Abstract
Objective
Emerging evidence suggests that genetic components contribute significantly to cartilage degeneration in osteoarthritis pathophysiology but little evidence is available on genetics of cartilage regeneration. Therefore, we investigated cartilage regeneration in genetic murine models using common inbred strains and a set of recombinant inbred lines generated from LG/J (healer of ear-wounds) and SM/J (non-healer) inbred strains.
Methods
An acute full-thickness cartilage injury was introduced through microsurgery in the trochlear groove of 8-weeks old mice (N=265). Knee joints were sagittally sectioned and stained with toluidine blue to evaluate regeneration. For ear-wound phenotype, a bilateral 2-mm through-and-through puncture was made (N=229) at 6-weeks and healing outcomes measured after 30-days. Broad-sense heritability and genetic correlations were calculated for both phenotypes.
Results
Time-course studies from recombinant inbred lines show no significant regeneration until 16-weeks post-surgery; at that time, the strains can be segregated into three categories: good, intermediate and poor healers. Heritability (H2) showed that both cartilage regeneration (H2=26%; p=0.006) and ear-wound closure (H2=53%; p<0.00001) are significantly heritable. The genetic correlations between the two healing phenotypes for common inbred strains (r=0.92) and recombinant inbred lines (r=0.86) were found to be extremely high.
Conclusion
We report that i) articular cartilage regeneration is heritable, ii) the differences between the lines being due to genetic differences and iii) a strong genetic correlation between the two phenotypes exists indicating that they plausibly share a common genetic basis. We, therefore, surmise that LG/J by SM/J intercross can be used to dissect the genetic basis of variation in cartilage regeneration.
Keywords: articular cartilage, ear-wound, regeneration, genetics, heritability
Tissue regeneration is a property generally attributed to organisms other than mammals. For example, hydra and planaria can regenerate their entire bodies from a small fraction of their tissues (1, 2). Similarly, vertebrates including newts and salamanders can regenerate limbs or other structures after amputation (3, 4). In contrast, the regenerative capacity in mammals is extremely limited (5) and is generally confined to shedding and re-growth of antlers in deer (6) and moose (7), and ear-wound closure in rabbits (8). Despite this limited regeneration in adult mammals, supporting evidence suggests that certain mouse strains possess unusual abilities to regenerate/repair tissues even into adulthood. For instance, MRL/MpJ (9–12), and LG/J (11, 13) strains display 2-millimeter through-and-through ear-hole closure within 4-weeks.
Articular cartilage is generally thought to have limited regeneration capacity, but there are sporadic reports of full-thickness articular cartilage healing in rats (14), rabbits (15), dogs (16) and horses (17). Spontaneous regeneration of cartilage is possible only when the defect penetrates through the cartilage extending through the subchondral bone to the bone-cavity to presumably access mesenchymal stem cells and recruit them to the site of damage (18, 19). In this case, the newly formed tissue is typically fibrocartilage, which is mechanically weak and degenerates over time (15, 20). In partial-thickness defects no effective repair response occurs (10, 21) and attempts to restore/repair cartilage injuries have met with unpredictable and variable outcomes (22, 23).
Lack of self-regeneration in articular cartilage, mainly because of its intrinsic avascularity, contributes to cartilage degeneration, predisposing an individual to the development of osteoarthritis (OA), leading to prosthetic joint replacement and disability (22–24). While it is well-appreciated that OA is widespread, over 50% of individuals are protected from post-traumatic OA despite suffering from cartilage damage (25). The exact cause for this remains unknown and can be attributed to a greater intrinsic ability to regenerate cartilage (26). Our long-term goal is to understand why some people can repair or regenerate cartilage while insult or aging results in degeneration in others.
Recently, two mouse strains, MRL/MpJ and DBA/1, were reported to regenerate full-thickness articular cartilage defects in contrast to a failure of healing in age-matched C57BL/6 mice. The regenerated cartilage is hyaline-like in nature and has abundant chondrocytes, an extracellular matrix rich in proteoglycan, persistent types II and VI collagen neo-deposition, and cell proliferation within the repair tissue (10, 21). MRL/MpJ and LG/J, two closely-related mouse strains sharing 75% of their genomes identical-by-descent (2), can completely heal ear-wounds including ear cartilage (9, 13, 27). This raises the possibility that, like MRL/MpJ, LG/J may also heal articular cartilage. To investigate this possibility, we have examined the extent of cartilage regeneration in a set of common inbred strains, including both healers and non-healers, and in a set of recombinant inbred (RI) lines formed from the intercross of the LG/J (healer) and SM/J (non-healer) inbred mouse strains. The conceptual starting point for RI lines is that any differential phenotype will be attributed to a restricted set of genes according to the way they have been inherited from parental strains. It is estimated that 30–75% of the variation in OA is due to genetic differences (28, 29). While OA in humans can be studied using cartilage from donors, usually at the time of total joint replacement, it is impossible to directly investigate cartilage repair in humans. Therefore, we took the approach of investigating cartilage regeneration in genetic mouse models.
In this study we aim to (1) investigate heritability for articular cartilage regeneration in common inbred strains and RI lines and (2) to determine the correlation between ear-wound healing and articular cartilage regeneration. We found that cartilage regeneration is indeed a heritable trait in the LG/J by SM/J intercross so that this intercross can be used to dissect the genetic basis of variation in articular cartilage regeneration. Furthermore, a strong genetic correlation between the ability to heal ear-wounds and the ability to regenerate articular cartilage suggests a common underlying mechanism.
MATERIALS AND METHODS
All procedures were approved by the Animal Studies Committee at Washington University in St. Louis.
Mice
MRL/MpJ, DBA/1J, DBA/2J, C57BL/6J, LG/J and SM/J mice were obtained from The Jackson Laboratory (Bar Harbor ME). LGXSM RI lines were generated from the F2 intercross of LG/J females with the SM/J males. The detailed information and history of the RI lines has already been presented (30). We used nine RI lines namely LGXSM-4, LGXSM-5, LGXSM-6, LGXSM-18, LGXSM-19, LGXSM-33, LGXSM-35, LGXSM-46 and LGXSM-48 (Table-1).
Table 1.
Distribution of mice according to strain, time-point and phenotype. Least Squares Means ± Standard Error of the Mean (S.E.M.) are presented for articular cartilage regeneration score and filled ear-wound diameter at indicated time-points post-surgery.
