Abstract
Ecto-nucleotidases play a pivotal role in purinergic signal transmission. They hydrolyze extracellular nucleotides and thus can control their availability at purinergic P2 receptors. They generate extracellular nucleosides for cellular reuptake and salvage via nucleoside transporters of the plasma membrane. The extracellular adenosine formed acts as an agonist of purinergic P1 receptors. They also can produce and hydrolyze extracellular inorganic pyrophosphate that is of major relevance in the control of bone mineralization. This review discusses and compares four major groups of ecto-nucleotidases: the ecto-nucleoside triphosphate diphosphohydrolases, ecto-5′-nucleotidase, ecto-nucleotide pyrophosphatase/phosphodiesterases, and alkaline phosphatases. Only recently and based on crystal structures, detailed information regarding the spatial structures and catalytic mechanisms has become available for members of these four ecto-nucleotidase families. This permits detailed predictions of their catalytic mechanisms and a comparison between the individual enzyme groups. The review focuses on the principal biochemical, cell biological, catalytic, and structural properties of the enzymes and provides brief reference to tissue distribution, and physiological and pathophysiological functions.
Keywords: Alkaline phosphatase, Catalytic mechanism, 5′-Nucleotidase, Ecto-nucleotidase, NPP, NTPDase, Nucleoside triphosphate diphosphohydrolase, Nucleotide pyrophosphatase/phosphodiesterase
Introduction
Principal functional roles of ecto-nucleotidases
Cell surface-located enzymes hydrolyzing extracellular nucleotides have been described long before intercellular signaling via extracellular nucleotides was discovered as a major pathway for communication between cells [1–3]. Initially, the functional role of extracellular hydrolysis of nucleotides and in particular of ATP was not understood and measurement of “apparent” ecto-nucleotidase activity was often assumed to result from cell damage and access of substrates to cytosolic nucleotidases. To date, no functional processes energized by extracellular ATP have been identified. Depending on subtype, ecto-nucleotidases typically hydrolyze nucleoside tri-, di-, and monophosphates and dinucleoside polyphosphates and produce nucleoside diphosphates, nucleoside monophosphates, nucleosides, phosphate, and inorganic pyrophosphate (PPi). A major functional role of extracellular nucleotide hydrolysis and production of extracellular nucleoside would thus relate to the control of ligand availability at nucleotide (P2) receptors and at adenosine (P1) receptors. These processes are highly relevant for purinergic signal transmission and can result in prevention of receptor desensitization or termination of receptor activation by ligand hydrolysis or also in receptor activation by the hydrolysis products generated [4]. However, the evidence supporting a functional role of ecto-nucleotidases in purinergic signaling varies considerably between enzyme species. Furthermore, some ecto-nucleotidases are multifunctional proteins interacting with extracellular matrix proteins or they signal into the cell.
Nucleosides as final hydrolysis products can in turn be salvaged by cellular reuptake via specific transporters and rephosphorylation inside the cell [5]. In addition, the production of extracellular pyrophosphate from ATP is of major relevance in the control of bone mineralization [6] and vascular smooth muscle calcification [7]. In the digestive system, hydrolysis of extracellular nucleotides may furthermore serve in the absorption of their molecular constituents.
Substrates relevant for purinergic signaling
Major extracellular purine and pyrimidine compounds known to elicit cell surface receptor-mediated signals in mammalian cells include ATP, ADP, UTP, UDP, UDP-glucose, and some additional nucleotide sugars, some dinucleoside polyphosphates, and the nucleoside adenosine [1]. Whereas adenosine activates solely G protein-coupled receptors (the P1 receptors A1, A2A, A2B, and A3), nucleotides act via ionotropic (P2X) or G protein-coupled (P2Y) receptors. The homo- or hetero-trimeric P2X receptors (seven subtypes, P2X1 to P2X7) are activated by ATP and represent Na+-, K+-, and Ca2+-permeable ion channels. Ligand preferences (in brackets) of the eight human P2Y receptors are as follows: P2Y1 (ADP), P2Y2 (UTP=ATP), P2Y4 (UTP), P2Y6 (UDP), P2Y11 (ATP, NAD+), P2Y12 (ADP), P2Y13 (ADP), and P2Y14 (UDP, UDP-glucose and other nucleotide sugars). In addition, the P2Y-like receptor G protein-coupled receptor (GPR) 17 responds to both uracil nucleotides (such a UDP-glucose) and cysteinyl-leukotrienes [8]. Dinucleoside polyphosphates act on some P2X and P2Y receptors [9, 10]. Moreover, evidence has been provided that NAD+ [11–13] and ADP ribose [14] function as ligands at P2Y receptors.
ATP can also serve as a co-substrate of ecto-kinases in the phosphorylation of cell surface-located or extracellular proteins [15, 16]. In murine tissue, NAD+ can (in addition to acting as a P2Y receptor agonist) activate P2X7 receptors as a result of ADP ribosylation [17].
Four major groups of ecto-nucleotidases
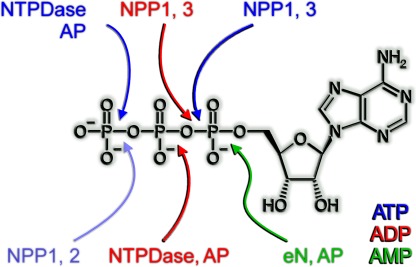
The four major groups of ecto-nucleotidases include the ecto-nucleoside triphosphate diphosphohydrolases (E-NTPDases), ecto-5′-nucleotidase (eN), ecto-nucleotide pyrophosphatase/phosphodiesterases (E-NPPs), and alkaline phosphatases (APs) (Table 1). The E-NTPDases are nucleotide-specific and hydrolyze nucleoside triphosphates and diphosphates with the nucleoside monophosphates as the final hydrolysis product. According to present knowledge, they represent the major nucleotide-hydrolyzing enzymes involved in purinergic signaling but they do not hydrolyze dinucleoside polyphosphates, ADP ribose, NAD+, or AMP. Similarly, the nucleoside monophosphate-hydrolyzing eN is nucleotide-specific and the major enzyme producing extracellular adenosine from AMP. The other two groups of enzymes act as ecto-nucleotidases but hydrolyze also other substrates. E-NPPs hydrolyze nucleoside triphosphates and diphosphates, dinucleoside polyphosphates, ADP ribose, NAD+, and a variety of artificial substrates but not AMP. Some members of this protein family hydrolyze phospholipids. APs finally hydrolyze nucleoside tri-, di-, and monophosphates, pyrophosphate, and a large variety of additional monoesters of phosphoric acid. As compared to E-NTPDases and eN, the examples for a role of E-NPPs and APs in the control of purinergic signaling are still scarce. Figure 1 depicts the different cleavage sites of members of the four enzyme families with ATP, ADP, or AMP as a substrate.
Table 1.
Overview of major human ecto-nucleotidases and some of their properties
| Family name and EC number | Protein name | Full name, aliases, historical names | Gene name | MA | # Cystine bridges | Major substrates | Final product | Substrate affinity (Km), μM |
|---|---|---|---|---|---|---|---|---|
| Ecto-nucleoside triphosphate diphosphohydrolase (EC 3.6.1.5) | NTPDase1 | CD39, ATPDase, ecto-apyrase | ENTPD1 | N, C | 5 | NTP, NDP | NMP, Pi | ATP ~10–200 |
| NTPDase2 | CD39L1, ecto-ATPase | ENTPD2 | N, C | 5 | NTP (NDP) | NMP, Pi | ATP ~70 | |
| NTPDase3 | CCD39L3, HB6 | ENTPD3 | N, C | 5 | NTP, NDP | NMP, Pi | ATP ~75 | |
| NTPDase4 | UDPase (hlALP70v), hlALP70 | ENTPD4 | N, C | 3 (a) | NTP, NDP, little ATP, ADP | NMP, Pi | UDP, GDP (b) ~200–500 | |
| NTPDase5 | CD39L4, PCPH, ER UDPase | ENTPD5 | N | 2 | UDP GDP,CDP, ADP, little ATP | NMP, Pi | GDP ~130 | |
| NTPDase6 | CD39L2 | ENTPD6 | N | 2 | GDP, IDP,UDP,CDP, no ATP | NMP, Pi | n.d. | |
| NTPDase7 | LALP1 | ENTPD7 | N, C | 3 (a) | UTP, GTP, CTP, little ATP | NMP, Pi | n.d. | |
| NTPDase8 | Liver canalicular ecto-ATPase, hATPDase | ENTPD8 | N, C | 5 | NTP (NDP) | NMP, Pi | ATP 81–226 | |
| Ecto-5′-nucleotidase (EC 3.1.3.5) | eN | CD73, ecto-5′-nucleotidase, eNT, low Km 5′-NT | 5NTE (ENT5) | GPI | 4 | NMP | Nucleoside, Pi | AMP 1–50 |
| NMN | NR, Pi | |||||||
| Ecto-nucleotide pyrophosphatase/phosphodiesterase (EC 3.6.1.9) (EC 3.1.4.1) | NPP1 | PC-1, NPPγ, PDNP1 | ENPP1 | N | 16 (a) | NPP1 to NPP3: | ||
| NTP, NDP | NMP, PPi, Pi | ATP ~50–500 | ||||||
| Dinucleoside polyphosphates | NMP + Npn-1 | ApnA ~1–20 | ||||||
| NAD+ | AMP + nicotinamide mononucleotide | – | ||||||
| NPP2 | Autotaxin, PD-1α, NPPα lysophospholipase-D, PDNP2 | ENPP2 | – | 15 | ADP-ribose | AMP + ribose-5-phosphate | ||
| UDP-glucose | UMP + glucose-6-phosphate | |||||||
| PPi | Pi | ~100 | ||||||
| In addition for NPP2: | ||||||||
| NPP3 | Gp130RB13-6, B10. PD1-β, NPPβ, PDNP3, CD203c | ENPP3 | N | 16 (a) | Lysophosphatidylcholine and sphingosylphosphorylcholine | Lysophosphatidic acid, sphingosine-1-phosphate | ~200 | |
| NPP4 | – | ENPP4 | C | ? | ||||
| NPP5 | – | ENPP5 | C | 0 (a) | ||||
| NPP6 | Choline-specific glycerophosphodiester phosphodiesterase | ENPP6 | C | 0 (a) | ||||
| NPP7 | Alk. sphingomyelinase | ENPP7 | C | 0 (a) | ||||
| Alkaline phosphatase (EC 3.1.3.1) | TNAP | Tissue nonspecific AP, liver–bone–kidney-type AP, TNSALP | ALPL | GPI | 2 | NTP, NDP, NMP (various monoesters of phosphoric acid) PPi | Nucleoside, Pi (dephospho-compound) Pi | Low ATP ~40, high ATP ~1,000 |
| PLAP | Placental AP, PLALP | ALPP | GPI | 2 | ||||
| GCAP | Germ cell AP, GCALP | ALPP2 | GPI | 2 | ||||
| IAP | Intestinal AP, IALP | ALPI | GPI | 2 | ||||
Cysteine bridges: (a) very certain, based on modeling; ? uncertain; (b) values for bovine enzyme. MA Membrane association either with N- and C-terminal or N- or C-terminal transmembrane domain or GPI (glycosylphosphatidylinositol) anchor
NMP nucleoside monophosphate, NDP nucleoside diphosphate, NTP nucleoside triphosphate, NMN nicotinamide mononucleotide, NR nicotinamide riboside
Fig. 1.
Overview of enzyme-specific cleavage sites of individual types of ecto-nucleotidases. Enzymes shown in blue, red, and green accept ATP, ADP and AMP, respectively, as substrates. The cleaved bond is marked by an arrow in the according color. NPPs cleave the same bond in ATP and ADP whereas NTPDases hydrolyze different bonds. The removal of the gamma-phosphate from ATP by NPP1 and NPP2 (pale blue) can be regarded a side reaction of this enzyme class caused by inverted binding of the nucleotide to the active site. See also Fig. 11
Additional nucleotide-metabolizing enzymes
Additional enzymes that can also hydrolyze certain nucleotides are not included in this review. Mammalian prostatic acid phosphatase (PAP) is expressed as a secreted or as a transmembrane protein [18]. At a pH of 7.0, the two mouse isoforms were found to dephosphorylate a large variety of compounds including AMP and to a minor extent also ADP. At acidic pH (pH 5.6), PAP dephosphorylates all purine nucleotides (AMP, ADP, ATP). This may have implications in inflammatory conditions where extracellular pH is reduced. By dephosphorylating extracellular AMP to adenosine and activating A1-adenosine receptors, the membrane-bound form of PAP (TM-PAP) is thought to exert antinociceptive effects in the dorsal spinal cord [19–21]. Similarly, mammalian tartrate-resistant acid phosphatase (TRAP) hydrolyzes a wide range of phosphate monoesters and anhydrides including nucleotides such as ATP, ADP, and (to a minor extent) AMP [22]. Among others, the enzyme is highly expressed in osteoclasts and osteoblasts and—due to its acid pH optimum—may be relevant for bone remodeling [23, 24]. Extracellular nucleoside diphosphates may be hydrolyzed also by soluble calcium-activated nucleotidase (CAN) [25]. Following recombinant expression, the human (but not the rodent) form of this enzyme is cleaved and released from cells. In addition, accumulating evidence has been provided that the sarcolemmal α-sarcoglycan is a Ca2+, Mg2+-dependent ecto-ATP diphosphohydrolase [26]. Ecto-ATPase activity has also been attributed to the neural cell adhesion molecule (NCAM) [27]. Finally, cell surface location of molecular components of mitochondrial F1Fo ATP synthase/F1 ATPase was described for several cell types. They have been identified as cell surface receptors for apparently unrelated ligands but generation of ATP or ADP has also been reported [28, 29]. Whether the observed nucleotidase activity was due to this ecto-protein requires further investigation [30].
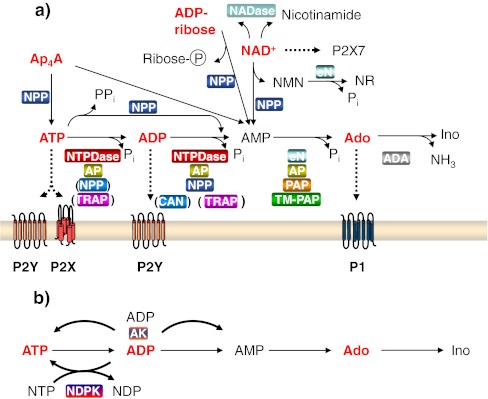
Additional ecto-enzymes such as ecto-nucleoside diphosphate kinase and ecto-adenylate kinase can interconvert extracellular nucleotides, others metabolize NAD+ (NAD glycohydrolases [NADases, CD38, CD157] and mono[ADPribosyl] transferases [ARTs]), or deaminate extracellular adenosine to inosine (adenosine deaminase) (ref. in [30–32]). In particular, ecto-nucleoside diphosphate kinase and ecto-adenylate kinase may in certain cell types significantly contribute to increase the pericellular concentrations of ATP via phosphotransfer reactions [30, 33, 34]. Figure 2 provides an overview of the cell surface-located metabolism of ATP (as an example for nucleoside triphosphates), Ap4A (as an example for dinucleoside polyphosphates), and NAD+ and its hydrolysis products and relates it to purinergic signaling.
Fig. 2.
Principal pathways of extracellular nucleotide metabolism. a Degradation of extracellular nucleotides and purinergic receptor activation with ATP, Ap4A, and NAD+ as examples. Compounds capable of receptor (P2X and P2Y nucleotide receptors or P1 adenosine receptors) activation are indicated in red. Enzymes capable of the specified catalytic reaction are highlighted with a colored background box. ADA ecto-adenosine deaminase, AP alkaline phosphatase, CAN soluble calcium-activated nucleotidase, eN ecto-5′-nucleotidase, NADase NAD-glycohydrolase, NPP ecto-nucleotide pyrophosphatase/phosphodiesterase, NTPDase ecto-nucleoside triphosphate diphosphohydrolase, PAP prostatic acid phosphatase, TM-PAP transmembrane-PAP, TRAP tartrate-resistant acid phosphatase. NAD+ can be hydrolyzed by NPPs to AMP and nicotinamide mononucleotide (NMN) and by NADase to nicotinamide and ADP-ribose. ADP-ribose can in turn be degraded by NPPs to AMP and ribose-5-phosphate. Nicotinamide mononucleotide can be dephosphorylated by eN to nicotinamide riboside (NR). Adenosine (Ado) or inosine (Ino) can be recycled into the cell via specific nucleoside transporters. NAD+ can function as a ligand of some P2 receptors (not indicated), and in murine tissue, it can activate P2X7 receptors as a result of ADP ribosylation. b Ecto-anabolism of nucleotides. At the surface of some cells, ATP can be synthesized extracellularly from ADP via ecto-nucleoside diphosphate kinase (NDPK), whereby another nucleoside triphosphate (NTP) serves as the phosphate donor. In contrast to NDPK, ecto-adenylate kinase (AK) is adenine nucleotide-specific. Depending on mass action, a phosphate can be transferred from one ADP molecule to another, resulting in the formation of ATP and AMP (or vice versa)
Structure and catalytic mechanism of ecto-nucleotidases
All four ecto-nucleotidases described in this review consist of relatively large chains of at least 500 amino acids and of at least two domains. Their crystal structures have been determined, although for eN, structural data are only available for homologues from bacteria and yeast. Complex structures with the reaction products or non-hydrolysable substrate analogues provide detailed insight into substrate binding and the catalytic mechanism. On the assumption that the modification of the nucleotide does not significantly perturb the catalytically competent substrate-binding mode, the relevant molecular interactions for base specificity or the specificity towards nucleoside triphosphates, diphosphates, or monophosphates can be characterized. Often also candidates for the nucleophilic water molecule attacking the phosphoryl group of the substrate can be visualized in these structures. The atomic structures allow in-depth comparisons of the catalytic mechanisms between the various ecto-nucleotidases and offer the possibility to design substances that interfere with the catalytic activity in an enzyme-specific manner.
This review discusses the four major groups of ecto-nucleotidases with an emphasis on the human enzymes. It focuses on the principal cell biological, catalytic and in particular also structural properties of the enzymes and provides only brief reference to tissue distribution, and physiological and pathophysiological functions.
Ecto-nucleoside triphosphate/diphosphohydrolases
General properties and functional role
Members of the ecto-nucleoside triphosphate diphosphohydrolase protein family are ecto-nucleotidases in the strict sense [30, 32, 35, 36]. These enzymes hydrolyze extracellular nucleotide tri- and diphosphates in the presence of millimolar concentrations of Ca2+ or Mg2+ with high rates at physiological extracellular pH between 7 and 8 (EC 3.6.1.5). Nucleoside monophosphates are the final hydrolysis product.
Eight paralogues have been identified in mammals. Four of these (NTPDase1, NTPDase2, NTPDase3, and NTPDase8) are typical cell surface-located enzymes. NTPDase4–7 share an intracellular organellar localization, but secreted forms of NTPDase5 and NTPDase6 have been reported (ref. in [36, 37]) (Fig. 3). NTPDase1, NTPDase2, NTPDase3, and NTPDase8 hydrolyze both nucleoside triphosphates and diphosphates but the range of substrates of the other enzymes is more restricted. Initially varying nomenclatures were in use for individual enzymes. Following a more detailed biochemical characterization and the availability of primary structures, the nomenclature has been revised [38, 39] (Table 1), but some of the original nomenclature is still used today. In particular, CD39 is often used in immunological contexts instead of NTPDase1.
Fig. 3.
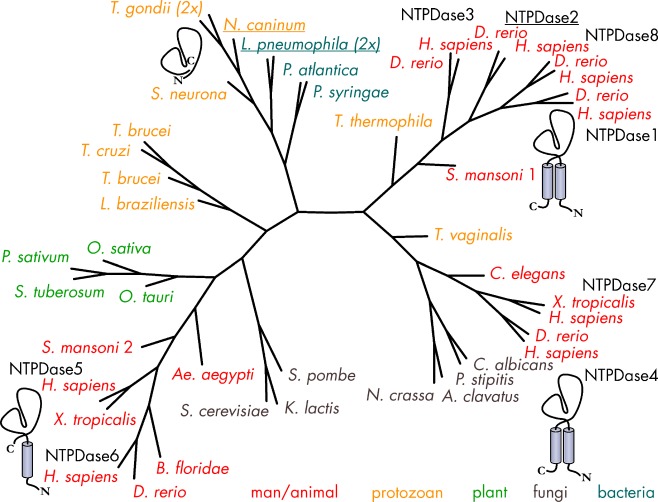
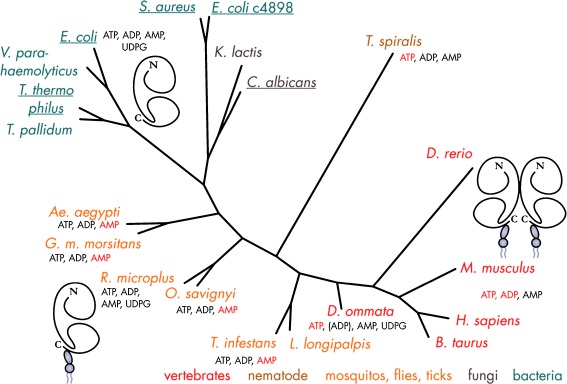
Radial phylogenetic tree of NTPDases. The tree highlights the clear segregation of vertebrate NTPDases into cell surface-located enzymes which are involved in purinergic signaling (NTPDase1–3 and NTPDase8) and the intracellularly located NTPDase4–7. Although NTPDases are ubiquitous in eukaryotes, cell-surface type forms are probably present in only a few non-vertebrate eukaryotes. The scarcity of bacterial NTPDase genes suggests that they have been acquired by horizontal gene transfer. Amino acid sequences have been aligned with Tcoffee [619]. The tree has been calculated with the program Protdist as included in Bioedit and the visualization was done with the program PhyloDraw. Proteins for which structural data are available have been underscored. GI Accession numbers—A. aegypti: 108878621; Aspergillus clavatus: 121713566; C. elegans: 17539006; Branchiostoma floridae: 210115619; C. albicans: 68480942; Danio rerio NTPDase1: 57525937, NTPDase2: 54261809, NTPDase3: 134133300, NTPDase4: 50539906, NTPDase6: 62955697, NTPDase8: 268837940; Homo sapiens NTPDase1: 1705710, NTPDase2: 45827719, NTPDase3: 4557425, NTPDase4: 3153211, NTPDase5: 3335102, HsNTPDase6: 3335098, NTPDase7: 9623384, NTPDase8: 158705943; Kluyveromyces lactis: 50311623; L. pneumophila: 81377241/gi 81377833; Leishmania braziliensis: 154333055; Neospora caninum: 3298332; Neurospora crassa: 85108997; Pisum sativum: 563612; Oryza sativa: 77548506; Ostreococcus tauri: 308802668; Pichia stipitis: 149389003; Pseudoalteromonas atlantica: 122971633; Pseudomonas syringae pv. tomato: 81730387; S. cerevisiae: 603637; Sarcocystis neurona: 32816824; Schistosoma mansoni 1: 33114187, 2: 114797038; Schizosaccharomyces pombe: 19114359; Solanum tuberosum: 2506931; Tetrahymena thermophila: 118383992; T. gondii: 2499220/gi2499221; Trichomonas vaginalis: 154413345; T. brucei: 72392821; Trypanosoma cruzi: 71414508; Xenopus tropicalis NTPDase5: 301618468, NTPDase7: 62859996
NTPDases are expressed in essentially every tissue [32, 36]. They reveal overlapping tissue distributions but in situ they are mostly expressed by different cell types [35]. Examples of cellular co-expression of NTPDases identified by immunocytochemistry include epithelial cells in a variety of tissues [40]. NTPDase1 is the most thoroughly investigated enzyme and considerable insight has been gained from the generation of knockout mice [41, 42]. NTPDase1 is a lymphocyte activation marker and found to be expressed on natural killer cells, monocytes, dendritic cells, and subsets of activated T cells. By modulating purinergic signaling, the enzyme plays a major role in the control of the cellular immune response [43–45]. In addition to other tissues, prominent expression of NTPDase1 is observed on vascular endothelium. By hydrolyzing prothrombotic ADP, NTPDase1 maintains vascular fluidity and plays an important role as a modulator of vascular inflammation and thrombosis [46, 47] as well as in cerebroprotection and cardioprotection [48]. It furthermore controls endothelial P2Y receptor-dependent vasorelaxation [49]. In addition, NTPDase1 is the major enzyme regulating nucleotide metabolism at the surface of vascular smooth muscle cells and thus contributes to the local regulation of vascular tone by nucleotides [49, 50].
NTPDase2 is expressed on the adventitial surface of blood vessels where it contributes to vascular hemostasis [46, 47]. It is co-expressed with NTPDase3 and eN in salivary cells and stratified epithelia of the gastrointestinal tract [40]. It is also found on taste buds [51] and was identified in a variety of tumor cells [37]. In the rodent brain, it is highly expressed by adult neural stem cells, where it has been implicated in the generation of new nerve cells, by non-myelinating Schwann cells of the peripheral nervous system, the satellite glia of dorsal root ganglia, and enteric glia [52–57]. In Xenopus laevis, NTPDase2 together with the P2Y1 receptor is essential for eye development [58].
NTPDase3 is expressed in subsets of neurons in the brain expressing the neuropeptide hypocretin-1/orexin-A [59], in a variety of epithelia including kidney, airways, reproductive and digestive systems [40], and in all Langerhans islet cell types [60]. NTPDase1 and NTPDase3 are expressed in airway epithelial surfaces, in addition to tissue nonspecific AP (TNAP) and E-NPPs [61, 62]. A recent immunocytochemical analysis in the mouse implicates NTPDase3 in the control of nociceptive nucleotide transmission [56]. The enzyme is highly expressed in dorsal root ganglion cells and their central projections. It was localized both in IB4 (isolectin B4)-binding and TRPV1 (transient receptor potential cation channel subfamily V member 1)-expressing sensory neurons and their axon terminals in lamina II of the dorsal horn. There was also extensive co-localization of NTPDase3 and eNT, suggesting that these two enzymes act (together with TM-PAP, [21]) in concert in nociceptive circuits to produce adenosine from extracellular ATP. NTPDase2 immunostaining was also apparent in a narrow band of intrinsic neurons in lamina II. Moreover, NTPDase3 was found to be present in large myelinated cutaneous axons and in specialized end organs that participate in tactile sensation. This points to the possibility that nucleotide signaling contributes to low-threshold mechanotransduction [56]. NTPDase8 has a more restricted tissue distribution. It is highly expressed in the liver, kidney, and intestine [32].
NTPDase4 is expressed in all tissues [63]. mRNA encoding the closely related NTPDase7 is similarly widely distributed, implicating that it exerts a general intracellular function [64]. Of the closely related NTPDase5 and NTPDase6, NTPDase5 has been reported in macrophages and in the liver, kidney, prostate, colon, and testis, while NTPDase6 mRNA appears to be expressed predominantly in the heart [65]. Both enzymes are expressed in the cochlea [66]. In humans, the gene ENTPD5 has been found to be identical to the PCPH proto-oncogene, and dysregulation of this gene was demonstrated in some human cancers [67, 68].
Substrates and catalytic properties
NTPDase1, NTPDase2, NTPDase3, and NTPDase8 hydrolyze both nucleoside triphosphates and diphosphates at physiological pH whereby they reveal rather broad substrate specificity towards purine and pyrimidine nucleotides (Table 1). Individual enzymes reveal, however, differences regarding pH sensitivity and also regarding preferences for Ca2+ and Mg2+. For example NTPDase2, NTPDase3, and NTPDase8 are considerably more active at acidic pH than NTPDase1 [69]. Purified cell surface NTPDases have a very high specific activity in the range of several hundred micromoles per minute per milligram [37]. Kinetic properties of the identical enzyme can vary between Ca2+ and Mg2+ salts of the nucleotide. Human NTPDase1 and NTPDase2 have a preference of adenine over uracil nucleotides. Furthermore, NTPDase1, NTPDase2, NTPDase3, and NTPDase8 hydrolyze ATP more rapidly than ADP. The ratio of the maximal rates for the hydrolysis of ATP and ADP can be taken as a signature of the enzyme paralogues [35, 36], whereby NTPDase2 has a particularly strong preference for the hydrolysis of ATP [70].
A comparative biochemical characterization has been performed with human and mouse NTPDase1 to NTPDase3 heterologously expressed in COS-7 cells [69]. Chelation of divalent cations with EDTA or EGTA abrogated catalytic activity. All enzymes exhibited Michaelis–Menten kinetics. The Km values for ATP of human NTPDase1, NTPDase2, and NTPDase3 were 17, 70, and 75 μM, respectively, suggesting that NTPDase1 is the enzyme with the highest affinity for ATP. The Km values for the mouse enzymes were somewhat lower. Recombinant human NTPDase8 hydrolyzed ATP and UTP about equally well and revealed a strong preference for nucleoside triphosphates [71, 72]. Km values varied to some extent between the two investigations and between the Ca2+ and Mg2+ salts of the nucleotides. For ATP, they ranged between 81 and 226 μM. Km values in the same range were obtained for ADP as a substrate. These values are in the same range as those obtained for the native or recombinant enzymes in other investigations [35, 36, 73].
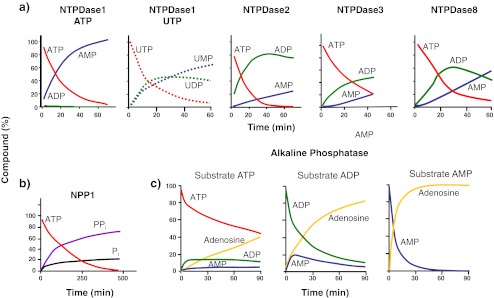
NTPDase2, NTPDase3, and NTPDase8 hydrolyze ATP to ADP, which is released from the enzyme and then further hydrolyzed to AMP. In the case of NTPDase2, considerable amounts of ADP accumulate before it is further hydrolyzed to AMP. In contrast, ATP is hydrolyzed by NTPDase1 directly to AMP, without significant amounts of ADP appearing as an intermediate product [69, 70, 74] (Fig. 4). However, hydrolysis of UTP by NTPDase1 leads to transient accumulation of free UDP [69]. Mammalian NTPDase2 (but not the other NTPDases) displays the unusual property of inactivation by substrate [75].
Fig. 4.
Idealized patterns of nucleotide hydrolysis and product formation by select members of the E-NTPDase, E-NPP, and AP families. The formation of ADP from ATP varies between NTPDases (a) and PPi is a major hydrolysis product of NPP1 (b) (the corresponding formation of AMP is not shown). Both types of enzymes produce AMP as final hydrolysis product. On hydrolysis of ATP, AP (c) produces a substrate pattern similar to NTPDase2, NTPDase3, and NTPDase8, except that hydrolysis proceeds to adenosine. Note that only minor amounts of ADP are released on hydrolysis of ATP by NTPDase1. In contrast, hydrolysis of UTP by the same enzyme is progressive with the formation of UDP as intermediate product. Unless indicated otherwise, the initial substrate was ATP. Hydrolysis of AMP by eN (not shown) would follow similar kinetics as those shown for AP. Graphs are modified from the following references: NTPDase1–3 (ATP): heterologous expression of the rat enzymes in CHO cells. Initial substrate concentration 250 or 500 μM [70, 192]; NTPDase1 (UTP), NTPDase8: heterologous expression of mouse enzymes in COS-7 cells. Initial substrate concentration 500 μM [69]. NPP1: Endogenous enzyme expressed by rat C6 glioma cells, presumably NPP1, application of 10 μM [γ-32P]ATP. The pattern of product formation varies, however, with the initial concentration of ATP [369]. The formation of Pi could in part have resulted from the presence of additional ecto-nucleotidases. AP: calf intestinal AP, commercial product from Fermentas Life Sciences, initial substrate concentration 500 μM (Peter Brendel, Frankfurt, unpublished)
The physiological relevance of the differential expression, the differences in substrate preference, and product formation or also pH dependence of the four ecto-forms is not well understood. This would require detailed information on the state of tissue and the expression of purinergic receptors in the immediate environment of the respective NTPDases.
Substrate preferences differ also for the intracellularly located NTPDases (Table 1). The two isoforms of human NTPDase4 hydrolyze nucleoside tri- and diphosphates, but ATP and ADP only to a minor extent [63, 76]. Both NTPDase5 and NTPDase6 reveal a high preference for nucleoside diphosphates whereby ADP is a poor substrate [39, 77–79]. NTPDase7 preferentially hydrolyses UTP, GTP, and CTP, but ATP and nucleoside diphosphates only to a minor extent [64].
The ATPase activity of NTPDase1, NTPDase2, NTPDase3, and NTPDase8 could potentially interfere with chaperone-assisted protein folding in the endoplasmic reticulum (ER) and phosphorylation in the Golgi apparatus as well as other reactions requiring intraorganellar ATP during the secretory pathway. Whereas it has been suggested that rat NTPDase1 is catalytically inactive intracellularly and becomes active only when fully glycosylated and cell surface-located [80], human NTPDase3 was found to acquire nucleotidase activity as soon as it is natively folded in the ER or in the Golgi intermediate compartment [81].
General molecular properties
The closely related NTPDase1, NTPDase2, NTPDase3, and NTPDase8 contain approximately 500 amino acid residues, and the apparent molecular mass of the glycosylated monomers is in the order of 70 to 80 kDa. They share approximately 40 % amino acid identity and important structural and functional features, including two transmembrane domains (TMDs) close to their N- and C-termini. The large extracellular loop contains the catalytic domain. This loop harbors five highly conserved sequence motifs, the “apyrase-conserved regions” (ACRs). The ACRs are a hallmark of the entire enzyme family and can also be found in the yeast enzymes [82–84]. In addition to the five canonical ACR motifs, a sixth region of high homology and functional importance (ACR6, see below) can be identified within subfamilies (Table 2). The four enzymes have in common four additional conserved regions as well as ten conserved cysteine residues [37].
Table 2.
Consensus sequences of the apyrase-conserved regions of NTPDase family proteins
| Apyrase-conserved region | Consensus sequence |
|---|---|
| ACR1 | (V/I) (V/I/M) X D A G S (S/T) (G/H/S) (T/S) |
| ACR2 | (A/S) T A G (M/L/V) R (L/D/M) (L/F/I) |
| ACR3 | G X X E G X (Y/F) X (W/F/Y) X X X N |
| ACR4 | (G/A/S) X X (D/E) X G G (A/G) S X Q |
| ACR5 | (W/R) (T/A/P/C) (L/D) G X X (L/I/V) |
| ACR6 (vertebrate NTPDase1–3, NTPDase8) | F X A (F/Y) (S/A) X (F/Y) (Y/F) (Y/F/W) |
The consensus sequence of ACR1–5 was generated from analysis of the profile Hidden Markov Model (pHMM logo [627]) generated for the GDA1_CD39 protein family. Unconserved positions are marked with an X. For positions with partial conservation, the respective alternatives are given in brackets. Boldface letters indicate strongly preferred residues. The newly described ACR6 exhibits greater sequence variability when all NTPDase proteins are compared and the shown consensus sequence is for vertebrate cell surface NTPDases only
Unraveling the molecular identity of the surface-located mammalian nucleotide-hydrolyzing enzymes turned out to be a tedious task, since the enzymes lose catalytic activity on detergent solubilization. Human NTPDase1 was first cloned as the CD39 lymphoid cell activation antigen (510 aa) [85], a glycoprotein of unknown function that played a role in B cell adhesion [86]. Considerable homology to an intracellular guanosine diphosphatase from yeast was recognized. In parallel, NTPDase1 was purified to homogeneity from human placenta [87]. Partial sequence identification was obtained but the CD39 sequence had not yet been published. Cloning and expression of the apyrase from potato tubers [82] finally provided the link to the CD39 sequence and demonstration of its ecto-nucleotidase activity [88]. At the same time, the vascular NTPDase was identified as NTPDase1 [89]. A catalytically active human variant with an N-terminal sequence of 11 amino acids differing from CD39 was cloned from a human placenta cDNA library, whereas a truncated form lacking ACR5 was catalytically inactive [90, 91]. The cell biology and function of these forms has not been further elucidated. Interestingly, limited tryptic digestion of human NTPDase1 resulted in the formation of two noncovalently membrane-associated fragments of 56 and 27 kDa that substantially augmented ATPase activity. The underlying molecular mechanisms are not understood [83].
Human NTPDase1 is constitutively palmitoylated at a cysteine within the N-terminal region. The covalent lipid modification of this region of the protein was important both in plasma membrane association and in targeting NTPDase1 to caveolae, where a co-localization of P2Y1 receptor and NTPDase1 was observed [92]. It was suggested that palmitoylation could modulate the function of NTPDase1 in regulating cellular signal transduction pathways [93, 94]. This notion is further supported by the observation that cholesterol-dependent lipid assemblies regulate the activity of NTPDase1 [95]. Removing of cholesterol from membranes reduced ATPase activity which is restored on readdition of cholesterol. NTPDase3 carries putative palmitoylation sites at both its N- and C-terminal cytoplasmic domains [93], but palmitoylation has not been shown experimentally. Digitonin, a detergent that preferentially binds to membrane cholesterol, has a marked stimulatory effect on human NTPDase2, suggesting that NTPDase2 may also be located in rafts [96].
Continued cloning efforts successfully unraveled additional enzymes with related catalytic activities. Mammalian NTPDase2 was first cloned and characterized from rat [97] and then from human sources [98]. Of the three human splice variants (NTPDase2α–γ) [99], only the NTPDase2α variant (495 aa), corresponding to the enzyme cloned from rat [97], was catalytically active. The region missing in the shorter human splice variants 2β and 2γ was found to be essential for correct folding, trafficking, and enzymatic activity. A specific residue (Cys399) which was absent in the shorter isoforms, accounted for the inactive intracellular phenotype shown for the shorter forms. It was suggested that this Cys residue is involved in an intra-chain disulfide bond that is essential for acquisition of tertiary structure and consequently for enzymatic activity. Structural data (see below) have now shown that Cys399 is part of an intrachain disulfide bridge and a central element of the fold of the C-terminal domain.
An additional splice variant also exists for rat NTPDase2 (NTPDase2α and β) [100]. NTPDase2β (545 aa) has an extended cytosolic C-terminus and was found to be localized both at the plasma membrane and at intracellular membranes. Both forms were expressed in a range of rat tissues, differed in their catalytic properties, and were differentially regulated [101, 102]. Their potential cell-specific function has not been determined.
Human genome mapping permitted the prediction of additional human members of the protein family [103]. The numbering (CD39-like, CD29L1-4, comp. Table 1) was unfortunate since these proteins are no cluster of differentiation (CD) proteins. NTPDase3 has subsequently been cloned from human (529 aa) [104] and additional mammalian sources. A splice variant (NTPDase3β) lacking the C-terminus including ACR5 was found to be targeted to the plasma membrane but was catalytically inactive. Since co-expression of both NTPDase3α and NTPDase3β reduced the amount of NTPDase3α targeted to the plasma membrane, the authors speculated that NTPDase3β could function as a possible modulator of nucleotidase activity and purinergic signaling [105]. NTPDase8 (495 aa) was cloned from liver cDNA libraries, heterologously expressed and functionally characterized [71, 72].
The intracellular NTPDase4–7 share the five canonical ACRs with the ecto-forms but their extracellular domains contain only four to six (NTPDase4, NTPDase7) cysteine residues. Whereas NTPDase4 and NTPDase7 also have two TMDs, NTPDase5 and NTPDase6 contain only one N-terminal TMD and are putative type II membrane proteins [37, 39] (Fig. 4). Two closely related forms of human NTPDase4 with different cellular localizations have been identified. Following cell transfection, they have been allocated to the Golgi apparatus (UDPase) [76] and to lysosomal/autophagic vacuoles (LALP70) [106], respectively. The two enzymes have an apparent molecular mass of approximately 70 kDa and differ by only eight amino acid residues, whereby the Golgi-located form represents a splice variant of the lysosomal/autophagic form [63]. The two variants differ in nucleotide preference and also in divalent cation dependence. The lysosomal/autophagic form has the highest preference for UTP and TTP, whereas CTP and UDP are the best substrates of the Golgi-located form. The closely related human NTPDase7 (604 aa, approximately 70 kDa) [64] has been allocated to intracellular organelles of unknown function. The functional roles of NTPDase4 and NTPDase7 are not known.
The glycosylated NTPDase5 and NTPDase6 have an apparent molecular mass of approximately 60 kDa. Expression of human NTPDase5 in COS-7 cells resulted in a secreted and soluble form [107]. A soluble ER-located form of NTPDase5 purified from bovine liver (ER-UDPase) has been suggested to promote reglycosylation reactions involved in glycoprotein folding and quality control in the ER [77]. Similarly, transfection of human NTPDase6 into COS cells resulted in soluble secreted and membrane-bound forms with the secreted form predominating [65, 108]. N-terminal amino acid sequencing of soluble NTPDase6 indicated cleavage of the signal peptide. Similar results were obtained with rat NTPDase6 whereby (similar to NTPDase4) the intracellular form was located in the Golgi apparatus [78]. Both enzymes were strongly activated by either Ca2+ or Mg2+. It is noteworthy that yeast (Saccharomyces cerevisiae) contains only two NTPDase orthologues (Ynd1/Apy1p and yeast GDPase) that are related to the equally Golgi-located NTPDase4 and to NTPDase5/6. Double deletion of the two yeast genes revealed that the two enzymes were required for Golgi glycosylation and cell wall integrity [109]. Similarly, the single homologous gene in the Drosophila genome encodes an intracellular enzyme, homologous to NTPDase6 [110].
The generation of an Entpd5 knockout mouse unraveled novel insight into the function of NTPDase5. Genetic inactivation resulted in two major histopathologic lesions: hepatopathy and aspermia. In addition, loss of Entpd5 promoted hepatocellular neoplasia [111]. The cellular mechanisms underlying these changes are not fully understood, but it is of interest that ENTPD5 has also been identified as the PCPH proto-oncogene [67]. The PCPH oncogene is a truncated form of ENTPD5 [112]. Furthermore, expression of the normal NTPDase5 protein is deregulated or lost in some human cancers and in a wide variety of malignant cells, consistent with a role as a tumor suppressor [111]. A recent study confirms the role of NTPDase5 in promoting N-glycosylation and protein folding in the ER. NTPDase5 was identified as an important link in the PI3K/PTEN/AKT signaling loop [68]. PI3K and PTEN (tumor suppressor protein phosphatase and tensin homologue deleted on chromosome ten) lipid phosphatase control the level of cellular phosphatidylinositol (3,4,5)-trisphosphate, an activator of AKT kinases that promotes cell growth and survival. It is suggested that NTPDase5 upregulation is important for AKT-activated cells to cope with elevated translational activity that generates more nascent polypeptide chains destined for the ER. NTPDase5 thus seems to mediate many of the observed cancer-related phenotypes associated with AKT activation. The physiological role of NTPDase6 is unknown. Whether NTPDase5 and NTPDase6 are released into body fluids in situ and what could be the functional consequences is not known.
A major question concerns the mechanisms underlying the difference in substrate specificity between the paralogues. Numerous mutations have been created to depict amino acid residues particularly within the ACRs responsible for determining catalytic properties [83, 84, 113–117]. In addition, five chimeric cDNAs were constructed in which N-terminal domains of increasing length of rat NTPDase1 were replaced by the corresponding sequences of NTPDase2 and vice versa and expressed in Chinese hamster ovary (CHO) cells [118]. An analysis of the catalytic activities of the expressed enzymes revealed that the sequences of NTPDase1 and NTPDase2 were sufficiently related to form functionally active protein chimeras. Amino acid residues between ACR3 and ACR5 and the cysteine-rich region between ACR4 and ACR5 had a particularly strong influence in conferring a wild-type phenotype to the respective chimera. The data implied that protein structure rather than the amino acid sequence in conserved ACRs per se may be of major relevance for determining differences in the catalytic properties between the two related wild-type enzymes. Chimeras of human NTPDase1 and NTPDase2 in which only the N-terminal halves of the proteins were swapped led to the conclusion that the N-terminal half of the protein (containing ACR1–4) regulates nucleotidase specificity [119]. In addition, in chimeras of NTPDase1 and NTPDase2, the TMDs have been identified of conferring substrate specificities on each enzyme [120].
Membrane topology
NTPDase1, NTPDase2, NTPDase3, NTPDase4, NTPDase7, and NTPDase8 share their general membrane topology with two TMDs at the N- and C-terminus, respectively, with P2X receptors. Otherwise, only epithelial Na+ channels and the acid-sensing ion channels reveal a similar membrane topology [121, 122]. Such a membrane topology is unusual for ecto-enzymes. These are typically attached to the membrane by a single protein or lipid link [123]. Considerable evidence has been accumulated demonstrating that the two TMDs play a role in the function and regulation of the enzymes in addition to anchoring the protein in the membrane. However, individual paralogues differ in their TMD sequences resulting in variable outcome of membrane perturbing experiments [124, 125].
In human NTPDase1 and NTPDase2 [75, 83, 120, 125] and also in chicken NTPDase8 [126], the two TMDs are important for maintaining catalytic activity and substrate specificity. Truncation of both TMDs in NTPDase1 or NTPDase2 resulted in the formation of monomers and 90 % loss of enzyme activity, equal to detergent solubilization [120, 123]. Furthermore, the TMDs of human NTPDase1 are important for folding and sorting as well as for optimal enzymatic activity and activation by cholesterol [127]. While NTPDase1 has been found to be localized in the apical as well as basolateral surface [95, 128–130], when expressed in the polarized Madin–Darby canine kidney (MDCK) cells, it is targeted to the apical side of the membrane. The N-terminal TMD contains an apical targeting signal; however, neither the N- nor the C-terminal domain was required for apical targeting, suggesting that an additional targeting signal is located at the ecto-domain. Folding, release from the ER, and transport to the plasma membrane relied only on the C-terminal TMD, whereas the N-terminus determined the proper orientation of the protein on the membrane. Both TMDs were required for activation of NTPDase1 by cholesterol.
The TMDs also impact on substrate specificity. Removal or disruption of TMDs of NTPDase1 and NTPDase2 abrogated the distinction between the two enzymes regarding substrate preference for ATP versus ADP and progressive cleavage of ATP [120, 131]. This would support the idea that the extracellular loops of the two enzymes have the same intrinsic substrate specificity and that the difference in catalytic properties is affected by their TMDs.
Similarly, membrane-associated NTPDase1 hydrolyzed ATP to completion without release of ADP as an intermediate [70, 74], whereas soluble NTPDase1 lacking both TMDs exhibited intermediate ADP release [74, 131]. The TMDs may also undergo coordinated motions during the process of nucleotide binding and hydrolysis [125, 132, 133], which could impact on the structure of the catalytic domain. Alterations in quaternary structure and subunit interactions may further affect the impact or interaction of ACRs involved in substrate binding and hydrolysis [125]. Whether posttranslational modifications such as protein phosphorylation contribute to this dynamic behavior remains to be investigated. It thus appears that protein structure rather than specific sequence requirements strongly influences catalytic properties. The notion that alterations in protein structure can impact on catalytic properties is now supported by structural analysis (see below).
Oligomeric structure
Using cross-linking of proteins in membrane fractions derived from transfected mammalian cells, members of the E-NTPDase family were shown to form oligomeric complexes. Oligomeric forms reveal increased catalytic activity [134, 135] and the state of oligomerization can affect catalytic properties [114, 136]. The formation of monomers has been held responsible for the strong reduction in catalytic activity following Triton X-100 solubilization. Similarly, mutants lacking one or both TMDs tend to form monomers and have low enzymatic activity [131, 135].
The apparent state of oligomerization varied between individual enzymes and may have depended on cross-linking and solubilization conditions. NTPDase1 to NTPDase3 were found as dimers to tetramers [83, 114, 123, 134, 136–140]. Oligomer formation of the cell surface-located pool of the enzymes could be verified by surface iodination [136]. When applying blue native gel polyacrylamide gel electrophoresis for the analysis of native complex formation, heterologously expressed NTPDase1 was found to consist of monomeric, dimeric, and trimeric forms, with the dimeric form predominating. NTPDase2 revealed monomeric to tetrameric forms, with the monomeric form being the least abundant. This suggests that different oligomeric forms coexist in the plasma membrane. Mutation experiments suggest that multimer formation in rat NTPDase1 and human NTPDase2 is governed by their TMDs [120, 135]. Previous evidence that oligomers are not linked by disulfide bridges [83, 136] has been substantiated by a recent analysis of NTPDase3 and of the crystal structure of the NTPDase2 ecto-domain (see below). Human NTPDase3 was found to preferentially form dimers [138]. Hydrogen bonding involving a conserved glutamine located in TMD1 near the extracellular surface was implicated in dimer formation [141]. The variable oligomeric structure of NTPDases from monomeric to oligomeric forms is in stark contrast to P2X receptors that share similar membrane topology [142]. P2X receptors form a single homooligomeric complex corresponding to a trimer, in the absence of tetramers, dimers, or monomers.
The similarity of the membrane topology of the surface-located NTPDase1, NTPDase2, NTPDase3, and NTPDase8 with P2X receptors and their ability to form oligomers makes them potential candidates for channel formation. It has been suggested that NTPDase1 could function as an ATP release channel. Release of ATP from NTPDase1-transfected Xenopus oocytes could be induced by hyperpolarizing pulses and required functional ecto-ATPase activity [143].
