RNA-binding proteins using binding residues and evolutionary information. J Mol Recognit. 2011;24:303–313. doi: 10.1002/jmr.1061. [DOI] [PubMed] [Google Scholar]
- 13.Shazman S, Mandel-Gutfreund Y. Classifying RNA-Binding Proteins Based on Electrostatic Properties. PLOS Comput Biol. 2008;4:1000146. doi: 10.1371/journal.pcbi.1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.UniProt Consortium, author. The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:190–195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeong E, Chung I, Miyano S. A neural network method for identification of RNA-interacting residues in protein. Genome Inform. 2004;15:105–116. [PubMed] [Google Scholar]
- 16.Wang L, Brown SJ. BindN: a web-based tool for efficient prediction of DNA and RNA binding sites in amino acid sequences. Nucleic Acids Res. 2006;34:243–248. doi: 10.1093/nar/gkl298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terribilini M, Lee JH, Yan C, Jernigan RL, Honavar V, Dobbs D. Prediction of RNA binding sites in proteins from amino acid sequence. RNA. 2006;12:1450–1462. doi: 10.1261/rna.2197306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Terribilini M, Sander JD, Lee JH, Zaback P, Jernigan RL, Honavar V, et al. RNABindR: a server for analyzing and predicting RNA-binding sites in proteins. Nucleic Acids Res. 2007;35:578–584. doi: 10.1093/nar/gkm294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Xue Z, Shen G, Xu J. PRINTR: Prediction of RNA binding sites in proteins using SVM and profiles. Amino Acids. 2008;35:295–302. doi: 10.1007/s00726-007-0634-9. [DOI] [PubMed] [Google Scholar]
- 20.Cheng CW, Su ECY, Hwang JK, Sung TY, Hsu WL. Predicting RNA-binding sites of proteins using support vector machines and evolutionary information. BMC Bioinformatics. 2008;9:6. doi: 10.1186/1471-2105-9-S12-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar M, Gromiha AM, Raghava GPS. Prediction of RNA binding sites in a protein using SVM and PSSM profile. Proteins. 2008;71:189–194. doi: 10.1002/prot.21677. [DOI] [PubMed] [Google Scholar]
- 22.Tong J, Jiang P, Lu Zh. RISP: A web-based server for prediction of RNA-binding sites in proteins. Comput Methods Programs Biomed. 2008;90:148–153. doi: 10.1016/j.cmpb.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 23.Chen YC, Lim C. Predicting RNA-binding sites from the protein structure based on electrostatics, evolution and geometry. Nucleic Acids Res. 2008;36:29. doi: 10.1093/nar/gkn008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maetschke SR, Yuan Z. Exploiting structural and topological information to improve prediction of RNA-protein binding sites. BMC Bioinformatics. 2009;10:341. doi: 10.1186/1471-2105-10-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Q, Cao Z, Liu H. Improve the prediction of RNA-binding residues using structural neighbours. Protein Pept Lett. 2010;17:287–296. doi: 10.2174/092986610790780279. [DOI] [PubMed] [Google Scholar]
- 26.Liu ZP, Wu LY, Wang Y, Zhang XS, Chen L. Prediction of protein-RNA binding sites by a random forest method with combined features. Bioinformatics. 2010;26:1616–1622. doi: 10.1093/bioinformatics/btq253. [DOI] [PubMed] [Google Scholar]
- 27.Perez-Cano L, Fernandez-Recio J. Optimal Protein-RNA Area, OPRA: A propensity-based method to identify RNA-binding sites on proteins. Proteins. 2010;78:25–35. doi: 10.1002/prot.22527. [DOI] [PubMed] [Google Scholar]
- 28.Zhao H, Yang Y, Zhou Y. Structure-based prediction of DNA binding proteins. Bioinformatics. 2010;26:1857–1863. doi: 10.1093/bioinformatics/btq295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao H, Yang Y, Zhou Y. Structure-based prediction of RNA-binding domains and RNA-binding sites and application to structural genomics targets. Nucleic Acids Res. 2011;39:3017–3025. doi: 10.1093/nar/gkq1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao M, Skolnick J. A threading-based method for the prediction of DNA-binding proteins with application to the human genome. PLOS Comput Biol. 2009;5:1000567. doi: 10.1371/journal.pcbi.1000567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Y, Faraggi E, Zhao H, Zhou Y. Improving protein fold recognition and template-based modelling by employing probabilisic-based matching between predicted one-dimensional structural properties of the query and corresponding native properties of templates. Bioinformatics. 2011;27:2076–2082. doi: 10.1093/bioinformatics/btr350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou H, Zhou Y. SPARKS 2 and SP3 servers in CASP 6. Proteins. 2005;61:152–156. doi: 10.1002/prot.20732. [DOI] [PubMed] [Google Scholar]
- 33.Liu S, Zhang C, Liang S, Zhou Y. Fold recognition by concurrent use of solvent accessibility and residue depth. Proteins. 2007;68:636–645. doi: 10.1002/prot.21459. [DOI] [PubMed] [Google Scholar]
- 34.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:244–248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Faraggi E, Yang Y, Zhang S, Zhou Y. Predicting continuous local structure and the effect of its substitution for secondary structure in fragment-free protein structure prediction. Structure. 2009;17:1515–1527. doi: 10.1016/j.str.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou H, Zhou Y. Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Sci. 2002;11:2714–2726. doi: 10.1110/ps.0217002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 39.Zhou H, Xue B, Zhou Y. DDOMAIN: Dividing structures into domains using a normalized domain-domain interaction profile. Protein Sci. 2007;16:947–955. doi: 10.1110/ps.062597307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hildebrand A, Remmert M, Biegert A, So ding J. Fast and accurate automatic structure prediction with HHpred. Proteins. 2009;77:128–132. doi: 10.1002/prot.22499. [DOI] [PubMed] [Google Scholar]
- 41.Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for annotation, visualization and integrated discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- 42.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ha SC, Lokanath NK, Quyen DV, Wu CA, Lowenhaupt K, Rich A, et al. A poxvirus protein forms a complex with left-handed Z-DNA: crystal structure of a Yatapoxvirus Zalpha bound to DNA. Proc Natl Acad Sci USA. 2004;101:14367–14372. doi: 10.1073/pnas.0405586101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herbert A, Alfken J, Kim YG, Mian IS, Nishikura K, Rich A. A Z-DNA binding domain present in the human editing enzyme, double-stranded RNA adenosine deaminase. Proc Natl Acad Sci USA. 1997;94:8421–8426. doi: 10.1073/pnas.94.16.8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Placido D, Brown BA, Lowenhaupt K, Rich A, Athanasiadis A. A left-handed RNA double helix bound by the Zalpha domain of the RNA-editing enzyme ADAR1. Structure. 2007;15:395–404. doi: 10.1016/j.str.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang Y, Guest JR. Direct evidence for mRNA binding and post-transcriptional regulation by Escherichia coli aconitases. Microbiology. 1999;145:3069–3079. doi: 10.1099/00221287-145-11-3069. [DOI] [PubMed] [Google Scholar]
- 47.Santulli-Marotto S, Nair SK, Rusconi C, Sullenger B, Gilboa E. Multivalent RNA aptamers that inhibit CTLA-4 and enhance tumor immunity. Cancer Res. 2003;63:7483–7489. [PubMed] [Google Scholar]