RNase P is a key tRNA maturation enzyme, and its enzymatic activity is one of the few performed, even today, by a catalytic RNA. Giegé and colleagues now show that Arabidopsis has evolved to entirely replace RNase P ribonucleoproteins with the protein-only PRORPs, which carry out all RNase P activity in the nucleus as well as organelles. This work has significant implications for discussion of the primordial RNA world's evolution to protein-dominated biology.
Keywords: RNase P, tRNA maturation, RNA processing, pentatricopeptide repeat, plant
Abstract
RNase P is an essential enzyme that cleaves the 5′ leader sequence of tRNA precursors. RNase Ps were believed until now to occur universally as ribonucleoproteins in organisms performing RNase P activity. Here we find that protein-only RNase P enzymes called PRORP (for proteinaceous RNase P) support RNase P activity in vivo in both organelles and the nucleus in Arabidopsis. Beyond tRNA, PRORP proteins are involved in the maturation of small nucleolar RNA (snoRNA) and mRNA. Finally, ribonucleoprotein RNase MRP is not involved in tRNA maturation in plants. Altogether, our results indicate that ribonucleoprotein enzymes have been entirely replaced by proteins for RNase P activity in plants.
RNase P is a virtually universal enzyme involved in the maturation of tRNAs, as it cleaves the 5′ leader sequence of tRNA precursors. It is thus essential to obtain functional tRNAs and is therefore pivotal for translation (Lai et al. 2010; Reiter et al. 2010). RNase P activities from all phyla of life were assumed to be universally performed by ribonucleoprotein enzymes whose catalytic activities are held by ribozymes (Altman 2007). This concept was first challenged with the proposition that spinach chloroplast and human mitochondria RNase Ps would not contain any RNA moiety (Wang et al. 1988; Rossmanith and Karwan 1998). More recently, protein-only RNase P enzymes called PRORP (for proteinaceous RNase P) have been characterized at the molecular level in endosymbiotic organelles in both humans and Arabidopsis (Holzmann et al. 2008; Gobert et al. 2010). Still, the dogma remained that RNase P enzymes would nonetheless universally occur as ribonucleoproteins in living organisms performing RNase P activity, with protein-only RNase Ps being marginal exceptions restricted to only some organelles (e.g., Esakova and Krasilnikov 2010).
The putative PRORP RNase P enzymes are characterized by the occurrence of a conserved “NYN” metallonuclease domain (Anantharaman and Aravind 2006). PRORP proteins also belong to the huge pentatricopeptide repeat (PPR) protein family. These proteins, typically from eukaryotes, are involved in a wide variety of post-transcriptional mechanisms (Schmitz-Linneweber and Small 2008). However, no functional information was available for PRORP proteins. We previously established that Arabidopsis PRORP1, a protein localized in organelles, can act in vitro as an RNase P enzyme that is a single protein (Gobert et al. 2010), although its function remained elusive in planta. We also used localization experiments (YFP fusions and immunodetections) to determine that PRORP2 and PRORP3, two paralogs of PRORP1, were present in Arabidopsis nuclei (Gobert et al. 2010). In addition, the fast-growing amount of genomic data has revealed that some important groups of eukaryotes, such as land plants and kinetoplastids, do not encode any recognizable genes for RNase P RNA or for proteins specific for ribonucleoprotein RNase P (Hartmann and Hartmann 2003). This has led to the hypothesis that some organisms, such as plants, might have entirely replaced ribonucleoprotein RNase P by another type of RNase P enzyme.
However, despite the absence of RNase P RNA genes in plant genomes, another ribonucleoprotein, called RNase MRP, is present in plant nuclei (Kiss et al. 1992). RNase MRP RNA is evolutionarily related to, although clearly distinct from, RNase P RNA. RNase MRP was found to be involved in ribosomal RNA maturation in several model systems (Esakova and Krasilnikov 2010). Still, it could not be excluded that RNase MRP might have acquired an additional RNase P function in plant nuclei.
Here we provide evidence to show that this is not the case and that the former hypothesis is accurate; i.e., that plants have entirely replaced ribonucleoproteins by PRORP enzymes for RNase P activity. We found that the down-regulation of essential RNase MRP subunits does not result in tRNA level changes, whereas the down-regulation of respective PRORP proteins results in decreased RNase P activities in both organelles and the nucleus. Furthermore, we provide evidence to show that, beyond tRNAs, PRORP enzymes, similar to ribonucleoprotein RNase Ps (Lai et al. 2010), are required in vivo for the maturation of other types of RNA species; i.e., small nucleolar RNA (snoRNA) and mRNA in the nucleus and mitochondria, respectively.
