Autophagy is a lysosomal degradation pathway that converts macromolecules into substrates for energy production during nutrient-scarce conditions. In this study, Reed and colleagues demonstrate that an ER membrane protein, Bax inhibitor-1 (BI-1), is a novel modulator of autophagy. The authors show that suppression of BI-1 expression inhibits autophagy and impairs tumors in xenograft studies. In addition, the authors uncover the mechanism by which BI-1 promotes autophagy. BI-1 interacts with IRE-1, which modulates the unfolded protein response (UPR) and apoptosis. Thus, this study provides novel insights into the role of BI-1 in the regulation of autophagy and tumorigenesis.
Keywords: BI-1, autophagy, ER stress, Ca2+, bioenergetics
Abstract
Autophagy is a lysosomal degradation pathway that converts macromolecules into substrates for energy production during nutrient-scarce conditions such as those encountered in tumor microenvironments. Constitutive mitochondrial uptake of endoplasmic reticulum (ER) Ca2+ mediated by inositol triphosphate receptors (IP3Rs) maintains cellular bioenergetics, thus suppressing autophagy. We show that the ER membrane protein Bax inhibitor-1 (BI-1) promotes autophagy in an IP3R-dependent manner. By reducing steady-state levels of ER Ca2+ via IP3Rs, BI-1 influences mitochondrial bioenergetics, reducing oxygen consumption, impacting cellular ATP levels, and stimulating autophagy. Furthermore, BI-1-deficient mice show reduced basal autophagy, and experimentally reducing BI-1 expression impairs tumor xenograft growth in vivo. BI-1's ability to promote autophagy could be dissociated from its known function as a modulator of IRE1 signaling in the context of ER stress. The results reveal BI-1 as a novel autophagy regulator that bridges Ca2+ signaling between ER and mitochondria, reducing cellular oxygen consumption and contributing to cellular resilience in the face of metabolic stress.
Autophagy is a catabolic process characterized by the formation of double-membraned vesicles that deliver intracellular components to lysosomes for degradation (Klionsky 2007, 2010; Mizushima 2010). Under basal conditions, autophagy provides a mechanism for the disposal of senescent proteins and organelles, performing a “housekeeping” function. However, during metabolic stresses, such as nutrient deprivation and hypoxia, autophagy is also employed to catabolize macromolecules and thereby generate substrates for production of ATP (Kroemer et al. 2010; Rabinowitz and White 2010). Dysregulation of autophagy has been implicated in aging and numerous diseases, including neurodegeneration, inflammation, host–pathogen interactions, cardiovascular diseases, and cancer (Mizushima et al. 2008). In the context of cancer, autophagy is hypothesized to play various roles in tumor development, progression, and responses to therapy (Kimmelman 2011). Among these potential roles of autophagy is promoting cancer cell survival under conditions where rapidly growing tumors outstrip their vascular supply, becoming malnourished and hypoxic (Boya et al. 2005; Degenhardt et al. 2006; Tsuchihara et al. 2009).
Inducers of autophagy include endoplasmic reticulum (ER) stress, a condition precipitated by hypoxia, oxidative stress, fluctuations in glucose, and other conditions, which causes accumulation of unfolded proteins in the ER lumen, thus triggering the unfolded protein response (UPR). The UPR is an adaptive response that aims to restore homeostasis in the ER by limiting new protein translation, increasing ER protein-folding capacity through the induction of various ER chaperones, and promoting degradation of unfolded ER proteins (Malhotra and Kaufman 2007). Many tumors show evidence of UPR activation, suggesting that ER stress is common in tumor microenvironments (Tsai and Weissman 2010; Mahadevan and Zanetti 2011).
Recent studies suggest that ER Ca2+ participates in the regulation of autophagy (Hoyer-Hansen et al. 2007). Proposed mechanisms for linking ER Ca2+ to autophagy include (1) modulation of Ca2+-dependent protein kinases (e.g., Ca2+/calmodulin kinase kinase-β) and (2) inositol triphosphate receptors (IP3Rs), which are responsible for transporting Ca2+ from ER into mitochondria to promote bioenergetics (Criollo et al. 2007). IP3Rs are also regulated by members of the Bcl-2 family, with anti-apoptotic proteins Bcl-2 and Bcl-XL promoting IP3-independent leakage of Ca2+ from the ER via physical interactions with IP3Rs (Kuo et al. 1998; Oakes et al. 2005; Rong et al. 2009). Interestingly, mitochondria and ER form interconnected membrane networks that influence various cellular processes, including metabolism and cell death (Csordas et al. 2006). In this regard, mitochondria preferentially accumulate Ca2+ in microdomains where ER and mitochondria are found in close apposition, termed mitochondria-associated microdomains (MAMs) (Rizzuto et al. 1998; Decuypere et al. 2011). The physiological significance of the Ca2+-rich microdomains at the sites of ER–mitochondria contact has recently been elucidated. For instance, elegant work from Foskett and colleagues (Cardenas et al. 2010) demonstrated that IP3R knockdown or IP3R inhibition reduces Ca2+ transport between ER and mitochondria (mitochondrial bioenergetics), lowering cellular ATP and consequently activating autophagy via an AMPK-dependent and mTOR-independent mechanism. Thus, apart from their role in second messenger (IP3)-mediated intracellular signaling, IP3Rs function as facilitators of mitochondrial bioenergetics.
BI-1 (Bax inhibitor-1) is an anti-apoptotic protein that was first discovered by functional screening of cDNA libraries for inhibitors of yeast cell death induced by ectopic expression of mammalian Bax (Xu and Reed 1998). BI-1 is overexpressed in several types of human cancers, and survival in culture of some tumor cell lines is BI-1-dependent (Reimers et al. 2008). BI-1 associates with the anti-apoptotic proteins Bcl-2 and Bcl-XL in ER membranes and operates downstream from Bcl-2 family proteins to control ER Ca2+ homeostasis (Chae et al. 2004; Xu et al. 2008). BI-1 also associates with IRE-1α complexes, suppressing IRE1α's intrinsic endoribonuclease activity (responsible for production of transcription factor XBP-1) and blunting IRE1α-mediated activation of stress kinases (Lisbona et al. 2009; Bailly-Maitre et al. 2010). Studies of genetically engineered mice (BI-1 transgenics and knockouts) have documented protective roles for BI-1 in several diseases where ER stress makes important contributions (Bailly-Maitre et al. 2006, 2010; Hunsberger et al. 2011; Krajewska et al. 2011). However, the roles of BI-1 in cancer remain poorly understood, and the mechanisms by which BI-1 impacts cellular processes that determine tumor cell survival in harsh microenvironments are undefined. Here, we document the importance of BI-1 for tumorigenesis and report a novel role for BI-1 as a regulator of IP3R-dependent Ca2+ transfer from ER to mitochondria, thereby impacting mitochondrial bioenergetics and promoting autophagy. Together with its previously identified function as a modulator of UPR signaling, the ability of BI-1 to reduce dependence on oxidative phosphorylation suggests that BI-1 contributes to tumorigenesis by promoting cellular resilience during metabolic stress.
Results
Tissues of BI-1-deficient mice show changes in autophagy markers
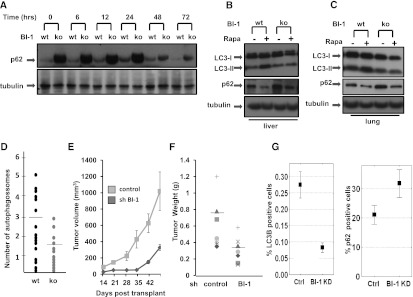
We compared levels of the autophagy marker protein p62 in tissues of wild-type and age-matched BI-1 knockout mice (littermates of the same sex) with and without in vivo treatment with autophagy inducer rapamycin (1 mg/kg) for various times (6–72 h). Due to its degradation by the autophagy lysosome system (Ichimura et al. 2008), conditions that suppress autophagy cause accumulation of p62, resulting in the formation of p62-positive inclusions in cells (Komatsu and Ichimura 2010). Elevations of p62 were seen at baseline in the hearts, livers, and lungs of BI-1 knockout mice (as measured by immunoblotting after normalization for total protein content), suggesting that basal autophagy is impaired in BI-1-deficient mice (Fig. 1). Treatment with rapamycin resulted in decreases in p62 in both wild-type and BI-1 knockout mice, which were more significant for BI-1-deficient mice due to the starting high levels of p62. In contrast to p62 protein, similar levels of p62 mRNA were observed in both wild-type and BI-1 knockout mice, regardless of rapamycin treatment (Supplemental Fig. S1), suggesting that p62 protein accumulation is not due to elevated transcriptional activity. Furthermore, numerous p62 inclusions were found in BI-1 knockout mouse tissues, including the kidneys and brains of BI-1 knockout mice (Supplemental Fig. 2).
Figure 1.