Glycosylation
The glycan moieties of glycoproteins can play important roles in maintaining polypeptide conformation and solubility, for protection of the polypeptide chain from proteolytic degradation, for signals for intracellular sorting and externalization, for cell adhesion, or also for catalytic activity [144]. Members of the E-NTPDase family are N-glycosylated. The degree of glycosylation varies between ecto-forms and intracellular forms. The number of predicted N-glycosylation sites amounts to 6, 6, 7, and 8 for NTPDase1–3 and NTPDase8, respectively, and 2, 3, 2, and 3, for NPTDase4–7, respectively. In contrast to other ecto-nucleotidases, glycan structures of NTPDases have not been determined. Experiments with endoglycosidase H digestion, which specifically recognizes and hydrolyzes the high-mannose N-glycans (that are characteristic for immature proteins resident in the ER) but not highly processed complex oligosaccharides, suggest that most of the N-linked glycosylation sites in human NTPDase3 are processed to complex oligosaccharides. But at least one site was high mannose or hybrid in structure [145]. Neuraminidase treatment of B cell-derived NTPDase1 revealed a small shift in molecular mass, suggesting the presence of sialic acid [86].
Only one of the putative N-glycosylation sites (N81 in NTPDase3), which is located near ACR1, is invariant in the cell surface-located human NTPDases [146]. Glycosylation does not appear to be required for enzymatic activity of NTPDase5 [147, 148] and NTPDase6 [79, 146]. This is shown by bacterial expression of the non-glycosylated proteins or by heterologous expression in the presence of tunicamycin (NTPDase5, [147]).
The results obtained for the ecto-forms varied between experimental conditions and the paralogue investigated. Deglycosylation of isolated NTPDase1 with peptide N-glycosidase F, which removes all N-glycans, resulted in a 30 % reduction in apparent molecular mass without loss of catalytic activity [83, 149, 150]. In contrast, inhibition of N-glycosylation with tunicamycin in COS-7 cells [80] or MDCK cells [127] expressing human NTPDase1 or mutation of putative N-glycosylation sites in rat NTPDase1 [151] impaired or abrogated catalytic activity. Possibly, removal of N-glycans from fully folded proteins no longer impairs their catalytic activity [150]. However, deglycosylation of NTPDase3 with peptide N-glycosidase F reduced catalytic activity [138]. In addition, mutation of N-glycosylation sites in NTPDase2 [99] or NTPDase3 [146] reduced enzymatic activity with the protein retained in the ER. Surface biotinylation analysis furthermore revealed that surface-expressed NTPDase1 receives a higher degree of N-glycosylation than the intracellular forms. Inhibition of N-glycosylation in the ER prevented its plasma membrane localization while terminal modification in the Golgi apparatus was less important [80, 127, 150]. Glycosylation was found to be essential for folding, trafficking, and activity of NTPDase1 and NTPDase2 [99].
Soluble forms
Minor amounts of soluble NTPDase activity were reported to constitutively circulate in human bloodstream [152]. But the truly soluble nature of the enzyme(s) has not been verified by phase partitioning and the molecular identity has not been determined. According to biochemical evidence, truly soluble ecto-forms of the NTPDase family would lose most of their catalytic activity. Shedding of undefined “ATPases” has been observed from endothelial cells from human umbilical vein under conditions of shear stress [153] and from cultured endothelial cells and astrocytes following oxygen–glucose deprivation [154]. There is evidence that catalytically active NTPDase1 can be shed in membrane-bound form from plasma membranes of NTPDase1-expressing cells. The hydrophobic form of NTPDase1 has been found in particulate secretions of rat pancreas under resting conditions [155] and as a result of stimulation with cholecystokininoctapeptide-8 where it is thought to regulate intraluminal ATP concentrations within the ductal tree [129, 156]. However, ecto-ATPase appears to be absent from guinea pig and human pancreatic secretions, implicating species-specific differences [157]. NTPDase1 was also found to be incorporated in microparticles of human and mouse plasma, where it may play a role in the exchange of regulatory signals between leucocytes and vascular cells [158] and in exosomes from diverse cancer cell types [159]. Release of soluble nucleotidases from stimulated sympathetic nerves innervating the guinea pig vas deferens nerve endings has previously been reported but the nature of the enzyme has not been determined [160, 161].
Protein interactions
Co-immunoprecipitation data suggest that NTPDase can be associated with cystic fibrosis transmembrane regulator in human and mouse red blood cell membranes [162]. The interaction between the two proteins might positively affect ATP transport and extracellular hydrolysis [163]. In addition to homooligomers, NTPDases may also form heterooligomers and engage in complexes with purine receptors. Using FRET microscopy of heterologously expressed fluorescence-tagged proteins, close molecular interaction, indicating complex formation, was observed between rat NTPDase1 and NTPDase2 and between NTPDase1 and a variety of P2Y nucleotide and P1 adenosine receptors [164]. The close interaction between NTPDases and P2Y receptors would have a severe impact on the availability of nucleotide agonists at their receptor, as has been implicated from the analysis of an NTPDase1/P2Y1 receptor fusion protein [165].
Downstream signaling
Surface-located NTPDases may have functions additional to their enzymatic activity. In a yeast two-hybrid system, the N-terminus of human NTPDase1 has been shown to interact with truncated Ran Binding Protein M (RanBPM) [166]. RanBPM provides a platform for the interaction of a variety of signaling proteins, including cell surface receptors, nuclear receptors, nuclear transcription factors, and cytosolic kinases [167]. NTPDase1 co-imunoprecipitated with RanBPM in B lymphocytes. Enzymatic activity of NTPDase1 was specifically downregulated after the binding of RanBPM to the N-terminal cytoplasmic domain of NTPDase1. It has been suggested [166] that NTPDase1 might be a bifunctional molecule with ecto-nucleotidase activity located in the large ecto-domain and the ability to bind to RanBPM expressed by the N-terminal cytoplasmic domain. As RanBPM may in turn modulate the ecto-enzymatic functions of NTPDase1, this intermolecular interaction may have important implications for the regulation of extracellular nucleotide signaling pathways.
Downstream signaling, nucleotides
The surface-located members of the NTPDase protein family have a strong influence on purine receptor-mediated signaling. In contrast to APs and E-NPPs, multiple examples have been provided demonstrating that endogenous (rather than added) NTPDases directly impact on the effective agonist concentrations at P2 and (together with AMP-hydrolyzing ecto-enzymes) on P1 adenosine receptors [36, 37, 168]. By hydrolyzing nucleoside triphosphates, they remove agonists for NTP-responsive P2 receptors. At the same time, they produce nucleoside diphosphates as agonist of nucleoside diphosphate-sensitive P2 receptors, which in turn can be inactivated by further hydrolysis to the nucleoside monophosphates. Only few examples are highlighted here.
In vitro, the impact of NTPDases on P2 receptor activation was demonstrated by co-expression of NTPDase1 or NTPDase2 with (ATP- and ADP-sensitive) P2Y1 receptors in human 1321N1 astrocytoma cells. These cells do not express P2 receptors and reveal low ecto-nucleotidase activity [169]. Ecto-nucleotidases selectively modulated the effective agonist concentration at P2Y1 receptors on identical or also neighboring cells, either by degrading ATP or by generating ADP from ATP. In addition, co-localized ecto-nucleotidases, by reducing levels of constitutively released nucleotide, reduced receptor desensitization. Preventing receptor desensitization following tonic or acute nucleotide release may thus be an additional important function of ecto-nucleotidases.
Further approaches to demonstrate the impact of endogenous ecto-NTPDases on purinergic signaling include the application of specific NTPDase inhibitors and knockout, knockdown, or overexpression of the enzymes. There are multiple examples showing that application of inhibitors such as the nucleotide analogue ARL 67156 potentiate nucleotide-mediated neurotransmission [170–173]. Deletion of NTPDase1 in mice revealed a key role of this enzyme in the prevention of thrombosis [41, 42], in purine signaling in angiogenesis [174], vascular permeability [175, 176], vascular relaxation [49], in the control of macrophage function [177], or also in promoting tumor growth [178]. Accordingly, transgenic mice expressing human NTPDase1 exhibited impaired platelet aggregation and were protected from thrombosis in a transplantation setting [179]. Moreover, adenovirus-mediated gene transfer of human placental NTPDase1 [91] into vascular smooth muscle cells was found to suppress thrombus formation and subsequent neointimal growth [180]. Similarly, gene transfer of this enzyme via cationic gelatin-coated stents inhibited subacute in-stent thrombosis and in addition suppressed neointimal hyperplasia and inflammation [181].
Knockdown of NTPDase2 abrogated the inhibitory effect of portal fibroblasts on P2Y receptor-mediated bile ductular proliferation in liver [182]. These and other studies unanimously demonstrate that NTPDases function to constitutively attenuate the impact of nucleotides on their receptors, an effect abolished by application of inhibitors or by deletion of the enzyme. At the same time, NTPDases promote the formation of adenosine from extracellular ATP. The formation of extracellular adenosine resulting from the tandem activity of NTPDase1 and eN has been particularly well documented [183, 184].
Phylogenetic relationship
Homologues of the mammalian ecto-forms of the E-NTPDase family are found in Xenopus [185] and zebrafish [186]. Interestingly, these enzymes are missing in the Drosophila and Caenorhabditis elegans genome. The apparent lack of P2X receptors in these organisms [187] further supports the notion that expression of ecto-NTPDases is a hallmark of purinergic signaling. In contrast, members of the protein family relating to the intracellular forms are present throughout the animal kingdom and also in plants and fungi [37, 188]. Surprisingly, members of the ecto-NTPDase family have been identified at the surface of several pathogenic protozoans and in no more but a few bacteria, including Legionella pneumophila, the causative agent of Legionaires’ disease [37, 189]. The limited occurrence of NTPDase genes in the bacterial kingdom strongly suggests an acquisition by horizontal gene transfer. In plants, NTPDases are thought to be involved in growth and symbiosis between plants and microbes. Even though extracellular ATP exhibits specific physiological effects, purinergic receptors have not been identified in plants [188]. The family of proteins related to NTPDase1 (CD39) and yeast GDPase (GAD1) are also referred to as GAD1_CD39 superfamily [37]. Figure 3 presents the phylogenetic tree of members of the enzyme superfamily with select examples from men, frog, fish, yeast, plant, and bacteria.
NTPDases share two common sequence motifs with members of the ASKHA (acetate and sugar kinases/Hsc70/actin) superfamily of phosphotransferases [190]. They contain the actin-HSP 70-hexokinase β- and γ-phosphate-binding motif [(I/L/V)X(I/L/V/C)DXG(T/S/G)(T/S/G)XX(R/K/C)] [37, 82, 97, 191], in which the DXG sequence is strictly conserved. These motifs are present in both ACR1 and ACR4 (Table 2). Members of this protein superfamily have ATP phosphotransferase or hydrolase activity, depend on divalent metal ion, and tend to form oligomeric structures. Striking similarities have previously been observed in the secondary structure of members of the actin/HSP70/sugar kinase superfamily [79, 192, 193].
Protein structure
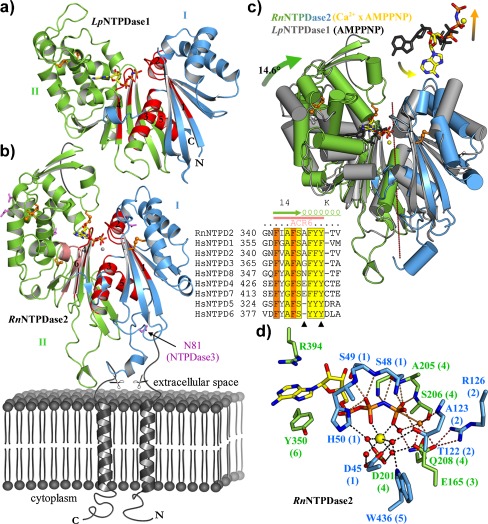
In spite of negligible global sequence identity, enzymes of the ASKHA superfamily share the principal structure of two major domains of similar folds on either side of a large cleft. Homology modeling of the NTPDase3 sequence revealed a high degree of structural fold similarity with a bacterial exopolyphosphatase (PDB 1T6C) that further refined structural predictions for members of the E-NTPDase family [145, 193, 194]. These similarities have now been substantiated by the determination of the crystal structures of the extracellular domain (ECD) of rat NTPDase2 [195] and the soluble NTPDase of the pathogenic bacterium L. pneumophila (LpNTPDase1), which is secreted into the replicated vacuole [196] (Table 3). Both enzymes share a sequence identity of approximately 20 %, and they both contain the five canonical ACR regions. The crystal structures reveal the pseudo-symmetrical arrangement of two extended RNaseH fold repeats that is also found in other members of the actin structural superfamily. Two structural domains are formed which are characterized by a central mixed β-sheet and a peripheral layer of mainly α helices. The terminal α-helices of the RNaseH fold repeats cross each other and interact more strongly with elements of the respective other repeat (Fig. 5).
Table 3.
Reported crystal structures of NTPDases
| Enzyme | Structure | Ligands | Resolution (Å) | PDB ID | Reference |
|---|---|---|---|---|---|
| Rat NTPDase2 | Apo form | – | 1.70 | 3CJ1 | [195] |
| Substrate analogue | AMPPNP, Ca2+ | 2.10 | 3CJA | [195] | |
| Product bound | AMP | 1.80 | 3CJ7 | [195] | |
| Product bound | AMP, phosphate, Ca2+ | 1.80 | 3CJ9 | [195] | |
| Legionella pneumophila NTPDase1 | Apo form | – | 1.60 | 3AAP | [196] |
| Substrate analogue | AMPPNP | 1.65 | 3AAR | [196] | |
| Inhibitor bound | ARL67156 | 2.00 | 3AAQ | [196] | |
| Rat NTPDase1 | Apo form | – | 2.00 | 3ZX3 | [198] |
| Complex | Decavanadate | 2.10 | 3ZX2 | [198] | |
| Complex | Heptamolybdate | 2.70 | 3ZX0 | [198] | |
| Toxoplasma gondii NTPDase3 | Apo form | – | 2.00 | 4A57 | [199] |
| Product bound | AMP | 2.20 | 4A59 | [199] | |
| Substrate analoguea | AMPPNP, Mg2+ | 2.85 | 4A5A | [199] | |
| Toxoplasma gondii NTPDase1 | Apo forma | – | 2.50 | 4A5B | [199] |
| Neospora caninum NTPDase | Apo form | – | 2.80 | 3AGR | [644] |
aPermanently active mutant
Fig. 5.
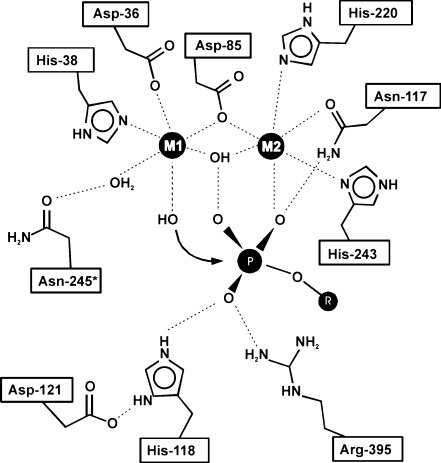
Structure and mechanism of NTPDases. a Crystal structure of an NTPDase from L. pneumophila (LpNTPDase1) in complex with the substrate analogue AMPPNP. The five ACR regions (numbered) are colored in red. b Model of a membrane-bound NTPDase (RnNTPDase2) based on the complex structure of the rat NTPDase2 ecto-domain with Ca2+ and AMPPNP. Protein regions missing from the crystal structure (due to flexibility in the crystal or being absent in the expression construct [borders indicated by scissors]) are depicted in gray. In addition to the five canonical ACR regions, a sixth region of high sequence conservation among mammalian NTPDases is shown in salmon (numbered 6). This region is symmetry-related to ACR2 and involved in nucleoside binding. A corresponding partial sequence alignment is shown as inset with residues involved in nucleoside binding indicated with triangles. c Superposition of rat NTPDase2 and the NTPDase from L. pneumophila based on domain I (blue). The bacterial NTPDase (gray) represents an open, the mammalian structure (colored) a closed conformation. The relative orientation of the two domains differs by a 14.6° rotation around the axis indicated in purple. In the open state, the substrate analogue adopts a considerably different conformation and does not chelate a metal ion. d Close-up view of the active site of NTPDase2. An extensive network of hydrogen bonds from residues of all six ACRs (numbers in brackets, see text and Table 2 for explanation on the sixth ACR region) functions to orient the substrate and the metal cofactor (yellow sphere) for catalysis. Hydrolysis proceeds via activation of a water molecule (orange) by the catalytic base E165
The bacterial enzyme is approximately 10 kDa smaller than the NTPDase2 ecto-domain. It can be described as a minimal construct lacking many of the structural elements of mammalian NTPDases, especially in the periphery of the second domain. Two adjacent, buried disulfide bridges are found within the core of the domain II of both NTPDases. These bridges are strictly conserved among all NTPDases but are missing in bacterial exopolyphosphatases, the next structural relatives of NTPDases. They can therefore be classified as a structural hallmark of NTPDases. NTPDase5 and NTPDase6 are lacking additional disulfide bonds, but homology modeling in our groups does strongly suggest a third disulfide bond to link ACR1 and ACR2 in NTPDase4 and NTPDase7 (Table 1).
Since the rat NTPDase2 structure was determined using protein refolded from bacterial inclusion bodies, no crystallographic insight into the glycosylation pattern was obtained. However, all potential N-glycosylation sites are well surface-exposed. The single site that is conserved among all cell surface NTPDases and required for full activity of NTPDase3 (N81 in NTPDase3, [146]) is found to be located very close to the membrane [195] (Fig. 5b).
In the solved complex structures of rat NTPDase2 ECD and the enzyme from L. pneumophila, the non-hydrolysable ATP analogue β,γ-imidoadenosine 5′-triphosphate (AMPPNP) is bound in the cleft between the two domains. In LpNTPDase1, the divalent metal ion required for catalysis is absent and the electron density for the substrate is indicative of multiple conformations of the ribose and the phosphate tail and only the most highly occupied state was modeled. Together with the observation that the enzyme appears to be in an open conformation (see below), this leads to the conclusion that the complex structure is likely to represent a non-productive binding mode. In the rat NTPDase2 structure, however, a fully occupied complex of the AMPPNP substrate analogue and a Ca2+ ion is bound in the interdomain cleft, engaging into a multitude of interactions with residues from all six ACR regions (Fig. 5b, d).
Noteworthy and in stark contrast to the dimetal center of E-NPPs, APs, and eN, the divalent metal ion is not directly bound to the protein. Rather, the metal ion is bound in a bidentate complex with the nucleotide, the complex representing the true substrate. The octahedral coordination sphere is further completed by four water molecules, which are oriented by the protein (Fig. 5d).
Active site and catalytic mechanism
The ACRs cluster around the interdomain cleft and participate in the formation of the active site. Determination of the structure of rat NTPDase2 ECD in complex with the nucleotide analogue AMPPNP and Ca2+ in a productive binding mode allowed the proposal of the catalytic mechanism. Furthermore, discrete functions can be assigned to individual ACRs and the background of a multitude of mutagenesis effects can now be explained (Table 4). ACR1 and ACR4 as well as ACR3 and ACR5 are related by the pseudo-twofold symmetry axis between the two domains. ACR1 and ACR4 are the loops formed by the first two β-strands of the two RNase H folding motifs. The respective residues are involved in water-mediated coordination of the metal ion (D45 and D201), binding of the substrate’s phosphate tail (S48-H50, G204-S206), and positioning of the nucleophilic water (S206). ACR3 and ACR5 are the two α-helices connecting domain I and II. ACR3 provides the catalytic base E165. E165 is also involved in water-mediated metal ion binding as is W436 of ACR5. ACR2 corresponds to the end of β-strand 4 and α-helix E. Residues involved in substrate and cofactor binding (T122), positioning of the nucleophilic water and the catalytic base (A123, R126), are recruited from this motif. As for the ACR pairs 1/4 and 3/5, a symmetry-related region is also found for ACR2. This sixth ACR region of NTPDase2 does also show high conservation (albeit not overall NTPDases) and participates in the formation of a sandwich-type base-binding pocket (Fig. 5b).
Table 4.
Explanation of the observed effects of previously reported active site mutations of NTPDases on activity
| Residuea | Function | Mutanta | Effect | Explanation | Reference |
|---|---|---|---|---|---|
| D45 | Involved in cofactor binding | D45A (3) | Inactive | Improper cofactor coordination | [84] |
| D45A (1) | More active but reduced kcat/Km in soluble form | [628] | |||
| G47 | Start of first phosphate-binding loop | G47A (3) | Inactive | Side chain would clash with penultimate phosphate | [84] |
| S48 | Hydrogen bond to bridging oxygen of cleaved bond | S48A (1) | Loss of activity in membrane-bound form | Depending on membrane anchorage side chain may be dispensable; influence on Km and kcat/Km unclear | [73] |
| S48A (1) | Increased ADPase, unaffected ATPase in soluble form | [628] | |||
| H50 | Binding of penultimate phosphate | H50G,S (1) | Reduced activity in membrane-bound form, increased activity in soluble form | Conservative substitution possible; depending on membrane anchorage side chain may be dispensable; influence on Km and kcat/Km unclear | [114] |
| H50A (1) | Increased activity in soluble form | [628] | |||
| Note: H50 is substituted for R in wild-type NTPDase3 | H50G (2) | Reduced activity in membrane-bound form, soluble form unaffected | [114] | ||
| R50G,H (3) | Higher activity | [116, 629] | |||
| R126 | Salt bridge to catalytic base | R126A (3) | Inactive | Conservative substitution possible; salt bridge R126-E165 may be less important in soluble form of NTPDase1 | [116] |
| R126K (3) | Active | [116] | |||
| R126A (1) | Slightly reduced activity in soluble form | [628] | |||
| E165 | Catalytic base; activation of nucleophilic water | E165D,Q (3) | Inactive | Activation of nucleophilic water Not possible | [116] |
| E165A (1) | Inactive | [628] | |||
| D201 | Cofactor binding | D201A (3) | Inactive | Improper cofactor coordination | [84] |
| D201A (1) | More active but reduced kcat/Km in soluble form | [628] | |||
| D201E (3) | Increased ATPase, unaffected ADPase | [113] | |||
| D201E (1) | Reduced activity, reduced kcat/Km | [73] | |||
| G203 | Start of second phosphate-binding loop | G203A (3) | Inactive | Side chain would clash with terminal phosphate | [84] |
| G204 | Hydrogen bond to bridging oxygen of cleaved bond | G204A (3) | 90 % loss of activity | Adverse steric effects | [630] |
| S206 | Positioning of nucleophilic water | S206A (3) | Inactive | Incorrect or insufficient positioning of nucleophilic water | [116] |
| S206A (1) | 90 % loss of activity in soluble form | [628] | |||
| Q208 | Positioning of nucleophilic water | Q208A (3) | Inactive | Incorrect positioning of nucleophilic water | [116] |
| W436 | Cofactor binding | W436A (3) | Increased ATPase and ADPase | Altered cofactor binding | [113] |
aNTPDase2 residue numbering is used. Numbers in brackets refer to the NTPDase enzyme used in the study
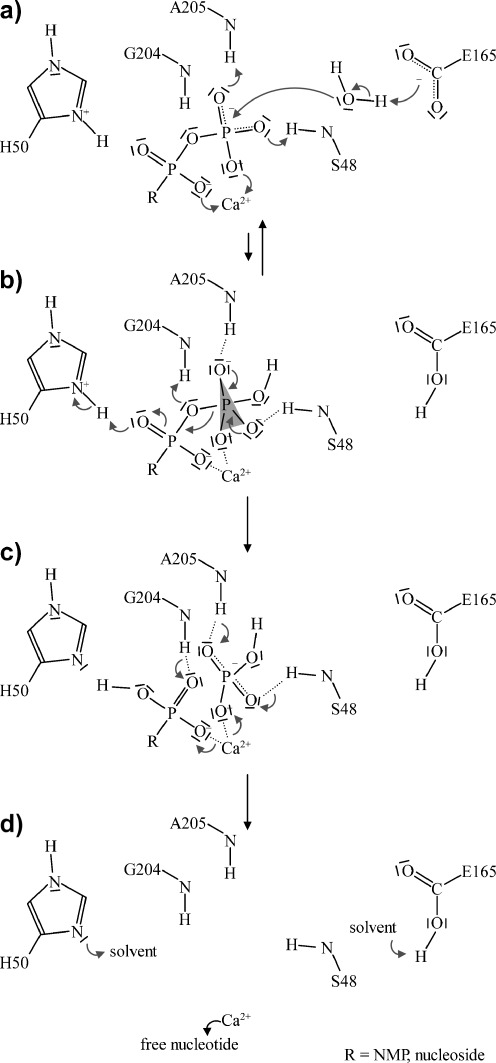
Hydrolysis proceeds via an inline nucleophilic attack of a nucleophilic water (Figs. 5d and 6) on the terminal phosphate. The water molecule is well positioned between the phosphorus atom of the γ-phosphate and the side chain of Glu165, which acts as the catalytic base. The partial positive charge of the phosphorus is increased by coordination of the metal ion. Upon nucleophilic attack, a trigonal planar transition state is thought to be formed. The backbone amides and side chain hydroxyls of the two phosphate-binding loops (i.e., ACR1 and 4) provide proton-donating hydrogen bonds to stabilize the accumulating negative charge of the transition state.
Fig. 6.
Schematic representation of the postulated reaction mechanism of NTPDase-mediated NTP (R = NMP) or NDP (R = nucleoside) hydrolysis. a Activation of the nucleophilic water by E165 and subsequent inline nucleophilic attack on the terminal phosphate. The negative charge of the phosphate groups is reduced by complexation of the divalent metal cation. A positively charged H50 may additionally draw partial negative charge from the terminal to the penultimate phosphate group, lowering the activation barrier. b Collapse of the trigonal bipyramidal transition state. The negative charge of the transition state is stabilized by proton-donating hydrogen bonds from the phosphate-binding loops (e.g., S48, A205). Additional hydrogen bonds may exist. c Product release. H50 may be responsible for protonation of the leaving group. d Reconstitution of the active site
No complex structure has yet been published for a nucleoside diphosphate (NDP)-bound NTPDase. However, the comparison of substrate analogue- and product AMP-bound complex structures of rat NTPDase2 is suggestive of a binding mode in which the binding site of the third (α in ATP) binding site is skipped. Potentially, NTPDases can achieve ADP/ATP discrimination by adjusting the distance between the base binding pocket and the catalytic base.
In addition to eN, the NTPDases seem to represent a family of ecto-nucleotidases in which a dynamic domain rearrangement is coupled to activity. Members of the ASKHA structural superfamily are renowned for especially large conformational transitions coupled to function (e.g., F-actin versus G-actin). A multitude of experiments have shown that a strong linkage exists between the active site and transmembrane helix interactions [71, 73, 125, 132, 133, 141, 197]. Oxidative cross-linking experiments carried out with cysteine mutants of membrane-bound variants of NTPDase1 indicate a rather dynamic coupling of coordinated movements of the transmembrane helices to spatial rearrangements of the active site [125, 132, 133]. The observation of the bacterial NTPDase in an open and of the rat enzyme in a closed state (Fig. 5c) supports the idea that the active site of NTPDases is subject to dynamic rearrangements. In the two crystal structures, the relative orientation of the two structural domains differ by a 14.6° rotation around an axis parallel to the active site cleft. If such a rotational domain closure movement occurs in solution, then it may help in catalysis by switching between open states with less steric constraints for easier substrate binding and product release and closed states for hydrolysis. This notion is substantiated by our recent observation of the NTPDase1 ecto-domain with relative domain orientation differing by up to 7.4° [198]. In the NTPDases of the protozoan parasite Toxoplasma gondii, a restriction of a 12° domain closure motion is used to retain the enzyme in an inactive form. Reduction of a regulatory disulfide bridge initiates a domain closure and leads to activity [199].
Similarly, the domain motion may have an impact on catalytic specificities such as the processivity of NTPDase1. The transmembrane helices might influence catalysis by changing the frequency or magnitude of the domain motion. Mutational studies suggest that polar residues at the extracellular side of the transmembrane helices and proline-rich linker regions between the ECD and the transmembrane helices participate in the coupling process [132, 141, 197].
Ecto-5′-nucleotidase
General properties and functional role
Vertebrate ecto-5'-nucleotidase (eN, EC 3.1.3.5) hydrolyzes ribo- and deoxyribonucleoside 5′-monophosphates including AMP, CMP, UMP, IMP, and GMP, whereby AMP generally is the most effectively hydrolyzed nucleotide. It has much lower activity with deoxyribonucleotides as substrates. Recently, it was reported that heterologously expressed human eN can in addition hydrolyze nicotinamide mononucleotide to nicotinamide riboside + Pi and to a minor extent also NAD+ to adenosine and/or nicotinamide riboside. However, the turnover rates were much lower than with AMP as a substrate [200]. The physiological relevance of these catalytic reactions needs to be further investigated (Table 1).
EN is a Zn2+-binding glycosylphosphatidylinositol (GPI)-anchored homodimeric protein, with its catalytic domain facing the extracellular medium. It is found both in membrane-anchored and soluble forms. Reported Km values for AMP range between 1 and 50 μM (ref. in [201, 202]). The enzyme has a broad tissue distribution, and it is expressed by subpopulations of human T and B lymphocytes and also by a considerable variety of tumor cells [203]. Catalytic activity varies considerably between individual tissues [204]. The existence of both catalytically active and inactive forms of eN has been described which may result from differential posttranslational processing [205–208] or also from the interaction of the enzyme with extracellular matrix proteins (see below).
The production of extracellular adenosine from extracellular AMP is considered to be a major function of eN. eN would thus act as a control point for the extracellular provision of this signal molecule. Changes in turnover and expression of the enzyme would—together with the provision of the substrate AMP—control the availability of adenosine. Importantly, ATP and ADP are competitive inhibitors of vertebrate eN with Ki values in the low micromolar range. These nucleotides apparently still bind to the catalytic site but without being hydrolyzed [209, 210]. Cellular release of ATP/ADP would thus result in feed-forward inhibition of eN and delay of extracellular adenosine formation until their extracellular levels have been reduced to low micromolar levels by other ecto-nucleotidases [211]. Additional functions of eN include the provision of adenosine for cellular reuptake and purine salvage [5] and, as lined out below, potential roles in specific protein (cell) interactions and downstream cellular signaling.
The adenosine formed in turn activates specific G protein-coupled adenosine (P1) receptors (A1, A2A, A2B, A3) [212]. These have a very broad tissue distribution and are involved in a multiplicity of physiological and pathological functions. In this respect, the functional impact of eN directly relates to that of the extracellular adenosine. This notion is supported by studies on eN knockout mice. eN has been deleted in at least three different laboratories. Knockout animals are healthy and display normal behavior [213–215]. In-depth experimental analyses of these animals underpinned the pathophysiological impact of eN and demonstrated that in the tissues investigated a significant amount of extracellular adenosine is derived from released nucleotide. Interestingly, certain 2-substituted AMP derivatives, including 2-hexylthio-AMP, 2-cyclopentylthio-AMP, 2-cyclohexylmethylthio-AMP, and 2-cyclohexylethylthio-AMP were shown to be substrates of eN and readily converted to the corresponding 2-substituted adenosine derivatives. Such compounds may serve as new lead structures for the development of potent prodrugs of adenosine A2A receptors that are preferentially activated in tissues with elevated eN expression such as in inflammatory conditions [216].
Several overviews have covered the multiple pathophysiological roles of eN. They reveal novel insight into its functional involvement in endothelial and fluid transport; endothelial barrier function; adaption to hypoxia and ischemic preconditioning; the cardiovascular system; lung, liver, and kidney function [184, 217–221]; the airways [222]; immunity and inflammation [223, 224]; leucocyte trafficking [225]; the nervous system [226, 227], including a novel role of eN in inhibiting nociception [228]; or the immune control of cancer [229–232]. eN is a component of the multiresistance machinery of tumor cells against immune surveillance and/or anticancer therapy [233]. eN-deficient mice have increased antitumor immunity and are resistant to experimental metastasis [234]. Depending on the pathophysiological context, therapeutic targeting of the eN pathway can thus involve targeting of adenosine receptors equally as well as targeting of eN via inhibitors or targeted application of soluble forms of eN [218].
Ecto-5′-nucleotidase versus soluble 5′-nucleotidases
The presence of both soluble eN and intracellular nucleoside 5′-monophosphate-hydrolyzing enzymes has initially created some confusion regarding the identity of 5′-nucleotidase-hydrolyzing enzymes. This initiated the development of a unifying nomenclature to differentiate between the various forms [201]. Sequence analysis has subsequently allowed clearly differentiating eN from intracellular 5′-nucleotidases. Of the seven human 5′-nucleotidases, six have an exclusively intracellular location. Ecto-5′-nucleotidase is now referred to as eN, with aliases including eNT, low Km 5′-NT or CD73, the latter abundantly used in immunological contexts. The intracellular and phylogenetically unrelated 5′-nucleotidases are either cytosolic (cN-IA, cN-IB, cN-II, cN-II, cdN) or have a mitochondrial localization (mdN) [235, 236].
General molecular properties
EN was first cloned from rat [237], human placenta [238], and the electric ray Torpedo electric organ [239], followed by identification of the cDNA sequence of a considerable variety of mammalian species [202, 240]. The human cDNA encodes a precursor of 574 residues that is converted to the mature form of 523 residues after cleavage of the signal peptide and the hydrophobic domain at the C-terminus, which is replaced with the GPI anchor, similar to the enzymes cloned from rat [237] and mouse [204]. The nucleotide sequence of murine eN cDNA is 86 and 92 % identical to that of the human and rat cDNAs, respectively. The reported apparent molecular mass of the glycosylated GPI-anchored mammalian eN amounts to 60–80 kDa for the monomer and 160 kDa for the dimer [201, 241]. The MALDI-MS analysis of eN from bull seminal plasma indicated a mass of 65.568 Da, implicating that the posttranslational modification of the protein contributed about 6,000 Da [242]. Mass spectrometric analyses of bull seminal plasma eN [242] revealed that all eight cysteines form intramolecular S–S bonds, suggesting that the subunits of the dimeric enzyme are linked by non-covalent bonds [241]. Recombinant soluble forms of rat [243] and mouse [244] eN lacking the GPI anchor have become available.
Glycosylation
Human and mouse eN contain four and rat eN contains five potential N-linked glycosylation sites. For bovine liver eN, which contains four putative N-linked glycosylation sites [245], all Asn residue candidates were shown to be modified, containing complex mixtures of glycans [242]. Using mass spectrometry, the N-linked glycosidic structures could be determined. Three of the Asn residues contained high-mannose oligosaccharide chains, six to nine mannose residues in length. One was modified only partially, showing a heterogeneous mixture of glycans including high-mannose, complex and hybrid structures. eN is a sialoglycoprotein and can be separated by two-dimensional electrophoresis into up to 13 isoforms. The differences in pI presumably relate to differing contents in sialic acid residues (ref. in [201, 246]). In addition, in Torpedo electric organ [246] and in developing kitten visual cortex [247], the HNK-1 epitope, a sulfated, glucuronic acid-containing carbohydrate epitope, that has been found in a variety of cell adhesion molecules, has been identified. The glycosylation pattern presumably varies not only between species but also between tissues. The lectin (carbohydrate-binding protein) concanavalin A binds to eN and is an effective non-competitive inhibitor of catalytic activity [201]. Deglycosylation of mouse eN did not alter its catalytic activity [248].
The GPI anchor
GPI-anchoring of eN could be demonstrated by release of the enzyme by phosphatidylinositol-specific phospholipase C from Bacillus thuringiensis. The degree of eN release from the cell surface varied between cell types, implicating acylation of the inositol ring in the GPI anchor which renders GPI anchors resistant to phosphatidylinositol-specific phospholipase C (ref. in [249–252]). GPI anchors can confer a variety of functional properties to proteins. This includes rapid lateral motility, cellular sorting to the apical membrane surface of polarized cells, clustering in lipid rafts and also transmembrane signaling [250, 252]. Accordingly, eN is sorted to the apical membrane of hepatic cells [253, 254].
Using sequence information from isolated C-terminal fragments of the mature rat [255], human [238] and bovine [245] enzyme, it was shown that the GPI anchor is linked to the conserved Ser523 residue. The hydrophobic C-terminal extension is cleaved and replaced by GPI. Mutational analyses using bovine liver eN determined 13 amino acids as the minimum domain of the C-terminal hydrophobic region for GPI modification [256]. Deletion of the whole hydrophobic domain (17 amino acids) or of the entire cleaved-off domain (25 amino acids) resulted in secretion of the protein. The structures of the GPI anchors from bovine liver [257, 258] and from bull seminal plasma [242] eN were characterized. They concurred with the glycan and lipid structures of mammalian GPI-anchored proteins [259].
Soluble forms
Soluble eN have been described in several tissues, including serum [260], and synovial fluid [261]. The serum form presumably is derived from the liver [262]. Forms of released eN may have different cellular origins. They can be derived by proteolytic cleavage devoid of the GPI anchor, as shown for the soluble eN in bull seminal plasma [263]. In this case, enzyme activity of the soluble and truncated eN was different from that of the GPI-anchored enzyme and AMP was not the preferred substrate. Furthermore, there were differences in the glycan moieties, and the Km values for the substrates tested were 5–10 times higher than those observed previously for the GPI-anchored enzyme. On the other hand, soluble forms of eN from the electric ray Torpedo and from bovine cerebral cortex can be derived from the GPI-anchored enzyme [264] by endogenous phospholipase C cleavage. This was shown using an antibody specific for the inositol 1,2-(cyclic)monophosphate that is formed on phospholipase C cleavage of the intact GPI anchor. Membrane-bound and soluble forms had very similar Km values for AMP and were both inhibited by micromolar concentrations of ATP.
Furthermore, eN can be released in GPI-anchored form via microvesicles or exosomes from cells [159, 265] or in snake venom [266], and it can serve the transfer of the enzyme between cells, as exemplified for adipocytes [267]. It can also be released into pancreatic juice [156]. Binding of monoclonal antibodies to lymphocyte eN but not to endothelial eN caused shedding of the protein [249]. In principle, soluble forms of the enzyme might be able to diffuse between cells and confer activity of eN to sites distant from eN expression.
Protein interactions
EN functions as a lymphocyte adhesion protein. It is involved in lymphocyte binding to the vascular endothelium. Binding of peripheral blood lymphocytes to cultured endothelial cells could be inhibited by a monoclonal antibody against human eN, implicating the involvement of protein–protein interactions [268, 269]. It also mediated adhesion between B cells and follicular dendritic cells [270]. Several studies suggest that eN can interact with components of the extracellular matrix. Antibodies against chicken eN inhibited spreading of chicken fibroblasts and myoblasts after their attachment to a laminin substrate [271, 272]. Furthermore, eN was found to directly bind to laminin/nidogen and fibronectin, interactions that did not involve the active site of the enzyme or the carbohydrate moieties [273, 274]. These two extracellular matrix proteins are involved in a variety of biological processes, including cell adhesion, spreading, growth, and migration. Antibodies directed against eN or laminin specifically perturbed the association of the two proteins. Laminin stimulated and fibronectin inhibited catalytic activity [275, 276]. Moreover, laminin and fragments of fibronectin were found to activate eN on BCS-TC2 human colon adenocarcinoma cells. Thus, a role of eN as a cell receptor for extracellular matrix proteins was proposed [275]. In support of this, a monoclonal antibody against eN from a human pancreatic tumor cell line (PaTu 8902) reduced the attachment of these cells to a fibronectin substratum. The antibody had no effect on cells lacking membrane-bound AMPase activity [277].
The concept that eN can interact with extracellular matrix proteins was recently revived in a study on human breast cancer cells [278]. eN was found to strongly and specifically interact with tenascin C, a non-structural extracellular matrix protein involved in cell adhesion and migration. This interaction led to an inhibition of eN catalytic activity, increased cell adhesion, and decreased cell migration on a tenascin C substrate. Cancer cells expressing reduced eN revealed weaker attachment and migration on tenascin C. There was, however, relatively weak interaction with fibronectin and no interaction with laminin with human eN. These data indicate that eN might function as a receptor for tenascin C. This interaction could influence cell adhesion and migration and in addition reduce the generation of local adenosine.
Downstream signaling
Additional functions of eN independent of catalytic activity have been described. eN can function as a co-stimulatory molecule in human T cells. Tyrosine phosphorylation and dephosphorylation of several proteins has been demonstrated on application of monoclonal anti-eN antibodies to lymphocytes—but not to endothelial cells [249, 279]. Antibody application together with submitogenic concentrations of PMA stimulated T-cell proliferation and secretion of interleukin-2 as well as expression of interleukin-2 receptors [280, 281]. Activation of downstream signaling mediated by antibody ligation has also been observed for other GPI-anchored proteins. Yet, neither the GPI anchor [282] nor catalytic activity [283] appeared to be essential for signal transmission through eN. At present, the mechanisms and physiological implications of these phenomena are poorly understood. It remains an open question whether there is a natural in situ ligand that could mimic the action of antibody ligation [284–286]. Induction of cellular signaling events via cross-linking may also involve cleavage of the GPI anchor [287, 288].
Phylogenetic relationship
Phylogenetically, eN can be grouped into the calcineurin superfamily of dinuclear metallophosphatases with multiple members in prokaryotes, invertebrates, and vertebrates. They are characterized by two sequence motifs: motif A, DXH(X)∼25-GDXXD(X)∼25GNH[D/E], and a shorter motif B, GH-(X) ∼50GHX[H/X]. In mammals, these include in addition to eN the Ser/Thr protein phosphatases and tartrate-resistant acid phosphatase [289, 290]. Molecular cloning revealed phylogenetic relationships between vertebrate eN and functionally related enzymes in invertebrates, yeasts/fungi, eubacteria, and archaea but seem to be lacking in plants [201, 202, 240]. The bacterial enzymes reveal a broader spectrum of substrate specificity. For example, the periplasmic 5′-nucleotidase encoded by the ushA gene of Escherichia coli hydrolyzes not only nucleoside monophosphates but also nucleoside di- and triphosphates as well as other 5´-ribo- and 5´-deoxyribonucleotides, and nucleotide diphosphate sugars [291, 292]. The periplasmic 5′-nucleotidase of Haemophilus influenzae termed NadN hydrolyzes NAD+ into AMP and nicotinamide ribonucleoside 5′-diphosphate, both of which can further be dephosphorylated [200].
A number of blood-feeding arthropods encode soluble proteins related to mammalian eN that are secreted in their saliva. These enzymes rather display apyrase activity, hydrolyzing ATP and ADP and lack the hydrophobic C-terminal region essential for GPI-anchoring, an exception being the eN from the cattle tick Rhipicephalus microplus (formerly Boophilus microplus). Hydrolysis of ATP and ADP prevents platelet aggregation and thus facilitates feeding. Examples of insect salivary apyrases include the mosquito Aedes aegypti [293], Anopheles gambiae [294], the triatomine bug Triatoma infestans [295], the sand fly Lutzomyia longipalpis [296], or a variety of ticks [297]. Figure 7 depicts the phylogenetic relationship of select members of the 5′-nucleotidase family and their substrate specificity.
Fig. 7.
Radial phylogenetic tree and substrate specificity of members of the 5′-nucleotidase family. Availability of structural data is indicated by underscores. Functional data are available for some purified nucleotidases. Accepted substrates are shown in black. No turnover could be detected with substances shown in red. Mammalian eN occurs in dimeric form, is membrane-attached via a GPI anchor. Bacterial 5′-nucleotidases are monomeric. Membrane attachment via a lipid anchor has been reported for Vibrio parahaemolyticus [620] and the 5′-nucleotidase from R. microplus seems to be membrane-bound via a GPI anchor [621]. No functional data are available for 5′-nucleotidases from yeasts and fungi which are phylogenetically distantly related to those of bacteria and animals. Amino acid sequences have been aligned with ClustalW and the tree was calculated with ProML. GI accession numbers and references for functional studies—A. aegypti (mosquito): gi556272 [293]; Bos taurus: gi285642; C. albicans: 68477828; D. rerio: 32766695; Discopyge ommata (electric ray): 62772; E. coli (ushA): 2506086; E. coli CFT073 (c4898): 26250712; Glossina morsitans morsitans (Tse tse fly): 14488055 [622]; Homo sapiens: 4505467; Kluyveromyces lactis: 50303513; L. longipalpis (sandfly): 4887100; Mus musculus: 6754900; Ornithodoros savignyi (soft tick): 152207619 [623]; R. microplus (cattle tick): 1737443; S. aureus: 88193920; T. thermophilus: 55981297; Treponema pallidum: 5902689; T. infestans: 34481604 [295]; Trichinella spiralis: 22656349 [624]; Vibrio parahaemolyticus: 217191. UDPG, UDP-glucose
Protein structure
Information on the spatial structure of eN is currently available only from crystal structures of the homologous bacterial periplasmic 5′-nucleotidases (5NT) and of 5NT from the yeast Candida albicans (Table 5). The sequence identity of human eN to 5NTs of known spatial structure is 39 % (Thermus thermophilus 5NT, Tt5NT), 30 % (H. influenzae NAD nucleotidase, HiNadN), 26 % (E. coli c4898), 25 % (E. coli 5NT, Ec5NT), 22 % (Staphylococcus aureus 5NT), and 20 % (C. albicans 5NT). The structure of Ec5NT has been studied most intensively (Table 5); however, due to the significantly higher sequence identity, it is clear that Tt5NT currently provides the best model for the structure of eN. In agreement with its function in the salvage of nucleotides of the growth medium, Ec5NT is a rather unspecific enzyme [192, 291]. Unfortunately, no information is available concerning the specificity or activity of Tt5NT which might help to pinpoint the structural factors responsible for substrate specificity. HiNadN cleaves AMP with about fivefold higher specific activity compared to NAD [200]. Nevertheless, the enzyme has been designated as a NAD nucleotidase due to the importance of the uptake of NAD metabolites for the growth of H. influenzae. Interestingly, AMP is hydrolyzed by HiNadN with 16- and 15-fold higher specific activity than ATP and ADP, respectively. Thus, the higher sequence identity between eN and HiNadN compared to Ec5HT is also reflected in a higher selectivity towards AMP.
Table 5.
Crystal structures of bacterial and yeast 5′-nucleotidases related to eN
| Crystal form | Conformation | Ligands | Resolution (Å) | PDB ID | References |
|---|---|---|---|---|---|
| E. coli 5NT | |||||
| I | Open | CO2−3, SO3−4, Zn²+ | 1.73 | 1USH | [300] |
| I | Open | ATP, CO2−3, SO3−4, Zn²+ | 1.70 | 1HP1 | [301] |
| II | Open | WO2−4, Zn²+ | 2.22 | 2USH | [300] |
| III | Closed | Adenosine, PO3−4, Zn²+ | 2.10 | 1HO5 | [301] |
| IV | Closed | AMPCP, Mn²+ | 1.85 | 1HPU | [301] |
| Va | Open | Ni²+ | 2.10 | 1OID | [303] |
| VIa | Open | Ni²+ | 2.33 | 1OIE | [303] |
| VIIb | Intermediate | CO2−3, SO3−4, Mn²+ | 2.10 | 1OI8 | [303] |
| E. coli c4898c | |||||
| I | Closed | Uridine, Mn²+, Fe²+ | 1.90 | 3IVD | Ramagopal et al., unpublishedd |
| II | Closed | Cytidine, Mg²+, Fe²+ | 1.70 | 3IVE | Ramagopal et al., unpublishedd |
| T. thermophilus 5NT | |||||
| I | Closed | Thymidine, PO3−4, Zn²+ | 1.75 | 2Z1A | Nakakaga et al., unpublishedd |
| S. aureus 5NT | |||||
| I | Closed | α-Ketoglutarate, Mn2+, Zn2+ | 2.05 | 3QFK | Minasov et al., unpublishedd |
| H. influenzae NadN | |||||
| I | Open | Adenosine | 1.3 | 3ZTV | [200] |
| II | Open and closed | AMPCP, PO3−4 | 2.0 | 3ZU0 | [200] |
| C. albicans 5NT | |||||
| I | Closed | Zn2+ | 1.90 | 3C9F | Patskovski et al., unpublishedd |
aCrystal forms V and VI were generated using a disulfide-trapped (S228C, P513C) E. coli 5NT mutant
bCrystal form VII was generated using a disulfide-trapped (P90C, L424C) E. coli 5NT mutant
cE. coli c4898 is a putative E. coli 5NT. To avoid confusion with E. coli 5NT, this putative 5NT enzyme is described by its gene locus (c4898)
dCrystal structures of E. coli c4898, T. thermophilus 5NT, S. aureus 5NT, and C. albicans 5NT were determined as part of structural genomics projects and no accompanying publications are currently available for these structures
On the basis of short sequence motifs, it became clear that the N-terminal domain of eN belongs to a large superfamily of distantly related metallophosphoesterases acting on diverse substrates such as Ser/Thr phosphoproteins, various nucleotides, sphingomyelin, as well as RNA and DNA [298]. The first structure of an enzyme of this superfamily was determined for a plant purple acid phosphatase [299], which is homologous to mammalian tartrate-resistant acid phosphatases. The core fold of the catalytic domain of these enzymes is a four-layered αββα structure. Two βαβαβ motifs are sandwiched on top of each other and the dimetal site is coordinated by loops at the C-terminal ends of the parallel β-strands of this motif.
5NT consists of two domains, which are linked by a long α-helix (Fig. 8a) [300]. The N-terminal domain (residues 25 to 342 for Ec5NT) is related to the calcineurin superfamily of metallophosphatases and binds two metal ions. This domain confers the phosphohydrolase activity to the enzyme. The C-terminal domain (residues 362 to 550) has a unique structure which has not been described in other proteins so far. This domain is responsible for the substrate specificity as it binds the base and ribose moieties of the nucleotide substrates [301]. Thus, the active site is located in a cleft between the two domains (Fig. 8).
Fig. 8.