Results and Discussion
Since RNase P activity is essential to obtain functional tRNAs and thus for translation in both organelles and the nucleus, the PRORP genes encoding putative RNase P enzymes were predicted to be essential genes. We had already observed that PRORP1 knockout mutation results in lethality and we thus expected the nuclear-localized PRORP2 and PRORP3 (Gobert et al. 2010) to be essential proteins as well if they were the only RNase P enzymes operating in plant nuclei. Two independent mutant lines of PRORP2 and PRORP3 did not show lethality or macroscopic phenotypes. However, homozygous double mutant prorp2 × prorp3 could not be obtained from the genotyping of >500 plants. In addition, the siliques of plants homozygous for one mutated gene but heterozygous for the second showed 50% of positions where seeds were completely absent (Supplemental Fig. S1). This result shows that PRORP2 and PRORP3 have redundant functions in the nucleus and that this function is essential.
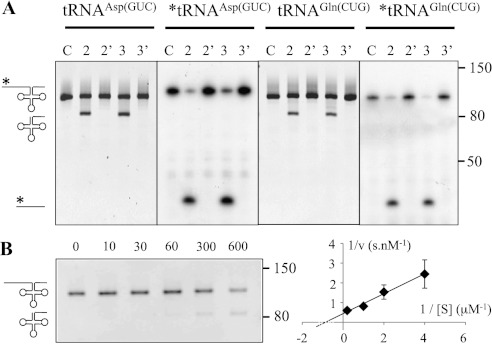
Because the predicted function of PRORP2 and PRORP3 is to perform RNase P activity in plant nuclei, we first had to establish that PRORP2 and PRORP3 could hold RNase P activity as single-subunit protein-only RNase P enzymes. For this, we performed in vitro cleavage assays with recombinant PRORP2 and PRORP3 and transcripts representing precursors for the representative Arabidopsis nuclear tRNAAsp(GUC) and tRNAGln(CUG). In the presence of PRORP2 or of PRORP3, the tRNA leader sequences were clipped off (Fig. 1). The cleavage products were characterized by circular RT–PCR cloning and sequencing, which revealed that maturation had occurred immediately upstream of position +1, as expected for canonical RNase P activity. Moreover, we generated catalytic mutants of PRORP2 and PRORP3 by introducing two aspartate-to-alanine point mutations in the catalytic sites of the respective proteins. No activity was observed when transcripts were put in the presence of the PRORP catalytic mutants (Fig. 1A). We also verified by an established procedure (Gobert et al. 2010) that the purified recombinant PRORP proteins were not contaminated with bacterial RNase P RNA (Supplemental Fig. S2). In order to get first insights into the mode of action of PRORP enzymes, we performed a kinetic analysis of PRORP activity. Reactions were performed with 100 nM exemplary chosen PRORP3 and 0.25–5 μM tRNAGln precursor for 10–600 sec. An apparent KM of 0.3 ± 0.09 μM and a vmax of 2 ± 0.15 nM × s−1 were deduced (Fig. 1B). These parameters are comparable with those observed for eukaryotic ribonucleoprotein RNase P (Hsieh et al. 2009). Altogether, with these results, we found that PRORP proteins whose localization is strictly restricted to nuclei in Arabidopsis have canonical RNase P activity as single-protein enzymes.
Figure 1.
The nuclear proteins PRORP2 and PRORP3 have RNase P activity. (A) The activity was assayed using in vitro transcripts representing the precursors of nuclear tRNAAsp(GUC) and tRNAGln(CUG). Precursor transcripts are 101 nt and 104 nt long, respectively. RNase P endonucleolytic cleavage results in the release of 5′ leader sequences 17 nt and 19 nt long, respectively. (Lanes C) Precursor transcripts alone. (Lanes 2,3) Precursor transcripts incubated with PRORP2 and PRORP3. (Lanes 2′,3′) Precursor transcripts incubated with the catalytic mutant of PRORP2 and PRORP3. After the reactions, RNA molecules were separated on 8% polyacrylamide gels and visualized by ethidium bromide staining and autoradiography. Stars show 5′ radiolabeled RNA molecules and autoradiographed gels. Molecular weights are given in nucleotides. (B) Kinetic analysis of tRNAGln cleavage by PRORP3. Representative experiment performed with 1 μM substrate for five reaction times (in seconds). Similar experiments were performed with four substrate concentrations. Average values from triplicate experiments are shown in a Lineweaver-Burk plot that was used to derive KM and vmax values.