BI-1-deficient mice have reduced autophagy. (A) Protein levels of p62 were assessed by immunoblot analysis of hearts from wild-type (wt) and BI-1 knockout (ko) mice treated with rapamycin for various times as indicated (in hours). Lysates were normalized for total protein content and analyzed by SDS-PAGE/immunoblotting using anti-p62 or anti-tubulin antibodies. Wild-type (wt) and BI-1 knockout (ko) livers (B) and lungs (C) were collected after 24 h of injection with rapamycin. Tissues lysates were analyzed by immunoblotting as above using anti-LC3, anti-p62, and anti-tubulin antibodies. (D) Wild-type (wt) and BI-1 knockout (ko) mice were injected with rapamycin. After 24 h, hearts were dissected, processed, and examined by transmission electron microscopy. From 20 images each for wild-type (wt) and knockout specimens, the total number of autophagosomes was determined using ImageJ software. Mice were injected with 1 mg of rapamycin per kilogram of body weight. Horizontal bars indicate mean. Data are statistically significant by t-test (P = 0.009). (E) Female BALB/c nu/nu mice were injected subcutaneously with H322M cells (5 × 106) containing scrambled control or BI-1 shRNA vectors. Tumor volumes were measured over time (mean ± SD; n = 10). (F) Seven weeks post-transplantation, animals were sacrificed, and tumors were excised and weighed (mean ± SD; n = 10 animals per group). (G) Tumor sections were analyzed for LC3 and p62 staining by quantitative immunohistochemistry. The overall low percentage of cells showing punctate LC3 immunostaining may reflect the antibody conditions employed, which were designed to avoid detection of diffuse cytosolic staining of nonautophagic cells.
LC3 (Atg8 ortholog) is a major constituent of autophagosomes (Kabeya et al. 2000). The proteolytically processed cytoplasmic form of LC3 (LC3-I, 16 kDa) becomes lipid conjugated to generate LC3-II (14 kDa), which is inserted into autophagosome membranes. After fusion of autophagosomes with lysosomes, those LC3-II molecules that are bound to the inner membrane of autophagosomes are degraded by lysosomal proteases. Tissues of BI-1 knockout mice showed reduced LC3-II levels at baseline and after rapamycin treatment (Fig. 1B,C).
Finally, BI-1's impact on autophagy in vivo was also documented by quantification of autophagic vesicles (AVs) using electron microscopy to analyze the heart tissue of wild-type and BI-1 knockout mice. After 24 h of treatment with rapamycin (1 mg/kg), AVs, mainly in the form of autophagolysosomes with partially degraded material in their lumen, were detected throughout the cytoplasm of wild-type mice. In contrast, significantly fewer AVs were observed in BI-1-deficient hearts (Fig. 1D; see Supplemental Fig. S3 for representative images). We conclude, therefore, that BI-1 deficiency alters autophagy in vivo.
BI-1 modulates autophagy in tumor xenografts
To assess the role of BI-1 on autophagy in the context of tumorigenesis, we employed a xenograft model in which the lung cancer cell line H322M (which contains high endogenous levels of BI-1 mRNA, consistent with prior reports for non-small-cell lung cancers) (Tanaka et al. 2006) was stably transduced with control or BI-1 shRNA knockdown vectors and injected subcutaneously into immunocompromised (nu/nu) mice. Whereas cells infected with the shRNA control rapidly generated large palpable tumors, H322M cells containing shRNA BI-1 produced much smaller tumors (Fig. 1E,F). Tumors were processed for immunohistochemical evaluation of autophagy markers LC3 and p62, showing reduced percentages of tumor cells with punctate LC3 staining and elevated percentages with p62 staining, suggesting reduced autophagy in BI-1 knockdown tumors (Fig. 1G; Supplemental Fig. S4).
BI-1 deficiency impairs autophagy in cultured cells
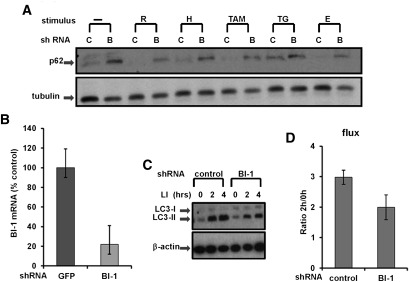
In addition to evaluating the effects of BI-1 deficiency in vivo, we used shRNA vectors to knock down BI-1 expression in cultured tumor cell lines and then monitored effects on autophagy markers in vitro (Fig. 2). Using the same genetically modified H322M lung cancer cells employed for in vivo tumor xenografts in which endogenous BI-1 mRNA was reduced by >80% by shRNA (Fig. 2B), levels of p62 were assessed before and after various stressors. At basal conditions, BI-1 knockdown H322M cells showed markedly elevated levels of p62, suggesting that autophagy was impaired. Challenging the stably transduced control (green fluorescent protein [GFP] shRNA) and BI-1 shRNA cells with various inducers of autophagy resulted in decreases in p62, suggesting that although BI-1-deficient cells have reduced autophagy, they still remain responsive to autophagy induction by the classical pathways that sense nutrient availability (Fig. 2A).
Figure 2.
BI-1 knockdown reduces autophagic flux. (A) Stably transduced H322M cells were cultured in standard rich medium or with serum- and glucose-deficient medium ([H] HBSS; [E] EBSS) alone or with various agents, including rapamycin (R; 25 μg/mL), thapsigargin (TG; 5 μM), and tamoxifen (TAM; 10 μM), for 16 h. Lysates were prepared, normalized for total protein content, and analyzed by SDS-PAGE/immunoblotting using anti-p62 and anti-tubulin antibodies. (B) H322M cells were stably transduced with recombinant shRNA lentiviruses targeting BI-1 or GFP. Relative levels of endogenous BI-1 mRNA were assessed by quantitative RT–PCR (expressing data as a percentage of control relative to cells transduced with GPF shRNA control vector). (C) H322M cells that were stably transduced with recombinant shRNA lentiviruses targeting BI-1 or GFP (control) were treated with LIs (20 mM NH4Cl and 100 μM leupeptin) for either 2 h or 4 h, as indicated. Levels of LC3-I and LC3-II were analyzed by immunoblotting. Blots were reprobed with anti-β-actin antibody as a loading control. (D) Bands were quantified by densitometry, and measurements were used to calculate LC3 flux (mean ± SD; n = 3 experiments; P = 0.017).
To translate the results found in mammalian cells to other organisms, we knocked down BI-1 in Drosophila S2 cells and monitored p62 levels after 2 h of starvation (glucose deprivation). Once again, experimentally reducing BI-1 expression caused an accumulation of p62 protein (Supplemental Fig. S5A). Comparable elevations in p62 were found in BI-1 knockdown S2 cells and in cells where the essential autophagy gene Atg1 was silenced, thus serving as a positive control.
To determine LC3 flux, we cultured control and BI-1 knockdown H322M cells in the absence or presence of the lysosomal inhibitors (LIs) NH4Cl and leupeptin for various times to measure autophagic flux (Martinez-Vicente et al. 2011). Specifically, the ratio of LC3-II levels at 2 h versus 0 h of exposure to LIs was calculated as a representation of LC3 flux (Martinez-Vicente et al. 2011). H322M cells in which BI-1 was knocked down showed reduced levels of LC3 flux, indicating decreased autophagy (Fig. 2C,D). Moreover, comparison of changes in LC3-II at two different times upon blockage of degradation (which can be used as a direct measurement of autophagosome formation) revealed a lower net increase in LC3-II content in the BI-1-deficient cells. Thus, the decrease in autophagic flux seen in BI-1-deficient cells is probably not due to compromised clearance of autophagosomes, but rather is a reflection of reduced autophagosome formation.
BI-1 modulates antibacterial autophagy
Autophagy plays an important role in host defense by eradicating intracellular bacteria through lysosomal destruction (Noda and Yoshimori 2010). We used primary macrophages infected with group A Streptococcus (GAS) as a model for functional assessment of the effects of BI-1 deficiency on autophagy. Accordingly, macrophage cultures were prepared from wild-type versus BI-1 knockout mice and infected with GAS, and the numbers of intracellular viable bacteria were enumerated at various times thereafter. Compared with wild-type cells, the BI-1-deficient macrophages contained significantly higher numbers of GAS (Supplemental Fig. 6), thus providing further evidence that BI-1 deficiency impairs autophagy.
BI-1 overexpression promotes autophagy in cancer cells
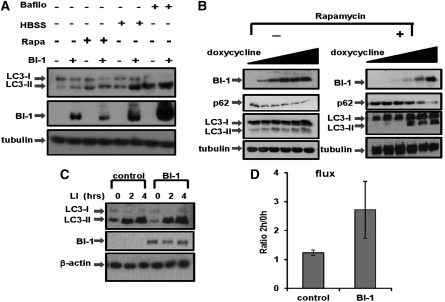
In addition to the studies of BI-1 deficiency, we also ascertained the effects of BI-1 overexpression using cultured cancer cell lines. For these experiments, 293T cells (Fig. 3A) were transiently transfected with either empty vector or pDNA3-BI-1-HA and then treated with various autophagy inducers. Overexpressing BI-1 increased the steady-state levels of LC3-II, in agreement with higher autophagic activity in these cells (also supported by higher rates of conversion of LC3-I to LC3-II in these cells). This increase in basal autophagic activity was also observed upon autophagy induction by culturing cells without nutrients (Hank's buffered salt solution [HBSS]) and, to a lesser extent, by rapamycin.
Figure 3.
BI-1 overexpression increases autophagic flux. (A) The relative levels of LC3-I and LC3-II were assessed by immunoblotting using lysates prepared from 293T cells transiently transfected with empty vector or BI-1-HA vector. Where indicated, cells were pretreated with bafilomycin (bafilo; 200 nM) or rapamycin (rapa; 25 μg/mL) for 16 h, or HBSS for 4 h. (B) The levels of p62 and LC3 were measured in HeLa cells in which BI-1 expression was conditionally driven using a doxycycline-inducible system. Cells were cultured with various concentrations (0, 50, 100, 250, 500, and 1000 ng/mL) for 12 h. (Left) Untreated cell. (Right) Rapamycin-treated cells (25 μg/mL for 12 h). Cell extracts were normalized for total protein content before analysis by SDS-PAGE/immunoblotting using anti-HA (to detect HA-BI-1), anti-p62, and anti-LC3 antibodies. Tubulin was used to verify equal protein loading. (C) HeLa cells in which BI-1 expression was conditionally driven using a doxycycline-inducible system (1 μg/mL) were treated with LIs (20 mM NH4Cl and 100 μM leupeptin) for either 2 h or 4 h. Levels of LC3-I and LC3-II were analyzed by immunoblotting of cell lysates. β-Actin served as a loading control. (D) LC3-II bands were quantified by densitometry, and measurements were used to calculate LC3 flux. LC3 flux was quantified by dividing levels of LC3-II after 2 h of LI treatment per level of LC3-II without LI (mean ± SD; n = 3 independent experiments; P = 0.026).
