Abstract
Bruno protein binds to multiple sites—BREs—in the oskar mRNA 3′ UTR, thereby controlling oskar mRNA translation. Bruno also binds and regulates other mRNAs, although the binding sites have not yet been defined. Bruno has three RRM type RNA binding motifs, two near the amino terminus and an extended RRM at the C terminus. Two domains of Bruno—the first two RRMs (RRM1+2), and the extended RRM (RRM3+)—can each bind with specificity to the oskar mRNA regulatory regions; these and Bruno were used for in vitro selections. Anti-RRM3+ aptamers include long, highly constrained motifs, including one corresponding to the previously identified BRe. Anti-RRM1+2 aptamers lack constrained motifs, but are biased towards classes of short and variable sequences. Bruno itself selects for several motifs, including some of those bound by RRM3+. We propose that the different RNA binding domains allow for combinatorial binding, with extended Bruno binding sites assembled from sequences bound by the individual domains. Examples of such sites were identified in known targets of Bruno, and shown to confer Bruno-dependent translational repression in vivo. Other proteins with multiple RRMs may employ combinatorial binding to achieve high levels of specificity and affinity.
Key words: RNA binding, translational regulation, RRM, Bruno, combinatorial binding
Introduction
Recognition of RNA sequences or structures by proteins is crucial for many aspects of RNA function. For gene expression, proteins bind to mRNAs in the nucleus to mediate their processing, splicing and nucleocytoplasmic transport. In the cytoplasm RNA binding proteins are required for translation, and contribute to a wide range of post-transcriptional control events including subcellular localization, translational repression and activation, and mRNA decay and stabilization. The selective nature of these regulatory events—not all mRNAs are treated the same—demands a means for the specific recognition of individual mRNAs. Each such mRNA is expected to contain appropriate cis-acting regulatory elements, with regulatory proteins binding with specificity to the elements.1 In practice, while there are examples of proteins binding with high specificity to well-defined elements, many RNA binding proteins appear to have relatively broad specificity. For example, polypyrimidine tract binding protein (PTB) recognizes sequences rich in U and C, but with no strict requirement for a specific sequence.2,3 Similarly, hnRNP proteins typically recognize sequences with only moderately constrained sequences.4 For proteins with broad binding specificity, the ability to selectively regulate a small fraction of the large population of cellular mRNAs presents a challenge.
RNA binding is mediated by a variety of protein domains.5,6 The most common is the RNA recognition motif or RRM. This domain is found in many proteins, with about 2% of human proteins containing one or more RRMs.6,7 RRM domains typically bind single stranded RNA, although there are examples of binding to single stranded DNA or proteins. The RRM consists of four anti-parallel beta strands and two alpha helices, arranged in an alpha/beta sandwich. Contacts with RNA occur along the beta sheet, with side chains stacking with the bases. Interactions between the core RRM and RNA typically span up to four nucleotides, providing only limited specificity in binding.8 Many RRMs rely on additional structural features to expand the RNA binding surface and increase specificity and affinity. These additional features vary considerably among different proteins, and involve additions to the N- or C-termini or expansion of loops connecting the secondary structure elements.9
Often, proteins contain two of more copies of the RRM. The multiple RRMs can be adjacent to one another, joined by short linkers or separated from one another. The presence of multiple RRMs raises questions about how they contribute to RNA binding. Structural studies with tandem RRMs revealed that the use of two RRMs increases the size of the RNA sequence recognized, with two themes for how this is accomplished. In the first, the two RRMs interact with a single region of RNA. Tandem RRMs of the Sex lethal and HuD proteins create a cleft in which the RNA lies, the RRMs of nucleolin form a sandwich with the RNA in the middle, and two RRMs of the poly(A) binding protein (PABP) form an extended binding platform.10–13 Alternatively, tandem RRMs bind to sequences separated from one another in the mRNA. This option is displayed by the two carboxyl terminal RRMs (RRMs 3 and 4) of polypyrimidine tract binding protein (PTB). Interactions between the two RRMs position the RNA binding surfaces apart from one another, and the RNA substrate must contain a spacer between its binding sites for high affinity binding.14 In the case of proteins with RRMs separated from one another, it remains unclear how the different RRMs bind in the context of the intact protein.
Bruno (Bru) is an RRM protein from D. melanogaster.15 Bru binds to the oskar (osk) mRNA to repress its translation. Bru binds primarily to two regions of the osk 3′ UTR, the AB and C regions. Sequence alignment and mutagenesis studies identified consensus Bru binding sites, BREs (Bru Response Elements), within these regions. Mutation of all BREs in the osk mRNA greatly reduces Bru binding in vitro and disrupts translational repression in vivo.16 Recently, a subset of the BREs—only those in the C region—were found to act not only in repression, but also in translational activation. Bru is a strong candidate to mediate activation, but this role may well require other factors or nearby regulatory elements since repression appears to be the default role of Bru binding sites.17–19
Bru contains three RRMs, organized in a manner shared by the closely related protein CUG-BP1 as well as several families of RRM proteins: there are two tandem RRMs near the amino terminus of the protein, and a third RRM positioned at the C terminus.15 None of the isolated RRMs of Bru individually displays high affinity RNA binding, but two larger domains of Bru each bind to the osk AB and C regions. One domain (RRM1+2) consists of RRMs 1 and 2, and the other (RRM3+) consists of RRM3 plus the final 42 amino acids of the spacer linking RRM2 and RRM3 (Fig. 1A). Both domains show specific binding to the osk regulatory regions, but binding of the RRM3+ protein is more sensitive to mutation of the BREs and thus has a higher degree of specificity for the BREs.20,21 Bru binds in vitro to the 3′ UTRs of other regulatory targets, including the gurken (grk), cyclin A (cycA), Sex lethal (Sxl) and germ cell-less (gcl) mRNAs. For these mRNAs no individual Bru binding sites have been identified, and there has been only limited delineation of the regions to which Bru binds. Only the cycA 3′ UTR has a perfect match to the consensus BRE sequence.22–26 Therefore, it seems clear that the BRE consensus sequence provides an incomplete picture of the Bru binding site or sites.
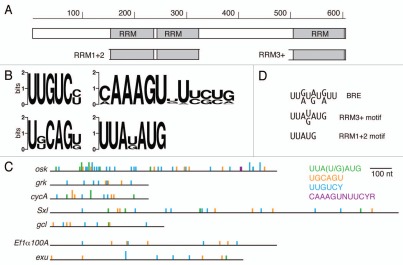
Figure 1.
Bru protein and binding motifs. (A) Organization of Bru protein. The structure is shown schematically, to scale, with the three RRM RNA binding domains indicated. The subdomains of Bru used for selections, RRM1+2 and RRM3+, are shown. (B) Graphical representations of preferred binding motifs identified by in vitro selections. The height of each stack represents the information content at each nucleotide of the motif in bits. At top left is the predominant motif identified from the Bru selection. The other motifs were identified from the RRM3+ selection. (C) comparison of the BRe consensus sequence and similar motifs from the selections. (D) The 3′ UTRs of the indicated mRNAs are shown schematically, with motifs from the aptamer selections indicated. The full height bars are perfect matches to the motifs, while the half height bars have a single mismatch. The top five 3′ UTRs are of Bru target mRNAs, while the bottom two 3′ UTRs are from other mRNAs not known to be regulated by Bru.
To obtain a more complete understanding of Bru binding sites, and to begin to ask how the different RNA binding domains of Bru contribute to specificity of binding, we used in vitro selection methods to identify preferred binding sequences for Bru and for the isolated RNA binding domains of Bru. The results indicate that the RRM3+ domain is a major determinant of binding specificity, with one of the motifs identified by the selection being effectively equivalent to the BRE. By contrast, RRM1+2 has lower specificity and selects aptamers enriched in short sequences, some of which are highly overrepresented in the osk AB and C regions to which Bru binds. We suggest that Bru recognizes extended combinatorial binding sites consisting of both highly constrained RRM3+ binding motifs and nearby sequences showing a preference for RRM1+2 binding. From the results of the selections, candidate Bru binding sites were identified in the grk and cycA 3′ UTRs. These sites conferred Brudependent translational repression on reporter mRNAs in the ovary, as did sites from the osk 3′ UTR.
Results
In vitro selection of aptamers.
In vitro selection of binding sites (SELEX; reviewed in ref. 27) was performed with Bru proteins. The proteins used for this analysis were full length Bru, as well as the isolated Bru RRM1+2 and RRM3+ domains. Although the known Bru binding sites define a short consensus sequence (7–9 nt; Fig. 1C), the random sequence RNA for the selections was longer (50 nt) to ensure that larger sequence motifs, or motifs that must be presented within structures, could be bound and recovered. Progress of the selections was monitored by testing naive and selected RNA pools for binding. Each selection led to an increase in affinity, with larger increases for the RRM3+ and Bru selections (Fig. S1). After 11 rounds of selection, bound RNAs from the final round were converted to DNA and sequenced. Each family of aptamers (Tables S1–3) was evaluated for overrepresented primary sequences using pattern searches and MEME,28 and for secondary structures using MFOLD.29,30 The highest degree of specificity was found for Bru and RRM3+, and these results are presented first.