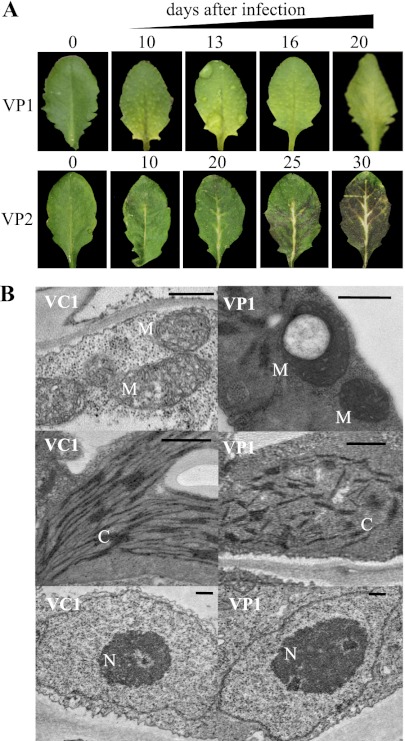
Nevertheless, the question remained regarding whether PRORP enzymes would indeed be responsible for tRNA 5′ maturation in both organelles and the nucleus in planta. In order to answer this question, we had to explore the molecular phenotypes of PRORP mutations in vivo. Since both the single-knockout mutation of PRORP1 and the double-knockout mutation of PRORP2 and PRORP3 result in lethality, we used virus-induced gene silencing (VIGS) (Burch-Smith et al. 2006) to down-regulate PRORP proteins in vivo. We infected wild-type plants with viral vectors targeting PRORP1 to obtain plants where the PRORP1 level was decreased. Then, we used prorp3 homozygous knockout plants with viral vectors targeting PRORP2 to obtain plants where PRORP3 was absent and PRORP2 level was decreased (later called PRORP2/3 plants for simplicity). This resulted in macroscopic symptoms; i.e., yellow chlorotic patches for PRORP1, and white veins followed by rapid leaf senescence for PRORP2/3 (Fig. 2A). These phenotypes correlated with decreases of PRORP1 and PRORP2 mRNA levels (Fig. 3A,C) as well as with decreased PRORP1 and PRORP2 protein levels in purified organelles or nuclei, respectively (Fig. 3A,C). At the cellular level, PRORP1 down-regulation resulted in altered structures of mitochondria and chloroplasts; i.e., very dense mitochondria with disorganized cristae and sometimes containing vacuoles and chloroplasts with completely disorganized thylakoid structures. However, nuclei were unaffected in PRORP1 down-regulation plants (Fig. 2B). PRORP2 down-regulation did not result in any obvious cellular structural change 20 d after viral infection; i.e., both nuclei and organelles did not show apparent structural alteration (Supplemental Fig. S3).
Figure 2.
Macroscopic phenotypes of plants infected by viral constructs targeting PRORP sequences. (A) Arabidopsis leaves not infected (0) and 10–30 d after viral infection. In wild-type plants infected by viral constructs targeting PRORP1 (VP1), the macroscopic phenotype appeared after 10 d and peaked at 20 d. In prorp3 homozygous knockout plants infected by viral constructs targeting PRORP2 (VP2), white veins appeared after 10 d and were followed by complete leaf senescence after 30 d. Samples were collected at 20 d for the molecular analysis. (B) The phenotypes of infected leaves were investigated at the cellular level after 20 d by electron microscopy. No obvious phenotype was visible in green tissue from VP2 cells (Supplemental Fig. S3); however, in VP1 cells, altered structures of mitochondria (M) and chloroplasts (C) could be observed; i.e., with disorganized thylakoid structures and dense mitochondrial structures sometimes containing vacuoles. No structural alteration of nuclei (N) was visible in VP1 cells. (VC1) Control plants infected with viral empty vectors. Bars, 500 nm.
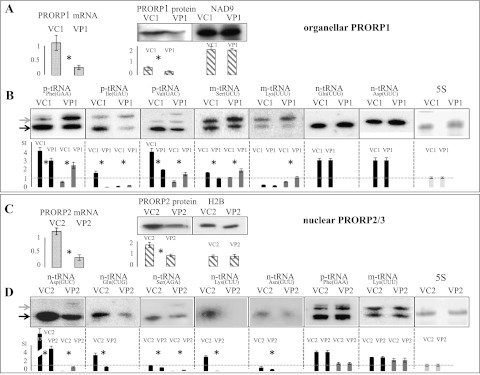
Figure 3.
