Abstract
Flavonoids are specialized compounds widely distributed and with diverse functions throughout the plant kingdom and with several benefits for human health. In particular, flavonols, synthesized by flavonol synthase (FLS), protect plants against UV-B radiation and are essential for male fertility in maize and other plants. We have recently characterized a UV-B inducible ZmFLS1, corresponding to the first to be described in monocot plants. Interestingly, the new assembly of the B73 maize genome revealed the presence of a second putative FLS gene (ZmFLS2), with very high identity with ZmFLS1. ZmFLSs expression was analyzed in different maize tissues, and by combining electrophoretic mobility shift assays and transient expression experiments, we show that both genes are direct targets of anthocyanin (C1/PL1 + R/B) and 3-deoxy flavonoid (P1) transcriptional regulators. ZmFLS expression analyses show higher levels of both transcripts in high altitude landraces than inbred lines, and both genes are regulated by UV-B radiation in all lines analyzed. Moreover, the high sequence conservation of the ZmFLS promoters between maize lines suggests that the differences observed in ZmFLS expression are due to allelic variations in the transcription factors that regulate their activities. Finally, we generated pFLS1::FLS1-RFP transgenic plants and analyzed ZmFLS1 expression in different maize tissues; we found that this enzyme is localized in the ER and the perinuclear region.
Keywords: UV-B, duplication, grasses, natural variation, maize
Introduction
Flavonoids are widely distributed plant metabolites with diverse biological functions. There is considerable evidence showing a role for flavonoids in contributing to the human health associated to their antioxidant, anti-proliferative and anti-inflammatory properties, and consequently, their indication in prevention against cancer and cardiovascular disease (Knekt et al., 2000; Hirvonen et al., 2001; Mak et al., 2006; Vargo et al., 2006; Geleijnse and Hollman, 2008; Kaur et al., 2008; Kang et al., 2009). Flavonols, the most abundant and widespread subgroup of flavonoids, play important functions in plant physiology, growth, and development, including the modulation of basipetal auxin transport in Arabidopsis (Brown et al., 2001; Peer et al., 2004; Kuhn et al., 2011; Lewis et al., 2011), attraction of and defense against insects (Gronquist et al., 2001), pollen fertility (Mo et al., 1992; Taylor and Jorgensen, 1992; Ylstra et al., 1994; Taylor and Hepler, 1997), and UV-B protection (Solovchenko and Schmitz-Eiberger, 2003; Jaakola et al., 2004; Stracke et al., 2007, 2010a; Kusano et al., 2011). Moreover, flavonols have recently been shown to participate in the ethylene-signaling pathway (Lewis et al., 2011).
The biosynthesis of flavonols from dihydroflavonols is catalyzed by the enzyme flavonol synthase (FLS), a soluble 2-oxoglutarate-dependent dioxygenase (2-ODD). FLS cDNAs were cloned from a large number of dicot plants, and they were functionally expressed in bacteria, yeast, and plants (Martens et al., 2010). However, the regulation of flavonol biosynthesis has only been well studied in Arabidopsis thaliana. In this species, both the regulators and the biosynthetic genes are mainly regulated at the level of transcription (Quattrocchio et al., 2006; Jenkins, 2008). One subgroup of R2R3-type MYB proteins, called PFG family for Production of Flavonol Glucosides, is constituted of PFG1/MYB12, PFG2/MYB11, and PFG3/MYB111, which exhibit differential spatial expression patterns and regulate flavonol accumulation in a tissue- and developmental-specific manner (Stracke et al., 2007). However, a PFG1-3-independent flavonol accumulation occurs in pollen and siliques/seeds (Stracke et al., 2010b). Moreover, it has been demonstrated that the bZIP transcription factor long HYpocotyl5 (HY5), an important participant in the UV-B-induced signal transduction cascade mediated by UVR8, regulates the expression of PFG1/MYB12 under UV-B radiation (Stracke et al., 2010a). Thus, AtFLS1 and other genes encoding enzymes involved in flavonol biosynthesis are targets of these regulators (Mehrtens et al., 2005; Stracke et al., 2007, 2010a).
We recently characterized an FLS enzyme from maize, ZmFLS1, which converts dihydroflavonols to the corresponding flavonols, partially complementing the flavonol deficiency of the Arabidopsis fls1 mutant. In addition, we showed that the ZmFLS1 transcript level is increased by UV-B radiation, induction that is at least in part mediated by the activation of the regulators P1 and C1/PL1 + R/B (Falcone Ferreyra et al., 2010). The most recent release (version 5b.60) of the maize genome (inbred B73) allowed us to identify a second ZmFLS gene with very high identity to ZmFLS1. Thus, the aim of this work is to investigate these two maize FLS genes, and comparatively evaluate this paralogous pair in evolutionarily closely related grasses. Here, we show the ZmFLSs expression pattern in different tissues of the B73 maize line, and demonstrate that both genes are regulated by the P1 and C1/PL1 + R/B regulators. We further analyze ZmFLS expression in different maize inbreds and landraces from high altitudes, and we find that ZmFLSs are induced by UV-B exposure in all lines analyzed. Higher transcript levels of both ZmFLS genes were found in high altitude lines compared to inbred lines in the absence of UV-B, indicating that higher flavonol levels may be a constitutive mechanism of protection against high irradiance in these landraces. Moreover, the high sequence conservation of the ZmFLS promoters between maize lines suggests that different expression levels are probably a result from allelic variations in the trans-acting regulatory machinery.
Results
Identification and analyses of flavonol synthases in maize and other grasses
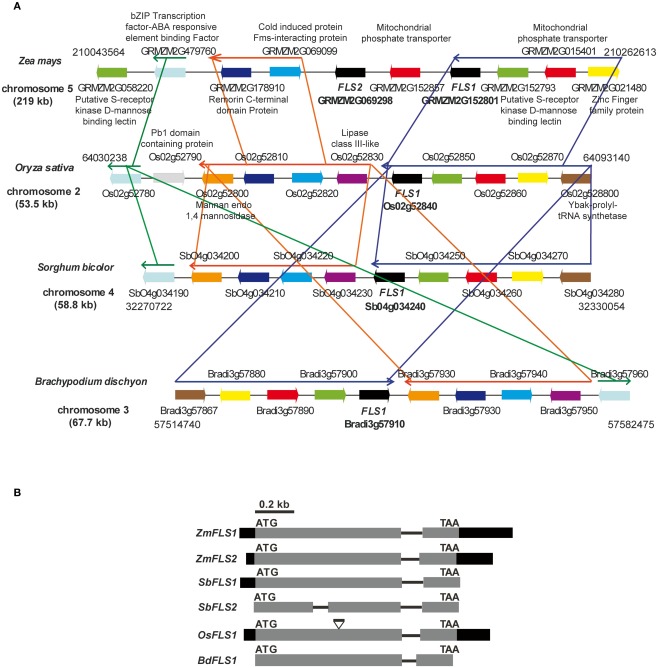
We recently described ZmFLS1 (GRMZM2G152801), encoding the first monocot flavonol synthase enzyme (Falcone Ferreyra et al., 2010). Interestingly, the new assembly of the B73 genome (release 5b.60) revealed the presence of a second putative FLS gene (GRMZM2G069298, ZmFLS2) with very high identity to ZmFLS1. Both genes are located in chromosome 5 separated by ∼50 kb (Figure 1A) and have identical structure, with two exons separated by an intron (Figure 1B). ZmFLS1 and ZmFLS2 share 96% identity, with higher identity in the coding regions (99%), and major differences between the respective 3′UTR regions. The predicted amino acid sequences exhibit 99% identity, with only two differences at positions 49 and 184, corresponding to isoleucine (I) and glycine (G) to methionine (M) and serine (S) for ZmFLS1 and ZmFLS2, respectively. Their putative upstream regulatory regions (arbitrarily defined here as 1.5 kb upstream of the start codon) exhibit 61% identity (Figure A1 in Appendix) while 95% identity is observed for the region from −350 to the start codon. In comparison with FLS genes from other grasses with completed sequenced genomes, like sorghum (Sorghum bicolor), rice (Oryza sativa), and Brachypodium distachyon, a high percentage of identity is observed at the nucleotide level (70–89%) with the highest homology found with one of the FLS genes in sorghum (SbFLS1). Based on the available genome sequences, Brachypodium and rice contain only one FLS gene (Bradi3g57910 and Os02g52840, respectively), while two FLS genes are present in S. bicolor (BTx623 line; Figure 1B). But unlike what is found in maize, the two sorghum FLS genes are located in different chromosomes [4 and 3 for SbFLS1 (Sb04g034240) and SbFLS2 (Sb03g002040), respectively] and SbFLS1 is more closely related to ZmFLSs (87–89% identity) than to SbFLS2 (72% identity), which is consistent with synteny analyses (see below). The length and the structure of the FLS genes are conserved between ZmFLSs, SbFLS1, and BdFLS1, each having two exons and one intron (Figure 1B), while OsFLS1 has three exons and two introns, a structure similar to AtFLS1 (Stracke et al., 2009). Although the annotation of SbFLS2 suggests the presence of a second intron, this region includes a region that, in all FLSs from other grasses, encodes a part of the FLS protein. Thus, the deduced protein sequence of SbFLS2 lacks 27 amino acids. In addition, the second intron of OsFLS1, SbFLS2, and the only one for ZmFLSs and SbFLS1 are placed at the same position in all these genes (Figure 1B).
Figure 1.
Regional synteny (A) and structure of FLS genes (B) from grasses. (A) Genomic arrangement of FLS and neighboring genes in Zea mays, Sorghum bicolor, Brachypodium distachyon, and Oryza sativa. The numbers on each side indicate the localization of the regions in the chromosomes, each gene in the different species is indicated with the same color. The length of chromosomes displayed is indicated in kb. Thin arrows above the genes indicate conserved regions between species. The graph was constructed using CoGe’s Genome Evolution Analysis tool (CoGe team software) at http://synteny.cnr.berkeley.edu/CoGe/GEvo.p. (B) Exons are represented by boxes, gray and black boxes indicate coding and UTR regions, respectively, and black lines represent non-coding regions. Start and stop codons are indicated.
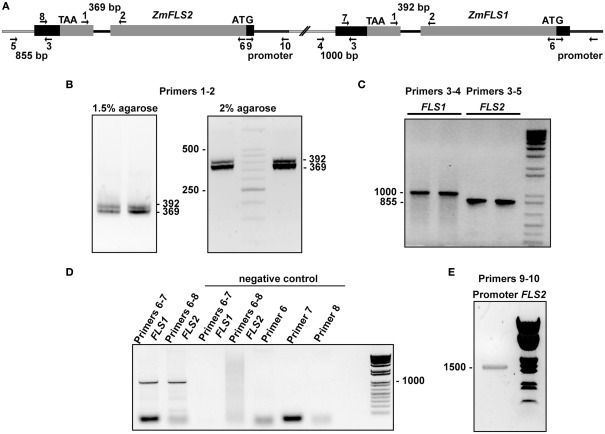
To verify the presence of the two tandemly arranged ZmFLS genes in the B73 genome, we designed primers to amplify fragments downstream of the 3′UTR of each gene, using the same forward primer that hybridizes in the 3′UTR of both genes, and specific reverse primers for each gene hybridizing elsewhere proximal in the chromosome. In addition, we amplified the intron of both ZmFLS genes using primers that hybridize in exons 1 and 2 of each gene (Figures A2B,C in Appendix). Moreover, to verify that both ZmFLS genes are localized in chromosome 5, we were able to amplify both genes using the same BAC clone (c0247D19) as a template (Figure A2D in Appendix). After sequencing the PCR products, we confirmed that the two different ZmFLS genes are present in chromosome 5 of the B73 maize inbred.