From the selection for RNAs that bind to full length Bru the predominant motif is UUGUCY (Fig. 1B). In the 90 clones sequenced, there are 92 instances of UUGUCY and 173 additional copies with a single mismatch. Additional motifs identified from the selection with RRM3+ [UUA(U/G)AUG and UGCAGU, below] are present at lower frequencies. There are many copies of a BRE-like UUA(U/G)AUG motif: 3 are perfect copies and an additional 21 have a single mismatch. There are no perfect matches to the UGCAGU motif, but there are 6 examples with a single mismatch.
The selection against RRM3+ identified several constrained motifs. One is a long sequence motif, 5′-CAAAGUNUUCYR (Y, pyrimidine; R, purine). From the 32 sequenced clones of the final round of selection, 16 had the consensus sequence or a close variant (Fig. 1B). The appearance of such a long motif was surprising: the core RRM domain typically binds to three nucleotides, and RRMs with structural features that extend the core typically recognize sequences no longer than six or seven nucleotides.5,31 The RRM3+ domain has an additional structural element that is important for binding,21 but recognition of a 12 nucleotide sequence by a single RRM would be unprecedented (it is also possible that this sequence is recognized by a dimer or multimer of RRM3+). One plausible explanation is that a portion of the sequence is required indirectly for correct presentation of the actual binding site, as is the case for recognition of U1 snRNA by the N terminal RRM of the U1A protein.32 However, there is no structure predicted to be formed by the isolated CAAAGUNUUCYR sequence, nor by the sequence in the context of the anti-RRM3+ aptamers. Notably, this sequence did not appear among the anti-Bru aptamers. Therefore, it is possible that recognition of CAAAGUNUUCYR relies in part or whole on an unconventional form of RNA binding by RRM3+, perhaps relying on surfaces normally not accessible or folded differently in the context of Bru.
Two other motifs identified among anti-RRM3+ aptamers are UGCAGU and UUA(U/G)AUG. The UGCAGU motif appeared 4 times, with 18 additional copies with a single mismatch (Fig. 1B). The UUA(U/G)AUG motif appeared 9 times, with 6 additional copies with a single mismatch (Fig. 1B). A notable feature of the UUA(U/G)AUG motif is that it can be superimposed on the BRE (Fig. 1C). The BRE was identified as a consensus sequence, but the importance of each nucleotide was not tested. The UUA(U/G)AUG motif serves as an updated and better definition of the BRE.
In contrast to the results with RRM3+ and Bru, selections against RRM1+2 did not produce highly overrepresented sequence motifs detected by MEME analysis. Following the approach taken by others in analysis of aptamers selected by proteins related to Bru [mouse Etr-3 protein;33 human CUG-BP1 34], the frequencies of tri- and tetra-nucleotides were determined, and pattern searches tested whether the most abundant tetranucleotides were core sequences of longer motifs. No single long and highly overrepresented motif was discovered. Instead, we identified a number of frequently appearing tetranucleotides (Table 1 and S4).
Table 1.
Highly enriched tetranucleotides in the RRM1+2 aptamers
| Tetranucleotide | n | Copies in osk AB (127 nt) | Copies in osk C (76 nt) | Copies in osk 3′ UTR (1041 nt) | Copies in anti-Bru aptamers | Rank among anti-Bru aptamer tetranucleotides |
| U/purine rich (only U or purine) | ||||||
| UUAU | 37 | 5 | 3 | 11 | 74 | 12 |
| UAUG | 24 | 5 | 2 | 9 | 23 | 55 |
| UGGA | 19 | - | - | 3 | 1 | 218 |
| UGUU | 17 | 7 | 1 | 14 | 103 | 5 |
| UUUA | 16 | 4 | 2 | 16 | 57 | 18 |
| C/A rich (at least 3 of the 4 nt are A or C) | ||||||
| AUCA | 27 | - | 1 | 8 | 11 | 90 |
| CAAA | 23 | - | - | 15 | 20 | 147 |
| UCAA | 20 | - | - | 9 | 15 | 74 |
| UCAC | 20 | - | - | 2 | 11 | 90 |
| AAAA | 18 | 3 | 181 | |||
| AAAG | 17 | - | - | 5 | 3 | 181 |
| CAUA | 16 | - | - | 2 | 17 | 71 |
| Other (don't fit in either group above) | ||||||
| UAUC | 22 | - | - | 3 | 35 | 32 |
| UUCU | 19 | 2 | - | 11 | 66 | 15 |
| AGCU | 18 | - | 1 | 5 | 3 | 181 |
| UUCA | 17 | - | - | 2 | 10 | 95 |
| UCUG | 16 | 1 | - | 7 | 40 | 25 |
| UCUA | 16 | 1 | 2 | 5 | 38 | 28 |
| UCUU | 15 | 1 | 1 | 6 | 92 | 7 |
| GUCU | 15 | 1 | 1 | 11 | 81 | 9 |
Tetranucleotides appearing at least 15 times in the RRM 1+2 aptamers are shown.
One subset of the tetranucleotides consists only of U and purines (U/R-rich); some of these form the core of the most common pentanucleotide, UUAUG, which appears 19 times in the aptamers. This pentanucleotide can be superimposed on the 5′ portion of the BRE consensus sequence (Fig. 1C). The U/R-rich tetranucleotides appear frequently within the AB and C regions of the osk 3′ UTR, the two regions implicated in Bru-mediated translational control (Table 1), suggesting that enrichment of these short sequences in the anti-RRM1+2 aptamers may have biological relevance.
Another subset of tetranucleotides is enriched in C and A (at least three of the four nucleotides); some of these form the core of the second most common pentanucleotide, UCA AA, which appears 18 times. The C/A-rich tetranucleotides are not closely related to the BREs, and are almost completely absent from the osk 3′ UTR AB and C regions (Table 1). Thus, there is no prediction about their biological relevance.
For comparison of anti-RRM1+2 and anti-Bru aptamers, the frequencies of tetranucleotides were also determined in the anti-Bru aptamers (Table S4). Not surprisingly, tetranucleotides from the predominant Bru motif (UUG UCY) are among the most highly represented. In addition, four of the six most common tetranucleotides contain 3 U's and one G, a trend similar to the enrichment of U/R-rich tetranucleotides in the anti-RRM1+2 aptamers.
The absence of one or even a few dramatically overrepresented sequence motifs in the anti-RRM1+2 aptamers suggests that the specificity of RRM1+2 is low. There is some degree of specificity, since binding of RRM1+2 to the osk AB region RNA is diminished by mutation of the BREs.20 Consistent with at least weak specificity, the pool of RNAs selected by RRM1+2 binds better than the naive pool (Fig. S1). The specificity for the osk AB region may well involve binding to the many U/purine rich sequences in that region, some of which are altered by mutation of the BREs.16
While the enrichment of particular sequence motifs is one outcome of the selections, there were also changes in nucleotide composition which were most striking for the anti-Bru aptamers. The initial template pool of random DNA sequences was synthesized with equal amounts of each nucleotide. Following selection with Bru, the residue U was enriched from 25% to 50%, while the residues A and G were reduced in the population to less than 15% each (Table 2). This level of U frequency is similar to that of the osk AB region, which has the strongest Bru binding of the two osk regulatory regions.16 The anti-Bru aptamers differ from the osk AB region for frequency of C residues: the osk AB region is only 10% C, but C is the second most abundant nucleotide (23%) in the anti-Bru aptamers. Less extreme changes in nucleotide composition occurred in the other selections. For both anti-RRM1+2 and anti-RRM3+ aptamers the frequency of U increased slightly, but not to the same extent as for the anti-Bru aptamers. Unlike the anti-Bru aptamers, there was no reduction in the frequency of A in the anti-RRM1+2 and anti-RRM3+ aptamers (Table 2).
Table 2.
Nucleotide composition of aptamers and osk 3′ UTR regions
| Nucleotide composition | ||||
| RNAs | A | C | G | U |
| osk 3′ UTR | 29% | 18% | 18% | 36% |
| osk AB region | 18% | 10% | 17% | 54% |
| osk C region | 28% | 9% | 16% | 47% |
| anti-RRM1+2 aptamers | 28% | 21% | 19% | 32% |
| anti-RRM3+ aptamers | 27% | 21% | 19% | 33% |
| anti-Bru aptamers | 14% | 23% | 13% | 50% |
The results of the selections reveal a complex picture of Bru binding specificity, with different domains displaying different patterns of specificity. One domain, RRM3+, appears to provide a high degree of binding specificity, as highly constrained motifs of 6 nt or longer were identified. A second binding domain, RRM1+2, does not have a high degree of specificity but may bind preferentially to regions enriched in certain types of sequences. Since anti-Bru aptamers can contain the well defined RRM3+ motifs and at least some of the shorter sequences identified from the RRM1+2 selection, it appears that Bru may recognize combinatorial sites in which the different domains of Bru bind independently to different motifs (see Discussion). An additional binding specificity, for the highly constrained UUGUCY motif, is detected only using full length Bru. Recognition of this motif could involve one or both of the RRM binding domains, but with a novel specificity imposed by the organization or folding of the domains in the context of Bru. Alternatively, UUGUCY binding could rely on a separate and previously unrecognized RNA binding domain.