| Strain | Time-point (Articular cartilage) |
N | Articular cartilage |
Ear-wound (30-day) |
|---|---|---|---|---|
| Time-course study | ||||
| LGXSM-6 | 4 wks | 12 | 3.62±0.63 | |
| LGXSM-6 | 8 wks | 09 | 3.43±0.73 | |
| LGXSM-6 | 12 wks | 10 | 2.82±0.69 | |
| LGXSM-6 | 16 wks | 18 | 5.70±0.52 | |
| LGXSM-33 | 4 wks | 06 | 2.54±0.91 | |
| LGXSM-33 | 8 wks | 09 | 3.65±0.73 | |
| LGXSM-33 | 12 wks | 11 | 2.80±0.67 | |
| LGXSM-33 | 16 wks | 07 | 3.26±0.84 | |
| Genetic study | ||||
| LGXSM-4 | 12 wks | 15 | 1.99±0.36 | 0.97±0.09 |
| LGXSM-5 | 12 wks | 12 | 3.70±0.40 | 0.96±0.10 |
| LGXSM-5 | 16 wks | 08 | 4.02±1.08 | 1.24±0.11 |
| LGXSM-6 | 12 wks | 07 | 3.16±0.52 | 1.23±0.13 |
| LGXSM-6 | 16 wks | 16 | 5.71±0.77 | 1.43±0.08 |
| LGXSM-18 | 12 wks | 10 | 2.30±0.43 | 0.89±0.11 |
| LGXSM-19 | 12 wks | 12 | 2.67±0.40 | 1.29±0.10 |
| LGXSM-33 | 12 wks | 11 | 2.84±0.42 | 0.49±0.10 |
| LGXSM-33 | 16 wks | 07 | 2.66±1.65 | 0.86±0.17 |
| LGXSM-35 | 12 wks | 15 | 2.93±036 | 1.05±0.09 |
| LGXSM-35 | 16 wks | 06 | 5.45±1.25 | 1.20±0.13 |
| LGXSM-46 | 12 wks | 13 | 2.76±0.38 | 1.42±0.09 |
| LGXSM-48 | 12 wks | 12 | 2.60±0.40 | 0.86±0.10 |
| LG/J | 12 wks | 08 | 3.34±0.49 | 1.68±0.12 |
| LG/J | 16 wks | 05 | 9.03±1.39 | 1.18±0.14 |
| SM/J | 12 wks | 16 | 2.81±0.35 | 0.53±0.09 |
| SM/J | 16 wks | 06 | 2.23±1.25 | 0.51±0.13 |
| MRL/MpJ | 12 wks | 11 | 5.29±0.77 | 1.19±0.07 |
| MRL/MpJ | 16 wks | 06 | 5.79±0.95 | 1.51±0.08 |
| DBA/1J | 12 wks | 04 | 2.22±1.27 | 0.48±0.11 |
| DBA/1J | 16 wks | 04 | 2.69±1.17 | 0.54±0.09 |
| DBA/2J | 12 wks | 09 | 4.28±1.42 | 0.43±0.07 |
| DBA/2J | 16 wks | 07 | 2.27±0.88 | 0.54±0.07 |
| C57BL/6J | 16 wks | 09 | 2.62±0.78 | 0.55±0.06 |
Full-thickness cartilage injury (Phenotyping)
At 8-weeks of age, bilateral full-thickness articular cartilage defects were created through microsurgery in the knee joints of both hind limbs (10). Briefly, mice were anesthetized by intraperitoneal injection (0.5–1.0-ml/kg body-weight) of rodent-cocktail comprised of ketamine (100-mg/ml), xylazine (20-mg/ml) and acepromazine (10-mg/ml). A small (0.5–1.0-cm) medial parapatellar skin-incision was made, the joint capsule opened and the patella luxated laterally to expose the trochlear groove articulating surface. A full-thickness lesion was made by creating a circular defect in the cartilage with a sterile 27Gx½″ needle using a circular motion until the subchondral bone was reached (Fig. 1A). Penetration of the subchondral bone was confirmed by the appearance of a blood droplet following removal of the needle. The joint capsule was closed with 6-0 absorbable polypropylene suture in a simple continuous pattern. Subcutaneous tissue was apposed with a single mattress suture and Vetclose skin-glue was applied on the skin to bridge over the closed edges. The mice were fully weight-bearing within 2–3 hours of surgery and showed no signs of lameness or systemic effects for the duration of the experiment.
Fig. 1.
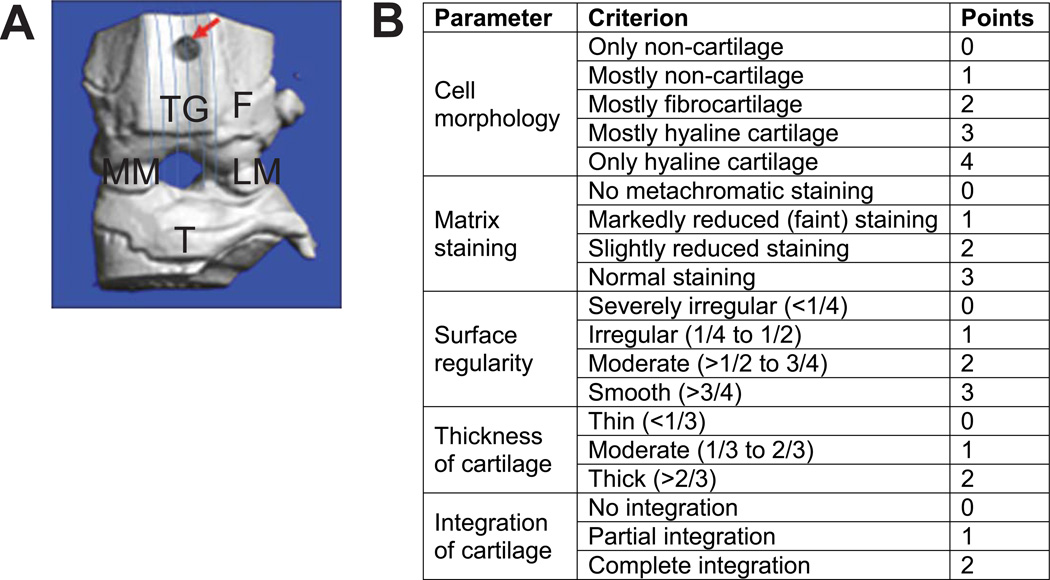
(A) Anatomical location of full-thickness articular cartilage lesion on the trochlear groove. Arrow indicates site of defect at zero time, lines indicate the direction of serial sagittal sections at 5-micron interval all through the trochlear groove, F = femur, TG = trochlear groove, MM = medial meniscus, LM = lateral meniscus, T = tibia. (B) A sketch of scoring sheet to grade histological sections for articualr cartilage regeneration.
Tissue harvesting and decalcification
Mice were sacrificed at indicated time-points (Table-1) using CO2 chambers. Both knee joints were dissected, fixed in 10% neutral buffered-formalin and decalcified using 10% formic acid in 5% formaldehyde solution for 48 hours on a rocking platform at 4°C. Following two washes with water, joints were incubated in 0.01M ethylenediaminetetraacetic acid in phosphate-buffered saline (PBS) for 4–6 hours at room-temperature. Finally the samples were washed four-times with water for 15 min each and subjected to a series of washes with increasing concentrations of ethanol for 30 min each. Samples were paraffin embedded, mounted in paraffin blocks and sagittally sectioned at 5-micron intervals extending through the entire trochlear groove using standard methods. The sections were mounted on polylysine-coated slides (Fischer Scientific) before staining.
Toluidine blue staining
After de-waxing, every third slide (at 15-micron intervals) was subjected to toluidine blue staining. Briefly, slides were immersed for 5 min in 0.04% toluidine blue solution prepared in 0.1M sodium acetate (pH 4.0). Then the slides were washed thrice with tap water, twice with absolute ethanol and finally twice with xylene before being cover-slipped.
Microscopy and histological scoring
Stained sections were viewed using 4X and 10X objectives on a bright-field microscope (Eclipse E800; Nikon) and images captured by a mounted camera (2000R Fast1394; Retiga) and QCapturePro (QImaging) software. Scoring of three selected sections was done by three observers (MFR, SH, EEJ) blinded to sample identity. Specimens were graded by accepted methodology (10) with slight modifications for a score ranging from 0 (no-healing) to 14 (full-healing) (Fig. 1B). The defect site was either easily recognizable by itself or was identified by a well-appreciated penetration mark of the needle through the growth plate which served as a persistent landmark to locate the defect.
Immunofluorescence for collagens
Paraffin sections were baked at 60°C for 2–4 hours followed by deparaffinization in xylene twice for 5 min each. Slides were then rehydrated in descending concentration of ethanol with final washes with distilled water and PBS. For antigen unmasking the slides were digested with 1% hyaluronidase in PBS for 30 min at 37°C in a customized humidified chamber. After washing with PBS, slides were blocked in 10% normal goat serum in PBS for 1 hour at room-temperature. After draining the blocking buffer off, slides were incubated with rabbit polyclonal antibody to collagen type I (ab34710, Abcam Inc.) or with antibody IIF (that recognizes the triple-helical domain of type II collagen protein) (31) at 1:200 dilutions in 1.5% goat serum in PBS and incubated at 4°C for overnight followed by three washes with PBS. The secondary antibodies used were goat anti-rabbit Alexa Fluor 488 (Invitrogen) and goat anti-rat Alexa Fluor 546 (Invitrogen) respectively at dilutions of 1:250 in 1.5% goat serum in PBS for 30 min at room-temperature. Finally, the slides were rinsed with distilled water before being sealed with a water-based mount. Sections were viewed using a 4X objective on a fluorescence microscope (Eclipse E800; Nikon) and images captured as described above.