In order to analyze the orthologous FLS genes in detail, we explored the organization of the corresponding genes in the chromosomes. The analysis shows that FLS neighboring genes are conserved in maize (ZmFLS1, ZmFLS2), rice (OsFLS1), Brachypodium (BdFLS1), and sorghum (SbFLS1, with the exception of SbFLS2 gene); however, some re-arrangements among blocks of linked genes are observed as well as inversions, duplication, and deletions of certain genes, particularly in maize (Figure 1A). Interestingly, synteny is spread over a region of 220 kb in maize while in other grasses this region is only 53–68 kb, as it was previously described for different regions of maize chromosomes (Li and Gill, 2002; Ilic et al., 2003; Bruggmann et al., 2006; Goette and Messing, 2009; Wei et al., 2009).
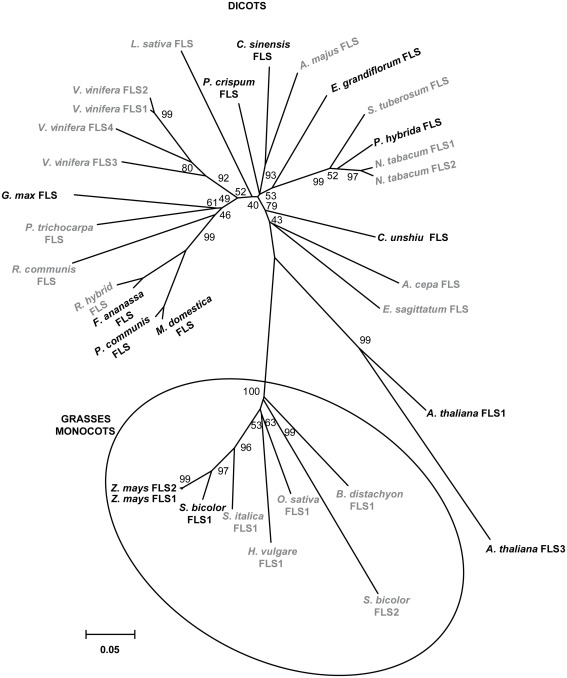
A phylogenetic tree generated with the amino acid sequences of plant FLS enzymes with demonstrated and predicted functionality showed a marked separation between enzymes from dicotyledonous and those from grasses monocotyledonous plants (Figure 2).
Figure 2.
Phylogeny of selected FLS proteins. The numbers indicate bootstrap values (10,000 replicates). Bar = 0.1 amino acid substitutions per site. FLSs from monocot grass plants are clustered in circles. Enzymes with demonstrated and predicted activities are indicated in black and gray, respectively.
Expression analysis and regulation of ZmFLSs by P1 and C1 + R
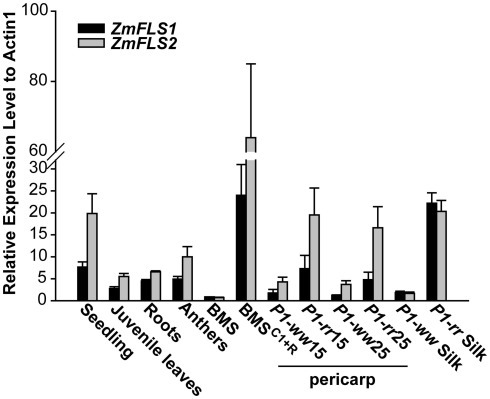
To analyze the expression of ZmFLS1 and ZmFLS2, we conducted quantitative RT-PCR (RT-quantitative PCR, qPCR) on RNA extracted from 7-day-old seedlings and juvenile leaves, roots, anthers (before anthesis), silks, 14 and 25 days after pollination (DAP) pericarps lacking (P1-ww) or accumulating (P1-rr) the phlobaphene pigments controlled by the maize P1 gene (Grotewold et al., 1994), Black Mexican Sweet (BMS) maize cells, ectopically expressing the C1 + R anthocyanin regulators (BMSC1+R, Grotewold et al., 1998), and untransformed control cells (BMS). Transcripts for ZmFLS1 and ZmFLS2 were detected in all the tissues analyzed, with the highest levels found in young seedlings (Figure 3). It is worth mentioning that the expression patterns described are consistent with the microarray database from a genome-wide atlas of transcription in different tissues and developmental stage of maize (Sekhon et al., 2011).
Figure 3.
Expression analysis of ZmFLS genes. ZmFLS expression evaluated by RT-qPCR in different tissues of the maize B73 inbred line: seedling (7-day-old plants), juvenile leaves (21-day-old plants), roots (21-day-old plants), and anthers; 15 and 25 days-old pericarps and silks expressing or not P1 gene and Black Mexican Sweet (BMS) maize cells ectopically expressing the C1 + R regulators [BMSC1+R] and untransformed controls (BMS). Each reaction was normalized using the Ct values corresponding to the actin1 mRNA (J01238). The means of the results obtained from biological triplicates are shown; the error bars indicate the SD of the samples.
For both genes, transcripts were present at significantly higher levels in P1-rr, compared to P1-ww pericarps and silks, indicating that both genes are regulated by P1. Similarly, both ZmFLS1 and ZmFLS2 showed higher expression in BMSC1+R than in BMS cells, suggesting that both genes are also under the control of the C1 and R anthocyanin regulators (Figure 3).
Previously, we showed by transient expression experiments in BMS cells that ZmFLS1 is positively regulated by P1 and C1 + R (Falcone Ferreyra et al., 2010). To investigate whether ZmFLS2 is similarly regulated, we carried out transient co-transfection experiments of maize protoplast cells with the regulators driven by the 35S promoter (p35S::P1 or p35S::C1 + p35S::R) and ZmFLS2 promoter-luciferase reporter fusion (pZmFLS2::Luc; 1.5 kb upstream of the start codon). In addition, given that the proximal promoter regions (from −350 to the start codon) of ZmFLS genes are highly similar, we investigated to what extent the respective regions of ZmFLS1 and ZmFLS2 were sufficient for regulation by P1 and C1 + R.
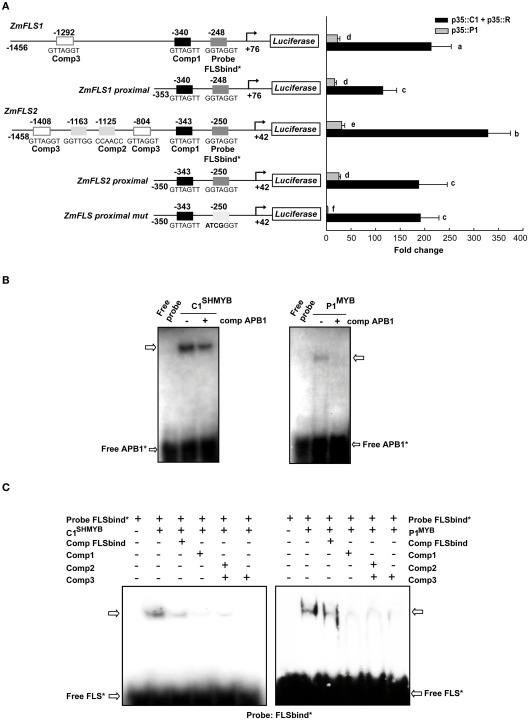
Both ZmFLS1 and ZmFLS2 are robustly activated by C1 + R, with a significantly higher induction of ZmFLS2, compared to ZmFLS1 (P < 0.05, Figure 4A). The analysis of candidate P1/C1 binding sites present in the ZmFLS promoters show additional candidate motifs in the distal region of the ZmFLS2 promoter (Figure 4A). To investigate whether these binding sites could contribute to a major activation of ZmFLS2 by C1 + R, we tested the proximal promoter regions up to −350 bp from the translation start codon; and, in this case, both genes were similarly activated, in accordance to the high identity of both ZmFLS proximal promoters (95%). For both genes, the 5′ deletion of the promoter caused a significant reduction in their activation, indicating that the additional binding sites present in the distal promoter regions are important for C1 + R regulation. Additionally, when the most proximal binding site in both promoters was mutated (Figure 4A), regulation of both promoter::luciferase constructs by C1 + R remained the same, suggesting that this binding site is not the main contributor to C1 + R regulation. Thus, for ZmFLS genes, it is possible that the binding of C1 + R to DNA involves only one cis-regulatory element in the promoter (at −340 and −343 bp in ZmFLS1 and ZmFLS2, respectively) or alternatively, another element could be involved in the binding and regulation, such as an E-box that is present in both promoters (at −296 and −293 bp, respectively), as it was suggested for the Bz1 promoter (Roth et al., 1991).
Figure 4.
Regulation of ZmFLS expression by P1 and C1 + R. (A) Transient expression following the co-transfection of maize protoplasts with p35S::P1 or p35S::C1 + p35S::R along with the constructs shown on the left. Putative p1/C1 binding sites are indicated with boxes and the sequences of the oligonucleotide probes used for DNA-binding experiments are shown below the boxes. For each construct analyzed and for each regulator, different letters indicate a significant difference at P < 0.05. (B) Binding of purified C1SHMYB and P1MYB proteins to 32P-labeled APB1 probe containing the haPBS of A1 promoter analyzed by electrophoretic mobility shift assays (EMSA; Hernandez et al., 2004). (C) Gel mobility retardation analyses with purified C1SHMYB and P1MYB proteins and FLSbind as probe. Free probe is indicated with an arrow.+ and − indicate the presence or absence of different competitors, respectively.
Both ZmFLS genes were activated by P1, with higher levels of activation for ZmFLS2 than for ZmFLS1. When the proximal promoter regions were analyzed, ZmFLS1 showed similar activation as when the full promoter was used, while a decreased activation was observed for ZmFLS2, suggesting that the additional P1-binding sites, absent in ZmFLS1, can contribute to ZmFLS2 regulation by P1. Finally, when the most proximal binding site was mutated, the ZmFLS activation by P1 was almost completely lost, indicating an important and major contribution of this site in P1 regulation of ZmFLS genes (Figure 4A).