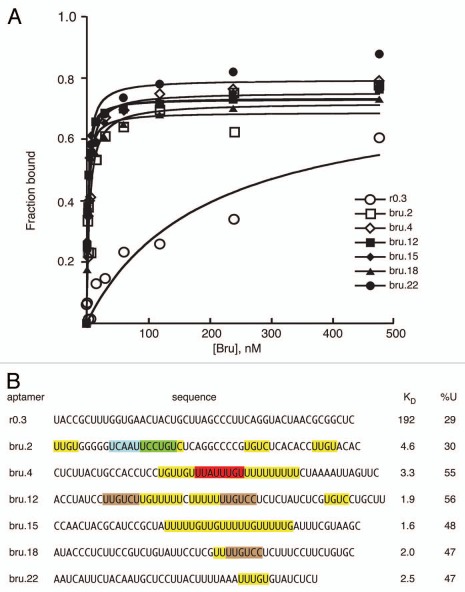
Binding affinities were determined for examples of anti-Bru aptamers (Fig. 2). All bound Bru with much higher affinities (KD of 1.6–4.6 nM) than a control RNA from the starting pool (KD 192 nM). Inspection of the aptamer sequences reveals the characteristics described above. All contain at least one perfect or near perfect copy of the predominant anti-Bru UUGUCY motif, but the number of such motifs varies substantially: at the extremes, one aptamer has one copy of a singly mismatched UUGUCY motif, while one aptamer has two perfect copies and three singly mismatched copies. All but one of the anti-Bru aptamers has U content approaching or above 50%. All aptamers have U/purine rich regions, including but not limited to the UUGUCY motif. Most aptamers also have A/C rich regions, although these are less abundant than the U/purine rich regions. These properties suggest that it is a combination of multiple features that underlies strong Bru binding, rather than the presence of a single critical motif. Functional studies using RNAs with either two or four copies of individual candidate binding sites (below) support this view.
Figure 2.
Bru binding to Anti-Bru aptamers. (A) Results of filter binding assays with recombinant Bru (0.9–477 nM) and anti-Bru aptamers. (B) Aptamer sequences (excluding the sequences from the cloning vector that are common to all) are shown, with KD values obtained from the data of part A. Motifs identified from the in vitro selections are shaded. The predominant UUGUCY motif from the Bru selection is shaded brown, with only perfect copies indicated. All of the anti-Bru aptamers have at least one copy of a singly-mismatched version of the motif (these are not shaded), while the control RNA obtained from the pool prior to selection (r0.3) has none. Examples of the motifs from the RRM3+ selection that also appeared in the Bru selection are shaded in green and red. Instances of the 5 tetranucleotides that appear most frequently in the anti-Bru aptamers (UUGU, UUUU, UGUC, UUUG and UGUU; Sup. Table 4) are shaded in yellow (except where the sequences are within another shaded region). The proportion of U, which is highly enriched in the Bru selection, is given for each RNA.
Candidate Bru binding motifs in known targets of Bru regulation.
Five genes have been reported to be subject to regulation by Bru. The best characterized example of such regulation is osk, which contains BREs in the AB and C regions of its 3′ UTR. Mutation of the BREs substantially reduces Bru binding in vitro, and leads to translational defects in vivo.16,17 The BRE consensus sequence closely resembles the UUA(U/G)AUG motif selected by RRM3+. Perfect copies of the predominant motif selected by Bru (UUGUCY) are present once in the AB region and twice in the C region, and a sequence that differs at only one position relative to the CAAAGUNUUCYR motif selected by RRM3+ is present in the C region. Notably, mutation of each type of Bru binding site in the osk mRNA leads to defects in translational regulation in vivo.17
The other reported targets of Bru translational repression are the cycA, grk, Sxl and gcl mRNAs.22–26 Bru is assumed to act directly to repress translation of these mRNAs, as the 3′ UTRs of each mRNA can bind to Bru in vitro. Bru binding sites have not been precisely mapped in any of the mRNAs, and the number of binding sites is not known. However, based on competition binding experiments, the gcl and grk 3′ UTRs bind Bru with much lower affinity than the osk 3′ UTR16,26 and therefore should have fewer or weaker binding sites (no such experiments have been reported for the other targets).
The 3′ UTRs of the Bru-regulated mRNAs were searched for sequences corresponding to each of the longer motifs from the RRM3+ and Bru selections (Fig. 1D). The only perfect matches (other than in osk) were in the cycA mRNA, with one copy of the BRE-like UUA(U/G)AUG motif and one copy of the UGCAGU motif (both from the RRM3+ selection). Both motifs are adjacent to short sequences similar to the A/C-rich tetranucleotides identified among the anti-RRM1+2 aptamers, consistent with the notion of Bru binding to combinatorial sites (see Discussion). None of the other known targets of Bru have perfect matches to any of the identified motifs. However, there are multiple copies of one or more of the motifs when a single mismatch is allowed (Fig. 1D). Bru binds to the gcl 3′ UTR primarily in the first 130 nt,26 and this region contains four motifs with single mismatches.
Translational repression by Bru binding motifs.
To determine if candidate Bru regulatory sites confer translational repression, we used GFP mRNA reporters. Reporter transcripts were expressed using the UAS/GAL4 system in the germ line cells of the ovary,35 where Bru is present.15 The control transgene mRNA consists of the GFP coding region, a portion of the SV40 3′ UTR, and a portion of the fs(1)K10 3′ UTR including the polyadenylation site. To determine if the reporter mRNAs are translation-ally regulated, we compared mRNA and GFP protein levels by quantitative real time PCR (qRT-PCR) and confocal microscopy, respectively. The control transgene shows strong GFP expression in the germ line cells of the egg chamber (Fig. 3B), consistent with the absence of translational repression. Derivatives of this transgene were constructed by insertion of copies of candidate regulatory sites from the known Bru targets.
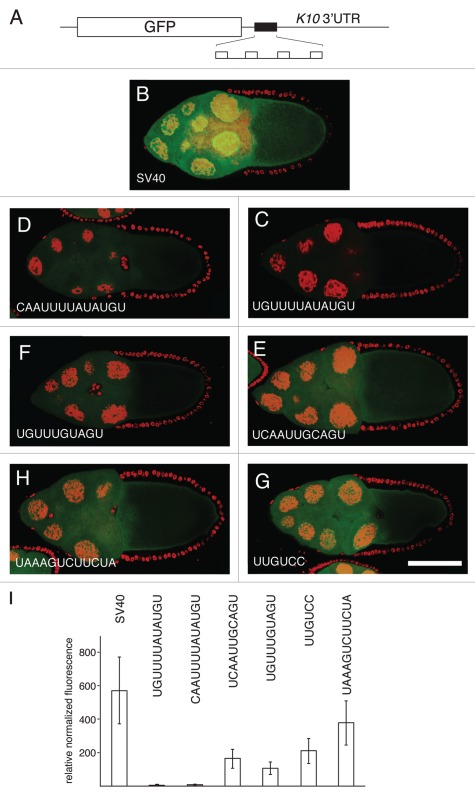
Figure 3.
Translational repression by candidate regulatory sites. (A) schematic diagram of reporter mRNAs with the variable region indicated by a filled box. For the SV40 reporter, the variable region is only SV40 sequences. For the remaining reporter mRNAs the variable region has four copies of a candidate Bru regulatory site embedded in SV40 sequences. (B–H) examples of GFP levels in stage 10A egg chambers expressing GFP reporter transgenes with the matα4-GAL-VP16 driver. All confocal images were taken on the same day at the same settings. The scale bar represents 75 µm. (B) control GFP transgene with no anti-Bru aptamer binding motifs. The remaining image parts are for GFP transgenes with the Bru binding motifs indicated in the figure and described below. (C) UGUUUUAUAUGU is from the osk AB region, and consists of a BRE-like motif adjacent to a short U/G rich motif (like those from the RRM1+2 selection). (D) CAAUUUUAUAUGU is from the cycA 3′UTR, and consists of a BRE-like sequence adjacent to a short C/A rich motif (like those from the RRM1+2 selection). (E) UCAAUUGCAGU is from the cycA 3′ UTR, and consists of a copy of the UGCAGU motif (from the RRM3+ selection) adjacent to a short C/A rich motif (like those from the RRM1+2 selection). (F) UGUUUGUAGU is from the grk 3′UTR, and consists of the UGCAGU motif (from the RRM3+ selection but with a single mismatch) adjacent to a short U/G rich motif (like those from the RRM1+2 selection). (G) UUGUCc is the type II Bru binding site, which appears three times in the AB and C regions of the osk 3′UTR. (H) UAAAGUCUUCUA is from the osk C region, and is a type III Bru binding site with a single mismatch relative to the longest aptamer motif from the RRM3+ selection. (I) Relative GFP levels in the nurse cell cytoplasm of stage 9/10 egg chambers for each of the reporter transgenes. GFP levels (obtained from 45 measurements for each transgene) were normalized to the RNA levels (from 3 measurements for each transgene). Transgene RNA levels were normalized relative to rp49 RNA levels. All of the reporter transgene mRNAs with candidate regulatory sites show reductions in GFP levels that are significantly lower than for the control (p < 0.0001 by the Tukey-Kramer method).