PRORP proteins are involved in tRNA 5′ maturation in vivo. Genes were down-regulated by VIGS. Wild-type plants were infected with viral empty vector (VC1) or viral vectors expressing siRNA targeting PRORP1 (VP1) and analyzed 20 d after infection. prorp3 homozygous knockout plants were transformed with viral empty vector (VC2) and a viral construct targeting PRORP2 (VP2), thus resulting in plants where PRORP3 is absent and PRORP2 is down-regulated. (A) Levels of PRORP1 mRNA and proteins were analyzed in response to VIGS treatment. mRNA levels were monitored by real-time quantitative RT–PCR. Error bars represent the standard deviation for three biological replicates. Protein levels were analyzed by Western blot on purified mitochondria for PRORP1, and equal loading was controlled with antibodies specific for the mitochondrial NAD9. Error bars represent the standard deviation for three biological replicates. (B) RNA levels in PRORP1 down-regulation plants were investigated by Northern blot hybridizations with tRNA-specific probes. (p-tRNA) Plastidial tRNA; (m-tRNA) mitochondrial tRNA; (n-tRNA) nuclear-encoded tRNAs. Black arrows indicate mature tRNAs, whereas gray arrows show precursor molecules. Histograms show the average quantifications for three replicate hybridization for each RNA species investigated. Black bars indicate mature RNA levels, whereas gray bars show precursor molecule levels. Expression levels are given in signal intensities (SI), normalized so that 1 corresponds to 5S RNA level (indicated by horizontal lines). (*) Experiments where RNA levels were significantly different. Equal loading was verified with 5S rRNA and blot staining. (C) Levels of both PRORP2 mRNA and proteins were analyzed in response to VIGS treatment. For PRORP2 protein, Western blot was performed on purified nuclei, and equal loading was controlled with antibodies specific for nuclear histone 2B (H2B). (D) RNA levels in PRORP2/3 plants were investigated by Northern blot hybridizations.
At the molecular level, the analysis of tRNAs in PRORP1 down-regulation mutants revealed that mitochondrial and plastidial mature tRNAs had decreased levels in the mutant; i.e., organelle tRNA levels in the mutant were, on average, 43% that of tRNA levels in control plants, with a standard deviation (SD) of 12 for five tRNAs. On the other hand, precursor molecules containing the 5′ leader sequences had increased levels; i.e., control plant precursor levels were, on average, 36% (SD = 7 for five tRNAs) that of precursor levels in the mutant, on average. The molecules detected during Northern hybridization were characterized by circular RT–PCR cloning and sequencing to confirm that they correspond to both mature and 5′ precursor molecules. Nuclear-encoded tRNAs had unaffected levels in PRORP1 mutants (Fig. 3B). In contrast, in PRORP2/3 VIGS plants 20 d after infection, organellar tRNAs were unaffected, whereas nuclear-encoded tRNAs had decreased levels; i.e., nuclear tRNA levels in PRORP2/3 plants were, on average, 31% (SD = 8 for five tRNAs) that of tRNA levels in control plants (Fig. 3D). tRNA precursor molecules containing 5′ leaders could also be detected (for tRNAAsp(GUC) and tRNASer(AGA)) (Fig. 3D), but to a lesser extent as compared with PRORP1 mutants. This might be caused by degradation mechanisms and/or quality control processes that would target unprocessed nonfunctional nuclear tRNAs in plant nuclei (Houseley and Tollervey 2009). The effects of PRORP protein level decreases were huge for some tRNAs, whereas they were minor for others. These differences in tRNA level variations might be explained by differences in turnover rates of individual tRNAs. It is becoming increasingly evident that multiple pathways coexist for the turnover of tRNAs; e.g., for normal tRNAs upon cellular stress and for damaged or unprocessed tRNAs (Phizicky and Hopper 2010). This brings the expectation that different turnover rates might exist for individual tRNAs. It is then imaginable that upon PRORP down-regulation, specific tRNAs—e.g., with comparatively faster turnover rates—might accumulate unprocessed precursor molecules faster and thus lead to stronger molecular phenotypes. Alternatively, the differences in tRNA level variations might be explained by differences in cleavage kinetic parameters of individual tRNA precursors by PRORP enzymes. Nonetheless, taken together, we found that the down-regulation of PRORP proteins results in the decrease of tRNA 5′ maturation; i.e., of RNase P activity in the respective compartments where PRORP proteins are localized.