Crystal structure of Ec5NT. a Open form of the enzyme. The N-terminal domain (yellow) binds the two metal ions (light blue). An ATP molecule (blue) is complexed to the substrate-binding pocket of the C-terminal domain (red). b Closed form of the enzyme in complex with AMPCP. The non-hydrolysable ADP analogue binds between the two domains. The nucleoside moiety of the substrate analogue is bound to the same binding pocket as in a, which is rotated together with the C-terminal domain by ∼96° around the axis shown in green. The bending residues, i.e., the region of the protein which accommodates the rotation of the rigid domains, are depicted in green. c Binding mode of the substrate analogue inhibitor α,β-methylene-ADP to the closed form of Ec5NT (stereo view). The residues which are not conserved in eN are depicted in broken lines. The residue labels refer to the human enzyme. The loop shown in green is predicted to be shorter in the ecto-enzymes. The coloring of the protein domains is the same as in a. d Scheme of the interactions involved in inhibitor binding. The upper label for each residue refers to human eN and the bottom label to Ec5NT
Domain movement
Crystallization of Ec5NT in different crystal forms showed that the enzyme undergoes a large and unique domain motion in catalysis (Fig. 8a) [302]. In the open form, the nucleotide binding pocket is well accessible for substrate binding and product leaving. However, the substrate ATP binds at a distance of about 25 Å away from the dimetal site [300]. In the closed conformation, the ADP analogue α,β-methylene-ADP (AMPCP) binds close to the two metal ions (Fig. 8). In both complex structures, the adenosine moiety of the substrate or substrate analogue binds to the same binding site of the C-terminal domain. eN shows a unique type of hinge-bending domain movement which resembles a ball-and-socket motion. The C-terminal domain is the ball which rotates approximately around its center. As a result, the residues at the domain interface (where the active site is located) move predominantly along the interface. This is in contrast to classical domain closure motions, where a cleft between two domains opens up or closes with movement of the residues mainly perpendicular to the domain interface. The rotation axis describing the conformational switch between the open and closed forms passes through the bending residues, which accommodate the rotational transition between the two connected rigid domains. In Ec5NT, residues 352–364 are the bending residues. They are located at the C-terminal end of the long α-helix connecting the two domains. In the closed conformation, the helix is broken in the region of residues K355 and G356. At these two residues, also the principal main chain torsion angle changes take place. The influence of mutations of these residues has not yet been explored; however, the two residues are not strongly conserved. The sliding domain movement is facilitated by the almost complete lack of direct contacts between the two domains. Instead, a layer of water molecules is present between the domains at the interface. Trapping of Ec5NT in the open and closed states via engineered disulfide bridges indicated that the domain motion is necessary for catalysis, presumably since binding and release of large substrates can only occur via the open conformation [303, 304]. Open and closed forms resembling those of Ec5NT have also been observed for HiNadN [200]. The domains rotate by 87° relative to each other between the two conformations. Interestingly, in crystal form II of HiNadN, which was obtained in the presence of AMPCP and phosphate (from the hydrolysis of AMPCPP), one monomer in the asymmetric unit is present in the open conformation and a second monomer is present in the closed form. This finding indicates that both forms exist in equilibrium in solution in the presence of these ligands and that the packing forces of the crystal stabilized both forms in one crystal during the crystallization process.
Active site and catalytic mechanism
The complex structure of AMPCP (α,β-methylene-ADP) with Ec5NT in the closed form provided the first detailed structural insight into substrate binding and catalysis of these enzymes [301]. Since then, further product complexes of other bacterial 5NTs have been determined (Table 5). The two metal ions in the active site are approximately 3.3 Å apart. Different divalent metal ions can activate Ec5NT, with Co2+ conferring the highest activity, followed by Mn2+ [305]. However, purified Ec5NT contains at least one tightly bound zinc ion and it appears likely that the periplasmic enzyme can bind different metal ions and presumably forms heterodinuclear centers for sufficient activation [305–307]. Metal-binding site 2 (Fig. 8c, d) has a significantly higher affinity for Mn2+ as binding site 1 [301]. The residues of the coordination sphere of the site 2 metal ion are conserved between Ec5NT and human eN, whereas Q254 of the E. coli enzyme coordinating the metal ion 1 is replaced by an asparagine residue (N245) in the human enzyme. Most likely, the shorter Asn residue binds a water molecule which in turn is coordinated to the metal ion.
AMPCP binds to the active site at the interface between both domains such that the nucleoside moiety is bound by the C-terminal domain whereas the terminal phosphate group is coordinated to the N-terminal domain (Figs. 8c, d and 9) [301]. The base is bound by a hydrophobic stacking interaction between two phenylalanines and the N1 nitrogen of the adenine ring forms a hydrogen bond to the side chain of Asn431. In human eN, this residue is replaced by G419 and a water molecule might replace the missing side chain to interact with the N1 nitrogen. In the E. coli enzyme, Asp504, Gly458, and Arg410 coordinate the ribose group. Arg375 and Arg379 bind the α-phosphate group of the nucleotide whereas Arg410 is coordinated to the β-phosphate group.
Fig. 9.
Scheme of the dimetal center and proposed structure of the Michaelis complex for eN catalysis. The binding mode of the substrate’s terminal phosphate group is supported by the binding mode of the inhibitor α,β-methylene-ADP to Ec5NT. Residue labels refer to human eN. All residues except for N245 are conserved in the E. coli enzyme. Therefore, the coordination of N245 via a water molecule to metal ion 1 is hypothetical. In the bacterial enzyme, N245 is replaced by a glutamine, which is directly coordinated to the metal ion
These residues are conserved in human eN, with two exceptions: Asn431 is replaced by a glycine residue (Gly419) and Arg375 is replaced by a serine. There is no suitable residue nearby in human eN which could replace Asn431 (of Ec5NT) in coordinating the base directly. Instead, a water molecule might bind between the base and Gly419 in the human enzyme. It is not clear from these mutations how the specificity towards AMP is achieved in eN compared to the bacterial 5NTs. Possibly, the substrate specificity is achieved by a control of the domain rotation of the enzyme, which influences the distance between the base binding site and the catalytic metal ions.
In the Ec5NT × AMPCP structure (Fig. 8c, d), the terminal phosphate group of AMPCP is bound to the N-terminal domain of the enzyme via hydrogen bonds to Asn116, His117, and the metal-bridging water molecule. In addition, one oxygen atom of the terminal phosphate group is coordinated to the site 2 metal ion. These interactions polarize the phosphate group for attack of the nucleophile (Fig. 9). A likely candidate for the nucleophile is the water molecule terminally coordinated to the site 1 metal ion [301, 308]. It is located almost opposite of the leaving group, deviating by only 25° from a perfect inline arrangement. The distance between the water molecule and the phosphorus atom is 3.2 Å. Coordination to the metal ion lowers the pKa value of the water molecule such that it is more easily deprotonated to a hydroxide ion at neutral pH. The transition state is stabilized by the two metal ions, His117 and Arg410. No protein residue is appropriately positioned to protonate the leaving group. Protonation of the leaving group is probably not required for the phosphate oxygens leaving the ATP and ADP substrates, as these are stable in the deprotonated state and thus good leaving groups. For catalysis of AMP hydrolysis, however, protonation of the alkoxide leaving group would be beneficial. Perhaps a water molecule transfers a proton to the leaving group. The proposed mechanism for 5NTs resembles that suggested for the related TRAPs based on structural and many spectroscopic data [299, 309]. This includes a direct transfer of the phosphoryl group to the water nucleophile without the formation of a covalent intermediate. For the TRAPs, such a direct transfer mechanism is supported by the inversion of the configuration at the phosphorus atom [310].
Alternative mechanisms have been proposed for other enzymes of this superfamily of metallophosphoesterases (for a more comprehensive comparison of 5NTs to the related enzymes of this superfamily, see ref. [311]). The metal-bridging water molecule (or hydroxide ion) might be the nucleophile attacking the terminal phosphate group. Furthermore, a bidentate metal-bridging binding mode of the phosphate group has been proposed for substrate binding. This is supported by the binding mode of phosphate in some product complexes. For HiNadN, a co-crystal structure with the ADP analogue AMPCP and phosphate was obtained in the closed form [200]. The complex structure obtained after the hydrolysis of AMPCPP clearly resembles a product complex of cleavage of the ATP analogue. Only the phosphate ion is coordinated to the metal ions, and the phosphate groups of AMPCP are behind the phosphate ion bound to the C-terminal domain. In contrast to the product structure of Ec5NT with adenosine and phosphate [301], where the phosphate ion is coordinated only to the site 2 metal ion, one oxygen atom of the phosphate ion in the HiNadN × AMPCP × PO3−4 structure replaces the metal-bridging water ligand and is asymmetrically bridging the two metal ions (2.2 Å to site 1 and 2.04 Å to site 2).
A mutational analysis of active site residues has so far only been published for Ec5NT, mainly studying the influence of metal-binding residues (Table 6) [307]. The effects of the mutations are in good agreement with the proposed mechanism based on the co-crystal structures with substrates and products. The mutations of the metal ion coordination sphere are replacements with Asn or Gln, which should be able to bind the divalent metal ions but the partial charge of the metal ion is changed by the different nature of the replaced ligand. The results indicate that both metal ions participate in transition state stabilization or activation of the nucleophile. Interestingly, only mutations of M2 ligands also have a small effect on the Km value, in agreement with the presumed binding of the terminal phosphate group of the substrate to M2. The largest effect on the specific activity was observed for the replacement of the catalytic histidine (H117 in Ec5NT and H118 in human eN), demonstrating the importance of this residue for transition state stabilization.
Table 6.
Explanation of the observed effects of previously reported active site mutations of Ec5NT on activity
| Residuea | Function | Mutant | Effect | Explanation |
|---|---|---|---|---|
| D41/36 | M1 ligand | D41N | Activity 9.1 %, Km no effect | Effect of M1 ion on transition state stabilization or nucleophile activation |
| H43/38 | M1 ligand | H43N | Activity 0.8 %, Km no effect | Effect of M1 ion on transition state stabilization or nucleophile activation |
| D84/85 | M1/M2 ligand | D84N | Activity 0.1 %, Km increased 1.7-fold | Effect of both metal ions on transition state stabilization or nucleophile activation, M2 on substrate binding |
| H117/118 | Transition state stabilization | H117N | Activity 0.04 %, Km no effect | Protonated H117 is essential for transition state stabilization |
| E118/119 | Buried residue next to H117 | G118Q | Activity 6.1 %, Km increased 5-fold | Distortion of active site |
| H217/220 | M2 ligand | H217N | Activity 0.2 %, Km no effect | Effect of M2 ion on transition state stabilization |
| H252/243 | M2 ligand | H252N | Activity 21.9 %, Km increased 1.8-fold | Effect of M2 ion on transition state stabilization and substrate binding |
aThe first residue number refers to the E. coli enzyme and the second number to human eN. All mutations are reported in ref. [307]
For the NTPDases, the selectivity for NTPs and NDPs is understood to be based on different binding modes of the triphosphate and diphosphate chains of the substrate. For the 5′-nucleotidases, however, the structural basis of the unspecificity of Ec5NT with respect to the hydrolysis of ATP, ADP, and AMP compared to the selectivity of the ecto-enzymes for AMP is not clear. It is unlikely that the substrate specificity is directly influenced by active site residues involved in substrate binding since these are mostly conserved or their mutation does not change the unspecificity of the E. coli enzyme (our unpublished results). We hypothesize that the substrate specificity might be achieved via a control of the domain motion [308]. The orientation of the two domains relative to each other changes the distance between the dimetal center of the N-terminal domain and the base binding site of the C-terminal domain, which is likely to strongly influence the binding modes of NTPs, NDPs, and nucleoside monophosphates (NMPs). If the domain orientations that are optimal for a productive binding mode of NTPs and NDPs are disfavored (i.e., of too high energy) in the ecto-enzyme, these nucleotides are not turned over. Perhaps the dimeric structure of the ecto-enzymes contributes to substrate specificity via a control of the domain orientations.
Ecto-nucleotide pyrophosphatase/phosphodiesterases
General properties and functional role
To date, seven ecto-nucleotide pyrophosphatase/phosphodiesterase paralogues (E-NPPs) were found to be expressed in vertebrates, forming the E-NPP protein family (Fig. 10). They have been numbered NPP1–NPP7 according to their order of cloning but only three (NPP1–NPP3) can hydrolyze nucleotides [312]. Similar to APs, their spectrum of substrates is not restricted to nucleotides. NPP1 to NPP3 hydrolyze pyrophosphate or phosphodiester bonds in a wide variety of substrates. They were found to hydrolyze nucleoside triphosphates and diphosphates, NAD+, FAD, UDP sugars, and dinucleoside polyphosphates and also artificial substrates such as p-nitrophenyl thymidine monophosphate. The advent of molecular cloning resulted in the identification of individual enzyme species that have been named independently by several groups (Table 1). This has subsequently led to an adjustment of the nomenclature that is still used today [38, 39]. In addition to nucleotides, NPP2 hydrolyzes phospholipids, which in the light of recent structural and functional data seems to be its major catalytic function, and NPP6 and NPP7 hydrolyze phospholipids only. In this context, the name autotaxin (ATX) is generally used for NPP2. Potential catalytic properties of NPP4 and NPP5 remain unknown.
Fig. 10.
Radial phylogenetic tree and membrane topology of NPP-type ecto-enzymes. The tree highlights the separation between the multidomain NPP1–3 and NPP4-7, which consist of only the catalytic domain. Separation into the seven different NPPs has occurred early in evolution. Included is the bacterial NPP from X. axonopodis pv. citri, for which structural data are available. NPP-type enzymes are ubiquitous in nature. Amino acid sequences have been aligned with ClustalW and the tree was calculated with ProML. GI Accession numbers (clockwise): D. rerio NPP2: 82187289, NPP4: 62955749, NPP5: 113462025, NPP6: 61806626; H. sapiens NPP1: 23503088, NPP2: 290457674, NPP3: 206729860, NPP4: 172045555, NPP5: 50401201, NPP6: 108935979, NPP7: 134047772; X. axonopodis pv. citri: 21243551; X. laevis NPP3: 254946550; X. tropicalis NPP1: 254946558, NPP7: 296010817
NPP1–NPP3 have a wide tissue distribution. Their cell and tissue-specific expression can vary between rodents and humans [312]. NPP1–NPP3 can be co-expressed in the same tissue and even by the same cell, as for example in cultured hepatocytes [313] or osteoblasts [314]. Similar to APs, they are often expressed on epithelial surfaces. This includes airway epithelia [315], the lung [313], liver epithelia, kidney epithelia [316], and intestinal epithelia [317]. Thiamine pyrophosphate, a likely substrate of E-NPPs, has been used early on as a substrate for the enzyme histochemical detection of microglia and blood vessels of the brain [318, 319]. Autotaxin catalytic activity is present in various biological fluids including serum, seminal plasma [320], or cerebrospinal fluid [321]. The protein is highly expressed during development [322, 323].
The functional role of individual E-NPPs is very variable and depends on the specific catalytic properties and pattern of expression [312]. NPP1 is a marker of late stage in the differentiation of antibody-producing B cells [324, 325], it is expressed in T cells [326], and in astrocytic brain tumors [327]. A major function of NPP1 is in bone mineralization and also in vascular smooth muscle cell calcification [6, 7, 328]. By hydrolyzing extracellular ATP to AMP, it contributes to the formation of extracellular PPi together with the inorganic pyrophosphate transporter ANK-mediated PPi efflux. PPi can function as an inhibitor of calcification. It thus interplays with AP which in turn hydrolyzes extracellular PPi [329, 330]. Accordingly, knockout mice for NPP1 or a natural mutant of NPP1 (tiptoe walking mouse) reveal pathological calcification of ligaments and joint capsules [331]. Double knockouts for TNAP and NPP1 partially reverse the hypomineralization deficiencies and have a less severe phenotype. This underlines the opposing and reciprocal actions of NPP1 and TNAP in the production and hydrolysis of extracellular PPi, respectively [332, 333]. Furthermore, NPP1 has been linked to insulin resistance and type 2 diabetes (for review, see [334–336]).
NPP3 has been shown to promote differentiation and invasion of glial cells but the underlying mechanisms have not been identified [337]. Moreover, the basophil and mast cell-expressed cell surface antigen CD203c was identified as NPP3 [338]. This protein defines the basophil lineage in the bone marrow and shows an elevated distribution on the cell surface of activated basophils. It can serve as a marker for the estimation of hypersensitivity to allergens [339–342]. Recent experiments implicate a dual role of NPP3 in vascular smooth muscle calcification by producing (together with NPP1) PPi from ATP on the one hand and subsequently hydrolyzing (together with TNAP) the PPi produced [7].
The multiple biological functions of NPP2 (autotaxin) are now mainly attributed to its phospholipase D activity and include promotion of cancer cell invasion, cell migration, lymphocyte trafficking, and angiogenesis [343–345]. A brain-specific variant (PD-Iα/ATXγ) is found almost exclusively on the myelin forming oligodendrocytes of the brain [346]. It acts in the modulation of oligodendrocyte extracellular matrix interactions and oligodendrocyte remodeling, a property independent of its catalytic activity [347, 348].
Substrates and catalytic properties
NPP1–NPP3 are classified as both alkaline nucleotide pyrophosphatase (EC 3.6.1.9) and phosphodiesterase I (EC 3.1.4.1). Catalytic activity depends on divalent cations and the enzymes reveal an alkaline pH optimum between pH 9 and 10. Similar to APs, they are also active at pH 7.4 with about 20 % of maximal activity [349–352]. Enzyme activity is inhibited by EDTA. Much of the evidence regarding nucleotide substrate specificity relies on earlier literature with proteins whose isoform specificity was not defined. NPPs hydrolyze pyrophosphate bonds in nucleoside tri- and diphosphates, NAD+, FAD, diadenosine polyphosphates, and UDP sugars (Fig. 11). Recombinant NPP1 to NPP3 have been reported to hydrolyze ATP to AMP and PPi (Fig. 3) whereby NPP1 revealed the highest activity and that of NPP2 was barely detectable [353]. 3′,5′cyclic AMP was reported to be either no substrate [315, 350] or a (poor) substrate [354–357]. There is ample evidence for the hydrolysis of extracellular cAMP by intact cells but the responsible enzyme has not been clearly identified [358, 359]. Nucleoside monophosphates such as AMP or UMP are not hydrolyzed but exert competitive product inhibition of the NPP reaction [354, 356, 360, 361].
Fig. 11.
Substrate specificity of NPP-type ecto-enzymes. Catalysis is thought to occur by nucleophilic attack of the Thr alkoxide onto the boxed phosphorus atom, so that a covalent intermediate with the R′ group is formed. In a second step, the covalent intermediate is hydrolyzed by nucleophilic attack from a Zn2+-activated water. To some of the enzymes, the substrates may bind in both orientations and this is also reflected by their ability to hydrolyze dinucleotides (e.g., NPP1 and NPP3). NPP2 and NPP6/7 are active towards the same class of substrates. Due to swapped specificities in the R and R′ binding sites, the substrates bind in an inverted position resulting in the generation of different hydrolysis products. The scheme is based on data presented in the following papers—NPP1: [351, 353, 412, 453, 625]; NPP2: [351, 353, 377, 380, 403, 406]; NPP3: [351]; NPP6: [388]; NPP7: [389, 626]. GPC glycerophosphorylcholine, LPC lysophosphatidylcholine, pNPPC p-nitrophenyl phosphorylcholine, SPC sphingosylphosphorylcholine
Recently, it has been demonstrated that soluble heterologously expressed murine NPP1 [362, 363] or NPP3 [7] hydrolyzes (at pH 7.4) PPi in addition to ATP. This result corroborates earlier observations that NPP2 cleaves PPi into Pi [364]. This would implicate a considerable overlap in the catalytic properties of TNAP and NPPs. However, the specificity constant kcat/Km for PPi cleavage by NPP1 is unknown, and hence, the significance of this NPP1 activity remains unclear. In addition, E-NPPs hydrolyze phosphodiester bonds of artificial substrates such as p-nitrophenyl thymidine monophosphates or p-nitrophenyl-phenyl phosphonate and the sulfate–phosphate bond in the AMP derivative 3′-phosphoadenosine 5′-phosphosulfate [355, 365, 366]. The most popular artificial substrate used for characterizing catalytic activity of NPPs has been p-nitrophenyl thymidine monophosphate that is cleaved into AMP and the yellow p-nitrophenol.
Hydrolysis of nucleotides by E-NPPs typically releases nucleoside 5′-monophosphates. ATP is hydrolyzed by NPP1 into AMP and PPi (Figs. 1, 4, and 11). But it can also be hydrolyzed into ADP and Pi. It has been suggested that hydrolysis of ATP into AMP involves a covalent AMP intermediate whereas hydrolysis into ADP involves a phosphate-bound intermediate, depending on how the substrate approaches the catalytic site [312, 334]. The previously described autophosphorylation of NPP1 [366, 367] would then represent one of its covalent intermediates for ATP hydrolysis. Cleavage products of UDP-glucose are UMP and glucose 1-phosphate, NAD+ is hydrolyzed into AMP and nicotinamide mononucleotide, FAD into AMP and flavin mononucleotide, 3′,5′-cAMP into 5′-AMP, and dinucleoside polyphosphates are hydrolyzed asymmetrically (involving the α,β-pyrophosphate bond) into AMP and nucleoside 5′(n-1) phosphate (Fig. 11). NPP1 has been reported to even degrade antisense phosphorothionate oligonucleotides, resulting in the formation of a ladder of shorter products and the release of the mononucleoside 5′-phosphorothionate [368].
Reported Km values of E-NPPs for ATP range between 17 and 300 μM [354, 360, 365, 369]. For a nucleotide pyrophosphatase isolated from rat liver, Km values for UDP-glucose and NAD+ were found to be similar to those for ATP and UTP [360]. Km values for dinucleoside polyphosphates are lower than those for ATP [35]. Heterologously expressed NPP1, NPP2, and NPP3 hydrolyzed dinucleoside polyphosphates with a pH optimum between 8.5 and 9 and Km values for diadenosine 5′,5′″-P1,P3-triphosphate (Ap3A) were 5.1, 8.0, and 49.5 μM, respectively [351]. Km values in the 3–10-μM range have also been reported for the hydrolysis of diadenosine polyphosphates by a purified ecto-pyrophosphatase (presumably NPP1) from rat brain [352]. This agrees well with earlier reports on the hydrolysis of dinucleoside polyphosphates by chromaffin cells [370, 371] or human serum [372]. ATP acts as an inhibitor of diadenosine polyphosphate hydrolysis [373]. Interestingly, NPP1 and NPP3 contain in their catalytic domain the motif GXGXXG, which represents a consensus sequence for the binding of dinucleotides. The GXGXXG motif is not conserved in NPP2. Mutation of these three glycines into alanine abolished the nucleotide phosphodiesterase activity of NPP1 [366]. Yet, recombinant NPP2 prepared and isolated from a vaccinia virus lysate of BS-C-1 cells was found to hydrolyze both ATP [364] and dinucleoside polyphosphates [351].
An important step forward in understanding the functional role of NPP2 (autotaxin) was the discovery of its identity with lysophospholipase D in serum [374, 375]. It converts extracellular LPC to lysophosphatidic acid (Fig. 11). Domain swapping experiments between NPP1 and NPP2 suggested that the expression of lysophospholipase D activity depends on isoform-specific determinants in the N-terminal and the catalytic as well as the nuclease-like domains of NPP2. It was hypothesized that sequences in all three domains together form a lysophospholipid-binding site [376]. In addition, ATX has the capacity to hydrolyze several other lysophospholipids to produce lysophosphatidic acid (LPA), including lysophosphatidylserine, lysophosphatidylethanolamine, and lysophosphatidylinositol [377]. LPA in turn acts on at least six distinct G protein-coupled receptors (LPA1-6) and elicits a large variety of both short-term and long-term cellular responses, including promotion of cell proliferation, survival, and motility of cells [343]. NPP2 null mice are embryologically lethal, presumably due to deficient LPA-dependent angiogenesis [378, 379].
Furthermore, NPP2 was found to hydrolyze sphingosylphosphorylcholine to produce sphingosine-1-phosphate (S1P) which can regulate diverse cellular processes such as motility and angiogenesis via the S1P family of receptors [380]. NPP2 is regulated through product inhibition by both lysophosphatidic acid and S1P [381]. More recent evidence suggests that NPP2 can in addition form cyclic phosphatidic acid, an analogue of lysophosphatidic acid in which a ring closure is formed by phosphate [382]. This enters yet another dimension to the biological functions of this protein. Cyclic phosphatidic acid reveals several distinct biological activities, including anti-mitogenic effects, inhibition of tumor cell invasion and metastasis, or enhancement of neurite outgrowth and survival [383]. The cell motility-inducing activity of NPP2 depends on the production of bioactive lipids [384–386]. It is now thought that the major physiological functions of NPP2 result from its phospholipase activity [387].
NPP6 and NPP7 hydrolyze phosphodiester bonds in lysophospholipids or other choline phosphodiesters, displaying lysophospholipase C activity. NPP6 is a glycerophosphodiester phosphodiesterase with a marked preference for choline containing phospholipids. It hydrolyzes lysophosphatidic acid to form monoacylglycerol and phosphorylcholine but not lysophosphatidic acid, identifying it as a lysophospholipase C. It also hydrolyzes efficiently glycerophosphorylcholine and sphingosylphosphorylcholine [388]. NPP7 was shown to possess alkaline sphingomyelin phosphodiesterase (sphingomyelinase) activity, generating ceramide from sphingomyelin in the intestinal tract [389, 390]. Interestingly, its catalytic activity is inhibited by ATP but not by ADP or AMP. In addition, NPP7 was found to cleave the phosphocholine head group and thus to inactivate platelet-activating factor, a pro-inflammatory phospholipid involved in the pathogenesis of inflammatory bowel disease [391].
General molecular properties
Human NPP1–NPP3 contain 863–925 amino acid residues with an apparent molecular mass of approximately 115 to 125 kDa. They share 40–50 % identity at the protein level. NPP1 was first identified as a surface-located plasma cell differentiation antigen (PC-1) of mouse B lymphocytes [392] and subsequently further characterized [393] and cloned and sequenced from mouse [394, 395] and human cDNA libraries [396, 397]. NPP3 was identified as the target of the monoclonal antibody RB13-6 in rat brain glial precursor cells and cloned from rat [398] and a human prostate cDNA library [399].
NPP1 is targeted to the basolateral and NPP3 to the apical surface of hepatocytes [317]. An N-terminal dileucine-based signal (AAASLLAP) is involved in directing NPP1 to the basolateral membrane of polarized MDCK cells [400] or to matrix vesicles of mineralizing cells [314]. NPP3 lacks the cytosolic dileucine motif and is apically targeted in the polarized MDCK and Caco-2 cells and in airway epithelia [315]. NPP1 and NPP3 differ regarding their detergent solubility which may relate to their targeting. The basolateral protein NPP1 was fully soluble in various detergents, whereas the apical protein NPP3 was very poorly solubilized by Lubrol and resistant in part to Triton extraction, a property observed in many apically targeted proteins [401].
NPP2 was initially described as a 125-kDa autocrine motility factor in the conditioned medium of the human melanoma cell line A2058 [402]. It was named autotaxin and cloned from the same source [403]. The subsequently identified rat [404] and human [405] phosphodiesterase 1α (PD-1α) turned out to be a splice variant. Analysis of the human gene [406] further clarified the issue of splice variants. The ENPP2 gene contains 27 exons with a considerable number of potential alternative splicing sites. Three different isoforms have been characterized that differ in length and tissue distribution [344, 406]. The nomenclature of the isoforms varies between authors [343, 344, 348, 406]. The largest isoform (ATXm, ATXα), the human melanoma-derived autotaxin [403], comprises 915 aa and contains a highly basic insertion in the catalytic domain. The shortest form (863 aa), the human tetracarcinoma isoform (ATXt, ATXβ) [407], is identical to plasma lysophospholipase D. The “brain-specific” intermediate form (889 aa) (ATXγ, PD-1α) [408] contains a 25-residue insertion close to the nuclease-like domain. In many of the studies, the isoforms investigated have not been specified. Probably, most studies refer to the ATXt/ATXβ isoform [406]. Differences in catalytic properties or cellular localization between the isoforms have not been reported.
NPP1 and NPP3 are type II membrane proteins with an N-terminal transmembrane helix, a short cytoplasmic, and a large extracellular domain [312, 331]. In contrast, NPP2 is a secreted protein and synthesized as a pre-pro-enzyme (Fig. 10). The 27 N-terminal amino acid residues functioning as a signal peptide are removed co-translationally, followed by further cleavage of the resulting soluble pro-NPP2 by furine-type proteases, generating the mature, fully active enzyme [409, 410]. Secretion of NPP2 from adipocytes is driven by N-glycosylation and signal peptide cleavage [411]. Membrane-bound NPP1 and NPP3 are homodimeric proteins whereas NPP2 is monomeric. In contrast, NPP4 to NPP7 have an inverted ecto-domain. They contain a putative N-terminal signal peptide and a C-terminal TMD. After cleavage of the signal peptide, this is predicted to result in the formation of type-I membrane proteins with a C-terminal transmembrane helix. Their cytosolic domains are rather short (2–76 residues).
Domain structure
E-NPPs possess a striking modular structure (Fig. 12). Based on the domain organization of the ecto-domain, E-NPPs can be divided into two groups. The ecto-domain of NPP1–3 consists of two short somatomedin B-like repeats of 40 to 50 amino acids, the central catalytic domain of approximately 400 amino acids, and a C-terminal nuclease-like domain (NLD) of approximately 250 residues that bears structural similarity with the DNA- or RNA-non-specific endoribonucleases. In contrast, NPP4–7 consist of solely the catalytic domain. The catalytic domains of NPP1–7 share 24 to 60 % amino acid identity between the human isoforms [334, 367]. The nuclease-like domain itself reveals no catalytic activity, but it bears isoform-specific determinants for catalysis. An incomplete nuclease domain of NPP1 was found to result in the loss of catalytic activity and swapping of the nuclease-like domains of NPP2 and NPP1 abrogated catalytic activity in NPP2. In contrast, NPP1 with the nuclease-like domain of NPP2 was fully active [376, 412]. A C-terminal lysine (Lys852) was found to be essential for catalysis of NPP2 [413]. Furthermore, the nuclease-like domain is required for the translocation of NPP1 from the ER to the plasma membrane [412]. Residues 829–850 are necessary for secretion of NPP2 [413].
Fig. 12.
Structure of NPP-type enzymes. a Domain organization of NPPs: NPP1–3 are composed of two SMB domains, the catalytic domain and a nuclease-like domain. In contrast, NPP4–7 lack any additional non-catalytic domains. An NPP-specific insertion into the AP fold “CAP” is shown in a lighter color. The position of the NPP2-specific deletion required for formation of the two LPA-binding sites is indicated. b Ribbon/surface representation of mouse NPP2 and NPP from X. axonopodis pv. citri (Xac) with an identical view on the active site. The bound product molecules LPA and AMP are shown as sticks and metal ions (2 × Zn2+, Ca2+, Na+, K+) as spheres. The WPG loop of Xac NPP that is missing in NPP2 (see d) is shown in yellow. c Surface representation of NPP2 along the LPA channel showing the intricate arrangement of the four domains, the linker, and the long lasso regions. N-linked glycosylation sites are shown as sticks and the structurally important N524 glycan is labeled. The eye indicates the approximate view in b. d Surface representation of NPP2 with a longitudinal section along the hydrophobic LPA-binding channel. Only one of the Zn2+ ions is visible. One LPA molecule (LPA1) is bound to the active site and the hydrophobic pocket. The second LPA molecule (partial model, LPA2) binds in the hydrophobic channel more distal to the active site. In all other NPP-type enzymes, these two binding sites are occluded by an amino acid stretch consisting of a highly conserved WPG motif at the beginning and a tyrosine at the end that is involved in binding the base of nucleotide substrates. The corresponding loop region and active site-bound AMP of Xac NPP is shown in green and magenta sticks, respectively. LPA lysophosphatidic acid, SMB somatomedin B
In all three proteins, the extracellular nuclease-like domain harbors a putative “EF hand” Ca2+-binding motif (DXD/NXDGXXD) [414, 415]. This motif is essential for catalytic activity in NPP1 and NPP3 but has little effect on the catalytic activity of NPP2 [366, 416]. Although not a complete EF hand is found in NPPs [366], the crystal structure of mouse and rat NPP2 show clear density for a bound Ca2+ ion (see below). The catalytic and nuclease-like domains of NPP2 are linked by an essential disulfide bond [413]. NPP1–NPP3 further contain two cysteine-rich tandem repeats, the somatomedin B-like domains proximal to their N-terminal TMD. It is thought that these domains function as protein interaction domains, similar to the somatomedin B domain of vitronectin, a large glycoprotein found in blood and the extracellular matrix [334].
Glycosylation
N-glycosylation of E-NPPs accounts for up to 20 kDa of the molecular weights of NPP1–NPP3 [398, 417, 418]. N-glycosylation is not required for the apical targeting of NPP3 [419]. Carbohydrate side chains in NPP2 are exclusively N-linked [418] and required for catalytic activity [420]. The rat NPP2 sequence contains five predicted N-glycosylation sites, three of which are glycosylated and map to the catalytic domain [420]. Of the three sites, only glycosylation at Asn-524 is essential for the expression of the catalytic and motility-stimulating activities of NPP2, in contrast to earlier investigations that could not relate N-glycosylation of NPP2 to stimulation of motility [418]. The Asn-524 glycosylation site is conserved in NPP2 of several vertebrates and also in NPP1–6 but not in NPP7. Asn-524 contains an oligomannosidic side chain with the structure identified as Man8GlcNAc2 and Man9GlcNAc2 [420]. For rat NPP5, the presence of a high-mannose-type glycan has been reported [421].
Soluble forms
As mentioned above, NPP2 is a secreted protein and exists only in soluble form. Soluble forms have also been described for NPP1 [365, 417], NPP3 [422], NPP6 [388], and NPP7 [423]. E-NPPs are found in human serum [152, 424, 425]. The endogenous mechanisms for shedding the membrane-bound forms need to be further characterized. To date, reports that alkaline phosphodiesterase I of undefined molecular identity can be released from plasma membranes of eukaryotic cells by phosphatidylinositol-specific phospholipase C from B. thuringiensis [426–429] remain unexplained.
Protein interactions
Several interactions of NPP2 with other macromolecules have been described. NPP2 is capable of binding to glycosaminoglycans, major components of the extracellular matrix. Binding of heparin competitively inhibited catalytic activity and binding of NPP1 to the glycosaminoglycan was diminished in the presence of the enzyme substrate ATP. It has been speculated that, due to its binding of glycosaminoglycans, NPP1 may associate with the extracellular matrix. This interaction may be negatively regulated by the presence of its extracellular substrates [365].
NPP2 and NPP3 contain an RGD tripeptide motif in their second and first somatomedin B-like domain, respectively. This motif may mediate an interaction with integrins [366]. Plasma autotaxin was found to associate with platelets during aggregation, and activated platelets bound recombinant autotaxin in an integrin-dependent manner [430, 431].
A novel function has been attributed to the nuclease-like domain at the C-terminal end of autotaxin (ATXγ, PD-1α), here referred to as modulator of oligodendrocyte remodeling and focal adhesion organization (MORFO) domain [432, 433]. The domain functions independently of the enzymatic activity of autotaxin and antagonizes the adhesion of oligodendrocytes to extracellular matrix proteins such as fibronectin, vitronectin, and laminin-2/merosin. The MORFO domain facilitates the in vitro morphological maturation of postmigratory, premyelinating oligodendrocytes and mediates a reorganized assembly of focal adhesions. Process outgrowth was found to be mediated in part via the EF hand-like motif located at the far C-terminal end of the MORFO domain. It is hypothesized that the MORFO–EF site binds to a cell surface receptor [348, 434].
Downstream signaling, purinergic mechanisms
In spite of the proof in vitro and in situ in many cellular systems of NPP-mediated ecto-nucleotidase activity, stringent experimental evidence that the enzymes are involved in the canonical P2 receptor-mediated purinergic signaling is scarce. One of the limitations results from the fact that classical inhibitors of P2 receptors can also inhibit E-NPPs [351, 369]. In principle, E-NPPs could modulate nucleotide signaling by hydrolyzing purinoceptors agonists and together with APs or ecto-5′-nucleotidase produce adenosine. C6 glioma cells express a surface-located nucleotide pyrophosphatase (presumably NPP1) as the major ecto-nucleotidase [369] that can modulate purinergic receptor function. Furthermore, inhibition of NPPase activity in C6 glioma cells was found to potentiate the inhibitory effect of ATP and ADP on the isoproterenol-induced increase in intracellular cyclic AMP synthesis, corroborating a role of NPPs in modulating purinoceptor-mediated signal transduction [435]. E-NPPs presumably are the major enzymes responsible for the extracellular hydrolysis of dinucleoside polyphosphates, a widely distributed group of extracellular signal molecules. Since these nucleotides are asymmetrically cleaved, E-NPPs not only terminate the action of these molecules on their receptors [436]. They also can produce nucleotides as additional agonists of P2X and P2Y receptors [437]. For example, hydrolysis of Ap5A, Ap4A, or Ap3A yields adenosine tetraphosphate, ATP, and ADP, respectively [373]. This could in turn lead to the additional or also continued activation of P2 receptors, depending on available receptor subtype. Together with NAD-glycohydrolase (NADase), E-NPPs presumably are the major enzymes for the extracellular hydrolysis of NAD+. Whereas NADase catalyzes the formation of nicotinamide and ADP-ribose, AMP and nicotinamide mononucleotide are formed as a result of NAD+ cleavage by E-NPPs. AMP can then be further converted to the signal molecule adenosine (Fig. 1). Similarly, hydrolysis of ADP ribose by E-NPPs will result in AMP formation.
Phylogenetic relationship
Members of the E-NPP gene family are expressed also in invertebrates, yeast, plants, and bacteria, and the divergent development of seven different NPP-type enzymes must have occurred early in vertebrate evolution (Fig. 10). In Xenopus, each gene displays a distinct specific expression pattern during development and in the adult [438]. E-NPPs from the nematode C. elegans share between 18 and 33 % amino acid similarity with human E-NPPs. Longer forms of E-NPPs such as human NPP1–NPP3 appear to exist only in vertebrates. Zebrafish NPP2 shares 70 % amino acid similarity with the human form and also displays lysophospholipase D activity [367, 439]. Interestingly, the fowlpox virus genome encodes a protein similar to NPP1 [440]. Together with APs, E-NPPs belong to a superfamily of phospho-/sulfo-coordinating metalloenzymes [367, 441, 442], also referred to as the AP superfamily [443, 444]. This relationship is based on primary structure, the similar fold of the catalytic domain, the conserved location of the amino acid residues that coordinate two metals in the catalytic site and their spatial arrangement as well as the similar reaction mechanisms. It is further strengthened by the recent analysis of the crystal structure of mouse [445] and rat [431] NPP2 and human placental AP [446, 447].
Protein structure
Structural information is available from crystal structures of mouse and rat NPP2 [431, 445], the somatomedin B1 (SMB1) domain of human NPP1 (PDBID 2YS0, unpublished), and of the bacterial NPP enzyme from Xanthomonas axonopodis pv. citri (Xac) [441], which corresponds to the catalytic domain alone (Table 7). The single phosphodiesterase (PDE) domain of NPP4–7 has a sequence identity of 30 % to the Xac NPP and the PDE domain (catalytic domain) of NPP2.
Table 7.
Reported crystal structures of NPPs
| Structure | Ligand | Resolution (Å) | PDB ID | References |
|---|---|---|---|---|
| X. axonopodis pv. citri NPP | ||||
| Apo-form | – | 1.75 | 2GSN | [441] |
| Transition state analogue | Vanadate | 1.30 | 2GSO | [441] |
| Product bound | AMP | 2.00 | 2GSU | [441] |
| Product bound | AMP | 1.45 | 2RH6 | [441] |
| Mouse NPP2 | ||||
| Apo-form | Sulfate | 2.00 | 3NKM | [445] |
| Product bound | LPA | 1.70–1.80 | 3NKN, 3NKO, 3NKP, 3NKQ, 3NKR | [445] |
| Rat NPP2 | ||||
| Apo-form | Phosphate | 2.05 | 2XR9 | [431] |
| Inhibitor bound | HA155 | 3.20 | 2XRG | [431] |
As predicted from early modeling studies [367], the catalytic domain common to all seven vertebrate NPP-type ecto-enzymes is homologous to the family of APs. As in the AP, two Zn2+ ions are tightly bound in the active site by a set of six conserved Asp/His residues. In addition to the α/β/α core of AP family members, the PDE domain contains a smaller α/β cap domain which partially shields the active site from solvent. This cap domain is specific to E-NPPs and phosphonoacetate hydrolases of proteobacteria [448].
In NPP1–3, the PDE domain is connected to a catalytically inactive nuclease-like domain by a long lasso-like linker that wraps around the NLD to enter it from the distal side (Fig. 12). The lasso-like loop binds to the NLD by hydrophobic interactions, hydrogen bonds, and two cysteine bonds. Our modeling studies indicate that in NPP1 and NPP3, a third cysteine bridge tethers the lasso to the NLD. The NLD domain has a mixed α/β fold with a central six-stranded antiparallel β-sheet. The PDE and NLD domains bind to each other by multiple modes of interaction, including an interdomain disulfide link (C413–C805, rat NPP2 numbering) [413], several salt and hydrogen bonds, and a high-mannose glycan chain at Asn-524 [420] that packs against a depression at the domain interface. Although a direct influence of the NLD onto the PDE domain is not obvious from the crystal structures, the structural and functional importance of the domain–domain interaction is highlighted by the fact that treatments such as disruption of the interdomain disulfide bond or removal of the Asn-524 glycan lead to variants with no or greatly reduced activity in NPP2 [413, 420].
Interestingly, despite the critical importance of the glycan chain at Asn-524 (Asn-585 in human numbering) for structure and function, successful production of active NPP2 in a bacterial expression system as well as crystallization and diffraction data collection has been reported for human NPP2 [449]. The NLD also contains an EF hand-like motif, which is absent in the closest homologue with known structure, the nuclease from Anabaena sp. Although not flanked by helices, the EF motifs of both NPP2 structures show binding to a Ca2+ ion. An intact EF consensus sequence was found to be important for trafficking, structure, and/or activity of NPP1–3 [414, 415, 431], most likely because it participates in the PDE–NLD interaction via a highly conserved salt bridge (K430-D739, rat NPP2 numbering). Additional sites for Na+ and K+ ions were found in the crystal structure of mouse NPP2, but these sites were not present in the rat orthologue.
Distal to the NLD, the PDE domain of NPP1–3 is flanked by the two N-terminal SMB domains. The SMB domains are connected to the PDE domain by a linker that itself is reinforced by two cysteine bridges to the PDE domain. Both SMB domains share the same knotted pattern of four conserved disulfide bridges that is also found in vitronectin. In NPP1 and NPP3, the SMB domains have been reported to mediate a (non-quantitative) homodimerization of the protein. Unfortunately, the crystal structure of NPP2 offers no insight into the mechanism of this dimerization as all cysteines of the SMB domains are involved in intrachain disulfides and NPP1 and NPP3 lack additional cysteine residues. However, studies on the somatomedin domain of vitronectin show that a heterogeneous disulfide pattern is compatible with biological activity [450]. The SMB domain pair of NPP2 was recently identified responsible for the ability of NPP2 to bind to activated platelets in an integrin-dependent fashion [430, 431, 451]. An RGD consensus motif for integrin-binding in SMB2 was found to be largely dispensable for this association [431]. Instead, the interaction mode of the SMB domains was suggested to be more reminiscent of complex structures of vitronectin with its interaction partners plasminogen activator inhibitor-1 and urokinase-type plasminogen activator. In case of SMB1, the potential binding interface contacts the PDE domain, rendering it unavailable for protein–protein association, but in SMB2, it is oriented to the solvent. Additional mutagenesis experiments identified residues H119, D122, and D129 as involved in the integrin-binding interface [431]. Whether NPP1 and NPP3 do bind to adapter proteins via their SMB domains is currently unclear. However, it is noteworthy that the K173Q (or K121Q, uncertain start codon) polymorphism of human NPP1 which was reported to be associated with increased insulin resistance caused by an increase in binding affinity to the insulin receptor [336] does also map to SMB2.
Both cysteine-rich SMB domains form extensive interactions with the catalytic PDE domain. In the NPP2 structures, a channel is formed between the SMB domains and the PDE domain that constitutes a binding site for LPA. It was suggested that this second LPA-binding site allows NPP2 to function also as a lipid carrier molecule and to shuttle the LPA product directly from the active site to the cell surface LPA receptors. In NPP1 and NPP3, this channel is occupied by an insertion loop that is found in all other E-NPPs including the bacterial Xac NPP enzyme (Fig. 12c, d).
Active site and catalytic mechanism
As stated above, E-NPPs belong to the functionally diverse AP superfamily. The catalyzed reaction therefore proceeds in a similar manner as in the well-studied family of AP via a two step mechanism with a covalent intermediate (Fig. 13). Identification of the nucleophilic group was initially achieved by incubating the enzymes with radioactive substrates, quenching the reaction by addition of phenol or a pH shift and subsequent sequencing of radioactive peptides [356, 452, 453]. The nucleophile in E-NPPs is mostly a Thr alkoxide (NPP1–5 and NPP7). However, in NPP6 it is a Ser alkoxide as in APs.
Fig. 13.
Comparison of the active site structures of human placental AP (PLAP) and human NPP1. The active site structure of human NPP1 differs from PLAP mainly by the absence of the magnesium ion binding site, the replacement of Arg-166 by Asn-277, and the presence of a substrate specificity site formed by Tyr-371, Phe-321, Phe-257, and Leu-290
In contrast to APs, E-NPPs are more specific for phosphodiester versus phosphomonoester substrates. Considerable insight into NPP activity and substrate specificity has been gained from kinetic studies and the crystal structures of Xac NPP in complex with the product AMP and the transition state analogue vanadate, and the mouse NPP2 structure in complex with LPA products of differing chain length [441, 445] (Fig. 12). The structural data available establish or confirm the importance of several active site residues for catalysis (Table 8).
Table 8.
Explanation of the observed effects of previously reported active site mutations of NPPs on activity
| Residuea | Function | Mutanta | Effect | Explanation | References |
|---|---|---|---|---|---|
| D218 | Metal 2 binding | D218N (1) | Inactive | Improper cofactor coordination | [367] |
| K255 | Start of first phosphate-binding loop | K255A (1) | Reduced activity | Slight distortion of active site structure, influence on Km and kcat/Km unclear | [367] |
| T256 | Zn2+-activated Thr alkoxide acts as nucleophile | T256A (1) | Complete loss of activity | Nucleophilic attack on substrate not possible | [367] |
| T256A (2) | [384, 431] | ||||
| T256A (7) | [631] | ||||
| T256A (wheat NPP) | [454] | ||||
| T256S (1) | Almost complete loss of activity, trapping of covalent intermediate | Nucleophilic attack on substrate still possible but distorted active site geometry with potentially larger effect on 2nd catalytic step | [367] | ||
| T256S (wheat NPP) | Complete loss of PDE activity but large gain in activity towards monoester substrates | Thr/Ser alkoxide as major determinant for diesterase vs. monoesterase activity | [454] | ||
| F257 | Binding of nucleobase/lipid | F257A (1) | Reduced activity | Most likely reduced substrate affinity | [367] |
| F257A (2) | Reduced activity | [445] | |||
| N277 | Phosphate binding, stabilization of transition state | N277A | Complete loss of activity | Missing stabilization of transition state | [445] |
| N277A | Strong reduction in activity | [431] | |||
| N277R (wheat NPP) | Complete loss of PDE activity but large gain in activity towards monoester substrates | Positively charged side chain allows monoester cleavage like in APs | [454] | ||
| L290 | Binding of nucleobase | L290A (2) | Reduced activity | Most likely reduced substrate affinity | [445] |
| F321 | Binding of nucleobase | F321Q (2) | Partial loss of activity | Exact role for nucleotide binding remains unclear | [431] |
| F321S (wheat NPP) | Reduction of activity | [454] | |||
| F321A (2) | Reduced PDE activity, unaltered LysoPLD activity | [445] | |||
| Y371 | Nucleoside binding | Y371A (2) | Reduced activity | Distortion in nucleoside/lipid-binding pocket | [445] |
| Y371Q (2) | Reduced activity | [431] | |||
| D376 | Metal 1 binding | D376N (1) | Largely inactive, trapping of covalent intermediate, slightly rescued by high Zn2+ | Reduced metal affinity, missing activation of nucleophilic water for second catalytic step | [367] |
| H380 | Metal 1 binding | H380Q (2) | Inactive | Reduced metal affinity | [384] |
| H380Q (1) | Largely inactive, trapping of covalent intermediate, rescued by high Zn2+ | [367] | |||
| D423 | Metal 2 binding | D423N (1) | Inactive, slightly rescued by high Zn2+ | Reduced metal affinity | [367] |
| H424 | Metal 2 binding | H424Q (2) | Inactive | [384] | |
| H535 | Metal 1 binding | H535Q (1) | Inactive, rescued by high Zn2+ | Reduced metal affinity | [367] |
| H535Q (2) | Inactive | [384] |
aHuman NPP1 residue numbering is used. Numbers in brackets refer to the NPP enzyme used in the study
As in APs, two Zn2+ ions are bound in the active site and the metal coordinating residues are conserved (Fig. 13). The third metal-binding site for Mg2+ in AP is not present in E-NPPs. Mg2+, which is required for positioning of a Zn2+-binding Asp residue, is functionally replaced by a Tyr OH group in E-NPPs. One additional key difference in the active sites is the functional substitution of R166 in AP (E. coli numbering) by N277 in NPP (human NPP1 numbering). R166 stabilizes the trigonal bipyramidal transition state in AP-catalyzed monoester cleavage by a bidentate interaction with two equatorial oxygens, the third equatorial oxygen being bound between the Zn2+ ions. In contrast, the uncharged side chain amide of N277 of E-NPPs forms only a monodentate interaction with one of the equatorial oxygens. The phosphate oxygen connected to the R′ substituent, which is part of the covalent intermediate formed after attack of the threonine nucleophile (Figs. 11 and 13), is not involved in any hydrogen bonding interactions or metal coordination. This reflects the adaptation to the smaller amount of negative charge accumulation in the transition state of phosphodiester compared to phosphomonoester cleavage [441].
