S. cerevisiae mating proteins are often primed before mitotic completion, allowing for the adverse scenario of attempting to both fuse and divide simultaneously. Kim and Rose delineate an elegant mechanism that avoids this. The key mating determinant, Fus2p, integrates two signaling pathways: a Cdk-dependent pathway that promotes Fus2p nuclear localization and the pheromone signaling pathway that drives its export. Careful balance between these two pathways keeps Fus2p sequestered in the nucleus until mitotic completion, preventing its inappropriate activation of mating fusion. This study provides remarkable insight into how cells coordinate differentiation and mitosis.
Keywords: cell fusion, Cla4p, Fus2p, Fus3p, mating differentiation, nucleo–cytoplasmic trafficking
Abstract
Yeast cells induce the genes required for mating prior to the completion of mitosis. To ensure proper cell cycle progression prior to mating differentiation, a key cytoplasmic regulator of cell fusion, Fus2p, is sequestered in the nucleus by cyclin-dependent kinase (Cdk). In response to pheromone signaling, the mitogen-activated protein kinase Fus3p phosphorylates Ser 84 in Fus2p to drive nuclear export. We found that Fus3p becomes active and phosphorylates S84 as early as S phase, raising the question of how Cdk prevents inappropriate activation of Fus2p. Countering Fus3p, Cdk and a p21-activated kinase, Cla4p, maintain Fus2p's nuclear localization by phosphorylating Ser 67, which drives nuclear import and inhibits nuclear export. When Cdk and Cla4p activities drop after cell division, Fus3p promotes Fus2p export both via S84 phosphorylation and by down-regulating S67 phosphorylation. Thus, potential premature activation of Fus2p in mitosis is prevented by cell cycle-dependent phosphorylation that overrides the mating pheromone-induced phosphorylation that drives nuclear export.
Cellular proliferation and differentiation are, in general, mutually exclusive processes, which are carefully regulated for proper development in higher eukaryotes. When cells start to differentiate, they lose proliferative capacity by transcriptional repression of cell cycle genes and coordinate expression of specialized sets of genes, leading to dramatic changes in cellular structures and functions. In many systems, key cell cycle regulators participate in controlling differentiation. Conversely, cell differentiation decisions often directly regulate cell cycle progression (Zhu and Skoultchi 2001; Galderisi et al. 2003). Thus, differentiation must be temporally coordinated with cell cycle exit to prevent both premature differentiation and re-entry into the cell cycle.
In the budding yeast Saccharomyces cerevisiae, pheromone from the opposite mating type stimulates vegetatively growing haploid cells to arrest in G1, prior to entry into the next cell cycle. Arrested cells differentiate into specialized cells capable of cell and nuclear fusion to produce diploid cells (White and Rose 2001). Mating requires transcriptional activation of mating-responsive genes and reorganization of the cytoskeleton, allowing polarized cell growth to form a mating projection, often called a “shmoo” (Kurjan 1993; Elion 2000).
At the molecular level, mating differentiation is initiated by pheromone binding to its specific receptor (Ste2p or Ste3p) on the cell surface. The receptor is coupled to a heterotrimeric G protein (Gα/Gpa1p, Gβ/Ste4p, and Gγ/Ste18p) and a downstream mitogen-activated protein (MAP) kinase cascade. Upon activation, the Gβγ complex is released from the Gα subunit and recruits the scaffold protein Ste5p to the plasma membrane. The MAP kinase kinase kinase Ste11p is activated by a p21-activated kinase (PAK), Ste20p, an effector protein of the GTPase Cdc42p. Ste11p then phosphorylates and activates a MAP kinase kinase, Ste7p, which in turn phosphorylates and activates two partially redundant MAP kinases, Fus3p and Kss1p. Either kinase is sufficient to activate the transcription factor Ste12p responsible for mating-specific gene expression, although genetic studies indicate that Fus3p has specificity for the mating pathway and cell fusion; Kss1p is primarily involved in the filamentation response to starvation (Madhani et al. 1997).
Differentiation into mating cells requires cell cycle arrest in G1 to maintain the integrity of the genome following nuclear fusion. Fus3p phosphorylates the cyclin-dependent kinase (Cdk) inhibitor Far1p (Chang and Herskowitz 1992), which physically interacts with and inhibits the G1 cyclin/Cdk required for the initiation of the cell cycle (START), ultimately leading to arrest in G1 (Peter and Herskowitz 1994). Once past START, cells must complete an additional cell division cycle before being able to mate. The G1 cyclins inhibit pheromone signaling and downstream cellular response by promoting the phosphorylation and degradation of both Far1p (McKinney et al. 1993; Henchoz et al. 1997) and Ste5p, the MAP kinase cascade scaffold protein (Garrenton et al. 2009) whose membrane association is also disrupted to prevent signaling (Strickfaden et al. 2007). However, as cells progress through mitosis, mating signaling becomes active, leading to transcriptional induction of pheromone-responsive genes (Strickfaden et al. 2007; Ydenberg and Rose 2009). Thus, mitotic cells must prevent the premature activation of mating differentiation because inappropriate activation of the mating pathway disrupts cell cycle progression and is detrimental to cell survival (Strickfaden et al. 2007; Ydenberg and Rose 2009).