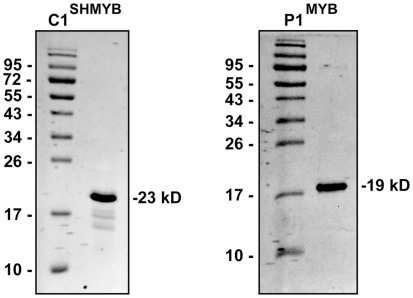
To verify which P1/C1 binding sites identified in the ZmFLS promoters are recognized by these transcription factors, we conducted electrophoretic mobility shift assays (EMSA). For these experiments, we expressed and affinity-purified from E. coli as N-terminal histidine-tagged fusions the P1 MYB domain (P1MYB), as well as a version of the MYB domain of C1, C1SHMYB, that binds DNA with higher affinity than C1MYB and with comparable affinity to P1MYB (Hernandez et al., 2004; N6His-ZmP1MYB and N6His-ZmC1SHMYB), obtaining 90–95% pure proteins (Figure A3 in Appendix). C1SHMYB and P1MYB proteins bind to the APB1 probe, the positive control in our experiments that contains the haPBS present in the A1 promoter (Figure 4B), which was previously demonstrated to bind both proteins with high affinity (Hernandez et al., 2004). C1SHMYB and P1MYB proteins can also bind effectively FLSbind (Figure 4C), the labeled probe that contains the most proximal binding site present in both ZmFLS promoters at −248 and −250 bp for ZmFLS1 and ZmFLS2, respectively (Figure 4A). Oligonucleotides containing other sites (comp 1–3) competed with the binding to FLSbind (Figure 4C), as FLSbind does itself. These results indicate that C1 and P1 can bind to all P1/C1 binding sites identified in the ZmFLS1 and ZmFLS2 promoters.
In vivo localization of ZmFLS1
To confirm the in vivo expression pattern of ZmFLS1, and to investigate its subcellular localization, we generated transgenic maize plants expressing ZmFLS1 fused to the red fluorescent protein (RFP) under its own promoter (pZmFLS1::ZmFLS1-RFP). The 3′UTR region and introns can contain regulatory functions (Bailey-Serres and Dawe, 1996; Patel et al., 2004; Rose et al., 2008; Parra et al., 2011); thus, the construct included the endogenous 3′UTR. It is noteworthy that we only generated transgenic plants expressing ZmFLS1 fused to RFP, since at the time these transgenic plants were constructed, the B73 maize genome was being sequenced and only the sequence of ZmFLS1 was available in GenBank.
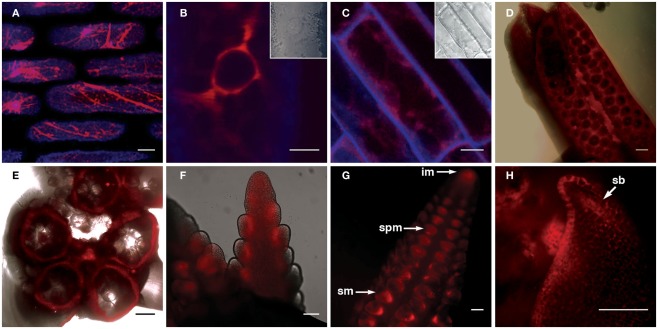
The FLS1-RFP fusion protein showed strong localization to the ER, as indicated by the “patchwork” pattern of fluorescence within leaf and root cells (Figures 5A,C), and was most easily detected in the perinuclear region in all cells in which fluorescence was detected (illustrated by Figure 5B). In developing inflorescence primordia, although expression was detected in all cells, higher levels of FLS1-RFP fluorescence could be observed in cells subtending spikelet pair (spm) and spikelet meristems (sp), as well as the L1 layer of suppressed bracts (sb; Figures 5F–H). In mature anthers, FLS1-RFP could be found in the degraded tapetum tissue surrounding pollen grains (Figures 5D–E), in accordance with the flavonol accumulation described in the later stages of maize anther development by Deboo et al. (1995). How the tissue and subcellular localization of the ZmFLS1 fusion protein relates to the function of the enzyme, remains to be determined.
Figure 5.
Transgenic expression of pZmFLS1::ZmFLS1-RFP. Confocal laser-scanning micrographs showing localization of ZmFLS1-RFP in maize leaf epidermal cells (A,B); root cells (C); anthers (D,E), tassel branch primordia (F), and ear primordia (G,H). FLS1-RFP was most easily detected in the ER (A) and perinuclear region (B) of cells. Red represents FLS1-RFP fluorescence, while blue represents cell wall autofluorescence. Brightfield images indicate the position of the nucleus and cell boundaries [(B–C) insets and (D–E) overlay]. Scale bars represent 5, 10, 20, 100 μm [(A–H), respectively]. White arrows indicate the inflorescence meristem (im), spikelet pair meristem (spm), spikelet meristem (sm), and suppressed bract (sb).
Expression of ZmFLSs in different maize lines
In order to investigate the evolution of ZmFLS genes, we amplified by PCR ZmFLS1 and ZmFLS2 genes in different maize lines, covering from position +739 (exon 1) to the 3′UTR using specific reverse primers for each gene. A high conservation at nucleotide level was observed between maize lines for both genes, showing minor differences mainly in the intron and in the 3′UTR (Figure A4 in Appendix). The phylogenetic tree generated from these sequences showed that ZmFLS1 and ZmFLS2 genes group in different clusters in all lines analyzed (Figure A5 in Appendix) indicating that paralogous genes arose by duplication before maize lines divergence.
Previously, we showed that the combined expression of ZmFLS1 and ZmFLS2 is induced by UV-B radiation in the B73 inbred, as well as in the W23 inbred line with (W23B PL) and without (W23b pl) the dominant alleles for the B and PL1 anthocyanin regulators. We examined ZmFLS1 and ZmFLS2 expression levels in different maize inbred lines (A619, Mo17, W22, B73, and W23 with or without the dominant alleles for the B and PL1 regulators) and in five maize landraces from high altitudes (Arrocillo Amarillo, Cacahuacintle, Conico, and Confite Puneño) by RT-qPCR using specific primers for each FLS gene covering the regions previously sequenced. Under control conditions without UV-B radiation, higher transcript levels of ZmFLS1 and ZmFLS2 were observed in 28-day-old seedlings from high altitude lines, compared to levels in the low-altitudes inbreds (Figures 6A,B). When the plants were irradiated with UV-B, ZmFLS transcripts increased in all lines, showing greater induction in the low-altitudes inbreds (Figure 6C). However, it is important to emphasize that despite the greater increase in ZmFLS expression by UV-B in low-altitudes inbreds, transcript levels, particularly for ZmFLS1 after the UV-B treatment, are in general lower than those in high altitude plants.
Figure 6.
Regulation of ZmFLS expression by UV-B radiation. ZmFLS1 (A) and ZmFLS2 (B) transcript levels under control conditions in the absence of UV-B (no UV-B) and after an 8-h-UV-B-treatment (UV-B) analyzed by RT-qPCR. Adult leaf samples from maize inbred lines (A619, Mo17, W22, B73, and W23 with or without the dominant alleles for the B PL1 regulators) and from five maize landraces from high altitudes (Mishca, Conico, Confite, Cacahuacintle, and Arrocillo) were analyzed. (C) Induction of ZmFLS genes expression by UV-B radiation. Each reaction was normalized using the Ct values corresponding to the thioredoxin-like transcript (AW927774). The means of the results obtained from biological triplicates are shown; the error bars indicate the SD of the samples.
Conservation of ZmFLS promoters between maize lines
To study if the differences observed in ZmFLS1 and ZmFLS2 expression levels between the maize inbred lines and the landraces from high altitudes (Figure 6) are a consequence of allelic variations in their promoters, we amplified the proximal promoters of ZmFLS genes from these maize lines. A high similarity of sequence was found in both promoters in all lines, with the conservation of the C1/P1-binding sites present in the B73 line, showing only short insertions or deletions at the 3′ promoter end, downstream of the putative TATA box (Figure A6 in Appendix). Consequently, the increased expression of ZmFLS genes in the maize landraces from high altitudes could be explained by differences in the activities and/or expression levels of C1 + R and P1 regulators in these landraces.
Discussion
The recently completed maize genome sequence (release 5b.60) shows that two FLS genes are present in the B73 line, resulting from complete genome duplication during evolution (Schnable et al., 2009). Consequently, this study was aimed at characterizing both ZmFLS genes and their patterns of expression and regulation, in order to analyze if duplication/divergence resulted in them being expressed under different conditions with functional specificity, or if there is functional redundancy between these genes. The analysis of both genes showed that there is an extremely high degree of identity between them, both at the nucleotide and amino acid levels, and also in their promoters. In addition, we found that there is a very high level of conservation in the sequences and genomic synteny with FLS genes from other grasses like rice, Brachypodium, and sorghum (SbFLS1, Figure 1). Although two FLS genes are also present in S. bicolor (SbFLS1 and SbFLS2), unlike in O. sativa and B. distachyon which have only one FLS gene, SbFLS1 is more closely related to the maize counterparts than to SbFLS2, and the annotated amino acid sequence of SbFLS2 lacks 27 amino acids. It is possible that SbFLS2 is a pseudo-gene that has not yet been lost, perhaps a result of a whole-genome duplication of the common ancestor of the grasses (Salse et al., 2008; Paterson et al., 2009). This hypothesis is consistent with the fact that no matching transcript could be found in the sorghum EST collections, and no expression of SbFLS2 could be observed under standard growth conditions, or upon exposure to salt stress or UV-B radiation in contrast to SbFLS1 (unpublished data). Moreover, according to the evolution of grass genomes from a common ancestor (50–70 mya), maize, and sorghum diverged from each other ∼12 mya, and maize underwent a complete genome duplication (∼5 mya) followed by re-arrangements leading to the current chromosomes (Salse et al., 2008; Buell, 2009; Paterson et al., 2009). Based on this, we can hypothesize that ZmFLS genes resulted from gene duplication ∼5 mya.
It is important to mention that, although we had previously reported an analysis of ZmFLS1 expression by RT-qPCR (Falcone Ferreyra et al., 2010); because there was no evidence of a second closely linked gene with very high identity to ZmFLS1, the primers used recognized both transcripts. The two ZmFLS genes studied here are expressed in all maize tissues analyzed, showing higher levels of expression in tissues where the P1 and C1 + R regulators are present, indicating that both genes are regulated by these transcription factors (Figure 3). To validate this hypothesis, we combined in vitro DNA-binding experiments with in vivo transient co-expression studies in maize protoplasts, showing that ZmFLS2 is more activated by the two regulators than ZmFLS1, probably due to the presence of additional P1/C1 binding sites in the distal region of its promoter (Figure 4).
Differences in regulatory elements present in ZmFLS-3′UTRs genes may contribute to their differential expression regulation. In humans, C. elegans and yeasts, regulatory elements present in the 5′ and 3′UTRs affect mRNA stability and translation rate; however, information of this type of regulation in plants is scarce (Zubiaga et al., 1995; Yang et al., 2003; Wilusz and Wilusz, 2004; Shalgi et al., 2005; Merritt et al., 2008). The role of mRNA sequence determinants in posttranscriptional regulation of the adh1 gene was investigated in maize protoplasts under hypoxia, demonstrating that the 5′and 3′UTRs are required for the regulation of the expression of this gene under this stress condition (Bailey-Serres and Dawe, 1996). Moreover, the 5′and 3′UTRs of the amaranth RbcS gene function as translational enhancers in different tissues (Patel et al., 2004). RNA binding proteins that complex with 5′ or 3′UTR are involved in the stability and translation of chloroplast mRNA (Bruick and Mayfield, 1999). However, additional studies are needed to investigate the contribution of the ZmFLS UTRs to gene regulation, for example by transient experiments with constructs containing the 3′UTR of each ZmFLS gene. Furthermore, it is well-known that mRNA levels do not necessarily correlate with protein levels in plant cells (Kawaguchi et al., 2004; Branco-Price et al., 2008; Mustroph et al., 2009). It has been reported that mRNA translation state is perturbed by stressful conditions like hypoxia, dehydration, light availability, cadmium intoxication, elevated temperature and high salinity in Arabidopsis (Kawaguchi et al., 2004; Mustroph et al., 2009; Matsuura et al., 2010; Sormani et al., 2011; Juntawong and Bailey-Serres, 2012). Therefore, we cannot rule out that ZmFLS mRNAs could be differentially translated.