To further evaluate the role of BI-1 in autophagy regulation, we used HeLa cells, which we previously engineered to inducibly express BI-1 under control of a tetracycline/doxycycline-regulated promoter (Wang et al. 2004). Upon addition to cultures of doxycycline at various concentrations, BI-1 protein expression was induced (Fig. 3B). We compared the levels of autophagy biomarkers p62 and LC3-I/II in these cells before and after inducing BI-1 at basal conditions (Fig. 3B, left) and upon inducing autophagy with rapamycin (Fig. 3B, right). Inducing BI-1 resulted in a decline in p62 in both the basal conditions and following autophagy induction with rapamycin. Overexpressing BI-I increased the steady-state levels of LC3-II, in agreement with higher autophagic activity in these cells (also supported by higher rates of conversion of LC3-I to LC3-II in these cells) (Fig. 3B). Similar to results seen in human cancer cells, when either Drosophila or human BI-1 protein was expressed in fly S2 cells, a reduction in p62 levels was observed (Supplemental Fig. S5B).
In addition to the end-point studies, we also ascertained the effects of BI-1 overexpression on autophagy flux. Similar to experiments performed in BI-1 knockdown cells, control and BI-1-overexpressing cells were cultured in the absence or presence of the LIs NH4Cl and leupeptin (Fig. 3C). Quantitative analysis of LC3-II levels demonstrated that BI-1-overexpressing cells have increased LC3 flux (Fig. 3D).
As another measure of autophagy, we generated a stable tumor cell line overexpressing GFP-LC3 combined with BI-1 under control of a tetracycline-inducible promoter. Localization of LC3 was assessed by high-content imaging using GFP-tagged protein. During autophagy, GFP-LC3 distribution changes from cytosolic diffuse to punctate, reflecting its attachment to AVs (Kabeya et al. 2000). Using a high-throughput microscopy system, we quantified both the number of “spots” of GFP-LC3 and the “spot” intensity per cell. We observed that BI-1 induction significantly increased both the average number of spots per cell and the average spot intensity per cell, with similar results obtained using both live-cell imaging and an analysis of fixed cells (Supplemental Fig. S7A–D). We conclude, therefore, that overexpression of BI-1 increases autophagy.
BI-1 does not require autophagy for cytoprotection
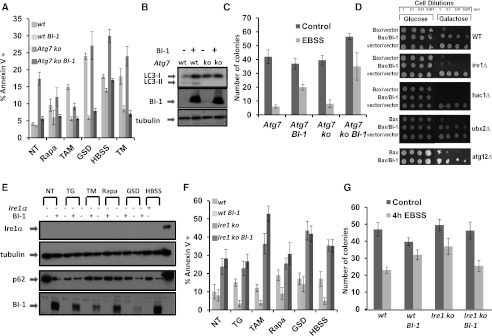
Previous studies have shown that BI-1 is cytoprotective against apoptosis, particularly triggers of ER stress (Chae et al. 2004). Given that BI-1 modulates autophagy, we next addressed whether this property might account for BI-1's cytoprotective activity. For this purpose, we stably transduced BI-1 into autophagy-deficient Atg7−/− mouse embryonic fibroblasts (MEFs) using recombinant lentivirus (Fig. 4B). After 48 h, cells were treated with apoptosis inducers, and then the percentage of annexin V-positive (dead and dying) cells was determined by flow cytometry. The percentage of annexin V-positive cells in cultures of wild-type and Atg7−/− knockout cells overexpressing BI-1 were reduced comparably, suggesting that BI-1 does not require autophagy for its cytoprotective activity (Fig. 4A). Similar results were obtained measuring cell death based on propidium iodide (PI) staining (Supplemental Fig. S8).
Figure 4.
BI-1 requires IRE1α but not autophagy for cytoprotective activity. (A) Wild-type (wt) and Atg7−/− (knockout [ko]) MEFs were stably transduced with empty or BI-1-encoding lentiviruses. Cells were cultured under basal conditions (NT) or for 24 h with various cell stress agents ([TAM] 20 μM tamoxifen; [Rapa] 35 μg/mL rapamycin; [GSD] glucose and serum [FBS] deprivation; [TM] 10 μg/mL tunicamycin) in complete medium or nutrient-depleted medium (HBSS) for 8 h. Cells were then stained with annexin V–FITC and analyzed by FACS. Results are expressed as percentage of annexin V+ cells. (B) To verify defective autophagy in Atg7−/− MEFs, LC3 levels were analyzed by immunoblotting. Tubulin was used as a loading control. (C) A clonogenic survival assay was performed using Atg7−/− (ko) cells stably transduced with empty or BI-1-encoding lentivirus. Cells were cultured (200 cells per six-well plate) in nutrient-depleted medium (EBSS) for 4 h, then washed and cultured for an additional 10 d in complete medium. Cells were stained with crystal violet, and the number of colonies (>1 mm) per well was counted (mean ± SD; n = 3; P = 0.001 for EBSS-treated control vs. BI-1). (D) Saturated overnight cultures of BY4741 wild-type (wt) yeast or the indicated gene deletion strains containing a galactose-inducible Bax expression vector and either BI-1 or control plasmids were normalized to OD600 = 1.0 and then serially diluted in increments of 1:10. Yeast cultures were spotted onto glucose (Bax-off) or galactose (Bax-on) plates and grown for 3 d at 30°C. (E) Ire1α−/− (−) cells were stably transduced with either GFP or BI-1 lentiviruses. Cells were cultured for 24 h with various cell stress agents ([TG] 5 μM thapsigargin; [TM] 10 μM tunicamcyin; [Rapa] 35 μg/mL rapamycin; [GSD] glucose and FBS deprivation) or in nutrient-depleted medium (HBSS) for 8 h. Cell lysates were normalized for total protein content and subjected to immunoblot analysis to detect p62, Ire1, and tubulin (loading control). (F) Ire1α−/− (knockout [ko]) or Ire1α+/+ (wild-type [wt]) MEFs were stably transduced with either GFP or BI-1 lentiviruses and cultured with cell stress agents as in E. To assess cell death, cells were stained with annexin V–FITC, enumerating the percentage of annexin V+ (dead) cells by flow cytometry (mean ± SD; n = 3 independent experiments). (G) Clonogenic survival assays were performed using Ire1α−/− MEFs stably transduced with GFP control or BI-1 lentiviruses. Cells were seeded (200 cells per six-well plate) in nutrient-depleted medium (EBSS) for 4 h and then cultured for an additional 10 d in nutrient-rich medium. Cells were stained with crystal violet, and the number of colonies (>1 mm) per well was counted (mean ± SD; n = 3 independent experiments). Differences between control and BI-1-overexpressing cells were not significantly different (P = 0.09 by t-test).
Clonogenic survival assays were also performed, wherein cells were subjected to the stress of culture for 4 h in nutrient-depleted medium (Eagle's balanced salt solution [EBSS]) and then cultured for 10 d in complete medium before counting surviving colonies (Fig. 4C). In agreement with the annexin V staining, BI-1-overexpressing Atg7−/− MEFs showed significantly increased numbers of colonies compared with control Atg7−/− MEFs (P = 0.013), indicating that BI-1 retains the ability to promote cell survival even in autophagy-defective cells.
Extensive cross-talk occurs between the mechanisms regulating apoptosis, ER stress, and autophagy (Kang et al. 2011; Rubinstein et al. 2011; Verma and Datta 2012). We tested whether minimal medium elicits an ER stress response in Atg7−/− MEFs. Using BiP (Grp78) as a marker of UPR signaling, we observed that Atg7−/− cells are more sensitive to ER stress induction by nutrient deprivation compared with wild-type cells (Supplemental Fig. S9). Interestingly, BI-1 overexpression suppressed UPR marker expression (BiP) in wild-type but not Atg7−/− cells, implying that autophagy influences the ability of BI-1 to impact UPR signaling—even though autophagy is not required for BI-1-mediated protection from ER stress-induced cell death.
BI-1 requires IRE1α for cytoprotection but not for autophagy modulation
Because BI-1 is reported to bind to and suppress IRE1 signaling (Lisbona et al. 2009; Bailly-Maitre et al. 2010), we explored the impact of IRE1 on BI-1-mediated cytoprotection and autophagy. We conducted initial experiments in yeast, where the cytoprotective activity of human BI-1 was first demonstrated by showing that it blocks yeast cell death induced by ectopic expression of the mammalian proapoptotic protein Bax (Xu and Reed 1998). In yeast, only the IRE1 component of the UPR machinery is present (unlike mammalian cells). We observed that yeast IRE1 binds human BI-1 by coimmunoprecipitation (co-IP) experiments (data not shown), indicating that human BI-1 interacts with both yeast and human IRE1. Using various gene deletion mutants, we determined that IRE1 and its downstream target, HAC (yeast ortholog of XBP1), are required for BI-1-mediated protection from Bax in yeast, but not the ERAD component UBX2 or the autophagy gene ATG12 (Fig. 4D). Thus, the IRE1/HAC pathway is required for BI-1 to protect against Bax in yeast, while autophagy is not.