Three of the candidate regulatory sites were from the osk mRNA. One site consists of a BRE and flanking U/G-rich sequence, UGUUUUAUAUGU. This site is expected to mediate translational repression, as it contains the BRE which has been shown to have this activity.16,18,19 A second site is the UUGUCY motif (type II Bru binding site) from the anti-Bru aptamers, which appears once in the osk AB region and twice in the osk C region. The third site from osk is UAAAGUCUUCUA (the type III Bru binding site), which differs at only one position from consensus for the CAAAGUNUUCYR motif of the anti-RRM3+ aptamers. Mutation of each of these sites in the osk mRNA does interfere with translational regulation.17 Testing these regulatory elements in the reporter assay allows us to ask, with an mRNA whose regulation is much simpler than osk, if they are sufficient to confer repression.
Three other candidate regulatory sites, consisting of motifs defined by the selections, were from the cycA and grk 3′ UTRs. The two cycA sites are those mentioned in the previous section, and are comprised of an RRM3+ motif adjacent to an A/C rich region: CAAUUUUAUAUGU and UCAAUUGCAGU. Within the grk mRNA there are no perfect matches to any of the anti-RRM3+ or anti-Bru aptamer motifs. However, there are three copies of the UGCAGU RRM3+ motif with a single mismatch, and each is positioned close to U/G- or A/C-rich sequences. One of these candidate regulatory sites, UGUUUGUAGU, was chosen for analysis.
In an initial round of experiments we tested reporter trans-genes bearing two copies of candidate regulatory sites, one at each end of 88 nt of SV40 sequences (the control transgene also has SV40 sequences in its 3′ UTR). None of these transgenes showed a large degree of translational repression (data not shown). By contrast, a similar GFP reporter with the osk AB region is dramatically repressed.17 One key difference between the osk AB and candidate site reporters is the density of binding sites: the reporters tested here have two sites in ∼100 nt, while the osk AB region is similar in size but has more Bru binding sites (most of the region consists of sequences that either match one of the motifs from the Bru and RRM3+ selections, or are enriched in the simpler sequences from the RRM1+2 selection).
In a second round of experiments we modified the reporter transgenes to include a total of four candidate Bru regulatory sites, with the two additional copies evenly spaced within the central SV40 sequences (Fig. 3A). Notably, each of the reporter mRNAs with candidate Bru regulatory sites had reduced levels of GFP. For the two reporter mRNAs with the strongest repression (more than 10 fold reduction), the regulatory sites (from the osk and cycA mRNAs) consist of a BRE-like sequence flanked by a short sequence similar to the enriched short motifs from the RRM1+2 selections (Fig. 3C, D and I). Intermediate levels of repression were conferred by three sites. One was the type II Bru binding site (the UUGUCY motif from the Bru selection) (Fig. 3G and I). The others were from the cycA and grk mRNAs, and consisted of a UGCAGU motif (from the RRM3+ selection) adjacent to one of the short motifs (like those from the RRM1+2 selection). The lowest level of repression was provided by the type III Bru binding site.
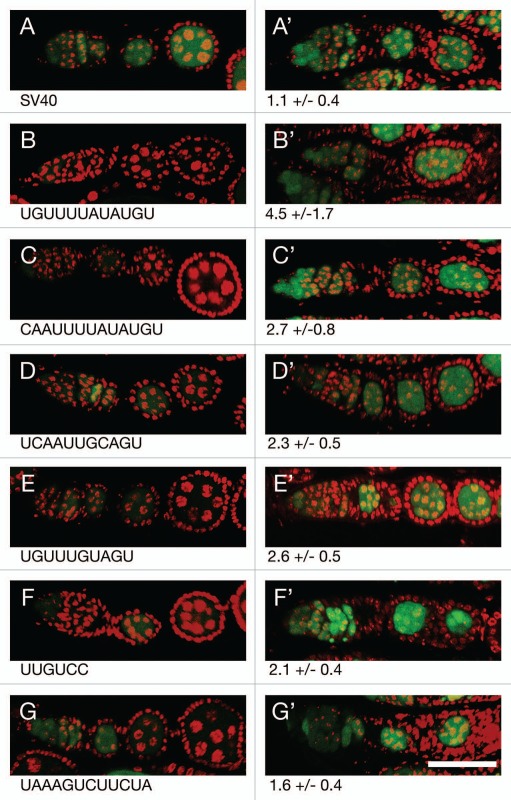
Repression by each of the regulatory sites is presumably due to the action of Bru. To confirm this expectation, the level of GFP produced by each reporter was compared between flies with Bru activity (aret/+ heterozygotes; aret is the gene that encodes Bru) and flies with substantially reduced Bru activity (aret/aret transheterozygotes) (Fig. 4). For the control reporter mRNA there was very little difference in GFP level when Bru activity was reduced (Fig. 4A). However, for each of the reporters showing repression, reducing Bru activity resulted in an increase in GFP (Fig. 4B–G). Not surprisingly, the largest increase in GFP came from the reporters that are most strongly repressed.
Figure 4.
Translational repression of reporter mRNAs requires Bru. (A–G) The pairs of parts show GFP expressed from a reporter mRNA in aret−/+ ovaries (A–G, left) or aret−/aret− ovaries (A′–G′, right). The identity of the Bru regulatory sites is shown below, with the relative increase in GFP level from mutation of aret indicated beneath the mutant parts. The scale bar represents 50 µm. In all cases the driver was nosGAL4VP16, which is active at early stages of oogenesis (the aret mutant ovaries arrest oogenesis and do not progress to the stage shown in Figure). The transgenes are the same as those in Figure, with four copies of a particular Bru binding motif as indicated.
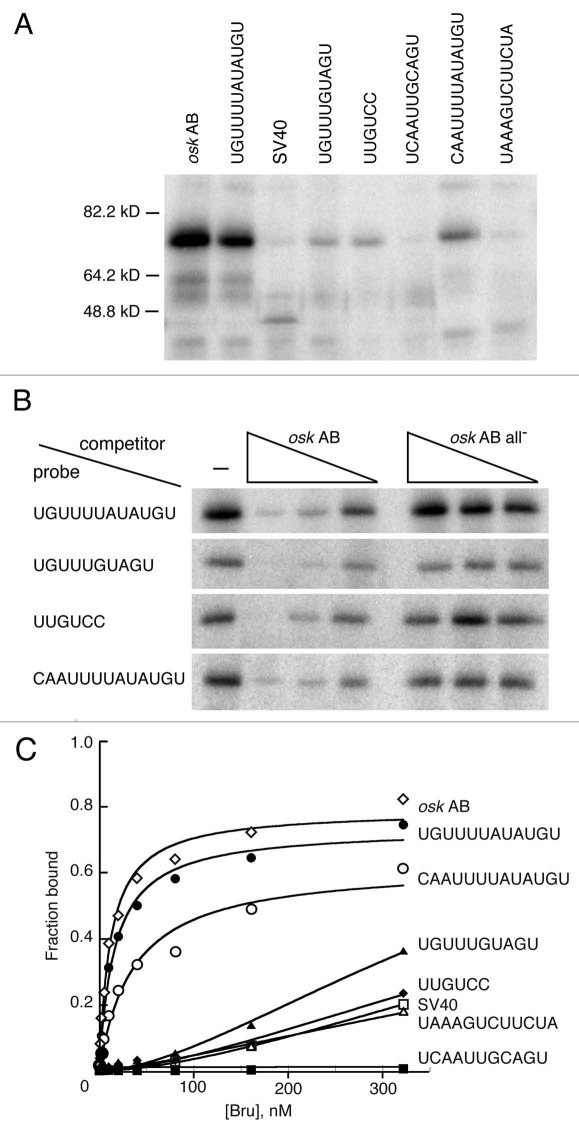
Binding of Bru in ovary extracts to each of the multimerized elements was tested in a UV crosslinking assay. The portion of each transgene bearing the multimerized elements was transcribed and radiolabeled in vitro, incubated with ovarian extract, UV irradiated, and the adducts displayed by denaturing electrophoresis and phosphorimaging. In this assay the RNA with the multiple copies of the combinatorial UGUUUUAUAUGU site (corresponding to a sequence from the osk AB region) bound most strongly (Fig. 5A). Binding to two other combinatorial sites and the UUGUCY site was also detected, but was weaker. The other sites did not bind detectably in this assay. To confirm that the observed binding was due to Bru, competition binding assays were performed (Fig. 5B). The presence of unlabeled osk AB RNA with known Bru binding sites greatly reduced binding, confirming that the crosslinked protein is Bru.