Zalatan et al. [441] found that the R166→N substitution in E-NPPs is not sufficient to explain the discrimination between mono-diester substrates. However, recently a N277R mutation in a plant NPP [454] was found to gain a significant increase in phosphatase activity towards the small phosphomonoester compound p-nitrophenylphosphate. Mutational data also suggest that the Thr/Ser substitution in E-NPPs versus APs favors PDE over phosphatase activity (Table 8). A third contribution comes from the formation of specific R′ binding pockets for the substrate moiety representing the covalent intermediate. In the AMP product-bound form of Xac NPP, the nucleobase is sandwiched between F91 (edge-on-plane) and Y174 (plane-on-plane). In the nucleotide-specific vertebrate E-NPPs, NPP1 and NPP3, these residues are preserved. Y174 is absent in NPP2, which is far more active towards lysophospholipids. It is the last residue of a 19 amino acid long “WPG loop” deletion (156WPG…Y174) from the catalytic core of the PDE domain that is missing exclusively in NPP2. This region is deleted from the fold of the NPP2 PDE domain to generate the space required for formation of the R′ binding pocket for the long fatty acid chain of lysophospholipids (Fig. 12d). Noteworthy, the lipid-binding pocket is narrow enough to exclude diacyl-phospholipids from binding [431]. As mentioned above, the deletion is thought to also serve the formation of a “backdoor” channel for direct shuttling of LPA to GPCRs. Because the WPG loop is present in all other E-NPPs, a different mode for lysophosphatidyl and sphingosyl binding must exist in NPP6 and NPP7. Compared to the lysophospholipase D activity of NPP2, these enzymes exhibit lysophospholipase C activity [388, 391]. In addition, NPP6 shows high specificity for choline head groups. Hence, it is straightforward to assume that in NPP6 and NPP7, the substrate binds in an inverse orientation with the R′ site—potentially formed from residues of the WPG loop—being specific for the choline lipid head group.
Alkaline phosphatases
General properties and functional role
APs belong to the best-studied enzymes. They served as prototypes for GPI-anchored proteins and marker enzymes for the plasma membrane [455–457] as well as for a wide variety of enzymes that use two metal ions to catalyze phosphoryl transfer reactions [458]. APs are widely expressed in prokaryotes and eukaryotes. They are found in essentially every mammalian tissue, including in a variety of tumors [459, 460] and in serum [461]. They are often (but by far not exclusively) located at exchange surfaces such as the endothelium, the apical brush border of enterocytes, kidney tubules [462, 463], the biliary epithelium, or the mucosal surface of airways [464]. They are also expressed by embryonic stem cells [465], primordial stem cells [466], in neural stem cells [55, 467], in the vascular endothelium and the neuropile of the brain [319, 468, 469], or in hair follicles [470]. Multiple technical approaches for the heterologous expression of APs have been elaborated [471].
Several AP paralogues are expressed in vertebrates (Table 1, Fig. 14). Their number and specificity vary between species. The human genome encodes four AP genes. The names of the four corresponding APs reflect their predominant (but not exclusive) tissue distribution. TNAP reveals high expression in liver, bone, and kidney (also liver bone and kidney AP) and in other tissues. Placental AP (PLAP), germ cell AP (GCAP), and intestinal AP (IAP) reveal a more restricted tissue distribution. The corresponding genes are referred to as ALPL, ALPP, ALPP2, and ALPI, respectively. For comparison, the mouse genome harbors genes encoding TNAP (Akp2), IAP (Akp3), EAP (embryonic AP) (Akp5), a recently discovered additional “global” IAP (Akp6), and the pseudogene Akp-ps1 [472]. The mouse EAP isoenzyme is related to both the human PLAP and GCAP isoenzymes. In the rat, TNAP (Alp1) and two isoenzymes of IAP (IAPI and IAPII) (Alpi and Alpi2) have been identified (ref. in [473, 474]).
Fig. 14.
Radial phylogenetic tree and substrate specificity of APs. Genes for canonical APs are ubiquitous but have not been cloned from plants. The tree highlights the closer relation of tissue nonspecific APs (TNAP) to those found in bacteria, archaea, and invertebrates. The divergence of the other human APs has occurred rather late in evolution. Availability of structural data is indicated by underscores. Amino acid sequences have been aligned with ClustalW and the tree was calculated with ProML. GI accession numbers—Antarctic bacterium TAB5: 7327837; D. rerio AP: 41055949; intestinal AP: 62122905, intestinal AP2: 68448521, E. coli: 581187; H. salinarum: 167728700; H. sapiens germ cell AP (GCAP): 110347479, intestinal AP (IAP): 157266292, placental AP (PLAP): 94721246, TNAP: 116734717; Mus musculus embryonic AP (EAP): 7327837, TNAP: 160333226, intestinal AP (IAP): 110347479; Pandalus borealis (shrimp): 13539555; Shewanella sp.: 19071967; Vibrio sp.: 243065523
APs typically are homodimeric proteins with apparent subunit sizes of about 80 kDa but heterodimers have been reported between IAP, GCAP, and PLAP [475]. They behave as non-cooperative allosteric enzymes in which the stability and the catalytic properties of each monomer are controlled by the conformation of the second subunit [476]. A dimeric structure has also been suggested after incorporation of the protein into supported lipid bilayers and analysis by atomic force microscopy [477]. Each catalytic site contains three metal ions, two Zn2+, and one Mg2+ that are necessary for catalytic activity. Zn2+ is loaded onto APs in the early secretory pathway that contains specific zinc transporters (ZnTs). Activation of human TNAP requires the presence of ZnT5/ZnT6 heterodimers and ZnT7 homooligomers [478, 479]. It is thought that the activation takes place in two steps. First, the transporters stabilize the apo-form of TNAP independently of their Zn2+ transport activity. In a second step, Zn2+ is loaded onto the protein, converting it from the apo- to the holo-form. The catalytically active holo-form is then trafficked to the plasma membrane [480]. In the absence of ZnT5/ZnT6 heterodimers, the apo-form is degraded [481]. In contrast, the placental enzyme (PLAP) is constitutively expressed in the apo-form and trafficked to the plasma membrane in ZnT5/ZnT6-deficient cells [478, 479], implicating that the activation processes of the two enzymes differ.
Substrates and catalytic properties
All mammalian APs reveal broad substrate specificity. They catalyze the hydrolysis of monoesters of phosphoric acid, releasing phosphate and also catalyze transphosphorylation reactions in the presence of high concentrations of phosphate acceptors [474]. E. coli AP displays low phosphodiesterase activity [482] and there is also evidence for phosphodiesterase activity of mammalian TNAP, including hydrolysis of cAMP [483]. The broad substrate specificity qualified APs to be widely used tools as in vitro and in vivo reporters and as molecular biology and diagnostic reagents [473, 484].
Proof of physiologically relevant substrates has been more difficult to achieve. Three phosphocompounds accumulate endogenously in hypophosphatasia: phosphoethanolamine, PPi, and pyridoxal 5′-phosphate (PLP), indicating that these are natural substrates for TNAP [485]. TNAP has been shown to hydrolyze the mineralization inhibitor PPi thereby maintaining proper bone mineralization [330]. Accordingly, mutations in the TNAP gene that cause impairment or loss of functional activity lead to hypophosphatasia in humans [486, 487]. Similarly, TNAP knockout mice (Akp2−/−) accumulate extracellular PPi, reveal hypomineralization [488–491], and can serve as a model for the infantile form of hypophosphatasia [492]. The defect can be eliminated by subcutaneous injections of soluble TNAP targeted to mineralizing tissue [493, 494] or by lentiviral application of a bone-targeted form of TNAP [495].
PLP has been identified as another TNAP substrate [496–498]. Abnormalities in PLP metabolism can be indirectly involved in the generation of seizures as exemplified for TNAP knockout mice [489, 499]. Also, the bacterial endotoxin lipopolysaccharide (LPS) is a substrate for AP [500]. LPA is detoxified by dephosphorylation of its lipid A moiety. Recent evidence suggests that IAP expressed on apical surfaces of microvillus membranes of enterocytes detoxifies LPS in the intestine, thereby preventing inflammation, sustaining barrier function, and maintaining homeostasis within intestinal flora [501–504]. This hypothesis is supported by the analysis of IAP knockout mice [505, 506]. In addition, LPS-dephosphorylating activity of gut-derived IAP has been suggested to be part of the endogenous hepatocyte defense system against LPS [507]. Furthermore, intestinal AP has been implicated in fatty acid transport in enterocytes [463]. Moreover, AP can dephosphorylate extracellular proteins at physiological pH [508–513]. Calf IAP is widely used in molecular biology to dephosphorylate nucleic acids.
Of relevance in the present context, APs can hydrolyze extracellular ATP via ADP and AMP to adenosine [362, 363, 514] (Fig. 4). This qualifies the enzymes as ecto-nucleotidases. APs are the only ecto-nucleotidases that can sequentially dephosphorylate nucleoside triphosphates to the nucleoside. They thus have the potential to inactivate agonists of P2 receptors and to produce adenosine as a P1 receptor agonist and to serve purine salvage through cellular reuptake of the nucleoside. To date, the functional involvement of APs in purinergic signaling has received rather little attention. An involvement in purinergic signaling has been implicated for TNAP in NG108-15 neuronal cells [515], in human airways [464, 516], in bone remodeling [24], and in the control of axon outgrowth in cultured hippocampal neurons [517].
APs have a pH optimum in the high alkaline range (>9). An alkaline pH is typical for the duodenal brush border surface and possibly also the osteoclast surface in the bone remodeling compartments [24]. In other settings, where the extracellular pH is close to neutral, AP activity is expected to be reduced. At a pH of 8.0, Km values for a variety of compounds are in the order of 1 mM and below (Table 1). The synthetic drug p-nitrophenylphosphate has become the most commonly used substrate for kinetic studies on APs because of the simple spectrophotometric assay [473, 474].
Few detailed studies exist on the kinetics of nucleotide hydrolysis. Their outcome is influenced by the isolation procedure employed and the microenvironment of the enzyme. Maximal activity is obtained at millimolar concentrations of Ca2+ and/or Mg2+. Rat osseous plate AP solubilized by phosphatidylinositol-specific phospholipase C revealed a pH optimum of 9.4 with the activity decreasing to about 25 % at pH 7.4 [518]. Similarly, extracellular hydrolysis of ATP to adenosine by TNAP-expressing NG108-15 cells at pH 7.4 was about 20 % of that at pH 8.5, but sufficient to elicit a physiological P1 receptor-mediated cellular response from extracellular ATP [515]. In a study on rat osseous plate AP (TNAP), complex pH-dependent kinetics for ATP hydrolysis were observed [519]. At a pH of 9.4 the Km value for ATP was 154 μM, whereas at a pH of 7.5, apparent high and low affinity dissociation constants of 82 μM and 1.3 mM, respectively, were observed. In another study, detergent-solubilized AP from the same source revealed Km values at pH 9.4 for ATP of approximately 10 μM and 1 mM for high affinity and low affinity sites, respectively, whereas at pH 7.5, the Km value was 8.1 μM [520]. As compared to that, a Km value of 1.5 mM was determined for a membrane-bound fraction of the enzyme [521]. Analyzing TNAP-, NPP1-, or POSPHO1-deficient mice, a recent study addressed the hydrolysis of ATP and ADP in isolated osteoblast-derived membrane-limited matrix vesicles and compared it to recombinant enzymes (all at pH 7.4) [362]. It identified TNAP as the major ATP- and ADP-hydrolyzing enzyme of the mouse matrix vesicles. Furthermore, purified recombinant human TNAP yielded Km values for ATP and ADP of 30 and 40 μM, respectively, with comparable maximal rates. A kinetic analysis of AMP hydrolysis by TNAP at the mucosal surface of human airway epithelium also revealed high affinity and low affinity activity sites. The Km values of the high affinity activity at pH 7.4 was 36 μM, that for the low affinity 717 μM. Vmax was about duplicated at low affinity Km [464]. These data clearly suggest that APs can hydrolyze extracellular nucleotides at low micromolar concentrations at physiological pH in vivo.
General molecular properties
The sequences of the cDNAs for the human APs were published in the 1980s (PLAP [522–524], IAP [525, 526], TNAP [527, 528], and GCAP[529, 530]). This was followed by the cloning and characterization of the corresponding genes. The predicted proteins contain 507 (TNAP), 509 (IAP), and 513 (GCAP, PLAP) amino acid residues. PLAP exhibits a considerable number of allelic variants (>20) [531]. The protein sequences of the tissue-specific forms PLAP, GCAP, and IAP are more closely related: The positional identity of the very closely related GCAP and PLAP is 98 %, that of PLAP and IAP 87 %, and that of the tissue-specific APs with TNAP between 50 and 60 % (Fig. 14). All APs contain a signal sequence of 17 to 21 amino acid residues and a C-terminal hydrophobic domain that is replaced by a GPI anchor. All mammalian APs have five cysteine residues per subunit, four of which are engaged in forming disulfide bonds [532].
Of the 13 exons of the TNAP gene, the first two exons (Ia, Ib) are noncoding. Each exon has its own promoter sequence, and thus, the presence of exons Ia and Ib in the mRNAs is mutually exclusive. The mRNAs have different 5′-untranslated sequences but the resulting protein sequences are identical [533–535]. In humans and rats, the upstream promoter Ia is preferentially utilized by osteoblasts and the downstream promoter Ib in kidney [536]. In the mouse, the first promoter is active in embryo-derived cells, whereas the second promoter is silent in basal conditions but it is activated by dibutyryl cAMP in fibroblastic cells. In the whole animal, the transcript driven by the first promoter is found in most tissues albeit at different levels, while the one driven by the second promoter is specifically expressed at high levels only in the heart [537].
Glycosylation
The number of predicted N-linked glycosylation sites is five for TNAP, three for IAP, and two for IGCAP and PLAP [531, 538]. Glycosylation of APs can vary between tissues, between isoforms of the same enzyme, and also between APs in serum and the respective tissue of origin [539]. An analysis of the AP isoforms of TNAP from human kidney, bone, and liver confirmed N-glycosylation for all three isoforms. In contrast to the liver isoenzyme, bone and kidney isoforms were also O-glycosylated [538]. The number of N-linked sites has not been determined but the contribution of N-linked carbohydrates to the molecular mass of human liver AP was estimated to be about 30 % [540]. N-linked sugars are essential for full activity of the TNAP isoforms [538, 541] but not for the germ cell (GCAP) [542], the placental (PLAP), and intestinal (IAP) enzymes [538, 543]. Structural differences in posttranslational glycosylation of bone AP isoforms in serum could be associated with different catalytic properties [544]. Human bone AP isoforms in serum separated by high-performance liquid chromatography also differed in the number of sialic acid residues attached to their glycan moieties [545].
The structure of the N-linked glycan has been determined for human PLAP [546]. It was concluded that an enzyme monomer contains a single N-linked sugar chain. The sugar chains of PLAP have typical complex-type biantennary structures, containing either Manα1→6(Manα1→3)Manβ1→4GlcNAcβ1→4GlcNAc or Manα1→6(Manα1→3)Manβ1→4GlcNAcβ1→4(Fucα1→6)GlcNAc as the core portion. Variation was caused by the numbers of N-acetylneuraminic acid in their outer chains. All the sialic acid residues of the sugar chains occurred as the NeuAcα2→3Gal group. Evidence for N-linked glycosylation on both Asn residues (Asn-122 and Asn-249) was later provided from an analysis of the crystal structure of human PLAP [446]. An analysis of recombinant human PLAP secreted from CHO cells revealed that the glycosylation pattern varies with culture conditions [547]. Using site-directed mutagenesis and protein expression in MDCK cells, it could be shown that N-glycosylation of human PLAP is neither necessary for oligomerization nor for apical cell sorting. Interestingly, when cells were depleted of cholesterol and treated with the inhibitor of N-acetylglucosamine transferase tunicamycin, PLAP was not able to oligomerize and was missorted to the basolateral surface. This would support an indirect role of N-glycosylation, possibly mediated by a raft-associated glycosylated interactor [548].
In GCAP, the N-linked carbohydrates were estimated to only contribute 11 % of total protein mass. It was suggested that both N-linked glycosylation sites were utilized during GCAP processing [542]. Finally, a detailed structural analysis was performed of the oligosaccharides of the tumor-derived IAP-like Kasahara isoenzyme purified from FL amnion cells [549]. It identified sialylated mono-, bi-, tri-, and tetraantennary complex-type sugar chains with the Galβ1→GlcNAc β1 outerchains. Both fucosylated and nonfucosylated trimannosyl cores were found in the sugar chains. The structure of these sugar chains was thus quite different from that of human PLAP that contained biantennary complex-type oligosaccharides only [546]. Since the two enzymes show almost 90 % sequence identity, this provides further evidence for cell-specific patterns of AP glycosylation.
The GPI anchor
That AP can be released from membranes was first shown using phospholipases from Bacillus cereus and a rabbit kidney cortex homogenate [550]. Later, human placental AP was the first protein identified to be fixed to the plasma membrane by a GPI anchor and to be released by bacterial phosphatidylinositol-specific phospholipase C (PI-PLC) [551, 552]. Yet, depending on the system investigated, various percentages of the enzyme proved to be PI-PLC-resistant, presumably as a result of inositol acylation [553, 554]. Since then, placental AP has been widely used as a model system for investigating C-terminal processing of GPI-anchored proteins. The N-terminal signal peptide is specifically removed and the C-terminal of the nascent pre-pro-PLAP provides the signal for processing, during which a largely hydrophobic 29-residue C-terminal peptide is removed. The phosphatidylinositol-glycan moiety is then added to the newly exposed Asp-484 terminus [555, 556]. Phosphatidylinositol-glycan tailing is a prerequisite for transport from the ER and for PLAP enzyme activity. Otherwise, proteins are retained in the ER in an inactive conformation [555]. Conversely, interruption of the C-terminal 29-amino acid hydrophobic stretch by a charged residue resulted in protein secretion into the medium [557]. Processing of PLAP can also be demonstrated in cell-free systems [558]. The residues required for proper processing of the nascent protein into the mature GPI-anchored protein were analyzed using a “mini PLAP” of 29 kDa, in which the N-terminus and C-terminus were retained but most of the interior of the PLAP sequence was deleted. Processing of the nascent mutant protein occurred only when a small amino acid was located at the site of cleavage and phosphatidylinositol-glycan attachment, the ω site [559]. The structure of the GPI anchor of human PLAP was determined by mass spectrometry and gas chromatography–mass spectrometry [560, 561].
GPI-anchored proteins have the ability to spontaneously insert into the lipid bilayer of cells and liposomes. Human PLAP [477, 643] and also bovine IAP [562] serve as model systems for GPI anchor targeting into rafts and for raft partitioning in model membranes. These studies further detail the mechanisms that organize membranes in functional platforms or microenvironments, enriched in sphingolipids and cholesterol. The acyl and alkyl chain composition of GPI anchors influences the association with the ordered domains. The activity of the GPI-anchored PLAP may in turn be modulated by features of the membrane environment such as lipid rafts and membrane curvature [563]. GPI-anchored AP molecules inserted in supported model membranes can be imaged by atomic force microscopy, in physiological buffer. The enzymes are generally observed in the most ordered domains. This direct access to the membrane structure at a mesoscopic scale allowed establishing the GPI protein-induced changes in microdomain size and provided direct evidence for the temperature-dependent distribution of GPI proteins between fluid and ordered membrane domains [456].
Soluble forms
AP is abundant in serum. Approximately 95 % of the circulating total AP in healthy humans is derived from bone and liver sources [545]. Studies in mice suggest that serum AP levels are genetically determined [564]. Numerous studies support the clinical utility of APs as a marker for various disorders including bone and liver disease [565, 566]. Bone AP circulates as a variable mixture of anchorless isoforms, which can be separated by high-performance liquid chromatography [545, 567]. It may initially be released from osteoblasts in an anchor-intact form, possibly associated with membrane vesicles, which are then susceptible to circulating GPI-specific phospholipase D activity, which is high in serum [566, 568, 569]. In contrast, in bile, where GPI-specific phospholipase D is absent, AP is released in an aggregating form. This aggregated form of AP is low in the serum of healthy individuals but is increased in hepatobiliary diseases, especially choleostasis. It presumably includes both enzyme–lipid complexes and membrane fragments to which AP is still attached [570, 571]. The physiological role of serum AP remains an open question. Furthermore, after ingestion of a high-fat diet, IAP in the intestinal luminal content increases by more than 10-fold. The process by which ingested fatty acids mediate the IAP release into the intestinal lumen involves production of lipoprotein particles secreted from the enterocytes [572].
Protein interactions
Similar to eN and NPPs, APs can bind extracellular matrix components. Initial experiments using affinity chromatography, identified collagen type I, II, and X as binding partners of TNAP [573–575]. The interaction of collagens with TNAP is thought to be important as a nucleator for the process of skeletal calcification. Both are constituents of matrix vesicles, cell-derived microstructures present in the extracellular matrix that have been shown to initiate mineral deposition in calcifying cartilage and other tissues. A putative protein–protein interaction domain in APs has been identified that bears homology with that of other proteins known to interact with collagen such as von Willebrand factor, complement factor B, or cartilage matrix protein [576]. This domain is in part responsible for the isozyme-specific binding of TNAP to collagen [577] and located within a top flexible extracellular loop of the enzyme, the crown domain [446] (see below). Within this crown domain, a site for collagen attachment has been localized between aa 405 and 435 [476]. Molecular modeling of TNAP shows that this domain is located at the very tip of the crown and highly accessible. Its functional relevance has been confirmed by mutational analysis [578]. In addition, PLAP has the capacity to bind the Fc portion of human IgG, acting as a placental IgG receptor. PLAP would thus be involved in the internalization of IgGs and transfer of IgG molecules from the maternal circulation to the fetus during pregnancy [579]. This view is compatible with the finding that large amounts of PLAP are contained in clathrin-coated vesicles prepared from placenta [580]. More recently was TNAP identified as an interaction partner of the (equally GPI-anchored and laminin-interacting) cellular prion protein (PrPC). This interaction takes place in lipid rafts of cultured neuroepithelial cell lines (1C11) that can be differentiated towards a serotonergic (1C115-HT) or noradrenergic (1C11NE) phenotype. Since TNAP can dephosphorylate laminin, it is hypothesized that TNAP acts as a functional protagonist in the PrPC interplay [511].
Several protein interactions have also been reported for IAP. In vitro and in vivo studies showed that exogenous IAP quickly binds to the asialoglycoprotein receptor on hepatocytes. Since the liver is the major LPS-removing organ, the enzyme may constitute part of the endogenous defense system against LPS [507]. Bovine IAP can bind purified chick intestinal calcium-binding protein calbindin in a calcium-dependent manner [581] and to serum immunoglobulin G that also enhances catalytic activity [582].
Downstream signaling
APs potentially can participate in downstream purinergic signaling pathways. Whereas eN hydrolyzes AMP to promote adenosine-mediated P1 receptor signaling, APs would be involved in both inactivation of agonists of P2X and P2Y receptors and activation of P1 receptors. In case of the co-expression of both eN and AP, eN may predominate at low and APs may predominate at high extracellular AMP concentrations [464]. Evidence for AP-mediated hydrolysis of extracellular ATP and subsequent P1 receptor activation has been derived from studies on NG108-15 cells [515] and fetal fibroblasts [583]. Recently, it was shown that TNAP is co-localized at growth cones of cultured hippocampal cells with ionotropic P2X7 receptors, whose activation inhibited axonal growth. TNAP facilitated axonal growth by hydrolyzing ATP in the immediate environment of the receptor and preventing P2X7 receptor activation. Furthermore, inhibiting the P2X7 receptor reduced TNAP expression, whereas the addition of AP enhanced P2X7 receptor expression. The results suggested that TNAP, by regulating both ligand availability and protein expression of P2X7 receptors, is important for axonal development [517]. TNAP was also found to be essential for the differentiation of cultured adult neural stem cells into neurons or oligodendrocytes. However, the involvement of purinergic mechanisms was not ascertained [584]. Activation of downstream signaling mediated by antibody ligation as observed for eN has not been described for APs. The functional implications of the ecto-nucleotidase activity of APs certainly deserve further investigation.
Phylogenetic relationship
Genes for canonical APs are ubiquitous in vertebrates, invertebrates, bacteria, and archaea but have not been cloned from plants (Fig. 14). Much progress has been made in understanding the catalytic mechanisms of APs by studying the bacterial enzymes. There is a high degree of similarity between the eukaryotic APs and E. coli AP(EcAP) [585]. Their active site contains a binuclear cluster of Zn2+ ions, which is involved in phosphate binding, and a conserved Ser residue, which is phosphorylated in the course of catalysis [586]. The dimeric EcAP is exported into the periplasmic space from where it can be secreted to the cell exterior [587, 588]. Several APs have been identified in Bacillus subtilis [589]. Mammalian APs are 20–30-fold more active than the corresponding bacterial enzymes. Their amino acid sequences are 25–30 % conserved [590].
Sequence data including the conservation of metal-binding motifs, structural analysis, and deduced catalytic mechanisms place APs into a superfamily of phospho-/sulfo-coordinating metalloenzymes [367, 441, 442], also referred to as AP superfamily [443, 444]. These include the families of arylsulfatases, phosphopentomutases, 2,3-bisphosphoglycerate-independent phosphoglycerate mutases, and the NPP family. Members of the AP superfamily possess conserved Ser, Thr, or Cys residues that have been found to be phosphorylated or sulfated. On the basis of the sequence and structural conservation, it has been proposed that catalytic cycles of all the members of this superfamily include phosphorylation (or sulfation, or phosphonation) of these conserved Ser/Thr/Cys residues. Thus, all enzymes of this superfamily share the same reaction scheme that was originally proposed for APs [444, 586].
Protein structure
Information on the spatial structure and enzyme mechanism of AP was first obtained for the enzyme from E. coli [586]. Since then, crystal structures have been obtained for several bacterial APs and for shrimp AP (Table 9). Of the four human isoenzymes of AP (Table 1), the X-ray structure is available for human placental AP (PLAP) (Fig. 15a) [446]. The homodimeric enzyme has dimensions of 100 × 72 × 59 Å. Each monomer consists of 479 residues and four metal ions. In comparison to the structure of AP from E. coli, the following three additional structural elements are present in the human enzyme. (1) The N-terminal residues (1–25) including an α-helix (9–25) interact with the neighboring monomer and might thus stabilize the dimer; 555 Å2 are buried upon interaction of the N-terminal α-helix with the neighboring monomer (Fig. 15a). (2) An insertion of 60 residues forms the “crown domain” close to the dimer interface, thus forming interactions between the two monomers. The residues of this domain thus interact with each other at the dimer interface. The domain consists of two three-stranded parallel β-sheets. A crown domain is also present in the APs from Halobacterium salinarum [591] and Vibrio sp. [592], but it has a different fold in the bacterial enzymes. (3) Finally, a metal-binding domain consisting of 76 residues (209–285) is present at the distal ends of the longest axis of the dimer. The metal-binding domain contains two β-strands extending the central β-sheet of the core AP domain and two α-helices, one on each side of this β-sheet. Thus, the core β-sheet of each monomer contains 10 β-strands, of which nine are parallel. Several α-helices are packed against both sides of the β-sheet. The metal-binding domain contains an additional non-catalytic metal ion which is coordinated by E216, E270, D285, the backbone carbonyl group of F269, and a water molecule. It is conserved in all human and mouse APs, but its functional or structural role remains to be established [473, 474]. The binding site appears to be occupied by a magnesium or calcium ion in the crystal structure [446] and was later identified as a Ca2+ ion in the purified enzyme [578].
Table 9.
Selected crystal structures of APs in the protein data bank
| Structure | Ligand | Resolution (Å) | PDB ID | References |
|---|---|---|---|---|
| Homo sapiens (PLAP) | ||||
| Native structure | Phosphate | 1.8 | 1EW2 | [446] |
| Complex with l-Phe, S92 is phosphorylated | l-Phe | 1.9 | 3MK2 (1ZEF) | [594, 632] |
| Shrimp (Pandalus borealis) AP | ||||
| Native structure | 1.92 | 1K7H | [633] | |
| Phosphate complex | Phosphate | 2.15 | 1SHN | [634] |
| Escherichia coli AP | ||||
| Phosphorylated intermediate of the S102T mutant | Phosphorylated nucleophile | 2.2 | 2GA3 | [600] |
| Vanadate complex as model for the transition state | Vanadate | 1.9 | 1B8J | [598] |
| Halobacterium salinarum AP | Phosphate | 1.7 | 2X98 | [591] |
| Shewanella sp. AP | Sulfate | 2.2 | 3A52 | [635] |
| Vibrio sp. AP | Sulfate | 1.4 | 3E2D | [592] |
| AP from TAB5 (Antarctic bacterium) | 1.95 | 2IUC | [636] |
Fig. 15.
Crystal structure of human placental AP (PLAP). a Stereo view of the fold of PLAP. The view is perpendicular to the twofold molecular axis relating the two monomers of the dimeric protein. Monomer A is colored in red and the other monomer B in beige. These regions correspond to the core part of the monomer that is conserved with the bacterial APs. Additional domains in the mammalian protein are colored as follows. The crown domain is shown in light blue (monomer A) and blue (monomer B). The metal-binding domain is indicated in yellow. The N-terminal helices (green, monomer A and orange, monomer B) form contacts to the neighboring monomer. The zinc ions of the active site are shown in green and the magnesium ions in white. The glycosylation sites are indicated in cyan. The C-terminal residue (479) of the X-ray structure is shown in red (large spheres) to indicate a position close to Asp484, to which the GPI anchor is attached for immobilization of the protein to the cell membrane. A phenylalanine ligand bound to a peripheral binding site is shown in red sticks (hidden behind the protein in monomer A). Generated from pdb ids 1EW2 and 1ZEF. b Stereo view of the active site of PLAP. The zinc ions are shown in green and the magnesium ion is depicted as a gray sphere. The coordination bonds are shown as broken lines. Tyr367, depicted with yellow carbon atoms, belongs to the neighboring subunit of the dimeric protein. The interactions between Arg166 and the bound phosphate ion are indicated as broken lines. Generated from PDB id 1EW2. c Inhibitory binding mode of l-Phe in the active site of PLAP. The zinc ions are shown in green and the magnesium ion is depicted as a gray sphere. Coordination bonds and hydrogen bonding interactions are shown as broken lines. The bound phenylalanine inhibitor is depicted with yellow carbon atoms. Generated from PDB id 3MK2. See the electronic form of the article for a colored version of this figure
Interestingly, all three regions differ in sequence between the different AP isoforms, and they have been proposed to be involved in isozyme-specific properties including the uncompetitive inhibition and allosteric behavior [474]. The crown domain as well as the metal-binding domain is unique in fold as no similar folds could be found by a comparison to structures in the Protein Data Bank. Mammalian APs all contain two disulfide bridges, corresponding to C121–C183 and C467–C474 in PLAP and a free cysteine (C101). The glycosylation sites N122 and N249 in human PLAP, the disulfide bridges, and the protein residues coordinating the fourth metal ion in hpAP are conserved in all four human AP isoenzymes.
Using the structure of PLAP, a homology model for the human tissue nonspecific AP (TNAP) was constructed [578]. Based on this model, mutations associated with hypophosphatasia could be rationalized. The mutations are clustered within five regions: the active site and its vicinity, the active site valley, the homodimeric interface, the crown domain, and the fourth metal-binding site. Several severe mutations occur around this Ca2+ binding site, demonstrating the importance of this additional metal site for the eukaryotic APs.
Active site structure
The core catalytic residues are conserved between (human) PLAP and bacterial APs: the three metal ions and the coordinating ligands, the serine nucleophile, and an arginine residue (Fig. 15b). The two zinc ions are 4.0 Å apart and the Mg2+ ion binds at 4.7 Å distance to Zn2. A unique feature of the active site of the mammalian enzyme in comparison to EcAP is the presence of Y367 at a distance of 6.1 Å to the phosphate ion. This tyrosine, which protrudes from the neighboring subunit into the active site, has been shown to be involved in the uncompetitive inhibition of mammalian APs [532, 593].
To characterize the structural basis of the allosteric behavior and uncompetitive inhibition of PLAP, co-crystal structures with various ligands influencing the catalytic activity of the enzyme have been determined [594]. Mammalian APs show an uncompetitive inhibition by some l-amino acids. A complex structure with l-Phe demonstrated the binding site for these inhibitors in the active site hydrophobic pocket (Fig. 15c). The catalytic serine residue (S92) is phosphorylated in this structure. The amino group of the phenylalanine inhibitor is coordinated to Zn1 and interacts with the phosphoryl group, in agreement with the uncompetitive nature of the inhibition, i.e., the inhibitor binds to the intermediate but not to the free enzyme. The l-Phe molecule, as well as AMP and p-nitrophenyl-phosphonate in further co-crystal structures, also binds to a second binding site at a distance of 28 Å from the active site (Fig. 15a, shown as red sticks in monomer B). The functional relevance of this obviously quite unspecific binding site between two helices remains unclear.
Catalytic mechanism
The work on the reaction mechanism of AP is mostly based on structural, kinetic, and mutational studies on the enzyme from E. coli [458, 595]. In contrast to many other metallophosphatases, which activate the water molecule for a direct attack on the substrate, the reaction mechanism of AP is special in that it proceeds via a covalent intermediate [596] (Fig. 16). A serine residue (S102 in EcAP, S92 in hpAP) is phosphorylated during the reaction. S92 is coordinated to Zn2 [586, 597], which lowers the pKa of the alcohol group such that it is more easily deprotonated to an alcoholate group for attack on the phosphate group. The phosphate group is coordinated to both zinc ions. This binding mode has been observed in crystal structures in complex with phosphate ions [446, 586]. The first transition state is stabilized by the two zinc ions and R166. A crystallographic model for the transition state has been obtained in the form of a co-crystal structure of EcAP with vanadate [598]. The vanadate is covalently bound to the serine nucleophile and adopts a trigonal bipyramidal geometry. The interactions with the metal ions and the arginine side chain resemble those shown in Fig. 16(f). In addition, structures of EcAP with the transition state analogue AlF3 have been determined [599]. Expulsion of the negatively charged leaving group is facilitated by coordination to Zn1. The phosphorylated serine intermediate is coordinated in a similar way to both zinc ions as the substrates phosphate group, but in a reverse orientation. Structures of the phosphorylated intermediate could be determined for PLAP in the co-crystal structure with the uncompetitive inhibitor l-Phe [594] and for EcAP by exchange of the zinc ions with cadmium [586], by mutation of the serine nucleophile to a threonine [600] and by a D153G–D330N double mutant [599].
Fig. 16.
Proposed reaction mechanism of AP [586]. a Scheme of the active site in the unliganded state. b–d Michaelis complex, transition state, and product complex, respectively, of the formation of the phosphorylated serine intermediate. e–g Dephosphorylation of the intermediate. The residue numbers refer to human placental AP (PLAP)
For efficient catalysis and to avoid substrate saturation and product inhibition, enzymes in general need to strongly discriminate between transition state and ground state structures. For EcAP, it was recently demonstrated that destabilization of the enzyme × substrate ground state complex is achieved by an electrostatic repulsion between the deprotonated Ser102 and the negatively charged phosphate ester substrates [601]. Molecular dynamics simulations and hybrid quantum mechanics/molecular mechanics calculation indicated that substrate promiscuity with respect to different leaving groups results from stabilization of different charge distributions of the leaving groups via different interactions involving the zinc ions and other active site residues such as Lys328 [602].
The second part of the mechanism is essentially very similar to the formation of the covalent intermediate; however, the roles of the two zinc ions are exchanged. At Zn1, the leaving group is replaced by the water nucleophile. Zn1 now activates the water nucleophile and Zn2 the leaving group. The pentaphosphate transition state is stabilized by the same interactions as the first transition state. As a result of the two inline steps, the overall reaction proceeds with a retention of the configuration at the phosphorus atom [603].
The equilibrium between the non-covalent phosphate complex and the phosphoseryl intermediate has also been studied by nuclear magnetic resonance (NMR) [604–608]. Via 31P NMR, it was shown that the hydrolysis of the substrate (the first step) is rate-limiting at alkaline pH, whereas the cleavage of the phosphoseryl intermediate is rate-limiting below pH 6 since the hydroxide ion nucleophile coordinated to Zn1 becomes protonated [604, 608]. Replacement of Zn2+ by the much softer Cd2+ in metal-binding site 1 shifts the pKa of the coordinated water to alkaline pH such that the phosphoseryl intermediate forms upon incubation of the enzyme with phosphate.
The function of the conserved magnesium ion has been investigated by kinetic and mutational studies (Table 10). Mutagenesis of the ligands of the magnesium ion has large detrimental effects on catalysis [609, 610]. A hydroxide ion coordinated to the Mg2+ ion has been proposed to act as a general base to accept the proton from the serine nucleophile [611]. In the dephosphorylation step, this water molecule coordinated to the Mg2+ ion might protonate the leaving Ser alcoholate. Interestingly, the third metal ion is not present in the NPP enzymes, which otherwise share a conserved dizinc site and probably quite similar mechanism. AP preferentially hydrolyzes phosphate monoesters compared to diesters whereas NPP has a much higher activity towards phosphate diesters. The preferences of these enzymes for phosphate monoesters and diesters are reversed by ∼1015-fold [441, 482]. The removal of the magnesium binding site from EcAP reduced the activity towards phosphate monoesters up to 106-fold whereas the hydrolysis of phosphate diesters was affected only ∼2-fold. The authors concluded that the role of the magnesium ion was likely not in the deprotonation of the nucleophile or protonation of the leaving group, which should affect the hydrolysis of monoesters and diesters likewise [442]. Instead, the magnesium ion might stabilize the transition state by interaction with the transferred phosphoryl group via a coordinated water molecule.
Table 10.
Explanation of the observed effects of selected previously reported active site mutations of E. coli AP on activity
| Residue | Function | Mutant | Effect | Explanation | References |
|---|---|---|---|---|---|
| S102 | Nucleophile | S102A, S102G, S102L | ~103–104-fold reduction in kcat | Side chain cannot act as nucleophile, direct attack of water? | [613, 615] |
| S102C | 100-fold reduction of kcat | Cysteine chain might takeover the role of the nucleophile but the environment is optimized for Ser | [612, 613] | ||
| R166 | Binds to substrates phosphate group | R166Q, R166S, R166A | Little effect on kcat, Km increases over 50-fold | R166 is not essential for catalysis, it is mainly involved in substrate binding but not in transition state stabilization | [616, 637] |
| D101 | Binds to R166 | D101A | Increase in kcat in the presence of a phosphate acceptor | Higher flexibility of R166, change in rate-limiting steps and faster phosphate release | [638–640] |
| D101S | |||||
| D153 | Binds to R166 and to Mg-coordinated water | D153G | 5-fold higher kcat, no change in Km | Faster phosphate release | [641] |
| K328 | Binds to D153 and phosphate ion via a water molecule | K328A | 6-fold increase in activity at pH 10.3 | Faster phosphate release | [642] |
Replacement of the serine nucleophile (S102) reduces kcat about 103–104-fold; however, the hydrolytic rate is still ∼105–107-fold higher than that of the non-enzymatic reaction [612, 613]. In the S102A or S102G variants, the reaction might proceed via a direct attack of a Zn2-coordinated water nucleophile on the substrate phosphate group, similar to mechanisms in other dinuclear metallohydrolases [614]. Interestingly, mutation of R166 (R166Q, R166S, and R166A) increases Km 50-fold but has only minor effects on kcat [615, 616]. Thus, the role of the arginine appears to be mainly in substrate binding and not in the stabilization of the transition state, which may be predominantly achieved by the two metal ions. Mutation of the active site residues D153 and K328 of AP results in an increase of the release rate of the product phosphate ion at alkaline pH and thus in an increase of the rate constant. However, these residues are replaced by histidine residues in the mammalian enzymes.
Synopsis
There exists a surprising multiplicity of ecto-nucleotidases, including several protein families with several members each. The physiological implications of this diversity are only partially understood. Not all of these enzymes hydrolyze only nucleotides. This applies to all APs and members of the NPP protein family (NPP2, NPP4–7). Conversely, not all members of a given protein family can hydrolyze nucleotides. Whereas NPP1–3 can hydrolyze nucleotides, NPP4–7 hydrolyze phospholipids only. Not all members of a given protein family may be cell surface-located. They can be associated with specific intracellular organelles, as for NTPDase4–7. Almost all of these enzymes primarily exist in membrane-bound form but they differ regarding their mode of membrane anchorage. eN and all APs are GPI-anchored whereas E-NTPDases and E-NPPs (with the exception of NPP2) possess transmembrane helices. Moreover, soluble forms devoid of a membrane anchor have been described (eN, APs, NPP2). In some cases, membrane-bound forms are shed from the cell surface in the form of microvesicles (eN, APs, NTPDase1). Membrane anchorage restricts the impact of ecto-nucleotidases to cell surface-located catalysis and autocrine and paracrine signaling. Soluble forms considerably broaden the reach of the enzymes by diffusion within a tissue or by their distribution within tissue fluids. The truly soluble forms may also be referred to as exo-nucleotidases.
For all ecto-nucleotidases, the availability of the extracellular nucleotide substrates governs catalytic activity whereas required metal ions such as Ca2+ or Mg2+ are continuously available in millimolar concentrations in the extracellular space. eN, NPPs, and AP require in addition the binding of Zn2+ ions which presumably are loaded onto the enzymes during their passage through the secretory pathway. Since extracellular Zn2+ concentrations are very low, the affinity for Zn2+ of the proteins must be high. Km values for ATP typically are in the low micromolar range. This would suggest that at resting extracellular ATP concentrations in the nanomolar range, the velocity of nucleotide hydrolysis would be low. Stimulated release and an increase in extracellular ATP to micromolar concentrations [30] would, however, strongly accelerate the catalytic rate of ecto-nucleotidases—whereby they compete with nucleotide receptors for nucleotide binding. Ecto-nucleotidases differ regarding substrate specificity and pH optima. Alkaline pH optima (as for APs and E-NPPs) may not appear optimal for catalytic activity in most tissues but may represent specific adaptive features, e.g., at the surface of HCO–3-secreting tissues such as duodenal epithelium or in the bone remodeling compartment [24]. Similarly, an acidic pH optimum (as for PAPs and TRAP) or sustained activity at acidic pH (as for some E-NTPDases) may be an adaption to conditions of inflammation, ischemia, and cancer growth, where a fall in extracellular pH is observed [617, 618].
Whereas previous biochemical analyses had led to the identification and characterization of prototypic members of individual enzyme families, only the advent of molecular cloning and recombinant expression revealed the presence of multiple paralogues (E-NTPDases, APs, E-NPPs) and also of multiple orthologues throughout the animal kingdom, plants, fungi, and bacteria. It permitted the unequivocal identification of their catalytic and additional biochemical properties and important initial studies on their regulation and pathways of synthesis and their physiological and pathophysiological roles. Whereas impressive evidence has been compiled for the (patho)physiological relevance of nucleotide hydrolysis by eN and E-NTPDases in purinergic signaling, the examples for E-NPPs and APs are still rare. But E-NPPs and APs are of considerable importance in tissue calcification by producing and hydrolyzing PPi.
Mutagenesis studies had provided important insight into the requirements of protein structure and essential amino acid residues for enzymatic activity. Only recently and based on crystal structures, detailed information regarding the spatial structures and catalytic mechanisms have become available for members of all four ecto-nucleotidase families reviewed here. This allows detailed predictions and comparison of their catalytic mechanisms.
Alkaline phosphatase is probably the best characterized enzyme, to a great part, however, based on the work on the E. coli homologue, which has been studied intensely by kinetic, spectroscopic, crystallographic, and mutational studies. The AP enzyme mechanism is unique in its use of two catalytic zinc ions and a protein nucleophile. In most metallohydrolases, the water nucleophile directly attacks the scissile bond, without the formation of a protein-bound intermediate. In AP, the phosphoryl group of the substrate is first transferred to a serine nucleophile under the formation of a covalent phosphoryl-seryl intermediate, which is hydrolyzed in a second step by attack of a water nucleophile. This ping-pong double inline displacement mechanism is mostly identical in the E-NPPs, which are structurally related, i.e., they share a common fold. The main differences are the use of a threonine nucleophile, the absence of the magnesium ion found in the AP active site, and the presence of further residues conferring the substrate specificity of the E-NPPs.
The other two ecto-nucleotidases discussed in this review directly transfer the substrate’s phosphoryl group to water. Like AP, eN contains two divalent metal ions in the active site, most likely a zinc ion and another divalent ion such as calcium. The fold of eN is not related to AP, NPP, or NTPDase. However, the N-terminal domain with the two metal ions belongs to the same superfamily of dimetal phosphatases as TRAP. Also, the catalytic mechanism of eN is unique, one metal ion binds and activates the water nucleophile whereas the other metal ion binds and polarizes the terminal phosphate group of the substrate. NTPDases also have a unique enzyme fold and catalytic mechanism compared to the other ecto-nucleotidases. A divalent metal ion binds the two terminal phosphate groups of the substrate. In contrast to the other three enzymes, a metal ion is not involved in the activation of the water nucleophile. Instead, a glutamate side chains acts as a general base to deprotonate the water molecule. All four ecto-nucleotidases are thus dependent on divalent metal ions for activity. In AP, NPP, and eN, the two metal ions are bound to the enzyme by several protein side chains also in the absence of bound substrate, whereas in the NTPDases, the metal ion only binds with the substrate and it is coordinated via four water molecules.
NTPDases and eN consist of two domains and the active center is located at the domain interface. For both enzymes, a domain motion is involved in enzyme catalysis. eN undergoes a large conformational change of about 96° between the open and closed forms of the enzyme, most likely to enable substrate binding and product leaving in the open conformation. For the NTPDases, the domain motion is much smaller and appears to be involved in the significant influence of the membrane attachment of the enzyme on catalytic activity.
Ecto-nucleotidases represent important potential therapeutic targets for interfering with P2 or P1 receptor-mediated cellular signaling pathways or the control of extracellular PPi formation. Unraveling their atomic structures will facilitate the further development of ecto-nucleotidase inhibitors or also of substances that may be employed as prodrugs for the tissue-specific generation of purinergic receptor agonists or possibly also antagonists.
Acknowledgments
The research work of the authors was supported by the Deutsche Forschungsgemeinschaft (to HZ: 140/17-4; Zi 140/18-1 and to NS: Str 477/11, Str 477/12, Str 477/13).