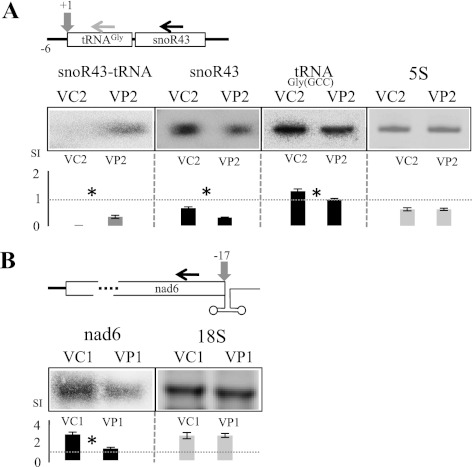
RNase P activity has been initially defined as the activity responsible for the 5′ maturation of tRNAs (Furdon et al. 1983; Altman 2007). However, it has become evident that ribonucleoprotein RNase P enzymes in both prokaryotes and eukaryotes are involved in the maturation of a wide array of substrates; e.g., 4.5S RNA, tmRNA, viral RNAs, mRNAs, and riboswitches (e.g., Altman et al. 2005; Evans et al. 2006; Lai et al. 2010). Experiments performed in yeast have suggested that RNase P could also be involved in the splicing-independent maturation of C/D-box snoRNAs (Coughlin et al. 2008). In plants, tRNA–snoRNA dicistronic transcripts can be found. In particular, in Arabidopsis, a tRNAGly–C/D-box snoRNA (snoR43) gene family is present (Kruszka et al. 2003; Michaud et al. 2011). Previous work has already suggested the involvement of both RNase P and RNase Z (the enzyme that performs the 3′ maturation of tRNAs) in the maturation of these transcripts because 5′ mature transcripts ends were required for the cleavage of tRNAGly–snoR43 by RNase Z in vitro (Barbezier et al. 2009). We thus investigated the involvement of nuclear PRORP enzymes in this maturation in vivo. For this, we monitored the accumulation of snoR43 in the PRORP2/3 VIGS plants. We found that this C/D-box snoRNA has decreased levels in the mutant as compared with control plants; i.e., levels in PRORP2/3 plants were, on average, 38% (SD = 1.5) that of control plant levels. We also observed an accumulation of the tRNAGly–snoR43 precursors in the mutant (Fig. 4A). This shows that snoR43 accumulation is PRORP-dependant. This suggests that PRORP RNase P cleavage is first required for the subsequent downstream release of snoR43. This is consistent with observations in other eukaryotes where RNase P cleavage is required before 3′ processing can be achieved (Wolin and Matera 1999) and in opposition to the occurrence of alternative maturation pathways where 3′ cleavage occurs before RNase P 5′ cleavage (Barbezier et al. 2009; Bayfield et al. 2010).
Figure 4.
PRORP proteins are required for the maturation of snoRNA and mRNA in the nucleus and mitochondria. (A) Nuclear PRORP proteins are required for the accumulation of the C/D-box snoRNA snoR43, present downstream from tRNAGly. The two genes are expressed as a dicistronic transcript, transcription being initiated 6 nt upstream of tRNAGly. RNA levels were monitored by Northern hybridizations with a tRNAGly-specific probe (horizontal gray arrow) and a snoR43-specific probe (horizontal black arrow) in PRORP2/3 VIGS plants (VP2) and control plants (VC2). Images were acquired by PhosphorImaging for 72 h for the tRNA–snoR43 precursor (top panel), for 24 h for snoR43 (middle panel), and for 1 h for tRNAGly (bottom panel). The complete tRNAGly and snoR43 hybridized blots exposed for 72 h are shown in Supplemental Figure S4. (B) The organellar PRORP1 is required for the accumulation of nad6 mRNA in mitochondria. RNA levels were monitored by Northern hybridization with a nad6-specific probe in PRORP1 VIGS plants (VP1) and control plants (VC1). Vertical arrows show the position of RNase P cleavages required for the transcript maturations, relative to tRNAGly start and nad6 termination codon, respectively. Histograms show quantifications as described for Figure 3.
Similarly, PRORP enzymes could be involved in mRNA maturation. Indeed, endonucleolytic cleavages that could correspond to RNase P and RNase Z activities have been mapped at transcript ends of mRNAs in Arabidopsis mitochondria at the level of tRNA-like structures called t-elements (Forner et al. 2007). In particular, a processing event that could correspond to RNase P cleavage has been mapped 17 nucleotides (nt) upstream of the nad6 termination codon (Forner et al. 2007). We previously showed that PRORP1 is able to cleave in vitro a transcript representing the t-element present at the 3′ end of nad6 (Gobert et al. 2010). We thus investigated the accumulation of nad6 in PRORP1 VIGS plants and found that nad6 has decreased levels in the mutant; i.e., levels in PRORP1 plants that were, on average, 32% (SD = 1.2) that of control plant levels. (Fig. 4B). This shows that correct 3′ transcript processing by PRORP1 is required for the accumulation of nad6 mRNA in vivo.
