Abstract
Small interfering RNAs (siRNAs) have been used extensively in reverse genetic research, and many have made their way into clinical trials. The most widely used siRNA structure consists of double-stranded RNA with 19 base pairs and 2-nucleotide overhangs at the 3’-end of both strands (19+2). Although widely used, this symmetric structure bears inherent disadvantages in both research and clinical applications. One of the most common caveats is the off-target effect leading to adverse effects in clinical application. In the current study, using C-C chemokine receptor (CCR5) as a target, we have shown that 19+2 siRNA could still cause considerable global off-target effects regardless of rational design based on its thermodynamic asymmetry. However, we demonstrated that structurally asymmetric siRNA targeting CCR5 could be adopted to improve the strand specificity and greatly reduce the off-target effects without significantly compromising its on-target effects. Data from microarray analysis suggest that an unidentified mechanism resulting in global gene down-regulation might be avoided through strand shortening. Taken together, our work suggested a promising and simple way to improve strand specificity and overcome the off-target gene-expression effects without introducing more complications while retaining the efficacy of siRNA.
In this study, Yuan and colleagues demonstrate that asymmetrically shortened siRNAs can silence their targets as efficiently as their traditional 19 + 2 counterparts in vitro. Moreover, the authors suggest that shortening siRNAs from their 3′ end improves the loading specificity of the RNA-induced silencing complex and results in fewer off-target gene expression effects.
Introduction
Small interfering RNAs (siRNAs) have been widely used in research for over a decade since the effector complex RISC (RNA-induced silencing complex) was discovered and characterized (Lingel et al., 2004; Liu et al., 2004; Meister et al., 2004; Sontheimer, 2005; Ameres et al., 2007; MacRae et al., 2007; Czech and Hannon, 2011; Sakurai et al., 2011). In recent years, many siRNAs have been investigated for their clinical potential, and some have demonstrated their potential therapeutic value for various diseases, including the infection of human immunodeficiency virus type 1 (HIV-1) (Capodici et al., 2002; Coburn and Cullen, 2002; Novina et al., 2002; Soutschek et al., 2004; Chang et al., 2005; Fu et al., 2005; Kim and Rossi, 2007). One of the main obstacles for the clinical use of siRNA is unintended off-target effects (Castanotto and Rossi, 2009), i.e., the down-regulation or up-regulation of genes other than the target. To address this issue, a variety of modifications have been introduced to siRNA (Soutschek et al., 2004; Bramsen et al., 2009; Vaish et al., 2011), most of which took advantage of chemical substitutions or modifications. However, it has also been reported that siRNAs with a shortened passenger strand could similarly function well and reduce off-target gene-expression effects (Sano et al., 2008; Sun et al., 2008; Chang et al., 2009).
The mechanisms behind off-target gene-expression effects are still not fully understood; however, there are some known factors that have been shown to contribute significantly to these effects. For example, unintended incorporation of the passenger strand into RISC is a major cause of unintended changes in gene-expression patterns. The thermodynamic properties of the ends of a given siRNA determine which strand will be recognized as the guide strand for RISC (Ameres et al., 2007), and the strand with a relatively less stable 5’-end is more likely to guide the silencing. As such, it was postulated that the rational designing of siRNAs with appropriate thermodynamic asymmetry could circumvent this problem (Reynolds et al., 2004; Tomari et al., 2004). However, thermodynamic asymmetry is not the only determinant of which strand submits to RISC selection, as independent research conducted by two groups showed that asymmetric antisense 3’-overhang could also guide the strand selection (Sano et al., 2008; Chang et al., 2009).
Besides unintended gene silencing caused by passenger strand, other factors such as RISC saturation, microRNA (miRNA)-like effects, and innate immunity activation may contribute to the global off-target effects of siRNA. Furthermore, it is conceivable that additional unknown mechanisms may promote these therapeutically negative factors.
To address these problems, in our current report on siRNAs targeting CCR5, a chemokine receptor and a coreceptor for HIV-1 infection, one strand at a time of an siRNA was shortened to give rise to a series of asymmetric structures, and the shortened forms of siRNA were thoroughly investigated for their potency, strand specificity, and off-target effects. We constructed two reporter plasmids to measure the loading efficiency of each strand of the siRNAs into RISC. Then mRNA microarray analysis was performed to provide insights into the differences of overall off-target effects between asymmetric siRNAs and their 19+2 counterpart [double-stranded RNA with 19 base pairs (bp) and 2-nucleotide overhangs at the 3’-end of both strands].
Materials and Methods
siRNA preparation
All siRNAs were chemically synthesized by GenePharma (Shanghai, China) using standard phosphoramidate chemistry, annealed, and purified by HPLC to a purity of >97%. Lyophilized siRNAs were dissolved in diethyl pyrocarbonate (DEPC)-treated water to a final concentration of 20 nM.
Cell culture and medium
GHOST-R5/X4 and GHOST-X4 cells, from the National Institutes of Health AIDS Research and Reference Reagent Program, were cultured in high-glucose Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 500 μg/ml G418, 100 μg/ml hygromycin, and 1 μg/ml puromycin. HEK293T cells were cultured in high-glucose DMEM supplemented with 10% FBS. All cells were incubated in a humidified atmosphere with 5% CO2 at 37°C in a water-jacketed CO2 incubator (Thermo Electron Corp., Waltham, MA).
Nucleic acid transfection
Transfection of GHOST-R5/X4 cells was performed with X-tremeGENE siRNA Transfection Reagents (Roche, Mannheim, Germany) following standard protocols provided by the manufacturer. In brief, GHOST-R5/X4 cells (4×104/well) were plated in a 24-well plate and transfected with 20 pmol of siRNA after 24 hr of culture. HEK293T cells and GHOST-X4 cells were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. HEK293T cells (1.8×105/well) or GHOST-X4 cells (8×104/well) were plated in a 24-well plate 24 hr before transfection, and cells were cotransfected with 200 ng of plasmids and either 4 pmol or 20 pmol of siRNA. For mRNA microarray analysis, GHOST-R5/X4 cells were plated in a six-well plate at a density of 1.2×105 cells/well and transfected with 1 μmol of siRNA. For cell viability analysis, GHOST-R5/X4 cells were plated in a 96-well plate at a density of 5×103 cells/well and transfected with 4 pmol of siRNA.