We also show that both ZmFLS genes are regulated by UV-B radiation in both high altitude landraces and low-altitudes inbreds. However, under control conditions in the absence of UV-B radiation, higher transcript levels for both genes are present in high altitude plants compared with levels in the low-altitudes inbreds (Figure 6).
Maize lines growing at high altitudes have developed mechanisms to prevent damage caused by high UV-B exposure, such as the accumulation of C-glycosyl flavones in leaves, maysin, and its biosynthetic precursor rhamnosylisoorientin, flavones commonly found in silks (Snook et al., 1993; Casati and Walbot, 2005). Previously, we also showed that B, PL, and P1 are up-regulated by UV-B in W23 and B73 lines, while in high altitude landraces these regulators are also expressed in leaves at high levels (Casati and Walbot, 2005). Thus, given the protective role of flavonols to UV-B radiation (Stafford, 1990; Ryan et al., 2001, 2002; Solovchenko and Schmitz-Eiberger, 2003; Jaakola et al., 2004; Stracke et al., 2010a; Kusano et al., 2011; Pollastri and Tattini, 2011), it is possible that the high transcript levels of both ZmFLS genes may also contribute to the adaptation to this stress condition with higher UV-B fluxes.
The analysis of the ZmFLS proximal promoters in high altitude landraces and low-altitude inbreds showed a high degree of conservation in the distribution of cis-regulatory elements, perhaps suggesting allelic variations in the trans-regulating machinery, either provided by differences in the activities and/or in the expression levels of the transcription factors that regulate the expression of both ZmFLS genes. Previous reports have shown that flavonols are essential for pollen germination and conditional male fertility in maize and petunia (Mo et al., 1992; Taylor and Jorgensen, 1992; Ylstra et al., 1994; Taylor and Hepler, 1997), but not in Arabidopsis (Burbulis et al., 1996; Ylstra et al., 1996). However, maize plants lacking the P1 and R/B + C1/PL1 anthocyanin regulators are fertile (Coe and Neuffer, 1988; Dooner et al., 1991; Neuffer et al., 1997), suggesting that additional regulators, not yet identified, are involved in the regulation of ZmFLS genes in anthers.
The FLS1-RFP fusion protein was detected in all tissues analyzed in agreement with transcript pattern established by RT-qPCR (Figures 3 and 5). At the subcellular level, this enzyme was localized at the ER and the perinuclear region, but not in the nucleus, contrary to what was recently reported for FLS1 in Arabidopsis (Kuhn et al., 2011), and previously for other flavonoid enzymes (Saslowsky et al., 2005). Interestingly, FLS1-RFP was observed in tapetum tissue of mature anthers, suggesting a role in late pollen development. Future experiments of localization will be necessary to investigate the specific participation of FLS1 and FLS2 proteins in maize anther development and pollen viability by analyzing maize fls mutant plants and complementation. While we cannot guarantee the functionality of the fusion protein, additional experiments such as the complementation of A. thaliana mutant plants in the fls1 gene with the construct pFLS1::FLS1-RFP would allow to demonstrate its functionality in planta. Initial supporting evidence demonstrated that 35S:FLS1-GFP was able to complement A. thaliana fls1 mutant plants, restoring the levels of anthocyanins and flavonols (Falcone Ferreyra et al., 2010). Finally, based on the high identity between ZmFLS1 and ZmFLS2, we can speculate that ZmFLS2 would be able to complement the A. thaliana fls1 mutants similarly as ZmFLS1. These proteins show only two differences in their amino acid sequences at positions not participating in 2-oxoglutarate and substrate binding, or the coordination of the ferrous iron necessary for the enzyme activity.
In conclusion, although it is expected that paralogous genes may be expressed in different tissues, at different levels and also regulated by distinct factors, our results indicate that it is not the case for duplicated ZmFLS genes. Moreover, while pseudogenization process eliminates unnecessary genes, it is notably that none of the ZmFLS genes turned non-functional or were removed from the genome. It is possible that ZmFLS genes are still diverging to specific functions. However, we cannot rule out that unidentified regulators can be involved in the differential regulation of ZmFLS genes to meet the physiological needs of the plants under specific conditions not investigated in our study.
Materials and Methods
Plant material, growth conditions, and UV treatments
The Zea mays highland lines Confite Puneño, Mischa, Conico, Arrocillo Amarillo, and Cacahuacintle were obtained from the Germplasm Resources Information Network (GRIN, http://www.ars-grin.gov/cgi-bin/npgs/acc/).
The two near isogenic maize (Z. mays) lines that differ in flavonoid content from the genotype W23 (W23B PL and W23b pl) correspond to those previously described (Casati and Walbot, 2003). B73 seeds were obtained from the Instituto Nacional de Tecnología Agropecuaria (INTA, Pergamino, Buenos Aires, Argentina) while A619, W22, Mo17 seeds were maintained as laboratory stocks by self-pollination. The generation and analysis of the BMS cells expressing p35S::C1 and p35S::R were previously described (Grotewold et al., 1998).
Maize plants were grown in greenhouse conditions with supplemental visible lighting to 1000 μE m−2 s−1 with 15 h of light and 9 h of dark without UV-B for 28 days. UV-B treatments were performed by illuminating plants with UV-B lamps for 8 h using fixtures mounted 30 cm above the plants (TL 20 W/12; Phillips) at a UV-B intensity of 2 W m−2 and a UV-A intensity of 0.65 W m−2. The bulbs were covered with cellulose acetate filters (100 mm extra clear cellulose acetate plastic, Tap Plastics, Mountain View, CA, USA); the cellulose acetate sheeting does not remove any UV-B radiation from the spectrum but excludes wavelengths lower than 280 nm. No UV-B-treated plants (control) were exposed for the same period of time under the same lamps covered with polyester filters (100 mm clear polyester plastic; Tap Plastics, 0.04 W/m−2, UV-A, 0.4 W/m−2), which absorbs both UV-B and wavelengths lower than 280 nm. Lamp outputs were recorded using a UV-B/UV-A radiometer (UV203 AÞB radiometer; Macam Photometrics) to ensure that bulbs and filters provided the designated UV light dosage in all treatments. Adult leaf samples (leaf 9 or 10) were collected immediately after irradiation and stored at −80°C. The UV-B treatment experiments were repeated at least three times.
For gene expression analyses in tissues of the B73 maize line, plants were grown in greenhouse conditions as described above and samples were collected from anthers, roots (21-day-old plants), seedlings (7-day-old plants), and juvenile leaves (21-day-old plants). Pericarps and silks were obtained from maize A619P-rr and A619P-ww plants grown in the field under natural sunlight conditions.
Gene expression analyzes by RT-qPCR
Tissues from three independent biological replicates were frozen in liquid nitrogen and stored at −80°C. Total RNA was extracted using the RNeasy Plant Mini kit with optional DNase treatment (Qiagen) or alternatively with Trizol Protocol (Invitrogen) followed by DNase treatment (Promega). cDNAs were synthesized from 4 μg of total RNA using Superscript Reverse Transcription Enzyme II (Invitrogen) with oligo-dT as a primer. The resulting cDNAs were used as templates for qPCR amplification in a iCycler iQ detection system with the Optical System Software version 3.0a (Bio-Rad), using the intercalation dye SYBR Green I (Invitrogen) as a fluorescent reporter and Platinum Taq Polymerase (Invitrogen). Primers were designed to generate unique 150–250 bp-fragments using the PRIMER3 software (Rozen and Skaletsky, 2000). Three biological replicates were used for each sample plus negative control (reaction without reverse transcriptase). To normalize the UV treatment data, primers for thioredoxin-like transcript (AW927774) were used, while for tissue dependent expression studies, primers for Actin1 (J01238) were used for maize species (Table A1 in Appendix). Amplification conditions were as follows: 2 min denaturation at 94°C; 40–45 cycles at 94°C for 10 s, 57°C for 15 s, and 72°C for 20 s, followed by 5 min at 72°C. Melting curves for each PCR product were determined by measuring the decrease of fluorescence with increasing temperature (from 65 to 95°C). To confirm the size of the PCR products and to check that they corresponded to a unique and expected PCR product, the final PCR products were separated on a 2% (w/v) agarose gel and also sequenced. Primers used for ZmFLS1 and ZmFLS2 are listed in Table A1 in Appendix (ZmFLS-RT-forward, ZmFLS1-RT-reverse, and ZmFLS2-RT-reverse, respectively).
Amplification of ZmFLS genes and promoters, cloning, and mutagenesis
To verify the presence of two ZmFLS genes in B73, primers were designed to amplify fragments downstream of the 3′UTR of each gene using the same forward primer (ZmFLS-3′UTR-forward, ZmFLS1-crom5-reverse, and ZmFLS2-crom5-reverse, Table A1 in Appendix). PCR reactions were made with Platinum Pfx Polymerase (Invitrogen) under the following conditions: 1× Pfx buffer, 1× enhancer, 2 mM MgSO4, 0.5 mM of each dNTP, 0.5 μM of each primer, 0.3 U Platinum Pfx Polymerase, and sterile water added to obtain a volume of 20 μl. Cycling conditions were as follows: 5 min denaturation at 95°C, 35 cycles at 20 s denaturation at 95°C, 30 s annealing at 50°C, 90 s amplification at 68°C.
The BAC clone (c0247D19) was obtained from Arizona Genomics Institute (AGI, Tucson, USA). ZmFLS genes were amplified from the BAC clone by PCR using the same forward primer (ZmFLS-cds-forward) and specific primers for each gene (ZmFLS1-RT-reverse and ZmFLS2-RT-reverse, Table A1 in Appendix) that hybridize in the 3′UTR. PCR reactions were performed with Platinum Pfx Polymerase (Invitrogen) as described above but with annealing at 60°C. The PCR products were purified from the gels and sequenced.
To amplify the introns of the ZmFLS genes from B73, primers were designed to hybridize in the exon 1 and 2 of genes (Table A1 in Appendix). PCR reaction were performed with GoTaq (Promega) and Pfu Polymerases (Invitrogen; 10:1) under the following conditions: 1× buffer, 2 mM MgCl2, 0.5 μM of each primer, 0.5 mM of each dNTP and 0.5 U of enzyme, in 25 μl of final volume under the following cycling condition: 5 min denaturation at 94°C; 35 cycles at 94°C for 20 s, 57°C for 25 s, and 72°C for 40 s, followed by 7 min at 72°C.
To amplify ZmFLS1 and ZmFLS2 genes in different maize lines, PCR reaction were performed as described above for B73 using primers that hybridize in the exon 1 and 3′UTR of each gene (ZmFLS-intron2-forward, ZmFLS1-RT-reverse, and ZmFLS2-RT-reverse, respectively, Table A1 in Appendix).
