Abstract
Tinnitus is a common and often incapacitating hearing disorder marked by the perception of phantom sounds. Susceptibility factors remain largely unknown but GABAB receptor signaling has long been implicated in the response to treatment and, putatively, in the etiology of the disorder. We hypothesized that variation in KCTD12, the gene encoding an auxiliary subunit of GABAB receptors, could help to predict the risk of developing tinnitus. Ninety-five Caucasian outpatients with a diagnosis of chronic tinnitus were systematically screened for mutations in the KCTD12 open reading frame and the adjacent 3′ untranslated region by Sanger sequencing. Allele frequencies were determined for 14 known variants of which three (rs73237446, rs34544607, and rs41287030) were polymorphic. When allele frequencies were compared to data from a large reference population of European ancestry, rs34544607 was associated with tinnitus (p = 0.04). However, KCTD12 genotype did not predict tinnitus severity (p = 0.52) and the association with rs34544607 was weakened after screening 50 additional cases (p = 0.07). Pending replication in a larger cohort, KCTD12 may act as a risk modifier in chronic tinnitus. Issues that are yet to be addressed include the effects of neighboring variants, e.g., in the KCTD12 gene regulatory region, plus interactions with variants of GABAB1 and GABAB2.
Keywords: KCTD12, association analysis, tinnitus, cortical inhibition
Introduction
Tinnitus is an unpleasant and often agonizing condition marked by the phantom perception of sound. According to recent epidemiological estimates, 25% of the US general populations are affected with one in three subjects reporting daily tinnitus (Shargorodsky et al., 2010). While there is evidence of a genetic susceptibility to tinnitus (Sand, 2011), a complex mode of inheritance suggests the presence of multiple risk genes, and comparatively small effect sizes for any single risk allele. Heritability estimates vary from 0.11 to 0.39 (Petersen et al., 2002; Kvestad et al., 2010). Owing to the lack of linkage studies, the search for candidate genes in primary tinnitus is hypothesis-driven. A well-established theoretical framework for tinnitus has been provided by the disruption of GABAB receptor signaling in animal models (Szczepaniak and Møller, 1995, 1996) that may explain altered cortical inhibition in patients (Eichhammer et al., 2004). More recently, a key role for GABAB receptors has been confirmed by the effects of receptor agonists on tinnitus symptomatology (Zheng et al., 2012), renewing the interest in controlled clinical trials (Westerberg et al., 1996). In the light of these developments, further characterization of the GABAB receptor complex is advocated, including the genes encoding the respective receptor structures.
An auxiliary subunit that associates tightly with the carboxy terminus of GABAB2 receptors is KCTD12 (also known as PFET1 or BTB/POZ domain-containing protein), a potassium channel tetramerization domain-containing protein (Bartoi et al., 2010). Coassembly of KCTD12 and GABAB2 changes the properties of the GABAB(1,2) core receptor by increasing agonist potency, by altering G-protein signaling, and by promoting desensitization (Schwenk et al., 2010). Effects on the pharmacology and the kinetics of GABAB receptors occur in various cochlear cell classes, e.g., in type I fibrocytes of the spiral ligament and in type I vestibular hair cells (Resendes et al., 2004). Knockdown of the KCTD12 ortholog right on leads to improper neuronal differentiation in the zebrafish auditory pathway (Kuo, 2005). Unlike other KCTD proteins, however, KCTD12 is also widely expressed in the adult mammalian brain (Metz et al., 2011) and, therefore, likely to act beyond early periods of maturation. KCTD12 is encoded by an intronless gene on human chr13q21 for which only limited data are presently available in hearing disorders. To determine the impact of this candidate gene on chronic tinnitus, we systematically screened the entire open reading frame for genetic variants, and compared observed allele frequencies to published reference data.
Materials and methods
In 95 German outpatients (67 men and 28 women, age 50.6 ± 12.1 years, mean ± SD) consulting for chronic tinnitus, the diagnosis was confirmed by a detailed neurootological examination including otoscopy, stapedius reflexes, middle ear pressure measurements, and pure tone audiometry. For the present study, only patients with subjective tinnitus were included. Tinnitus severity was assessed by the Tinnitus Questionnnaire (TQ) (Goebel and Hiller, 1994). An additional 50 subjects with chronic tinnitus (40 men and 10 women, age 49.3 ± 11.3 years, mean ± SD) formed an extension sample and underwent the same diagnostic workup as outlined above. All participants were Cauacasians and a majority originated from the Upper Palatinate region of Bavaria. All provided informed consent and the study was approved by the local ethics committee at the University of Regensburg.