RT-qPCR analysis
After transfection, total RNA was extracted from GHOST-R5/X4 cells with TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Isolated RNA was dissolved and diluted in DEPC-treated water and reverse-transcribed using a ReverTra Ace qPCR RT Kit (Toyobo, Osaka, Japan); the resulting cDNA was subjected to quantitative PCR (qPCR) analysis. qPCR was performed using SYBR Green Realtime PCR Master Mix (Toyobo) in an ABI 7300 Real-Time PCR system (Applied Biosystem, Carlsbad, CA). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal control for relative quantification of CCR5. Primer sequences were as follows: CCR5 forward, 5’-AAGCACATTGCCAAACGC; CCR5 reverse, 5’-TGTCACAAGCCCACAGAT; GAPDH forward, 5’-ATGGCACCGTCAAGGCTG AG; GAPDH reverse, 5’-GCTAAG CAGTTGGTGGTGCA. Relative expression was calculated by using the ΔΔCt method. Primer concentrations and annealing temperature were optimized in independent experiments for the amplification efficiency to reach 90–105%, which is the prerequisite for relative quantification.
Flow cytometry analysis
For the evaluation of CCR5 surface expression, cells were trypsinized and stained with phycoerythrin (PE)-conjugated mouse anti-human CD195 (CCR5) monoclonal antibody or PE-conjugated mouse IgG2a,κ as isotype control (BD Biosciences, San Jose, CA). After staining, the cells were fixed with 2% paraformaldehyde. For detection of enhanced green fluorescent protein (EGFP) fluorescence, the cells were trypsinized, washed, and fixed directly with 2% paraformaldehyde. Cells were analyzed on a BD FACS Arial Flow Cytometer, and the data were collected using BD FACSDiva software (BD Biosciences).
Pseudovirus preparation and infection
Luciferase reporter pseudovirus was prepared as previously described (Montefiori, 2009). In brief, HEK293T cells were cotransfected with plasmid expressing the envelope proteins of JR-FL and pNL4-3, which had been mutated to inactivate env gene and carried the sequences encoding Renilla luciferase that was under the control of the HIV-1 long terminal repeat (LTR). After cotransfection, the culture supernatant was collected and cleared, and the 50% tissue culture infective dose (TCID50) was determined.
For infection, GHOST-R5/X4 cells were cultured in a 24-well plate and transfected with various siRNAs. Forty-eight hours after transfection, when cells were grown to confluence, 1,000 TCID50 virus suspension containing 5 μg/ml DEAE was added to each well and incubated for 3 hr. Unbound virus was removed by washing, and the cell culture was continued for 48 hr before the cells were lysed. Luciferase activity was determined on a GloMax 96 Microplate Luminometer (Promega, Madison, WI) using the Bright-Glo Luciferase Kit (Promega).
RNA stability
siRNA (2 μl, 40 pmol) was mixed with 8 μl of FBS and incubated at 37°C for 1, 2, 4, 6, or 8 hr. siRNA diluted in DEPC-treated water was used as control. The mixtures were separated on a 4% ethidium bromide (EB)-prestained agarose gel and analyzed by a Tanon 3500 Gel Image System (Tanon Science & Technology, Shanghai, China).
ELISA
ELISA was carried out using the READY-SET-GO! cytokine ELISA kit (eBioscience, San Diego, CA) according to the manufacturer's instructions. Cells were cultured and transfected with siRNAs as described earlier. Twenty-four hours after transfection, supernatant was collected, and the cells were lysed by RIPA buffer (Santa Cruz Biotechnology, Santa Cruz, CA). Cytokine levels were determined by ELISA, and the concentrations were calculated based on standard curves derived from experiments performed at the same time on standard samples.
Cell viability analysis
Cells were transfected with 40 nM siRNA and incubated for 48 hr; then the supernatant was discarded and cells were incubated in DMEM/10% fetal calf serum supplemented with 10% CCK-8 solution (DOJINDO Laboratories, Kumamoto, Japan) at 37°C for 1 hr. The OD450 was measured using an Infinite M200 (Tecan, Männedorf, Switzerland).
mRNA microarray analysis
Cells were cultured and transfected with siRNAs as described in the section on nucleic acid transfection. Forty-eight hours after transfection, cells were lysed with TRIzol for total RNA extraction and mRNA microarray analysis. Microarray analysis was performed by LC Bio Co. Ltd. (Hangzhou, China) using Affymetrix U133 plus 2.0 array chips (Affymetrix, Santa Clara, CA). Data were scanned and processed using GCOS1.4 software.
Construction of EGFP reporter plasmid
pEGFP-C1 plasmid was amplified by PCR using PrimeSTAR HS DNA Polymerase (Takara, Japan) to introduce siRNA target sequence. The primers used were as follows: sense target forward, 5’-GGATCCACCGGATCTAGATAAAAGTGTCAAGTCCAATCTAT GTTCTGATCATAATCAGCCATACCAC; sense target reverse, 5’-GTGGTATGGCTGATT ATGATCAGAACATAGATTGGACTTGACACTTTTATCTAGATCCGGTGGATCC; antisense target forward, 5’-GGATCCACCGGATCTAGATAAAACATAGATTGGAC TTGACACTTCTGATCATAATCAGCCATACCAC; antisense target reverse, 5’-GTGGTATGGCTGA TTATGATCAGAAGTGTCAAGTCCAATCTATGTTTTATCTAGATCCGGTGGATCC. Then PCR products were subjected to digestion by DpnI restriction enzyme (NEB, Hitchin, Herts, UK) to remove methylated DNA templates so that only mutated DNA was retained. The DpnI-treated DNA was used to transform E. coli, and the clones were verified by sequencing and plasmids isolated and purified using PureYield Plasmid Midiprep System (Promega).