To clone the promoter of ZmFLS2 from the B73 genotype, primers were designed to amplify a 1.5-kb fragment upstream of the start codon, as predicted from www.maizesequence.com release 5b.60. Restriction sites NotI and KpnI were included in the forward and reverse primers, respectively (NotI-ZmFLS2-prom-forward and KpnI-ZmFLS2-prom-reverse, Table A1 in Appendix). Genomic DNA was isolated from leaf tissue using a DNA isolation kit (Qiagen). PCR reactions were performed with Platinum Taq Polymerase (Invitrogen) under the following condition: 1× buffer, 0.3% DMSO, 2 mM MgCl2, 0.5 μM of each primer, 0.5 mM of each dNTP, 100 ng genomic DNA, and 0.3 U Platinum Taq Polymerase in a volume of 25 μl. Cycling conditions were as follows: 30 s denaturation at 95°C, 30 s annealing at 68°C, 2 min amplification at 72°C, with a 1°C decrement of annealing temperature in each cycle until it reached 58°C, followed by 25 cycles of 30 s denaturation at 95°C, 30 s annealing at 58°C, 2 min amplification at 72°C. The PCR products were purified from the gels, digested with the corresponding restriction enzymes and purified. The pZmFLS1::Luc construct (pMSZ011; Falcone Ferreyra et al., 2010) was restricted with NotI and KpnI and the ZmFLS1 promoter was replaced by the ZmFLS2 promoter, resulting in the pZmFLS2::Luc construct. Proximal promoters of ZmFLS1 and ZmFLS2 from B73 were amplified by PCR using the pZmFLS1::Luc and pZmFLS2::Luc constructs as templates with Platinum Taq Polymerase and Pfu Polymerases (Invitrogen; 10:1) under the following conditions: 1× buffer, 2 mM MgCl2, 0.5 μM of each primer, 0.5 mM of dNTPs and 0.5 U of enzyme, in 25 μl of final volume under the following cycling condition: 5 min denaturation at 94°C; 35 cycles at 94°C for 20 s, 59°C for 30 s, and 72°C for 50 s, followed by 7 min at 72°C. The PCR products were purified from the gels, digested with the corresponding restriction enzymes and purified, and the ZmFLS promoters (1.5 kb) in pMSZ011 were replaced by the ZmFLS proximal promoters. For ZmFLS proximal promoter analysis from different maize lines, DNA was extracted from leaves using the CTAB Method, and PCR reactions were performed with Platinum Taq Polymerase under the following conditions: 1× buffer, 2 mM MgCl2, 0.4% DMSO, 0.5 μM of each primer, 0.5 mM of each dNTP and 0.5 U of enzyme, in 25 μl final volume. Amplification conditions were as follows: 5 min denaturation at 94°C; 35 cycles at 94°C for 25 s, 59°C for 35 s, and 72°C for 50 s, followed by 5 min at 72°C. PCR products were purified, cloned into pGEM®-T-Easy vector (Promega) and sequenced.
Mutagenesis of the P1/C1 binding site at −250 bp in ZmFLS promoters was made by PCR using the same forward primer described above for amplification of proximal promoters but using a reverse oligonucleotide with the mutated binding site (ZmFLSprom-mut-reverse; Table A1 in Appendix). PCR reactions were performed with GoTaq (Promega) and Pfu Polymerases (Invitrogen; 10:1) under the following conditions: 1× buffer, 2 mM MgCl2, 0.5 μM of each primer, 0.5 mM of each dNTP, and 0.5 U of enzyme, in 25 μl final volume under the following cycling condition: 5 min denaturation at 94°C; 35 cycles at 94°C for 20 s, 50°C for 35 s, and 72°C for 50 s, followed by 7 min at 72°C. The PCR product (mega primer) was purified from the gel and then used as a forward primer (0.16 μM) in a new PCR with the reverse primer (1 μM) described above for proximal promoters (Table A1 in Appendix). The PCR products were purified from the gels, digested with the corresponding restriction enzymes and purified, and the ZmFLS1 proximal promoter in pMSZ011 was replaced by the ZmFLS mutated proximal promoter. Finally, mutations in the P1/C1 binding site were confirmed by sequencing.
Cloning, expression and purification of recombinant proteins
The MYB domain of P1 (P1MYB) cloned in pET19b (Invitrogen) was used for its expression in E. coli (Williams and Grotewold, 1997). The plasmid for the expression of the MYB domain of C1SH was obtained by subcloning from pTYB2 vector (Hernandez et al., 2004) to pET19b. Thus, the C1SH MYB domain was cut with NdeI and XhoI restriction enzymes and inserted in pET19b previously digested with the same enzymes. BL21(DE3)pLys cells were transformed with the clones described; P1MYB expression was achieved by induction of the cell culture (250 ml) with 1 mM IPTG at 37°C for 3 h, while C1SHMYB expression was done at 30°C with 0.5 mM IPTG for 6 h. Protein purification was essentially done as described by Williams and Grotewold (1997) with the following modifications. After binding the proteins to a Ni-NTA resin (Invitrogen) by rocking at 4°C for 1 h, the resin was loaded onto a column, washed three times with 15 volumes of binding buffer (50 mM sodium phosphate, pH 8.0, 500 mM NaCl, 20 mM imidazole, 5% glycerol and 1 mM phenylmethylsulfonyl fluoride), followed by three washes with seven volumes of washing buffer (50 mM sodium phosphate, pH 8.0, 500 mM NaCl, 5% glycerol, 50 mM imidazole). Elution was carried out by seven sequential additions of 1.5 ml of elution buffer (50 mM sodium phosphate, pH 8.0, 500 mM NaCl, 5% glycerol, and 200 mM imidazole). Finally, recombinant proteins were desalted in desalting buffer (10 mM Tris-HCl pH 7.5, 50 mM NaCl, 1 mM EDTA, 5% glycerol, and 1 mM DTT) by four cycles of concentration and dilution using Amicon Ultra-15 3K (Millipore) and stored at −80°C. Protein levels were estimated both by comparison with dilution series of bovine serum albumin on a Coomassie Blue-stained SDS-PAGE and also using the Bradford reagent (Bio-Rad, Bradford, 1976). The yield of 90–95% pure recombinant proteins obtained in these conditions was 4 and 6 mg/L of culture for P1MYB and C1SHMYB, respectively.
Electrophoretic mobility shift assays
End labeling of synthetic oligonucleotide probes (APB10, FLSbind-forward, comp1-forward, comp2-forward, and comp3-forward, Table A1 in Appendix) was carried out using T4 polynucleotide kinase (Invitrogen) in the presence of a 2-M excess of [γ-32P]ATP (>8,000 Ci/mmol). The labeled oligonucleotides were then annealed to equal amounts of complementary oligonucleotides (APB01, FLSbind-reverse, comp1-reverse, comp2-reverse, and comp3-reverse) by heating to 95°C and slowly cooling down to room temperature to generate APB1, FLSbind, comp1, comp2, and comp3 probes, respectively. A fraction of the double-stranded labeled oligonucleotides was precipitated on glass filters for quantification by scintillation of the radiation incorporated. The probe used as positive control (APB1) contains the high affinity P1-binding sites from the a1 gene promoter, Protein-DNA incubations were performed essentially as described previously (Heine et al., 2004) with the following modifications. Approximately 2 μg of purified proteins were incubated in a buffer containing 10 mM Tris, pH 7.5, 50 mM NaCl, 1 mM EDTA, 5% glycerol, 2 μg of salmon sperm DNA, 1 mM DTT with 1 ng of end-labeled, double-stranded DNA probes, for 1 h at 4°C in the presence or absence of competitors as indicated in the Figures. Protein-DNA complexes were resolved on 8% polyacrylamide gels (30:0.8 acrylamide:bis-acrylamide) in 0.25× Tris-borate/EDTA (22.5 mM Tris-Borate and 0.5 mM EDTA) at 100 V for 90 min at 4°C. The gels were then dried onto Whatman paper and subjected to autoradiography at −80°C overnight.
Transient expression experiments in maize protoplasts
The p35S::C1 + p35S::R, p35S::P1, p35S::Renilla, and p35S::BAR and p35S::GFP plasmids have all been previously described (Grotewold et al., 1994; Sainz et al., 1997; Hernandez et al., 2004, 2007).
Protoplasts from 11- to 13-day-old etiolated maize seedlings were obtained from kernels of B73xMo17 plant. After chopping second or third leaves into small pieces, leaf stripes were digested in 3% cellulase RS, 0.6% macerozyme R10 (both from Yakult Honsha Co., Japan), 0.6 M mannitol, 10 mM MES (pH5.7), 5 mM CaCl2, and 0.1% (w/v) BSA for 15 min under vacuum followed by 2:30 h gentle shaking (40 rpm) at 25°C in the dark. After releasing the protoplasts at 80g, the protoplasts were filtered through a 35-μm nylon mesh and collected by centrifugation at 150g for 1 min. The protoplasts were washed in ES buffer (0.6 M mannitol, 5 mM MES, pH 5.7, 10 mM KCl) and counted with a hemocytometer. Electroporation was carried out on ∼105 protoplasts with 40 μg of total DNA per transformation, using 100 V/cm, 10 ms, and one pulse with a BTX Electro-Square-Porator T820. After electroporation, protoplasts were incubated for 18–22 h in the dark at RT before performing the luciferase reaction. Transformation efficiency was estimated following GFP expression by fluorescence microscopy. Transient expression assays for Luciferase and Renilla were performed essentially as previously described (Sheen, 1991; Feller et al., 2006; Hernandez et al., 2007).
Generation of maize transgenic plants expressing pZmFLS1::ZmFLS1-mRFP
Construct was generated using the MultiSite Gateway® Pro 3.0 system (Invitrogen).
Two sets of primers (P1/P2, P3/P4; Table A1 in Appendix) were designed for the amplification of two genomic fragments. The first set of primers amplified a fragment (P1–P2) that extends 2.483 kb upstream of the start codon to the tag insertion site within the coding sequence of ZmFLS1 (just before the stop codon). The second set of primers amplified a fragment (P3–P4) from the tag insertion site to 2.1 kb downstream of the gene to include 3′ UTR and regulatory sequences. P1 and P4 contained, in addition to gene-specific sequences, sequences partially overlapping the attB1 and attB2 Gateway forward and reverse primers, respectively (used for a second PCR). P2 and P3 contained sequences partially overlapping the RFP primers. A second PCR reaction, designated triple-template PCR (TT-PCR), utilized two primers containing the complete attB1 and attB2 Gateway sequences and partially overlapping the P1 and P4 primers. Thus, the RFP fluorescent tag was introduced into the ZmFLS1 gene (C-terminal) and resulted in an internally tagged full-length gene sequence flanked by attB1 and attB2 sites ready for Gateway recombination cloning. The construct was recombined into the maize pTF101.1 binary vector, tagging ZmFLS1 with mRFP1 at the C-terminus. Agrobacterium-mediated transformation of maize HiII was performed by the Iowa State University Plant Transformation Facility. T0s were crossed to the inbred line B73 and T1s imaged live using a Zeiss 510 or 710 confocal laser-scanning microscope. Construct sequences and image metadata are available at http://maize.jcvi.org/cellgenomics/index.shtml.