Genomic DNA was extracted from lymphocytes using standard procedures prior to amplification of the KCTD12 open reading frame and adjacent 3′ sequence by PCR. Briefly, two overlapping amplicons of 438 bp (a) and 819 bp (b) were generated using the following primer pairs: 5′-CGG TTG CAG CTC CTG AGT-3′ (forward, a), 5′-AGC TCT GGC AGC TCG AAG TA-3′ (reverse, a), 5′-CTC GTG CTG CCC GAC TAC TT-3′ (forward, b) and 5′-GAC AGG TCT CAC CCA GCT AC-3′ (reverse, b). PCR products were purified with ExoSAP-IT (GE Healthcare, Freiburg, Germany) for Sanger sequencing, and for the identification of variants against the human genome reference (Genome Reference Consortium Build 37, February 2009 release). In the extension sample, only amplicon b was sequenced. Multiple sequence alignments were conducted with DNA Dynamo 1.0 (Blue Tractor Software, UK). Linkage disequilibrium and conformity with Hardy–Weinberg equilibrium was measured with HaploView 4.2 (Barrett et al., 2005). PS V2.1.15 (Dupont and Plummer, 1990) was used for power simulations. KCTD12 allele frequencies from a large reference population of European ancestry (NHLBI GO Exome Sequencing Project, ESP) were retrieved with the Exome Variant Server (URL: http://evs.gs.washington.edu/EVS/). ESP allele frequency data were compared to the frequencies observed in tinnitus using Fisher's exact tests. T-tests were employed to compare self-reported tinnitus severity in carriers and non-carriers of the minor KCTD12 alleles. STATA 8.0 (Stata Corporation, College Station, TX, USA) was used for descriptive statistics. The Shapiro–Wilk statistic served to test the null hypothesis of normally distributed TQ scores. The level of statistical significance was set at p < 0.05. All p-values are uncorrected for multiple testing.
For estimating the functionality of confirmed sequence variants, evolutionary conservation in primates was assessed with a phylogenetic hidden Markov model-based method, phastCons, that describes the process of DNA substitution at each site in a genome and the way this process changes from one site to the next (Siepel et al., 2005). Computational annotations of SNP function (Xu and Taylor, 2009) were obtained from the SNPinfo WebServer (URL: http://snpinfo.niehs.nih.gov/snpfunc.htm, accessed Dec. 2011). In silico predictions of structural effects at the amino acid level were based on information from homologous proteins using metaPrDOS at default parameters (Ishida and Kinoshita, 2008).
Results
We confirmed the existence of two coding variants, F87F (rs73237446) and T178T (rs34544607), plus one previously described, non-coding variant in the gene's 3′ UTR (rs41287030) at heterozygosities of 0.01, 0.10, and 0.02, respectively. All genotype distributions conformed to the Hardy–Weinberg equilibrium (p > 0.75). No novel sequence variants emerged and 11 KCTD12 variants listed in dbSNP were absent from our sample (rs141180437, rs116710456, rs143013358, rs694997, rs146434030, rs141477426, rs144225285, rs139291676, rs151278314, rs142368706, and rs140689403, Table 1). When allele frequencies in subjects with chronic tinnitus were compared to reference frequencies from a large control population of European ancestry, an increased prevalence of the minor allele was noted for T178T (0.0494 vs. 0.0263, p = 0.04). To put this finding into perspective, the original screening sample was augmented by 100 chromosomes from a second set of patients, whereupon the MAF in cases dropped to 0.0458 for rs34544607, weakening the association with tinnitus (p = 0.07). Power simulations, based on the entire sample of patients diagnosed with chronic tinnitus and on ESP control data, indicated that we should expect a statistical power of >80% to detect a susceptibility factor with an allelic relative risk of >1.77 for the T178T variant. The number of tinnitus cases needed to reach this power was estimated at 363.