Because IRE1α reportedly modulates autophagy (Ogata et al. 2006), we next explored the role of IRE1α for BI-1's proautophagic and cytoprotective activities by using Ire1α−/− MEFs that had been stably transduced with BI-1 versus control lentiviruses. Levels of LC3 and p62 were evaluated by immunoblot analysis in basal conditions and following treatment with various ER stress and autophagy inducers (Fig. 4E). Reduced p62 levels in BI-1-overexpressing Ire1α−/− cells suggested that IRE1α is not necessary for BI-1 to induce autophagy. BI-1 also retained its proautophagic activity in Perk−/− cells (Supplemental Fig. S10), further distinguishing components of the ER stress response from BI-1's autophagic activity.
To test the cytoprotective function of BI-1 in Ire1α−/− cells, we treated cells with various autophagy inducers (rapamycin, tamoxifen, glucose, and FBS deprivation, or HBSS) or the ER stress inducer thapsigargin and then measured cell viability by annexin V staining (Fig. 4F). Under these conditions, most of the cell death probably occurred via apoptosis, based on costaining FITC–annexin V plus PI costaining analysis (<3% PI-positive cells) (data not shown). While BI-1 overexpression increased the percentages of surviving Ire1+/+ cells, it did not rescue Ire1−/− cells. Similar results were obtained using a clonogenic survival assay where nutrient deprivation was applied as a cellular stress (Fig. 4G). Thus, while other mechanisms may make contributions, we conclude that BI-1's cytoprotective activity is largely dependent on IRE1α.
BI-1 modulates ER Ca2+ and mitochondria bioenergetics
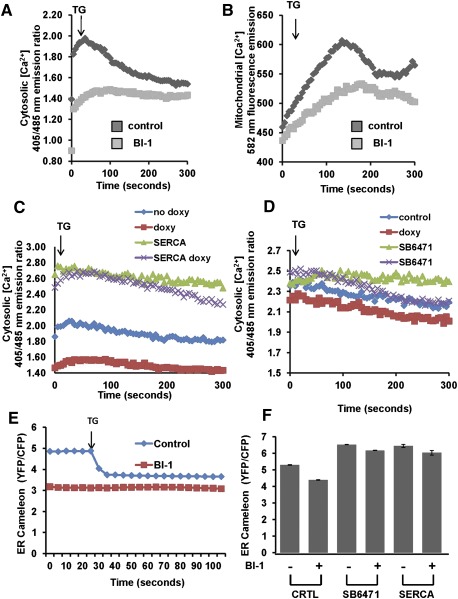
BI-1 is known to impact ER Ca2+ in a manner that phenocopies anti-apoptotic proteins Bcl-2 and Bcl-XL, causing reductions in resting steady-state levels of free Ca2+ in the ER lumen (Xu et al. 2008; Weston and Puthalakath 2010; Hunsberger et al. 2011; Rodriguez et al. 2011). We therefore considered that BI-1's ability to regulate ER Ca2+ might account for its proautophagic activity. To lay a foundation for these studies, we compared the pools of ER Ca2+ in HeLa cells containing tetracycline/doxycycline-inducible BI-1 (with vs. without BI-1 induction) using the irreversible ER Ca2+ ATPase inhibitor thapsigargin. Cells were loaded with the cytosolic Ca2+-sensing dye Indo1 or the mitochondrial Ca2+-sensing dye Rhod2 and then placed into Ca2+-free medium to assess intracellular Ca2+ fluxes upon treatment with thapsigargin. Inducing BI-1 expression by addition of the tetracycline analog doxycycline resulted in reduced levels of ER (thapsigargin-releasable) Ca2+ (Fig. 5A) and, correspondingly, reduced mitochondrial Ca2+ uptake (Fig. 5B). These data are consistent with prior reports showing reduced sequestration of Ca2+ in the ER of BI-1-overexpressing cells, such that release of ER Ca2+ by thapsigargin treatment results in less Ca2+ efflux into the cytosol and therefore less uptake into mitochondria (Xu et al. 2008).
Figure 5.
SERCA nullifies BI-1-mediated reduction of ER Ca2+. HeLa cells in which BI-1 expression was conditionally driven using a doxycycline-inducible system were cultured with (BI-1) or without (control [C]) 1 μg/mL doxycycline for 12 h. Cells were then loaded with the cytosolic Ca2+ probe Indo-1 AM (A) or the mitochondrial Ca2+ probe Rhod 2-AM (B) for 30 min at 37°C. After transfer to Ca2+-free PBS, cells were stimulated with thapsigargin (5 μM) and immediately analyzed using a microplate reader. Average fluorescent intensity was captured every 5 sec for a total time of 300 sec. We used a 405/485-nm emission ratio and 582 nm to measure Indo-1AM and Rhod-2 AM, respectively. (C,D) BI-1-inducible HeLa cells were either transiently transfected with control versus SERCA plasmids (C) or treated with DMSO versus SERCA agonist compound SB-6471 (10 μM) (D). Cells were then cultured with (to induce BI-1) or without doxycycline for 12 h prior to Indo1-AM loading. After transfer to Ca2+-free medium, thapsigargin (TG; 5 μM) was added (arrow), and the levels of cytosolic Ca2+ were measured every 5 sec for a total time of 300 sec. (E) Control and BI-1-overexpressing cells (BI-1) were transfected with the ER Ca2+ cameleon plasmid YC4.3ER for 48 h prior to analysis of ER Ca2+. (F) BI-1-inducible HeLa cells were either transiently transfected with control versus SERCA plasmids or treated with DMSO versus SERCA agonist compound SB-6471 (10 μM). Cells were then cultured with (to induce BI-1) or without doxycycline for 12 h prior to ER cameleon transfection and ER Ca2+ analysis. Results are expressed as emission ratio YFP/CFP.
To assess the role of altered ER Ca2+ in the cellular phenotypes of BI-1, we re-established ER Ca2+ in BI-1-overexpressing cells by either overexpressing sarco/endoplasmic reticulum Ca2+ ATPase (SERCA) (ER Ca2+ pump) or stimulating SERCA pharmacologically with a chemical agonist (Supplemental Fig. S11). By overexpressing SERCA or stimulating endogenous SERCA, our goal was to overcome the Ca2+ leakage from ER created by BI-1, thus re-establishing ER Ca2+ levels toward normal. Indeed, SERCA overexpression by gene transfer and endogenous SERCA activation by pharmacological agonist treatment re-established pools of thapsigargin-releasable (ER) Ca2+ to normal levels in BI-1-overexpressing cells, as measured by Indo1 fluorescence in cells cultured in Ca2+-free medium (Fig. 5C,D). These results were independently confirmed using an ER targeted Ca2+-sensing fluorescent protein (“cameleon”) (Fig. 5E) previously used as a probe for assessing the impact of BI-1 and Bcl-2 family proteins on ER Ca2+ (Xu and Reed 1998; Palmer et al. 2004). Both SERCA overexpression and pharmacological stimulation of endogenous SERCA overcame the effect of BI-1, elevating ER CA2+ (Fig. 5F).
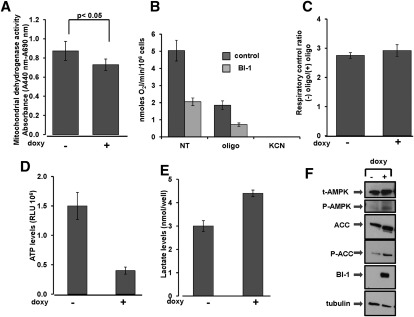
Mitochondria require Ca2+ to maintain the tricarboxylic acid (TCA) cycle, which provides reducing equivalents to support oxidative phosphorylation and ATP production. Given that BI-1-overexpressing cells showed reduced levels of mitochondrial Ca2+, we assessed whether mitochondrial metabolism was affected. To this end, we first measured levels of total mitochondrial dehydrogenases that are normally activated by Ca2+. Of note, cells overexpressing BI-1 showed a significant reduction in the activity of these enzymes, suggesting a reduction of the TCA cycle activity (Fig. 6A). Isocitrate dehydrogenase (IDH), a mitochondrial matrix protein that represents a component of the TCA cycle, is responsible for catalyzing the reversible conversion of isocitrate to α-ketoglutarate and CO2 in a two-step reaction. Because the activity of this enzyme is regulated by changes in mitochondrial matrix Ca2+ ion concentration, we measured its activity in BI-1-overexpressing cells. Activity of IDH was significantly lower in BI-1-overexpressing cells compared with control cells (Supplemental Fig. S12), a result that was in agreement with lower levels of mitochondrial Ca2+. Consistent with reduced activity of IDH (Qi et al. 2008), levels of NADH were also reduced in BI-1-overexpressing cells (data not shown), and levels of mitochondrial NADPH were increased (Supplemental Fig. S13).
Figure 6.