Glossary
- ACR
Apyrase-conserved regions
- AMPCP
α,β-Methylene-ADP
- AMPPNP
β,γ-Imidoadenosine 5′-triphosphate
- AP
Alkaline phosphatases
- ATX
Autotaxin
- CAN
Calcium-activated nucleotidase
- CHO
Chinese hamster ovary
- ECD
Extracellular domain
- eN
5′-Nucleotidase
- ER
Endoplasmic reticulum
- FRET
Fluorescence resonance energy transfer
- GCAP
Germ cell AP
- GPC
Glycerophosphorylcholine
- GPI
Glycosylphosphatidylinositol
- IAP
Intestinal AP
- LPA
Lysophosphatidic acid
- LPC
Lysophosphatidylcholine
- LPS
Lipopolysaccharide
- MALDI
Matrix-assisted laser desorption/ionization
- MDCK
Madin–Darby canine kidney
- MS
Mass spectrometry
- NDP
Nucleoside diphosphate
- NLD
Nuclease-like domain
- NMP
Nucleoside monophosphate
- NMN
Nicotinamide mononucleotide
- NMR
Nuclear magnetic resonance
- NPP
Nucleotide pyrophosphatase/phosphodiesterase
- NR
Nicotinamide riboside
- NTP
Nucleoside triphosphate
- NTPDase
Nucleoside triphosphate diphosphohydrolase
- PAP
Prostatic acid phosphatase
- PDB
Protein Data Bank
- PDE
Phosphodiesterase
- PI-PLC
Phosphatidylinositol-specific phospholipase C
- PLAP
Placental AP
- PLP
Pyridoxal 5′-phosphate
- PMA
Phorbol myristate acetate
- pNPPC
p-Nitrophenyl phosphorylcholine
- PPi
Pyrophosphate
- S1P
Sphingosine-1-phosphate
- SMB
Somatomedin B
- SPC
Sphingosylphosphorylcholine
- RanBPM
Ran Binding Protein M
- TMD
Transmembrane domain
- TNAP
Tissue nonspecific AP
- TRAP
Tartrate-resistant acid phosphatase
References
- 1.Burnstock G. Physiology and pathophysiology of purinergic neurotransmission. Physiol Rev. 2007;87:659–797. doi: 10.1152/physrev.00043.2006. [DOI] [PubMed] [Google Scholar]
- 2.Zimmermann H, Mishra SK, Shukla V, Langer D, Gampe K, Grimm I, Delic J, Braun N. Ecto-nucleotidases, molecular properties and functional impact. An R Acad Nac Farm. 2007;73:537–566. [Google Scholar]
- 3.Zimmermann H. ATP and acetylcholine, equal brethren. Neurochem Int. 2008;52:634–648. doi: 10.1016/j.neuint.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 4.Burnstock G. Pathophysiology and therapeutic potential of purinergic signaling. Pharmacol Rev. 2006;58:58–86. doi: 10.1124/pr.58.1.5. [DOI] [PubMed] [Google Scholar]
- 5.King AE, Ackley MA, Cass CE, Young JD, Baldwin SA. Nucleoside transporters: from scavengers to novel therapeutic targets. Trends Pharm Sci. 2006;27:416–425. doi: 10.1016/j.tips.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 6.Johnson K, Polewski M, Etten D, Terkeltaub R. Chondrogenesis mediated by PPi depletion promotes spontaneous aortic calcification in NPP1−/− mice. Arterioscler Thromb Vasc Biol. 2005;25:686–691. doi: 10.1161/01.ATV.0000154774.71187.f0. [DOI] [PubMed] [Google Scholar]
- 7.Villa-Bellosta R, Wang XN, Millán JL, Dubyak GR, ONeill WC. Extracellular pyrophosphate metabolism and calcification in vascular smooth muscle. Amer J Physiol Heart Circ Phy. 2011;301:H61–H68. doi: 10.1152/ajpheart.01020.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciana P, Fumagalli M, Trincavelli ML, Verderio C, Rosa P, Lecca D, Ferrario S, Parravicini C, Capra V, Gelosa P, Guerrini U, Belcredito S, Cimino M, Sironi L, Tremoli E, Rovati GE, Martini C, Abbracchio MP. The orphan receptor GPR17 identified as a new dual uracil nucleotides/cysteinyl-leukotrienes receptor. EMBO J. 2006;25:4615–4627. doi: 10.1038/sj.emboj.7601341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abbracchio MP, Burnstock G, Boeynaems JM, Barnard EA, Boyer JL, Kennedy C, Knight GE, Fumagalli M, Gachet C, Jacobson KA, Weisman GA. International Union of Pharmacology LVIII: update on the P2Y G protein-coupled nucleotide receptors: from molecular mechanisms and pathophysiology to therapy. Pharmacol Rev. 2006;58:281–341. doi: 10.1124/pr.58.3.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harden TK, Sesma JI, Fricks IP, Lazarowski ER. Signalling and pharmacological properties of the P2Y14 receptor. Acta Physiol. 2010;199:149–160. doi: 10.1111/j.1748-1716.2010.02116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mutafova-Yambolieva VN, Hwang SJ, Hao X, Chen H, Zhu MX, Wood JD, Ward SM, Sanders KM. β-Nicotinamide adenine dinucleotide is an inhibitory neurotransmitter in visceral smooth muscle. Proc Natl Acad Sci USA. 2007;104:16359–16364. doi: 10.1073/pnas.0705510104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hwang SJ, Durnin L, Dwyer L, Rhee PL, Ward SM, Koh SD, Sanders KM, Mutafova-Yambolieva VN. β-Nicotinamide adenine dinucleotide is an enteric inhibitory neurotransmitter in human and nonhuman primate colons. Gastroenterology. 2011;140:608–U347. doi: 10.1053/j.gastro.2010.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallego D, Gil V, Aleu J, Martinez-Cutillas M, Clave P, Jimenez M. Pharmacological characterization of purinergic inhibitory neuromuscular transmission in the human colon. Neurogastroenterol Motil. 2011;23:792–e338. doi: 10.1111/j.1365-2982.2011.01725.x. [DOI] [PubMed] [Google Scholar]
- 14.Gustafsson AJ, Muraro L, Dahlberg C, Migaud M, Chevallier O, Khanh HN, Krishnan K, Li NL, Islam MS. ADP ribose is an endogenous ligand for the purinergic P2Y1 receptor. Mol Cell Endocrinol. 2011;337:122–123. doi: 10.1016/j.mce.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 15.Ehrlich YH, Hogan MV, Pawlowska Z, Wieraszko A, Katz E, Sobocki T, Babinska A, Kornecki E. Surface protein phosphorylation by ecto-protein kinases. Role in neuronal development and synaptic plasticity. Adv Exp Med Biol. 1998;446:51–71. doi: 10.1007/978-1-4615-4869-0_4. [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez FA, Contreras C, Bolanos-Garcia V, Allende JE. Protein kinase CK2 as an ectokinase: the role of the regulatory CK2beta subunit. Proc Natl Acad Sci USA. 2008;105:5693–5698. doi: 10.1073/pnas.0802065105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seman M, Adriouch S, Scheuplein F, Krebs C, Freese D, Glowacki G, Deterre P, Haag F, Koch-Nolte F. NAD-induced T cell death: ADP-ribosylation of cell surface proteins by ART2 activates the cytolytic P2X7 purinoceptor. Immunity. 2003;19:571–582. doi: 10.1016/S1074-7613(03)00266-8. [DOI] [PubMed] [Google Scholar]
- 18.Quintero IB, Araujo CL, Pulkka AE, Wirkkala RS, Herrala AM, Eskelinen EL, Jokitalo E, Hellström PA, Tuominen HJ, Hirvikoski PP, Vihko PT. Prostatic acid phosphatase is not a prostate specific target. Cancer Res. 2007;67:6549–6554. doi: 10.1158/0008-5472.CAN-07-1651. [DOI] [PubMed] [Google Scholar]
- 19.Zylka MJ, Sowa NA, Taylor-Blake B, Twomey MA, Herrala A, Voikar V, Vihko P. Prostatic acid phosphatase is an ectonucleotidase and suppresses pain by generating adenosine. Neuron. 2008;60:111–122. doi: 10.1016/j.neuron.2008.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sowa NA, Vadakkan KI, Zylka MJ. Recombinant mouse PAP has pH-dependent ectonucleotidase activity and acts through A1-adenosine receptors to mediate antinociception. PLoS One. 2009;4:e4248. doi: 10.1371/journal.pone.0004248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zylka MJ. Pain-relieving prospects for adenosine receptors and ectonucleotidases. Trends Mol Med. 2011;17:188–196. doi: 10.1016/j.molmed.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitic N, Valizadeh M, Leung EW, de JJ, Hamilton S, Hume DA, Cassady AI, Schenk G. Human tartrate-resistant acid phosphatase becomes an effective ATPase upon proteolytic activation. Arch Biochem Biophys. 2005;439:154–164. doi: 10.1016/j.abb.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 23.Oddie GW, Schenk G, Angel NZ, Walsh N, Guddat LW, Jersey J, Cassady AI, Hamilton SE, Hume DA. Structure, function, and regulation of tartrate-resistant acid phosphatase. Bone. 2000;27:575–584. doi: 10.1016/S8756-3282(00)00368-9. [DOI] [PubMed] [Google Scholar]
- 24.Kaunitz JD, Yamaguchi DT. TNAP, TrAP, ecto-purinergic signaling, and bone remodeling. J Cell Biochem. 2008;105:655–662. doi: 10.1002/jcb.21885. [DOI] [PubMed] [Google Scholar]
- 25.Smith TM, Kirley TL. The calcium activated nucleotidases: a diverse family of soluble and membrane associated nucleotide hydrolyzing enzymes. Purinergic Signal. 2006;2:327–333. doi: 10.1007/s11302-005-5300-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sandona D, Gastaldello S, Martinello T, Betto R. Characterization of the ATP-hydrolysing activity of alpha-sarcoglycan. Biochem J. 2004;381:105–112. doi: 10.1042/BJ20031644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dzhandzhugazyan K, Bock E. Demonstration of (Ca2+-Mg2+)-ATPase activity of the neural cell adhesion molecule. FEBS Lett. 1993;336:279–283. doi: 10.1016/0014-5793(93)80820-K. [DOI] [PubMed] [Google Scholar]
- 28.Champagne E, Martinez LO, Collet X, Barbaras R. Ecto-F1F0 ATP synthase/F1 ATPase: metabolic and immunological functions. Curr Opin Lipidol. 2006;17:279–284. doi: 10.1097/01.mol.0000226120.27931.76. [DOI] [PubMed] [Google Scholar]
- 29.Fu Y, Hou YJ, Fu CL, Gu MX, Li CH, Kong W, Wang XA, Shyy JYJ, Zhu Y. A novel mechanism of γ/δ T-lymphocyte and endothelial activation by shear stress the role of ecto-ATP synthase β chain. Circ Res. 2011;108:410–417. doi: 10.1161/CIRCRESAHA.110.230151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yegutkin GG. Nucleotide- and nucleoside-converting ectoenzymes: important modulators of purinergic signalling cascade. BBA Mol Cell Res. 2008;1783:673–694. doi: 10.1016/j.bbamcr.2008.01.024. [DOI] [PubMed] [Google Scholar]
- 31.Deaglio S, Malavasi F. The CD38/CD157 mammalian gene family: an evolutionary paradigm for other leucocyte surface enzymes. Purinergic Signal. 2006;2:431–441. doi: 10.1007/s11302-006-9002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kukulski F, Levesque SA, Sévigny J. Impact of ectoenzymes on P2 and P1 receptor signaling. Adv Pharmacol. 2011;61:263–299. doi: 10.1016/B978-0-12-385526-8.00009-6. [DOI] [PubMed] [Google Scholar]
- 33.Yegutkin GG, Henttinen T, Jalkanen S. Extracellular ATP formation on vascular endothelial cells is mediated by ecto-nucleotide kinase activities via phosphotransfer reactions. FASEB J. 2001;15:251–260. doi: 10.1096/fj.00-0268com. [DOI] [PubMed] [Google Scholar]
- 34.Yegutkin GG, Mikhailov A, Samburski SS, Jalkanen S. The detection of micromolar pericellular ATP pool on lymphocyte surface by using lymphoid ecto-adenylate kinase as intrinsic ATP sensor. Mol Biol Cell. 2006;17:3378–3385. doi: 10.1091/mbc.E05-10-0993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zimmermann H. Ecto-nucleotidases. In: Abbracchio MP, Williams M, editors. Handbook of experimental pharmacology. Purinergic and pyrimidergic signalling. Heidelberg: Springer; 2001. pp. 209–250. [Google Scholar]
- 36.Robson SC, Sévigny J, Zimmermann H. The E-NTPDase family of ectonucleotidases: structure function relationships and pathophysiological significance. Purinergic Signal. 2006;2:409–430. doi: 10.1007/s11302-006-9003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knowles AF. The GDA1_CD39 superfamily: NTPDases with diverse functions. Purinergic Signal. 2011;7:21–45. doi: 10.1007/s11302-010-9214-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zimmermann H, Beaudoin AR, Bollen M, Goding JW, Guidotti G, Kirley TL, Robson SC, Sano K. Proposed nomenclature for two novel nucleotide hydrolyzing enzyme families expressed on the cell surface. In: Lemmens R, vanDuffel L, editors. Ecto-ATPases and related ectonucleotidases. Maastricht: Shaker; 2000. pp. 1–8. [Google Scholar]
- 39.Zimmermann H. Ectonucleotidases: some recent developments and a note on nomenclature. Drug Dev Res. 2001;52:44–56. doi: 10.1002/ddr.1097. [DOI] [Google Scholar]
- 40.Lavoie ÉG, Gulbransen BD, Martín-Satué M, Aliagas E, Sharkey KA, Sévigny J. Ectonucleotidases in the digestive system: focus on NTPDase3 localization. Amer J Physiol Gastrointest L. 2011;300:G608–G620. doi: 10.1152/ajpgi.00207.2010. [DOI] [PubMed] [Google Scholar]
- 41.Enjyoji K, Sévigny J, Lin Y, Frenette P, Christie PD, Schulte am Esch J, Imai M, Edelberger JM, Rayburn H, Lech M, Beeler DM, Csizmadia E, Wagner DD, Robson SC, Rosenberg RD. Targeted disruption of cd39/ATP diphosphohydrolase results in disordered hemostasis and thromboregulation. Nature Med. 1999;5:1010–1017. doi: 10.1038/12447. [DOI] [PubMed] [Google Scholar]
- 42.Pinsky DJ, Broekman MJ, Peschon JJ, Stocking KL, Fujita T, Ramasamy R, Connolly ES, Huang J, Kiss S, Zhang Y, Choudhri TF, McTaggart RA, Liao H, Drospoulos JHF, Price VL, Marcus AJ, Maliszewski CR. Elucidation of the thromboregulatory role of CD39/ectoapyrase in the ischemic brain. J Clin Invest. 2002;109:1031–1040. doi: 10.1172/JCI10649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mizumoto N, Kumamoto T, Robson SC, Sévigny J, Matsue H, Enjyoji K, Takashima A. CD39 is the dominant Langerhans cell associated ecto-NTPDase: modulatory roles in inflammation and immune responsiveness. Nature Med. 2002;8:358–365. doi: 10.1038/nm0402-358. [DOI] [PubMed] [Google Scholar]
- 44.Dwyer KM, Deaglio S, Gao W, Friedman D, Strom TB, Robson SC. CD39 and control of cellular immune responses. Purinergic Signal. 2007;3:171–180. doi: 10.1007/s11302-006-9050-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deaglio S, Robson SC. Ectonucleotidases as regulators of purinergic signaling in thrombosis, inflammation, and immunity. Adv Pharmacol. 2011;61:301–332. doi: 10.1016/B978-0-12-385526-8.00010-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sévigny J, Sundberg C, Braun N, Guckelberger O, Csizmadia E, Qawi I, Imai M, Zimmermann H, Robson SC. Differential catalytic properties and vascular topography of murine nucleoside triphosphate diphosphohydrolase 1 (NTPDase1) and NTPDase2 have implications for thromboregulation. Blood. 2002;99:2801–2809. doi: 10.1182/blood.V99.8.2801. [DOI] [PubMed] [Google Scholar]
- 47.Robson SC, Wu Y, Sun XF, Knosalla C, Dwyer K, Enjyoji K. Ectonucleotidases of CD39 family modulate vascular inflammation and thrombosis in transplantation. Semin Thromb Hemostasis. 2005;31:217–233. doi: 10.1055/s-2005-869527. [DOI] [PubMed] [Google Scholar]
- 48.Marcus AJ, Broekman MJ, Drosopoulos JHF, Olson KE, Islam N, Pinsky DJ, Levi R. Role of CD39 (NTPDase-1) in thromboregulation, cerebroprotection, and cardioprotection. Semin Thromb Hemostasis. 2005;31:234–246. doi: 10.1055/s-2005-869528. [DOI] [PubMed] [Google Scholar]
- 49.Kauffenstein G, Fürstenau CR, D'Orleans-Juste P, Sévigny J. The ecto-nucleotidase NTPDase1 differentially regulates P2Y1 and P2Y2 receptor-dependent vasorelaxation. Brit J Pharmacol. 2010;159:576–585. doi: 10.1111/j.1476-5381.2009.00566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kauffenstein G, Drouin A, Thorin-Trescases N, Bachelard H, Robaye B, D'Orleans-Juste P, Marceau F, Thorin E, Sévigny J. NTPDase1 (CD39) controls nucleotide-dependent vasoconstriction in mouse. Cardiovasc Res. 2010;85:204–213. doi: 10.1093/cvr/cvp265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bartel DL, Sullivan SL, Lavoie ÉG, Sévigny J, Finger TE. Nucleoside triphosphate diphosphohydrolase-2 is the ecto-ATPase of type I cells in taste buds. J Comp Neurol. 2006;497:1–12. doi: 10.1002/cne.20954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Braun N, Sévigny J, Mishra S, Robson SC, Barth SW, Gerstberger R, Hammer K, Zimmermann H. Expression of the ecto-ATPase NTPDase2 in the germinal zones of the developing and adult rat brain. Eur J Neurosci. 2003;17:1355–1364. doi: 10.1046/j.1460-9568.2003.02567.x. [DOI] [PubMed] [Google Scholar]
- 53.Braun N, Sévigny J, Robson SC, Hammer K, Hanani M, Zimmermann H. Association of the ecto-ATPase NTPDase2 with glial cells of the peripheral nervous system. Glia. 2004;45:124–132. doi: 10.1002/glia.10309. [DOI] [PubMed] [Google Scholar]
- 54.Shukla V, Zimmermann H, Wang L, Kettenmann H, Raab S, Hammer K, Sévigny J, Robson SC, Braun N. Functional expression of the ecto-ATPase NTPDase2 and of nucleotide receptors by neuronal progenitor cells in the adult murine hippocampus. J Neurosci Res. 2005;80:600–610. doi: 10.1002/jnr.20508. [DOI] [PubMed] [Google Scholar]
- 55.Mishra SK, Braun N, Shukla V, Füllgrabe M, Schomerus C, Korf H-W, Gachet C, Ikehara Y, Sévigny J, Robson SC, Zimmermann H. Extracellular nucleotide signaling in adult neural stem cells: synergism with growth factor-mediated cellular proliferation. Development. 2006;133:675–684. doi: 10.1242/dev.02233. [DOI] [PubMed] [Google Scholar]
- 56.Vongtau HO, Lavoie ÉG, Sévigny J, Molliver DC. Distribution of ecto-nucleotidases in mouse sensory circuits suggests roles for nucleoside triphosphate diphosphohydrolyse-3 in nociception and mechanoreception. Neuroscience. 2011;193:387–398. doi: 10.1016/j.neuroscience.2011.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zimmermann H. Purinergic signaling in neural development. Semin Cell Dev Biol. 2011;22:194–204. doi: 10.1016/j.semcdb.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 58.Massé K, Bhamra S, Eason R, Dale N, Jones EA. Purine-mediated signalling triggers eye development. Nature. 2007;449:1058–1062. doi: 10.1038/nature06189. [DOI] [PubMed] [Google Scholar]
- 59.Belcher SM, Zsarnovzky A, Crawford PA, Hemani H, Spurling L, Kirley TL. Immunolocalization of ecto-nucleoside triphosphate diphosphohydrolase 3 in rat brain: implications for modulation of multiple homeostatic systems including feeding and sleep wake behaviors. Neuroscience. 2006;137:1331–1346. doi: 10.1016/j.neuroscience.2005.08.086. [DOI] [PubMed] [Google Scholar]
- 60.Lavoie ÉG, Fausther M, Kauffenstein G, Kukulski F, Kunzli BM, Friess H, Sévigny J. Identification of the ectonucleotidases expressed in mouse, rat, and human Langerhans islets: potential role of NTPDase3 in insulin secretion. Amer J Physiol Endocrinol Met. 2010;299:F647–F656. doi: 10.1152/ajpendo.00126.2010. [DOI] [PubMed] [Google Scholar]
- 61.Burch LH, Picher M. E-NTPDases in human airways: regulation and relevance for chronic lung diseases. Purinergic Signalling. 2006;2:399–408. doi: 10.1007/s11302-006-9001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fausther M, Pelletier J, Ribeiro CM, Sévigny J, Picher M. Cystic fibrosis remodels the regulation of purinergic signaling by NTPDase1 (CD39) and NTPDase3. Amer J Physiol Lung Cell M Ph. 2010;298:L804–L818. doi: 10.1152/ajplung.00019.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Biederbick A, Kosan C, Kunz J, Elsässer HP. First apyrase splice variants have different enzymatic properties. J Biol Chem. 2000;275:19018–19024. doi: 10.1074/jbc.M001245200. [DOI] [PubMed] [Google Scholar]
- 64.Shi JD, Kukar T, Wang CY, Li QZ, Cruz PE, Davoodi-Semiromi A, Yang P, Gu YR, Lian W, Wu DH, She JX. Molecular cloning and characterization of a novel mammalian endo-apyrase (LALP1) J Biol Chem. 2001;276:17474–17478. doi: 10.1074/jbc.M011569200. [DOI] [PubMed] [Google Scholar]
- 65.Yeung G, Mulero JJ, McGowan DW, Bajwa SS, Ford JE. CD39L2, a gene encoding a human nucleoside diphosphatase, predominantly expressed in the heart. Biochemistry USA. 2000;39:12916–12923. doi: 10.1021/bi000959z. [DOI] [PubMed] [Google Scholar]
- 66.O'Keeffe MG, Thorne PR, Housley GD, Robson SC, Vlajkovic SM. Developmentally regulated expression of ectonucleotidases NTPDase5 and NTPDase6 and UDP-responsive P2Y receptors in the rat cochlea. Histochemistry Cell Biol. 2010;133:425–436. doi: 10.1007/s00418-010-0682-1. [DOI] [PubMed] [Google Scholar]
- 67.Paéz JG, Recio JA, Rouzaut A, Notario V. Identity between the PCPH proto-oncogene and the CD39L4 (ENTPD5) ectonucleoside triphosphate diphosphohydrolase gene. Int J Oncol. 2001;19:1249–1254. doi: 10.3892/ijo.19.6.1249. [DOI] [PubMed] [Google Scholar]
- 68.Fang M, Shen Z, Huang S, Zhao L, Chen S, Mak TW, Wang X. The ER UDPase ENTPD5 promotes protein N-glycosylation, the Warburg effect, and proliferation in the PTEN pathway. Cell. 2010;143:711–724. doi: 10.1016/j.cell.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 69.Kukulski F, Lévesque SA, Lavoie ÉG, Lecka J, Bigonnesse F, Knowles AF, Robson SC, Kirley TL, Sévigny J. Comparative hydrolysis of P2 receptor agonists by NTPDase 1, 2, 3 and 8. Purinergic Signal. 2005;1:193–204. doi: 10.1007/s11302-005-6217-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Heine P, Braun N, Zimmermann H. Functional characterization of rat ecto-ATPase and ecto-ATP diphosphohydrolase after heterologous expression in CHO cells. Eur J Biochem. 1999;262:102–107. doi: 10.1046/j.1432-1327.1999.00347.x. [DOI] [PubMed] [Google Scholar]
- 71.Knowles AF, Li C. Molecular cloning and characterization of expressed human ecto-nucleoside triphosphate diphosphohydrolase 8 (E-NTPDase 8) and its soluble extracellular domain. Biochemistry USA. 2006;45:7323–7333. doi: 10.1021/bi052268e. [DOI] [PubMed] [Google Scholar]
- 72.Fausther M, Lecka J, Kukulski F, Levesque SA, Pelletier J, Zimmermann H, Dranoff JA, Sévigny J. Cloning, purification, and identification of the liver canalicular ecto-ATPase as NTPDase8. Amer J Physiol Gastrointest L. 2007;292:G785–G795. doi: 10.1152/ajpgi.00293.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Musi E, Islam N, Drosopoulos JHF. Constraints imposed by transmembrane domains affect enzymatic activity of membrane-associated human CD39/NTPDase1 mutants. Arch Biochem Biophys. 2007;461:30–39. doi: 10.1016/j.abb.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 74.Chen W, Guidotti G. Soluble apyrases release ADP during ATP hydrolysis. Biochem Biophys Res Commun. 2001;282:90–95. doi: 10.1006/bbrc.2001.4555. [DOI] [PubMed] [Google Scholar]
- 75.Chiang WC, Knowles AF. Transmembrane domain interactions affect the stability of the extracellular domain of the human NTPDase 2. Arch Biochem Biophys. 2008;472:89–99. doi: 10.1016/j.abb.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 76.Wang TF, Guidotti G. Golgi localization and functional expression of human uridine diphosphatase. J Biol Chem. 1998;273:11392–11399. doi: 10.1074/jbc.273.18.11392. [DOI] [PubMed] [Google Scholar]
- 77.Trombetta ES, Helenius A. Glycoprotein reglycosylation and nucleotide sugar utilization in the secretory pathway: identification of a nucleoside diphosphatase in the endoplasmic reticulum. EMBO J. 1999;18:3282–3292. doi: 10.1093/emboj/18.12.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Braun N, Fengler S, Ebeling C, Servos J, Zimmermann H. Sequencing, functional expression and characterization of NTPDase6, a nucleoside diphosphatase and novel member of the ecto-nucleoside triphosphate diphosphohydrolase family. Biochem J. 2000;351:639–647. doi: 10.1042/0264-6021:3510639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ivanenkov VV, Murphy-Piedmonte DM, Kirley TL. Bacterial expression, characterization, and disulfide bond determination of soluble human NTPDase6 (CD39L2) nucleotidase: implications for structure and function. Biochemistry USA. 2003;42:11726–11735. doi: 10.1021/bi035137r. [DOI] [PubMed] [Google Scholar]
- 80.Zhong XT, Malhotra R, Woodruff R, Guidotti G. Mammalian plasma membrane ecto-nucleoside triphosphate diphosphohydrolase 1, CD39, is not active intracellularly—the N-glycosylation state of CD39 correlates with surface activity and localization. J Biol Chem. 2001;276:41518–41525. doi: 10.1074/jbc.M104415200. [DOI] [PubMed] [Google Scholar]
- 81.Ivanenkov VV, Sévigny J, Kirley TL. Trafficking and intracellular ATPase activity of human ecto-nucleotidase NTPDase3 and the effect of ER-targeted NTPDase3 on protein folding. Biochemistry USA. 2008;47:9184–9197. doi: 10.1021/bi800402q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Handa M, Guidotti G. Purification and cloning of a soluble ATP-diphosphohydrolase (apyrase) from potato tubers (Solanum tuberosum) Biochem Biophys Res Commun. 1996;218:916–923. doi: 10.1006/bbrc.1996.0162. [DOI] [PubMed] [Google Scholar]
- 83.Schulte am Esch J, Sévigny J, Kaczmarek E, Siegel JB, Imai M, Koziak K, Beaudoin AR, Robson SC. Structural elements and limited proteolysis of CD39 influence ATP diphosphohydrolase activity. Biochemistry USA. 1999;38:2248–2258. doi: 10.1021/bi982426k. [DOI] [PubMed] [Google Scholar]
- 84.Smith TM, Kirley TL. Site-directed mutagenesis of a human brain ecto-apyrase: evidence that the E-type ATPases are related to the actin/heat shock 70/sugar kinase superfamily. Biochemistry USA. 1999;38:321–328. doi: 10.1021/bi9820457. [DOI] [PubMed] [Google Scholar]
- 85.Maliszewski CR, DeLepesse GJT, Schoenborn MA, Armitage RJ, Fanslow WC, Nakajima T, Baker E, Sutherland GR, Poindexter K, Birks C, Alpert A, Friend D, Gimpel SD, Gayle RB. The CD39 lymphoid cell activation antigen: molecular cloning and structural characterization. J Immunol. 1994;153:3574–3583. [PubMed] [Google Scholar]
- 86.Kansas GS, Wood GS, Tedder TF. Expression, distribution, and biochemistry of human CD39: role in activation-associated homotypic adhesion of lymphocytes. J Immunol. 1991;146:2235–2244. [PubMed] [Google Scholar]
- 87.Christoforidis S, Papamarcaki T, Galaris D, Kellner R, Tsolas O. Purification and properties of human placental ATP diphosphohydrolase. Eur J Biochem. 1995;234:66–74. doi: 10.1111/j.1432-1033.1995.066_c.x. [DOI] [PubMed] [Google Scholar]
- 88.Wang TF, Guidotti G. CD39 is an ecto-(Ca2+, Mg2+)-apyrase. J Biol Chem. 1996;271:9898–9901. doi: 10.1074/jbc.271.17.9898. [DOI] [PubMed] [Google Scholar]
- 89.Kaczmarek E, Siegel JB, Sévigny J, Koziak K, Hancock WW, Beaudoin A, Bach FH, Robson SC. Vascular ATP diphosphohydrolase (CD39/ATPDase) In: Plesner L, Kirley TL, Knowles AF, editors. Ecto-ATPases. Recent progress on structure and function. New York: Plenum; 1997. pp. 171–185. [Google Scholar]
- 90.Makita K, Shimoyama T, Sakurai Y, Yagi H, Matsumoto M, Narita N, Sakamoto Y, Saito S, Ikeda Y, Suzuki M, Titani K, Fujimura Y. Placental ecto-ATP diphosphohydrolase: its structural feature distinct from CD39, localization and inhibition on shear-induced platelet aggregation. Int J Hematol. 1998;68:297–310. doi: 10.1016/S0925-5710(98)00080-2. [DOI] [PubMed] [Google Scholar]
- 91.Matsumoto M, Sakurai Y, Kokubo T, Yagi H, Makita K, Matsui T, Titani K, Fujimura Y, Narita N. The cDNA cloning of human placental ecto-ATP diphosphohydrolases I and II. FEBS Lett. 1999;453:335–340. doi: 10.1016/S0014-5793(99)00751-6. [DOI] [PubMed] [Google Scholar]
- 92.Kittel A, Kaczmarek E, Sévigny J, Lengyel K, Csizmadia E, Robson SC. CD39 as a caveolar-associated ectonucleotidase. Biochem Biophys Res Commun. 1999;262:596–599. doi: 10.1006/bbrc.1999.1254. [DOI] [PubMed] [Google Scholar]
- 93.Koziak K, Kaczmarek E, Kittel A, Sévigny J, Blusztajn JK, Schulte am Esch J, Imai M, Guckelberger O, Goepfert C, Qawi I, Robson SC. Palmitoylation targets CD39/endothelial ATP diphosphohydrolase to caveolae. J Biol Chem. 2000;275:2057–2062. doi: 10.1074/jbc.275.3.2057. [DOI] [PubMed] [Google Scholar]
- 94.Kittel A, Csapo Z, Csizmadia E, Jackson SW, Robson SC. Co-localization of P2Y1 receptor and NTPDase1/CD39 within caveolae in human placenta. Eur J Histochem. 2004;48:253–259. [PubMed] [Google Scholar]
- 95.Papanikolaou A, Papafotika A, Murphy C, Papamarcaki T, Tsolas O, Drab M, Kurzchalia TV, Kasper M, Christoforidis S. Cholesterol-dependent lipid assemblies regulate the activity of the ecto-nucleotidase CD39. J Biol Chem. 2005;280:26406–26414. doi: 10.1074/jbc.M413927200. [DOI] [PubMed] [Google Scholar]
- 96.Knowles AF, Chiang WC. Enzymatic and transcriptional regulation of human ecto-ATPase/E-NTPDase2. Arch Biochem Biophys. 2003;418:217–227. doi: 10.1016/j.abb.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 97.Kegel B, Braun N, Heine P, Maliszewski CR, Zimmermann H. An ecto-ATPase and an ecto-ATP diphosphohydrolase are expressed in rat brain. Neuropharmacology. 1997;36:1189–1200. doi: 10.1016/S0028-3908(97)00115-9. [DOI] [PubMed] [Google Scholar]
- 98.Mateo J, Harden TK, Boyer JL. Functional expression of a cDNA encoding a human ecto-ATPase. Brit J Pharmacol. 1999;128:396–402. doi: 10.1038/sj.bjp.0702805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mateo J, Kreda S, Henry CE, Harden TK, Boyer JL. Requirement of Cys(399) for processing of the human ecto-ATPase (NTPDase2) and its implications for determination of the activities of splice variants of the enzyme. J Biol Chem. 2003;278:39960–39968. doi: 10.1074/jbc.M307854200. [DOI] [PubMed] [Google Scholar]
- 100.Vlajkovic SM, Housley GD, Greenwood D, Thorne PR. Evidence for alternative splicing of ecto-ATPase associated with termination of purinergic transmission. Mol Brain Res. 1999;73:85–92. doi: 10.1016/S0169-328X(99)00244-2. [DOI] [PubMed] [Google Scholar]
- 101.Wang CJF, Vlajkovic SM, Housley GD, Braun N, Zimmermann H, Robson SC, Sévigny J, Soeller C, Thorne PR. C-terminal splicing of NTPDase2 provides distinctive catalytic properties, cellular distribution and enzyme regulation. Biochem J. 2005;385:729–736. doi: 10.1042/BJ20040852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vlajkovic SM, Wang CJH, Soeller C, Zimmermann H, Thorne PR, Housley GD. Activation-dependent trafficking of NTPDase2 in Chinese hamster ovary cells. Int J Biochem Cell Biol. 2007;39:810–817. doi: 10.1016/j.biocel.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 103.Chadwick BP, Frischauf AM. The CD39-like gene family: identification of three new human members (CD39L2, CD39L3, and CD39L4), their murine homologues, and a member of the gene family from Drosophila melanogaster. Genomics. 1998;50:357–367. doi: 10.1006/geno.1998.5317. [DOI] [PubMed] [Google Scholar]
- 104.Smith TM, Kirley TL. Cloning, sequencing, and expression of a human brain ecto-apyrase related to both the ecto-ATPases and CD39 ecto-apyrases. Biochim Biophys Acta. 1998;1386:65–78. doi: 10.1016/S0167-4838(98)00063-6. [DOI] [PubMed] [Google Scholar]
- 105.Crawford PA, Gaddie KJ, Smith TM, Kirley TL. Characterization of an alternative splice variant of human nucleoside triphosphate diphosphohydrolase 3 (NTPDase3): a possible modulator of nucleotidase activity and purinergic signaling. Arch Biochem Biophys. 2007;457:7–15. doi: 10.1016/j.abb.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Biederbick A, Rose S, Elsässer HP. A human intracellular apyrase-like protein, LALP70, localizes to lysosomal/autophagic vacuoles. J Cell Sci. 1999;112:2473–2484. doi: 10.1242/jcs.112.15.2473. [DOI] [PubMed] [Google Scholar]
- 107.Mulero JJ, Yeung G, Nelken ST, Ford JE. CD39-L4 is a secreted human apyrase, specific for the hydrolysis of nucleoside diphosphates. J Biol Chem. 1999;29:20064–20067. doi: 10.1074/jbc.274.29.20064. [DOI] [PubMed] [Google Scholar]
- 108.Hicks-Berger CA, Chadwick BP, Frischauf AM, Kirley TL. Expression and characterization of soluble and membrane-bound human nucleoside triphosphate diphosphohydrolase 6 (CD39L2) J Biol Chem. 2000;275:34041–34045. doi: 10.1074/jbc.M004723200. [DOI] [PubMed] [Google Scholar]
- 109.Gao XD, Kaigorodov V, Jigami Y. YND1, a homologue of GDA1, encodes membrane-bound apyrase required for Golgi N- and O-glycosylation in Saccharomyces cerevisiae. J Biol Chem. 1999;274:21450–21456. doi: 10.1074/jbc.274.30.21450. [DOI] [PubMed] [Google Scholar]
- 110.Knowles AF. The single NTPase gene of Drosophila melanogaster encodes an intracellular nucleoside triphosphate diphosphohydrolase 6 (NTPDase6) Arch Biochem Biophys. 2009;484:70–79. doi: 10.1016/j.abb.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 111.Read R, Hansen G, Kramer J, Finch R, Li L, Vogel P. Ectonucleoside triphosphate diphosphohydrolase type 5 (Entpd5)-deficient mice develop progressive hepatopathy, hepatocellular tumors, and spermatogenic arrest. Vet Pathol. 2009;46:491–504. doi: 10.1354/vp.08-VP-0201-R-AM. [DOI] [PubMed] [Google Scholar]
- 112.Velasco JA, Avila MA, Notario V. The product of the cph oncogene is a truncated, nucleotide-binding protein that enhances cellular survival to stress. Oncogene. 1999;18:689–701. doi: 10.1038/sj.onc.1202324. [DOI] [PubMed] [Google Scholar]
- 113.Smith TM, Carl SAL, Kirley TL. Mutagenesis of two conserved tryptophan residues of the E-type ATPases: inactivation and conversion of an ecto-apyrase to an ecto-NTPase. Biochemistry USA. 1999;38:5849–5857. doi: 10.1021/bi990171k. [DOI] [PubMed] [Google Scholar]
- 114.Grinthal A, Guidotti G. Substitution of His59 converts CD39 apyrase into an ADPase in a quaternary structure dependent manner. Biochemistry USA. 2000;39:9–16. doi: 10.1021/bi991751k. [DOI] [PubMed] [Google Scholar]
- 115.Drosopoulos JHF, Broekman MJ, Islam N, Maliszewski CR, Gayle RB, Marcus AJ. Site-directed mutagenesis of human endothelial cell ecto-ADPase/soluble CD39: requirement of glutamate 174 and serine 218 for enzyme activity and inhibition of platelet recruitment. Biochemistry USA. 2000;39:6936–6943. doi: 10.1021/bi992581e. [DOI] [PubMed] [Google Scholar]
- 116.Yang F, Hicks-Berger CA, Smith TM, Kirley TL. Site-directed mutagenesis of human nucleoside triphosphate diphosphohydrolase 3: the importance of residues in the apyrase conserved regions. Biochemistry USA. 2001;40:3943–3950. doi: 10.1021/bi002711f. [DOI] [PubMed] [Google Scholar]
- 117.Javed R, Yarimizu K, Pelletier N, Li C, Knowles AF. Mutagenesis of lysine 62, asparagine 64, and conserved region 1 reduces the activity of human ecto-ATPase (NTPDase 2) Biochemistry USA. 2007;46:6617–6627. doi: 10.1021/bi700036e. [DOI] [PubMed] [Google Scholar]
- 118.Heine P, Braun N, Sévigny J, Robson SC, Servos J, Zimmermann H. The C-terminal cysteine-rich region dictates specific catalytic properties in chimeras of the ecto-nucleotidases NTPDase1 and NTPDase2. Eur J Biochem. 2001;262:102–107. doi: 10.1046/j.1432-1327.1999.00347.x. [DOI] [PubMed] [Google Scholar]
- 119.Hicks-Berger CA, Kirley TL. Expression and characterization of human ecto-ATPase and chimeras with CD39 ecto-apyrase. IUBMB Life. 2000;50:43–50. doi: 10.1080/15216540050176584. [DOI] [PubMed] [Google Scholar]
- 120.Grinthal A, Guidotti G. Transmembrane domains confer different substrate specificities and adenosine diphosphate hydrolysis mechanisms on CD39, CD39L1, and chimeras. Biochemistry USA. 2002;41:1947–1956. doi: 10.1021/bi015563h. [DOI] [PubMed] [Google Scholar]
- 121.Gonzales EB, Kawate T, Gouaux E. Pore architecture and ion sites in acid-sensing ion channels and P2X receptors. Nature. 2009;460:599–U62. doi: 10.1038/nature08218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Browne LE, Jiang LH, North RA. New structure enlivens interest in P2X receptors. Trends Pharmacol Sci. 2010;31:229–237. doi: 10.1016/j.tips.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang TF, Handa M, Guidotti G. Structure and function of ectoapyrase (CD39) Drug Develop Res. 1998;45:245–252. doi: 10.1002/(SICI)1098-2299(199811/12)45:3/4<245::AID-DDR22>3.0.CO;2-U. [DOI] [Google Scholar]
- 124.Mukasa T, Lee Y, Knowles AF. Either the carboxyl- or the amino-terminal region of the human ecto-ATPase (E-NTPDase 2) confers detergent and temperature sensitivity to the chicken ecto-ATP-diphosphohydrolase (E-NTPDase 8) Biochemistry USA. 2005;44:11160–11170. doi: 10.1021/bi050019k. [DOI] [PubMed] [Google Scholar]
- 125.Grinthal A, Guidotti G. CD39, NTPDase1, is attached to the plasma membrane by two transmembrane domains. Why? Purinergic Signal. 2006;2:391–398. doi: 10.1007/s11302-005-5907-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Li CS, Lee Y, Knowles AF. The stability of chicken nucleoside triphosphate diphosphohydrolase 8 requires both of its transmembrane domains. Biochemistry USA. 2010;49:134–146. doi: 10.1021/bi901820c. [DOI] [PubMed] [Google Scholar]
- 127.Papanikolaou A, Papafotika A, Christoforidis S. CD39 reveals novel insights into the role of transmembrane domains in protein processing, apical targeting and activity. Traffic. 2011;12:1148–1165. doi: 10.1111/j.1600-0854.2011.01224.x. [DOI] [PubMed] [Google Scholar]
- 128.Lemmens R, Kupers L, Sévigny J, Beaudoin AR, Grondin G, Kittel A, Waelkens E, VanDuffel L. Purification, characterization, and localization of an ATP diphosphohydrolase in porcine kidney. Amer J Physiol Renal Physiol. 2000;278:F978–F988. doi: 10.1152/ajprenal.2000.278.6.F978. [DOI] [PubMed] [Google Scholar]
- 129.Sørensen CE, Amstrup J, Rasmussen HN, Ankorina-Stark I, Novak I. Rat pancreas secretes particulate ecto-nucleotidase CD39. J Physiol London. 2003;551:881–892. doi: 10.1113/jphysiol.2003.049411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kittel A, Pelletier J, Bigonnesse F, Guckelberger O, Kordas K, Braun N, Robson SC, Sévigny J. Localization of nucleoside triphosphate diphosphohydrolase-1 (NTPDase1) and NTPDase2 in pancreas and salivary gland. J Histochem Cytochem. 2004;52:861–871. doi: 10.1369/jhc.3A6167.2004. [DOI] [PubMed] [Google Scholar]
- 131.Zebisch M, Sträter N. Characterization of rat NTPDase1, -2, and -3 ectodomains refolded from bacterial inclusion bodies. Biochemistry USA. 2007;46:11945–11956. doi: 10.1021/bi701103y. [DOI] [PubMed] [Google Scholar]
- 132.Grinthal A, Guidotti G. Dynamic motions of CD39 transmembrane domains regulate and are regulated by the enzymatic active site. Biochemistry USA. 2004;43:13849–13858. doi: 10.1021/bi048644x. [DOI] [PubMed] [Google Scholar]
- 133.Grinthal A, Guidotti G. Bilayer mechanical properties regulate the transmembrane helix mobility and enzymatic state of CD39. Biochemistry USA. 2007;46:279–290. doi: 10.1021/bi061052p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Stout JG, Kirley TL. Control of cell membrane ecto-ATPase by oligomerization state: intermolecular cross-linking modulates ATPase activity. Biochemistry USA. 1996;35:8289–8298. doi: 10.1021/bi960563g. [DOI] [PubMed] [Google Scholar]
- 135.Wang TF, Ou Y, Guidotti G. The transmembrane domains of ectoapyrase (CD39) affect its enzymatic activity and quaternary structure. J Biol Chem. 1998;273:24814–24821. doi: 10.1074/jbc.273.38.24814. [DOI] [PubMed] [Google Scholar]
- 136.Failer BU, Aschrafi A, Schmalzing G, Zimmermann H. Determination of native oligomeric state and substrate specificity of rat NTPDase1 and NTPDase2 after heterologous expression in Xenopus oocytes. Eur J Biochem. 2003;270:1802–1809. doi: 10.1046/j.1432-1033.2003.03542.x. [DOI] [PubMed] [Google Scholar]
- 137.Carl SAL, Smith TM, Kirley TL. Cross-linking induces homodimer formation and inhibits enzymatic activity of chicken stomach ecto-apyrase. Biochem Mol Biol Int. 1998;44:463–470. doi: 10.1080/15216549800201482. [DOI] [PubMed] [Google Scholar]
- 138.Smith TM, Kirley TL. Glycosylation is essential for functional expression of a human brain ecto-apyrase. Biochemistry USA. 1999;38:1509–1516. doi: 10.1021/bi9821768. [DOI] [PubMed] [Google Scholar]
- 139.Murphy DM, Ivanenkov VV, Kirley TL. Identification of cysteine residues responsible for oxidative cross-linking and chemical inhibition of human nucleoside-triphosphate diphosphohydrolase 3. J Biol Chem. 2002;277:6162–6169. doi: 10.1074/jbc.M110105200. [DOI] [PubMed] [Google Scholar]
- 140.Chiang WC, Knowles AF. Inhibition of human NTPDase 2 by modification of an intramembrane cysteine by p-chloromercuriphenylsulfonate and oxidative cross-linking of the transmembrane domains. Biochemistry USA. 2008;47:8775–8785. doi: 10.1021/bi800633d. [DOI] [PubMed] [Google Scholar]
- 141.Gaddie KJ, Kirley TL. Conserved polar residues stabilize transmembrane domains and promote oligomerization in human nucleoside triphosphate diphosphohydrolase 3. Biochemistry USA. 2009;48:9437–9447. doi: 10.1021/bi900909g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Nicke A, Baumert HG, Rettinger J, Eichele A, Lambrecht G, Mutschler E, Schmalzing G. P2X1 and P2X3 receptors form stable trimers: a novel structural motif of ligand-gated ion channels. EMBO J. 1998;17:3016–3028. doi: 10.1093/emboj/17.11.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bodas E, Aleu J, Pujol G, Martin-Satué M, Marsal J, Solsona C. ATP crossing the cell plasma membrane generates an ionic current in Xenopus oocytes. J Biol Chem. 2000;275:20268–20273. doi: 10.1074/jbc.M000894200. [DOI] [PubMed] [Google Scholar]
- 144.Rademacher TW, Parekh RB, Dwek RA. Glycobiology. Annu Rev Biochem. 1988;57:785–838. doi: 10.1146/annurev.bi.57.070188.004033. [DOI] [PubMed] [Google Scholar]
- 145.Ivanenkov VV, Meller J, Kirley TL. Characterization of disulfide bonds in human nucleoside triphosphate diphosphohydrolase 3 (NTPDase3): implications for NTPDase structural modeling. Biochemistry USA. 2005;44:8998–9012. doi: 10.1021/bi047487z. [DOI] [PubMed] [Google Scholar]
- 146.Murphy DM, Kirley TL. Asparagine 81, an invariant glycosylation site near apyrase conserved region 1, is essential for full enzymatic activity of ecto-nucleoside triphosphate diphosphohydrolase. Arch Biochem Biophys. 2003;419:251–252. doi: 10.1016/j.abb.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 147.Mulero JJ, Yeung G, Nelken ST, Bright JM, McGowan DW, Ford JE. Biochemical characterization of CD39L4. Biochemistry USA. 2000;39:12924–12928. doi: 10.1021/bi000960y. [DOI] [PubMed] [Google Scholar]
- 148.Murphy-Piedmonte DM, Crawford PA, Kirley TL. Bacterial expression, folding, purification and characterization of soluble NTPDase5 (CD39L4) ecto-nucleotidase. BBA Proteins Proteomics. 2005;1747:251–259. doi: 10.1016/j.bbapap.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 149.Christoforidis S, Papamarcaki T, Tsolas O. Human placental ATP diphosphohydrolase is a highly N-glycosylated plasma membrane enzyme. BBA Biomem. 1996;1282:257–262. doi: 10.1016/0005-2736(96)00065-X. [DOI] [PubMed] [Google Scholar]
- 150.Zhong XT, Kriz R, Kumar R, Guidotti G. Distinctive roles of endoplasmic reticulum and Golgi glycosylation in functional surface expression of mammalian E-NTPDasel, CD39. BBA Gen Subjects. 2005;1723:143–150. doi: 10.1016/j.bbagen.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 151.Wu JJ, Choi LE, Guidotti G. N-linked oligosaccharides affect the enzymatic activity of CD39: diverse interactions between seven N-linked glycosylation sites. Mol Biol Cell. 2005;16:1661–1672. doi: 10.1091/mbc.E04-10-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yegutkin GG, Samburski SS, Mortensen SP, Jalkanen S, Gonzalez-Alonso J. Intravascular ADP and soluble nucleotidases contribute to acute prothrombotic state during vigorous exercise in humans. J Physiol. 2007;579:553–564. doi: 10.1113/jphysiol.2006.119453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yegutkin G, Bodin P, Burnstock G. Effect of shear stress on the release of soluble ecto-enzymes ATPase and 5′-nucleotidase along with endogenous ATP from vascular endothelial cells. Brit J Pharmacol. 2000;129:921–926. doi: 10.1038/sj.bjp.0703136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ceruti S, Colombo L, Magni G, Vigano F, Boccazzi M, Deli MA, Sperlagh B, Abbracchio MP, Kittel A. Oxygen-glucose deprivation increases the enzymatic activity and the microvesicle-mediated release of ectonucleotidases in the cells composing the blood–brain barrier. Neurochem Int. 2011;59:259–271. doi: 10.1016/j.neuint.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 155.Beaudoin AR, Vachereau A, Grondin G, St-Jean P, Rosenberg MD, Strobel R. Microvesicular secretion, a mode of cell secretion associated with the presence of an ATP-diphosphohydrolase. FEBS Lett. 1986;203:1–2. doi: 10.1016/0014-5793(86)81423-5. [DOI] [PubMed] [Google Scholar]
- 156.Yegutkin GG, Samburski SS, Jalkanen S, Novak I. ATP-consuming and ATP-generating enzymes secreted by pancreas. J Biol Chem. 2006;281:29441–29447. doi: 10.1074/jbc.M602480200. [DOI] [PubMed] [Google Scholar]