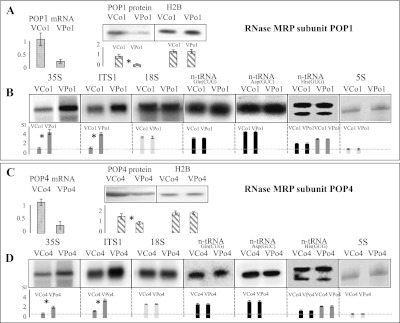
In the absence of genes encoding RNase P RNA in plant genomes, the possibility remained that RNase MRP, another ribonucleoprotein localized in plant nuclei (Kiss et al. 1992), might have acquired RNase P activity in plants. RNase MRP is essential for the maturation of cytosolic rRNA precursors, as it performs the endonucleolytic maturation at site A3 (Esakova and Krasilnikov 2010). Interestingly, in yeast and animals, some proteins—such as POP1 and POP4—that are essential components of RNase MRP are also essential subunits of ribonucleoprotein RNase P (Hartmann and Hartmann 2003). Because POP1 and POP4 are present in plant genomes, we used VIGS to down-regulate these proteins in vivo and thus investigate the potential involvement of RNase MRP in plant tRNA 5′ maturation. For this, we infected mtr4 knockout plants with viral constructs targeting POP1 and POP4. MTR4 is a nonessential putative RNA helicase, a cofactor of the exosome. In its absence, rRNA precursors and maturation intermediates or by-products accumulate and thus become detectable by Northern hybridization (Lange et al. 2011). We found that, as expected from functional analysis in other kingdoms (Esakova and Krasilnikov 2010), both POP1 and POP4 down-regulations resulted in the accumulation of unprocessed cytosolic rRNA precursors—in particular, the large 35S rRNA precursor—as well as the ITS1 (internal transcribed spacer 1) fragment corresponding to the region between the 18S and 5.8S rRNAs that contains the A3 cleavage sites (Fig. 5; Lange et al. 2011). The accumulations of both fragments are consistent with an A3 maturation defect. On the other hand, the down-regulation of both POP1 and POP4 did not result in any variation of nuclear tRNA levels (Fig. 5). This indicates that RNase MRP is indeed involved in rRNA maturation in plants and appears to perform the canonical RNase MRP maturation of rRNA at site A3. It indicates as well that RNase MRP does not appear to be involved in tRNA maturation in plants.
Figure 5.
POP1 and POP4, two essential subunits of RNase MRP, are involved in rRNA maturation but not tRNA maturation. Genes were down-regulated by VIGS. mtr4 knockout plants (Lange et al. 2011) were infected with viral empty vector (VCo1 and VCo4) or viral vectors expressing siRNA targeting POP1 (VPo1) and POP4 (VPo4) and analyzed 20 d after infection. (A) Levels of POP1 mRNA and proteins were analyzed in response to VIGS treatment. mRNA levels were monitored by real-time quantitative RT–PCR. Error bars represent the standard deviation for three biological replicates. For POP1 protein, Western blot was performed on purified nuclei, and equal loading was controlled with antibodies specific for nuclear histone 2B (H2B). Error bars represent the standard deviation for three biological replicates. (B) rRNA and tRNA levels were investigated by Northern blot hybridizations in POP1 down-regulation plants using rRNA fragment-specific or tRNA-specific probes. The 35S rRNA precursor and ITS1 fragments were analyzed to investigate canonical RNase MRP activity. Histograms show quantifications as described for Figure 3. Equal loading was verified with the mature 18S rRNA and blot staining. (C) Levels of POP4 mRNA and proteins were analyzed in response to VIGS treatment as described above. (D) rRNA and tRNA levels were investigated by Northern blot hybridizations in POP4 down-regulation plants as described above.
Here, we found that three PRORP proteins function as RNase P enzymes in the Arabidopsis nucleus and organelles in vivo. The three paralogs share high sequence identity and have the same conserved domains but divergent localization signals (Gobert et al. 2010). We thus propose that their specificities for tRNAs are achieved in vivo through precise subcellular localizations. However, as we began to find, PRORP enzymes also have, beyond tRNAs, a wider spectrum of substrates. In the absence of mechanistic data on the mode of action of PRORP enzymes, we are unable to predict whether the structures or minimal substrates recognized by individual PRORP enzymes are the same or whether specificities exist for some substrates. It cannot be theoretically excluded that another yet unidentified alternative pathway, using catalytic RNA or not, would have RNase P activity in plants. However, it seems unlikely because the putative enzyme(s) would be expected to rescue, at least partially, the knockout mutations of PRORP proteins. Altogether, in light of our results, Arabidopsis rather appears to be the first example of an organism that performs the maturation of tRNA 5′ leader sequences entirely with PRORP proteins in its three compartments where gene expression takes place. It thus becomes apparent that the distribution of RNase P enzymes in life is more complex than previously thought. Some organisms, such as yeast or bacteria, exclusively use ribonucleoproteins for RNase P activity. Other organisms, such as plants, appear to use only PRORP proteins, whereas other organisms, such as human, use both types of enzymes (Supplemental Fig. S5). In humans, several studies have suggested that RNase P RNA is imported into mitochondria (e.g., Mercer et al. 2011). This would suggest that both ribonucleoprotein RNase P and PRORP can coexist in mitochondria. In our view, this is unlikely because the protein subunits of human ribonucleoprotein RNase P are not found in human mitochondrial proteomes and do not possess mitochondrial targeting sequences (Rossmanith 2011). If present in mitochondria, human RNase P RNA might rather be involved in another yet unidentified function in this compartment. Accordingly, we assume that PRORP and ribonucleoprotein RNase Ps function in specialized compartments when they occur in the same organism. The incidence of organisms relying entirely on protein-only RNase P and their comparison with other systems where both PRORP and ribonucleprotein RNase P enzymes coexist or where ribonucleproteins are entirely responsible for RNase P activity will serve as a paradigm and offer a unique opportunity to understand how the transition was made from the prebiotic RNA world to the present day protein-dominated world.