BI-1 reduces mitochondria bioenergetics. (A) Activity of mitochondrial dehydrogenases was measured in control ([−] doxy) and BI-1 overexpressing ([+] doxy) HeLa cells. Values were normalized for the number of viable cells (trypan blue exclusion). Background absorbance was read at 690 nm and subtracted from readings at 440 nm (A440nm − A690nm). The difference between samples was significant (P < 0.05, by unpaired t-test). (B) Endogenous oxygen consumption rates in control (no doxy) and BI-1-overexpressing (doxy) cells were monitored by an oxymeter before (NT) and after exposure to oligomycin (2 μM) and FCCP (1 μM). Results are expressed as nanomoles of O2 per minute per 1 × 106 cells. Results for untreated control versus BI-1-expressing cells were statistically significant (P = 0.0015). (C) RCR in control ([−] doxy) and BI-1 overexpressing ([+] doxy) cells was calculated by dividing O2 consumption before and after addition of oligomycin. The efficiency of oxidative phosphorylation coupling was comparable in control and BI-1-overexpressing cells. (D) ATP levels were measured in control versus BI-1-overexpressing cells using a bioluminescence method. Data represent relative luminescence units (RLUs) per 106 viable cells (mean ± SD; n = 3; P = 0.002. (E) Levels of extracellular lactate were measured in control ([−] doxy) and BI-1-overexpressing ([+] doxy) cells. Results are represented as nanomoles of lactate per well (mean ± SD; n = 3; P = 0.021. (F) Lysates from control versus BI-1-overexpressing cells were normalized for total protein content and subjected to immunoblot analysis to detect AMPK, phospho-AMPK, acetylCoA carboxylase (ACC), and phospho-acetylCoA carboxylase (P-ACC). Anti-HA antibody was used to detect expression of BI-1-HA protein. Equivalent sample loading was shown by monitoring tubulin levels.
As a direct measurement of mitochondrial respiration, O2 consumption rates were assessed by an oxymeter. BI-1-overexpressing cells showed a marked reduction in O2 consumption (Fig. 6B) compared with control cells in which BI-1 expression was not induced. The reduction in mitochondrial O2 consumption in BI-1-overexpressing cells was observed under both coupled and uncoupled (+FCCP) conditions and does not appear to be due to an intrinsic defect in the oxidative phosphorylation machinery based on analysis of respiratory control ratio (RCR) values (Fig. 6C).
To assure that reduced oxidative phosphorylation was not due to reduced mitochondrial mass, we assessed levels of the mitochondrial proteins AIF, Hsp60, and Cyclophilin D (Supplemental Fig. S14). Induction of BI-1 expression for up to 72 h did not change levels of these mitochondrial proteins. To further evaluate mitochondrial mass, we also stained cells with nonylacridine orange (NAO), which binds to cardiolipin in mitochondrial membranes, finding that NAO staining was unaffected by BI-1 expression (Supplemental Fig. S15).
Since mitochondria generate the vast majority of cellular ATP via oxidative phosphorylation, we next measured ATP levels in control versus BI-1-overexpressing HeLa cells. When normalized for numbers of viable cells via trypan blue exclusion, cells overexpressing BI-1 contained reduced ATP concentrations compared with controls (Fig. 6D). In contrast, human H322M cells in which BI-1 was knocked down contained elevated ATP levels compared with cells with scrambled shRNA (Supplemental Fig. S16A). Moreover, ATP levels in Drosophila S2 cells were elevated in BI-1 siRNA-treated cells compared with control siRNA-treated cells in both nutrient-rich (Supplemental Fig. S16B) and nutrient-depleted (Supplemental Fig. S16C) cultures.
When oxygen is reduced, cells use anaerobic glycolysis as an alternative means of energy (ATP) production, wherein glucose is converted in lactate. HeLa cells overexpressing BI-1 showed increased levels of lactate production compared with control cells (Fig. 6E), suggesting a greater reliance of glycolysis for generating ATP.
If BI-1 overexpression causes a net decline in cellular ATP levels, we predicted that cellular sensors of ATP would be impacted, such as AMPK. Once activated, this kinase phosphorylates substrates that consequently limit anabolic processes and facilitate catabolic pathways to provide energy. BI-1-overexpressing cells contained increased levels of phospho-AMPK (indicative of kinase activation) and also showed increased phosphorylation of the AMPK substrate acetylCoA carboxylase (Fig. 6F). These findings confirm that BI-1 causes a net decline in cellular ATP by impacting mitochondrial bioenergetics.
IP3R is essential for BI-1-mediated autophagy
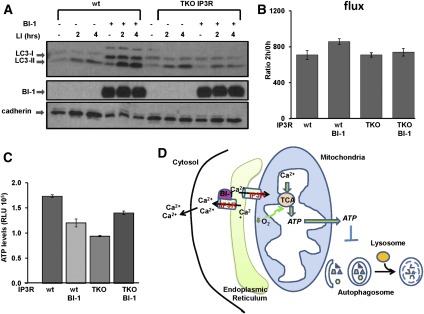
Because IP3Rs have been reported to enhance mitochondrial bioenergetics (Cardenas et al. 2010), we next explored whether BI-1's ability to modulate cellular ATP and promote autophagy is dependent on these Ca2+ channels. Because most vertebrate genomes contain three IP3R genes, we employed chicken DT40 lymphoma cells in which all three IP3R isoforms (triple-knockout [TKO] IP3R) were genetically ablated (Sugawara et al. 1997). Wild-type and TKO IP3R DT40 cells were stably transduced with BI-1 versus control lentiviruses, and expression of BI-1 was confirmed (Fig. 7A). Then, cells were treated with LIs to assess autophagic flux by measuring LC3 levels. Indeed, when expressed in TKO IP3R DT40 cells, BI-1 lost its ability to stimulate LC3 flux, whereas BI-1 retained its proautophagic activity when expressed in wild-type DT40 cells (Fig. 7A,B). To assess the impact of IP3R loss on mitochondrial bioenergetics, we measured levels of ATP in wild-type and TKO IP3R DT40 cells with or without overexpression of BI-1. When expressed in wild-type cells, BI-1 reduced ATP levels (as previously shown with other cell lines), whereas levels of ATP in IP3R DT40 cells overexpressing BI-1 were not changed or slightly increased (Fig. 7C). Altogether, these results indicate that BI-1 reduces cellular ATP and induces autophagy via a mechanism that requires IP3Rs (Fig. 7D).
Figure 7.
BI-1 requires IP3R to modulate ATP levels and induce autophagy. (A) Wild-type (wt) and TKO IP3R DT40 cells were stably infected with either control or BI-1 viruses. Where indicated, cells were treated with the LIs NH4Cl (20 mM) and leupeptin (10 μM) for either 2 h or 4 h. (B) Levels of LC3-II were quantified by densitometry to measure LC3 flux. (C) ATP levels were measured in wild-type (wt) and TKO IP3R DT40 cells stably transduced with either GFP or BI-1 lentiviruses. Data represent relative luminescence units (RLUs) per 106 viable cells (trypan blue exclusion) of three independent measurements. (D) By increasing ER Ca2+ leakage via the IP3R, BI-1 reduces the amount of Ca2+ at the microdomains where ER and mitochondria are in close proximity. Consequently, low mitochondrial Ca2+ levels reduce activity of the TCA cycle with reduction of O2 consumption and ATP production. Low ATP levels trigger a cascade of signal transduction events that activates autophagy.
SERCA reverses BI-1-mediated reduction in bioenergetics and BI-1-induced autophagy
To assess the role of altered ER Ca2+ in the cellular phenotypes mediated by BI-1, we restored ER Ca2+ in BI-1-overexpressing cells by overexpressing SERCA or stimulating SERCA with chemical agonists. Then, with ER Ca2+ levels sustained at high levels (Fig. 5), we tested the ability of BI-1 to promote autophagy and regulate cellular bioenergetics. SERCA overexpression reduced autophagy, which was evidenced by accumulation of p62 and reduced LC3-II levels (Supplemental Fig. S17A). Similarly, the SERCA chemical agonist SB-1163 also reversed BI-1-mediated autophagy, as evidenced by levels of the autophagy markers LC3 and p62 (Supplemental Fig. S17D). Furthermore, SERCA overexpression (Supplemental Fig. S17B,C) and pharmacological stimulation of SERCA (Supplemental Fig. S18E, F) partially restored ATP levels in BI-1-overexpressing cells while also either partially or fully restoring mitochondrial dehydrogenase activity (TCA cycle marker).
To ascertain the relevance of BI-1-mediated changes in ER Ca2+ to cytoprotection, we overexpressed SERCA in BI-1-inducible HeLa cells and challenged these cells with the ER stressors thapsigargin and tunicamycin. SERCA did not modify BI-1's ability to protect against ER stress-induced cell death (Supplemental Fig. S18A). SERCA also did not interfere with BI-1-mediated suppression of stress kinase activation induced by ER stress agents (Supplemental Fig. S18B). Taken together, these results establish a novel function for BI-1 whereby its ability to reduce ER Ca2+ suppresses mitochondrial ATP production to promote autophagy, independent of BI-1's regulation of ER stress responses and ER stress-induced cell death.
Discussion
BI-1 has previously been shown to have multiple functions that may contribute to its role in adaptation to cell stress and preservation of cell survival, including (1) interaction with and suppression of signaling by IRE1α (Lisbona et al. 2009), an initiator of UPR signaling linked to apoptosis, and (2) regulating ER Ca2+ homeostasis, possibly in association with anti-apoptotic Bcl-2 family proteins that are known to regulate IP3R function in ER membranes (Bcl-2 and Bcl-XL) (Xu et al. 2008). Additionally, we demonstrate here a role for BI-1 as a modulator of autophagy and attribute this function to its ability to regulate ER Ca2+. Altogether, our data are consistent with a model in which BI-1 reduces ER Ca2+ levels, resulting in diminished IP3R-mediated transport of Ca2+ from ER into mitochondria, causing a decline in mitochondrial bioenergetics that results in reduced cellular ATP and thus promoting autophagy.