Ear-wound punch (Phenotyping)
A through-and-through 2-mm in diameter hole was created bilaterally in the center of the cartilaginous part of external ear of 6-week old mice using a metal ear punch (Fisher Scientific) as previously described (32). The holes were measured at 30-days post-wounding by using a grid-etched reticle and healing area was calculated in millimeters by deducting the residual hole diameter from 2-mm.
Statistical analysis
Post-surgery time-course
All statistical analyses were performed in SYSTAT-12 (Systat Software, Inc.). We first examined a post-surgery time series for LGXSM-6 and LGXSM-33 as these two strains have been shown to vary substantially in ear-wound healing (27). Variation among strains, sexes and healing time-points was examined using ANOVA:
where Sex, Time post-surgery, and Strain are considered as fixed effects. A significant interaction term implies that there is significant variation in the timing of cartilage regeneration between the strains. As there were no significant sex differences, sex was removed from the model.
Genetic heritability
We used ANOVA to first test for significant differences between strains at each time-point for cartilage regeneration and ear-hole size using the following model:
with Sex as a fixed effect and Strain as a random effect. Sex was removed from the model because there were no significant sex differences. Since these are fully inbred strains, the broad-sense heritability of the traits was calculated by the following equation:
the variance among strains (σ2st) divided by the sum of the within (σ2r) and between (σ2st) strain variance. This includes all sources of genetic variation between strains. Genetic correlations were calculated using a similar procedure for the whole between strain variance/covariance matrices obtained from MANOVA including both cartilage regeneration and ear-wound healing.
Post-hoc pair-wise significance tests comparing specific strain pairs were performed using Tukey’s Honestly-Significant-Difference test to control for multiple comparisons.
RESULTS
Time-course study
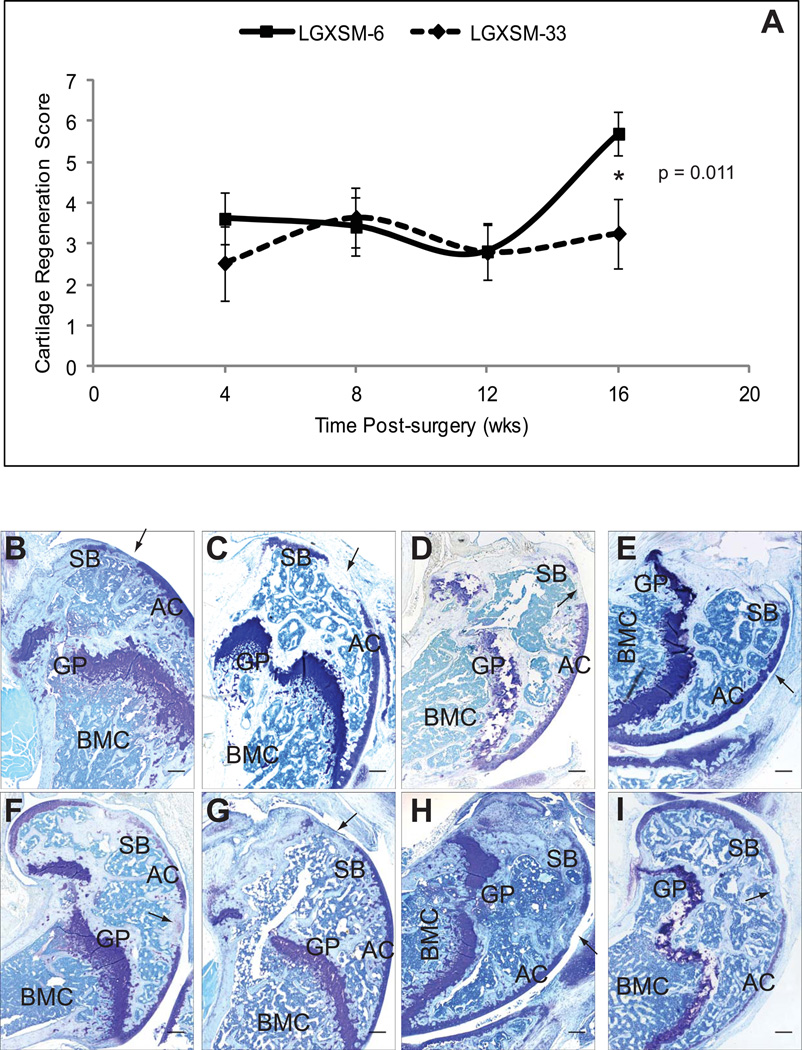
To examine the time-course patterns of articular cartilage regeneration, we first selected RI lines LGXSM-6 and LGXSM-33 and examined them at 4-, 8-, 12- and 16-weeks post-surgery. These strains were chosen because they respectively represent ear-wound healer and non-healer strains from the intercross of healer (LG/J) and non-healer (SM/J) parental lines (27). The time-course experiments revealed that there was no significant strain, sex, or strain by sex interaction effect on cartilage regeneration at 4-, 8-, or 12-weeks in these strains. However, when we compared healing at 16-weeks (Fig. 2A, Table-1), we found strain effects that were significantly (p<0.0003) stronger than those found at earlier ages (strain by post-surgery time-period interaction). At 16-weeks, LGXSM-6 regenerated cartilage significantly better than it had at 12-weeks (p<0.0002) and also significantly better than LGXSM-33 at both 12- (p<0.0004) and 16-weeks (p=0.011) post-injury. LGXSM-33 completely failed to regenerate its cartilage. The remarkable difference in cartilage regeneration observed between LGXSM-6 and LGXSM-33 was due to LGXSM-6 successfully producing a proteoglycan-positive extracellular matrix at the site of the defect by 16-weeks post-surgery, as indicated by toluidine blue staining (Fig. 2B–E), while LGXSM-33 never produced such a matrix (Fig. 2F–I). With our procedures and mice from the LG/J and SM/J intercross, significant healing does not occur until 16-weeks post-surgery. We also found no significant difference in healing between the left and right knees in either strain or at any time-point.
Fig. 2. Time-course of articular cartilage regeneration.
(A) A time-course analysis of articular cartilage regeneration in LGXSM-6 and LGXSM-33 lines showed a significant difference in regeneration only at 16-weeks post-surgery. (B–I) Representative sagittal sections of full-thickness articular cartilage lesions form LGXSM-6 (B–E) and LGXSM-33 (F–I) lines stained with toluidine blue for proteoglycan at 4-, 8-, 12- and 16-weeks post-surgery are shown. There was a significant proteoglycan deposition in LGXSM-6 (healer) compared to LGXSM-33 (non-healer) at 16-weeks time-point. At all other time-points, no significant regenerative response was observed. Asterisk (*) indicates statistical significance (p<0.011); arrow indicates site of defect; AC = articular cartilage, SB = subchondral bone, GP = growth plate, BMC = bone marrow cavity, bar (all panels) = 0.1 mm
Regeneration/repair in common inbred strains
The parental strains LG/J and SM/J, and several other inbred strains were also evaluated for cartilage regeneration and ear-wound healing including MRL/MpJ, DBA/1J, DBA/2J and C57BL/6J.