- 157.Kordas KS, Sperlagh B, Tihanyi T, Topa L, Steward MC, Varga G, Kittel A. ATP and ATPase secretion by exocrine pancreas in rat, guinea pig, and human. Pancreas. 2004;29:53–60. doi: 10.1097/00006676-200407000-00056. [DOI] [PubMed] [Google Scholar]
- 158.Banz Y, Beldi G, Wu Y, Atkinson B, Usheva A, Robson SC. CD39 is incorporated into plasma microparticles where it maintains functional properties and impacts endothelial activation. Brit J Haematol. 2008;142:627–637. doi: 10.1111/j.1365-2141.2008.07230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Clayton A, Al-Taei S, Webber J, Mason MD, Tabi Z. Cancer exosomes express CD39 and CD73, which suppress T cells through adenosine production. J Immunol. 2011;187:676–683. doi: 10.4049/jimmunol.1003884. [DOI] [PubMed] [Google Scholar]
- 160.Todorov LD, Mihaylova-Todorova S, Westfall TD, Sneddon P, Kennedy C, Bjur RA, Westfall DP. Neuronal release of soluble nucleotidases and their role in neurotransmitter inactivation. Nature. 1997;387:76–79. doi: 10.1038/387076a0. [DOI] [PubMed] [Google Scholar]
- 161.Westfall DP, Todorov LD, Mihaylova-Todorova ST. ATP as a cotransmitter in sympathetic nerves and its inactivation by releasable enzymes. J Pharmacol Exp Ther. 2002;303:439–444. doi: 10.1124/jpet.102.035113. [DOI] [PubMed] [Google Scholar]
- 162.Sterling KM, Shah S, Kim RJ, Johnston NIF, Salikhova AY, Abraham EH. Cystic fibrosis transmembrane conductance regulator in human and mouse red blood cell membranes and its interaction with ecto-apyrase. J Cell Biochem. 2004;91:1174–1182. doi: 10.1002/jcb.20017. [DOI] [PubMed] [Google Scholar]
- 163.Abraham EH, Sterling KM, Kim RJ, Salikhova AY, Huffman HB, Crockett MA, Johnston N, Parker HW, Boyle WE, Hartov A, Demidenko E, Efird J, Kahn J, Grubman SA, Jefferson DM, Robson SC, Thakar JH, Lorico A, Rappa G, Sartorelli AC, Okunieff P. Erythrocyte membrane ATP binding cassette (ABC) proteins: MRP1 and CFTR as well as CD39 (ecto-apyrase) involved in RBC ATP transport and elevated blood plasma ATP of cystic fibrosis. Blood Cells Molecules Dis. 2001;27:165–180. doi: 10.1006/bcmd.2000.0357. [DOI] [PubMed] [Google Scholar]
- 164.Schicker K, Hussl S, Chandaka GK, Kosenburger K, Yang JW, Waldhoer M, Sitte HH, Boehm S. A membrane network of receptors and enzymes for adenine nucleotides and nucleosides. BBA Mol Cell Res. 2009;1793:325–334. doi: 10.1016/j.bbamcr.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 165.Alvarado-Castillo C, Lozano-Zarain P, Mateo J, Harden TK, Boyer JL. A fusion protein of the human P2Y1 receptor and NTPDase1 exhibits functional activities of the native receptor and ectoenzyme and reduced signaling. Mol Pharmacol. 2002;62:521–528. doi: 10.1124/mol.62.3.521. [DOI] [PubMed] [Google Scholar]
- 166.Wu Y, Sun XF, Kaczmarek E, Dwyer KM, Bianchi E, Usheva A, Robson SC. RanBPM associates with CD39 and modulates ecto-nucleotidase activity. Biochem J. 2006;396:23–30. doi: 10.1042/BJ20051568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Murrin LC, Talbot JN. RanBPM, a scaffolding protein in the immune and nervous systems. J Neuroimmune Pharmacol. 2007;2:290–295. doi: 10.1007/s11481-007-9079-x. [DOI] [PubMed] [Google Scholar]
- 168.Zimmermann H. Ecto-nucleotidases in the nervous system. Novartis Found Symp. 2006;275:113–128. doi: 10.1002/9780470032244.ch10. [DOI] [PubMed] [Google Scholar]
- 169.Alvarado-Castillo C, Harden TK, Boyer JL. Regulation of P2Y1 receptor-mediated signaling by the ectonucleoside triphosphate diphosphohydrolase isozymes NTPDase1 and NTPDase2. Mol Pharmacol. 2005;67:114–122. doi: 10.1124/mol.104.006908. [DOI] [PubMed] [Google Scholar]
- 170.Kennedy C, Westfall TD, Sneddon P. Modulation of purinergic neurotransmission by ecto-ATPase. Semin Neurosci. 1996;8:195–199. doi: 10.1006/smns.1996.0025. [DOI] [Google Scholar]
- 171.Westfall TD, Kennedy C, Sneddon P. The ecto-ATPase inhibitor ARL 67156 enhances parasympathetic neurotransmission in the guinea-pig urinary bladder. Eur J Pharmacol. 1997;329:169–173. doi: 10.1016/S0014-2999(97)89178-9. [DOI] [PubMed] [Google Scholar]
- 172.Machida T, Heerdt PM, Reid AC, Schäfer U, Silver RB, Broekman MJ, Marcus AJ, Levi R. Ectonucleoside triphosphate diphosphohydrolase1/CD39, localized in neurons of human and porcine heart, modulates ATP-induced norepinephrine exocytosis. J Pharmacol Exp Ther. 2005;313:570–577. doi: 10.1124/jpet.104.081240. [DOI] [PubMed] [Google Scholar]
- 173.Ghildyal P, Palani D, Manchanda R. Post- and prejunctional consequences of ecto-ATPase inhibition: electrical and contractile studies in guinea-pig vas deferens. J Physiol London. 2006;575:469–480. doi: 10.1113/jphysiol.2006.109678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Goepfert C, Sundberg C, Sévigny J, Enjyoji K, Hoshi T, Csizmadia E, Robson SC. Disordered cellular migration and angiogenesis in cd39-null mice. Circulation. 2001;104:3109–3115. doi: 10.1161/hc5001.100663. [DOI] [PubMed] [Google Scholar]
- 175.Guckelberger O, Sun XF, Sévigny J, Imai M, Kaczmarek E, Enjyoji K, Kruskal JB, Robson SC. Beneficial effects of CD39/ecto-nucleoside triphosphate diphosphohydrolase-I in murine intestinal ischemia-reperfusion injury. Thromb Haemost. 2004;91:576–586. doi: 10.1160/TH03-06-0373. [DOI] [PubMed] [Google Scholar]
- 176.Sun XF, Cardenas A, Wu Y, Enjyoji K, Robson SC. Vascular stasis, intestinal hemorrhage, and heightened vascular permeability complicate acute portal hypertension in cd39-null mice. Amer J Physiol Gastrointest L. 2009;297:G306–G311. doi: 10.1152/ajpgi.90703.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Levesque SA, Kukulski F, Enjyoji K, Robson SC, Sévigny J. NTPDase1 governs P2X7-dependent functions in murine macrophages. Eur J Immunol. 2010;40:1473–1485. doi: 10.1002/eji.200939741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Feng LL, Sun XF, Csizmadia E, Han LH, Bian S, Murakami T, Wang X, Robson SC, Wu Y. Vascular CD39/ENTPD1 directly promotes tumor cell growth by scavenging extracellular adenosine triphosphate. Neoplasia. 2011;13:206–U34. doi: 10.1593/neo.101332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Dwyer KM, Robson SC, Nandurkar HH, Campbell DJ, Gock H, Murray-Segal LJ, Fisicaro N, Mysore TB, Kaczmarek E, Cowan PJ, D'Apice AJ. Thromboregulatory manifestations in human CD39 transgenic mice and the implications for thrombotic disease and transplantation. J Clin Invest. 2004;113:1440–1446. doi: 10.1172/JCI19560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Furukoji E, Matsumoto M, Yamashita A, Yagi H, Sakurai Y, Marutsuka K, Hatakeyama K, Morishita K, Fujimura Y, Tamura S, Asada Y. Adenovirus-mediated transfer of human placental ectonucleoside triphosphate diphosphohydrolase to vascular smooth muscle cells suppresses platelet aggregation in vitro and arterial thrombus formation in vivo. Circulation. 2005;111:808–815. doi: 10.1161/01.CIR.0000155239.46511.79. [DOI] [PubMed] [Google Scholar]
- 181.Takemoto Y, Kawata H, Soeda T, Imagawa K, Somekawa S, Takeda Y, Uemura S, Matsumoto M, Fujimura Y, Jo J, Kimura Y, Tabata Y, Saito Y. Human placental ectonucleoside triphosphate diphosphohydrolase gene transfer via gelatin-coated stents prevents in-stent thrombosis. Arterioscler Thromb Vasc Biol. 2009;29:857–862. doi: 10.1161/ATVBAHA.109.186429. [DOI] [PubMed] [Google Scholar]
- 182.Jhandier MN, Kruglov EA, Lavoie ÉG, Sévigny J, Dranoff JA. Portal fibroblasts regulate the proliferation of bile duct epithelia via expression of NTPDase2. J Biol Chem. 2005;280:22986–22992. doi: 10.1074/jbc.M412371200. [DOI] [PubMed] [Google Scholar]
- 183.Deaglio S, Dwyer KM, Gao W, Friedman D, Usheva A, Erat A, Chen JF, Enjyoji K, Linden J, Oukka M, Kuchroo VK, Strom TB, Robson SC. Adenosine generation catalyzed by CD39 and CD73 expressed on regulatory T cells mediates immune suppression. J Exp Med. 2007;204:1257–1265. doi: 10.1084/jem.20062512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Eltzschig HK. Adenosine: an old drug newly discovered. Anesthesiology. 2009;111:904–915. doi: 10.1097/ALN.0b013e3181b060f2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Massé K, Eason R, Bhamra S, Dale N, Jones EA. Comparative genomic and expression analysis of the conserved NTPDase gene family in Xenopus. Genomics. 2006;87:366–381. doi: 10.1016/j.ygeno.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 186.Rosemberg DB, Rico EP, Langoni AS, Spinelli JT, Pereira TC, Dias RD, Souza DO, Bonan CD, Bogo MR. NTPDase family in zebrafish: nucleotide hydrolysis, molecular identification and gene expression profiles in brain, liver and heart. Comp Biochem Physiol Pt B. 2010;155:230–240. doi: 10.1016/j.cbpb.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 187.Burnstock G, Verkhratsky A. Evolutionary origins of the purinergic signalling system. Acta Physiol (Oxf) 2009;195:415–447. doi: 10.1111/j.1748-1716.2009.01957.x. [DOI] [PubMed] [Google Scholar]
- 188.Tanaka K, Gilroy S, Jones AM, Stacey G. Extracellular ATP signaling in plants. Trends Cell Biol. 2010;20:601–608. doi: 10.1016/j.tcb.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Sansom FM, Robson SC, Hartland EL. Possible effects of microbial ecto-nucleoside triphosphate diphosphohydrolases on host–pathogen interactions. Microbiol Mol Biol Rev. 2008;72:765. doi: 10.1128/MMBR.00013-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Buss KA, Cooper DR, Ingram-Smith C, Ferry JG, Sanders DA, Hasson MS. Urkinase: structure of acetate kinase, a member of the ASKHA superfamily of phosphotransferases. J Bacteriol. 2001;183:680–686. doi: 10.1128/JB.183.2.680-686.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Flaherty KM, Mckay DB, Kabsch W, Holmes KC. Similarity of the three dimensional structures of actin and the ATPase fragment of a 70-kDa heat shock cognate protein. Proc Natl Acad Sci USA. 1991;88:5041–5045. doi: 10.1073/pnas.88.11.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Vorhoff T, Zimmermann H, Pelletier J, Sévigny J, Braun N. Cloning and characterization of the ecto-nucleotidase NTPDase3 from rat brain: predicted secondary structure and relation to members of the E-NTPDase family and actin. Purinergic Signal. 2005;1:259–270. doi: 10.1007/s11302-005-6314-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Kirley TL, Crawford PA, Smith TM. The structure of the nucleoside triphosphate diphosphohydrolases (NTPDases) as revealed by mutagenic and computational modeling analyses. Purinergic Signal. 2006;2:379–389. doi: 10.1007/s11302-005-5301-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ivanenkov VV, Crawford PA, Toyama A, Sévigny J, Kirley TL. Epitope mapping in cell surface proteins by site-directed masking: defining the structural elements of NTPDase3 inhibition by a monoclonal antibody. Protein Eng Des Sel. 2010;23:579–588. doi: 10.1093/protein/gzq027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Zebisch M, Sträter N. Structural insight into signal conversion and inactivation by NTPDase2 in purinergic signaling. Proc Nat Acad Sci USA. 2008;105:6882–6887. doi: 10.1073/pnas.0802535105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Vivian JP, Riedmaier P, Ge HH, Nours J, Sansom FM, Wilce MCJ, Byres E, Dias M, Schmidberger JW, Cowan PJ, d'Apice AJF, Hartland EL, Rossjohn J, Beddoe T. Crystal structure of a Legionella pneumophila ecto-triphosphate diphosphohydrolase, a structural and functional homolog of the eukaryotic NTPDases. Structure. 2010;18:228–238. doi: 10.1016/j.str.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 197.Gaddie KJ, Kirley TL. Proline residues link the active site to transmembrane domain movements in human nucleoside triphosphate diphosphohydrolase 3 (NTPDase3) Purinergic Signal. 2010;6:327–337. doi: 10.1007/s11302-010-9180-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Zebisch M, Schäfer P, Krauss M, Sträter N. Crystallographic evidence for a domain motion in rat nucleoside triphosphate diphosphohydrolase (NTPDase) 1. J Mol Biol. 2012;15:288–306. doi: 10.1016/j.jmb.2011.10.050. [DOI] [PubMed] [Google Scholar]
- 199.Krug U, Zebisch M, Krauss M, Sträter N. Structural insight into the activation mechanism of Toxoplasma gondii nucleoside triphosphate diphosphohydrolases by disulfide reduction. J Biol Chem. 2012;287:3051–66. doi: 10.1074/jbc.M111.294348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Garavaglia S, Bruzzone S, Cassani C, Canella L, Allegrone G, Sturla L, Mannino E, Millo E, De FA, Rizzi M. The high-resolution crystal structure of periplasmic Haemophilus influenzae NAD nucleotidase reveals a novel enzymatic function of human CD73 related to NAD metabolism. Biochem J Immediate Publication. 2011;441:131–141. doi: 10.1042/BJ20111263. [DOI] [PubMed] [Google Scholar]
- 201.Zimmermann H. 5′-Nucleotidase—molecular structure and functional aspects. Biochem J. 1992;285:345–365. doi: 10.1042/bj2850345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Hunsucker SA, Mitchell BS, Spychala J. The 5′-nucleotidases as regulators of nucleotide and drug metabolism. Pharmacol Ther. 2005;107:1–30. doi: 10.1016/j.pharmthera.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 203.Jin DC, Fan J, Wang L, Thompson LF, Liu AJ, Daniel BJ, Shin T, Curiel TJ, Zhang B. CD73 on tumor cells impairs antitumor T-cell responses: a novel mechanism of tumor-induced immune suppression. Cancer Res. 2010;70:2245–2255. doi: 10.1158/0008-5472.CAN-09-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Resta R, Hooker SW, Hansen KR, Laurent AB, Park JL, Blackburn MR, Knudsen TB, Thompson LF. Murine ecto-5′-nucleotidase (CD73)—cDNA cloning and tissue distribution. Gene. 1993;133:171–177. doi: 10.1016/0378-1119(93)90635-G. [DOI] [PubMed] [Google Scholar]
- 205.Christensen LD. No correlation between CD73 expression and ecto-5′-nucleotidase activity on blood mononuclear cells in vitro—evidence of CD73 (ecto-5′-nucleotidase) on blood mononuclear cells with distinct antigenic properties. APMIS. 1996;104:126–134. doi: 10.1111/j.1699-0463.1996.tb00697.x. [DOI] [PubMed] [Google Scholar]
- 206.Navarro JM, Olmo N, Turnay J, López-Conejo MT, Lizarbe MA. Ecto-5′-nucleotidase from a human colon adenocarcinoma cell line. Correlation between enzyme activity and levels in intact cells. Mol Cell Biochem. 1998;187:121–131. doi: 10.1023/A:1006808232059. [DOI] [PubMed] [Google Scholar]
- 207.Garcia-Ayllon MS, Campoy FJ, Vidal CJ, Muñoz-Delgado E. Identification of inactive ecto-5′-nucleotidase in normal mouse muscle and its increased activity in dystrophic Lama2(dy) mice. Neurosci Res. 2001;66:656–665. doi: 10.1002/jnr.10014. [DOI] [PubMed] [Google Scholar]
- 208.Morote-Garcia JC, Campo LFS, Campoy FJ, Vidal CJ, Muñoz-Delgado E. The increased ecto-5′-nucleotidase activity in muscle, heart and liver of laminin alpha 2-deficient mice is not caused by an elevation in the mRNA content. Int J Biochem Cell Biol. 2006;38:1092–1101. doi: 10.1016/j.biocel.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 209.Naito Y, Lowenstein JM. 5′-Nucleotidase from rat heart. Biochemistry USA. 1981;20:5188–5194. doi: 10.1021/bi00521a014. [DOI] [PubMed] [Google Scholar]
- 210.Grondal EJM, Zimmermann H. Purification, characterization and cellular localization of 5′-nucleotidase from Torpedo electric organ. Biochem J. 1987;245:805–810. doi: 10.1042/bj2450805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.James S, Richardson PJ. Production of adenosine from extracellular ATP at the striatal cholinergic synapse. J Neurochem. 1993;60:219–227. doi: 10.1111/j.1471-4159.1993.tb05841.x. [DOI] [PubMed] [Google Scholar]
- 212.Fredholm BB, Ijzerman AP, Jacobson KA, Klotz KN, Linden J. International Union of Pharmacology. XXV. Nomenclature and classification of adenosine receptors. Pharmacol Rev. 2001;53:527–552. [PMC free article] [PubMed] [Google Scholar]
- 213.Castrop H, Huang YN, Hashimoto S, Mizel D, Hansen P, Theilig F, Bachmann S, Deng CX, Briggs J, Schnermann J. Impairment of tubuloglomerular feedback regulation of GFR in ecto-5′-nucleotidase/CD73-deficient mice. J Clin Invest. 2004;114:634–642. doi: 10.1172/JCI21851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Thompson LF, Eltzschig HK, Ibla JC, Wiele CJ, Resta R, Morote-Garcila JC, Colgan SP. Crucial role for ecto-5′-nucleotidase (CD73) in vascular leakage during hypoxia. J Exp Med. 2004;200:1395–1405. doi: 10.1084/jem.20040915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Koszalka P, Ozuyaman B, Huo YQ, Zernecke A, Flogel U, Braun N, Buchheiser A, Decking UKM, Smith ML, Sévigny J, Gear A, Weber AA, Molojavyi A, Ding ZP, Weber C, Ley K, Zimmermann H, Gödecke A, Schrader J. Targeted disruption of cd73/ecto-5′-nucleotidase alters thromboregulation and augments vascular inflammatory response. Circ Res. 2004;95:814–821. doi: 10.1161/01.RES.0000144796.82787.6f. [DOI] [PubMed] [Google Scholar]
- 216.El-Tayeb A, Iqbal J, Behrenswerth A, Romio M, Schneider M, Zimmermann H, Schrader J, Müller CE. Nucleoside-5′-monophosphates as prodrugs of adenosine A2A receptor agonists activated by ecto-5′-nucleotidase. J Med Chem. 2009;52:7669–7677. doi: 10.1021/jm900538v. [DOI] [PubMed] [Google Scholar]
- 217.Kriz W. Adenosine and ATP: traffic regulators in the kidney. J Clin Invest. 2004;114:611–613. doi: 10.1172/JCI22669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Colgan SP, Eltzschig HK, Eckle T, Thompson LF. Physiological roles of ecto-5′-nucleotidase. Purinergic Signal. 2006;2:351–360. doi: 10.1007/s11302-005-5302-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Schetinger MR, Morsch VM, Bonan CD, Wyse AT. NTPDase and 5′-nucleotidase activities in physiological and disease conditions: new perspectives for human health. Biofactors. 2007;31:77–98. doi: 10.1002/biof.5520310205. [DOI] [PubMed] [Google Scholar]
- 220.Rajakumar SV, Lu B, Crikis S, Robson SC, d'Apice AJF, Cowan PJ, Dwyer KM. Deficiency or inhibition of CD73 protects in mild kidney ischemia-reperfusion injury. Transplantation. 2010;90:1260–1264. doi: 10.1097/TP.0b013e3182003d9b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Robson SC, Schuppan D. Adenosine: tipping the balance towards hepatic steatosis and fibrosis. J Hepatol. 2010;52:941–943. doi: 10.1016/j.jhep.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Lazarowski ER, Boucher RC. Purinergic receptors in airway epithelia. Curr Opin Pharmacol. 2009;9:262–267. doi: 10.1016/j.coph.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Bours MJ, Swennen EL, Di VF, Cronstein BN, Dagnelie PC. Adenosine 5′-triphosphate and adenosine as endogenous signaling molecules in immunity and inflammation. Pharmacol Ther. 2006;112:358–404. doi: 10.1016/j.pharmthera.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 224.Junger WG. Immune cell regulation by autocrine purinergic signaling. Nature Rev Immunol. 2011;11:201–212. doi: 10.1038/nri2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Salmi M, Jalkanen S. Cell-surface enzymes in control of leukocyte trafficking. Nature Rev Immunol. 2005;5:760–771. doi: 10.1038/nri1705. [DOI] [PubMed] [Google Scholar]
- 226.Dunwiddie TV, Masino SA. The role and regulation of adenosine in the central nervous system. Annu Rev Neurosci. 2001;24:31–55. doi: 10.1146/annurev.neuro.24.1.31. [DOI] [PubMed] [Google Scholar]
- 227.Dare E, Schulte G, Karovic O, Hammarberg C, Fredholm BB. Modulation of glial cell functions by adenosine receptors. Physiol Behav. 2007;92:15–20. doi: 10.1016/j.physbeh.2007.05.031. [DOI] [PubMed] [Google Scholar]
- 228.Sowa NA, Taylor-Blake B, Zylka MJ. Ecto-5′-nucleotidase (CD73) inhibits nociception by hydrolyzing AMP to adenosine in nociceptive circuits. J Neurosci. 2010;30:2235–2244. doi: 10.1523/JNEUROSCI.5324-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Spychala J. Tumor-promoting functions of adenosine. Pharmacol Ther. 2000;87:161–173. doi: 10.1016/S0163-7258(00)00053-X. [DOI] [PubMed] [Google Scholar]
- 230.Stagg J, Smyth MJ. Extracellular adenosine triphosphate and adenosine in cancer. Oncogene. 2010;29:5346–5358. doi: 10.1038/onc.2010.292. [DOI] [PubMed] [Google Scholar]
- 231.Zhang B. CD73: a novel target for cancer immunotherapy. Cancer Res. 2010;70:6407–6411. doi: 10.1158/0008-5472.CAN-10-1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Zhi XL, Wang YJ, Zhou XR, Yu J, Jian RR, Tang SX, Yin LH, Zhou P. RNAi-mediated CD73 suppression induces apoptosis and cell-cycle arrest in human breast cancer cells. Cancer Sci. 2010;101:2561–2569. doi: 10.1111/j.1349-7006.2010.01733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Mikhailov A, Sokolovskaya A, Yegutkin GG, Amdahl H, West A, Yagita H, Lahesmaa R, Thompson LF, Jalkanen S, Blokhin D, Eriksson JE. CD73 participates in cellular multiresistance program and protects against TRAIL-induced apoptosis. J Immunol. 2008;181:464–475. doi: 10.4049/jimmunol.181.1.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Stagg J, Divisekera U, Duret H, Sparwasser T, Teng MWL, Darcy PK, Smyth MJ. CD73-deficient mice have increased antitumor immunity and are resistant to experimental metastasis. Cancer Res. 2011;71:2892–2900. doi: 10.1158/0008-5472.CAN-10-4246. [DOI] [PubMed] [Google Scholar]
- 235.Bianchi V, Spychala J. Mammalian 5′-nucleotidases. J Biol Chem. 2003;278:46195–46198. doi: 10.1074/jbc.R300032200. [DOI] [PubMed] [Google Scholar]
- 236.Borowiec A, Lechward K, Tkacz-Stachowska K, Skladanowski AC. Adenosine as a metabolic regulator of tissue function: production of adenosine by cytoplasmic 5′-nucleotidases. Acta Biochim Pol. 2006;53:269–278. [PubMed] [Google Scholar]
- 237.Misumi Y, Ogata S, Hirose S, Ikehara Y. Primary structure of rat liver 5′-nucleotidase deduced from the cDNA. Presence of the COOH-terminal hydrophobic domain for possible post-translational modification by glycophospholipid. J Biol Chem. 1990;265:2178–2183. [PubMed] [Google Scholar]
- 238.Misumi Y, Ogata S, Ohkubo K, Hirose S, Ikehara Y. Primary structure of human placental 5′-nucleotidase and identification of the glycolipid anchor in the mature form. Eur J Biochem. 1990;191:563–569. doi: 10.1111/j.1432-1033.1990.tb19158.x. [DOI] [PubMed] [Google Scholar]
- 239.Volknandt W, Vogel M, Pevsner J, Misumi Y, Ikehara Y, Zimmermann H. 5′-Nucleotidase from the electric ray electric lobe: primary structure and relation to mammalian and procaryotic enzymes. Eur J Biochem. 1991;202:855–861. doi: 10.1111/j.1432-1033.1991.tb16443.x. [DOI] [PubMed] [Google Scholar]
- 240.Zimmermann H, Braun N. Ecto-nucleotidases: molecular structures, catalytic properties, and functional roles in the nervous system. Prog Brain Res. 1999;120:371–385. doi: 10.1016/S0079-6123(08)63570-0. [DOI] [PubMed] [Google Scholar]
- 241.Martinez-Martinez A, Muñoz-Delgado E, Campoy FJ, Flores-Flores C, Rodriguez-Lopez JN, Fini C, Vidal CJ. The ecto-5′-nucleotidase subunits in dimers are not linked by disulfide bridges but by non-covalent bonds. Biochim Biophys Acta. 2000;1478:300–308. doi: 10.1016/S0167-4838(00)00035-2. [DOI] [PubMed] [Google Scholar]
- 242.Fini C, Amoresano A, Andolfo A, D'Auria S, Floridi A, Paolini S, Pucci P. Mass spectrometry study of ecto-5′-nucleotidase from bull seminal plasma. Eur J Biochem. 2000;267:4978–4987. doi: 10.1046/j.1432-1327.2000.01545.x. [DOI] [PubMed] [Google Scholar]
- 243.Servos J, Reiländer H, Zimmermann H. Catalytically active soluble ecto-5′-nucleotidasse purified after heterologous expression as a tool for drug screening. Drug Develop Res. 1998;45:269–276. doi: 10.1002/(SICI)1098-2299(199811/12)45:3/4<269::AID-DDR25>3.0.CO;2-B. [DOI] [Google Scholar]
- 244.Sowa NA, Voss MK, Zylka MJ. Recombinant ecto-5′-nucleotidase (CD73) has long lasting antinociceptive effects that are dependent on adenosine A1 receptor activation. Mol Pain. 2010;6:20. doi: 10.1186/1744-8069-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Suzuki K, Furukawa Y, Tamura H, Ejiri N, Suematsu H, Taguchi R, Nakamura S, Suzuki Y, Ikezawa H. Purification and cDNA cloning of bovine liver 5′- nucleotidase, a GPI-anchored protein, and its expression in COS cells. J Biochem Tokyo. 1993;113:607–613. doi: 10.1093/oxfordjournals.jbchem.a124090. [DOI] [PubMed] [Google Scholar]
- 246.Vogel M, Kowalewski HJ, Zimmermann H, Janetzko A, Margolis RU, Wollny HE. Association of the HNK-1 epitope with 5′-nucleotidase from Torpedo marmorata (electric ray) electric organ. Biochem J. 1991;278:199–202. doi: 10.1042/bj2780199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Vogel M, Zimmermann H, Singer W. Transient association of the HNK-1 epitope with 5′-nucleotidase during development of the cat visual cortex. Eur J Neurosci. 1993;5:1423–1425. doi: 10.1111/j.1460-9568.1993.tb00209.x. [DOI] [PubMed] [Google Scholar]
- 248.Morote-Garcia JC, Garcia-Ayllón AS, Campoy FJ, Vidal CJ, Muñoz-Delgado E. Active and inactive ecto-5′-nucleotidase variants in liver of control and dystrophic Lama2dy mice. Int J Biochem Cell Biol. 2004;36:422–433. doi: 10.1016/S1357-2725(03)00266-8. [DOI] [PubMed] [Google Scholar]
- 249.Airas L, Niemela J, Salmi M, Puurunen T, Smith DJ, Jalkanen S. Differential regulation and function of CD73, a glycosyl- phosphatidylinositol-linked 70-kD adhesion molecule, on lymphocytes and endothelial cells. J Cell Biol. 1997;136:421–431. doi: 10.1083/jcb.136.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Chatterjee S, Mayor S. The GPI-anchor and protein sorting. Cell Mol Life Sci. 2001;58:1969–1987. doi: 10.1007/PL00000831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Sharom FJ, Lehto MT. Glycosylphosphatidylinositol-anchored proteins: structure, function, and cleavage by phosphatidylinositol-specific phospholipase C. Biochem Cell Biol. 2002;80:535–549. doi: 10.1139/o02-146. [DOI] [PubMed] [Google Scholar]
- 252.Sharom FJ. GPI-anchored proteins: biophysical behaviour and cleavage by PI-specific phospholipases. In: Dangerfield JA, Metzner C, editors. GPI membrane anchors—the much needed link. Oak Park: Bentham Science; 2010. pp. 34–52. [Google Scholar]
- 253.Ali N, Evans WH. Priority targeting of glycosyl-phosphatidylinositol-anchored proteins to the bile-canalicular (apical) plasma membrane of hepatocytes. Involvement of ‘late’ endosomes. Biochem J. 1990;271:193–199. doi: 10.1042/bj2710193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Ihrke G, Martin GV, Shanks MR, Schrader M, Schroer TA, Hubbard AL. Apical plasma membrane proteins and endolyn-78 travel through a subapical compartment in polarized WIF-B hepatocytes. J Cell Biol. 1998;141:115–133. doi: 10.1083/jcb.141.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Ogata S, Hayashi Y, Misumi Y, Ikehara Y. Membrane-anchoring domain of rat liver 5′-nucleotidase: identification of the COOH-terminal serine-523 covalently attached with a glycolipid. Biochemistry USA. 1990;29:7923–7927. doi: 10.1021/bi00486a021. [DOI] [PubMed] [Google Scholar]
- 256.Furukawa Y, Tamura H, Ikezawa H. Mutational analysis of the COOH-terminal hydrophobic domain of bovine liver 5′-nucleotidase as a signal for glycosylphosphatidylinositol (GPI) anchor attachment. BBA-Biomembranes. 1994;1190:273–278. doi: 10.1016/0005-2736(94)90084-1. [DOI] [PubMed] [Google Scholar]
- 257.Taguchi R, Hamakawa N, Haradanishida M, Fukui T, Nojima K, Ikezawa H. Microheterogeneity in glycosylphosphatidylinositol anchor structures of bovine liver 5′-nucleotidase. Biochemistry USA. 1994;33:1017–1022. doi: 10.1021/bi00170a021. [DOI] [PubMed] [Google Scholar]
- 258.Taguchi R, Hamakawa N, Maekawa N, Ikezawa H. Application of electrospray ionization MS/MS and matrix-assisted laser desorption/ionization-time of flight mass spectrometry to structural analysis of the glycosyl-phosphatidylinositol-anchored protein. J Biochem Tokyo. 1999;126:421–429. doi: 10.1093/oxfordjournals.jbchem.a022467. [DOI] [PubMed] [Google Scholar]
- 259.Kinoshita T, Fujita M, Maeda Y. Biosynthesis, remodelling and functions of mammalian GPI-anchored proteins: recent progress. J Biochem. 2008;144:287–294. doi: 10.1093/jb/mvn090. [DOI] [PubMed] [Google Scholar]
- 260.Sunderman FW., Jr The clinical biochemistry of 5′-nucleotidase. Annu Clin Lab Sci. 1990;20:123–139. [PubMed] [Google Scholar]
- 261.Johnson SM, Patel S, Bruckner FE, Collins DA. 5′-Nucleotidase as a marker of both general and local inflammation in rheumatoid arthritis patients. Rheumatology (Oxford) 1999;38:391–396. doi: 10.1093/rheumatology/38.5.391. [DOI] [PubMed] [Google Scholar]
- 262.Chuang NN, Newby AC, Luzio JP. Characterization of different molecular forms of 5′-nucleotidase in normal serum and in serum from cholestatic patients and bile-duct-ligated rats. Biochem J. 1984;224:689–695. doi: 10.1042/bj2240689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Fini C, Talamo F, Cherri S, Coli M, Floridi A, Ferrara L, Scaloni A. Biochemical and mass spectrometric characterization of soluble ecto-5′-nucleotidase from bull seminal plasma. Biochem J. 2003;372:443–451. doi: 10.1042/BJ20021687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Vogel M, Kowalewski H, Zimmermann H, Hooper NM, Turner AJ. Soluble low-Km 5′-nucleotidase from electric ray (Torpedo marmorata) electric organ and bovine cerebral cortex is derived from the glycosyl-phosphatidylinositol-anchored ectoenzyme by phospholipase C cleavage. Biochem J. 1992;284:621–624. doi: 10.1042/bj2840621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Trams EG, Lauter CJ, Salem N, Jr, Heine U. Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochim Biophys Acta. 1981;645:63–70. doi: 10.1016/0005-2736(81)90512-5. [DOI] [PubMed] [Google Scholar]
- 266.Ogawa Y, Murayama N, Yanoshita R. Molecular cloning and characterization of ecto-5′-nucleotidase from the venoms of Gloydius blomhoffi. Toxicon. 2009;54:408–412. doi: 10.1016/j.toxicon.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 267.Müller G, Jung C, Wied S, Biemer-Daub G, Frick W. Transfer of the glycosylphosphatidylinositol-anchored 5′-nucleotidase CD73 from adiposomes into rat adipocytes stimulates lipid synthesis. Brit J Pharmacol. 2010;160:878–891. doi: 10.1111/j.1476-5381.2010.00724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Airas L, Hellman J, Salmi M, Bono P, Puurunen T, Smith DJ, Jalkanen S. CD73 is involved in lymphocyte binding to the endothelium: characterization of lymphocyte vascular adhesion protein 2 identifies it as CD73. J Exp Med. 1995;182:1603–1608. doi: 10.1084/jem.182.5.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Airas L, Niemela J, Jalkanen S. CD73 engagement promotes lymphocyte binding to endothelial cells via a lymphocyte function-associated antigen-1-dependent mechanism. J Immunol. 2000;165:5411–5417. doi: 10.4049/jimmunol.165.10.5411. [DOI] [PubMed] [Google Scholar]
- 270.Airas L, Jalkanen S. CD73 mediates adhesion of B cells to follicular dendritic cells. Blood. 1996;88:1755–1764. [PubMed] [Google Scholar]
- 271.Codogno P, Doyennette-Moyne M-A, Aubery M, Dieckhoff J, Lietzke R, Mannherz HG. Polyclonal and monoclonal antibodies against chicken gizzard 5′-nucleotidase inhibit the spreading process of chicken embryonic fibroblasts on laminin substratum. Exptl Cell Res. 1988;174:344–354. doi: 10.1016/0014-4827(88)90305-9. [DOI] [PubMed] [Google Scholar]
- 272.Mehul B, Doyennette-Moyne MA, Aubery M, Mannherz HG, Codogno P. 5′-Nucleotidase is involved in chick embryo myoblast spreading on laminin. Cell Biol Int Rep. 1990;14:155–164. doi: 10.1016/0309-1651(90)90032-T. [DOI] [PubMed] [Google Scholar]
- 273.Stochaj U, Richter H, Mannherz HG. Chicken gizzard 5′-nucleotidase is a receptor for the extracellular matrix component fibronectin. Eur J Cell Biol. 1990;51:335–338. [PubMed] [Google Scholar]
- 274.Stochaj U, Mannherz HG. Chicken gizzard 5′-nucleotidase functions as a binding protein for the laminin/nidogen complex. Eur J Cell Biol. 1992;59:364–372. [PubMed] [Google Scholar]
- 275.Olmo N, Turnay J, Risse G, Deutzmann R, Mark K, Lizarbe A. Modulation of 5′-nucleotidase activity in plasma membranes and intact cells by the extracellular matrix proteins laminin and fibronectin. Biochem J. 1992;282:181–188. doi: 10.1042/bj2820181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Mehul B, Aubery M, Mannherz HG, Codogno P. Dual mechanism of laminin modulation of ecto-5′-nucleotidase activity. J Cell Biochem. 1993;52:266–274. doi: 10.1002/jcb.240520303. [DOI] [PubMed] [Google Scholar]
- 277.Flocke K, Lesch G, Elsässer HP, Bosslet K, Mannherz HG. Monoclonal antibodies against 5′-nucleotidase from a human pancreatic tumor cell line—their characterization and inhibitory capacity on tumor cell adhesion to fibronectin substratum. Eur J Cell Biol. 1992;58:62–70. [PubMed] [Google Scholar]
- 278.Sadej R, Inail K, Rajfur Z, Ostapkowicz A, Kohler J, Skladanowski AC, Mitchell BS, Spychala J. Tenascin C interacts with ecto-5′-nucleotidase (EN) and regulates adenosine generation in cancer cells. BBA Mol Basis Dis. 2008;1782:35–40. doi: 10.1016/j.bbadis.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 279.Dianzani U, Redoglia V, Bragardo M, Attisano C, Bianchi A, Difranco D, Ramenghi U, Wolff H, Thompson LF, Pileri A, Massaia M. Co-stimulatory signal delivered by CD73 molecule to human CD45RAhiCD45ROlo (naive) CD8+ T lymphocytes. J Immunol. 1993;151:3961–3970. [PubMed] [Google Scholar]
- 280.Thompson LF, Ruedi JM, Glass A, Low MG, Lucas AH. Antibodies to 5′-nucleotidase (CD73), a glycosyl-phosphatidylinositol-anchored protein, cause human peripheral blood T cells to proliferate. J Immunol. 1989;143:1815–1821. [PubMed] [Google Scholar]
- 281.Massaia M, Perrin L, Bianchi A, Ruedi J, Attisano C, Altieri D, Rijkers GT, Thompson LF. Human T cell activation. Synergy between CD73 (ecto-5′-nucleotidase) and signals delivered through CD3 and CD2 molecules. J Immunol. 1990;145:1664–1674. [PubMed] [Google Scholar]
- 282.Resta R, Hooker SW, Laurent AB, Shuck JK, Misumi Y, Ikehara Y, Koretzky GA, Thompson LF. Glycosyl phosphatidylinositol membrane anchor is not required for T cell activation through CD73. J Immunol. 1994;153:1046–1053. [PubMed] [Google Scholar]
- 283.Gutensohn W, Resta R, Misumi Y, Ikehara Y, Thompson LF. Ecto-5′-nucleotidase activity is not required for T cell activation through CD73. Cell Immunol. 1995;161:213–217. doi: 10.1006/cimm.1995.1029. [DOI] [PubMed] [Google Scholar]
- 284.Resta R, Thompson LF. T cell signalling through CD73. Cell Signal. 1997;9:131–139. doi: 10.1016/S0898-6568(96)00132-5. [DOI] [PubMed] [Google Scholar]
- 285.Airas L. CD73 and adhesion of B-cells to follicular dendritic cells. Leuk Lymphoma. 1998;29:37–47. doi: 10.3109/10428199809058380. [DOI] [PubMed] [Google Scholar]
- 286.Resta R, Yamashita Y, Thompson LF. Ecto-enzyme and signaling functions of lymphocyte CD73. Immunol Rev. 1998;161:95–109. doi: 10.1111/j.1600-065X.1998.tb01574.x. [DOI] [PubMed] [Google Scholar]
- 287.Sharom FJ, Radeva G. GPI-anchored protein cleavage in the regulation of transmembrane signals. In: Quinn PJ, editor. Membrane dynamics and domains. New York: Kluwer Academic/Plenum; 2004. pp. 295–315. [DOI] [PubMed] [Google Scholar]
- 288.Fujita M, Jigami Y. Lipid remodeling of GPI-anchored proteins and its function. Biochim Biophys Acta. 2008;1780:410–420. doi: 10.1016/j.bbagen.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 289.Zhuo S, Clemens JC, Stone RL, Dixon JE. Mutational analysis of a Ser/Thr phosphatase—identification of residues important in phosphoesterase substrate binding and catalysis. J Biol Chem. 1994;269:26234–26238. [PubMed] [Google Scholar]
- 290.Zimmermann H. Extracellular purine metabolism. Drug Develop Res. 1996;39:337–352. doi: 10.1002/(SICI)1098-2299(199611/12)39:3/4<337::AID-DDR15>3.0.CO;2-Z. [DOI] [Google Scholar]
- 291.Glaser L, Melo A, Paul R. Uridine diphosphate sugar hydrolase. Purification of enzyme and protein inhibitor. J Biol Chem. 1967;242:1944–1954. [PubMed] [Google Scholar]
- 292.Neu HC. The 5′-nucleotidases (uridine diphosphate sugar hydrolases) of the Enterobacteriaceae. Biochemistry USA. 1968;7:3766–3773. doi: 10.1021/bi00850a059. [DOI] [PubMed] [Google Scholar]
- 293.Champagne DE, Smartt CT, Ribeiro JMC, James AA. The salivary gland-specific apyrase of the mosquito Aedes aegypti is a member of the 5′-nucleotidase family. Proc Natl Acad Sci USA. 1995;92:694–698. doi: 10.1073/pnas.92.3.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Lombardo F, Cristina M, Spanos L, Louis C, Coluzzi M, Arca B. Promoter sequences of the putative Anopheles gambiae apyrase confer salivary gland expression in Drosophila melanogaster. J Biol Chem. 2000;275:23861–23868. doi: 10.1074/jbc.M909547199. [DOI] [PubMed] [Google Scholar]
- 295.Faudry E, Lozzi SP, Santana JM, D'Souza-Ault M, Kieffer S, Felix CR, Ricart CA, Sousa MV, Vernet T, Teixeira AR. Triatoma infestans apyrases belong to the 5′-nucleotidase family. J Biol Chem. 2004;279:19607–19613. doi: 10.1074/jbc.M401681200. [DOI] [PubMed] [Google Scholar]
- 296.Charlab R, Valenzuela JG, Rowton ED, Ribeiro JMC. Toward an understanding of the biochemical and pharmacological complexity of the saliva of a hematophagous sand fly Lutzomyia longipalpis. Proc Nat Acad Sci USA. 1999;96:15155–15160. doi: 10.1073/pnas.96.26.15155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Stutzer C, Mans BJ, Gaspar ARM, Neitz AWH, Maritz-Olivier C. Ornithodoros savignyi: soft tick apyrase belongs to the 5′-nucleotidase family. Exp Parasitol. 2009;122:318–327. doi: 10.1016/j.exppara.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 298.Koonin EV. Conserved sequence pattern in a wide variety of phosphoesterases. Protein Sci. 1994;3:356–358. doi: 10.1002/pro.5560030218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Sträter N, Klabunde T, Tucker P, Witzel H, Krebs B. Crystal structure of a purple acid phosphatase containing a dinuclear Fe(III)–Zn(II) active site. Science. 1995;268:1489–1492. doi: 10.1126/science.7770774. [DOI] [PubMed] [Google Scholar]
- 300.Knöfel T, Sträter N. X-ray structure of the Escherichia coli periplasmic 5′-nucleotidase containing a dimetal catalytic site. Nature Struct Biol. 1999;6:448–453. doi: 10.1038/8253. [DOI] [PubMed] [Google Scholar]
- 301.Knöfel T, Sträter N. Mechanism of hydrolysis of phosphate esters by the dimetal center of 5′-nucleotidase based on crystal structures. J Mol Biol. 2001;309:239–254. doi: 10.1006/jmbi.2001.4656. [DOI] [PubMed] [Google Scholar]
- 302.Knöfel T, Sträter N. E-coli 5′-nucleotidase undergoes a hinge-bending domain rotation resembling a ball-and-socket motion. J Mol Biol. 2001;309:255–266. doi: 10.1006/jmbi.2001.4657. [DOI] [PubMed] [Google Scholar]
- 303.Schultz-Heienbrok R, Maier T, Sträter N. Trapping a 96 degrees domain rotation in two distinct conformations by engineered disulfide bridges. Protein Sci. 2004;13:1811–1822. doi: 10.1110/ps.04629604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Schultz-Heienbrok R, Maier T, Sträter N. A large hinge bending domain rotation is necessary for the catalytic function of Escherichia coli 5′-nucleotidase. Biochemistry USA. 2005;44:2244–2252. doi: 10.1021/bi047989c. [DOI] [PubMed] [Google Scholar]
- 305.Neu HC. The 5′-nucleotidase of Escherichia coli. I. Purification and properties. J Biol Chem. 1967;242:3896–3904. [PubMed] [Google Scholar]
- 306.Dvorak HF, Heppel LA. Metallo-enzymes released from Escherichia coli by osmotic shock. II. Evidence that 5′-nucleotidase and cyclic phosphodiesterase are zinc metallo-enzymes. J Biol Chem. 1968;243:2647–2653. [PubMed] [Google Scholar]
- 307.McMillen L, Beacham IR, Burns DM. Cobalt activation of Escherichia coli 5′-nucleotidase is due to zinc ion displacement at only one of two metal-ion-binding sites. Biochem J. 2003;372:625–630. doi: 10.1042/BJ20021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Sträter N. Ecto-5′-nucleotidase: structure function relationships. Purinergic Signal. 2006;2:343–350. doi: 10.1007/s11302-006-9000-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Klabunde T, Sträter N, Fröhlich R, Witzel H, Krebs B. Mechanism of Fe(III)–Zn(II) purple acid phosphatase based on crystal structures. J Mol Biol. 1996;259:737–748. doi: 10.1006/jmbi.1996.0354. [DOI] [PubMed] [Google Scholar]
- 310.Mueller EG, Crowder MA, Averill BA, Knowles JR. Purple acid phosphatase: a diiron enzyme that catalyzes a direct phospho group transfer to water. J Am Chem Soc. 1993;115:2974–2975. doi: 10.1021/ja00060a055. [DOI] [Google Scholar]
- 311.Sträter N. 5′-Nucleotidase. In: Messerschmidt A, Bode W, Cygler M, editors. Handbook of metalloproteins. Chichester: Wiley; 2004. pp. 62–70. [Google Scholar]
- 312.Stefan C, Jansen S, Bollen M. Modulation of purinergic signaling by NNP-type ectophosphodiesterases. Purinergic Signal. 2006;2:361–370. doi: 10.1007/s11302-005-5303-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Stefan C, Gijsbers R, Stalmans W, Bollen M. Differential regulation of the expression of nucleotide pyrophosphatases/phosphodiesterases in rat liver. Biochim Biophys Acta. 1999;1450:45–52. doi: 10.1016/S0167-4889(99)00031-2. [DOI] [PubMed] [Google Scholar]
- 314.Vaingankar SM, Fitzpatrick TA, Johnson K, Goding JW, Maurice M, Terkeltaub R. Subcellular targeting and function of osteoblast nucleotide pyrophosphatase phosphodiesterase 1. Amer J Physiol Cell Physiol. 2004;286:C1177–C1187. doi: 10.1152/ajpcell.00320.2003. [DOI] [PubMed] [Google Scholar]
- 315.Picher M, Boucher RC. Biochemical evidence for an ecto-alkaline phosphodiesterase I in human airways. Amer J Respir Cell Molec Biol. 2000;23:255–261. doi: 10.1165/ajrcmb.23.2.4088. [DOI] [PubMed] [Google Scholar]
- 316.Vekaria RM, Shirley DG, Sévigny J, Unwin RJ. Immunolocalization of ectonucleotidases along the rat nephron. Amer J Physiol Renal Physiol. 2006;290:F550–F560. doi: 10.1152/ajprenal.00151.2005. [DOI] [PubMed] [Google Scholar]
- 317.Scott LJ, Delautier D, Meerson NR, Trugnan G, Goding JW, Maurice M. Biochemical and molecular identification of distinct forms of alkaline phosphodiesterase I expressed on the apical and basolateral plasma membrane surfaces of rat hepatocytes. Hepatol. 1997;25:995–1002. doi: 10.1002/hep.510250434. [DOI] [PubMed] [Google Scholar]
- 318.Murabe Y, Sano Y. Thiaminepyrophosphatase activity in the plasma membrane of microglia. Histochemistry. 1981;71:45–52. doi: 10.1007/BF00592569. [DOI] [PubMed] [Google Scholar]
- 319.Langer D, Hammer K, Koszalka P, Schrader J, Robson S, Zimmermann H. Distribution of ectonucleotidases in the rodent brain revisited. Cell Tissue Res. 2008;334:199–217. doi: 10.1007/s00441-008-0681-x. [DOI] [PubMed] [Google Scholar]
- 320.Aoki J. Mechanisms of lysophosphatidic acid production. Semin Cell Dev Biol. 2004;15:477–489. doi: 10.1016/j.semcdb.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 321.Sato K, Malchinkhuu E, Muraki T, Ishikawa K, Hayashi K, Tosaka M, Mochiduki A, Inoue K, Tomura H, Mogi C, Nochi H, Tamoto K, Okajima F. Identification of autotaxin as a neurite retraction-inducing factor of PC12 cells in cerebrospinal fluid and its possible sources. J Neurochem. 2005;92:904–914. doi: 10.1111/j.1471-4159.2004.02933.x. [DOI] [PubMed] [Google Scholar]
- 322.Ohuchi H, Hayashibara Y, Matsuda H, Onoi M, Mitsumori M, Tanaka M, Aoki J, Arai H, Noji S. Diversified expression patterns of autotaxin, a gene for phospholipid-generating enzyme during mouse and chicken development. Develop Dynam. 2007;236:1134–1143. doi: 10.1002/dvdy.21119. [DOI] [PubMed] [Google Scholar]
- 323.Koike S, Yutoh Y, Keino-Masu K, Noji S, Masu M, Ohuchi H. Autotaxin is required for the cranial neural tube closure and establishment of the midbrain-hindbrain boundary during mouse development. Develop Dynam. 2011;240:413–421. doi: 10.1002/dvdy.22543. [DOI] [PubMed] [Google Scholar]