The ability of BI-1 to modulate autophagy was demonstrated here using multiple cell lines involving both constitutive and conditional BI-1 overexpression and using various experimental methods for BI-1 gene silencing or gene knockout. Additionally, the findings were translated from mammalian cells (human and mouse) to avian cells (DT40) and insect cells (Drosophila S2 cells). The role of BI-1 in modulating autophagy was also demonstrated in vivo by analysis of tissues of BI-1−/− mice and analysis of BI-1-deficient tumor xenografts. In contrast to our results, Hetz and colleagues (Castillo et al. 2011) have recently reported that BI-1 inhibits autophagy through a mechanism involving IRE1. They observed that BI-1-mediated suppression of JNK activation downstream from IRE1 inhibits the release of the essential autophagy protein Beclin from Bcl-2, an event that requires JNK-mediated phosphorylation of Bcl-2 (Pattingre et al. 2005). Thus, it is possible that BI-1 can modulate autophagy via two independent mechanisms having opposing effects, including (1) suppressing IRE1-mediated JNK activation in the context of ER stress, thus reducing autophagy, and (2) reducing Ca2+/IP3R-dependent mitochondrial bioenergetics, resulting in lower cellular ATP and thus stimulating autophagy under basal conditions. The relative contributions of these two opposing mechanisms to net autophagic flux may vary with cell type and cell context. Regardless, our findings elucidate a novel mechanism of autophagy regulation by BI-1, showing the existence of a IP3R-dependent, IRE1-independent pathway by which BI-1 promotes autophagy. The alterations in oxygen consumption and ATP levels observed in human tumor cell lines here, however, are inconsistent with circumstances that would result in autophagy suppression. Our analysis of BI-1 actions in IRE1 knockout cells also do not support a prominent role for this UPR signaling protein, in contrast to observations derived from IP3R knockout cells.
Recent studies have shown the existence of specific domains in which ER and mitochondria juxtapose. These MAMs of the ER have very specific functions that permit Ca2+ transfer between these two organelles (Szabadkai et al. 2006a,b). Moreover, it was recently reported that IP3Rs in ER membranes are required for maintaining mitochondrial bioenergetics by mediating the transfer of Ca2+ from ER to mitochondria, thereby supporting the activity of various dehydrogenases required for the TCA cycle (Cardenas et al. 2010). We suspected a possible connection between BI-1 and IP3Rs for three reasons. First, BI-1 causes a passive leak of Ca2+ from the ER, resulting in reduced resting levels of free Ca2+ in the ER lumen (Chae et al. 2004), a phenotype that closely resembles the effects of anti-apoptotic Bcl-2 and Bcl-XL proteins in ER membranes (He et al. 1997). Second, BI-1 appears to be required for Bcl-XL-mediated regulation of ER Ca2+, as demonstrated by experiments using BI-1−/− cells (Xu et al. 2008). Third, Bcl-2 and Bcl-XL associate with and modulate the function of IP3Rs in ER membranes, and BI-1 appears to associate with Bcl-2 and Bcl-XL based on co-IP experiments (Xu and Reed 1998). The discovery reported here that BI-1 modulates cellular ATP levels, lactate production, and mitochondrial O2 consumption (without impacting mitochondrial mass) points to a role for BI-1 in controlling mitochondrial bioenergetics. Thus, BI-1, from its location in ER membranes, impacts mitochondrial bioenergetics via a mechanism that requires IP3Rs. We therefore propose that BI-1 reduces the efficiency with which IP3Rs at MAMs transfer Ca2+ from ER into mitochondria. Furthermore, our data are consistent with the concept that the downstream consequences of this reduced efficiency of IP3R-mediated transfer of Ca2+ from ER to mitochondria are reduced cellular ATP levels and increased basal levels of autophagic flux.
The connection between ER Ca2+ and BI-1's ability to promote autophagy was firmly established by experimentally manipulating SERCA, an ER membrane ATPase that actively pumps Ca2+ into the lumen of the ER. While BI-1 causes a passive Ca2+ leak from the ER, SERCA actively pumps Ca2+ into the ER. Using SERCA overexpression as well as a pharmacological agonist of SERCA, we were able to overcome the effects of BI-1 on ER Ca2+, showing that when ER Ca2+ is restored to normal, BI-1 fails to modulate autophagy or cellular ATP levels. Thus, for the first time, a specific cellular function has been attributed to BI-1's activity as a regulator of ER Ca2+, demonstrating a linkage to mitochondrial bioenergetics and autophagy. It will be interesting to explore whether Bcl-2 and Bcl-XL, through their association with BI-1 in ER membranes, have a similar impact on mitochondrial bioenergetics and autophagy that is distinct from other mechanisms previously identified (Kroemer et al. 2010). While BI-1's ability to modulate intracellular Ca2+ homeostasis conceivably could impact autophagy through effects on Ca2+-dependent kinases previously implicated in autophagy regulation, such as CaMKKII and DAPK1 (Hoyer-Hansen et al. 2007; Zalckvar et al. 2009), we were unable to demonstrated an impact of either of these kinases on BI-1's ability to modulate autophagy.
Controversy has abounded over the question of whether autophagy is a cell survival or cell death mechanism (Shimizu et al. 2004; Codogno and Meijer 2005; Levine and Kroemer 2008; Maiuri et al. 2009; Kroemer and White 2010). We found that BI-1-mediated cytoprotection does not depend on the autophagy machinery, based on experiments using Atg7−/− cells. In contrast, BI-1's ability to protect against cell death was determined to be IRE1α-dependent, based on experiments using Ire1α−/− cells. Furthermore, the role of IRE1α appears to be specific and not a manifestation of a general perturbation in UPR signaling, inasmuch as BI-1 retained its cytoprotective activity in Perk−/− cells. Previously, it has been suggested that BI-1 is an inhibitor of IRE1, a property that may protect against ER stress-induced cell death in mammalian cells by limiting downstream activation of stress kinases that promote apoptosis (Rubio et al. 2011). However, IRE1's ability to activate XBP1 via its ribonuclease activity has been implicated in cell adaptation and survival following ER stress (Tirasophon et al. 2000). Together with other data implying a need for delicate control of IRE activity (Schroder et al. 2003), our observations in yeast showing that the IRE1/XBP1 (HAC) axis is required for BI-1-mediated protection from Bax suggest that BI-1 may collaborate with IRE1 to promote cell survival. The phenotype of BI-1 thus combines suppression of cell death (apoptosis) with promotion of autophagy and stands apart from other dual regulators of apoptosis and autophagy, such as Bcl-2 and Bcl-XL, which suppress both apoptosis and autophagy. Altogether, the distinct and functional separable phenotypes of BI-1 provide further evidence that BI-1 is a multifunctional protein, with at least one function defined by an ability to bind IRE1α and suppress ER stress-induced cell death and another function defined by an ability to modulate ER Ca2+ to impact mitochondrial bioenergetics and autophagy. The net outcome of these two distinct and separable functions in terms of cell life and death may vary with context, but heretofore, studies with BI-1 have uniformly demonstrated an overall survival benefit.
Although tumor-associated elevations in BI-1 expression have been reported in many human cancers (Reimers et al. 2008), the tumor xenograft data provide the first functional evidence that BI-1 can be an important promoter of tumorigenesis in vivo. In this regard, BI-1's ability to promote autophagy and anaerobic glycolysis may be an advantage for tumor cells in hypoxic microenvironments, along with its ability to prevent ER stress-induced apoptosis that also tends to accompany harsh microenvironments. Also, given that BI-1 has been shown to provide protection against ischemia reperfusion injury in mice (Bailly-Maitre et al. 2006), it will be interesting to elucidate the relative roles of the IRE1α-dependent (ER stress) versus the IP3R-dependent (autophagy/bioenergetics) mechanisms of BI-1 in that context. Altogether, the new findings reported here suggest that BI-1 may serve an especially important role to aid cellular resilience and ensure cellular survival under circumstances in which oxygen and nutrient deprivation are encountered, such as the microenvironment of some tumors.
Materials and methods
Immunoblotting analysis
Cells were homogenized in RIPA buffer (50 mM Tris-HCl at pH 7.4, 1% NP-40, 0.25% Na-deoxycholate, 150 mM Tris-HCl, 1 mM EDTA, 1 mM PMSF, 1 μg/mL leupeptin, aprotinin, pepstatin, 1 mM NaF, Na4VO3). Lysates were centrifuged at 10,000g for 5 min at 4°C, and total protein content was quantified by BCA assay (Pierce). Proteins were analyzed by SDS-PAGE and immunoblotting after transfer to nitrocellulose membranes (Millipore). Antibodies used include anti-LC3 (Sigma), anti-p62 (BD Biosciences), anti-HA (Roche), anti-tubulin (Roche), anti-SERCA (BD Biosciences), anti-AMPK (Cell Signaling Technologies), anti-p-AMPK (Cell Signaling Technologies), anti-acetylCoA carboxylase (Cell Signaling), and anti p-acetylCoA carboxylase (Cell Signaling Technologies). An enhanced chemiluminescence (ECL) method (Pierce) was used for detection.