There was no significant difference among strains in cartilage regeneration at 12-weeks (p=0.088; H2=0.11). Although MRL/MpJ showed evidence of healing (data not shown); it only healed significantly better than SM/J (p=0.015). In contrast, there was highly significant genetic variation in ear-wound closure at 30-days (p<1×10−15; H2=0.84). The phenotypic correlation between cartilage regeneration and ear-wound healing was low (r=0.17) at 12-weeks. We were unable to calculate a genetic correlation because of the lack of genetic variation in articular cartilage regeneration at 12-weeks among these strains.
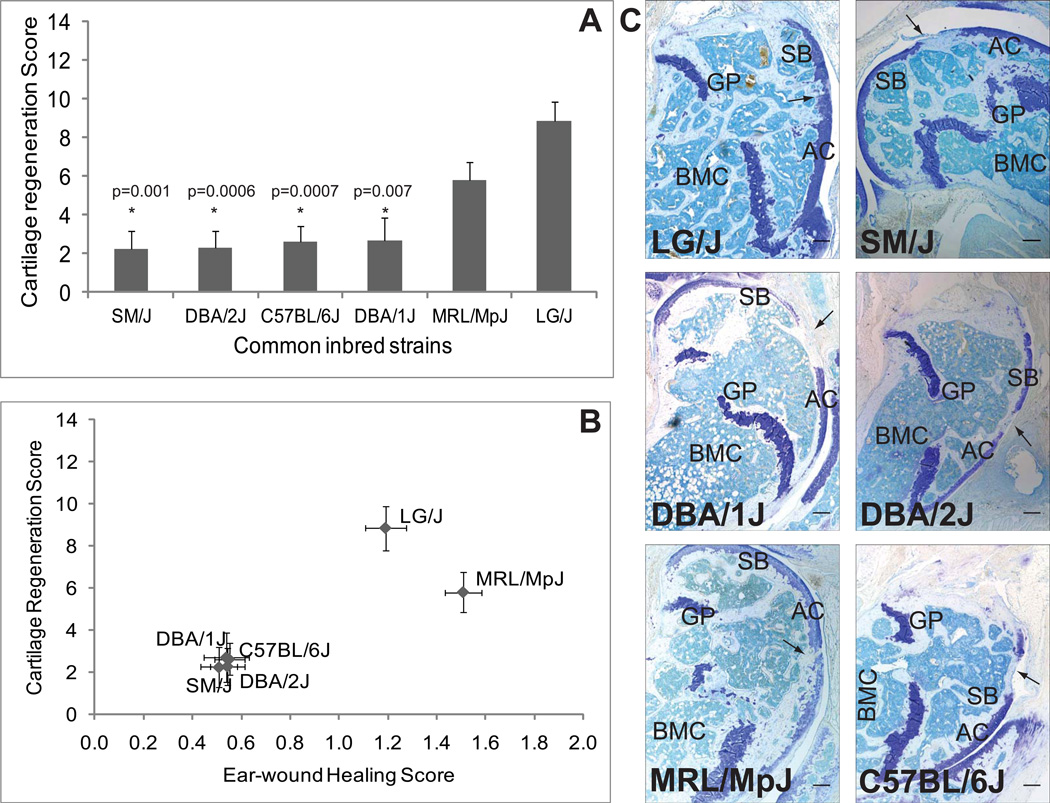
At 16-weeks post-injury of articular cartilage (Fig. 3A; Table-1) and 30-day after ear-punch, both articular cartilage regeneration and ear-hole closure are significantly heritable, with a heritability of 51% (p<0.0001) for articular cartilage regeneration and 84% (p<1×10−11) for ear-hole closure. The genetic correlation between the two different healing sites is extremely high (r=0.92, p<0.005; Fig. 3B) indicating that strains that heal wounds in the ear also regenerate articular cartilage. Post-hoc comparisons indicate that LG/J heals articular cartilage significantly better than C57BL/6J (p=0.0007), DBA/1J (p=0.007), DBA/2J (p=0.0006), and SM/J (p=0.001). MRL/MpJ has an intermediate level of articular cartilage regeneration, not significantly less than LG/J but not significantly more than the other strains. For ear-hole closure, LG/J and MRL/MpJ heal significantly better than C57BL/6J (p<0.0001; p<0.00001), DBA/1J (p<0.0001; p<0.00001), DBA/2J (p<0.0001, p<0.00001), and SM/J (p<0.0001, p<0.00001). Again, there is no significant difference in ear-wound healing between LG/J and MRL/MpJ. LG/J and MRL/MpJ are healers for both articular cartilage and ear-wounds while C57BL/6J, DBA/1J, DBA/2J, and SM/J are non-healers.
Fig. 3. Articular cartilage regeneration and ear-wound healing in common inbred strains.
(A) Articular cartilage regeneration in common inbred strains showed no healing response in SM/J, DBA/2J, C57BL/6J and DBA/1J strains while it showed better healing ability in MRL/MpJ and LG/J strains. Asterisk (*) with respective p values indicates statistically significant difference from LG/J strain. (B) Correlation between articular cartilage regeneration and ear-wound healing showed that the two phenotypes are strongly correlated in common inbred strains. (C) Representative sagittal sections of full-thickness articular cartilage lesions from common inbred strains namely LG/J, SM/J, MRL/MpJ, DBA/1J, DBA/2J and C57BL/6J showed that only LG/J and MRL/MpJ could show proteoglycan deposition indicating a better regenerative response compared to SM/J, DBA/1J, DBA72J and C57BL/6J strains. Arrow indicates site of defect; AC = articular cartilage, SB = subchondral bone, GP = growth plate, BMC = bone marrow cavity; bar (all panels) = 0.1 mm
The defect area in the non-healers showed mostly fibrous tissue with no cartilage matrix staining as determined by toluidine blue. Both MRL/MpJ and LG/J could regenerate cartilage and the defect site appeared to be hyaline cartilage as it showed proteoglycan staining similar to neighboring undisturbed cartilage. It also appeared smooth, of similar thickness compared to adjacent cartilage, and was better integrated with the neighboring cartilage tissue (Fig. 3C).
Regeneration/repair in the LGXSM RI lines
The comparison of RI lines, including the parental strains, at 12-weeks showed no statistically significant strain (p=0.26), sex (p=0.06), or strain by sex (p=0.51) interaction effects on articular cartilage regeneration. The average histological score for these strains was low at 12-weeks indicating a uniformly weak healing response (Table-1). Furthermore, the strains failed to produce a proteoglycan-positive extracellular matrix at the site of defect as indicated by the absence of toluidine blue staining. Also the defect remained unfilled (no hyaline cartilage) and contained fewer chondrocytes at the wound site (not shown).