- 324.Anderson KC, Bates MP, Slaughenhoupt B, Schlossman SF, Nadler LM. A monoclonal antibody with reactivity restricted to normal and neoplastic plasma cells. J Immunol. 1984;132:3172–3179. [PubMed] [Google Scholar]
- 325.Anderson KC, Bates MP, Slaughenhoupt BL, Pinkus GS, Schlossman SF, Nadler LM. Expression of human B cell-associated antigens on leukemias and lymphomas: a model of human B cell differentiation. Blood. 1984;63:1424–1433. [PubMed] [Google Scholar]
- 326.Deterre P, Gelman L, Gary-Gouy H, Arrieumerlou C, Berthelier V, Tixier EM, Ktorza S, Goding L, Schmitt C, Bismuth G. Coordinated regulation in human T cells of nucleotide-hydrolyzing ecto-enzymatic activities, including CD38 and PC-1—possible role in the recycling of nicotinamide adenine dinucleotide metabolites. J Immunol. 1996;157:1381–1388. [PubMed] [Google Scholar]
- 327.Aerts I, Martin JJ, Deyn PP, Ginniken C, Ostade X, Kockx M, Dua G, Slegers H. The expression of ecto-nucleotide pyrophosphatase/phosphodiesterase 1 (E-NPP1) is correlated with astrocytic tumor grade. Clin Neurol Neurosurg. 2011;113:224–229. doi: 10.1016/j.clineuro.2010.11.018. [DOI] [PubMed] [Google Scholar]
- 328.Prosdocimo DA, Wyler SC, Romani AM, ONeill WC, Dubyak GR. Regulation of vascular smooth muscle cell calcification by extracellular pyrophosphate homeostasis: synergistic modulation by cyclic AMP and hyperphosphatemia. Amer J Physiol Cell Physiol. 2010;298:C702–C713. doi: 10.1152/ajpcell.00419.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Johnson K, Terkeltaub R. Inorganic pyrophosphate (PPI) in pathologic calcification of articular cartilage. Front Biosci. 2005;10:988–997. doi: 10.2741/1593. [DOI] [PubMed] [Google Scholar]
- 330.Orimo H. The mechanism of mineralization and the role of alkaline phosphatase in health and disease. J Nippon Med Sch. 2010;77:4–12. doi: 10.1272/jnms.77.4. [DOI] [PubMed] [Google Scholar]
- 331.Goding JW, Grobben B, Slegers H. Physiological and pathophysiological functions of the ecto-nucleotide pyrophosphatase/phosphodiesterase family. BBA Mol Basis Dis. 2003;1638:1–19. doi: 10.1016/S0925-4439(03)00058-9. [DOI] [PubMed] [Google Scholar]
- 332.Anderson HC, Harmey D, Camacho NP, Garimella R, Sipe JB, Tague S, Bi XH, Johnson K, Terkeltaub R, Millán JL. Sustained osteomalacia of long bones despite major improvement in other hypophosphatasia-related mineral deficits in tissue nonspecific alkaline phosphatase/nucleotide pyrophosphatase phosphodiesterase 1 double-deficient mice. Amer J Pathol. 2005;166:1711–1720. doi: 10.1016/S0002-9440(10)62481-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Terkeltaub R. Physiologic and pathologic functions of the NPP nucleotide pyrophosphatase/phosphodiesterase family focusing in calcification. Purinergic Signal. 2006;2:371–377. doi: 10.1007/s11302-005-5304-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Stefan C, Jansen S, Bollen M. NPP-type ectophosphodiesterases: unity in diversity. Trends Biochem Sci. 2005;30:542–550. doi: 10.1016/j.tibs.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 335.Abate N, Chandalia M, Di PR, Foster DW, Grundy SM, Trischitta V. Mechanisms of disease: ectonucleotide pyrophosphatase phosphodiesterase 1 as a ‘gatekeeper’ of insulin receptors. Nat Clin Pract Endocrinol Metab. 2006;2:694–701. doi: 10.1038/ncpendmet0367. [DOI] [PubMed] [Google Scholar]
- 336.Goldfine ID, Maddux BA, Youngren JF, Reaven G, Accili D, Trischitta V, Vigneri R, Frittitta L. The role of membrane glycoprotein plasma cell antigen 1 ectonucleotide pyrophosphatase phosphodiesterase 1 in the pathogenesis of insulin resistance and related abnormalities. Endocrine Rev. 2008;29:62–75. doi: 10.1210/er.2007-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Deissler H, Blass-Kampmann S, Bruyneel E, Mareel M, Rajewsky MF. Neural cell surface differentiation antigen gp130RB13-6 induces fibroblasts and glioma cells to express astroglial proteins and invasive properties. FASEB J. 1999;13:657–666. doi: 10.1096/fasebj.13.6.657. [DOI] [PubMed] [Google Scholar]
- 338.Buhring HJ, Seiffert M, Giesert C, Marxer A, Kanz L, Valent P, Sano K. The basophil activation marker defined by antibody 97A6 is identical to the ectonucleotide pyrophosphatase/phosphodiesterase. Blood. 2001;97:3303–3305. doi: 10.1182/blood.V97.10.3303. [DOI] [PubMed] [Google Scholar]
- 339.Buhring HJ, Streble A, Valent P. The basophil-specific ectoenzyme E-NPP3 (CD203c) as a marker for cell activation and allergy diagnosis. Int Arch Allergy Immunol. 2004;133:317–329. doi: 10.1159/000077351. [DOI] [PubMed] [Google Scholar]
- 340.Hauswirth AW, Sonneck K, Florian S, Krauth MT, Bohm A, Sperr WR, Valenta R, Schernthaner GH, Printz D, Fritsch G, Buhring HJ, Valent P. Interleukin-3 promotes the expression of E-NPP3/CD203C on human blood basophils in healthy subjects and in patients with birch pollen allergy. Int J Immunopathol Pharmacol. 2007;20:267–278. doi: 10.1177/039463200702000207. [DOI] [PubMed] [Google Scholar]
- 341.Hauswirth AW, Escribano L, Prados A, Nunez R, Mirkina I, Kneidinger M, Florian S, Sonneck K, Vales A, Schernthaner GH, Sanchez-Munoz L, Sperr WR, Buhring HJ, Orfao A, Valent P. CD203c is overexpressed on neoplastic mast cells in systemic mastocytosis and is upregulated upon IgE receptor cross-linking. Int J Immunopathol Pharmacol. 2008;21:797–806. doi: 10.1177/039463200802100404. [DOI] [PubMed] [Google Scholar]
- 342.Hausmann OV, Gentinetta T, Bridts CH, Ebo DG. The basophil activation test in immediate-type drug allergy. Immunol Allergy Clin North Am. 2009;29:555–566. doi: 10.1016/j.iac.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 343.Meeteren LA, Moolenaar WH. Regulation and biological activities of the autotaxin-LPA axis. Prog Lipid Res. 2007;46:145–160. doi: 10.1016/j.plipres.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 344.Boutin JA, Ferry G. Autotaxin. Cell Mol Life Sci. 2009;66:3009–3021. doi: 10.1007/s00018-009-0056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Moolenaar WH, Perrakis A. Insights into autotaxin: how to produce and present a lipid mediator. Nat Rev Mol Cell Biol. 2011;12:674–679. doi: 10.1038/nrm3188. [DOI] [PubMed] [Google Scholar]
- 346.Fuss B, Baba H, Phan T, Tuohy VK, Macklin WB. Phosphodiesterase I, a novel adhesion molecule and/or cytokine involved in oligodendrocyte function. J Neurosci. 1997;17:9095–9103. doi: 10.1523/JNEUROSCI.17-23-09095.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Dennis J, Nogaroli L, Fuss B. Phosphodiesterase-Iα/autotaxin (PD-Iα/ATX): a multifunctional protein involved in central nervous system development. J Neurosci Res. 2005;82:737–742. doi: 10.1002/jnr.20686. [DOI] [PubMed] [Google Scholar]
- 348.Yuelling LM, Fuss B. Autotaxin (ADX): a multi-functional and multi-modular protein possessing enzymatic lysoPLD activity and matricellular properties. BBA Mol Cell Biol Lipids. 2008;1781:525–530. doi: 10.1016/j.bbalip.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Sela BA, Lis H, Sachs L. Enzymatic hydrolysis of uridine diphosphate-N-acetyl-D-galactosamine and uridine diphospate-N-acetyl-d-glucosamine by normal cells, and blocks in this hydrolysis in transformed cells and their revertants. J Biol Chem. 1972;247:7585–7590. [PubMed] [Google Scholar]
- 350.Evans WH, Hood DO, Gurd JW. Purification and properties of a mouse liver plasma-membrane glycoprotein hydrolysing nucleotide pyrophosphate and phosphodiester bonds. Biochem J. 1973;135:819–826. doi: 10.1042/bj1350819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Vollmayer P, Clair T, Goding JW, Sano K, Servos J, Zimmermann H. Hydrolysis of diadenosine polyphosphates by nucleotide pyrophosphatases/phosphodiesterases. Eur J Biochem. 2003;270:2971–2978. doi: 10.1046/j.1432-1033.2003.03674.x. [DOI] [PubMed] [Google Scholar]
- 352.Asensio AC, Rodríguez-Ferrera CR, Castañeyra-Perdomo A, Oaknin S, Rotllán P. Biochemical analysis of ecto-nucleotide pyrophosphatase phosphodiesterase activity in brain membranes indicates involvement of NPP1 isoenzyme in extracellular hydrolysis of diadenosine polyphosphates in central nervous system. Neurochem Int. 2007;50:581–590. doi: 10.1016/j.neuint.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 353.Gijsbers R, Aoki J, Arai H, Bollen M. The hydrolysis of lysophospholipids and nucleotides by autotaxin (NPP2) involves a single catalytic site. FEBS Lett. 2003;538:60–64. doi: 10.1016/S0014-5793(03)00133-9. [DOI] [PubMed] [Google Scholar]
- 354.Kelly SJ, Dardinger DE, Butler LG. Hydrolysis of phosphonate esters catalyzed by 5′-nucleotide phosphodiesterase. Biochemistry USA. 1975;14:4983–4988. doi: 10.1021/bi00693a030. [DOI] [PubMed] [Google Scholar]
- 355.Kelly SJ, Butler LG. Enzymic hydrolysis of phosphonate esters. Reaction mechanism of intestinal 5′-nucleotide phosphodiesterase. Biochemistry USA. 1977;16:1102–1104. doi: 10.1021/bi00625a011. [DOI] [PubMed] [Google Scholar]
- 356.Landt M, Butler LG. 5′-Nucleotide phosphodiesterase: isolation of covalently bound 5′-adenosine monophosphate, an intermediate in the catalytic mechanism. Biochemistry USA. 1978;17:4130–4135. doi: 10.1021/bi00613a004. [DOI] [PubMed] [Google Scholar]
- 357.Landt M, Everard RA, Butler LG. 5′-Nucleotide phosphodiesterase: features of the substrate binding site as deduced from specificity and kinetics of some novel substrates. Biochemistry USA. 1980;19:138–143. doi: 10.1021/bi00542a021. [DOI] [PubMed] [Google Scholar]
- 358.Chiavegatti T, Costa VL, Araujo MS, Godinho RO. Skeletal muscle expresses the extracellular cyclic AMP–adenosine pathway. Brit J Pharmacol. 2008;153:1331–1340. doi: 10.1038/sj.bjp.0707648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Giron MC, Bin A, Brun P, Etteri S, Bolego C, Florio C, Gaion RM. Cyclic AMP in rat ileum: evidence for the presence of an extracellular cyclic AMP–adenosine pathway. Gastroenterology. 2008;134:1116–1126. doi: 10.1053/j.gastro.2008.01.030. [DOI] [PubMed] [Google Scholar]
- 360.Bischoff E, Tran-Thi TA, Decker KF. Nucleotide pyrophosphatase of rat liver. A comparative study on the enzymes solubilized and purified from plasma membrane and endoplasmic reticulum. Eur J Biochem. 1975;51:353–361. doi: 10.1111/j.1432-1033.1975.tb03935.x. [DOI] [PubMed] [Google Scholar]
- 361.Moe OA, Jr, Butler LG. The catalytic mechanism of bovine intestinal 5′-nucleotide phosphodiesterase. pH and inhibition studies. J Biol Chem. 1983;258:6941–6946. [PubMed] [Google Scholar]
- 362.Ciancaglini P, Yadav MC, Simão AMS, Narisawa S, Pizauro JM, Farquharson C, Hoylaerts MF, Millán JL. Kinetic analysis of substrate utilization by native and TNAP-, NPP1-, or PHOSPHO1-deficient matrix vesicles. J Bone Miner Res. 2010;25:716–723. doi: 10.1359/jbmr.091023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Simão AM, Yadav MC, Narisawa S, Bolean M, Pizauro JM, Hoylaerts MF, Ciancaglini P, Millán JL. Proteoliposomes harboring alkaline phosphatase and nucleotide pyrophosphatase as matrix vesicle biomimetics. J Biol Chem. 2010;285:7598–7609. doi: 10.1074/jbc.M109.079830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Clair T, Lee HY, Liotta LA, Stracke ML. Autotaxin is an exoenzyme possessing 5′-nucleotide phosphodiesterase/ATP pyrophosphatase and ATPase activities. J Biol Chem. 1997;272:996–1001. doi: 10.1074/jbc.272.46.29347. [DOI] [PubMed] [Google Scholar]
- 365.Hosoda N, Hoshino S, Kanda Y, Katada T. Inhibition of phosphodiesterase/pyrophosphatase activity of PC-1 by its association with glycosaminoglycans. Eur J Biochem. 1999;265:763–770. doi: 10.1046/j.1432-1327.1999.00779.x. [DOI] [PubMed] [Google Scholar]
- 366.Bollen M, Gijsbers R, Ceulemans H, Stalmans W, Stefan C. Nucleotide pyrophosphatases/phosphodiesterases on the move. Crit Rev Biochem Molec Biol. 2000;35:393–432. doi: 10.1080/10409230091169249. [DOI] [PubMed] [Google Scholar]
- 367.Gijsbers R, Ceulemans H, Stalmans W, Bollen M. Structural and catalytic similarities between nucleotide pyrophosphatases/phosphodiesterases and alkaline phosphatases. J Biol Chem. 2001;276:1361–1368. doi: 10.1074/jbc.M007552200. [DOI] [PubMed] [Google Scholar]
- 368.Wojcik M, Cieslak M, Stec WJ, Goding JW, Koziolkiewicz M. Nucleotide pyrophosphatase/phosphodiesterase 1 is responsible for degradation of antisense phosphorothioate oligonucleotides. Oligonucleotides. 2007;17:134–145. doi: 10.1089/oli.2007.0021. [DOI] [PubMed] [Google Scholar]
- 369.Grobben B, Anciaux K, Roymans D, Stefan C, Bollen M, Esmans EL, Slegers H. An ecto-nucleotide pyrophosphatase is one of the main enzymes involved in the extracellular metabolism of ATP in rat C6 glioma. J Neurochem. 1999;72:826–834. doi: 10.1046/j.1471-4159.1999.0720826.x. [DOI] [PubMed] [Google Scholar]
- 370.Ramos A, Pintor J, Miras-Portugal MT, Rotllán P. Use of fluorogenic substrates for detection and investigation of ectoenzymatic hydrolysis of diadenosine polyphosphates: a fluorometric study on chromaffin cells. Anal Biochem. 1995;228:74–82. doi: 10.1006/abio.1995.1317. [DOI] [PubMed] [Google Scholar]
- 371.Gasmi L, Cartwright JL, McLennan AG. The hydrolytic activity of bovine adrenal medullary plasma membranes towards diadenosine polyphosphates is due to alkaline phosphodiesterase-I. Biochim Biophys Acta. 1998;1405:121–127. doi: 10.1016/S0167-4889(98)00097-4. [DOI] [PubMed] [Google Scholar]
- 372.Lüthje J, Ogilvie A. Catabolism of AP4A and Ap3A in human serum. Identification of isoenzymes and their partial characterization. Eur J Biochem. 1987;169:385–388. doi: 10.1111/j.1432-1033.1987.tb13624.x. [DOI] [PubMed] [Google Scholar]
- 373.Rotllán P, Asensio AC, Ramos A, Rodríguez-Ferrer CR, Oaknin S (2002) Ectoenzymatic hydrolysis of the signaling nucleotides diadenosine polyphoaphates. In: Recent research developments in Biochemistry, vol. 3. Trivandrum, India: Research Signpos, pp 191–209
- 374.Tokumura A, Majima E, Kariya Y, Tominaga K, Kogure K, Yasuda K, Fukuzawa K. Identification of human plasma lysophospholipase D, a lysophosphatidic acid-producing enzyme, as autotaxin, a multifunctional phosphodiesterase. J Biol Chem. 2002;277:39436–39442. doi: 10.1074/jbc.M205623200. [DOI] [PubMed] [Google Scholar]
- 375.Umezu-Goto M, Kishi Y, Taira A, Hama K, Dohmae N, Takio K, Yamori T, Mills GB, Inoue K, Aoki J, Arai H. Autotaxin has lysophospholipase D activity leading to tumor cell growth and motility by lysophosphatidic acid production. J Cell Biol. 2002;158:227–233. doi: 10.1083/jcb.200204026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Cimpean A, Stefan C, Gijsbers R, Stalmans W, Bollen M. Substrate-specifying determinants of the nucleotide pyrophosphatases/phosphodiesterases NPP1 and NPP2. Biochem J. 2004;381:71–77. doi: 10.1042/BJ20040465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Aoki J, Taira A, Takanezawa Y, Kishi Y, Hama K, Kishimoto T, Mizuno K, Saku K, Taguchi R, Arai H. Serum lysophosphatidic acid is produced through diverse phospholipase pathways. J Biol Chem. 2002;277:48737–48744. doi: 10.1074/jbc.M206812200. [DOI] [PubMed] [Google Scholar]
- 378.Tanaka M, Okudaira S, Kishi Y, Ohkawa R, Iseki S, Ota M, Noji S, Yatomi Y, Aoki J, Arai H. Autotaxin stabilizes blood vessels and is required for embryonic vasculature by producing lysophosphatidic acid. J Biol Chem. 2006;281:25822–25830. doi: 10.1074/jbc.M605142200. [DOI] [PubMed] [Google Scholar]
- 379.vanMeeteren LA, Ruurs P, Stortelers C, Bouwman P, vanRooijen MA, Pradere JP, Pettit TR, Wakelam MJO, SaulnierBlache JS, Mummery CL, Moolenaar WH, Jonkers J. Autotaxin, a secreted lysophospholipase D, is essential for blood vessel formation during development. Mol Cell Biol. 2006;26:5015–5022. doi: 10.1128/MCB.02419-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Clair T, Aoki J, Koh E, Bandle RW, Nam SW, Ptaszynska MM, Mills GB, Schiffmann E, Liotta LA, Stracke ML. Autotaxin hydrolyzes sphingosylphosphorylcholine to produce the regulator of migration, sphingosine-1-phosphate. Cancer Res. 2003;63:5446–5453. [PubMed] [Google Scholar]
- 381.Meeteren LA, Ruurs P, Christodoulou E, Goding JW, Takakusa H, Kikuchi K, Perrakis A, Nagano T, Moolenaar WH. Inhibition of autotaxin by lysophosphatidic acid and sphingosine 1-phosphate. J Biol Chem. 2005;280:21155–21161. doi: 10.1074/jbc.M413183200. [DOI] [PubMed] [Google Scholar]
- 382.Tsuda S, Okudaira S, Moriya-Ito K, Shimamoto C, Tanaka M, Aoki J, Arai H, Murakami-Murofushi K, Kobayashi T. Cyclic phosphatidic acid is produced by autotaxin in blood. J Biol Chem. 2006;281:26081–26088. doi: 10.1074/jbc.M602925200. [DOI] [PubMed] [Google Scholar]
- 383.Fujiwara Y. Cyclic phosphatidic acid—a unique bioactive phospholipid. Biochim Biophys Acta. 2008;1781:519–524. doi: 10.1016/j.bbalip.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Koh E, Clair T, Woodhouse EC, Schiffmann E, Liotta L, Stracke M. Site-directed mutations in the tumor-associated cytokine, autotaxin, eliminate nucleotide phosphodiesterase, lysophospholipase D, and motogenic activities. Cancer Res. 2003;63:2042–2045. [PubMed] [Google Scholar]
- 385.Hama K, Aoki J, Fukaya M, Kishi Y, Sakai T, Suzuki R. Lysophosphatidic acid and autotaxin stimulate cell motility of neoplastic and non-neoplastic cells through LPA(1) J Biol Chem. 2004;279:17634–17639. doi: 10.1074/jbc.M313927200. [DOI] [PubMed] [Google Scholar]
- 386.Kishi Y, Okudaira S, Tanaka M, Hama K, Shida D, Kitayama J, Yamori T, Aoki J, Fujimaki T, Arai H. Autotaxin is overexpressed in glioblastoma multiforme and contributes to cell motility of glioblastoma by converting lysophosphatidylcholine to lysophosphatidic acid. J Biol Chem. 2006;281:17492–17500. doi: 10.1074/jbc.M601803200. [DOI] [PubMed] [Google Scholar]
- 387.Luquain C, Sciorra VA, Morris AJ. Lysophosphatidic acid signaling: how a small lipid does big things. Trends Biochem Sci. 2003;28:377–383. doi: 10.1016/S0968-0004(03)00139-7. [DOI] [PubMed] [Google Scholar]
- 388.Sakagami H, Aoki J, Natori Y, Nishikawa K, Kakehi Y, Arai H. Biochemical and molecular characterization of a novel choline-specific glycerophosphodiester phosphodiesterase belonging to the nucleotide pyrophosphatase/phosphodiesterase family. J Biol Chem. 2005;280:23084–23093. doi: 10.1074/jbc.M413438200. [DOI] [PubMed] [Google Scholar]
- 389.Duan RD, Bergman T, Xu N, Wu J, Cheng Y, Duan JX, Nelander S, Palmberg C, Nilsson A. Identification of human intestinal alkaline sphingomyelinase as a novel ecto-enzyme related to the nucleotide phosphodiesterase family. J Biol Chem. 2003;278:38528–38536. doi: 10.1074/jbc.M305437200. [DOI] [PubMed] [Google Scholar]
- 390.Zhang Y, Cheng YJ, Hansen GH, Niels-Christiansen LL, Koentgen F, Ohlsson L, Nilsson A, Duan RD. Crucial role of alkaline sphingomyelinase in sphingomyelin digestion: a study on enzyme knockout mice. J Lipid Res. 2011;52:771–781. doi: 10.1194/jlr.M012880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Wu J, Nilsson A, Jonsson BA, Stenstad H, Agace W, Cheng Y, Duan RD. Intestinal alkaline sphingomyelinase hydrolyses and inactivates platelet-activating factor by a phospholipase C activity. Biochem J. 2006;394:299–308. doi: 10.1042/BJ20051121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Takahashi T, Old LJ, Boyse EA. Surface alloantigens of plasma cells. J Exp Med. 1970;131:1325–1341. doi: 10.1084/jem.131.6.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Goding JW, Shen FW. Structure of the murine plasma cell alloantigen PC-1: comparison with the receptor for transferring. J Immunol. 1982;129:2636–2640. [PubMed] [Google Scholar]
- 394.Driel IR, Wilks AF, Pietersz GA, Goding JW. Murine plasma cell membrane antigen PC-1: molecular cloning of cDNA and analysis of expression. Proc Natl Acad Sci USA. 1985;82:8619–8623. doi: 10.1073/pnas.82.24.8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Driel IR, Goding JW. Plasma cell membrane gylcoprotein PC-1: primary structure deduced from cDNA clones. J Biol Chem. 1987;262:4882–4887. [PubMed] [Google Scholar]
- 396.Buckley MF, Loveland KA, McKinstry WJ, Garson OM, Goding JW. Plasma membrane glycoprotein PC-1: cDNA cloning of the human molecule, amino acid sequence and chromosomal location. J Biol Chem. 1990;265:17506–17511. [PubMed] [Google Scholar]
- 397.Belli SI, Goding JW. Biochemical characterization of human PC-1, an enzyme possessing alkaline phosphodiesterase I and nucleotide pyrophosphate activities. Eur J Biochem. 1994;226:433–443. doi: 10.1111/j.1432-1033.1994.tb20068.x. [DOI] [PubMed] [Google Scholar]
- 398.Deissler H, Lottspeich F, Rajewsky MF. Affinity purification and cDNA cloning of rat neural differentiation and tumor cell surface antigen gp130RB13-6 reveals relationship to human and murine PC-1. J Biol Chem. 1995;270:9849–9855. doi: 10.1074/jbc.270.17.9849. [DOI] [PubMed] [Google Scholar]
- 399.Jin-Hua P, Goding JW, Nakamura H, Sano K. Molecular cloning and chromosomal localization of PD-Iβ (PDNP3), a new member of the human phosphodiesterase I genes. Genomics. 1997;45:412–415. doi: 10.1006/geno.1997.4949. [DOI] [PubMed] [Google Scholar]
- 400.Bello V, Goding JW, Greengrass V, Sali A, Dubljevic V, Lenoir C, Trugnan G, Maurice M. Characterization of a di-leucine-based signal in the cytoplasmic tail of the nucleotide-pyrophosphatase NPP1 that mediates basolateral targeting but not endocytosis. Mol Biol Cell. 2001;12:3004–3015. doi: 10.1091/mbc.12.10.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Delaunay JL, Breton M, Goding JW, Trugnan G, Maurice M. Differential detergent resistance of the apical and basolateral NPPases: relationship with polarized targeting. J Cell Sci. 2007;120:1009–1016. doi: 10.1242/jcs.002717. [DOI] [PubMed] [Google Scholar]
- 402.Stracke ML, Krutzsch HC, Unsworth EJ, Arestad AA, Cioce V, Schiffmann E, Liotta LA. Identification, purification, and partial sequence analysis of autotaxin, a novel motility-stimulating protein. J Biol Chem. 1992;267:2524–2529. [PubMed] [Google Scholar]
- 403.Murata J, Lee HJ, Clair T, Krutzsch HC, Arestad AA, Sobel ME, Liotta LA, Stracke ML. cDNA cloning of the human motility-stimulating protein, autotaxin, reveals a homology with phosphodiesterases. J Biol Chem. 1994;269:30479–30484. [PubMed] [Google Scholar]
- 404.Narita M, Goji J, Nakamura H, Sano K. Molecular cloning, expression, and localization of a brain-specific phosphodiesterase I/nucleotide pyrophosphatase (PD-Iα) from rat brain. J Biol Chem. 1994;269:28235–28242. [PubMed] [Google Scholar]
- 405.Kawagoe H, Soma O, Goji J, Nishimura N, Narita M, Inazawa J, Nakamura H, Sano K. Molecular cloning and chromosomal assignment of the human brain-type phosphodiesterase I/nucleotide pyrophosphatase gene (PDNP2) Genomics. 1995;30:380–384. doi: 10.1006/geno.1995.0036. [DOI] [PubMed] [Google Scholar]
- 406.Giganti A, Rodriguez M, Fould B, Moulharat N, Coge F, Chomarat P, Galizzi JP, Valet P, Saulnier-Blache JS, Boutin JA, Ferry G. Murine and human autotaxin α, β, and γ isoforms—gene organization, tissue distribution, and biochemical characterization. J Biol Chem. 2008;283:7776–7789. doi: 10.1074/jbc.M708705200. [DOI] [PubMed] [Google Scholar]
- 407.Lee HY, Murata J, Clair T, Polymeropoulos MH, Torres R, Manrow RE, Liotta LA, Stracke ML. Cloning chromosomal localization, and tissue expression of autotaxin from human tetracarcinoma cells. Biochem Biophys Res Commun. 1996;218:714–719. doi: 10.1006/bbrc.1996.0127. [DOI] [PubMed] [Google Scholar]
- 408.Kawagoe H, Stracke ML, Nakamura H, Sano K. Expression and transcriptional regulation of the PD-Iα/autotaxin gene in neuroblastoma. Cancer Res. 1997;57:2516–2521. [PubMed] [Google Scholar]
- 409.Jansen S, Stefan C, Creemers JWM, Waelkens E, VanEynde A, Stalmans W, Bollen M. Proteolytic maturation and activation of autotaxin (NPP2), a secreted metastasis-enhancing lysophospholipase D. J Cell Sci. 2005;118:3081–3089. doi: 10.1242/jcs.02438. [DOI] [PubMed] [Google Scholar]
- 410.Koike S, Keino-Masu K, Ohto T, Masu M. The N-terminal hydrophobic sequence of autotaxin (ENPP2) functions as a signal peptide. Genes Cells. 2006;11:133–142. doi: 10.1111/j.1365-2443.2006.00924.x. [DOI] [PubMed] [Google Scholar]
- 411.Pradere JP, Tarnus E, Gres S, Valet P, Saulnier-Blache JS. Secretion and lysophospholipase D activity of autotaxin by adipocytes are controlled by N-glycosylation and signal peptidase. BBA Mol Cell Biol Lipids. 2007;1771:93–102. doi: 10.1016/j.bbalip.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 412.Gijsbers R, Ceulemans H, Bollen M. Functional characterization of the non-catalytic ectodomains of the nucleotide pyrophosphatase/phosphodiesterase NPP1. Biochem J. 2003;371:321–330. doi: 10.1042/BJ20021943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Jansen S, Andries M, Derua R, Waelkens E, Bollen M. Domain interplay mediated by an essential disulfide linkage is critical for the activity and secretion of the metastasis-promoting enzyme autotaxin. J Biol Chem. 2009;284:14296–14302. doi: 10.1074/jbc.M900790200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Belli SI, Sali A, Goding JW. Divalent cations stabilize the conformation of plasma cell membrane glycoprotein PC-1 (alkaline phosphodiesterase I) Biochem J. 1994;304:75–80. doi: 10.1042/bj3040075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Andoh K, JinHua P, Terashima K, Nakamura H, Sano K. Genomic structure and promoter analysis of the ecto-phosphodiesterase I gene (PDNP3) expressed in glial cells. BBA Gene Struct Express. 1999;1446:213–224. doi: 10.1016/S0167-4781(99)00090-1. [DOI] [PubMed] [Google Scholar]
- 416.Lee J, Jung ID, Nam SW, Clair T, Jeong EM, Hong SY, Han JW, Lee HW, Stracke ML, Lee HY. Enzymatic activation of autotaxin by divalent cations without EF-hand loop region involvement. Biochem Pharmacol. 2001;62:219–224. doi: 10.1016/S0006-2952(01)00658-X. [DOI] [PubMed] [Google Scholar]
- 417.Belli SI, Driel IR, Goding JW. Identification and characterization of soluble form of the plasma cell membrane glycoprotein PC-1 (5′-nucleotide phosphodiesterase) Eur J Biochem. 1993;217:421–428. doi: 10.1111/j.1432-1033.1993.tb18261.x. [DOI] [PubMed] [Google Scholar]
- 418.Stracke ML, Arestad A, Levine M, Krutzsch HC, Liotta LA. Autotaxin is an N-linked glycoprotein but the sugar moieties are not needed for its stimulation of cellular motility. Melanoma Res. 1995;5:203–209. doi: 10.1097/00008390-199508000-00001. [DOI] [PubMed] [Google Scholar]
- 419.Meerson NR, Bello V, Delaunay JL, Slimane TA, Delautier D, Lenoir C, Trugnan G, Maurice M. Intracellular traffic of the ecto-nucleotide pyrophosphatase/phosphodiesterase NPP3 to the epical plasma membrane of MDCK and Caco-2 cells: apical targeting occurs in the absence of N-glycosylation. J Cell Sci. 2000;113:4193–4202. doi: 10.1242/jcs.113.23.4193. [DOI] [PubMed] [Google Scholar]
- 420.Jansen S, Callewaert N, Dewerte I, Andries M, Ceulemans H, Bollen M. An essential oligomannosidic glycan chain in the catalytic domain of autotaxin, a secreted lysophospholipase-D. J Biol Chem. 2007;282:11084–11091. doi: 10.1074/jbc.M611503200. [DOI] [PubMed] [Google Scholar]
- 421.Ohe Y, Ohnishi H, Okazawa H, Tomizawa K, Kobayashi H, Okawa K, Matozaki T. Characterization of nucleotide pyrophosphatase-5 as an oligomannosidic glycoprotein in rat brain. Biochem Biophys Res Commun. 2003;308:719–725. doi: 10.1016/S0006-291X(03)01454-2. [DOI] [PubMed] [Google Scholar]
- 422.Meerson NR, Delautier D, Durand-Schneider AM, Moreau A, Schilsky ML, Sternlieb I, Feldmann G, Maurice M. Identification of B10, an alkaline phosphodiesterase of the apical plasma membrane of hepatocytes and biliary cells, in rat serum: increased levels following bile duct ligation and during the development of cholangiocarcinoma. Hepatology. 1998;27:563–568. doi: 10.1002/hep.510270234. [DOI] [PubMed] [Google Scholar]
- 423.Wu J, Liu F, Nilsson A, Duan RD. Pancreatic trypsin cleaves intestinal alkaline sphingomyelinase from mucosa and enhances the sphingomyelinase activity. Am J Physiol Gastrointest Liver Physiol. 2004;287:G967–G973. doi: 10.1152/ajpgi.00190.2004. [DOI] [PubMed] [Google Scholar]
- 424.Yegutkin GG, Samburski SS, Jalkanen S. Soluble purine-converting enzymes circulate in human blood and regulate extracellular ATP level via counteracting pyrophosphatase and phosphotransfer reactions. FASEB J. 2003;17:U15–U34. doi: 10.1096/fj.02-1136fje. [DOI] [PubMed] [Google Scholar]
- 425.Laketa D, Bjelobaba I, Savic J, Lavrnja I, Stojiljkovic M, Rakic L, Nedeljkovic N. Biochemical characterization of soluble nucleotide pyrophosphatase/phosphodiesterase activity in rat serum. Mol Cell Biochem. 2010;339:99–106. doi: 10.1007/s11010-009-0373-1. [DOI] [PubMed] [Google Scholar]
- 426.Nakabayashi T, Ikezawa H. Alkaline phosphodiesterase I release from eucaryotic plasma membranes by phophatidylinositol-specific phospholipase C. I. The release from rat organs. J Biochem. 1986;99:703–712. doi: 10.1093/oxfordjournals.jbchem.a135529. [DOI] [PubMed] [Google Scholar]
- 427.Nakabayashi T, Ikezawa H. Alkaline phosphodiesterase I release from eucaryotic plasma membranes by phosphatidylinositol-specific phospholipase C. II. The release from brush border membranes of porcine intestine. Toxicon. 1986;24:975–984. doi: 10.1016/0041-0101(86)90003-6. [DOI] [PubMed] [Google Scholar]
- 428.Nakabayashi T, Matsuoka Y, Ikezawa H, Kimura Y. Alkaline phosphodiesterase I release from eucaryotic plasma membranes by phosphatidylinositol-specific phospholipase C—IV. The release from Cacia porcellus organs. Int J Biochem. 1994;26:171–179. doi: 10.1016/0020-711X(94)90142-2. [DOI] [PubMed] [Google Scholar]
- 429.Itami C, Taguchi R, Ikezawa H, Nakabayashi T. Release of ectoenzymes from small intestine brush border membranes of mice by phospholipases. Biosci Biotechnol Biochem. 1997;61:336–340. doi: 10.1271/bbb.61.336. [DOI] [PubMed] [Google Scholar]
- 430.Pamuklar Z, Federico L, Liu SY, Umezu-Goto M, Dong AP, Panchatcharam M, Fulerson Z, Berdyshev E, Natarajan V, Fang XJ, Meeteren LA, Moolenaar WH, Mills GB, Morris AJ, Smyth SS. Autotaxin/lysopholipase D and lysophosphatidic acid regulate murine hemostasis and thrombosis. J Biol Chem. 2009;284:7385–7394. doi: 10.1074/jbc.M807820200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Hausmann J, Kamtekar S, Christodoulou E, Day JE, Wu T, Fulkerson Z, Albers HM, Meeteren LA, Houben AJ, Zeijl L, Jansen S, Andries M, Hall T, Pegg LE, Benson TE, Kasiem M, Harlos K, Kooi CW, Smyth SS, Ovaa H, Bollen M, Morris AJ, Moolenaar WH, Perrakis A. Structural basis of substrate discrimination and integrin binding by autotaxin. Nat Struct Mol Biol. 2011;18:198–U262. doi: 10.1038/nsmb.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Fox MA, Colello RJ, Macklin WB, Fuss B. Phosphodiesterase-I α/autotaxin: a counteradhesive protein expressed by oligodendrocytes during onset of myelination. Mol Cell Neurosci. 2003;23:507–519. doi: 10.1016/S1044-7431(03)00073-3. [DOI] [PubMed] [Google Scholar]
- 433.Fox MA, Alexander JK, Afshari FS, Colello RJ, Fuss B. Phosphodieterase-I α/autotaxin and IFAK phosphorylation during controls cytoskeletal organization myelination. Mol Cell Neurosci. 2004;27:140–150. doi: 10.1016/j.mcn.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 434.Dennis J, White MA, Forrest AD, Yuelling LM, Nogaroli L, Afshari FS, Fox MA, Fuss B. Phosphodiesterase-I alpha/autotaxin’s MORFO domain regulates oligodendroglial process network formation and focal adhesion organization. Mol Cell Neurosci. 2008;37:412–424. doi: 10.1016/j.mcn.2007.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Grobben B, Claes P, Roymans D, Esmans EL, Onckelen H, Slegers H. Ecto-nucleotide pyrophosphatase modulates the purinoceptor-mediated signal transduction and is inhibited by purinoceptor antagonists. Brit J Pharmacol. 2000;130:139–145. doi: 10.1038/sj.bjp.0703289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Delicado EG, Miras-Portugal MT, Carrasquero LMG, León D, Pérez-Sen R, Gualix J. Dinucleoside polyphosphates and their interaction with other nucleotide signaling pathways. Pflugers Arch Eur J Physiol. 2006;452:563–572. doi: 10.1007/s00424-006-0066-5. [DOI] [PubMed] [Google Scholar]
- 437.Hoyle CHV, Hilderman RH, Pintor JJ, Schlüter H, King BF. Diadenosine polyphosphates as extracellular signal molecules. Drug Develop Res. 2001;52:260–273. doi: 10.1002/ddr.1123. [DOI] [Google Scholar]
- 438.Massé K, Bhamra S, Allsop G, Dale N, Jones EA. Ectophosphodiesterase/nucleotide phosphohydrolase (Enpp) nucleotidases: cloning, conservation and developmental restriction. Int J Dev Biol. 2010;54:181–193. doi: 10.1387/ijdb.092879km. [DOI] [PubMed] [Google Scholar]
- 439.Nakanaga K, Hama K, Aoki J. Autotaxin—an LPA producing enzyme with diverse functions. J Biochem. 2010;148:13–24. doi: 10.1093/jb/mvq052. [DOI] [PubMed] [Google Scholar]
- 440.Laidlaw SM, Anwar MA, Thomas W, Green P, Shaw K, Skinner MA. Fowlpox virus encodes nonessential homologs of cellular alpha-SNAP, PC-1, and an orphan human homolog of a secreted nematode protein. J Virol. 1998;72:6742–6751. doi: 10.1128/jvi.72.8.6742-6751.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Zalatan JG, Fenn TD, Brunger AT, Herschlag D. Structural and functional comparisons of nucleotide pyrophosphatase/phosphodiesterase and alkaline phosphatase: implications for mechanism and evolution. Biochemistry USA. 2006;45:9788–9803. doi: 10.1021/bi060847t. [DOI] [PubMed] [Google Scholar]
- 442.Zalatan JG, Fenn TD, Herschlag D. Comparative enzymology in the alkaline phosphatase superfamily to determine the catalytic role of an active-site metal ion. J Mol Biol. 2008;384:1174–1189. doi: 10.1016/j.jmb.2008.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Galperin MY, Bairoch A, Koonin EV. A superfamily of metalloenzymes unifies phosphopentomutase and cofactor-independent phosphoglycerate mutase with alkaline phosphatases and sulfatases. Protein Sci. 1998;7:1829–1835. doi: 10.1002/pro.5560070819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Galperin MY, Jedrzejas MJ. Conserved core structure and active site residues in alkaline phosphatase superfamily enzymes. Protein Struct Funct Genet. 2001;45:318–324. doi: 10.1002/prot.1152. [DOI] [PubMed] [Google Scholar]
- 445.Nishimasu H, Okudaira S, Hama K, Mihara E, Dohmae N, Inoue A, Ishitani R, Takagi J, Aoki J, Nureki O. Crystal structure of autotaxin and insight into GPCR activation by lipid mediators. Nat Struct Mol Biol. 2011;18:205–U271. doi: 10.1038/nsmb.1998. [DOI] [PubMed] [Google Scholar]
- 446.Du MH, Stigbrand T, Taussig MJ, Menez A, Stura EA. Crystal structure of alkaline phosphatase from human placenta at 1.8 A resolution—implication for a substrate specificity. J Biol Chem. 2001;276:9158–9165. doi: 10.1074/jbc.M009250200. [DOI] [PubMed] [Google Scholar]
- 447.Hoylaerts MF, Ding L, Narisawa S, Kerckhoven S, Millán JL. Mammalian alkaline phosphatase catalysis requires active site structure stabilization via the N-terminal amino acid microenvironment. Biochemistry USA. 2006;45:9756–9766. doi: 10.1021/bi052471+. [DOI] [PubMed] [Google Scholar]
- 448.Kim A, Benning MM, OkLee S, Quinn J, Martin BM, Holden HM, Dunaway-Mariano D. Divergence of chemical function in the alkaline phosphatase superfamily: structure and mechanism of the P-C bond cleaving enzyme phosphonoacetate hydrolases. Biochemistry USA. 2011;50:3481–3494. doi: 10.1021/bi200165h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Inoue K, Tanaka N, Haga A, Yamasaki K, Umeda T, Kusakabe Y, Sakamoto Y, Nonaka T, Deyashiki Y, Nakamura KT. Crystallization and preliminary X-ray crystallographic analysis of human autotaxin. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011;67:450–453. doi: 10.1107/S174430911005311X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Kamikubo Y, De GR, Kroon G, Curriden S, Neels JG, Churchill MJ, Dawson P, Oldziej S, Jagielska A, Scheraga HA, Loskutoff DJ, Dyson HJ. Disulfide bonding arrangements in active forms of the somatomedin B domain of human vitronectin. Biochemistry USA. 2004;43:6519–6534. doi: 10.1021/bi049647c. [DOI] [PubMed] [Google Scholar]
- 451.Kanda H, Newton R, Klein R, Morita Y, Gunn MD, Rosen SD. Autotaxin, an ectoenzyme that produces lysophosphatidic acid, promotes the entry of lymphocytes into secondary lymphoid organs. Nat Immunol. 2008;9:415–423. doi: 10.1038/ni1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Culp JS, Blytt HJ, Hermodson M, Butler LG. Amino acid sequence of the active site peptide of bovine intestinal 5′-nucleotide phosphodiesterase and identification of the active site residue as threonine. J Biol Chem. 1985;260:8320–8324. [PubMed] [Google Scholar]
- 453.Stefan C, Stalmans W, Bollen M. Threonine autophosphorylation and nucleotidylation of the hepatic membrane protein PC-1. Eur J Biochem. 1996;241:338–342. doi: 10.1111/j.1432-1033.1996.00338.x. [DOI] [PubMed] [Google Scholar]
- 454.Joye IJ, Belien T, Brijs K, Soetaert W, Delcour JA. Mutational analysis of wheat (Triticum aestivum L.) nucleotide pyrophosphatase/phosphodiesterase shows the role of six amino acids in the catalytic mechanism. Appl Microbiol Biotechnol. 2011;90:173–180. doi: 10.1007/s00253-010-2962-z. [DOI] [PubMed] [Google Scholar]
- 455.Eichholz A, Crane RK. Isolation of plasma membranes from intestinal brush borders. Methods Enzymol. 1974;31:123–134. doi: 10.1016/0076-6879(74)31012-9. [DOI] [PubMed] [Google Scholar]
- 456.Giocondi MC, Seantier B, Dosset P, Milhiet PE, Le GC. Characterizing the interactions between GPI-anchored alkaline phosphatases and membrane domains by AFM. Pflugers Arch. 2008;456:179–188. doi: 10.1007/s00424-007-0409-x. [DOI] [PubMed] [Google Scholar]
- 457.Gouridis G, Karamanou S, Koukaki M, Economou A. In vitro assays to analyze translocation of the model secretory preprotein alkaline phosphatase. Methods Mol Biol. 2010;619:157–172. doi: 10.1007/978-1-60327-412-8_10. [DOI] [PubMed] [Google Scholar]
- 458.Coleman JE. Structure and mechanism of alkaline phosphatase. Ann Rev Biophys Biomol Struct. 1992;21:441–483. doi: 10.1146/annurev.bb.21.060192.002301. [DOI] [PubMed] [Google Scholar]
- 459.McComb RB, Bowers GN, Posen S. Alkaline phosphatase. New York: Plenum; 1979. [Google Scholar]
- 460.Millán JL. Alkaline phosphatase as a reporter of cancerous transformation. Clin Chim Acta. 1992;209:123–129. doi: 10.1016/0009-8981(92)90343-O. [DOI] [PubMed] [Google Scholar]
- 461.Epstein E, Kiechle FL, Artiss JD, Zak B. The clinical use of alkaline phosphatase enzymes. Clin Lab Med. 1986;6:491–505. [PubMed] [Google Scholar]
- 462.Shirley DG, Vekaria RM, Sévigny J. Ectonucleotidases in the kidney. Purinergic Signal. 2009;5:501–511. doi: 10.1007/s11302-009-9152-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Lynes MD, Widmaier EP. Involvement of CD36 and intestinal alkaline phosphatases in fatty acid transport in enterocytes, and the response to a high-fat diet. Life Sci. 2011;88:384–391. doi: 10.1016/j.lfs.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 464.Picher M, Burch LH, Hirsh AJ, Spychala J, Boucher RC. Ecto-5′-nucleotidase and nonspecific alkaline phosphatase—two AMP-hydrolyzing ectoenzymes with distinct roles in human airways. J Biol Chem. 2003;278:13468–13479. doi: 10.1074/jbc.M300569200. [DOI] [PubMed] [Google Scholar]
- 465.Pease S, Braghetta P, Gearing D, Grail D, Williams RL. Isolation of embryonic stem (ES) cells in media supplemented with recombinant leukemia inhibitory factor (LIF) Dev Biol. 1990;141:344–352. doi: 10.1016/0012-1606(90)90390-5. [DOI] [PubMed] [Google Scholar]
- 466.Heath JK. Characterization of a xenogeneic antiserum raised against the fetal germ cells of the mouse: cross-reactivity with embryonal carcinoma cells. Cell. 1978;15:299–306. doi: 10.1016/0092-8674(78)90105-8. [DOI] [PubMed] [Google Scholar]
- 467.Langer D, Ikehara Y, Takebayashi H, Hawkes R, Zimmermann H. The ectonucleotidases alkaline phosphatase and nucleoside triphosphate diphosphohydrolase 2 are associated with subsets of progenitor cell populations in the mouse embryonic, postnatal and adult neurogenic zones. Neuroscience. 2007;150:863–879. doi: 10.1016/j.neuroscience.2007.07.064. [DOI] [PubMed] [Google Scholar]
- 468.Fonta C, Negyessy L, Renaud L, Barone P. Postnatal development of alkaline phosphatase activity correlates with the maturation of neurotransmission in the cerebral cortex. J Comp Neurol. 2005;486:179–196. doi: 10.1002/cne.20524. [DOI] [PubMed] [Google Scholar]
- 469.Negyessy L, Xiao J, Kantor O, Kovacs GG, Palkovits M, Doczi TP, Renaud L, Baksa G, Glasz T, Ashaber M, Barone P, Fonta C. Layer-specific activity of tissue non-specific alkaline phosphatase in the human neocortex. Neuroscience. 2011;172:406–418. doi: 10.1016/j.neuroscience.2010.10.049. [DOI] [PubMed] [Google Scholar]
- 470.Iida M, Ihara S, Matsuzaki T. Hair cycle-dependent changes of alkaline phosphatase activity in the mesenchyme and epithelium in mouse vibrissal follicles. Develop Growth Differ. 2007;49:185–195. doi: 10.1111/j.1440-169X.2007.00907.x. [DOI] [PubMed] [Google Scholar]
- 471.Aldag I, Bockau U, Rossdorf J, Laarmann S, Raaben W, Herrmann L, Weide T, Hartmann MW. Expression, secretion and surface display of a human alkaline phosphatase by the ciliate Tetrahymena thermophila. BMC Biotechnol. 2011;11:11. doi: 10.1186/1472-6750-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Narisawa S, Hoylaerts MF, Doctor KS, Fukuda MN, Alpers DH, Millán JL. A novel phosphatase upregulated in Akp3 knockout mice. Amer J Physiol Gastrointest L. 2007;293:G1068–G1077. doi: 10.1152/ajpgi.00073.2007. [DOI] [PubMed] [Google Scholar]
- 473.Millán JL. Mammalian alkaline phosphatase. From biology to applications in medicine and biotechnology. Weinheim: Wiley; 2006. [Google Scholar]
- 474.Millán JL. Alkaline phosphatases: structure, substrate specificity and functional relatedness to other members of a large superfamily of enzymes. Purinergic Signal. 2006;2:335–341. doi: 10.1007/s11302-005-5435-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Du MH, Millán JL. Structural evidence of functional divergence in human alkaline phosphatases. J Biol Chem. 2002;277:49808–49814. doi: 10.1074/jbc.M207394200. [DOI] [PubMed] [Google Scholar]
- 476.Hoylaerts MF, Manes T, Millán JL. Mammalian alkaline phosphatases are allosteric enzymes. J Biol Chem. 1997;272:22781–22787. doi: 10.1074/jbc.272.36.22781. [DOI] [PubMed] [Google Scholar]
- 477.Saslowsky DE, Lawrence J, Ren XY, Brown DA, Henderson RM, Edwardson JM. Placental alkaline phosphatase is efficiently targeted to rafts in supported lipid bilayers. J Biol Chem. 2002;277:26966–26970. doi: 10.1074/jbc.M204669200. [DOI] [PubMed] [Google Scholar]
- 478.Suzuki T, Ishihara K, Migaki H, Matsuura W, Kohda A, Okumura K, Nagao M, Yamaguchi-Iwai Y, Kambe T. Zinc transporters, ZnT5 and ZnT7, are required for the activation of alkaline phosphatases, zinc-requiring enzymes that are glycosylphosphatidylinositol-anchored to the cytoplasmic membrane. J Biol Chem. 2005;280:637–643. doi: 10.1074/jbc.M411247200. [DOI] [PubMed] [Google Scholar]
- 479.Suzuki T, Ishihara K, Migaki H, Ishihara K, Nagao M, Yamaguchi-Iwai Y, Kambe T. Two different zinc transport complexes of cation diffusion facilitator proteins localized in the secretory pathway operate to activate alkaline phosphatases in vertebrate cells. J Biol Chem. 2005;280:30956–30962. doi: 10.1074/jbc.M506902200. [DOI] [PubMed] [Google Scholar]