Phylogenetic analysis
The trees were constructed using MEGA 4.0 Software with the Neighbor-Joining method based on ClustalW multiple alignments (Tamura et al., 2007). For analysis of FLS proteins from monocot and dicot plants the following sequences were analyzed: Pyrus communis FLS (ABB70118), Fragaria x ananassa FLS (ABH07784.1), Rosa hybrid FLS (BAC66468), Ricinus communis FLS (XP_002513774), Populus trichocarpa (XP_002301003.1), Glycine max FLS (AB246668.1), Vitis vinifera FLS1 (XP_002285838.1), V. vinifera FLS2 (BAE75809.1), V. vinifera FLS3 (XP_002284410.1), V. vinifera FLS4 (XP_002285839.1), Lactuca sativa FLS (BAG12186.1), Petroselinum crispum FLS (AAP57395.1), Camellia sinensis FLS (ACL98052), Antirrhinum majus FLS (ABB53382.1), Eustoma grandiflorum FLS (BAD34463), Solanum tuberosum FLS (ACN81826.1), Petunia x hybrida FLS (Q07512.1), Nicotiana tabacum FLS1 (ABE28017.1), N. tabacum FLS2 (BAF96939.1), Citrus unshiu FLS (BAA36554.1), Allium cepa FLS (AAO63023.1), Epimedium sagittatum FLS (ABY63659.1), Hordeum vulgare FLS (BAJ98444.1), Setaria italica FLS (Si017742m), S. bicolor FLS1 (Sb04g034240), S. bicolor FLS2 (Sb03g002040), B. distachyon FLS1 (Bd5g57910), O. sativa FLS1 (Os02g52840).
Statistical analysis
Data presented were analyzed using one-way analysis of variance (ANOVA). Minimum significant differences were calculated by the Bonferroni, Holm–Sidak, Dunett, and Duncan tests (P < 0.05) using the Statgraphics Plus 5.0 Software.
Accession numbers
Sequence data from this article can be found in the maize genome sequence (version 3b.60 at maizesequence.org), sorghum genome sequence (release Sbi1.4 at gramene.org), TIGR Rice Genome Annotation Project (release 7 at rice.plantbiology.msu.edu), Brachypodium genome sequence (GBrowse v1.0 at brachypodium.org), and GenBank databases under the following accession numbers: ZmFLS1 (GRMZM2G152801); ZmFLS2 (GRMZM2G069298); SbFLS1 (Sb04g034240); SbFLS2 (Sb03g002040); OsFLS1 (Os02g52840); BdFLS1 (Bradi3g57910); Z. mays thioredoxin-like, AW927774; Z. mays actin1, J01238.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by grant USDA 2010-65115-20408 to Erich Grotewold, by FONCyT grant PICT-2006-00957 and PICT-2010-00105 to Paula Casati and Erich Grotewold and Excellence in Plant Molecular Biology and Biotechnology Graduate Fellowship to María Isabel Casas.
Appendix
Table A1.
Primers used for cloning, sequencing, RT-qPCR generation of maize transgenic plants, and EMSA.
| Name | Sequence |
|---|---|
| ZmFLS-3′UTR-forward | 5′TGTAAGGGCACTAATACATG3′ |
| ZmFLS-cds-forward | 5′ATGGGGGGCGAGACGCACCTGAG3′ |
| ZmFLS1-crom5-reverse | 5′GAAAAGGCTCACTCGGTCTA3′ |
| ZmFLS2-crom5-reverse | 5′GCCCTGCAGTTCAGTAACTT3′ |
| ZmFLS-intron2-forward | 5′GTCAATGGTACGAGGCCAAG3′ |
| ZmFLS-intron2-reverse | 5′CATGGGGAGCTTGTTGATCT3′ |
| NotI-ZmFLS2prom-forward | 5′CACCGCGGCCGCAGCCATGAAAAATGACAGT3′ |
| KpnI-ZmFLS2prom-reverse | 5′CTATGGTACCCGCAGCAGGAAGCAGGACC′3 |
| Not-ZmFLSprom-int-forward | 5′CACCGCGGCCGCGCTATTACTCGTTAGTTGG′3 |
| ZmFLSprom-mut-reverse | 5′GGCCGTACGACGCACCCGATAGAAACTCGTTTA′3 |
| KpnI-ZmFLS1prom-reverse | 5′CTATGGTACCCGCAGCAGGAAGCAGGGCC′3 |
| ZmFLS-RT-forward | 5′GACGGTGAACAAGGAGAAGAC′3 |
| ZmFLS1-RT-reverse | 5′GGCCATGCATGCGACTGGAAT′3 |
| ZmFLS2-RT-reverse | 5′ACAGAAGCGGGTGCACACTGC′3 |
| ZmActine1-forward | 5′CTTCGAATGCCCAGCAAT3′ |
| ZmActine1-reverse | 5′CGGAGAATAGCATGAGGAAG3′ |
| ZmThioredoxine-like-forward | 5′GGACCAGAAGATTGCAGAAG3′ |
| ZmThioredoxine-like-reverse | 5′ACGGATGTCCCATGAAGA3′ |
| APB10 | 5′GATCCGGGTCAGTGTACCTACCAACCTTAAACAC3′ |
| APB01 | 5′GATCGTGTTTAAGGTTGGTAGGTACACTGACCCG3′ |
| FLSbind-forward | 5′GAGTTTCTGGTAGGTGCGTCGTACGGCCAG3′ |
| FLSbind-reverse | 5′CTGGCCGTACGACGCACCTACCAGAAACTC3′ |
| Comp1-forward | 5′CTATTACTCGTTAGTTGGATTTTAGTTT3′ |
| Comp1-reverse | 5′AAACTAAAATCCAACTAACGAGTAATAG3′ |
| Comp2-forward | 5′CCAAGTCAAAATCCAACCATTATCTCTTGA3′ |
| Comp2-reverse | 5′TCAAGAGATAATGGTTGGATTTTGACTTGG3′ |
| Comp3-forward | 5′TAGATGTGATTCGTTAGGTGTGTTTTTATAA3′ |
| Comp3-reverse | 5′TTATAAAAACACACCTAACGAATCACATCTA3′ |
| P1 | 5′ggggacaagtttgtacaaaaaagcaggcttaGCGACGAATCCATTCTTGA3′ |
| P2 | 5′ggggacaactttgtatagaaaagttggggtgCATGGGGAGCTTGTTGATCTTGC3′ |
| P3 | 5′ ggggacaactttgtataataaagttgcaTAATTATGTAGCTCGGGTTC3′ |
| P4 | 5′ggggaccactttgtacaagaaagctgggtaTATTGCTGGCGTGAATAACC3′ |
Figure A1.
Alignment of ZmFLS promoters of B73 maize.
Figure A2.
Two ZmFLS genes are present in the B73 maize line. (A) Organization of ZmFLS genes in the chromosome 5. Primers used for amplification are indicated by arrows, and the predicted PCR product size is shown with numbers above them. Amplification of ZmFLS introns (B), 3′UTR regions downstream of ZmFLS genes (C), ZmFLS1 and ZmFLS2 genes from the BAC clone (D), and ZmFLS2 promoter (1.5 kb upstream the start codon) (E).
Figure A3.
SDS-PAGE analysis (15%) of the purified MYB domains of P1 and C1SH. The numbers on the sides of the gels indicate the molecular mass of standard proteins in kD.
Figure A4.
Sequence comparison of partial ZmFLS1 (A) and ZmFLS2 (B) genes in different maize lines. The regions of ZmFLS1 and ZmFLS2 genes range from +740 bp (exon 1) to +1218 and +1143 bp, respectively (3′UTR). The introns in ZmFLS genes are highlighted in gray, different nucleotides and insertions or deletions are highlighted in yellow. Stop codons are indicated in bold-underlined letters. Primers used for RT-qPCR are in bold-underlined letters on B73 sequences.
Figure A5.
Phylogenetic tree based on nucleotide sequences of ZmFLS1 and ZmFLS2 genes in maize inbred lines and landraces from high altitudes. The numbers indicate bootstrap values (10,000 replicates). Bar = 0.1 amino acid substitutions per site. OsFLS1 was defined as the out-group.
Figure A6.
Alignment of ZmFLS1 (A) and ZmFLS2 (B) proximal promoters of maize inbred lines and landraces from high altitudes. C1/P1-binding sites are highlighted in different colors. Insertions or deletions in promoters are highlighted in gray, while differences in nucleotides from B73 are highlighted in yellow. The putative TATA boxes are indicated in bold-underlined letters.