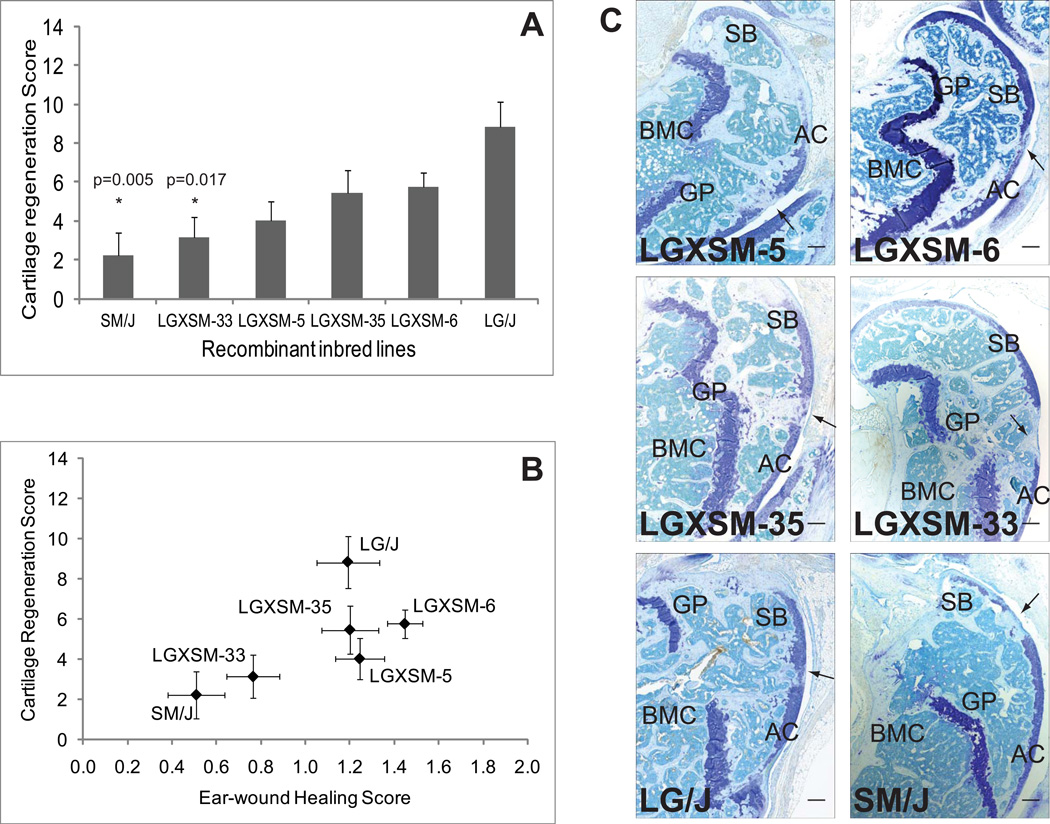
Significant strain differences were observed at 16-weeks (Fig. 4A; Table-1) with a significant strain by post-injury time effect on cartilage regeneration (p=0.0004). By 16-weeks the level of variation between strains had increased, with a broad-sense heritability of 26% (p<0.006). At 16-weeks LG/J showed the best cartilage regeneration, being significantly better healed than SM/J (p=0.005), LGXSM-33 (p=0.017), and better than LGXSM-5 (p=0.054) at a borderline significance level. None of the other pair-wise differences were significant when controlling for multiple comparisons, although after LG/J, LGXSM-6 and LGXSM-35 show moderate healing, and LGXSM-5, LGXSM-33, and SM/J show poor healing. The differences between LGXSM-6 and LGXSM-33 are not statistically significant here although they were statistically significant in the time-course analysis because here we adjusted the probability for multiple comparisons. However, taken alone, the strains at 16-weeks show the same differences they showed in time-course analysis. The heritability of 30-day ear-wound healing for the LGXSM RI lines is 53% (p<0.00001). Several strains heal significantly better than SM/J, including LG/J (p<0.01), LGXSM-35 (p<0.006), LGXSM-5 (p<0.001), and LGXSM-6 (p<0.00001), and LGXSM-6 heals significantly better than LGXSM-33 (p=0.0003). The genetic correlation for the RI line set is quite high (r=0.86; p=0.014) (Fig. 4B), especially given the environmental correlation of −0.12 which is not significantly different from zero.
Fig. 4. Articular cartilage regeneration and ear-wound healing in recombinant inbred lines.
(A) Articular cartilage regeneration in recombinant inbred lines showed no healing response in SM/J, LGXSM-5 and LGXSM-33 strains while intermediate to good healing responses were observed in LGXSM-6, LGXSM-35 and LG/J strains. Asterisk (*) with p given values indicates statistically significant difference from parental strain LG/J. (B) Correlation between knee articular cartilage regeneration and ear-wound healing showed that the two phenotypes are strongly correlated in the recombinant inbred lines. (C) Representative sagittal sections of full-thickness articular cartilage lesions from recombinant inbred lines LGXSM-5, LGXSM-6, LGXSM-33 and LGXSM-35 and parental strains LG/J and SM/J show that except for strains LGXSM-33 and SM/J, all other strains regenerated their articular cartilage and showed a deposition of proteoglycan indicating regenerative response. Arrow indicates site of defect; AC = articular cartilage, SB = subchondral bone, GP = growth plate, BMC = bone marrow cavity; bar (all panels) = 0.1 mm
Histologically, the site of articular cartilage defect in RI lines showed mostly fibrous tissue with no cartilage matrix staining in non-healers. In regenerating strains the injured area had hyaline cartilage based on proteoglycan staining similar to adjacent intact cartilage. The cartilage also appeared relatively smooth, of almost similar thickness compared to adjacent cartilage, and was better integrated with the neighboring cartilage tissue (Fig. 4C).
The full-thickness articular cartilage injuries in these mice involved the penetration of the needle through the growth plate. While articular cartilage showed regeneration in healer strains, no significant healing of growth plate cartilage was noted (not shown). It is known that growth plate does not have the ability to regenerate on its own after an injury. We made the injury in mature mice so we did not see any regeneration of growth plate in these mice.
Collagen staining
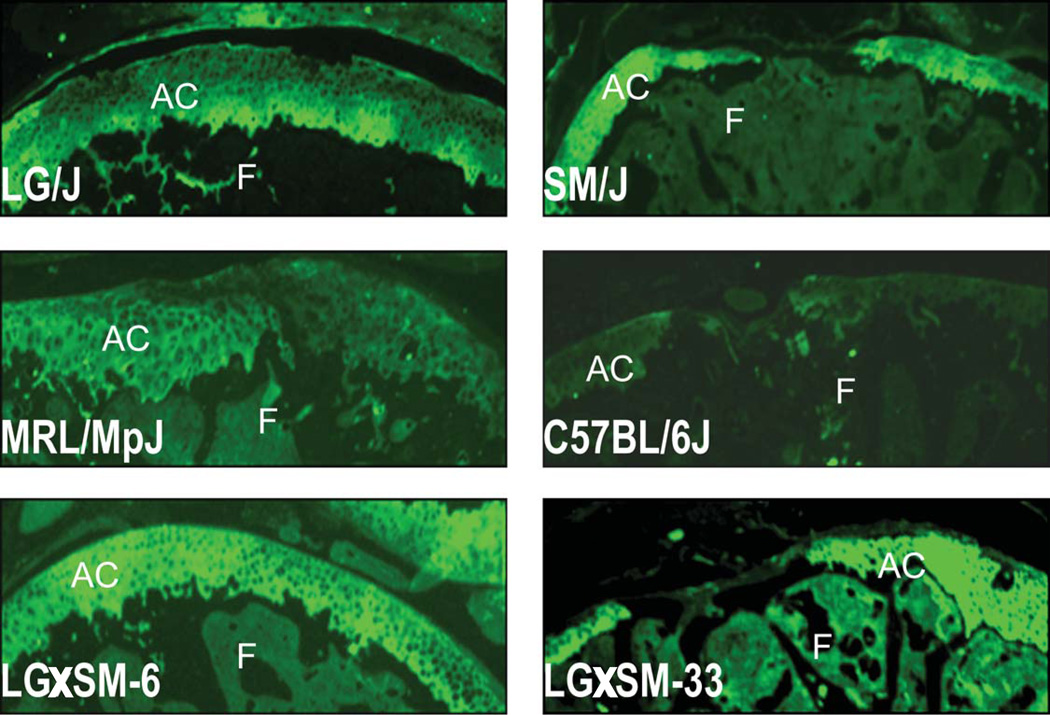
There was no collagen type I deposition on the regenerated or non-regenerated defect site or on the native healthy cartilage, however, bone showed positive signals for collagen type I. On the other hand, healthy cartilage as well as regenerated cartilage in healer strains showed collagen type II deposition. In the non-regenerating strains, the defect site did not stain for this type of collagen as compared to neighboring undamaged cartilage (Fig. 5).
Fig. 5. Collagen staining.