Figure 5.
Bru binding to regulatory sites. (A) UV crosslinking assay with ovarian protein and RNAs bearing four copies of the Bru binding sites embedded in SV40 sequences. The RNA probes are indicated at top by the identity of the binding site. The SV40 probe is SV40 RNA alone, and the osk AB probe is the AB region of the osk 3′ UTR. (B) competition binding assay. Crosslinking assays of the type shown in (A) were repeated, with or without the presence of unlabeled competitor RNAs. The competitors are osk AB RNA, or the same RNA (all-) with point mutations in BREs and type II Bru binding sites.17 exposures of the different rows are not equivalent, but were chosen to have similar binding signals in the absence of competitor. (C) Filter binding assay with recombinant Bru (1.26–322 nM) and the RNAs used in (A and B).
The absence of detectable binding of some of the RNAs was surprising, given their demonstrated effects on translation. Therefore, a quantitative filter binding assay was also used with recombinant Bru (Fig. 5C); the results are essentially identical to those from the crosslinking assay. The RNA with the UGUUUUAUAUGU sites shows the strongest binding, just as in the crosslinking assay, and binds Bru nearly as well as the osk AB RNA. Not surprisingly, this RNA mediates strong repression in vivo. The other combinatorial site with a BRE-like sequence, CAAUUUAUAUGU, also shows strong binding and strong repression. The RNA with the UGUUUGUAGU sites shows weaker binding and is less effective at translational repression. Two of the RNAs (those with UUGUCC or UAAAGUGUUCUA sites) do not bind Bru any more effectively than the SV40 control in this assay, and one RNA (with the UCAAUUGCAGU sites) does not bind detectably at all. Nevertheless, each of these RNAs confers some degree of translational repression in vivo, and the repression requires Bru (Figs. 3 and 4).
The likely explanation of this conundrum is that the short RNAs used for binding assays are folded into structures that completely or partially mask the binding sites. Structural predictions very strongly support this view. Notably, the RNA with UCAAUUGCAGU sites which fails to bind (and thus binds less well than the SV40 control) is predicted to adopt an extensively and stably base paired structure. The presence of each copy of the UCAAUUGCAGU site in a double stranded region with multiple G-C base pairs (Table S5) would very likely prevent specific Bru binding in vitro, as none of the sites would be in a single stranded conformation with the binding site exposed. Furthermore, even non-specific binding to single stranded RNA would be inhibited, with very little of the RNA in that conformation. However, when the segment of RNA is embedded within a reporter mRNA in vivo, in the presence of many other RNA binding proteins, the structure may not form or may be unstable, allowing Bru to bind and repress translation.
None of the other RNAs with different sites appear to have structures that would completely mask the binding sites, but most of the weakly binding RNAs have either a subset of the sites in strongly base paired helices, or all of the sites in more weakly based paired helices. By contrast, the two RNAs that bind strongly are predicted to have structures with the binding sites in regions that are only partially base paired, with primarily A–U and G–U base pairs. The exception to this trend is the RNA with the weakly repressing UAAAGUCUUCUA sites. Three of the four sites in the RNA are not expected to be in stable helices, and thus likely available for binding. However, this is the site that was recovered in the RRM3+ selection and not from the Bru selection, and does not make a substantial contribution to Bru binding to the osk mRNA in vitro.17
Discussion
How individual RRMs interact with RNA has been studied in depth, revealing fundamental features of binding of the core RRM, as well as variations allowed by modification or extension of the core RRM. What is understood in much less detail is how proteins with multiple RRMs, and in particular those with RRMs widely separated in the polypeptide chain, make use of the individual domains to achieve their binding specificities.
The CELF or Bruno-like proteins have three RRMs, with two tandem copies near the amino terminus and an isolated RRM at the carboxyl terminus. Proteins of this class play important roles in many levels of post-transcriptional regulation of gene expression. Studies on RNA binding by these proteins have focused largely on short repeat sequences, in part because the one member (CUG-BP1) was initially found to bind to repeated CUG,36,37 and in part because in vitro selections with intact proteins revealed a preference for short repeats of U and G such as UGU.33,34 Structural studies of members of this class of proteins have made use of isolated RNA binding domains bound to substrates with the short binding motifs.38,39 These studies provide a detailed picture of the molecular contacts between the individual domains and the bound RNAs, but do not address the question of how the different domains contribute to binding in the context of the intact protein. Furthermore, binding specificity is not limited to the short U/G rich repeats, as revealed by characterization of CUG-BP1 binding to the tumor necrosis factor (TFN) mRNA.40 This novel binding specificity might involve one of the RRMs, or a combination of different RRMs perhaps recognizing different motifs.
Bru is also a member of the CELF/Bruno-like protein family. We previously found that two different Bru RNA binding domains, RRM1+2 and RRM3+, could each bind to BRE-containing regulatory regions of the osk mRNA 3′ UTR. Mutation of the BREs reduced binding by both domains. This suggested that both domains had some specificity for BREs, and that each contributed to strong binding by Bru. Here, using selections with the Bru RNA binding domains and full length protein, we provide a more complete picture of the specificity of Bru RNA binding and how it is achieved. There are four main findings, with at least some of these relevant to the class of proteins related to Bru.
First, the Bru RRM1+2 domain displays a preference for short, low complexity sequences. This fits well with the properties of the same domain of CUG-BP1, including the preference for U/G rich sequences. However, for Bru RRM1+2 it is not only U/G rich sequences that are recovered from the SELEX, but also A/C rich sequences. We do not know if this binding preference is specific for Bru, or is shared by CUG-BP1 but not yet detected. The latter is possible, as SELEX experiments have not been performed with this domain of CUG-BP1. Similarly, no binding experiments with this class of substrate have been reported.
Second, the Bru RRM3+ domain binds to more complex sequence motifs, including one very closely related to the BRE motif previously implicated in translational repression by Bru in vivo. Because the core RRM recognizes only a few nucleotides, the more complex binding motifs must require additional structural elements. Indeed, the RRM3+ domain of Bru is an extended RRM with an amino terminal addition.21 Notably, RRM3 of CUG-BP1 has a similar amino terminal addition.38 Binding experiments with RRM3 of CUG-BP1 have only been performed with the simple U/G rich motifs,38 and no SELEX experiments have been reported. Therefore, it is possible that the RRM3 domain of CUG-BP1 has, as for Bru, the ability to bind more complex motifs. Such a binding activity might account for the more complex binding specificity revealed in studies with the TFN mRNA.40
Third, we found that full length Bru selects for aptamers enriched in a motif not recovered in selections with the isolated domains. Better understanding of this binding specificity, and whether it relies on a binding domain assembled from the known RNA binding domains or from a novel type of domain, will be a future goal. No similar binding specificity was detected in SELEX with other CELF/Bru-like proteins.33,34 Since Bru is similar to these proteins only in the RRMs, it is possible that the binding activity comes from the structural elements unique to Bru.
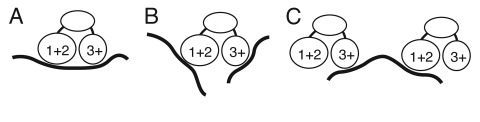
Fourth, our results suggest that the multiple RNA binding domains of Bru are used in recognition of combinatorial sites, with different domains binding to different parts of an extended binding site. Combinatorial binding would enhance specificity and affinity by increasing the number of contacts (Fig. 6A). Two types of evidence support this interpretation. First, mutation of either the RRM1+2 domain or the RRM3+ domain interferes with optimal binding to osk mRNA regulatory sites in vitro, and impairs translational control of osk mRNA in vivo.20 Thus, both domains make contributions to binding to osk mRNA, consistent with combinatorial binding. Based on the SELEX results, RRM3+ would provide a high degree of binding specificity, while RRM1+2 would provide less specificity but would enhance binding affinity. Support for combinatorial binding also comes from the makeup of the regulatory regions within mRNAs that mediate Bru-dependent translational repression. The best characterized such regions, the osk AB and C regions, contain motifs identified in each of the three selections. Our understanding of the regulatory elements in the other Bru targets is more limited. Nevertheless, it is notable that translational repression of a reporter mRNA is conferred by sequences from the cycA and grk mRNAs containing one of the highly constrained RRM3+ motifs close to an example of the short, lower complexity motifs selected by RRM1+2. We propose that the multiple RNA binding domains of Bru serve primarily in allowing combinatorial binding to extended binding sites. Combinatorial binding could be a general property of this class of proteins with three RRMs, and might explain the presence of a novel type of CUG-BP1 site in TFN mRNA.