Measurement of respiratory activity
Cultured cells were gently detached from the dish by trypsinization, washed in PBS, harvested by centrifugation at 500g for 5 min, and immediately assessed for O2 consumption. The rate of oxygen consumption was measured polarographically with a Clark-type oxygen electrode (Hansatech Instruments Ltd.) in a thermostatically controlled chamber equipped with a magnetic stirring device and a gas-tight stopper fitted with a narrow port for additions via a Hamilton microsyringe. Aliquots of 5 × 106 viable cells per milliliter were assayed in 200 mM sucrose, 1 g/L bovine serum albumin, 10 mM KH2PO4, 2.7 mM KCl, 1 mM MgCl2, 20 mM HEPES, and 0.5 mM EGTA (pH 7.4) at 37°C; after attainment of a stationary endogenous substrate-sustained respiratory rate, 2 μM oligomycin was added. The rates of oxygen consumption were corrected for 5 mM KCN-insensitive respiration. The RCR was obtained by dividing the rates of oxygen consumption achieved before and after the addition of oligomycin.
ATP measurements
Cellular ATP concentrations were assessed using the CellTiter-Glo luminescent cell viability assay (Promega) as per the manufacturer's instructions. Luminescence was measured using the Luminoskan Ascent (Thermo Electron Corp.) at 1-sec integration time per sample). Readings were normalized to the number of viable cells (trypan blue exclusion).
Mitochondrial dehydrogenase activity (WST-1 assay)
Cells (in 96-well plates) were analyzed for mitochondrial dehydrogenase activity using the WST-1 assay according to the manufacturer's protocol. The WST-1 substrate 4-[3-(4-iodophenyl)-2-(4-nitrophenyl)-2H-5-tetrazolio]-1,3-benzene disulphonate was converted to a colored formazan compound by the mitochondrial enzymes. The color intensity is proportionally related to activity of mitochondrial dehydrogenases. Briefly, 10 μL of WST-1 reagent was added to 100 μL of medium. The cells were incubated with the WST-1 reagent for 1–2 h at room temperature. The formazan dye formed was measured at the wavelengths of 450 nm and 690 nm (background) using a FlexStation 3 microplate reader (Molecular Devices).
Ca2+ analysis
Ca2+ measurements were performed essentially as described previously (Palmer et al. 2004). Briefly, cells were plated in 96-well plates and loaded with 50 μM Indo-1 AM and F-127 Pluronic (0.02%) for 30 min at 37°C. Cells were then stimulated with 5 μM thapsigargin to empty ER Ca2+ stores. Total fluorescent intensity was measured every 5 sec for a total time of 300 sec. Results were expressed as 400 nm/475 nm (calcium-free/calcium-bound) emission ratio. For direct ER Ca2+ measurements, cells were plated in glass-bottomed dishes and transfected with 4 μg of ER cameleon (YC4.3ER) for 48 h prior to analysis. Cells were stimulated with 5 μM thapsigargin to empty ER Ca2+ stores and then imaged on an Inverted IX81 Olympus wide-field and fluorescence microscope (FRET) with a cooled charge-coupled device camera (Photmometrics, Inc.) controlled by MetaFluor 6.1 software (Universal Imaging). Emission ratio imaging of the cameleon was accomplished by using a 436DF20 excitation filter, a 450-nm dichroic mirror, and two emission filters (475/40 for enhanced CFP and 535/25 for citrine). Data represent the emission ratio YFP/CFP versus time, with each line representing an average of not less than six cells. Mitochondrial Ca2+ measurements were assessed by Rhod-2 AM (50 μM, Invitrogen). Briefly, cells were loaded with the calcium-sensitive dye Rhod-2 for 30 min at 37°C and 5% CO2, then washed with PBS and reincubated for 30 min before analysis. Measurements of the Rhod-2 fluorescence were performed using a microplate reader (FlexStation 3, Molecular Devices) at 544-nm excitation and 590-nm emission wavelengths.
Lactate measurements
Lactate levels were quantified using a L-lactate assay kit according to the manufacturer's instructions (BioVision). In this assay, lactate is oxidized by lactate dehydrogenase to generate a product that interacts with a probe to produce a color (λmax = 450 nm), which is proportional to the amount of L-lactate. Experiments used 2000 cells per well of 96-well plates, which were seeded without or with 1 μg/mL doxycycline for 24 h.
LC3 flux
Cells were cultured for 2 h and 4 h with the LIs NH4Cl (20 mM) and leupeptin (100 μM). Cells were then harvested, lysed, normalized for total protein content, and subjected to immunoblot analysis using anti-LC3 antibody (Sigma). LC3-II levels were quantified by densitometry and normalized for β-actin. LC3 flux was quantified by dividing levels of LC3-II after 2 h of LI treatment per level of LC3-II without LI.
Animals
All animal procedures were conducted in compliance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) of Sanford-Burnham and were in accordance with National Institutes of Health (NIH) guidelines. Mice with targeted disruption of the BI-1 gene (Chae et al. 2004) were used for experiments at 8–12 wk of age. When indicated, animals were intraperitoneally injected with rapamycin (1 mg/kg; Sigma), and tissues were harvested after 6 h, 12 h, 48 h, and 72 h post-injection. Before sacrificing, mice were anesthetized by intraperitoneal injection of avertin. Tissues were dissected and immediately frozen in liquid nitrogen.
In vivo tumor xenografts
All animal experiments were approved by the IACUC of Sanford-Burnham. Xenograft tumors were established by subcutaneous injection of 5 × 106 H322M cells into the flanks of 6- to 8-wk-old female nu/nu mice. Tumor growth was monitored weekly using a Vernier caliper to calculate tumor volumes according to the formula (length × width2)/2.
Statistical analysis
The data are expressed as mean ± standard deviation (SD) from a minimum of three determinations. Statistical significance of differences between various samples was determined by t-test. P < 0.05 was considered significant.
Acknowledgments
We thank Dr. Ana Maria Cuervo for help with experimental design, data interpretation, and manuscript review; Dr. Fumihiko Urano for providing Ire1α and PERK knockout MEFs; Dr. Noboru Mizushima for providing the GFP-LC3 plasmid; and Dr. Kevin Foskett for kindly providing TKO IP3R DT40 cells. We thank Santosh Hariharan and Susanne Heynen-Genel for high-content cell imaging, Edward Monosov for transmission electron microscopy analyzes, Xian-Shu Huang for immunohistochemistry analysis, Melanie Hanaii and Tessa Siegfried for manuscript preparation, and Yoav Altman and Amy Cortez for technical assistance with FACS analysis. This work was supported by grants from the NIH (AG-15393). R.S. is supported by the Tobacco-Related Disease Research Foundation (18FT-0179).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.184325.111.
References
- Bailly-Maitre B, Fondevila C, Kaldas F, Droin N, Luciano F, Ricci JE, Croxton R, Krajewska M, Zapata JM, Kupiec-Weglinski JW, et al. 2006. Cytoprotective gene bi-1 is required for intrinsic protection from endoplasmic reticulum stress and ischemia-reperfusion injury. Proc Natl Acad Sci 103: 2809–2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly-Maitre B, Belgardt BF, Jordan SD, Coornaert B, von Freyend MJ, Kleinridders A, Mauer J, Cuddy M, Kress CL, Willmes D, et al. 2010. Hepatic Bax inhibitor-1 inhibits IRE1α and protects from obesity-associated insulin resistance and glucose intolerance. J Biol Chem 285: 6198–6207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boya P, Gonzalez-Polo RA, Casares N, Perfettini JL, Dessen P, Larochette N, Metivier D, Meley D, Souquere S, Yoshimori T, et al. 2005. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol 25: 1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas C, Miller RA, Smith I, Bui T, Molgo J, Muller M, Vais H, Cheung KH, Yang J, Parker I, et al. 2010. Essential regulation of cell bioenergetics by constitutive InsP3 receptor Ca2+ transfer to mitochondria. Cell 142: 270–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo K, Rojas-Rivera D, Lisbona F, Caballero B, Nassif M, Court FA, Schuck S, Ibar C, Walter P, Sierralta J, et al. 2011. BAX inhibitor-1 regulates autophagy by controlling the IRE1α branch of the unfolded protein response. EMBO J 30: 4465–4478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae HJ, Kim HR, Xu C, Bailly-Maitre B, Krajewska M, Krajewski S, Banares S, Cui J, Digicaylioglu M, Ke N, et al. 2004. BI-1 regulates an apoptosis pathway linked to endoplasmic reticulum stress. Mol Cell 15: 355–366 [DOI] [PubMed] [Google Scholar]
- Codogno P, Meijer AJ 2005. Autophagy and signaling: Their role in cell survival and cell death. Cell Death Differ 12: 1509–1518 [DOI] [PubMed] [Google Scholar]
- Criollo A, Maiuri MC, Tasdemir E, Vitale I, Fiebig AA, Andrews D, Molgo J, Diaz J, Lavandero S, Harper F, et al. 2007. Regulation of autophagy by the inositol trisphosphate receptor. Cell Death Differ 14: 1029–1039 [DOI] [PubMed] [Google Scholar]
- Csordas G, Renken C, Varnai P, Walter L, Weaver D, Buttle KF, Balla T, Mannella CA, Hajnoczky G 2006. Structural and functional features and significance of the physical linkage between ER and mitochondria. J Cell Biol 174: 915–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decuypere JP, Monaco G, Bultynck G, Missiaen L, De Smedt H, Parys JB 2011. The IP3 receptor-mitochondria connection in apoptosis and autophagy. Biochim Biophys Acta 1813: 1003–1013 [DOI] [PubMed] [Google Scholar]
- Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, et al. 2006. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 10: 51–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Lam M, McCormick TS, Distelhorst CW 1997. Maintenance of calcium homeostasis in the endoplasmic reticulum by Bcl-2. J Cell Biol 138: 1219–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer-Hansen M, Bastholm L, Szyniarowski P, Campanella M, Szabadkai G, Farkas T, Bianchi K, Fehrenbacher N, Elling F, Rizzuto R, et al. 2007. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-β, and Bcl-2. Mol Cell 25: 193–205 [DOI] [PubMed] [Google Scholar]
- Hunsberger JG, Machado-Vieira R, Austin DR, Zarate C, Chuang DM, Chen G, Reed JC, Manji HK 2011. Bax inhibitor 1, a modulator of calcium homeostasis, confers affective resilience. Brain Res 1403: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y, Kominami E, Tanaka K, Komatsu M 2008. Selective turnover of p62/A170/SQSTM1 by autophagy. Autophagy 4: 1063–1066 [DOI] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19: 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R, Zeh HJ, Lotze MT, Tang D 2011. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ 18: 571–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmelman AC 2011. The dynamic nature of autophagy in cancer. Genes Dev 25: 1999–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ 2007. Autophagy: From phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol 8: 931–937 [DOI] [PubMed] [Google Scholar]
- Klionsky DJ 2010. The autophagy connection. Dev Cell 19: 11–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Ichimura Y 2010. Physiological significance of selective degradation of p62 by autophagy. FEBS Lett 584: 1374–1378 [DOI] [PubMed] [Google Scholar]
- Krajewska M, Xu L, Xu W, Krajewski S, Kress CL, Cui J, Yang L, Irie F, Yamaguchi Y, Lipton SA, et al. 2011. Endoplasmic reticulum protein BI-1 modulates unfolded protein response signaling and protects against stroke and traumatic brain injury. Brain Res 1370: 227–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, White E 2010. Autophagy for the avoidance of degenerative, inflammatory, infectious, and neoplastic disease. Curr Opin Cell Biol 22: 121–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Marino G, Levine B 2010. Autophagy and the integrated stress response. Mol Cell 40: 280–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo TH, Kim HR, Zhu L, Yu Y, Lin HM, Tsang W 1998. Modulation of endoplasmic reticulum calcium pump by Bcl-2. Oncogene 17: 1903–1910 [DOI] [PubMed] [Google Scholar]
- Levine B, Kroemer G 2008. Autophagy in the pathogenesis of disease. Cell 132: 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisbona F, Rojas-Rivera D, Thielen P, Zamorano S, Todd D, Martinon F, Glavic A, Kress C, Lin JH, Walter P, et al. 2009. BAX inhibitor-1 is a negative regulator of the ER stress sensor IRE1α. Mol Cell 33: 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan NR, Zanetti M 2011. Tumor stress inside out: Cell-extrinsic effects of the unfolded protein response in tumor cells modulate the immunological landscape of the tumor microenvironment. J Immunol 187: 4403–4409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri MC, Tasdemir E, Criollo A, Morselli E, Vicencio JM, Carnuccio R, Kroemer G 2009. Control of autophagy by oncogenes and tumor suppressor genes. Cell Death Differ 16: 87–93 [DOI] [PubMed] [Google Scholar]
- Malhotra JD, Kaufman RJ 2007. The endoplasmic reticulum and the unfolded protein response. Semin Cell Dev Biol 18: 716–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, de Vries R, Arias E, Harris S, Sulzer D, et al. 2011. Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat Neurosci 13: 567–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N 2010. Noboru Mizushima: All about autophagy. Interview by Caitlin Sedwick. J Cell Biol 190: 946–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ 2008. Autophagy fights disease through cellular self-digestion. Nature 451: 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Yoshimori T 2010. Between canonical and antibacterial autophagy: Rab7 is required for GAS-containing autophagosome-like vacuole formation. Autophagy 6: 419–420 [DOI] [PubMed] [Google Scholar]
- Oakes SA, Scorrano L, Opferman JT, Bassik MC, Nishino M, Pozzan T, Korsmeyer SJ 2005. Proapoptotic BAX and BAK regulate the type 1 inositol trisphosphate receptor and calcium leak from the endoplasmic reticulum. Proc Natl Acad Sci 102: 105–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, et al. 2006. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol 26: 9220–9231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer AE, Jin C, Reed JC, Tsien RY 2004. Bcl-2-mediated alterations in endoplasmic reticulum Ca2+ analyzed with an improved genetically encoded fluorescent sensor. Proc Natl Acad Sci 101: 17404–17409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B 2005. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122: 927–939 [DOI] [PubMed] [Google Scholar]
- Qi F, Chen X, Beard DA 2008. Detailed kinetics and regulation of mammalian NAD-linked isocitrate dehydrogenase. Biochim Biophys Acta 1784: 1641–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz JD, White E 2010. Autophagy and metabolism. Science 330: 1344–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimers K, Choi CY, Bucan V, Vogt PM 2008. The Bax inhibitor-1 (BI-1) family in apoptosis and tumorigenesis. Curr Mol Med 8: 148–156 [DOI] [PubMed] [Google Scholar]
- Rizzuto R, Pinton P, Carrington W, Fay FS, Fogarty KE, Lifshitz LM, Tuft RA, Pozzan T 1998. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science 280: 1763–1766 [DOI] [PubMed] [Google Scholar]
- Rodriguez D, Rojas-Rivera D, Hetz C 2011. Integrating stress signals at the endoplasmic reticulum: The BCL-2 protein family rheostat. Biochim Biophys Acta 1813: 564–574 [DOI] [PubMed] [Google Scholar]
- Rong YP, Barr P, Yee VC, Distelhorst CW 2009. Targeting Bcl-2 based on the interaction of its BH4 domain with the inositol 1,4,5-trisphosphate receptor. Biochim Biophys Acta 1793: 971–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein AD, Eisenstein M, Ber Y, Bialik S, Kimchi A 2011. The autophagy protein Atg12 associates with antiapoptotic Bcl-2 family members to promote mitochondrial apoptosis. Mol Cell 44: 698–709 [DOI] [PubMed] [Google Scholar]
- Rubio C, Pincus D, Korennykh A, Schuck S, El-Samad H, Walter P 2011. Homeostatic adaptation to endoplasmic reticulum stress depends on Ire1 kinase activity. J Cell Biol 193: 171–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder M, Clark R, Kaufman RJ 2003. IRE1- and HAC1-independent transcriptional regulation in the unfolded protein response of yeast. Mol Microbiol 49: 591–606 [DOI] [PubMed] [Google Scholar]
- Shimizu S, Kanaseki T, Mizushima N, Mizuta T, Arakawa-Kobayashi S, Thompson CB, Tsujimoto Y 2004. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol 6: 1221–1228 [DOI] [PubMed] [Google Scholar]
- Sugawara H, Kurosaki M, Takata M, Kurosaki T 1997. Genetic evidence for involvement of type 1, type 2 and type 3 inositol 1,4,5-trisphosphate receptors in signal transduction through the B-cell antigen receptor. EMBO J 16: 3078–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabadkai G, Bianchi K, Varnai P, De Stefani D, Wieckowski MR, Cavagna D, Nagy AI, Balla T, Rizzuto R 2006a. Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J Cell Biol 175: 901–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabadkai G, Simoni AM, Bianchi K, De Stefani D, Leo S, Wieckowski MR, Rizzuto R 2006b. Mitochondrial dynamics and Ca2+ signaling. Biochim Biophys Acta 1763: 442–449 [DOI] [PubMed] [Google Scholar]
- Tanaka R, Ishiyama T, Uchihara T, Inadome Y, Iijima T, Morishita Y, Kano J, Goya T, Noguchi M 2006. Expression of the Bax inhibitor-1 gene in pulmonary adenocarcinoma. Cancer 106: 648–653 [DOI] [PubMed] [Google Scholar]
- Tirasophon W, Lee K, Callaghan B, Welihinda A, Kaufman RJ 2000. The endoribonuclease activity of mammalian IRE1 autoregulates its mRNA and is required for the unfolded protein response. Genes Dev 14: 2725–2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai YC, Weissman AM 2010. The unfolded protein response, degradation from endoplasmic reticulum and cancer. Genes Cancer 1: 764–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchihara K, Fujii S, Esumi H 2009. Autophagy and cancer: Dynamism of the metabolism of tumor cells and tissues. Cancer Lett 278: 130–138 [DOI] [PubMed] [Google Scholar]
- Verma G, Datta M 2012. The critical role of JNK in the ER–mitochondrial crosstalk during apoptotic cell death. J Cell Physiol 227: 1791–1795 [DOI] [PubMed] [Google Scholar]
- Wang Z, Cuddy M, Samuel T, Welsh K, Schimmer A, Hanaii F, Houghten R, Pinilla C, Reed JC 2004. Cellular, biochemical, and genetic analysis of mechanism of small molecule IAP inhibitors. J Biol Chem 279: 48168–48176 [DOI] [PubMed] [Google Scholar]
- Weston RT, Puthalakath H 2010. Endoplasmic reticulum stress and BCL-2 family members. Adv Exp Med Biol 687: 65–77 [DOI] [PubMed] [Google Scholar]
- Xu Q, Reed JC 1998. Bax inhibitor-1, a mammalian apoptosis suppressor identified by functional screening in yeast. Mol Cell 1: 337–346 [DOI] [PubMed] [Google Scholar]
- Xu C, Xu W, Palmer AE, Reed JC 2008. BI-1 regulates endoplasmic reticulum Ca2+ homeostasis downstream of Bcl-2 family proteins. J Biol Chem 283: 11477–11484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalckvar E, Berissi H, Mizrachy L, Idelchuk Y, Koren I, Eisenstein M, Sabanay H, Pinkas-Kramarski R, Kimchi A 2009. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep 10: 285–292 [DOI] [PMC free article] [PubMed] [Google Scholar]