Results
siRNA with shortened passenger strand mediated effective and durable gene silencing against human CCR5
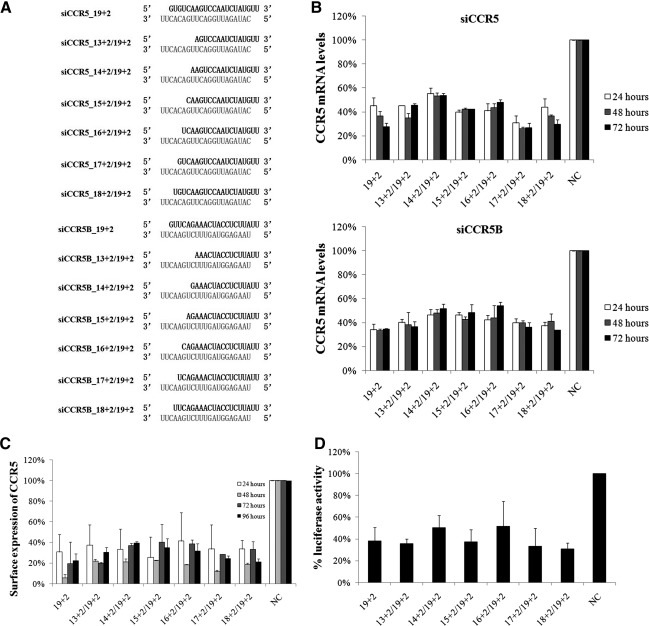
To investigate the effect of shortening passenger strand on the potency of siRNA, two previously reported siRNAs (Anderson and Akkina, 2007) targeting human CCR5, a chemokine receptor and an HIV-1 coreceptor, were used as the starting point. Each siRNA was shortened from the 5’-end of the passenger strand to generate a series of asymmetric siRNAs with double-stranded regions ranging from 13 to 18 bp (Fig. 1A). siRNAs at 40 nM were transfected into GHOST-R5/X4 cells, which are derived from human osteosarcoma cells stably transfected with human CCR5 and CXCR4. CCR5 mRNA was determined by RT-qPCR and its membrane expression detected by flow cytometry after transfection. Shortening passenger strand had little or no effect on the efficacy and durability of silencing. Most siRNAs achieved 40–70% inhibition of mRNA compared with the negative control (Fig. 1B) and 60–80% down-regulation of membrane protein expression (Fig. 1C). The relatively lower inhibition of mRNA might result from cleaved, but not completely degraded, mRNA of CCR5, which could be amplified by PCR primers, but not actually translated, as the primers used did not span the cleavage sites of siRNA targets. The similar results from two different siRNA sequences ruled out the possibility that this could be the result of sequence-specific effects. To validate functional knockdown of CCR5, an HIV-1 env-pseudotyped virus bearing gp120 of the JR-FL strain (a CCR5-using strain) and Renilla luciferase gene under the control of HIV-1 LTR was used to infect GHOST-R5/X4 cells at 48 hr after transfection of various siRNAs. Luciferase activities were suppressed to comparable levels by siRNAs with various lengths of passenger strand relative to conventional 19+2 siRNA (Fig. 1D), indicating that asymmetric siRNAs were at least as equally potent as their symmetric partners in vitro.
FIG. 1.
Efficient knockdown of CCR5 by passenger-strand shortened siRNAs. (A) Sequences of the two sets of siRNA targeting CCR5 used in the experiments. Guide strands are colored black, and sense strands are gray. (B) GHOST-R5/X4 cells were transfected with 40 nM siRNAs, and total RNAs were extracted for RT-qPCR analysis of CCR5 mRNA expression at the indicated time points after transfection. (C) GHOST-R5/X4 cells were transfected as in (B) and were subjected to flow cytometric analysis at the indicated time points after transfection. (D) Forty-eight hours after transfection of 40 nM siRNA, GHOST-R5/X4 cells were infected with 2,000 TCID50/ml HIV-1 JR-FL pseudovirus, and infectivity was determined by luciferase activity analysis. Data shown are the means of three independent experiments, and the error bars indicate the standard deviations. NC, negative control siRNA.
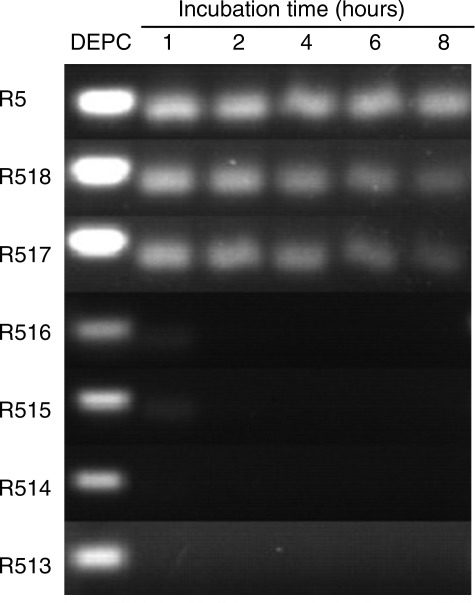
It was observed that trimming of siRNA was accompanied by decreasing of its stability. When incubated with serum, the longer RNAs could be detected after 8 hr, whereas the shorter ones degraded rapidly within 2 hr (Fig. 2). Despite the observed instability of the shortened siRNA, the CCR5 gene was effectively silenced for up to 96 hr by all of the asymmetric RNA tested here (Fig. 1C).
FIG. 2.
Stability of siRNA in serum was negatively correlated with the length of strand. Each siRNA was incubated in 80% FBS for 1–8 hr and electrophoresed on a 4% agarose gel, stained with EB, and visualized by UV irradiation. siRNA incubated in DEPC-treated water was used as loading control.