Table 1.
Allele frequencies for the KCTD12 sequence screened in subjects with chronic tinnitus as compared to frequencies in a large control population.
| dbSNP ID | chr13 position | Major>minor allelesa | Variant amino acid | MAF in chronic tinnitus (2N)b | MAF in controls (2N)c | p |
|---|---|---|---|---|---|---|
| rs141180437 | 77,460,118 | C>T | P56S | 0.0000 (190) | 0.0000 (6972) | n.s. |
| rs116710456 | 77,460,080 | G>A | Q68Q | 0.0000 (190) | 0.0003 (6858) | n.s. |
| rs143013358 | 77,460,078 | C>T | P69L | 0.0000 (190) | 0.0000 (6844) | n.s. |
| rs694997 | 77,460,068 | G>A | L72L | 0.0000 (190) | 0.0003 (6872) | n.s. |
| rs146434030 | 77,460,065 | C>A | A73A | 0.0000 (190) | 0.0000 (6894) | n.s. |
| rs73237446 | 77,460,023 | C>T | F87F | 0.0053 (190) | 0.0089 (6890) | n.s. |
| rs141477426 | 77,460,015 | G>A | R90H | 0.0000 (190) | 0.0001 (6858) | n.s. |
| rs144225285 | 77,459,981 | C>T | L101L | 0.0000 (190) | 0.0000 (6756) | n.s. |
| rs34544607 | 77,459,750 | G>C | T178T | 0.0458 (262) | 0.0263 (5058) | 0.07 |
| rs139291676 | 77,459,507 | C>T | P259P | 0.0000 (262) | 0.0000 (7018) | n.s. |
| rs151278314 | 77,459,394 | C>T | T297M | 0.0000 (262) | 0.0000 (7020) | n.s. |
| rs142368706 | 77,459,383 | G>A | A301T | 0.0000 (262) | 0.0000 (7020) | n.s. |
| rs140689403 | 77,459,359 | A>G | S309G | 0.0000 (262) | 0.0001 (7020) | n.s. |
| rs41287030 | 77,459,221 | C>T | – | 0.0076 (262) | – | – |
Fisher's exact tests were used to address allelic association.
Nucleobases on the transcribed strand.
Call rates of 85% were achieved in the first round of screening amplicon b.
Reference population of European ancestry from the NHLBI GO Exome Sequencing Project. Data retrieved with the Exome Variant Server (URL: http://evs.gs.washington.edu/EVS/), accessed December 2011.
MAF, minor allele frequency.
We next examined whether KCTD12 variants could serve as predictors of tinnitus severity. Overall, TQ scores followed a Gaussian distribution (Figure 1) and averaged 37.1 ± 16.3 (mean ± SD) out of 84 points (N = 144). By this measure, tinnitus was rated mild (0–30 points) in 55 subjects (38.2%), moderate (31–46 points) in 46 subjects (31.9%), severe (47–59 points) in 29 subjects (20.1%), and extreme (60–84 points) in 14 subjects (9.7%). There was no significant difference in mean TQ scores or in the degree of concomitant hearing loss between carriers and non-carriers of the minor allele at rs34544607 (p = 0.52 and p = 0.48, respectively, t-test, Figure 2). A positive family history of tinnitus in first-degree relatives did not predict rs34544607 genotype (p = 0.67, Fisher's exact test). As we encountered only one carrier of rs73237446, and only two carriers of rs41287030, the interplay of these substitutions with tinnitus severity, hearing loss, or with a family history of tinnitus could not be fully judged.
Figure 1.
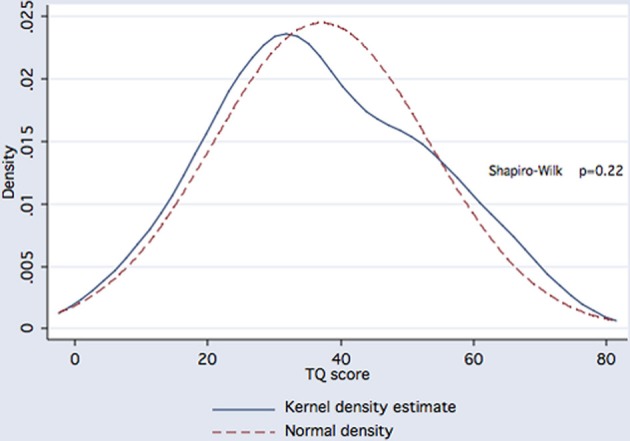
Distribution of TQ scores in 144 subjects with chronic tinnitus does not deviate from the expected Gaussian curve (p = 0.22).