Representative sagittal sections from selected healer and non-healer strains strained with type II collagen show that there was collagen type II positive staining at the site of injury after it has healed. In non-healer strains, there was no collagen type II staining at the injury site. AC = articular cartilage, F = femur
DISCUSSION
There are several reports on cartilage degeneration in OA pathophysiology but little evidence is available on genetics of cartilage regeneration. Here, we have demonstrated regeneration of full-thickness articular cartilage lesions in mouse strains each with a defined genetic composition. While genetic analyses have clearly shown that tissue regeneration is a complex multigenic trait (33), we hypothesize that any differential phenotype between the strains will be attributed to a restricted set of genomic regions.
The present study segregates RI lines into three distinct categories of articular cartilage regeneration: good, intermediate and poor healers suggesting that rearrangement of the alleles in the RI lines segregated genes that influence regeneration. The broad-sense heritability showed that articular cartilage regeneration is significantly heritable trait (p=0.006). Further, the genetic correlations between the two healing phenotypes (articular cartilage regeneration and ear-wound healing) were found to be extremely high indicating that the two processes share a common genetic basis.
MRL/MpJ and its close relative, LG/J, have unique healing and regenerative capabilities including the complete closure of through-and-through ear puncture wounds with normal tissue architecture without scarring over time and thus these strains display similar ear-wound healing phenotypes (9–13, 34, 35). The MRL/MpJ strain was originally generated from a series of crosses between AKR/J, C3H/HeDi, C57BL/6J and two final backcrosses to LG/J (2), so that MRL/MpJ and LG/J share 75% of their genomes identical-by-descent (9, 13, 35). Here we confirm the earlier ear-wound healing results and show that both MRL/MpJ, as in Fitzgerald et al. (10), and LG/J also regenerate articular cartilage over time. This is in contrast to C57BL/6J and SM/J stains, which are known non-healers for ear-wounds (10, 11, 13, 21) and articular cartilage injuries (10, 21). These observations indicate that this exceptional healing ability of LG/J and MRL/MpJ is likely due to their shared genetic composition. Furthermore, the high genetic correlation observed among common inbred strains between articular cartilage regeneration and ear-hole closure indicate that the same cellular and tissue mechanisms are likely to be responsible for healing in MRL/MpJ and LG/J.
Eltawil and colleagues (21) found that DBA/1 can regenerate articular cartilage under certain conditions. We were unable to confirm the wound-healing ability of this strain as it failed to regenerate/repair articular cartilage or ear-wounds. There could be multiple reasons for our failure to replicate. First, we did not follow the same protocol (21). While they produced a full-thickness cartilage defect void in the patellar groove, they did not pierce the underlying bone. Also, we tested DBA/1J in St. Louis (obtained from The Jackson Laboratory), but the origin of the DBA/1 animals used (21) in London is not clear. The DBA/1OlaHsd strain was separated from DBA/1J over 55 years ago while the DBA/1JBomTac strain was separated from DBA/1J nearly 20 years ago. New mutations or even unintended mixing may have occurred over these hundreds of generations. Thus, the rationale for the difference between our results and those of Eltawil et al. (21) in cartilage regeneration of DBA/1 mice remains unknown and may be attributed to genetic drift or differences in surgical procedure.
The ear-wounds of healer strains rapidly re-epithelialize with re-growth of both hair follicles and elastic cartilage while non-healer strains, including C57BL/6J, 129/SvJ, SM/J, DBA/1J and DBA/2J, form scar tissue around the edges of the wound (9–13, 35, 36). In addition, increased angiogenesis, cell proliferation, matrix formation and fibroblast migration occurs in tissue undergoing repair in the healer strains compared to the non-healer strains. MRL/MpJ, has also demonstrated a superior microvasculature response to skin wounds (37). Thus, the healing properties of MRL/MpJ, and likely LG/J, are a result of multiple pathways and not due to a single cause. We believe that the quality of repair in the ear-wound in our healer strains is similar to what has already been reported for MRL/MpJ (12, 36) and LG/J (9) with similar genes involved in the healing process (38). Therefore, in the presence of convincing data on the nature of healed ear-tissues we took it as exemplary for our study.
Several genetic linkage studies (32, 33, 39) have demonstrated that ear-wound healing is a complex genetic trait with contributions from approximately 20 quantitative trait loci (QTLs). The large number of QTLs involved in distinguishing healer and non-healer strains indicates plausible contributions from many genes involved in many different cellular- and tissue-level processes in the wound healing phenotype (38). Although a strong sexual dimorphism in the rate of ear-wound closure in the LGXSM cross exists, the QTLs have largely the same effect in both sexes (40). In our smaller study, we failed to detect sex differences in ear-would healing. Articular cartilage regeneration response in MRL/MpJ mice has been reported to be sexually dimorphic at 12-weeks albeit no significant sex differences were observed at any earlier time-point (10). Here, with larger sample size and more statistical power, we failed to detect sex difference for articular cartilage regeneration.
The specific reason for some strains being healers and others not remains unknown. However, healer strains MRL/MpJ, LG/J and LGXSM-6 have been shown to share some unique molecular features with the classical regenerators with respect to ear-wound healing. Healer strains have a distinct cell cycle phenotype i.e. a G2/M stage arrest and a heightened basal and wound-site DNA damage/repair response (27).
The LG/J and SM/J inbred strains differ for a wide variety of phenotypes, including growth (41), obesity and diabetes (42, 43), long bone growth (44–46), skeletal morphology (47), bone biomechanical properties (48), muscle-weight (49), and ear-wound healing (13). Further, each RI line genome is an independent isogenic line with genetic variance concentrated between lines and eliminated within lines. Previous work related to ear-wound healing has identified LGXSM-6 as a line that heals ear-wounds while LGXSM-33 fails to heal (27). For all these phenotypes, we have found that genetic variation is due to many genes of small effect and their interactions with the genetic background. The finding that LGXSM-5 has a relatively high ear-wound healing score and a relatively low cartilage regeneration score shows that the correlation between the traits is less than 1.00 (r=0.86) and suggests that there are a few genomic regions with differential effects on the two forms of healing.
We believe that this is the first study that characterizes and reports articular cartilage and ear-wound healing phenotypes from a genetics viewpoint in a segregating population. The key findings from our RI lines study are that (1) variation in ear-wound healing and articular cartilage regeneration are both heritable, differences between RI lines being due to genetic differences; (2) the genetic correlation between ear-wound healing and articular cartilage regeneration is very strong indicating that the two processes likely share a common genetic basis; (3) there is no correlation between ear-wound healing and articular cartilage regeneration due to environmental, non-genetic, factors; and (4) LGXSM-6 is a RI line that responds to injury in a fashion similar to LG/J while many other RI lines have intermediate healing phenotypes for both ear-wound healing and articular cartilage regeneration.
As a consequence of these findings, it is apparent that the LG/J by SM/J intercross segregates for multiple genetic factors affecting articular cartilage regeneration and that the genetic basis for this difference can be genetically mapped in the intercross population. In addition to the RI lines used here, there is also an advanced intercross line (45) in which fine-mapping studies can be performed. The previously mapped QTLs affecting ear-wound healing are also likely to affect cartilage regeneration in the knee, so that fine-mapped ear-wound healing loci are strong candidates for loci also affecting articular cartilage regeneration.
These studies may also shed light on protection of healer strains from developing OA. In a companion study (50) (unpublished data), we have demonstrated that a strain (LGXSM-6) shown to regenerate articular cartilage and heal ear-wound is relatively protected from post-traumatic OA compared to the strain LGXSM-33, which has more extensive OA and does not regenerate articular cartilage or ear-wounds. Thus, capacity to regenerate articular cartilage is inversely correlated with the development of post-traumatic OA as has been provisionally suggested (26).
The phenotypic differences observed among RI lines can be attributed to a restricted set of genes differentially inherited from the parental strains due to random segregation of genomic regions that influence regeneration. Taken together, this project creates a unique resource for the study of genes that contribute to tissue healing, cartilage regeneration and thus, perhaps, to protection from OA.