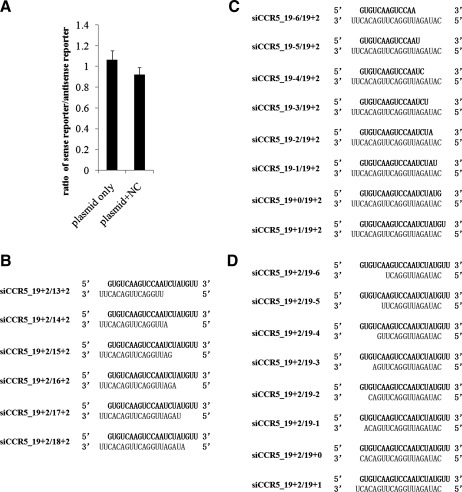
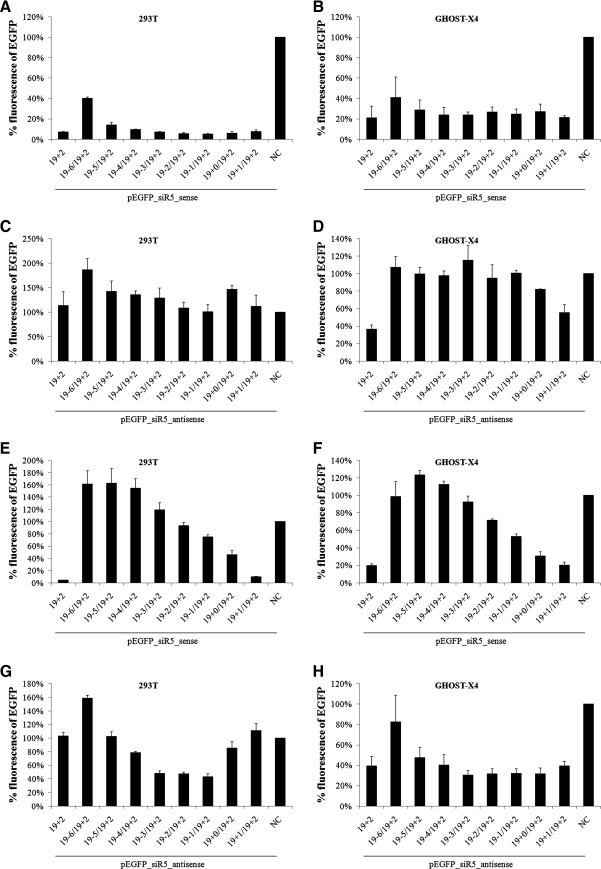
To further validate the specificity of the siRNA-induced knockdown, we constructed a pair of reporter plasmids based on pEGFP-C1, a widely used commercial plasmid expressing high levels of EGFP under the control of the cytomegalovirus immediate early promoter. Sequence complementary to siR5 siRNA guide strand (sense target) or passenger strand (antisense target) was inserted between the EGFP coding sequence and the 3’-untranslated region (UTR) by PCR-based mutagenesis. The plasmid inserted with sense target was designated as pEGFP_siR5_sense, and that inserted with antisense target was designated as pEGFP_siR5_antisense. The expression of EGFP did not differ between the two plasmids (Fig. 3A), whether transfected independently or cotransfected with control siRNA. Therefore, HEK293T cells could be cotransfected with either of the two reporter plasmids and various siRNAs for direct comparison of silencing efficiency. Two concentrations of siRNA (8 and 40 nM) were used to indicate if the effects of shortening were sensitive to siRNA concentration. The results showed that the patterns of relative inhibition by different siRNAs at two distinct concentrations were similar (Fig. 4A, C, E, and G). To confirm the results derived from HEK293T cells and allow direct comparison, cotransfection was repeated in GHOST-X4, a cell line identical to GHOST-R5/X4 except for the absence of CCR5 expression, thus eliminating the influence by endogenous CCR5.
FIG. 3.
siRNAs used for EGFP reporter experiments. (A) HEK293 cells were transfected with either of the two reporter plasmids with or without negative control siRNA (NC); the EGFP fluorescence in cells transfected with different plasmids was compared; and the ratio was plotted. Data shown are the means of three independent experiments, and the error bars indicate the standard deviations. (B) siRNA was trimmed from the 5’-end of the guide strand decrementally. Guide strands are colored black, and sense strands are colored gray. (C and D) siRNA was trimmed from the 3’-end decrementally. Guide strands are colored black, and sense strands are colored gray. NC, negative control siRNA.
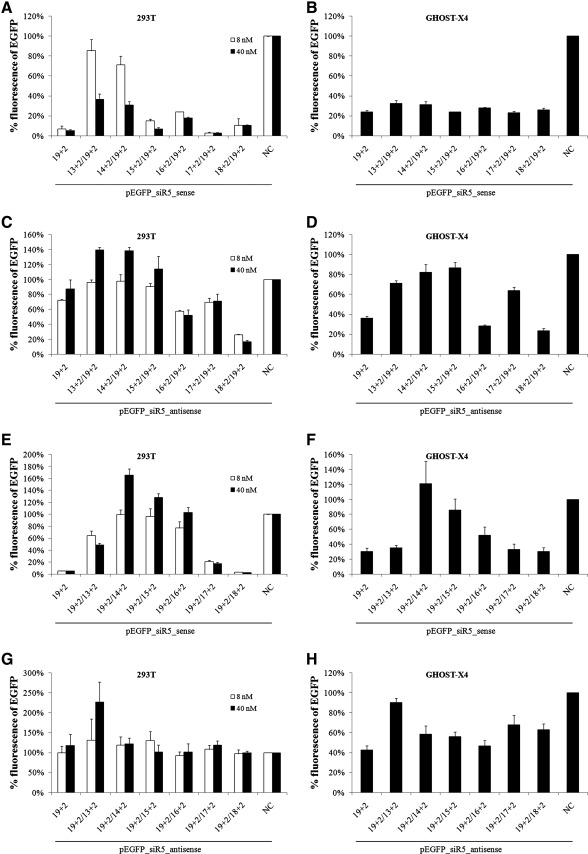
FIG. 4.
Sense silencing and antisense silencing by siRNAs shortened from 5’-end. EGFP reporter plasmids (200 ng) were cotransfected with the indicated siRNA into HEK293T cells (A, C, E, G) or GHOST-X4 cells (B, D, F, H). siRNA concentration was used at 8 nM (293T) or 40 nM (293T and GHOST-X4). Twenty-four hours after transfection, EGFP fluorescence was analyzed by flow cytometry. Data shown are the means of three independent experiments, and the error bars indicate the standard deviations. NC, negative control siRNA.
EGFP expression was efficiently inhibited by siRNAs with shortened passenger strand (Fig. 4A and B), confirming our results obtained from GHOST R5/X4 cells. siRNAs with shorter passenger strand failed to significantly inhibit EGFP at 8 nM (13+2/19+2 and 14+2/19+2 in Fig. 4A), probably due to extreme instability at low concentration and degradation before loading onto RISC. However, considerable inhibition was still achieved at 40 nM, even with the passenger strand shortened to 13+2 (∼64%).