- 480.Fukunaka A, Kurokawa Y, Teranishi F, Sekler I, Oda K, Ackland ML, Faundez V, Hiromura M, Masuda S, Nagao M, Enomoto S, Kambe T. Tissue nonspecific alkaline phosphatase is activated via a two-step mechanism by zinc transport complexes in the early secretory pathway. J Biol Chem. 2011;286:16363–16373. doi: 10.1074/jbc.M111.227173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Ishihara K, Yamazaki T, Ishida Y, Suzuki T, Oda K, Nagao M, Yamaguchi-Iwai Y, Kambe T. Zinc transport complexes contribute to the homeostatic maintenance of secretory pathway function in vertebrate cells. J Biol Chem. 2006;281:17743–17750. doi: 10.1074/jbc.M602470200. [DOI] [PubMed] [Google Scholar]
- 482.O'Brien PJ, Herschlag D. Functional interrelationships in the alkaline phosphatase superfamily: phosphodiesterase activity of Escherichia coli alkaline phosphatase. Biochemistry USA. 2001;40:5691–5699. doi: 10.1021/bi0028892. [DOI] [PubMed] [Google Scholar]
- 483.Rezende AA, Pizauro JM, Ciancaglini P, Leone FA. Phosphodiesterase activity is a novel property of alkaline phosphatase from osseous plate. Biochem J. 1994;301:517–522. doi: 10.1042/bj3010517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Jiang T, Xing B, Rao J. Recent developments of biological reporter technology for detecting gene expression. Biotechnol Genet Eng Rev. 2008;25:41–75. doi: 10.5661/bger-25-41. [DOI] [PubMed] [Google Scholar]
- 485.Whyte PP. Hypophosphatasia: natures window on alkaline phosphatase function in man. In: Bilezkian J, Raisz L, Rodan G, editors. Principles of bone biology. San Diego: Academic; 1996. pp. 951–968. [Google Scholar]
- 486.Mornet E. Hypophosphatasia. Orphanet J Rare Dis. 2007;2:40. doi: 10.1186/1750-1172-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Whyte MP. Physiological role of alkaline phosphatase explored in hypophosphatasia. Ann N Y Acad Sci. 2010;1192:190–200. doi: 10.1111/j.1749-6632.2010.05387.x. [DOI] [PubMed] [Google Scholar]
- 488.MacGregor GR, Zambrowicz BP, Soriano P. Tissue non-specific alkaline phosphatase is expressed in both embryonic and extraembryonic lineages during mouse embryogenesis but is not required for migration of primordial germ cells. Development. 1995;121:1487–1496. doi: 10.1242/dev.121.5.1487. [DOI] [PubMed] [Google Scholar]
- 489.Waymire JC, Mahuren JD, Jaje JM, Guilarte T, Coburn SP, MacGregor GR. Mice lacking tissue non-specific alkaline phosphatase die from seizures due to defective metabolism of vitamin B-6. Nature Genet. 1995;11:45–51. doi: 10.1038/ng0995-45. [DOI] [PubMed] [Google Scholar]
- 490.Narisawa S, Fröhlander N, Millán JL. Inactivation of two mouse alkaline phosphatase genes and establishment of a model of infantile hypophosphatasia. Develop Dynam. 1997;208:432–446. doi: 10.1002/(SICI)1097-0177(199703)208:3<432::AID-AJA13>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 491.Anderson HC, Sipe JB, Hessle L, Dhamyamraju R, Atti E, Camacho NP, Millán JL. Impaired calcification around matrix vesicles of growth plate and bone in alkaline phosphatase-deficient mice. Amer J Pathol. 2004;164:841–847. doi: 10.1016/S0002-9440(10)63172-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Fedde KN, Blair L, Silverstein J, Coburn SP, Ryan LM, Weinstein RS, Waymire K, Narisawa S, Millán JL, MacGregor GR, Whyte MP. Alkaline phosphatase knock-out mice recapitulate the metabolic and skeletal defects of infantile hypophosphatasia. J Bone Miner Res. 1999;14:2015–2026. doi: 10.1359/jbmr.1999.14.12.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493.Millán JL, Narisawa S, Lemire I, Loisel TP, Boileau G, Leonard P, Gramatikova S, Terkeltaub R, Camacho NP, Mckee MD, Crine P, Whyte MP. Enzyme replacement therapy for murine hypophosphatasia. J Bone Miner Res. 2008;23:777–787. doi: 10.1359/jbmr.071213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 494.Yadav MC, Lemire I, Leonard P, Boileau G, Blond L, Beliveau M, Cory E, Sah RL, Whyte MP, Crine P, Millán JL. Dose response of bone-targeted enzyme replacement for murine hypophosphatasia. Bone. 2011;49:250–256. doi: 10.1016/j.bone.2011.03.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 495.Yamamoto S, Orimo H, Matsumoto T, Iijima O, Narisawa S, Maeda T, Millán JL, Shimada T. Prolonged survival and phenotypic correction of Akp2(−/−) hypophosphatasia mice by lentiviral gene therapy. J Bone Miner Res. 2011;26:135–142. doi: 10.1002/jbmr.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496.Smith GP, Peters TJ. Subcellular localization and properties of pyridoxal phosphate phosphatases of human polymorphonuclear leukocytes and their relationship to acid and alkaline phosphatase. Biochim Biophys Acta. 1981;661:287–294. doi: 10.1016/0005-2744(81)90017-6. [DOI] [PubMed] [Google Scholar]
- 497.Wilson PD, Smith GP, Peters TJ. Pyridoxal 5′-phosphate: a possible physiological substrate for alkaline phosphatase in human neutrophils. Histochem J. 1983;15:257–264. doi: 10.1007/BF01006240. [DOI] [PubMed] [Google Scholar]
- 498.Whyte MP, Landt M, Ryan LM, Mulivor RA, Henthorn PS, Fedde KN, Mahuren JD, Coburn SP. Alkaline phosphatase: placental and tissue-nonspecific isoenzymes hydrolyze phosphoethanolamine, inorganic pyrophosphate, and pyridoxal 5′-phosphate—substrate accumulation in carriers of hypophosphatasia corrects during pregnancy. J Clin Invest. 1995;95:1440–1445. doi: 10.1172/JCI117814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 499.Narisawa S, Wennberg C, Millán JL. Abnormal vitamin B6 metabolism in alkaline phosphatase knock-out mice causes multiple abnormalities, but not the impaired bone mineralization. J Pathol. 2001;193:125–133. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH722>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 500.Poelstra K, Bakker WW, Klok PA, Kamps JA, Hardonk MJ, Meijer DK. Dephosphorylation of endotoxin by alkaline phosphatase in vivo. Am J Pathol. 1997;151:1163–1169. [PMC free article] [PubMed] [Google Scholar]
- 501.Bates JM, Akerlund J, Mittge E, Guillemin K. Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host Microbe. 2007;2:371–382. doi: 10.1016/j.chom.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Geddes K, Philpott DJ. A new role for intestinal alkaline phosphatase in gut barrier maintenance. Gastroenterology. 2008;135:8–12. doi: 10.1053/j.gastro.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 503.Goldberg RF, Austen WG, Jr, Zhang X, Munene G, Mostafa G, Biswas S, McCormack M, Eberlin KR, Nguyen JT, Tatlidede HS, Warren HS, Narisawa S, Millán JL, Hodin RA. Intestinal alkaline phosphatase is a gut mucosal defense factor maintained by enteral nutrition. Proc Natl Acad Sci USA. 2008;105:3551–3556. doi: 10.1073/pnas.0712140105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Lalles JP. Intestinal alkaline phosphatase: multiple biological roles in maintenance of intestinal homeostasis and modulation by diet. Nutr Rev. 2010;68:323–332. doi: 10.1111/j.1753-4887.2010.00292.x. [DOI] [PubMed] [Google Scholar]
- 505.Malo MS, Alam SN, Mostafa G, Zeller SJ, Johnson PV, Mohammad N, Chen KT, Moss AK, Ramasamy S, Faruqui A, Hodin S, Malo PS, Ebrahimi F, Biswas B, Narisawa S, Millán JL, Warren HS, Kaplan JB, Kitts CL, Hohmann EL, Hodin RA. Intestinal alkaline phosphatase preserves the normal homeostasis of gut microbiota. Gut. 2010;59:1476–1484. doi: 10.1136/gut.2010.211706. [DOI] [PubMed] [Google Scholar]
- 506.Chen KT, Malo MS, Beasley-Topliffe LK, Poelstra K, Millán JL, Mostafa G, Alam SN, Ramasamy S, Warren HS, Hohmann EL, Hodin RA. A role for intestinal alkaline phosphatase in the maintenance of local gut immunity. Dig Dis Sci. 2011;56:1020–1027. doi: 10.1007/s10620-010-1396-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Tuin A, Huizinga-Van der Vlag A, Loenen-Weemaes AM, Meijer DK, Poelstra K. On the role and fate of LPS-dephosphorylating activity in the rat liver. Am J Physiol Gastrointest Liver Physiol. 2006;290:G377–G385. doi: 10.1152/ajpgi.00147.2005. [DOI] [PubMed] [Google Scholar]
- 508.Swarup G, Cohen S, Garbers DL. Selective dephosphorylation of proteins containing phosphotyrosine by alkaline phosphatases. J Biol Chem. 1981;256:8197–8201. [PubMed] [Google Scholar]
- 509.Bütikofer P, Vassella E, Ruepp S, Boschung M, Civenni G, Seebeck T, Hemphill A, Mookherjee N, Pearson TW, Roditi I. Phosphorylation of a major GPI-anchored surface protein of Trypanosoma brucei during transport to the plasma membrane. J Cell Sci. 1999;112:1785–1795. doi: 10.1242/jcs.112.11.1785. [DOI] [PubMed] [Google Scholar]
- 510.Scheibe RJ, Kuehl H, Krautwald S, Meissner JD, Mueller WH. Ecto-alkaline phosphatase activity identified at physiological pH range on intact P19 and HL-60 cells is induced by retinoic acid. J Cell Biochem. 2000;76:420–436. doi: 10.1002/(SICI)1097-4644(20000301)76:3<420::AID-JCB10>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 511.Ermonval M, Baudry A, Baychelier F, Pradines E, Pietri M, Oda K, Schneider B, Mouillet-Richard S, Launay JM, Kellermann O. The cellular prion protein interacts with the tissue non-specific alkaline phosphatase in membrane microdomains of bioaminergic neuronal cells. PLoS One. 2009;4:e6497. doi: 10.1371/journal.pone.0006497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Kanshin E, Wang S, Ashmarina L, Fedjaev M, Nifant'ev I, Mitchell GA, Pshezhetsky AV. The stoichiometry of protein phosphorylation in adipocyte lipid droplets: analysis by N-terminal isotope tagging and enzymatic dephosphorylation. Proteomics. 2009;9:5067–5077. doi: 10.1002/pmic.200800861. [DOI] [PubMed] [Google Scholar]
- 513.Díaz-Hernández M, Gómez-Ramos A, Rubio A, Gómez-Villafuertes R, Naranjo JR, Miras-Portugal MT, Avila J. Tissue-nonspecific alkaline phosphatase promotes the neurotoxicity effect of extracellular tau. J Biol Chem. 2010;285:32539–32548. doi: 10.1074/jbc.M110.145003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Say JC, Ciuffi K, Furriel RP, Ciancaglini P, Leone FA. Alkaline phosphatase from rat osseous plates: purification and biochemical characterization of a soluble form. Biochim Biophys Acta. 1991;1074:256–262. doi: 10.1016/0304-4165(91)90161-9. [DOI] [PubMed] [Google Scholar]
- 515.Ohkubo S, Kimura J, Matsuoka I. Ecto-alkaline phosphatase in NG108-15 cells: a key enzyme mediating P1 antagonist-sensitive ATP response. Brit J Pharmacol. 2000;131:1667–1672. doi: 10.1038/sj.bjp.0703750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.Picher M, Burch LH, Boucher RC. Metabolism of P2 receptor agonists in human airways—implications for mucociliary clearance and cystic fibrosis. J Biol Chem. 2004;279:20234–20241. doi: 10.1074/jbc.M400305200. [DOI] [PubMed] [Google Scholar]
- 517.Díez-Zaera M, Díaz-Hernández JI, Hernández-Álvarez E, Zimmermann H, Díaz-Hernández M, Miras-Portugal MT. Tissue-nonspecific alkaline phosphatase promotes axonal growth of hippocampal neurons. Mol Biol Cell. 2011;22:1014–1024. doi: 10.1091/mbc.E10-09-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 518.Pizauro JM, Demenis MA, Ciancaglini P, Leone FA. Kinetic characterization of a membrane-specific ATPase from rat osseous plate and its possible significance on endochodral ossification. Biochim Biophys Acta. 1998;1368:108–114. doi: 10.1016/S0005-2736(97)00174-0. [DOI] [PubMed] [Google Scholar]
- 519.Pizauro JM, Ciancaglini P, Leone FA. Allosteric modulation by ATP, calcium and magnesium ions of rat osseous plate alkaline phosphatase. Biochim Biophys Acta. 1993;1202:22–28. doi: 10.1016/0167-4838(93)90058-Y. [DOI] [PubMed] [Google Scholar]
- 520.Demenis MA, Leone FA. Kinetic characteristics of ATP hydrolysis by a detergent-solubilized alkaline phosphatase from rat osseous plate. IUBMB Life. 2000;49:113–119. doi: 10.1080/15216540050022421. [DOI] [PubMed] [Google Scholar]
- 521.Simão AMS, Beloti MM, Cezarino RM, Rosa AL, Pizauro JM, Ciancaglini P. Membrane-bound alkaline phosphatase from ectopic mineralization and rat bone marrow cell culture. Comp Biochem Physiol Pt A. 2007;146:679–687. doi: 10.1016/j.cbpa.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 522.Kam W, Clauser EM, Kim YS, Kan YW, Rutter WJ. Cloning, sequencing, and chromosomal localization of human term placental alkaline phosphatase cDNA. Proc Natl Acad Sci USA. 1985;82:8715–8719. doi: 10.1073/pnas.82.24.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523.Millán JL. Molecular cloning and sequence analysis of human placental alkaline phosphatase. J Biol Chem. 1986;261:3112–3115. [PubMed] [Google Scholar]
- 524.Henthorn PS, Knoll BJ, Raducha M, Rothblum KN, Slaughter C, Weiss M, Lafferty MA, Fischer T, Harris H. Products of two common alleles at the locus for human placental alkaline phosphatase differ by seven amino acids. Proc Natl Acad Sci USA. 1986;83:5597–5601. doi: 10.1073/pnas.83.15.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 525.Berger J, Garattini E, Hua JC, Udenfriend S. Cloning and sequencing of human intestinal alkaline phosphatase cDNA. Proc Natl Acad Sci USA. 1987;84:695–698. doi: 10.1073/pnas.84.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 526.Henthorn PS, Raducha M, Edwards YH, Weiss MJ, Slaughter C, Lafferty MA, Harris H. Nucleotide and amino acid sequences of human intestinal alkaline phosphatase: close homology to placental alkaline phosphatase. Proc Natl Acad Sci USA. 1987;84:1234–1238. doi: 10.1073/pnas.84.5.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 527.Weiss MJ, Henthorn PS, Lafferty MA, Slaughter C, Raducha M, Harris H. Isolation and characterization of a cDNA encoding a human liver/bone/kidney-type alkaline phosphatase. Proc Natl Acad Sci USA. 1986;83:7182–7186. doi: 10.1073/pnas.83.19.7182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 528.Terao M, Mintz B. Cloning and characterization of a cDNA coding for mouse placental alkaline phosphatase. Proc Natl Acad Sci USA. 1987;84:7051–7055. doi: 10.1073/pnas.84.20.7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529.Knoll BJ, Rothblum KN, Longley M. Two gene duplication events in the evolution of the human heat-stable alkaline phosphatases. Gene. 1987;60:267–276. doi: 10.1016/0378-1119(87)90235-6. [DOI] [PubMed] [Google Scholar]
- 530.Millán JL, Manes T. Seminoma-derived Nagao isozyme is encoded by a germ-cell alkaline phosphatase gene. Proc Natl Acad Sci USA. 1988;85:3024–3028. doi: 10.1073/pnas.85.9.3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531.Fishman WH. Alkaline phosphatase isozymes: recent progress. Clin Biochem. 1990;23:99–104. doi: 10.1016/0009-9120(90)80019-F. [DOI] [PubMed] [Google Scholar]
- 532.Kozlenkov A, Manes T, Hoylaerts MF, Millán JL. Function assignment to conserved residues in mammalian alkaline phosphatases. J Biol Chem. 2002;277:22992–22999. doi: 10.1074/jbc.M202298200. [DOI] [PubMed] [Google Scholar]
- 533.Kishi F, Matsuura S, Kajii T. Nucleotide sequence of the human liver-type alkaline phosphatase cDNA. Nucleic Acids Res. 1989;17:2129. doi: 10.1093/nar/17.5.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 534.Toh Y, Yamamoto M, Endo H, Fujita A, Misumi Y, Ikehara Y. Sequence divergence of 5′ extremities in rat liver alkaline phosphatase mRNAs. J Biochem. 1989;105:61–65. doi: 10.1093/oxfordjournals.jbchem.a122620. [DOI] [PubMed] [Google Scholar]
- 535.Toh Y, Yamamoto M, Endo H, Misumi Y, Ikehara Y. Isolation and characterization of a rat liver alkaline phosphatase gene. A single gene with two promoters. Eur J Biochem. 1989;182:231–237. doi: 10.1111/j.1432-1033.1989.tb14822.x. [DOI] [PubMed] [Google Scholar]
- 536.Zernik J, Kream B, Twarog K. Tissue-specific and dexamethasone-inducible expression of alkaline phosphatase from alternative promoters of the rat bone/liver/kidney/placenta gene. Biochem Biophys Res Commun. 1991;176:1149–1156. doi: 10.1016/0006-291X(91)90405-V. [DOI] [PubMed] [Google Scholar]
- 537.Studer M, Terao M, Gianni M, Garattini E. Characterization of a second promoter for the mouse liver/bone/kidney-type alkaline phosphatase gene: cell and tissue specific expression. Biochem Biophys Res Commun. 1991;179:1352–1360. doi: 10.1016/0006-291X(91)91722-O. [DOI] [PubMed] [Google Scholar]
- 538.Nosjean O, Koyama I, Goseki M, Roux B, Komoda T. Human tissue non-specific alkaline phosphatases: sugar-moiety-induced enzymic and antigenic modulations and genetic aspects. Biochem J. 1997;321(Pt 2):297–303. doi: 10.1042/bj3210297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 539.Kuwana T, Sugita O, Yakata M. Sugar chain heterogeneity of bone and liver alkaline phosphatase in serum. Enzyme. 1991;45:63–66. doi: 10.1159/000468866. [DOI] [PubMed] [Google Scholar]
- 540.Garattini E, Hua JC, Pan YC, Udenfriend S. Human liver alkaline phosphatase, purification and partial sequencing: homology with the placental isozyme. Arch Biochem Biophys. 1986;245:331–337. doi: 10.1016/0003-9861(86)90223-7. [DOI] [PubMed] [Google Scholar]
- 541.Farley JR, Magnusson P. Effects of tunicamycin, mannosamine, and other inhibitors of glycoprotein processing on skeletal alkaline phosphatase in human osteoblast-like cells. Calcified Tissue Int. 2005;76:63–74. doi: 10.1007/s00223-004-0023-2. [DOI] [PubMed] [Google Scholar]
- 542.Pan CJ, Sartwell AD, Chou JY. Transcriptional regulation and the effects of sodium butyrate and glycosylation on catalytic activity of human germ cell alkaline phosphatase. Cancer Res. 1991;51:2058–2062. [PubMed] [Google Scholar]
- 543.Mueller WH, Kleefeld D, Khattab B, Meissner JD, Scheibe RJ. Effects of retinoic acid on N-glycosylation and mRNA stability of the liver/bone/kidney alkaline phosphatase in neuronal cells. J Cell Physiol. 2000;182:50–61. doi: 10.1002/(SICI)1097-4652(200001)182:1<50::AID-JCP6>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 544.Linder CH, Narisawa S, Millán JL, Magnusson P. Glycosylation differences contribute to distinct catalytic properties among bone alkaline phosphatase isoforms. Bone. 2009;45:987–993. doi: 10.1016/j.bone.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 545.Magnusson P, Farley JR. Differences in sialic acid residues among bone alkaline phosphatase isoforms: a physical, biochemical, and immunological characterization. Calcified Tissue Int. 2002;71:508–518. doi: 10.1007/s00223-001-1137-4. [DOI] [PubMed] [Google Scholar]
- 546.Endo T, Ohbayashi H, Hayashi Y, Ikehara Y, Kochibe N, Kobata A. Structural study on the carbohydrate moiety of human placental alkaline phosphatase. J Biochem. 1988;103:182–187. doi: 10.1093/oxfordjournals.jbchem.a122228. [DOI] [PubMed] [Google Scholar]
- 547.Nam JH, Zhang F, Ermonval M, Linhardt RJ, Sharfstein ST. The effects of culture conditions on the glycosylation of secreted human placental alkaline phosphatase produced in Chinese hamster ovary cells. Biotechnol Bioeng. 2008;100:1178–1192. doi: 10.1002/bit.21853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 548.Catino MA, Paladino S, Tivodar S, Pocard T, Zurzolo C. N- and O-glycans are not directly involved in the oligomerization and apical sorting of GPI proteins. Traffic. 2008;9:2141–2150. doi: 10.1111/j.1600-0854.2008.00826.x. [DOI] [PubMed] [Google Scholar]
- 549.Endo T, Higashino K, Hada T, Imanishi H, Muratani K, Kochibe N, Kobata A. Structures of the asparagine-linked oligosaccharides of an alkaline phosphatase, kasahara isozyme, purified from FL amnion cells. Cancer Res. 1990;50:1079–1084. [PubMed] [Google Scholar]
- 550.Slein MW, Logan GF. Characterization of the phospholipases of Bacillus cereus and their effects on erythrocytes, bone, and kidney cells. J Bacteriol. 1965;90:69–81. doi: 10.1128/jb.90.1.69-81.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 551.Ikezawa H, Yamanegi M, Taguchi R, Miyashita T, Ohyabu T. Studies on phosphatidylinositol phosphodiesterase (phospholipase C type) of Bacillus cereus. I. Purification, properties and phosphatase-releasing activity. Biochim Biophys Acta. 1976;450:154–164. [PubMed] [Google Scholar]
- 552.Low MG, Finean JB. Release of alkaline phosphatase from membranes by a phosphatidylinositol-specific phospholipase C. Biochem J. 1977;167:281–284. doi: 10.1042/bj1670281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 553.Wong YW, Low MG. Phospholipase resistance of the glycosyl-phosphatidylinositol membrane anchor on human alkaline phosphatase. Clin Chem. 1992;38:2517–2525. [PubMed] [Google Scholar]
- 554.Wong YW, Low MG. Biosynthesis of glycosylphosphatidylinositol-anchored human placental alkaline phosphatase: evidence for a phospholipase C-sensitive precursor and its post-attachment conversion into a phospholipase C-resistant form. Biochem J. 1994;301:205–209. doi: 10.1042/bj3010205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 555.Micanovic R, Gerber LD, Berger J, Kodukula K, Udenfriend S. Selectivity of the cleavage/attachment site of phosphatidylinositol-glycan-anchored membrane proteins determined by site-specific mutagenesis at Asp-484 of placental alkaline phosphatase. Proc Natl Acad Sci USA. 1990;87:157–161. doi: 10.1073/pnas.87.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 556.Ogata S, Hayashi Y, Takami N, Ikehara Y. Chemical characterization of the membrane-anchoring domain of human placental alkaline phosphatase. J Biol Chem. 1988;263:10489–10494. [PubMed] [Google Scholar]
- 557.Lowe ME. Site-specific mutations in the COOH-terminus of placental alkaline phosphatase: a single amino acid change converts a phosphatidylinositol-glycan-anchored protein to a secreted protein. J Cell Biol. 1992;116:799–807. doi: 10.1083/jcb.116.3.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 558.Bailey CA, Gerber L, Howard AD, Udenfriend S. Processing at the carboxyl terminus of nascent placental alkaline phosphatase in a cell-free system: evidence for specific cleavage of a signal peptide. Proc Natl Acad Sci USA. 1989;86:22–26. doi: 10.1073/pnas.86.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 559.Amthauer R, Kodukula K, Udenfriend S. Placental alkaline phosphatase: a model for studying COOH-terminal processing of phosphatidylinositol-glycan-anchored membrane proteins. Clin Chem. 1992;38:2510–2516. [PubMed] [Google Scholar]
- 560.Redman CA, Thomas-Oates JE, Ogata S, Ikehara Y, Ferguson MAJ. Structure of the glycosylphosphatidylinositol membrane anchor of human placental alkaline phosphatase. Biochem J. 1994;302:861–865. doi: 10.1042/bj3020861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 561.Fukushima K, Ikehara Y, Kanai M, Kochibe N, Kuroki M, Yamashita K. A β-N-acetylglucosaminyl phosphate diester residue is attached to the glycosylphosphatidylinositol anchor of human placental alkaline phosphatase—a target of the channel-forming toxin aerolysin. J Biol Chem. 2003;278:36296–36303. doi: 10.1074/jbc.M304341200. [DOI] [PubMed] [Google Scholar]
- 562.Giocondi MC, Besson F, Dosset P, Milhiet PE, Le GC. Remodeling of ordered membrane domains by GPI-anchored intestinal alkaline phosphatase. Langmuir. 2007;23:9358–9364. doi: 10.1021/la700892z. [DOI] [PubMed] [Google Scholar]
- 563.Sesana S, Re F, Bulbarelli A, Salerno D, Cazzaniga E, Masserini M. Membrane features and activity of GPI-anchored enzymes: alkaline phosphatase reconstituted in model membranes. Biochemistry USA. 2008;47:5433–5440. doi: 10.1021/bi800005s. [DOI] [PubMed] [Google Scholar]
- 564.Srivastava AK, Masinde G, Yu H, Baylink DJ, Mohan S. Mapping quantitative trait loci that influence blood levels of alkaline phosphatase in MRL/MpJ and SJL/J mice. Bone. 2004;35:1086–1094. doi: 10.1016/j.bone.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 565.Moss DW. Perspectives in alkaline phosphatase research. Clin Chem. 1992;38:2486–2492. [PubMed] [Google Scholar]
- 566.Moss DW. Release of membrane-bound enzymes from cells and the generation of isoforms. Clin Chim Acta. 1994;226:131–142. doi: 10.1016/0009-8981(94)90210-0. [DOI] [PubMed] [Google Scholar]
- 567.Magnusson P, Sharp CA, Farley JR. Different distributions of human bone alkaline phosphatase isoforms in serum and bone tissue extracts. Clin Chim Acta. 2002;325:59–70. doi: 10.1016/S0009-8981(02)00248-6. [DOI] [PubMed] [Google Scholar]
- 568.Low MG, Huang KS. Factors affecting the ability of glycosylphosphatidylinositol- specific phospholipase-D to degrade the membrane anchors of cell surface proteins. Biochem J. 1991;279:483–493. doi: 10.1042/bj2790483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 569.Anh DJ, Eden A, Farley JR. Quantitation of soluble and skeletal alkaline phosphatase, and insoluble alkaline phosphatase anchor-hydrolase activities in human serum. Clin Chim Acta. 2001;311:137–148. doi: 10.1016/S0009-8981(01)00584-8. [DOI] [PubMed] [Google Scholar]
- 570.Brocklehurst D, Wilde CE, Doar JW. The incidence and likely origins of serum particulate alkaline phosphatase and lipoprotein-X in liver disease. Clin Chim Acta. 1978;88:509–515. doi: 10.1016/0009-8981(78)90285-1. [DOI] [PubMed] [Google Scholar]
- 571.Broe ME, Roels F, Nouwen EJ, Claeys L, Wieme RJ. Liver plasma membrane: the source of high molecular weight alkaline phosphatase in human serum. Hepatol. 1985;5:118–128. doi: 10.1002/hep.1840050124. [DOI] [PubMed] [Google Scholar]
- 572.Nakano T, Inoue I, Alpers DH, Akiba Y, Katayama S, Shinozaki R, Kaunitz JD, Ohshima S, Akita M, Takahashi S, Koyama I, Matsushita M, Komoda T. Role of lysophosphatidylcholine in brush-border intestinal alkaline phosphatase release and restoration. Amer J Physiol Gastrointest L. 2009;297:G207–G214. doi: 10.1152/ajpgi.90590.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 573.Vittur F, Stagni N, Moro L, Bernhard B. Alkaline phosphatase binds to collagen; a hypothesis on the mechanism of extravesicular mineralization in epiphyseal cartilage. Experientia. 1984;40:836–837. doi: 10.1007/BF01951980. [DOI] [PubMed] [Google Scholar]
- 574.Wu LN, Genge BR, Lloyd GC, Wuthier RE. Collagen-binding proteins in collagenase-released matrix vesicles from cartilage. Interaction between matrix vesicle proteins and different types of collagen. J Biol Chem. 1991;266:1195–1203. [PubMed] [Google Scholar]
- 575.Wu LN, Genge BR, Wuthier RE. Evidence for specific interaction between matrix vesicle proteins and the connective tissue matrix. Bone Miner. 1992;17:247–252. doi: 10.1016/0169-6009(92)90745-Y. [DOI] [PubMed] [Google Scholar]
- 576.Tsonis PA, Argraves WS, Millán JL. A putative functional domain of human placental alkaline phosphatase predicted from sequence comparisons. Biochem J. 1988;254:623–624. doi: 10.1042/bj2540623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 577.Bossi M, Hoylaerts MF, Millán JL. Modifications in a flexible surface loop modulate the isozyme-specific properties of mammalian alkaline phosphatases. J Biol Chem. 1993;268:25409–25416. [PubMed] [Google Scholar]
- 578.Mornet E, Stura E, Lia-Baldini AS, Stigbrand T, Menez A, Du MH. Structural evidence for a functional role of human tissue nonspecific alkaline phosphatase in bone mineralization. J Biol Chem. 2001;276:31171–31178. doi: 10.1074/jbc.M102788200. [DOI] [PubMed] [Google Scholar]
- 579.Makiya R, Stigbrand T. Placental alkaline phosphatase has a binding site for the human immunoglobulin-G Fc portion. Eur J Biochem. 1992;205:341–345. doi: 10.1111/j.1432-1033.1992.tb16785.x. [DOI] [PubMed] [Google Scholar]
- 580.Makiya R, Thornell LE, Stigbrand T. Placental alkaline phosphatase, a GPI-anchored protein, is clustered in clathrin-coated vesicles. Biochem Biophys Res Commun. 1992;183:803–808. doi: 10.1016/0006-291X(92)90554-X. [DOI] [PubMed] [Google Scholar]
- 581.Leathers VL, Norman AW. Evidence for calcium mediated conformational changes in calbindin-D28K (the vitamin D-induced calcium binding protein) interactions with chick intestinal brush border membrane alkaline phosphatase as studied via photoaffinity labeling techniques. J Cell Biochem. 1993;52:243–252. doi: 10.1002/jcb.240520216. [DOI] [PubMed] [Google Scholar]
- 582.Shinozaki T, Watanabe H, Takagishi K, Pritzker KP. Allotype immunoglobulin enhances alkaline phosphatase activity: implications for the inflammatory response. J Lab Clin Med. 1998;132:320–328. doi: 10.1016/S0022-2143(98)90046-4. [DOI] [PubMed] [Google Scholar]
- 583.She QB, Mukherjee JJ, Chung T, Kiss Z. Placental alkaline phosphatase, insulin, and adenine nucleotides or adenosine synergistically promote long-term survival of serum-starved mouse embryo and human fetus fibroblasts. Cell Signal. 2000;12:659–665. doi: 10.1016/S0898-6568(00)00117-0. [DOI] [PubMed] [Google Scholar]
- 584.Kermer V, Ritter M, Albuquerque B, Leib C, Stanke M, Zimmermann H. Knockdown of tissue nonspecific alkaline phosphatase impairs neural stem cell proliferation and differentiation. Neurosci Lett. 2010;485:208–211. doi: 10.1016/j.neulet.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 585.Millán JL. Oncodevelopmental expression and structure of alkaline phosphatase genes. Anticancer Res. 1988;8:995–1004. [PubMed] [Google Scholar]
- 586.Kim EE, Wyckoff HW. Reaction mechanism of alkaline phosphatase based on crystal structures. Two-metal ion catalysis. J Mol Biol. 1991;218:449–464. doi: 10.1016/0022-2836(91)90724-K. [DOI] [PubMed] [Google Scholar]
- 587.Lazzaroni JC, Atlan D, Portalier RC. Excretion of alkaline phosphatase by Escherichia coli K-12 pho constitutive mutants transformed with plasmids carrying the alkaline phosphatase structural gene. J Bacteriol. 1985;164:1376–1380. doi: 10.1128/jb.164.3.1376-1380.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 588.Sone M, Kishigami S, Yoshihisa T, Ito K. Roles of disulfide bonds in bacterial alkaline phosphatase. J Biol Chem. 1997;272:6174–6178. doi: 10.1074/jbc.272.10.6174. [DOI] [PubMed] [Google Scholar]
- 589.Hulett FM, Kim EE, Bookstein C, Kapp NV, Edwards CW, Wyckoff HW. Bacillus subtilis alkaline phosphatases III and IV. Cloning, sequencing, and comparisons of deduced amino acid sequence with Escherichia coli alkaline phosphatase three-dimensional structure. J Biol Chem. 1991;266:1077–1084. [PubMed] [Google Scholar]
- 590.Murphy JE, Kantrowitz ER. Why are mammalian alkaline phosphatases much more active than bacterial alkaline phosphatases? Mol Microbiol. 1994;12:351–357. doi: 10.1111/j.1365-2958.1994.tb01024.x. [DOI] [PubMed] [Google Scholar]
- 591.Wende A, Johansson P, Vollrath R, Dyall-Smith M, Oesterhelt D, Grininger M. Structural and biochemical characterization of a halophilic archaeal alkaline phosphatase. J Mol Biol. 2010;400:52–62. doi: 10.1016/j.jmb.2010.04.057. [DOI] [PubMed] [Google Scholar]
- 592.Helland R, Larsen RL, Asgeirsson B. The 1.4 Å crystal structure of the large and cold-active Vibrio sp. alkaline phosphatase. Biochim Biophys Acta. 2009;1794:297–308. doi: 10.1016/j.bbapap.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 593.Kozlenkov A, Du MH, Cuniasse P, Ny T, Hoylaerts MF, Millán JL. Residues determining the binding specificity of uncompetitive inhibitors to tissue-nonspecific alkaline phosphatase. J Bone Miner Res. 2004;19:1862–1872. doi: 10.1359/JBMR.040608. [DOI] [PubMed] [Google Scholar]
- 594.Llinas P, Stura EA, Menez A, Kiss Z, Stigbrand T, Millán JL, Du MH. Structural studies of human placental alkaline phosphatase in complex with functional ligands. J Mol Biol. 2005;350:441–451. doi: 10.1016/j.jmb.2005.04.068. [DOI] [PubMed] [Google Scholar]
- 595.Holtz KM, Kantrowitz ER. The mechanism of the alkaline phosphatase reaction: insights from NMR, crystallography and site-specific mutagenesis. FEBS Lett. 1999;462:7–11. doi: 10.1016/S0014-5793(99)01448-9. [DOI] [PubMed] [Google Scholar]
- 596.Schwartz JH, Lipmann F. Phosphate incorporation into alkaline phosphatase of E. coli. Proc Natl Acad Sci USA. 1961;47:1996–2005. doi: 10.1073/pnas.47.12.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 597.Kim EE, Wyckoff HW. Structure of alkaline phosphatases. Clin Chim Acta. 1990;186:175–187. doi: 10.1016/0009-8981(90)90035-Q. [DOI] [PubMed] [Google Scholar]
- 598.Holtz KM, Stec B, Kantrowitz ER. A model of the transition state in the alkaline phosphatase reaction. J Biol Chem. 1999;274:8351–8354. doi: 10.1074/jbc.274.13.8351. [DOI] [PubMed] [Google Scholar]
- 599.Du MH, Lamoure C, Muller BH, Bulgakov OV, Lajeunesse E, Menez A, Boulain JC. Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase. J Mol Biol. 2002;316:941–953. doi: 10.1006/jmbi.2001.5384. [DOI] [PubMed] [Google Scholar]
- 600.Wang J, Kantrowitz ER. Trapping the tetrahedral intermediate in the alkaline phosphatase reaction by substitution of the active site serine with threonine. Protein Sci. 2006;15:2395–2401. doi: 10.1110/ps.062351506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 601.Andrews LD, Deng H, Herschlag D. Isotope-edited FTIR of alkaline phosphatase resolves paradoxical ligand binding properties and suggests a role for ground-state destabilization. J Am Chem Soc. 2011;133:11621–11631. doi: 10.1021/ja203370b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 602.López-Canut V, Roca M, Bertrán J, Moliner V, Tuñón I. Promiscuity in alkaline phosphatase superfamily. Unraveling evolution through molecular simulations. J Am Chem Soc. 2011;133:12050–12062. doi: 10.1021/ja2017575. [DOI] [PubMed] [Google Scholar]
- 603.Jones SR, Kindman LA, Knowles JR. Stereochemistry of phosphoryl group transfer using a chiral [16O, 17O, 18O] stereochemical course of alkaline phosphatase. Nature. 1978;275:564–565. doi: 10.1038/275564a0. [DOI] [PubMed] [Google Scholar]
- 604.Hull WE, Halford SE, Gutfreund H, Sykes BD. 31P nuclear magnetic resonance study of alkaline phosphatase: the role of inorganic phosphate in limiting the enzyme turnover rate at alkaline pH. Biochemistry USA. 1976;15:1547–1561. doi: 10.1021/bi00652a028. [DOI] [PubMed] [Google Scholar]
- 605.Otvos JD, Alger JR, Coleman JE, Armitage IM. 31P NMR of alkaline phosphatase. Saturation transfer and metal–phosphorus coupling. J Biol Chem. 1979;254:1778–1780. [PubMed] [Google Scholar]
- 606.Gettins P, Coleman JE. 31P nuclear magnetic resonance of phosphoenzyme intermediates of alkaline phosphatase. J Biol Chem. 1983;258:408–416. [PubMed] [Google Scholar]
- 607.Gettins P, Coleman JE. Zn(II)–113Cd(II) and Zn(II)–Mg(II) hybrids of alkaline phosphatase. 31P and 113Cd NMR. J Biol Chem. 1984;259:4991–4997. [PubMed] [Google Scholar]
- 608.Gettins P, Metzler M, Coleman JE. Alkaline phosphatase. 31P NMR probes of the mechanism. J Biol Chem. 1985;260:2875–2883. [PubMed] [Google Scholar]
- 609.Xu X, Kantrowitz ER. Binding of magnesium in a mutant Escherichia coli alkaline phosphatase changes the rate-determining step in the reaction mechanism. Biochemistry USA. 1993;32:10683–10691. doi: 10.1021/bi00091a019. [DOI] [PubMed] [Google Scholar]
- 610.Tibbitts TT, Murphy JE, Kantrowitz ER. Kinetic and structural consequences of replacing the aspartate bridge by asparagine in the catalytic metal triad of Escherichia coli alkaline phosphatase. J Mol Biol. 1996;257:700–715. doi: 10.1006/jmbi.1996.0195. [DOI] [PubMed] [Google Scholar]
- 611.Stec B, Holtz KM, Kantrowitz ER. A revised mechanism for the alkaline phosphatase reaction involving three metal ions. J Mol Biol. 2000;299:1303–1311. doi: 10.1006/jmbi.2000.3799. [DOI] [PubMed] [Google Scholar]
- 612.Han R, Coleman JE. Dependence of the phosphorylation of alkaline phosphatase by phosphate monoesters on the pKa of the leaving group. Biochemistry USA. 1995;34:4238–4245. doi: 10.1021/bi00013a013. [DOI] [PubMed] [Google Scholar]
- 613.Stec B, Hehir MJ, Brennan C, Nolte M, Kantrowitz ER. Kinetic and X-ray structural studies of three mutant E. coli alkaline phosphatases: insights into the catalytic mechanism without the nucleophile Ser102. J Mol Biol. 1998;277:647–662. doi: 10.1006/jmbi.1998.1635. [DOI] [PubMed] [Google Scholar]
- 614.Sträter N, Lipscomb WN, Klabunde T, Krebs B. Two-metal ion catalysis in enzymatic acyl- and phosphoryl-transfer reactions. Angew Chem Int Ed. 1996;35:2024–2055. doi: 10.1002/anie.199620241. [DOI] [Google Scholar]
- 615.Butler-Ransohoff JE, Rokita SE, Kendall D, Banzon JA, Carano KS, Kaiser ET, Matlin AR. Active-site mutagenesis of E. coli alkaline phosphatase: replacement of serine-102 with nonnucleophilic amino acids. J Org Chem. 1992;57:142–145. doi: 10.1021/jo00027a027. [DOI] [Google Scholar]
- 616.Chaidaroglou A, Brezinski DJ, Middleton SA, Kantrowitz ER. Function of arginine-166 in the active site of Escherichia coli alkaline phosphatase. Biochemistry USA. 1988;27:8338–8343. doi: 10.1021/bi00422a008. [DOI] [PubMed] [Google Scholar]
- 617.Stevens CR, Williams RB, Farrell AJ, Blake DR. Hypoxia and inflammatory synovitis: observations and speculation. Ann Rheum Dis. 1991;50:124–132. doi: 10.1136/ard.50.2.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 618.Bevan S, Geppetti P. Protons: small stimulants of capsaicin-sensitive sensory nerves. Trends Neurosci. 1994;17:509–512. doi: 10.1016/0166-2236(94)90149-X. [DOI] [PubMed] [Google Scholar]
- 619.Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 620.Tamao Y, Noguchi K, Sakai-Tomita Y, Hama H, Shimamoto T, Kanazawa H, Tsuda M, Tsuchiya T. Sequence analysis of nutA gene encoding membrane-bound Cl-dependent 5′-nucleotidase of Vibrio parahaemolyticus. J Biochem. 1991;109:24–29. doi: 10.1093/oxfordjournals.jbchem.a123345. [DOI] [PubMed] [Google Scholar]
- 621.Liyou N, Hamilton S, Elvin C, Willadsen P. Cloning and expression of ecto-5′-nucleotidase from the cattle tick Boophilus microplus. Insect Mol Biol. 1999;8:257–266. doi: 10.1046/j.1365-2583.1999.820257.x. [DOI] [PubMed] [Google Scholar]
- 622.Caljon G, Ridder K, Baetselier P, Coosemans M, Abbeele J. Identification of a tsetse fly salivary protein with dual inhibitory action on human platelet aggregation. PLoS One. 2010;5:e9671. doi: 10.1371/journal.pone.0009671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 623.Mans BJ, Gasper AR, Louw AI, Neitz AW. Purification and characterization of apyrase from the tick, Ornithodoros savignyi. Comp Biochem Physiol B Biochem Mol Biol. 1998;120:617–624. doi: 10.1016/S0305-0491(98)10061-5. [DOI] [PubMed] [Google Scholar]
- 624.Gounaris K, Selkirk ME, Sadeghi SJ. A nucleotidase with unique catalytic properties is secreted by Trichinella spiralis. Mol Biochem Parasitol. 2004;136:257–264. doi: 10.1016/j.molbiopara.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 625.Oda Y, Kuo MD, Huang SS, Huang JS. The plasma cell membrane glycoprotein, PC-1, is a threonine-specific protein kinase stimulated by acidic fibroblast growth factor. J Biol Chem. 1991;266:16791–16795. [PubMed] [Google Scholar]
- 626.North EJ, Howard AL, Wanjala IW, Pham TCT, Baker DL, Parrill AL. Pharmacophore development and application toward the identification of novel, small-molecule autotaxin inhibitors. J Med Chem. 2010;53:3095–3105. doi: 10.1021/jm901718z. [DOI] [PubMed] [Google Scholar]
- 627.Schuster-Böckler B, Schultz J, Rahmann S. HMM logos for visualization of protein families. BMC Bioinformatics. 2004;5:7. doi: 10.1186/1471-2105-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 628.Drosopoulos JHF. Roles of Asp54 and Asp213 in Ca2+ utilization by soluble human CD39/ecto-nucleotidase. Arch Biochem Biophys. 2002;406:85–95. doi: 10.1016/S0003-9861(02)00414-9. [DOI] [PubMed] [Google Scholar]
- 629.Hicks-Berger CA, Yang F, Smith TM, Kirley TL. The importance of histidine residues in human ecto-nucleoside triphosphate diphosphohydrolase-3 as determined by site-directed mutagenesis. BBA Protein Struct Mol Enzym. 2001;1547:72–81. doi: 10.1016/S0167-4838(01)00176-5. [DOI] [PubMed] [Google Scholar]
- 630.Kirley TL, Yang F, Ivanenkov VV. Site-directed mutagenesis of human nucleoside triphosphate diphosphohydrolase 3: the importance of conserved glycine residues and the identification of additional conserved protein motifs in eNTPDases. Arch Biochem Biophys. 2001;395:94–102. doi: 10.1006/abbi.2001.2570. [DOI] [PubMed] [Google Scholar]
- 631.Wu J, Hansen GH, Nilsson A, Duan RD. Functional studies of human intestinal alkaline sphingomyelinase by deglycosylation and mutagenesis. Biochem J. 2005;386:153–160. doi: 10.1042/BJ20041455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 632.Stec B, Cheltsov A, Millán JL. Refined structures of placental alkaline phosphatase show a consistent pattern of interactions at the peripheral site. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66:866–870. doi: 10.1107/S1744309110019767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 633.Backer M, McSweeney S, Rasmussen HB, Riise BW, Lindley P, Hough E. The 1.9 A crystal structure of heat-labile shrimp alkaline phosphatase. J Mol Biol. 2002;318:1265–1274. doi: 10.1016/S0022-2836(02)00035-9. [DOI] [PubMed] [Google Scholar]
- 634.Backer MME, McSweeney S, Lindley PF, Hough E. Ligand-binding and metal-exchange crystallographic studies on shrimp alkaline phosphatase. Acta Crystallogr D Biol Cryst. 2004;60:1555–1561. doi: 10.1107/S0907444904015628. [DOI] [PubMed] [Google Scholar]
- 635.Tsuruta H, Mikami B, Higashi T, Aizono Y. Crystal structure of cold-active alkaline phosphatase from the Psychrophile shewanella sp. Biosci Biotechnol Biochem. 2010;74:69–74. doi: 10.1271/bbb.90563. [DOI] [PubMed] [Google Scholar]
- 636.Wang E, Koutsioulis D, Leiros HK, Andersen OA, Bouriotis V, Hough E, Heikinheimo P. Crystal structure of alkaline phosphatase from the Antarctic bacterium TAB5. J Mol Biol. 2007;366:1318–1331. doi: 10.1016/j.jmb.2006.11.079. [DOI] [PubMed] [Google Scholar]
- 637.Butler-Ransohoff JE, Kendall DA, Kaiser ET. Use of site-directed mutagenesis to elucidate the role of arginine-166 in the catalytic mechanism of alkaline phosphatase. Proc Natl Acad Sci USA. 1988;85:4276–4278. doi: 10.1073/pnas.85.12.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 638.Chaidaroglou A, Kantrowitz ER. Alteration of aspartate 101 in the active site of Escherichia coli alkaline phosphatase enhances the catalytic activity. Protein Eng. 1989;3:127–132. doi: 10.1093/protein/3.2.127. [DOI] [PubMed] [Google Scholar]
- 639.Mandecki W, Shallcross MA, Sowadski J, Tomazic-Allen S. Mutagenesis of conserved residues within the active site of Escherichia coli alkaline phosphatase yields enzymes with increased kcat. Protein Eng. 1991;4:801–804. doi: 10.1093/protein/4.7.801. [DOI] [PubMed] [Google Scholar]
- 640.Chen L, Neidhart D, Kohlbrenner WM, Mandecki W, Bell S, Sowadski J, Abad-Zapatero C. 3-D structure of a mutant (Asp101→Ser) of E. coli alkaline phosphatase with higher catalytic activity. Protein Eng. 1992;5:605–610. doi: 10.1093/protein/5.7.605. [DOI] [PubMed] [Google Scholar]
- 641.Dealwis CG, Chen L, Brennan C, Mandecki W, Abad-Zapatero C. 3-D structure of the D153G mutant of Escherichia coli alkaline phosphatase: an enzyme with weaker magnesium binding and increased catalytic activity. Protein Eng. 1995;8:865–871. doi: 10.1093/protein/8.9.865. [DOI] [PubMed] [Google Scholar]
- 642.Xu X, Kantrowitz ER. A water-mediated salt link in the catalytic site of Escherichia coli alkaline phosphatase may influence activity. Biochemistry USA. 1991;30:7789–7796. doi: 10.1021/bi00245a018. [DOI] [PubMed] [Google Scholar]
- 643.Kahya N, Brown DA, Schwille P. Raft partitioning and dynamic behavior of human placental alkaline phosphatase in giant unilamellar vesicles. Biochemistry USA. 2005;44:7479–7489. doi: 10.1021/bi047429d. [DOI] [PubMed] [Google Scholar]
- 644.Matoba K, Shiba T, Takeuchi T, Sibley LD, Seiki M, Kikyo F, Horiuchi T, Asai T, Haradaa S (2010) Crystallization and preliminary X-ray structural analysis of nucleoside triphosphate hydrolases from Neospora caninum and Toxoplasma gondii. Acta Crystallogr Sect F Struct Biol Cryst Commun 66:1445–1448 [DOI] [PMC free article] [PubMed]