Figure 6.
Models for Bru binding. (A) combinatorial binding: different RNA binding domains of Bru interact with extended binding sites in the same substrate RNA. (B) Independent binding: different RNA binding domains interact with different substrate RNAs. (C) Bridging binding: different molecules of Bru bind to the same substrate RNA. For simplicity, the possible third Bru RNA binding domain identified from the Bru selection is not shown, as its position in the protein is unknown.
There are alternate but less plausible explanations for how multiple RNA binding domains are used. One option is that the different domains act in recognition of different substrate mRNAs, expanding the spectrum of potential regulatory targets (Fig. 6B). Bru does bind to multiple different mRNAs, but our evidence does not point to specialization of different binding domains for binding to different mRNAs. Furthermore, the low degree of specificity provided by RRM1+2 would not be effective in limiting Bru activity to a small proportion of all mRNAs. Another option is suggested by the ability of Bru to oligomerize RNAs bearing tandem copies of the osk AB region in vitro:41 the multiple RNA binding domains could bridge different molecules of target mRNAs, forming interconnected RNA/ protein particles (Fig. 6C). From kinetic considerations it is more likely that a single molecule of Bru would make multiple contacts with a single molecule of RNA, at least in situations in which appropriate binding sites are present and can be bound without conformational constraints. After initial binding of Bru to one RNA molecule via one domain, the other domains of Bru would most rapidly encounter any additional binding sites nearby in the same molecule. However, even if the initial binding of Bru to osk mRNA followed the kinetically favorable path, subsequent assembly of osk mRNA into particles could facilitate intermolecular binding of Bru: dissociation of individual Bru RNA binding domains from their substrate would allow binding to a different substrate positioned nearby. Such Bru dependent oligomerization of osk mRNA has thus far only been demonstrated in vitro with artificial RNAs, and the biological relevance of this form of binding remains uncertain. Although osk mRNA is assembled into particles in the ovary, assembly depends on Polypyrimidine Tract Binding Protein but apparently not Bru since BREs have been reported to be neither necessary nor sufficient for osk mRNA interaction in vivo.42
Multiple binding specificities of Bru RNA binding domains.
A notable feature of the in vitro selections with Bru or its sub-domains is the recovery of more than one type of binding motif from each selection. For the RRM1+2 domain it is possible that each of the component RRMs has its own rather limited binding specificity (although neither binds well in isolation20), and that these specificities are revealed by the selections. However, such an argument is not possible to explain the three very different binding motifs that were identified for RRM3+. Multiple binding specificities are not uncommon for RNA binding proteins, although different binding sites are sometimes recognized by different binding domains.43 For a single RNA binding domain, different binding specificities could be obtained through alternate conformations of the domain as observed for U2AF65,44,45 or structural reorganization upon binding as for NELF-E.46 In addition, changes in RNA structure have the potential to present a site differently to a binding protein.47 How the flexibility of Bru binding is achieved is not known, and will likely require structural studies with the protein or domains bound to different substrates for a complete understanding.
Achieving specificity in translational control.
Translational control of osk mRNA is essential. Defects in repression or activation from mutation of control elements disrupts development and is ultimately lethal.16,17,48 Although many factors are involved in translational control of osk mRNA, Bru appears to play the key role of selectively recognizing the mRNA. RRM-containing proteins, like Bru, often bind with modest specificity, which is not surprising given that the core RRM interacts with only 3–4 nucleotides of an RNA. Nevertheless, Bru effectively silences osk mRNA translation, and must do so with a high degree of specificity. Two strategies appear to be used to obtain this specificity. One is combinatorial binding via the multiple RNA binding domains of Bru. Second, the osk mRNA relies on a high density of Bru binding sites for regulation. We have observed that single Bru binding sites display only very weak binding to Bru or Bru subdomains. Similarly, in the reporter assays described here a high local density of Bru binding sites is required for substantial translational control. Two relatively distant copies (separated by a 88 nt spacer) of any of the candidate regulatory elements confer either weak or undetectable repression, while four copies in a region of the same length do provide repression. Thus, evidence that Bru can bind an mRNA does not by itself demonstrate that Bru will have a major effect on its translation. We suggest that it is the combination of moderately specific RNA binding by Bru and the presence of multiple Bru binding sites that together ensure that a target mRNA will be efficiently regulated.
Materials and Methods
Protein purification.
The RRM1+2 and RRM3+ Bru protein domains were expressed in E. coli using the pET3a vector for purification via the T7 tag. Pelleted cells from induced cultures were frozen at -80°C, thawed and resuspended in 1x T7 tag bind/wash buffer from a T7 purification kit (Novagen), and lysed by sonication. Debris was removed by centrifugation and the supernatent filtered with a 0.2 µm filter (Nalgene). Protein in the filtered supernatent was purified using the batch-wise method detailed in the manufacturer's protocol. A vivaspin spin column (Sartorius) was used to concentrate the protein in a final storage buffer of 50 mM HEPES pH 7.9, 100 mM KCl, 1 mM EDTA, 10% glycerol.
Bru protein was expressed in E. coli using the pET15b vector, which provides a 6xHis tag for purification. Pelleted cells from induced cultures were frozen at -80°C, thawed and resuspended in histag buffer (20 mM phosphate buffer pH 7.8, 500 mM NaCl, 20 mM imidazole, 10% glycerol), and lysed by sonication. Debris was removed by centrifugation and the supernatent filtered with a 0.2 µm filter (Nalgene). Protein was loaded onto ProBond resin (Invitrogen) and eluted with an increasing concentration of imidazole to a final concentration of 300 mM. Peak fractions were combined and concentrated by dialysis against PEG solution (25% PEG MW 15–20 K, 200 mM KCl, 1 mM EDTA). Additional dialysis was used to equilibrate in protein storage buffer (50 mM HEPES pH 7.9, 100 mM KCl, 1 mM EDTA, 10% glycerol).
Selection.
RNA for selection was prepared by transcription of a synthetic DNA template consisting of GAT AAT ACG ACT CAC TAT AGG GTT ACC TAG GTG TAG ATG CT (N)50 AAG TGA CGT CTG AAC TGC TTC GAA where the random segment was prepared with equimolar amounts of the four nucleotides. Transcripts were produced using the Ampliscribe T7 polymerase kit (epicentre), and gel purified.
Prior to incubation with the selective protein, 415 pmol (2.5 × 1014 unique molecules) of the RNA aptamer pool was passed through a nitrocellulose filter (Millipore, HAQP01300) secured by a syringe filter apparatus (Whatman, 420100). The aptamer pool was incubated with the selective protein (full length Bru, RRM1+2 or RRM3+) for 30 min at room temperature in 50 µl total volume of 1x SELEX binding buffer (20 mM HEPES, pH 7.9, 100 mM KCl and 2 mM MgCl2). For the first four rounds, equimolar amounts of RNA and protein (200–415 pmol) were used. For the final rounds of selection RNA was present at 5-fold molar excess. After each binding incubation, the reaction was again passed through a nitrocellulose filter and bound RNA was eluted by incubation for 5 min at 98°C in 200 µl of elution buffer (7 M Urea, 100 mM NaOAc, 3 mM EDTA). Eluted RNA was precipitated and resuspended in 20 µL of water. Ten microliters of the resuspended aptamer RNA was used for a cDNA reaction with M-MLV reverse transcriptase (Invitrogen) and the products were amplified by PCR (forward primer: GAT AAT ACG ACT CAC TAT AGG GTT ACC TAG GTG TAG ATG CT, reverse primer: TTC GAA GCA GTT CAG ACG TCA CTT). The PCR products were then used for a further round of transcription, binding, cDNA synthesis and amplification.
The selection process was monitored using a filter binding assay (described below) (Fig. S1). Every three rounds of selection, the binding of the selective protein to the current aptamer pool was compared to that of the initial aptamer pool as well as select previous aptamer pools. The selections were terminated after eleven rounds.
cDNAs from the selections were cloned using the TopoTA cloning kit (Invitrogen) and the inserts sequenced. The sequences were compared using the MEME program (http://meme.sdsc.edu/meme/meme.html or Bailey and Elkan, 1994). Tetranucleotide frequencies were determined using the search function of BBEdit 6.5 (Bare Bones Software), which identifies non-overlapping instances of the search string.
Transgenes.