Shortening the guide strand of siRNA from 5’-end did not change the preference for RISC loading
19+2 siRNA bears a symmetric structure, i.e., the two strands share the same topology. It has been shown that RISC could discriminate subtle thermodynamic differences between the two termini of siRNA, and the strand with the more thermodynamically unstable 5’-end would be chosen to guide silencing (Ameres et al., 2007). siR5 siRNA was analyzed on HPCDispatcher, developed in Dr. John J. Rossi's laboratory (Sakurai et al., 2011), to calculate the end free energy. The end free energy of the 5’-end of the guide strand was −7.7 kJ/mol, and that of the 3’-end was −10.1 kJ/mol. Thus, the potency and specificity of siR5 could be attributed to the significant differences in the end free energy of its two ends, i.e., its strict thermodynamic asymmetry.
To investigate how structural asymmetry of siRNA influences RISC loading preference, another series of siRNAs was synthesized and cotransfected with reporter plasmids into HEK293T cells. Instead of the passenger strand, the guide strand of siR5 siRNA was shortened (Fig. 3B). If RISC loading preference is subject to structural asymmetry, then antisense silencing should be enhanced when the guide strand is shortened. However, the passenger strand did not silence the antisense target more efficiently regardless of the guide-strand shortening (Fig. 4G and H). On the contrary, the sense target could still be significantly knocked down when the guide strand was not shortened too much (Fig. 4E and F), and passenger-strand shortening did not reduce the antisense silencing (Fig. 4C and D; 16+2/19+2 and 18+2/19+2). These results suggested that the preference of RISC loading could not be changed by shortening one strand of an siRNA from its 5’-end.
3’-end of siRNA is dispensable for its silencing activity, but contributes to strand selection of RISC
The duplex region formed between the 5’-portion of the guide strand and the 3’-portion of the passenger strand was thought to be critical for RNA interference (RNAi) activity, and previous studies have shown a critical role of the 3’-overhang for RISC loading (Sakurai et al., 2011); therefore, the siRNAs analyzed hitherto were all shortened from the 5’-end. However, it would also be interesting to know the effect of 3’-end shortening on siRNA activity. Two groups of siR5 siRNAs with the passenger strand or guide strand shortened from the 3’-end were synthesized (Fig. 3C and D). siRNAs shortened from the 3’-end of the passenger strand exhibited silencing activity comparable to that of the 19+2 prototype (Fig. 5A and B), similar to their 5’-shortened counterpart (Fig. 4A and B), whereas the activity of antisense silencing was completely abrogated by shortening the passenger strand (Fig. 5C and D), which was distinct from 5’-shortening (Fig. 4C and D). On the other hand, guide-strand shortening greatly compromised the ability of siRNA to silence sense target and enhanced the antisense silencing to various extents, depending on the cell type (Fig. 5E–H). When the 3’-overhang of guide strand was trimmed off, the efficacy of sense silencing was reduced by nearly 50%, and when the guide strand was further shortened, the siRNA completely lost its ability to suppress pEGFP_siR5_sense (Fig. 5E and F), but all of the shortened siRNAs showed antisense silencing no less than the 19+2 symmetric form (Fig. 5G and H). Thus, in contrast to 5’-shortening, when a strand was shortened from the 3’-end, the RISC preference would be biased by structural asymmetry and the silencing efficiency seemed more closely related to the extent of strand shortening rather than thermodynamic preference. Therefore, reducing the length of one strand of siRNA duplex from the 3’-end could significantly compromise its RISC loading while promoting the loading of the opposite strand, which may explain the observation by others that the 3’-overhang is essential for siRNA function (Sakurai et al., 2011).
FIG. 5.
Sense silencing and antisense silencing by siRNAs shortened from the 3’-end. EGFP reporter plasmids (200 ng) were cotransfected with the indicated siRNA into HEK293T cells (A, C, E, G) or GHOST-X4 cells (B, D, F, H). siRNA was used at 40 nM. Twenty-four hours after transfection, EGFP fluorescence was analyzed by flow cytometry. Data shown are the means of three independent experiments, and the error bars indicate the standard deviations. NC, negative control siRNA.
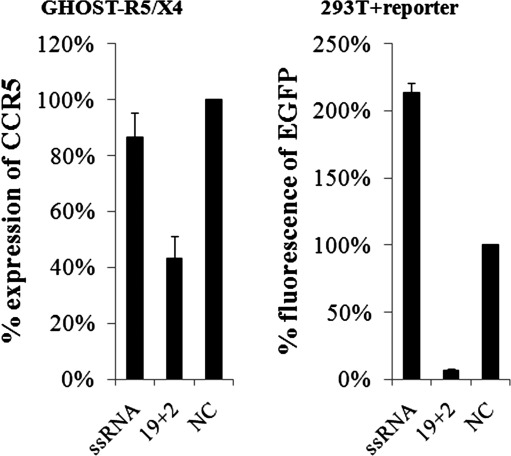
Notably, enhancement of EGFP expression by asymmetric siRNAs with shorter strand was consistently observed in our experiments in 293T cells. It was probably a consequence of target sequences inserted into the 3’-UTR, an important regulatory site in protein coding genes. When one strand of siRNA was shortened, the unpaired region of the opposite strand might pair with the inserted sequence in the 3’-UTR, which in turn could recruit transcriptional or translational factors and enhance the expression of EGFP. This process may be cell type–related, because it was not observed in GHOST-X4 cells. This was confirmed by the enhancement effect of unpaired siR5 guide-strand RNA observed in the EGFP reporter system, but not in GHOST-R5/X4 cells (Fig. 6).
FIG. 6.
Single-stranded RNA enhanced expression of EGFP reporter gene in HEK293T cells, but not genuine CCR5 gene in GHOST-R5/X4 cells. Data shown are the means of three independent experiments, and the error bars indicate the standard deviations. ssRNA, single-stranded antisense RNA of siR5; NC, negative control siRNA.
Taken together, both ends of siRNA could be shortened to give rise to a functional asymmetric structure. The 3’-end of the passenger strand was not necessary for the effector function of RISC, but may be a prerequisite for thermodynamic selection of the two strands by RISC. The strand lacking 3’-overhang seemed to be deselected, which is consistent with previous reports by others (Sano et al., 2008; Chang et al., 2009).