Materials and methods
Please see the Supplemental Material for detailed information.
RNase P cleavage assays
cDNAs representing nuclear tRNA precursors were designed with leader and trailer sequences of ∼20 nt and ∼10 nt, respectively; cloned in pUC19; and transcribed in vitro by T7 RNA polymerase, and RNase P cleavage assays were performed as previously described (Gobert et al. 2010).
Knockout mutants and VIGS analysis
T-DNA insertion lines were analyzed and crossed to obtain double mutants as described (Hammani et al. 2011). Genes were down-regulated by VIGS (Burch-Smith et al. 2006). Briefly, gene-specific sequences were cloned in tobacco rattle virus vector pTRV2 and transformed in Agrobacterium tumefaciens. Bacterial solutions were infiltrated in 3-wk-old Arabidopsis seedlings. Macroscopic and molecular phenotypes of infected plants were analyzed 20 d after agroinfiltration.
Subcellular fractionations and immunodetections
Nuclei and mitochondrial fractions were prepared as described previously (Giegé et al. 2003; Hammani et al. 2011), and Western blot analysis was performed with PRORP antibodies (Gobert et al. 2010) as well as POP1- and POP4-specific antibodies obtained from Dr. A. Schön (Leipzig, Germany).
Northern blot analysis
Northern blot analysis was performed as described previously with total RNA extracted from plant material (Giegé et al. 1998). Three biological replicates were performed for each RNA investigated. Signals were acquired with a FLA-7000 PhosphorImager and quantified with ImageGauge (Fujifilm). Values were normalized so that 1 corresponds to the signal obtained for mitochondrial 5S rRNA. Student t-tests were performed to show that the values measured for the different replicates were statistically similar (P < 0.05). Wilcoxon tests were performed to decide whether average values for the biological replicates were significantly different (P < 0.05) between control and mutant samples. Bands detected by Northern hybridizations were always characterized by cloning and sequencing.
Acknowledgments
We thank Dr. H. Lange for providing the mtr4 mutants. We thank M. Erhardt for assistance with electron microscopy. This work was supported by the French “Centre National de la Recherche Scientifique.” A.G. was supported by an ANR Blanc research grant “PRO-RNase P, ANR 11 BSV8 008 01” to P.G. P.G. was supported by the LabEx consortium “MitoCross.” B.G. was supported by a PhD grant from the University of Strasbourg.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.189514.112.
References
- Altman S 2007. A view of RNase P. Mol Biosyst 3: 604–607 [DOI] [PubMed] [Google Scholar]
- Altman S, Wesolowski D, Guerrier-Takada C, Li Y 2005. RNase P cleaves transient structures in some riboswitches. Proc Natl Acad Sci 102: 11284–11289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V, Aravind L 2006. The NYN domains: Novel predicted RNAses with a PIN domain-like fold. RNA Biol 3: 18–27 [DOI] [PubMed] [Google Scholar]
- Barbezier N, Canino G, Rodor J, Jobet E, Saez-Vasquez J, Marchfelder A, Echeverria M 2009. Processing of a dicistronic tRNA–snoRNA precursor: Combined analysis in vitro and in vivo reveals alternate pathways and coupling to assembly of snoRNP. Plant Physiol 150: 1598–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayfield MA, Yang R, Maraia RJ 2010. Conserved and divergent features of the structure and function of La and La-related proteins (LARPs). Biochim Biophys Acta 1799: 365–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch-Smith TM, Schiff M, Liu Y, Dinesh-Kumar SP 2006. Efficient virus-induced gene silencing in Arabidopsis. Plant Physiol 142: 21–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughlin DJ, Pleiss JA, Walker SC, Whitworth GB, Engelke DR 2008. Genome-wide search for yeast RNase P substrates reveals role in maturation of intron-encoded box C/D small nucleolar RNAs. Proc Natl Acad Sci 105: 12218–12223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esakova O, Krasilnikov AS 2010. Of proteins and RNA: The RNase P/MRP family. RNA 16: 1725–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D, Marquez SM, Pace NR 2006. RNase P: Interface of the RNA and protein worlds. Trends Biochem Sci 31: 333–341 [DOI] [PubMed] [Google Scholar]