References
- Bailey-Serres J., Dawe R. K. (1996). Both 5′ and 3′ sequences of maize adh7 mRNA are required for enhanced translation under low-oxygen conditions. Plant Physiol. 112, 685–695 10.1104/pp.112.2.685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- Branco-Price C., Kaiser K. A., Jang C. J., Larive C. K., Bailey-Serres J. (2008). Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabidopsis thaliana. Plant J. 56, 743–755 10.1111/j.1365-313X.2008.03642.x [DOI] [PubMed] [Google Scholar]
- Brown D. E., Rashotte A. M., Murphy A. S., Normanly J., Tague B. W., Peer W. A., Taiz L., Muday G. K. (2001). Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiol. 126, 524–535 10.1104/pp.126.2.524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggmann R., Bharti A. K., Gundlach H., Lai J., Young S., Pontaroli A. C., Wei F., Haberer G., Fuks G., Du C., Raymond C., Estep M. C., Liu R., Bennetzen J. L., Chan A. P., Rabinowicz P. D., Quackenbush J., Barbazuk W. B., Wing R. A., Birren B., Nusbaum C., Rounsley S., Mayer K. F., Messing J. (2006). Uneven chromosome contraction and expansion in the maize genome. Genome Res. 16, 1241–1251 10.1101/gr.5338906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruick R. K., Mayfield S. P. (1999). Light-activated translation of chloroplast mRNAs. Trends Plant Sci. 4, 190–195 10.1016/S1360-1385(99)01402-8 [DOI] [PubMed] [Google Scholar]
- Buell C. R. (2009). Poaceae genomes: going from unattainable to becoming a model clade for comparative plant genomics. Plant Physiol. 149, 111–116 10.1104/pp.108.128926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbulis I. E., Iacobucci M., Shirley B. W. (1996). A null mutation in the first enzyme of flavonoid biosynthesis does not affect male fertility in Arabidopsis. Plant Cell 8, 1013–1025 10.1105/tpc.8.6.1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati P., Walbot V. (2003). Gene expression profiling in response to ultraviolet radiation in maize genotypes with varying flavonoid content. Plant Physiol. 132, 1739–1754 10.1104/pp.103.022871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati P., Walbot V. (2005). Differential accumulation of maysin and rhamnosylorientin in leaves of high altitude landraces of maize after UV-B exposure. Plant Cell Environ. 28, 788–799 10.1111/j.1365-3040.2005.01329.x [DOI] [Google Scholar]
- Coe E. H., Neuffer M. G. (1988). “The genetics of corn,” in Corn and Corn Improvement, eds Sprague G. F., Dudley J. W. (Madison, WI: American Society of Agronomy; ), 81–258 [Google Scholar]
- Deboo G. B., Albertsen M. C., Taylor L. P. (1995). Flavanone 3-hydroxylase transcripts and flavonols accumulation are temporally coordinate in maize anther. Plant J. 7, 703–705 10.1046/j.1365-313X.1995.07050703.x [DOI] [PubMed] [Google Scholar]
- Dooner H. K., Robbins T. P., Jorgensen R. A. (1991). Genetic and developmental control of anthocyanin biosynthesis. Annu. Rev. Genet. 25, 173–199 10.1146/annurev.ge.25.120191.001133 [DOI] [PubMed] [Google Scholar]
- Falcone Ferreyra M. L., Rius S., Emiliani J., Pourcel L., Feller A., Morohashi K., Casati P., Grotewold E. (2010). Cloning and characterization of a UV-B inducible maize flavonol synthase. Plant J. 62, 77–91 10.1111/j.1365-313X.2010.04133.x [DOI] [PubMed] [Google Scholar]
- Feller A., Hernandez J. M., Grotewold E. (2006). An ACT-like domain participates in the dimerization of several plant bHLH transcription factors. J. Biol. Chem. 281, 28964–28974 10.1074/jbc.M605551200 [DOI] [PubMed] [Google Scholar]
- Geleijnse J. M., Hollman P. C. H. (2008). Flavonoids and cardiovascular health: which compounds, what mechanisms? Am. J. Clin. Nutr. 88, 12–13 [DOI] [PubMed] [Google Scholar]
- Goette W., Messing J. (2009). Change of gene structure and function by non-homologous end-joining, homologous recombination, and transposition of DNA. PLoS Genet. 5, e1000516. 10.1371/journal.pgen.1000516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronquist M., Bezzerides A., Attygalle A., Meinwald J., Eisner M., Eisner T. (2001). Attractive and defensive functions of the ultraviolet pigments of a flower (Hypericum calycinum). Proc. Natl. Acad. Sci. U.S.A. 98, 13745–13750 10.1073/pnas.231471698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E., Chamberlin M., Snook M., Siame B., Butler L., Swenson J., Maddock S., Clair G. S., Bowen B. (1998). Engineering secondary metabolism in maize cells by ectopic expression of transcription factors. Plant Cell 10, 721–740 10.1105/tpc.10.5.721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E., Drummond B. J., Bowen B., Peterson T. (1994). The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76, 543–553 10.1016/0092-8674(94)90117-1 [DOI] [PubMed] [Google Scholar]
- Heine G. F., Hernandez J. M., Grotewold E. (2004). Two cysteines in plant R2R3 MYB domains participate in REDOX-dependent DNA binding. J. Biol. Chem. 279, 37878–37885 10.1074/jbc.M405166200 [DOI] [PubMed] [Google Scholar]
- Hernandez J. M., Heine G. F., Irani N. G., Feller A., Kim M.-G., Matulnik T., Chandler V. L., Grotewold E. (2004). Different mechanisms participate in the R-dependent activity of the R2R3 MYB transcription factor C1. J. Biol. Chem. 279, 48205–48213 10.1074/jbc.M407845200 [DOI] [PubMed] [Google Scholar]
- Hernandez J. M., Feller A., Morohashi K., Frame K., Grotewold E. (2007). The basic helix loop helix domain of maize R links transcriptional regulation and histone modifications by recruitment of an EMSY-related factor. Proc. Natl. Acad. Sci. USA 104, 17222–17227 10.1073/pnas.0705629104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirvonen T., Pietinen P., Virtanen M., Ovaskainen M.-L., Häkkinen S., Albanes D. (2001). Intake of flavonols and flavones and risk of coronary heart disease in male smokers. Epidemiology 12, 62–67 10.1097/00001648-200101000-00011 [DOI] [PubMed] [Google Scholar]
- Ilic K., SanMiguel P. J., Bennetzen J. L. (2003). A complex history of rearrangement in an orthologous region of the maize, sorghum, and rice genomes. Proc. Natl. Acad. Sci. U.S.A. 100, 12265–12270 10.1073/pnas.1434476100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaakola L., Määttä-Riihinen K., Kärenlampi S., Hohtola A. (2004). Activation of flavonoid biosynthesis by solar radiation in bilberry (Vaccinium myrtillus L.) leaves. Planta 218, 721–728 10.1007/s00425-003-1161-x [DOI] [PubMed] [Google Scholar]
- Jenkins G. I. (2008). “Environmental regulation of flavonoid biosynthesis,” in Health Benefits of Organic Food: Effects of the Environment, eds Givens D. I., Minihane A. M., Shaw E. (Oxfordshire: CABI; ), 240–262 [Google Scholar]
- Juntawong P., Bailey-Serres J. (2012). Dynamic light regulation of translation status in Arabidopsis thaliana. Front. Plant Sci. 3:66. 10.3389/fpls.2012.00066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H.-K, Ecklund, D., Liu M., Datta S. (2009). Apigenin, a non-mutagenic dietary flavonoid, suppresses lupus by inhibiting autoantigen presentation for expansion of autoreactive Th1 and Th17 cells. Arthritis Res. Ther. 11, R59. 10.1186/ar2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur P., Shukla S., Gupta S. (2008). Plant flavonoid apigenin inactivates Akt to trigger apoptosis in human prostate cancer: an in vitro and in vivo study. Carcinogenesis 29, 2210–2217 10.1093/carcin/bgn201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi R., Girke T., Bray E. A., Bailey-Serres J. (2004). Differential mRNA translation contributes to gene regulation under non-stress and dehydration stress conditions in Arabidopsis thaliana. Plant J. 38, 823–839 10.1111/j.1365-313X.2004.02090.x [DOI] [PubMed] [Google Scholar]
- Knekt P., Isotupa S., Rissanen H., Heliövaara M., Järvinen R., Häkkinen S. (2000). Quercetin intake and the incidence of cerebrovascular disease. Eur. J. Clin. Nutr. 54, 415–417 10.1038/sj.ejcn.1600997 [DOI] [PubMed] [Google Scholar]
- Kuhn B. M., Geisler M., Bigler L., Ring C. (2011). Flavonols accumulate asymmetrically and affect auxin transport in Arabidopsis. Plant Physiol. 156, 585–595 10.1104/pp.111.175976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano M., Tohge T., Fukushima A., Kobayashi M., Hayashi N., Otsuki H., Kondou Y., Goto H., Kawashima M., Matsuda F., Niida R., Matsui M., Saito K., Fernie A. R. (2011). Metabolomics reveals comprehensive reprogramming involving two independent metabolic responses of Arabidopsis to UV-B light. Plant J. 67, 354–369 10.1111/j.1365-313X.2011.04599.x [DOI] [PubMed] [Google Scholar]
- Lewis D. R., Ramirez M. V., Miller N. D., Vallabhaneni P., Ray W. K., Helm R. F., Winkel B. S., Muday G. K. (2011). Auxin and ethylene induce flavonol accumulation through distinct transcriptional networks. Plant Physiol. 156, 144–164 10.1104/pp.111.172502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Gill B. S. (2002). The colinearity of the Sh2/A1 orthologous region in rice, sorghum and maize is interrupted and accompanied by genome expansion in the triticeae. Genetics 160, 1153–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak P., Leung Y.-K, Tang, W.-Y, Harwood, C., Ho S.-M. (2006). Apigenin suppresses cancer cell growth through Erb. Neoplasia 8, 896–890 10.1593/neo.06538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens S., Preuss A., Matern U. (2010). Multifunctional flavonoid dioxygenases: flavonol and anthocyanin biosynthesis in Arabidopsis thaliana L. Phytochemistry 71, 1040–1049 10.1016/j.phytochem.2010.04.016 [DOI] [PubMed] [Google Scholar]
- Matsuura H., Ishibashi Y., Shin-myo A., Kanaya S., Kato K. (2010). Genome-wide analyses of early translational responses to elevated temperature and high salinity in Arabidopsis thaliana. Plant Cell Physiol. 51, 448–462 10.1093/pcp/pcq010 [DOI] [PubMed] [Google Scholar]
- Mehrtens F., Kranz H., Bednarek P., Weisshaar B. (2005). The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol. 138, 1083–1096 10.1104/pp.104.058032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt C., Rasoloson D., Ko D., Seydoux G. (2008). 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr. Biol. 18, 1476–1482 10.1016/j.cub.2008.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo Y., Nagel C., Taylor L. P. (1992). Biochemical complementation of chalcone synthase mutants defines a role for flavonols in functional pollen. Proc. Natl. Acad. Sci. U.S.A. 89, 7213–7217 10.1073/pnas.89.15.7213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A., Zanetti M. E., Jang C. J. H., Holtan H. E., Repetti P. P., Galbraith T. G., Bailey-Serres J. (2009). Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 18843–18848 10.1073/pnas.0906131106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuffer M. G., Coe E. H., Wessler S. R. (1997). Mutants of Maize. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Parra G., Bradnam K., Rose A. B., Korf I. (2011). Comparative and functional analysis of intron-mediated enhancement signals reveals conserved features among plants. Nucleic Acids Res. 39, 5328–5337 10.1093/nar/gkr043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M., Corey A. C., Yin L.-Y., Ali S., Taylor W. C., Berry J. O. (2004). Untranslated regions from C4 amaranth AhRbcS1 mRNAs confer translational enhancement and preferential bundle sheath cell expression in transgenic C4 Flaveria bidentis. Plant Physiol. 136, 3550–3561 10.1104/pp.104.051508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A. H., Bower J. E., Bruggmann R., Dubchak I., Grimwood J., Gundlach H., Haberer G., Hellste U., Mitros T., Poliakov A., Schmutz J., Spannag M., Tang H., Wang X., Wicker T., Bharti A. K., Chapman J., Feltus F. A., Gowik U., Grigoriev I. V., Lyons E., Maher C. A., Martis M., Narechania A., Otillar R. P., Penning B. W., Salamov A. A., Wang Y., Zhang L., Carpita N. C., Freeling M., Gingle A. R., Hash C. T., Keller B., Klein P., Kresovich S., McCann M. C., Ming R., Peterson D. G., Rahman M., Ware D., Westhoff P., Mayer K. F. X., Messing J., Rokhsar D. S. (2009). The Sorghum bicolor genome and the diversification of grasses. Nature 457, 551–556 10.1038/nature07723 [DOI] [PubMed] [Google Scholar]