Reduced length of passenger strand correlated with reduced off-target gene silencing
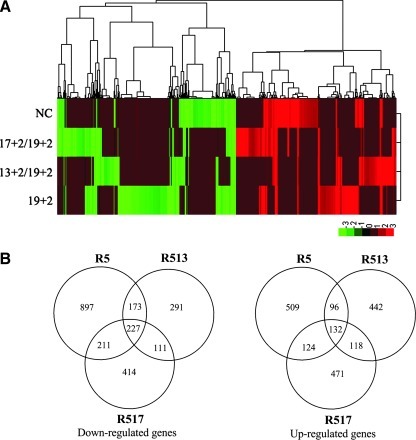
Microarray analysis was performed to investigate the overall off-target gene-expression effects of our siRNA constructs. It showed that considerably fewer genes were modulated by passenger-strand shortened siRNAs than 19+2 siRNA, and it was particularly evident in these down-regulated genes (Table 1, Figure 7A). Both asymmetric siRNAs (with 17 or 13 bp double-stranded region, respectively) showed significantly less off-target effects than 19+2 siRNA. Our observations suggested that shortening one strand of siRNA could greatly reduce the side effects.
Table 1.
Genes Down-Regulated or Up-Regulated by siRNA Transfection
| |
Numbers of genes that changed more than two fold |
Numbers of genes that changed more than three fold |
||
|---|---|---|---|---|
| siRNA | Up-regulation | Down-regulation | Up-regulation | Down-regulation |
| siR5_19+2 | 861 | 1,508 | 341 | 528 |
| siR5_17+2/19+2 | 845 | 963 | 347 | 383 |
| siR5_13+2/19+2 | 800 | 804 | 314 | 333 |
FIG. 7.
Off-target effects of siRNA. (A) Heat map of genes whose mRNA levels fluctuated by more than twofold. (B) Venn diagram showing the relationships between genes up-regulated or down-regulated by each siRNA.
siRNAs can also mimic miRNA when nucleotides 2–8 (core sequence) numbered from the 5’-end of the loaded strand completely pair with mRNA, whereas the other region is only partially complementary, thus targeting unintended mRNA sequences. Based on the microarray data, genes with more than twofold changes induced by siRNA treatment were selected. Their sequences were aligned to the core sequences of both strands of each siRNA, and the numbers of genes or sites fully complementary were counted to evaluate the correlation of miRNA-like off-target silencing. The absolute number of complementary genes was not markedly reduced along with the length of strand, and the number of complementary sites even doubled in genes down-regulated by siR5_13+2/19+2 (Table 2). These results indicated that most miRNA-like functions were not affected by strand shortening, which was consistent with the general knowledge that most miRNAs silence their targets by interfering with translation, rather than cleaving mRNA.
Table 2.
Sequence Complementarity Between Down-Regulated Genes and Core Sequence of siRNA
| Name | Core sequence | Complementary genes | Complementary sites |
|---|---|---|---|
| siR5_19+2/passenger | UGUCAAG | 100 | 133 |
| siR5_19+2/guide | AUAGAUU | 100 | 122 |
| siR5_17+2/19+2/passenger | UCAAGUC | 100 | 138 |
| siR5_17+2/19+2/guide | AUAGAUU | 100 | 134 |
| siR5_13+2/19+2/passenger | GUCCAAU | 79 | 193 |
| siR5_13+2/19+2/guide | AUAGAUU | 99 | 240 |
The differential off-target gene expression was not due to differences in innate immunity recognition and was independent of the RNAi pathway
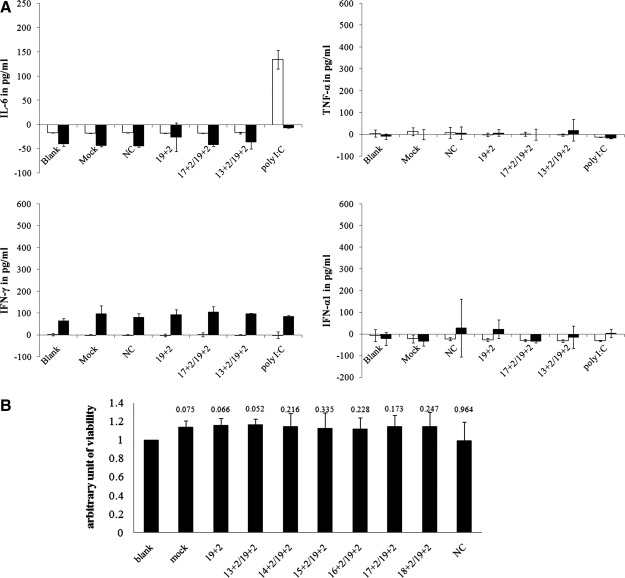
Introduction of exogenous RNA may activate many intracellular signaling pathways and disturb the homeostasis of cellular components, as evidenced by the modulation of gene-expression profiles (Liu et al., 2004). Pattern recognition receptors (PRRs), such as Toll-like receptors (TLRs) and RIG-I-like receptors (RLRs), can recognize exogenous nucleic acids and induce expression of key transcription factors for innate immune regulatory cytokines (Marques et al., 2006). To investigate if the reduction of side effects was the result of activated innate immunity, we measured interleukin-6 (IL-6), interferon-γ (IFN-γ), tumor necrosis factor-α (TNF-α), and IFN-α1, which are known to be up-regulated by exogenous RNA, in both the culture supernatants and the cell lysates 24 hr after transfection of siRNA. Our data showed that the siRNAs used did not induce expression of IL-6, whereas poly I:C, an analogue of double-stranded RNA, induced a high level of IL-6 secretion from GHOST-R5/X4 (Fig. 8A), and the induction of IFN-γ, TNF-α, and IFN-α1 was not observed even by poly I:C, which was consistent with previous observations that RNA duplex less than 30 bp did not trigger innate sensors for RNA (Sun et al., 2008). Additionally, no obvious cell-growth inhibition was detected during transfection of GHOST-R5/X4 cells (Fig. 8B). So the observed reduction of side effects was not a result of differential cytokine induction or cellular toxicity.
FIG. 8.
Cytokine production and cell viability were not influenced by siRNA. (A) GHOST-R5/X4 cells were transfected with 40 nM concentration of the indicated siRNA or poly I:C or treated with transfection reagent (mock) or medium (blank) alone. Twenty-four hours after transfection, cell supernatants were collected and cells lysed; supernatants and lysates were analyzed for the presence of IL-6, INF-γ, TNF-α, and IFN-α1. Experiments were performed in duplicate; values of cytokine concentrations were derived from standard curves accomplished simultaneously (see Supplementary Fig. S1; Supplementary Data are available online at www.liebertonline.com/hum). (B) GHOST-R5/X4 cells were transfected with 40 nM concentration of the indicated siRNA or treated with transfection reagent (mock) or medium (blank) alone. The numbers above each column indicate p values (compared with blank, paired t test). Data shown are the means of three independent experiments, and the error bars indicate the standard deviations. NC, negative control siRNA.