Transgenes with Bru binding motifs were all based on UAS-GFP.49 The UAS-GFP-osk AB transgene has been described in reference 17. For the initial set of reporter trans-genes the binding motifs were placed at the ends of an 88 nt segment from the SV40 3′ UTR, and cloned as BamHI-BglII fragments into the BamHI site of UAS-GFP, just after GFP. The final transgenes were further modified by replacing internal portions of the SV40 sequences with two additional binding motifs such that the four binding motifs were distributed at equal distances within the SV40 segment. The sequences from each of the fragments from the final clones, and from the SV40 control, are shown below. Bru binding motifs are underlined.
UAS-GFP-UGU UUU AUA UGU TGT TTT ATA TGT GAT GAG TTT GGG ACA AAC CAC ATG TTT TAT ATG TTG AAA AAA ATG CTT TAT TTG TTG TTT TAT ATG TGC TAT TGC TTC ATT TGT AAC CTG TTT TAT ATG T
UAS-GFP-UGU UUG UAG UTG TTT GTA GTG ATG AGT TTG GGA CAA ACC ACA ACT GTT TGT AGT TGA AAA AAA TGC TTT ATT TGT GTG TTT GTA GTT GCT ATT GCT TCA TTT GTA ACC TGT TTG TAG T
UAS-GFP-UUG UCC TTG TCC GAT GAG TTT GGA CAA CCA CAA CTA TTG TCC AGT GAA AAA AAT GCT TTA TTT GTG ATT GTC CTG ATG CTA TTG CTT CAT TTG TAA CCC TTG TCC
UAS-GFP-UCA AUU GCA GUT CAA TTG CAG TGA TGA GTT TGG GAC AAA CCA CAA TCA ATT GCA GTT GAA AAA AAT GCT TTA TTT GTT CAA TTG CAG TTG CTA TTG CTT CAT TTG TAA CCT CAA TTG CAG T
UAS-GFP-CAA UUU UAU AUG UCA ATT TTA TAT GTG ATG AGT TTG GGA CAA ACC ACC AAT TTT ATA TGT TGA AAA AAA TGC TTT ATT TGT CAA TTT TAT ATG TCT ATT GCT TCA TTT GTA ACC CAA TTT TAT ATG T
UAS-GFP-UAA AGU CUU CUA TAA AGT CTT CTA GAT GAG TTT GGG ACA AAC CAC ATA AAG TCT TCT ATG AAA AAA ATG CTT TAT TTG TTA AAG TCT TCT AGC TAT TGC TTC ATT TGT AAC CTA AAG TCT TCT A
UAS-GFP-SV40 CAG ACA TGA TGA TGA GTT TGG ACA AAC CAC AAC TAG AAT GCA GTG AAA AAA ATG CTT TAT TTG TGA AAT TTG TGA TGC TAT TGC TTT ATT TGT AAC CCA GAC ATG AT.
Confocal analysis.
Transgenic flies were grown at 25°C. 2- to 3-day-old flies were placed in well yeasted vials and incubated at 25°C for another 2 days. Ovaries were dissected in PBS and fixed in a solution of 1.2 mL of PBS and 150 µL 37% formaldehyde for twenty minutes with gentle mixing. The ovaries were then washed for one hour in four changes of PBT (1x PBS, 0.1% Tween 20). Quantitative immunofluorescence data were obtained using Leica software from images collected by confocal microscopy using a single plane of focus. The GFP signal from nurse cell cytoplasm was sampled from three different locations in the egg chamber in each of 15 stage 9 or 10 egg chambers. Samples to be imaged for figures were stained with Topro (Molecular Probes) to label nuclei.
RT-PCR.
Ovaries from 20 females, prepared as described above, were dissected in PBS and homogenized with a pestle. Total RNA from the ovaries was prepared using Tri Reagent-LS (Molecular Research Center, Inc.,) according to manufacturer instructions. The isolated RNA was reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). The cDNAs were used in a quantitative real-time PCR with primers for either the GFP coding region (GFP-F, TTT TCG TTG GGA TCT TTC GAA; GFP-R, ACG GCG GCG TGC AAC) or rp49 (rp49-F, GCG CAC CAA GCA CTT CAT C; rp49-R, GAC GCA CTC TGT TGT CGA TAC C). The quantitative real-time PCR was carried out using the Power SYBR® Green Master Mix (Applied Biosystems) per manufacturer instructions in a 7900HT Fast Real-Time PCR System (Applied Biosystems). The GFP cDNA sample levels were normalized using rp49 cDNA sample levels. The real-time PCR was quantitated using the SDS software v2.2 (Applied Biosystems).
Flies.
Transgenic fly stocks were established by standard methods. Expression of UAS transgenes was driven by the nos-GAL4VP16 driver50 or the matα4-GAL-VP16 driver,51 as indicated. The aret alleles used were aretZ2286 from M. Lilly22 and aretPA from Trudi Schüpbach.52
RNA binding.
UV crosslinking assays with ovarian extracts were performed as described in reference 16, using RNAs uniformly radiolabeled with alpha 32P-UTP. For the competition binding experiments the unlabeled competitor RNAs were present at 10-, 100- and 1,000-fold excess.
RNAs for filter binding assays were synthesized with a T7 polymerase kit (epicentre, AS3107) and gel purified. 5′ phosphates were removed with Shrimp Alkaline Phosphatase and the RNAs then labeled with gamma 32P-ATP and T4 polynucleotide kinase.
Nitrocellulose (Whatman, 10-401-196) and nylon (Amersham Biosciences, RPN119B) filters were incubated in binding buffer for 30 minutes, placed together in a dot blot apparatus (Whatman, 10-447-900), modified as in reference 53, and pre-washed with 100 µl of binding buffer. RNA (200 pM) was incubated with Bru for 30–120 min at room temperature in a 50 µl volume of 1x SELEX binding buffer (binding was found to be similar with the different incubation times, indicating that saturation was achieved, and 60 min incubations were used for the data shown). For the binding assays of Figure 2, tRNA (70 pg) was included as a non-specific competitor. For the binding assays of Figure 5, tRNA (10 ng) and heparin (20 ng) were included as non-specific competitors. The binding reactions were passed sequentially through the nitro-cellulose and nylon filters, followed by a wash with 350 µl of binding buffer. The filters were imaged with a BioRad phosphoimager (Molecular Imager PharosFX System). All assays were performed in triplicate. Radioactive signal intensities were measured and plotted for comparison using this equation: bound RNA/total RNA. The binding data for the osk AB RNA and the UGUUUUAUAUGU- and CAAUUUUAUAUG U-containing fragments were fit to a hyperbola: y = (ymax[Bru])/(Kd + [Bru]).
Acknowledgments
We thank Trudi Schüpbach and Mary Lilly for fly stocks, members of the Macdonald lab and Brad Hall for useful comments on the manuscript, and Brad Hall and Anna Szafranska for advice on the selections. Rick Russell provided valuable advice on RNA binding assays and their interpretation. Michael Mahometa from the UT Division of Statistics and Scientific Computation provided important guidance on statistical analysis.
Financial Support
This work was supported by grants from the NIH (GM54409, GM076536 and GM077040) and from the Welch Foundation (F-1654).