Acknowledgements
We acknowledge with thanks the important technical support by Crystal Idleburg, John Freeman, Elizabeth DeLassus, Patricia Keller (Late), Timothy Morris and Joseph Futhey.
Financial support
This project was supported by an ARRA Grand Opportunity Grant, Award Number RC2-AR058978 (LJS, JMC) and the histological analysis was supported by Musculoskeletal Research Center, Award Number P30-AR057235 from the National Institute of Arthritis, Musculoskeletal and Skin Diseases.
Footnotes
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Arthritis, Musculoskeletal and Skin Diseases or the National Institutes of Health.
Author contributions:
Designed research: LJS, JMC
Performed research: MFR, SH, EEJ, KLJ, JF
Contributed new reagents or analytic tools: JF, EHK
Analyzed data: MFR, LJS, SH, EEJ, JMC
Wrote the paper: MFR, LJS, JMC
References
- 1.Gierer A, Berking S, Bode H, David CN, Flick K, Hansmann G, et al. Regeneration of hydra from reaggregated cells. Nat New Biol. 1972;239(91):98–101. doi: 10.1038/newbio239098a0. [DOI] [PubMed] [Google Scholar]
- 2.Murphy ED, Roths JB. Autoimmunity and lymphproliferation: Induction by mutant gene lpr and acceleration by a male-associated factor in strain BXSB. In: Rose NR, Bigazzi PE, Warner NL, editors. Genetic Control of Autoimmune Disease. New York: Elsevier; 1979. pp. 207–220. [Google Scholar]
- 3.Stocum DL. The urodele limb regeneration blastema. Determination and organization of the morphogenetic field. Differentiation. 1984;27(1):13–28. doi: 10.1111/j.1432-0436.1984.tb01403.x. [DOI] [PubMed] [Google Scholar]
- 4.Tanaka EM. Regeneration: if they can do it, why can't we? Cell. 2003;113(5):559–562. doi: 10.1016/s0092-8674(03)00395-7. [DOI] [PubMed] [Google Scholar]
- 5.Harty M, Neff AW, King MW, Mescher AL. Regeneration or scarring: an immunologic perspective. Dev Dyn. 2003;226(2):268–279. doi: 10.1002/dvdy.10239. [DOI] [PubMed] [Google Scholar]
- 6.Kierdorf U, Kierdorf H. Deer antlers - a model of mammalian appendage regeneration: an extensive review. Gerontology. 2011;57(1):53–65. doi: 10.1159/000300565. [DOI] [PubMed] [Google Scholar]
- 7.Carlson BM. Some principles of regeneration in mammalian systems. Anat Rec B New Anat. 2005;287(1):4–13. doi: 10.1002/ar.b.20079. [DOI] [PubMed] [Google Scholar]
- 8.ten Koppel PG, van Osch GJ, Verwoerd CD, Verwoerd-Verhoef HL. A new in vivo model for testing cartilage grafts and biomaterials: the 'rabbit pinna punch-hole' model. Biomaterials. 2001;22(11):1407–1414. doi: 10.1016/s0142-9612(00)00298-2. [DOI] [PubMed] [Google Scholar]
- 9.Clark LD, Clark RK, Heber-Katz E. A new murine model for mammalian wound repair and regeneration. Clin Immunol Immunopathol. 1998;88(1):35–45. doi: 10.1006/clin.1998.4519. [DOI] [PubMed] [Google Scholar]
- 10.Fitzgerald J, Rich C, Burkhardt D, Allen J, Herzka AS, Little CB. Evidence for articular cartilage regeneration in MRL/MpJ mice. Osteoarthritis Cartilage. 2008;16(11):1319–1326. doi: 10.1016/j.joca.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 11.Kench JA, Russell DM, Fadok VA, Young SK, Worthen GS, Jones-Carson J, et al. Aberrant wound healing and TGF-beta production in the autoimmune-prone MRL/+ mouse. Clin Immunol. 1999;92(3):300–310. doi: 10.1006/clim.1999.4754. [DOI] [PubMed] [Google Scholar]
- 12.Heber-Katz E, Leferovich JM, Bedelbaeva K, Gourevitch D. Spallanzani's mouse: a model of restoration and regeneration. Curr Top Microbiol Immunol. 2004;280:165–189. doi: 10.1007/978-3-642-18846-6_5. [DOI] [PubMed] [Google Scholar]
- 13.Blankenhorn EP, Bryan G, Kossenkov AV, Clark LD, Zhang XM, Chang C, et al. Genetic loci that regulate healing and regeneration in LG/J and SM/J mice. Mamm Genome. 2009;20(11–12):720–733. doi: 10.1007/s00335-009-9216-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anraku Y, Mizuta H, Sei A, Kudo S, Nakamura E, Senba K, et al. The chondrogenic repair response of undifferentiated mesenchymal cells in rat full-thickness articular cartilage defects. Osteoarthritis Cartilage. 2008;16(8):961–964. doi: 10.1016/j.joca.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 15.Shapiro F, Koide S, Glimcher MJ. Cell origin and differentiation in the repair of full-thickness defects of articular cartilage. J Bone Joint Surg Am. 1993;75(4):532–553. doi: 10.2106/00004623-199304000-00009. [DOI] [PubMed] [Google Scholar]
- 16.Breinan HA, Hsu HP, Spector M. Chondral defects in animal models: effects of selected repair procedures in canines. Clin Orthop Relat Res. 2001;(391 Suppl):S219–S230. [PubMed] [Google Scholar]
- 17.Convery FR, Akeson WH, Keown GH. The repair of large osteochondral defects. An experimental study in horses. Clin Orthop Relat Res. 1972;82:253–262. [PubMed] [Google Scholar]
- 18.Wei X, Gao J, Messner K. Maturation-dependent repair of untreated osteochondral defects in the rabbit knee joint. J Biomed Mater Res. 1997;34(1):63–72. doi: 10.1002/(sici)1097-4636(199701)34:1<63::aid-jbm9>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 19.De Bari C, Dell'accio F. Mesenchymal stem cells in rheumatology: a regenerative approach to joint repair. Clin Sci (Lond) 2007;113(8):339–348. doi: 10.1042/CS20070126. [DOI] [PubMed] [Google Scholar]
- 20.Frisbie DD, Oxford JT, Southwood L, Trotter GW, Rodkey WG, Steadman JR, et al. Early events in cartilage repair after subchondral bone microfracture. Clin Orthop Relat Res. 2003;(407):215–227. doi: 10.1097/00003086-200302000-00031. [DOI] [PubMed] [Google Scholar]
- 21.Eltawil NM, De Bari C, Achan P, Pitzalis C, Dell'accio F. A novel in vivo murine model of cartilage regeneration. Age and strain-dependent outcome after joint surface injury. Osteoarthritis Cartilage. 2009;17(6):695–704. doi: 10.1016/j.joca.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buckwalter JA, Mankin HJ. Articular cartilage: degeneration and osteoarthritis, repair, regeneration, and transplantation. Instr Course Lect. 1998;47:487–504. [PubMed] [Google Scholar]
- 23.Hunziker EB. Articular cartilage repair: basic science and clinical progress. A review of the current status and prospects. Osteoarthritis Cartilage. 2002;10(6):432–463. doi: 10.1053/joca.2002.0801. [DOI] [PubMed] [Google Scholar]
- 24.Ding C, Cicuttini F, Jones G. How important is MRI for detecting early osteoarthritis? Nat Clin Pract Rheumatol. 2008;4(1):4–5. doi: 10.1038/ncprheum0676. [DOI] [PubMed] [Google Scholar]
- 25.Anderson DD, Chubinskaya S, Guilak F, Martin JA, Oegema TR, Olson SA, et al. Post-traumatic osteoarthritis: improved understanding and opportunities for early intervention. J Orthop Res. 2011;29(6):802–809. doi: 10.1002/jor.21359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ward BD, Furman BD, Huebner JL, Kraus VB, Guilak F, Olson SA. Absence of posttraumatic arthritis following intraarticular fracture in the MRL/MpJ mouse. Arthritis Rheum. 2008;58(3):744–753. doi: 10.1002/art.23288. [DOI] [PubMed] [Google Scholar]