A Venn diagram was generated to show the relationships between genes up-regulated or down-regulated by each siRNA (Fig. 7B). Surprisingly, most of the genes down-regulated by each siRNA were not overlapped, suggesting a mechanism independent of the RNAi pathway that underlay the observed off-target effects, because the RISC is predominantly loaded with the siR5 guide strand (Fig. 4A–D), which was the same for all three siRNAs tested.
Discussion
Although widely used in research, the application of siRNA as therapeutics is still far from reach. One of the major concerns is off-target gene-expression effects. A variety of modifications of siRNA chemistry or structure have been introduced to minimize the off-target effect (Martinez et al., 2002; Chiu and Rana, 2003; Kim et al., 2004; Siolas et al., 2004; Judge et al., 2005; Sun et al., 2008; Bramsen et al., 2009; Vaish et al., 2011). It has been recently reported that modifying siRNAs with unlocked nucleobase analog could markedly reduce the off-target effects (Vaish et al., 2011). However, chemical modifications at dispersed sites greatly increase the cost and complicate the purification of siRNA. In the current study, we modified siRNAs by shortening the passenger strand and demonstrated that the asymmetrically shortened siRNA could silence their targets as efficiently as their 19+2 counterpart, while improving the loading specificity of RISC when shortened from the 3′-end giving rise to less off-target gene expression effects. This strategy is simple and easy to perform, without additional cost added to its manufacturing.
siRNA exerts its function through incorporation into RISC, a complicated and delicately regulated ribonucleoprotein complex. However, in mammalian cells, RISC is mainly ready for loading of miRNA (Gregory et al., 2005). miRNA is a conserved regulatory small RNA species that plays vital roles in regulating many physiological activities, such as development, metabolism, and immunity (Bartel, 2004; He and Hannon, 2004; Biton et al., 2011). The reservoir of RISC components is not unlimited and is saturable by exogenous siRNA, thus affecting the normal function of miRNA. The concentrations of siRNA used in the current study were relatively low (8 and 40 nM) to avoid saturation of RISC. All of the shortened siRNAs tested in the current study showed comparable inhibition of CCR5 and suppression of HIV-1 pseudovirus infection at 40 nM and fivefold lower concentration. Furthermore, the shortened siRNAs showed equal knockdown potency despite their reduced stability. This may be a result of improved RISC loading efficiency or merely reflects the insulation effect of transfection reagents, which needs further investigation. Nevertheless, these results suggest that asymmetric siRNAs deserve further study for potential clinical application.
Two potential mechanisms resulting in off-target silencing have been elucidated (Vaish et al., 2011). One is accidental incorporation of passenger strand that could guide RISC to cleave the sequence complementary to its normal target. Although the detailed RISC loading mechanism remains to be illustrated, thermodynamic asymmetry of siRNA duplex was the only definitely known factor that determines the selection of siRNA strand by RISC. Sano and Chang reported that 3’-overhang could also guide RISC loading (Sano et al., 2008; Chang et al., 2009). Using the EGFP reporter system, our data indicated that 3’-overhang might have a higher priority than thermodynamic asymmetry in strand selection. The biased strand preference caused by asymmetric structure of siRNA suggested that strand shortening from the 3’-end of the passenger strand may be a promising and valuable approach to improve the specificity of RISC loading and aid the design of more potent and more specific siRNAs based on current guidelines (Reynolds et al., 2004).
Another cause for the off-target effect is miRNA-like silencing. This type of off-target gene-expression effects is sequence-dependent; it causes only gene down-regulation and can be caused by either strand of siRNA. Our microarray analysis showed that the number of down-regulated genes was influenced more profoundly than that of up-regulated genes by the length of the siRNA strand, which implicated an underlying gene-silencing mechanism that might be minimized by strand shortening. But it seemed that miRNA-like off-target effects were not influenced at all by strand shortening, because the number of miRNA targets did not change with strand length (Table 2). To obtain a comprehensive insight into the miRNA-like silencing during treatment with asymmetric siRNA, changes in protein levels need to be investigated, because most miRNAs do not influence the levels of target mRNAs (Bartel, 2004).
Exogenous RNA can stimulate various innate immune responses (Judge et al., 2005). Although reports have shown that RNA duplex less than 30 bp could not induce a protein kinase R-dependent type I IFN pathway (Marques et al., 2006), the exogenous nature of siRNA implied an inherent susceptibility of siRNA to a great deal of RNA sensors from the innate defense system, including TLRs, RLRs, and many other molecules that can bind RNA, and there are reports that in some cases siRNA could induce innate immune response (Alexopoulou et al., 2001; Kleinman et al., 2008; Yoneyama and Fujita, 2008; Kawai and Akira, 2010). In the current study, however, no such innate stimulation was observed.
Therefore, some unidentified mechanisms might exist that could result in global gene down-regulation by RNA, and this mechanism is sensitive to the length of RNA. Whether this is an unidentified intrinsic feature of small RNAs or the result of some unknown innate signaling pathway remains to be shown.
In summary, the current study demonstrated that the asymmetric design of siRNA is a promising approach to reduce unintended side effects due to off-target silencing, and the 3’-shortening is a good strategy to promote the strand specificity of siRNA without compromising its potency. The major caveat for the shortened structure of the RNA may be its instability. New technologies that make RNA more stable or novel delivery methods that provide better protection of the naked RNA will need to be considered for practical application of the siRNA.
Supplementary Material
Acknowledgments
We thank Ying Chu for technical support, Hongyong Song for pseudovirus preparation, and Min Qiu for advice. This work was supported by the National Science Foundation (grant no. 30870124), and an International Collaborative Research Grant from the Ministry of Science and Technology (grant no. 2009DFA31260). National Science and Technology Major Project (grant no. 2012ZX10001007-009).
Author Disclosure Statement
No competing financial interests exist.