Figure 2.
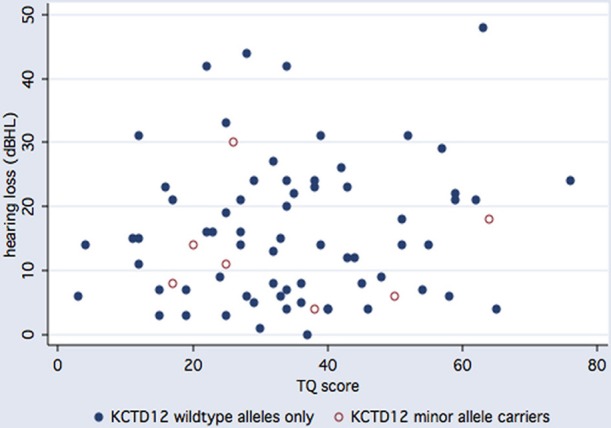
Stratification of TQ scores by hearing loss and KCTD12 minor allele carrier status. Open circles indicate T178T carriers, filled circles indicate homozygous carriers of wildtype alleles. The degree of hearing loss is expressed as the binaural pure tone average involving air conduction across seven test frequencies (0.125, 0.25, 0.5, 1, 2, 4, and 8 kHz).
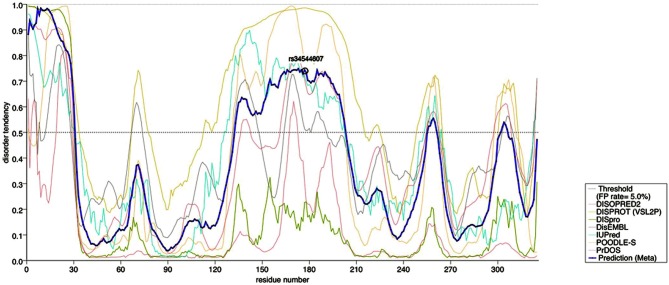
Using the degree of evolutionary conservation as a surrogate parameter of functionality, both rs73237446 and rs34544607 scored high on the comparative genomics scale (Figure 3). Further in silico analyses confirmed that rs73237446 maps to the potassium channel tetramerization domain (Figure 3) whereas residue 178, encoded by rs34544607, maps to a disordered region of KCTD12 (Figure 4) which may affect the molecular recognition of proteins and DNA. The non-coding variant rs41287030 is only poorly conserved among primates but could have acquired a functional role in the recent past. Thus, rs41287030 would appear to alter a micro RNA binding site and may thereby inhibit protein translation (see the corresponding SNPinfo entry for prediction results).
Figure 3.
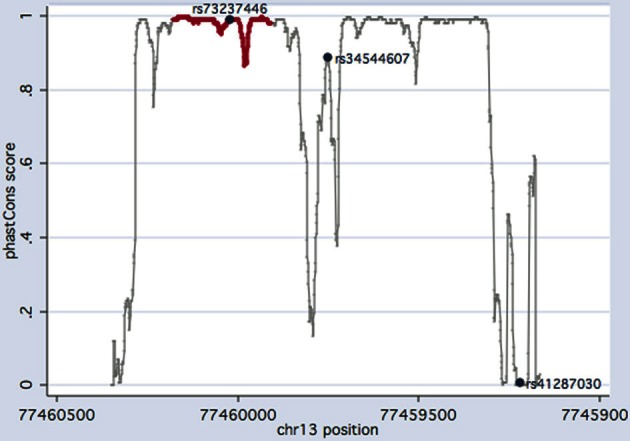
Comparative genomic analysis of the KCTD12 sequence screened. Only confirmed variants are shown. F87F (rs73237446) and T178T (rs34544607) map to regions (x-axis) highly conserved in primates. Conservation scores (y-axis) for the potassium channel tetramerization domain (delimited by residues 36 and 125) are plotted in red.