Supplementary Material
References
- 1.Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 2.Singh R, Valcárcel J, Green MR. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- 3.Pérez I, Lin CH, McAfee JG, Patton JG. Mutation of PTB binding sites causes misregulation of alternative 3′ splice site selection in vivo. RNA. 1997;3:764–778. [PMC free article] [PubMed] [Google Scholar]
- 4.Blanchette M, Green RE, MacArthur S, Brooks AN, Brenner SE, Eisen MB, Rio DC. Genome-wide analysis of alternative pre-mRNA splicing and RNA-binding specificities of the Drosophila hnRNP A/B family members. Mol Cell. 2009;33:438–449. doi: 10.1016/j.molcel.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y, Varani G. Protein families and RNA recognition. FEBS J. 2005;272:2088–2097. doi: 10.1111/j.1742-4658.2005.04650.x. [DOI] [PubMed] [Google Scholar]
- 6.Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Varani G, Nagai K. RNA recognition by RNP proteins during RNA processing. Annu Rev Biophys Biomol Struct. 1998;27:407–445. doi: 10.1146/annurev.biophys.27.1.407. [DOI] [PubMed] [Google Scholar]
- 8.Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 9.Cléry A, Blatter M, Allain FH. RNA recognition motifs: boring? Not quite. Curr Opin Struct Biol. 2008;18:290–298. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 10.Handa N, Nureki O, Kurimoto K, Kim I, Sakamoto H, Shimura Y, et al. Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature. 1999;398:579–585. doi: 10.1038/19242. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, Tanaka Hall TM. Structural basis for recognition of AU-rich element RNA by the HuD protein. Nat Struct Biol. 2001;8:141–145. doi: 10.1038/84131. [DOI] [PubMed] [Google Scholar]
- 12.Allain FH, Bouvet P, Dieckmann T, Feigon J. Molecular basis of sequence-specific recognition of pre-ribosomal RNA by nucleolin. EMBO J. 2000;19:6870–6881. doi: 10.1093/emboj/19.24.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deo RC, Bonanno JB, Sonenberg N, Burley SK. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell. 1999;98:835–845. doi: 10.1016/s0092-8674(00)81517-2. [DOI] [PubMed] [Google Scholar]
- 14.Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, et al. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- 15.Webster PJ, Liang L, Berg CA, Lasko P, Macdonald PM. Translational repressor bruno plays multiple roles in development and is widely conserved. Genes Dev. 1997;11:2510–2521. doi: 10.1101/gad.11.19.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim-Ha J, Kerr K, Macdonald PM. Translational regulation of oskar mRNA by bruno, an ovarian RNA-binding protein, is essential. Cell. 1995;81:403–412. doi: 10.1016/0092-8674(95)90393-3. [DOI] [PubMed] [Google Scholar]
- 17.Reveal B, Yan N, Snee MJ, Pai C, Gim Y, Macdonald PM. BREs Mediate Both Repression and Activation of oskar mRNA Translation and Act In trans. Dev Cell. 2010;18:496–502. doi: 10.1016/j.devcel.2009.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lie Y, Macdonald PM. Translational regulation of oskar mRNA occurs independent of the cap and poly(A) tail in Drosophila ovarian extracts. Development. 1999;126:4989–4996. doi: 10.1242/dev.126.22.4989. [DOI] [PubMed] [Google Scholar]
- 19.Castagnetti S, Hentze MW, Ephrussi A, Gebauer F. Control of oskar mRNA translation by Bruno in a novel cell-free system from Drosophila ovaries. Development. 2000;127:1063–1068. doi: 10.1242/dev.127.5.1063. [DOI] [PubMed] [Google Scholar]
- 20.Snee M, Benz D, Jen J, Macdonald PM. Two distinct domains of Bruno bind specifically to the oskar mRNA. RNA Biol. 2008:5. [PubMed] [Google Scholar]
- 21.Lyon A, Reveal B, Macdonald PM, Hoffman D. Bruno protein contains an expanded RNA recognition motif. Biochemistry. 2009;48:12202–12212. doi: 10.1021/bi900624j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sugimura I, Lilly MA. Bruno inhibits the expression of mitotic cyclins during the prophase I meiotic arrest of Drosophila oocytes. Dev Cell. 2006;10:127–135. doi: 10.1016/j.devcel.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 23.Filardo P, Ephrussi A. Bruno regulates gurken during Drosophila oogenesis. Mech Dev. 2003;120:289–297. doi: 10.1016/s0925-4773(02)00454-9. [DOI] [PubMed] [Google Scholar]
- 24.Yan N, Macdonald PM. Genetic interactions of Drosophila melanogaster arrest reveal roles for translational repressor Bruno in accumulation of Gurken and activity of Delta. Genetics. 2004;168:1433–1442. doi: 10.1534/genetics.104.033985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Z, Lin H. Sex-lethal is a target of Brunomediated translational repression in promoting the differentiation of stem cell progeny during Drosophila oogenesis. Dev Biol. 2007;302:160–168. doi: 10.1016/j.ydbio.2006.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore J, Han H, Lasko P. Bruno negatively regulates germ cell-less expression in a BRE-independent manner. Mech Dev. 2009;126:503–516. doi: 10.1016/j.mod.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 27.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 28.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 29.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 31.Auweter SD, Oberstrass FC, Allain FH. Sequencespecific binding of single-stranded RNA: is there a code for recognition? Nucleic Acids Res. 2006;34:4943–4959. doi: 10.1093/nar/gkl620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oubridge C, Ito N, Evans PR, Teo CH, Nagai K. Crystal structure at 1.92 A resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature. 1994;372:432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- 33.Faustino NA, Cooper TA. Identification of putative new splicing targets for ETR-3 using sequences identified by systematic evolution of ligands by exponential enrichment. Mol Cell Biol. 2005;25:879–887. doi: 10.1128/MCB.25.3.879-887.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marquis J, Paillard L, Audic Y, Cosson B, Danos O, Le Bec C, Osborne HB. CUG-BP1/CELF1 requires UGU-rich sequences for high-affinity binding. Biochem J. 2006;400:291–301. doi: 10.1042/BJ20060490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rorth P. Gal4 in the Drosophila female germline. Mech Dev. 1998;78:113–118. doi: 10.1016/s0925-4773(98)00157-9. [DOI] [PubMed] [Google Scholar]
- 36.Timchenko LT, Timchenko NA, Caskey CT, Roberts R. Novel proteins with binding specificity for DNA CTG repeats and RNA CUG repeats: implications for myotonic dystrophy. Hum Mol Genet. 1996;5:115–121. doi: 10.1093/hmg/5.1.115. [DOI] [PubMed] [Google Scholar]
- 37.Timchenko LT, Miller JW, Timchenko NA, DeVore DR, Datar KV, Lin L, et al. Identification of a (CUG) n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 1996;24:4407–4414. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsuda K, Kuwasako K, Takahashi M, Someya T, Inoue M, Terada T, et al. Structural basis for the sequence-specific RNA-recognition mechanism of human CUGBP1 RRM3. Nucleic Acids Res. 2009;37:5151–5166. doi: 10.1093/nar/gkp546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teplova M, Song J, Gaw HY, Teplov A, Patel DJ. Structural insights into RNA recognition by the alternate-splicing regulator CUG-binding protein 1. Structure. 2010;18:1364–1377. doi: 10.1016/j.str.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang L, Lee JE, Wilusz J, Wilusz CJ. The RNA-binding protein CUGBP1 regulates stability of tumor necrosis factor mRNA in muscle cells: implications for myotonic dystrophy. J Biol Chem. 2008;283:22457–22463. doi: 10.1074/jbc.M802803200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chekulaeva M, Hentze MW, Ephrussi A. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell. 2006;124:521–533. doi: 10.1016/j.cell.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 42.Besse F, Lopez de Quinto S, Marchand V, Trucco A, Ephrussi A. Drosophila PTB promotes formation of high-order RNP particles and represses oskar translation. Genes Dev. 2009;23:195–207. doi: 10.1101/gad.505709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abe R, Sakashita E, Yamamoto K, Sakamoto H. Two different RNA binding activities for the AU-rich element and the poly(A) sequence of the mouse neuronal protein mHuC. Nucleic Acids Res. 1996;24:4895–901. doi: 10.1093/nar/24.24.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sickmier EA, Frato KE, Shen H, Paranawithana SR, Green MR, Kielkopf CL. Structural basis for polypyrimidine tract recognition by the essential pre-mRNA splicing factor U2AF65. Mol Cell. 2006;23:49–59. doi: 10.1016/j.molcel.2006.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thickman KR, Sickmier EA, Kielkopf CL. Alternative conformations at the RNA-binding surface of the N-terminal U2AF(65) RNA recognition motif. J Mol Biol. 2007;366:703–710. doi: 10.1016/j.jmb.2006.11.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rao JN, Schweimer K, Wenzel S, Wöhrl BM, Rösch P. NELF-E RRM undergoes major structural changes in flexible protein regions on target RNA binding. Biochemistry. 2008;47:3756–3761. doi: 10.1021/bi702429m. [DOI] [PubMed] [Google Scholar]
- 47.Banerjee H, Rahn A, Davis W, Singh R. Sex lethal and U2 small nuclear ribonucleoprotein auxiliary factor (U2AF65) recognize polypyrimidine tracts using multiple modes of binding. RNA. 2003;9:88–99. doi: 10.1261/rna.2131603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Munro TP, Kwon S, Schnapp BJ, St Johnston D. A repeated IMP-binding motif controls oskar mRNA translation and anchoring independently of Drosophila melanogaster IMP. J Cell Biol. 2006;172:577–588. doi: 10.1083/jcb.200510044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reich J, Snee MJ, Macdonald PM. miRNA-dependent translational repression in the Drosophila ovary. PLoS ONE. 2009;4:4669. doi: 10.1371/journal.pone.0004669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Doren M, Williamson AL, Lehmann R. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr Biol. 1998;8:243–246. doi: 10.1016/s0960-9822(98)70091-0. [DOI] [PubMed] [Google Scholar]
- 51.Martin SG, St. Johnston D. A role for Drosophila LKB1 in anterior-posterior axis formation and epithelial polarity. Nature. 2003;421:379–384. doi: 10.1038/nature01296. [DOI] [PubMed] [Google Scholar]
- 52.Schüpbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics. 1991;129:1119–1136. doi: 10.1093/genetics/129.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wong I, Lohman TM. A double-filter method for nitrocellulose-filter binding: application to proteinnucleic acid interactions. Proc Natl Acad Sci USA. 1993;90:5428–5432. doi: 10.1073/pnas.90.12.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.