References
- Alexopoulou L. Holt A.C. Medzhitov R. Flavell R.A. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- Ameres S.L. Martinez J. Schroeder R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell. 2007;130:101–112. doi: 10.1016/j.cell.2007.04.037. [DOI] [PubMed] [Google Scholar]
- Anderson J. Akkina R. Complete knockdown of CCR5 by lentiviral vector-expressed siRNAs and protection of transgenic macrophages against HIV-1 infection. Gene Ther. 2007;14:1287–1297. doi: 10.1038/sj.gt.3302958. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Biton M. Levin A. Slyper M., et al. Epithelial microRNAs regulate gut mucosal immunity via epithelium–T cell crosstalk. Nat. Immunol. 2011;12:239–246. doi: 10.1038/ni.1994. [DOI] [PubMed] [Google Scholar]
- Bramsen J.B. Laursen M.B. Nielsen A.F., et al. A large-scale chemical modification screen identifies design rules to generate siRNAs with high activity, high stability and low toxicity. Nucleic Acids Res. 2009;37:2867–2881. doi: 10.1093/nar/gkp106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capodici J. Kariko K. Weissman D. Inhibition of HIV-1 infection by small interfering RNA-mediated RNA interference. J. Immunol. 2002;169:5196–5201. doi: 10.4049/jimmunol.169.9.5196. [DOI] [PubMed] [Google Scholar]
- Castanotto D. Rossi J.J. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.I. Yoo J.W. Hong S.W., et al. Asymmetric shorter-duplex siRNA structures trigger efficient gene silencing with reduced nonspecific effects. Mol. Ther. 2009;17:725–732. doi: 10.1038/mt.2008.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L.J. Liu X. He J. Lentiviral siRNAs targeting multiple highly conserved RNA sequences of human immunodeficiency virus type 1. Gene Ther. 2005;12:1133–1144. doi: 10.1038/sj.gt.3302509. [DOI] [PubMed] [Google Scholar]
- Chiu Y.L. Rana T.M. siRNA function in RNAi: a chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn G.A. Cullen B.R. Potent and specific inhibition of human immunodeficiency virus type 1 replication by RNA interference. J. Virol. 2002;76:9225–9231. doi: 10.1128/JVI.76.18.9225-9231.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B. Hannon G.J. Small RNA sorting: matchmaking for Argonautes. Nat. Rev. Genet. 2011;12:19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu G.F. Lin X.H. Han Q.W., et al. RNA interference remarkably suppresses bcl-2 gene expression in cancer cells in vitro and in vivo. Cancer Biol. Ther. 2005;4:822–829. doi: 10.4161/cbt.4.8.1889. [DOI] [PubMed] [Google Scholar]
- Gregory R.I. Chendrimada T.P. Cooch N. Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- He L. Hannon G.J. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Judge A.D. Sood V. Shaw J.R., et al. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat. Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- Kawai T. Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- Kim D.H. Rossi J.J. Strategies for silencing human disease using RNA interference. Nat. Rev. Genet. 2007;8:173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- Kim D.H. Behlke M.A. Rose S.D., et al. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat. Biotechnol. 2004;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- Kleinman M.E. Yamada K. Takeda A., et al. Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature. 2008;452:591–597. doi: 10.1038/nature06765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingel A. Simon B. Izaurralde E. Sattler M. Nucleic acid 3’-end recognition by the Argonaute2 PAZ domain. Nat. Struct. Mol. Biol. 2004;11:576–577. doi: 10.1038/nsmb777. [DOI] [PubMed] [Google Scholar]
- Liu J. Carmell M.A. Rivas F.V., et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- MacRae I.J. Zhou K. Doudna J.A. Structural determinants of RNA recognition and cleavage by Dicer. Nat. Struct. Mol. Biol. 2007;14:934–940. doi: 10.1038/nsmb1293. [DOI] [PubMed] [Google Scholar]
- Marques J.T. Devosse T. Wang D., et al. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat. Biotechnol. 2006;24:559–565. doi: 10.1038/nbt1205. [DOI] [PubMed] [Google Scholar]
- Martinez J. Patkaniowska A. Urlaub H., et al. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- Meister G. Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Montefiori D.C. Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol. Biol. 2009;485:395–405. doi: 10.1007/978-1-59745-170-3_26. [DOI] [PubMed] [Google Scholar]
- Novina C.D. Murray M.F. Dykxhoorn D.M., et al. siRNA-directed inhibition of HIV-1 infection. Nat. Med. 2002;8:681–686. doi: 10.1038/nm725. [DOI] [PubMed] [Google Scholar]
- Reynolds A. Leake D. Boese Q., et al. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- Sakurai K. Amarzguioui M. Kim D.H., et al. A role for human Dicer in pre-RISC loading of siRNAs. Nucleic Acids Res. 2011;39:1510–1525. doi: 10.1093/nar/gkq846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano M. Sierant M. Miyagishi M., et al. Effect of asymmetric terminal structures of short RNA duplexes on the RNA interference activity and strand selection. Nucleic Acids Res. 2008;36:5812–5821. doi: 10.1093/nar/gkn584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siolas D. Lerner C. Burchard J., et al. Synthetic shRNAs as potent RNAi triggers. Nat. Biotechnol. 2004;23:227–231. doi: 10.1038/nbt1052. [DOI] [PubMed] [Google Scholar]
- Sontheimer E.J. Assembly and function of RNA silencing complexes. Nat. Rev. Mol. Cell Biol. 2005;6:127–138. doi: 10.1038/nrm1568. [DOI] [PubMed] [Google Scholar]
- Soutschek J. Akinc A. Bramlage B., et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- Sun X. Rogoff H.A. Li C.J. Asymmetric RNA duplexes mediate RNA interference in mammalian cells. Nat. Biotechnol. 2008;26:1379–1382. doi: 10.1038/nbt.1512. [DOI] [PubMed] [Google Scholar]
- Tomari Y. Matranga C. Haley B., et al. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- Vaish N. Chen F. Seth S., et al. Improved specificity of gene silencing by siRNAs containing unlocked nucleobase analogs. Nucleic Acids Res. 2011;39:1823–1832. doi: 10.1093/nar/gkq961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M. Fujita T. RNA recognition and signal transduction by RIG-I-like receptors. Immunol. Rev. 2008;227:54–65. doi: 10.1111/j.1600-065X.2008.00727.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.