Figure 4.
In silico structural analysis of the KCTD12 protein. KCTD12 residues are plotted on the x-axis. Eight prediction scores on the y-axis (see legend) are combined using metaPrDOS to locate disordered regions based on homologies with other proteins (blue line). T178T (rs34544607) maps to a relative maximum, i.e., a region that may affect interactions with other proteins or DNA. A default false positive (FP) threshold of 5% was applied.
Discussion
Screening of the KCTD12 ORF in chronic tinnitus extends preliminary results on genomic variation as obtained from 88 subjects with congenital deafness (Kuo, 2005). As in the earlier study, no novel sequence variants were identified. However, a trend was observed for association of chronic tinnitus with a highly conserved, synonymous substitution, rs34544607. The relevance of this finding is unclear in view of the moderately sized sample and the use of an external reference population. It is conceivable that some control subjects from the ESP may have experienced mild, subclinical forms of tinnitus, increasing the likelihood of a type II error. Pending replication of this association trend at a larger scale, the mechanism by which rs34544607 can affect hearing also remains to be elucidated. Possible explanations for synonymous mutations' functionality are offered by interference with RNA processing, or by changes in translation kinetics that affect protein folding (Sauna and Kimchi-Sarfaty, 2011). Phenotypically, rs34544607 carriers may be indistinguishable from other subjects unless treated with baclofen or another GABAB receptor agonist. If rs34544607 truly impacts on GABAB signaling, we should expect electrophysiological measures of cortical inhibition to discriminate between carriers and non-carriers. Electrophysiological data (motor threshold, short-interval intracortical inhibition, intracortical facilitation, and cortical silent period) were available only in a subset of our sample and did not suggest a major effect. The degree of hearing loss did not predict rs34544607 carrier status but further stratification by etiology (noise-induced vs. congenital) is recommended in future studies. With regard to rs41287030, the current lack of publicly available control data and a MAF < 0.01 in tinnitus subjects call for a re-examination in a larger population of affecteds and controls in order to test for a possible association with the phenotype.
Taken together, the present results implicate genetic variation in a GABAB receptor auxiliary subunit as a possible risk modifier in chronic tinnitus. More research is also invited to address KCTD12 promoter variants, and to explore the interaction with variants in genes encoding other elements of the receptor complex, e.g., GABAB1 and GABAB2 proteins.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge support by the German Research Foundation and the Open Access Publishing Fund of the University of Regensburg.
References
- Barrett J. C., Fry B., Maller J., Daly M. J. (2005). Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265. 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- Bartoi T., Rigbolt K. T., Du D., Köhr G., Blagoev B., Kornau H. C. (2010). GABAB receptor constituents revealed by tandem affinity purification from transgenic mice. J. Biol. Chem. 285, 20625–20633. 10.1074/jbc.M109.049700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont W. D., Plummer W. D., Jr. (1990). Power and sample size calculations. A review and computer program. Control. Clin. Trials 11, 116–128. 10.1016/0197-2456(90)90005-M [DOI] [PubMed] [Google Scholar]
- Eichhammer P., Langguth B., Zowe M., Kleinjung T., Jacob P., Sand P., Hajak G. (2004). GABAB-associated neuropsychiatric disorders. Psych-iatr. Prax. 31(Suppl. 1), S44–S46. 10.1055/s-2004-828429 [DOI] [PubMed] [Google Scholar]
- Goebel G., Hiller W. (1994). The tinnitus questionnaire. A standard instrument for grading the degree of tinnitus. Results of a multicenter study with the tinnitus questionnaire. HNO 42, 166–172. [PubMed] [Google Scholar]
- Ishida T., Kinoshita K. (2008). Prediction of disordered regions in proteins based on the meta approach. Bioinformatics 24, 1344–1348. 10.1093/bioinformatics/btn195 [DOI] [PubMed] [Google Scholar]
- Kuo S. F. (2005). Characterization of KCTD12/PFET1, an Intronless Gene with Predominant Fetal Expression. PhD dissertation. Harvard-MIT Division of Health Sciences and Technology. 10.1007/s10162-003-4042-x [DOI] [PMC free article] [PubMed]
- Kvestad E., Czajkowski N., Engdahl B., Hoffman H. J., Tambs K. (2010). Low heritability of tinnitus: results from the second Nord-Trøndelag health study. Arch. Otolaryngol. Head Neck Surg. 136, 178–182. 10.1001/archoto.2009.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz M., Gassmann M., Fakler B., Schaeren-Wiemers N., Bettler B. (2011). Distribution of the auxiliary GABAB receptor subunits KCTD8 12, 12b, and 16 in the mouse brain. J. Comp. Neurol. 519, 1435–1454. 10.1002/cne.22610 [DOI] [PubMed] [Google Scholar]
- Petersen H. C., Andersen T., Frederiksen H., Hoffman H. J., Christensen K. (2002). The heritability of tinnitus: a twin study. Poster presented at: Nordic Epidemiology Congress; June 9–12, 2002. Aarhus, Denmark.