- Forner J, Weber B, Thuss S, Wildum S, Binder S 2007. Mapping of mitochondrial mRNA termini in Arabidopsis thaliana: t-elements contribute to 5′ and 3′ end formation. Nucleic Acids Res 35: 3676–3692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furdon PJ, Guerrier-Takada C, Altman S 1983. A G43 to U43 mutation in E. coli tRNAtyrsu3+ which affects processing by RNase P. Nucleic Acids Res 11: 1491–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegé P, Konthur Z, Walter G, Brennicke A 1998. An ordered Arabidopsis thaliana mitochondrial cDNA library on high-density filters allows rapid systematic analysis of plant gene expression: A pilot study. Plant J 15: 721–726 [DOI] [PubMed] [Google Scholar]
- Giegé P, Sweetlove L, Leaver C 2003. Identification of mitochondrial protein complexes in Arabidopsis using two-dimensional Blue-Native polyacrylamide gel electrophoresis. Plant Mol Biol Rep 21: 133–144 [Google Scholar]
- Gobert A, Gutmann B, Taschner A, Gößringer M, Holzmann J, Hartmann RK, Rossmanith W, Giegé P 2010. A single Arabidopsis organellar protein has RNase P activity. Nat Struct Mol Biol 17: 740–744 [DOI] [PubMed] [Google Scholar]
- Hammani K, Gobert A, Hleibieh K, Choulier L, Small I, Giegé P 2011. An Arabidopsis dual-localized pentatricopeptide repeat protein interacts with nuclear proteins involved in gene expression regulation. Plant Cell 23: 730–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann E, Hartmann RK 2003. The enigma of ribonuclease P evolution. Trends Genet 19: 561–569 [DOI] [PubMed] [Google Scholar]
- Holzmann J, Frank P, Loffler E, Bennett KL, Gerner C, Rossmanith W 2008. RNase P without RNA: Identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell 135: 462–474 [DOI] [PubMed] [Google Scholar]
- Houseley J, Tollervey D 2009. The many pathways of RNA degradation. Cell 136: 763–776 [DOI] [PubMed] [Google Scholar]
- Hsieh J, Walker SC, Fierke CA, Engelke DR 2009. Pre-tRNA turnover catalyzed by the yeast nuclear RNase P holoenzyme is limited by product release. RNA 15: 224–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T, Marshallsay C, Filipowicz W 1992. 7-2/MRP RNAs in plant and mammalian cells: Association with higher order structures in the nucleolus. EMBO J 11: 3737–3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruszka K, Barneche F, Guyot R, Aihas J, Meneau I, Shiffer S, Marchfelder A, Echeverria M 2003. Plant dicistronic tRNA–snoRNA genes: A new mode of expression of the small nucleolar RNAs processed by RNase Z. EMBO J 22: 621–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai LB, Vioque A, Kirsebom LA, Gopalan V 2010. Unexpected diversity of RNase P, an ancient tRNA processing enzyme: Challenges and prospects. FEBS Lett 584: 287–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange H, Sement FM, Gagliardi D 2011. MTR4, a putative RNA helicase and exosome co-factor, is required for proper rRNA biogenesis and development in Arabidopsis thaliana. Plant J 68: 51–63 [DOI] [PubMed] [Google Scholar]
- Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AM, Haugen E, Bracken CP, Rackham O, Stamatoyannopoulos JA, et al. 2011. The human mitochondrial transcriptome. Cell 146: 645–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud M, Cognat V, Duchêne AM, Maréchal-Drouard L 2011. A global picture of tRNA genes in plant genomes. Plant J 66: 80–93 [DOI] [PubMed] [Google Scholar]
- Phizicky EM, Hopper AK 2010. tRNA biology charges to the front. Genes Dev 24: 1832–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter NJ, Osterman A, Torres-Larios A, Swinger KK, Pan T, Mondragon A 2010. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature 468: 784–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmanith W 2011. Of P and Z: Mitochondrial tRNA processing enzymes. Biochim Biophys Acta. doi: 10.1016/bbagrm.2011.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmanith W, Karwan RM 1998. Characterization of human mitochondrial RNase P: Novel aspects in tRNA processing. Biochem Biophys Res Commun 247: 234–241 [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Small I 2008. Pentatricopeptide repeat proteins: A socket set for organelle gene expression. Trends Plant Sci 13: 663–670 [DOI] [PubMed] [Google Scholar]
- Wang MJ, Davis NW, Gegenheimer P 1988. Novel mechanisms for maturation of chloroplast transfer RNA precursors. EMBO J 7: 1567–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin SL, Matera AG 1999. The trials and travels of tRNA. Genes Dev 13: 1–10 [DOI] [PubMed] [Google Scholar]