- 27.Bedelbaeva K, Snyder A, Gourevitch D, Clark L, Zhang XM, Leferovich J, et al. Lack of p21 expression links cell cycle control and appendage regeneration in mice. Proc Natl Acad Sci U S A. 2010;107(13):5845–5850. doi: 10.1073/pnas.1000830107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hunter DJ, Snieder H, March L, Sambrook PN. Genetic contribution to cartilage volume in women: a classical twin study. Rheumatology (Oxford) 2003;42(12):1495–1500. doi: 10.1093/rheumatology/keg400. [DOI] [PubMed] [Google Scholar]
- 29.Spector TD, Cicuttini F, Baker J, Loughlin J, Hart D. Genetic influences on osteoarthritis in women: a twin study. BMJ. 1996;312(7036):940–943. doi: 10.1136/bmj.312.7036.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hrbek T, de Brito RA, Wang B, Pletscher LS, Cheverud JM. Genetic characterization of a new set of recombinant inbred lines (LGXSM) formed from the inter-cross of SM/J and LG/J inbred mouse strains. Mamm Genome. 2006;17(5):417–429. doi: 10.1007/s00335-005-0038-7. [DOI] [PubMed] [Google Scholar]
- 31.Zhu Y, Oganesian A, Keene DR, Sandell LJ. Type IIA procollagen containing the cysteine-rich amino propeptide is deposited in the extracellular matrix of prechondrogenic tissue and binds to TGF-beta1 and BMP-2. Journal of Cell Biology. 1999;144(5):1069–1080. doi: 10.1083/jcb.144.5.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McBrearty BA, Clark LD, Zhang XM, Blankenhorn EP, Heber-Katz E. Genetic analysis of a mammalian wound-healing trait. Proc Natl Acad Sci U S A. 1998;95(20):11792–11797. doi: 10.1073/pnas.95.20.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masinde GL, Li X, Gu W, Davidson H, Mohan S, Baylink DJ. Identification of wound healing/regeneration quantitative trait loci (QTL) at multiple time points that explain seventy percent of variance in (MRL/MpJ and SJL/J) mice F2 population. Genome Res. 2001;11(12):2027–2033. doi: 10.1101/gr.203701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chadwick RB, Bu L, Yu H, Hu Y, Wergedal JE, Mohan S, et al. Digit tip regrowth and differential gene expression in MRL/Mpj, DBA/2, and C57BL/6 mice. Wound Repair Regen. 2007;15(2):275–284. doi: 10.1111/j.1524-475X.2007.00216.x. [DOI] [PubMed] [Google Scholar]
- 35.Masinde GL, Li R, Nguyen B, Yu H, Srivastava AK, Edderkaoui B, et al. New quantitative trait loci that regulate wound healing in an intercross progeny from DBA/1J and 129 x 1/SvJ inbred strains of mice. Funct Integr Genomics. 2006;6(2):157–163. doi: 10.1007/s10142-005-0004-1. [DOI] [PubMed] [Google Scholar]
- 36.Gourevitch D, Clark L, Chen P, Seitz A, Samulewicz SJ, Heber-Katz E. Matrix metalloproteinase activity correlates with blastema formation in the regenerating MRL mouse ear hole model. Dev Dyn. 2003;226(2):377–387. doi: 10.1002/dvdy.10243. [DOI] [PubMed] [Google Scholar]
- 37.Liu F, Smith J, Zhang Z, Cole R, Herron BJ. Genetic heterogeneity of skin microvasculature. Dev Biol. 2010;340(2):480–489. doi: 10.1016/j.ydbio.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li X, Gu W, Masinde G, Hamilton-Ulland M, Xu S, Mohan S, et al. Genetic control of the rate of wound healing in mice. Heredity. 2001;86(Pt 6):668–674. doi: 10.1046/j.1365-2540.2001.00879.x. [DOI] [PubMed] [Google Scholar]
- 39.Heber-Katz E, Chen P, Clark L, Zhang XM, Troutman S, Blankenhorn EP. Regeneration in MRL mice: further genetic loci controlling the ear hole closure trait using MRL and M.m. Castaneus mice. Wound Repair Regen. 2004;12(3):384–392. doi: 10.1111/j.1067-1927.2004.012308.x. [DOI] [PubMed] [Google Scholar]
- 40.Cheverud JM, Lawson HA, Funk R, Zhou J, Blankenhorn EP, Heber-Katz E. Healing quantitative trait loci in a combined cross analysis using related mouse strain crosses. Heredity. 2011 doi: 10.1038/hdy.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaughn TT, Pletscher LS, Peripato A, King-Ellison K, Adams E, Erikson C, et al. Mapping quantitative trait loci for murine growth: a closer look at genetic architecture. Genet Res. 1999;74(3):313–322. doi: 10.1017/s0016672399004103. [DOI] [PubMed] [Google Scholar]
- 42.Cheverud JM, Ehrich TH, Hrbek T, Kenney JP, Pletscher LS, Semenkovich CF. Quantitative trait loci for obesity- and diabetes-related traits and their dietary responses to high-fat feeding in LGXSM recombinant inbred mouse strains. Diabetes. 2004;53(12):3328–3336. doi: 10.2337/diabetes.53.12.3328. [DOI] [PubMed] [Google Scholar]
- 43.Cheverud JM, Vaughn TT, Pletscher LS, Peripato AC, Adams ES, Erikson CF, et al. Genetic architecture of adiposity in the cross of LG/J and SM/J inbred mice. Mamm Genome. 2001;12(1):3–12. doi: 10.1007/s003350010218. [DOI] [PubMed] [Google Scholar]
- 44.Norgard EA, Roseman CC, Fawcett GL, Pavlicev M, Morgan CD, Pletscher LS, et al. Identification of quantitative trait loci affecting murine long bone length in a two-generation intercross of LG/J and SM/J Mice. J Bone Miner Res. 2008;23(6):887–895. doi: 10.1359/JBMR.080210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Norgard EA, Jarvis JP, Roseman CC, Maxwell TJ, Kenney-Hunt JP, Samocha KE, et al. Replication of long-bone length QTL in the F9-F10 LG,SM advanced intercross. Mamm Genome. 2009;20(4):224–235. doi: 10.1007/s00335-009-9174-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanger TJ, Norgard EA, Pletscher LS, Bevilacqua M, Brooks VR, Sandell LJ, et al. Developmental and genetic origins of murine long bone length variation. J Exp Zool B Mol Dev Evol. 2011;316B(2):146–161. doi: 10.1002/jez.b.21388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kenney-Hunt JP, Vaughn TT, Pletscher LS, Peripato A, Routman E, Cothran K, et al. Quantitative trait loci for body size components in mice. Mamm Genome. 2006;17(6):526–537. doi: 10.1007/s00335-005-0160-6. [DOI] [PubMed] [Google Scholar]
- 48.Reich MS, Jarvis JP, Silva MJ, Cheverud JM. Genetic relationships between obesity and osteoporosis in LGXSM recombinant inbred mice. Genet Res (Camb) 2008;90(5):433–444. doi: 10.1017/S0016672308009798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lionikas A, Cheng R, Lim JE, Palmer AA, Blizard DA. Fine-mapping of muscle weight QTL in LG/J and SM/J intercrosses. Physiol Genomics. 2010;42A(1):33–38. doi: 10.1152/physiolgenomics.00100.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hashimoto S, Rai MF, Janiszak KL, Cheverud JM, Sandell LJ. Development of OA and tissue regenration in genetic strains of mice. Osteoarthritis Cartilage. 2011 doi: 10.1016/j.joca.2012.01.022. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]