- Peer W. A., Bandyopadhyay A., Blakeslee J. J., Makam S. N., Chen R. J., Masson P. H., Murphy A. S. (2004). Variation in expression and protein localization of the PIN family of auxin efflux facilitator proteins in flavonoid mutants with altered auxin transport in Arabidopsis thaliana. Plant Cell 16, 1898–1911 10.1105/tpc.021501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollastri S., Tattini M. (2011). Flavonols: old compounds for old roles. Ann. Bot. 108, 1225–1233 10.1093/aob/mcr234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F., Baudry A., Lepiniec L., Grotewold E. (2006). “The regulation of flavonoid biosynthesis,” in The Science of Flavonoids, ed. Grotewold E. (New York: Springer Science+Business Media, Inc.), 97–122 [Google Scholar]
- Rose A. B., Elfersi T., Parra G., Korf I. (2008). Promoter-proximal introns in Arabidopsis thaliana are enriched in dispersed signals that elevate gene expression. Plant Cell 20, 543–551 10.1105/tpc.107.057190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth B. A., Goff S. A., Klein T. M., Fromm M. E. (1991). C1- and R-dependent expression of the maize Bz1 gene requires sequences with homology to mammalian MYB and MYC binding sites. Plant Cell 3, 317–325 10.2307/3869371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H. (2000). Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132, 365–386 [DOI] [PubMed] [Google Scholar]
- Ryan K. G., Swinny E. E., Winefield C., Markham K. R. (2001). Flavonoids and UV photoprotection in Arabidopsis mutants. Z. Naturforsch. 56, 745–754 [DOI] [PubMed] [Google Scholar]
- Ryan K. G., Swinny E. E., Markham K. R., Winefield C. (2002). Flavonoid gene expression and UV photoprotection in transgenic and mutant Petunia leaves. Phytochemistry, 59, 23–32 10.1016/S0031-9422(01)00404-6 [DOI] [PubMed] [Google Scholar]
- Sainz M. B., Grotewold E., Chandler V. L. (1997). Evidence for direct activation of an anthocyanin promoter by the maize C1 protein and comparison of DNA binding by related Myb domain proteins. Plant Cell 9, 611–625 10.1105/tpc.9.4.611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salse J., Bolot S., Throude M., Jouffe V., Piegu B., Quraishi U. M., Calcagno T., Cooke R., Delseny D., Feuilleta F. (2008). Identification and characterization of shared duplications between rice and wheat provide new insight into grass genome evolution. Plant Cell 20, 11–24 10.1105/tpc.107.056309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saslowsky D. E., Warek U., Winkel B. S. J. (2005). Nuclear localization of flavonoid enzymes in Arabidopsis. J. Biol. Chem. 280, 23735–23740 10.1074/jbc.M413506200 [DOI] [PubMed] [Google Scholar]
- Schnable P. S., Ware D., Fulton R. S., Stein J. C., Wei F., Pasternak S., Liang C., Zhang J., Fulton L., Graves T. A., Minx P., Reily A. D., Courtney L., Kruchowski S. S., Tomlinson C., Strong C., Delehaunty K., Fronick C., Courtney B., Rock S. M., Belter E., Du F., Im K., Abbott R. M., Cotton M., Levy A., Marchetto P., Ochoa K., Jackson S. M., Gillam B., Chen W., Yan L., Higginbotham J., Cardenas M., Waligorski J., Applebaum E., Phelps L., Falcone J., Kanchi K., Thane T., Scimone A., Thane N., Henke J., Wang T., Ruppert J., Shah N., Rotter K., Hodges J., Ingenthron E., Cordes M., Kohlberg S., Sgro J., Delgado B., Mead K., Chinwalla A., Leonard S., Crouse K., Collura K., Kudrna D., Currie J., He R., Angelova A., Rajasekar S., Mueller T., Lomeli R., Scara G., Ko A., Delaney K., Wissotski M., Lopez G., Campos D., Braidotti M., Ashley E., Golser W., Kim H., Lee S., Lin J., Dujmic Z., Kim W., Talag J., Zuccolo A., Fan C., Sebastian A., Kramer M., Spiegel L., Nascimento L., Zutavern T., Miller B., Ambroise C., Muller S., Spooner W., Narechania A., Ren L., Wei S., Kumari S., Faga B., Levy M. J., McMahan L., Van Buren P., Vaughn M. W., Ying K., Yeh C. T., Emrich S. J., Jia Y., Kalyanaraman A., Hsia A. P., Barbazuk W. B., Baucom R. S., Brutnell T. P., Carpita N. C., Chaparro C., Chia J. M., Deragon J. M., Estill J. C., Fu Y., Jeddeloh J. A., Han Y., Lee H., Li P., Lisch D. R., Liu S., Liu Z., Nagel D. H., McCann M. C., San-Miguel P., Myers A. M., Nettleton D., Nguyen J., Penning B. W., Ponnala L., Schneider K. L., Schwartz D. C., Sharma A., Soderlund C., Springer N. M., Sun Q., Wang H., Waterman M., Westerman R., Wolfgruber T. K., Yang L., Yu Y., Zhang L., Zhou S., Zhu Q., Bennetzen J. L., Dawe R. K., Jiang J., Jiang N., Presting G. G., Wessler S. R., Aluru S., Martienssen R. A., Clifton S. W., McCombie W. R., Wing R. A., Wilson R. K. (2009). The B73 maize genome: complexity, diversity, and dynamics. Science 326, 1112–1115 10.1126/science.1178534 [DOI] [PubMed] [Google Scholar]
- Sekhon R. S., Lin H., Childs K. L., Hansey C. N., Robin Buell C., De Leon N., Kaeppler S. M. (2011). Genome-wide atlas of transcription during maize development. Plant J. 66, 553–563 10.1111/j.1365-313X.2011.04527.x [DOI] [PubMed] [Google Scholar]
- Shalgi R., Lapidot M., Shami R., Pilpel Y. (2005). A catalog of stability-associated sequence elements in 3′ UTRs of yeast mRNAs. Genome Biol. 6, R86. 10.1186/gb-2005-6-10-r86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. (1991). Molecular mechanisms underlying the differential expression of maize pyruvate, orthophosphate dikinase genes. Plant Cell 3, 225–245 10.2307/3869364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snook M. E., Gueldner R. C., Widstrom N. W., Wiseman B. R., Himmelsbach D. S., Harwood J. S., Costello C. E. (1993). Levels of maysin and maysin analogues in silks of maize germplasm. J. Agric. Food Chem. 41, 1481–1485 10.1021/jf00033a024 [DOI] [Google Scholar]
- Solovchenko A., Schmitz-Eiberger M. (2003). Significance of skin flavonoids for UV-B-protection in apple fruits. J. Exp. Bot. 54, 1977–1984 10.1093/jxb/erg199 [DOI] [PubMed] [Google Scholar]
- Sormani R., Delannoy E., Lageix S., Bitton F., Lanet E., Saez-Vasquez J., Deragon J. M., Renou J. P., Robaglia C. (2011). Sublethal cadmium intoxication in Arabidopsis thaliana impacts translation at multiple levels. Plant Cell Physiol. 52, 436–447 10.1093/pcp/pcr001 [DOI] [PubMed] [Google Scholar]
- Stafford H. A. (1990). Flavonoid Metabolism. Boca Raton: CRC Press, Inc [Google Scholar]
- Stracke R., De Vos R. C., Bartelniewoehner L., Ishihara H., Sagasser M., Martens S., Weisshaar B. (2009). Metabolomic and genetic analyses of flavonol synthesis in Arabidopsis thaliana support the in vivo involvement of leucoanthocyanidin dioxygenase. Planta 229, 427–445 10.1007/s00425-008-0841-y [DOI] [PubMed] [Google Scholar]
- Stracke R., Favory J.-J, Gruber, H. E., Bartelniewoehner L., Bartels S., Binkert M., Funk M., Weisshaar B., Ulm R. (2010a). The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ. 33, 88–103 [DOI] [PubMed] [Google Scholar]
- Stracke R., Jahns O., Keck M., Tohge T., Niehaus K., Fernie A. R., Weisshaar B. (2010b). Analysis of PRODUCTION OF FLAVONOL GLYCOSIDES-dependent flavonol glycoside accumulation in Arabidopsis thaliana plants reveals MYB11-, MYB12- and MYB111-independent flavonols glycoside accumulation. New Phytol. 188, 985–1000 10.1111/j.1469-8137.2010.03421.x [DOI] [PubMed] [Google Scholar]
- Stracke R., Ishihara H., Huep G., Barsch A., Mehrtens F., Niehaus K., Weisshaar B. (2007). Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 50, 660–677 10.1111/j.1365-313X.2007.03078.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 10.1093/molbev/msm092 [DOI] [PubMed] [Google Scholar]
- Taylor L. P., Hepler P. K. (1997). Pollen germination and tube growth. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 461–491 10.1146/annurev.arplant.48.1.461 [DOI] [PubMed] [Google Scholar]
- Taylor L. P., Jorgensen R. (1992). Conditional male fertility in chalcone synthase-deficient petunia. J. Hered. 83, 11–17 [Google Scholar]
- Vargo M. A., Voss O. H., Poustka F., Cardounel A. J., Grotewold E., Doseff A. I. (2006). Apigenin-induced-apoptosis is mediated by the activation of PKCδ and caspases in leukemia cells. Biochem. Pharmacol. 72, 681–692 10.1016/j.bcp.2006.06.010 [DOI] [PubMed] [Google Scholar]
- Wei F., Stein J. C., Lian C., Zhang J., Fulton R. S., Baucom R. S., De Paoli E., Zhoum S., Yang L., Han Y., Pasternak S., Narechania A., Zhang L., Yeh C.-T, Ying, K., Nagel D. H., Collura K., Kudrna D., Currie J., Lin J., Kim H., Angelova A., Scara G., Wissotski M., Golser W., Courtney L., Kruchowski S., Graves T. A., Rock S. M., Adams S., Fulton L. A., Fronick C., Courtney W., Kramer M., Spiegel L., Nascimento L., Kalyanaraman A., Chaparro C., Deragon J.-M., San Miguel P., Jiang N., Wessler S. R., Green P. J., Yu Y., Schwartz D. C., Meyers B. C., Bennetzen J. L., Martienssen R. A., McCombie R., Aluru S., Clifton S. W., Schnable P. S., Ware D., Wilson R. K., Wing R. A. (2009). Detailed analysis of a contiguous 22-Mb region of the maize genome. PLoS Genet. 5, e1000728. 10.1371/journal.pgen.1000728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C. E., Grotewold E. (1997). Differences between plant and animal Myb domains are fundamental for DNA-binding, and chimeric Myb domains have novel DNA-binding specificities. J. Biol. Chem. 272, 563–571 10.1074/jbc.272.12.7759 [DOI] [PubMed] [Google Scholar]
- Wilusz C. J., Wilusz J. (2004). Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet. 20, 491–497 10.1016/j.tig.2004.07.011 [DOI] [PubMed] [Google Scholar]
- Yang E., van Nimwegen E., Zavolan M., Rajewsky N., Schroeder M., Magnasco M., Darnell J. E., Jr. (2003). Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Res. 13, 1863–1872 10.1101/gr.997703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylstra B., Busscher J., Franken J., Hollman P. C. H., Mol J. N. M., Van Tunen A. J. (1994). Flavonols and fertilization in Petunia hybrida: localization and mode of action during pollen tube growth. Plant J. 6, 201–212 10.1046/j.1365-313X.1994.6020201.x [DOI] [Google Scholar]
- Ylstra B., Muskens M., Tunen A. (1996). Flavonols are not essential for fertilization in Arabidopsis thaliana. Plant Mol. Biol. 32, 1155–1158 10.1007/BF00041399 [DOI] [PubMed] [Google Scholar]
- Zubiaga A. M., Belasco J. G., Greenberg M. E. (1995). The nonamer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol. Cell. Biol. 15, 2219–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]