- Resendes B. L., Kuo S. F., Robertson N. G., Giersch A. B., Honrubia D., Ohara O., Adams J. C., Morton C. C. (2004). Isolation from cochlea of a novel human intronless gene with predominant fetal expression. J. Assoc. Res. Otolaryngol. 5, 185–202. 10.1007/s10162-003-4042-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sand P. G. (2011). “Genetic risk factors in chronic tinnitus,” in Textbook of Tinnitus, eds Møller A. R., Langguth B., DeRidder D., Kleinjung T. (New York, NY: Springer; ), 47–50. [Google Scholar]
- Sauna Z. E., Kimchi-Sarfaty C. (2011). Understanding the contribution of synonymous mutations to human disease. Nat. Rev. Genet. 12, 683–691. 10.1038/nrg3051 [DOI] [PubMed] [Google Scholar]
- Schwenk J., Metz M., Zolles G., Turecek R., Fritzius T., Bildl W., Tarusawa E., Kulik A., Unger A., Ivankova K., Seddik R., Tiao J. Y., Rajalu M., Trojanova J., Rohde V., Gassmann M., Schulte U., Fakler B., Bettler B. (2010). Native GABA(B) receptors are heteromultimers with a family of auxiliary subunits. Nature 465, 231–235. 10.1038/nature08964 [DOI] [PubMed] [Google Scholar]
- Shargorodsky J., Curhan G. C., Farwell W. R. (2010). Prevalence and characteristics of tinnitus among US adults. Am. J. Med. 123, 711–718. 10.1016/j.amjmed.2010.02.015 [DOI] [PubMed] [Google Scholar]
- Siepel A., Bejerano G., Pedersen J. S., Hinrichs A. S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L. W., Richards S., Weinstock G. M., Wilson R. K., Gibbs R. A., Kent W. J., Miller W., Haussler D. (2005). Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034–1050. 10.1101/gr.3715005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepaniak W. S., Møller A. R. (1995). Effects of L-baclofen and D-baclofen on the auditory system: a study of click-evoked potentials from the inferior colliculus in the rat. Ann. Otol. Rhinol. Laryngol. 104, 399–404. [DOI] [PubMed] [Google Scholar]
- Szczepaniak W. S., Møller A. R. (1996). Effects of (-)-baclofen, clonazepam, and diazepam on tone exposure-induced hyperexcitability of the inferior colliculus in the rat: possible therapeutic implications for pharmacological management of tinnitus and hyperacusis. Hear. Res. 97, 46–53. [PubMed] [Google Scholar]
- Westerberg B. D., Roberson J. B., Jr., Stach B. A. (1996). A double-blind placebo-controlled trial of baclofen in the treatment of tinnitus. Am. J. Otol. 17, 896–903. [PubMed] [Google Scholar]
- Xu Z., Taylor J. A. (2009). SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 37, W600–W605. 10.1093/nar/gkp290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Vagal S., McNamara E., Darlington C. L., Smith P. F. (2012). A dose-response analysis of the effects of L-baclofen on chronic tinnitus caused by acoustic trauma in rats. Neuropharmacology 62, 940–946. 10.1016/j.neuropharm.2011.09.027 [DOI] [PubMed] [Google Scholar]