A key cytoplasmic mating regulator, Fus2p, is highly induced by pheromone and acts together with Rvs161p, a homolog of amphiphysin, to promote cell wall removal at the zone of cell fusion (Brizzio et al. 1998; Gammie et al. 1998). Mating signaling leads to interaction with Rvs161p and localization of the complex to a cortical site at the shmoo tip, dependent on actin; a type V myosin, Myo2p; and an integral membrane protein, Fus1p (Brizzio et al. 1998; Paterson et al. 2008; Sheltzer and Rose 2009). Because amphiphysins typically bind membranes and play important roles in membrane remodeling, such as endocytosis and vesicular trafficking (Dawson et al. 2006; Friesen et al. 2006), it is likely that Fus2p/Rvs161p is associated with vesicles clustered at the junction between the two mating cells (Gammie et al. 1998; Paterson et al. 2008). Fus2p contains a putative Rho-guanine nucleotide exchange factor (GEF) or Dbl homology domain, suggesting that cortical Fus2p interacts with a cortical Rho protein to regulate the removal of the intervening cell walls (Paterson et al. 2008).
When initially induced, Fus2p accumulates in the nucleus of mitotic cells and only exits the nucleus after cells complete mitosis (Ydenberg and Rose 2009). Fus2p retained in the nucleus is unable to activate cell fusion, whereas premature nuclear exit is toxic (Ydenberg and Rose 2009; E Andrianantoandro and MD Rose, in prep.). Considering its biological function, it seems likely that premature association with the cell cortex might disrupt cell wall integrity. Previous work showed that Fus2p export from the nucleus is positively regulated by the mating signaling MAP kinase Fus3p via phosphorylation of Fus2p S84 and negatively regulated by the yeast Cdk, Cdc28p (Ydenberg and Rose 2009). However, the previous study did not clarify how Cdk activity inhibits nuclear exit or establish how cells transition from the mitotic to the mating state. In this study, we describe the molecular mechanism by which mitotic cells override pheromone signaling to keep Fus2p in the nucleus for proper cell cycle progression prior to mating differentiation.
Results
Fus2p contains a bipartite nuclear localization signal (NLS) adjacent to a Crm1p-dependent nuclear export signal (NES)
The N-terminal 104 amino acids are both necessary and sufficient for regulated nucleo–cytoplasmic transport of Fus2p, whereas localization to the shmoo tip requires the C-terminal 105–677 residues (Fig. 1A; Supplemental Fig. S1A,B; Ydenberg and Rose 2009). To identify the sequences regulating transport, Fus2p1–104 deletions were generated in which the truncated proteins were fused to C-terminal triple GFP (GFP3, ∼80 kDa) because small proteins (<40 kDa) are able to passively enter the nucleus. (Davis 1995). GFP3 had little effect on Fus2p transport (data not shown). The deletions were ectopically expressed in mitotic cells under control of the GAL1 promoter to observe nuclear import, after which their expression was repressed by glucose and subsequently treated with α-factor for export.
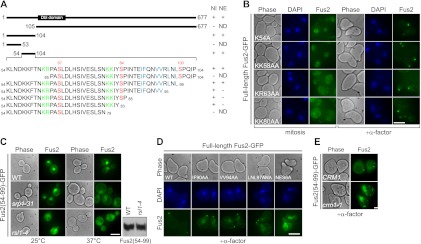
Figure 1.
Identification and characterization of Fus2p NLS and NES. (A) Schematic representation and summary of the functional sequences for Fus2p nuclear import (NI) and nuclear export (NE). (ND) Not determined. Essential sequences for nuclear import and export are highlighted in green and blue, respectively. Phosphorylation residues Ser 67, Ser 84, and Ser 100 are highlighted in red. A putative Dbl Rho-GEF domain is present at residues 116–321. (B) Fus2p point mutants (pMR6186, pMR6187, pMR6188, and pMR6189) were expressed in fus2Δ (MY10174) under the GAL1 promoter for 3 h (left), after which glucose and α-factor were added for 2 h (right). (C) Fus2p54–99 (pMR6159) was expressed under the GAL1 promoter in wild type (BY4741), srp1-31 (MY6457), and rsl1-4 (MY12131) for 30 min at 25°C; cells were then maintained at 25°C or shifted for 1 h to 37°C. Fus2p expression in rsl1-4 cells was shown by anti-GFP immunoblot. (D) Fus2p point mutants (pMR5469, pMR6192, pMR6191, pMR6190, and pMR6193) were expressed in fus2Δ (MY10174) under the GAL1 promoter for 3 h, after which glucose and α-factor were added for 2 h. (E) CRM1 (MY10239) and crm1-1 (MY10240) cells were arrested with α-factor for 90 min at 25°C, then cells were shifted to 37°C, and Fus2p54–99 (pMR6159) was expressed from the GAL1 promoter for 2 h. (B–E) Bar, 5 μm. (B,D) Cells were fixed and stained with DAPI. (C,E) GFP fluorescence was observed in living cells.
Whereas Fus2p1–53 was diffusely cytoplasmic, Fus2p54–104 accumulated in the nucleus and was exported to the cytoplasm by pheromone, like Fus2p1–104 (Fig. 1A; Supplemental Fig. S1A,B). When Fus2p54–104 was further dissected, Fus2p65–104 and Fus2p54–79, removing 54KLNDKKFTNKR64 or 80KKIY83, were found to be broadly dispersed in the cytoplasm (Fig. 1A; Supplemental Fig. S1A,B). Both sequences contain several basic residues, which are candidate NLSs. The basic residues were mutated to alanine in the context of full-length Fus2p, which carries a single internal GFP (Paterson et al. 2008). Mutations 54K and 58KK59 had little effect on nuclear localization, whereas mutations 63KR64 and 80KK81 caused diffuse cytoplasmic distribution (Fig. 1B), suggesting that they act as a bipartite NLS. All of the mutants localized normally to the shmoo tip by pheromone (Fig. 1B).
To identify the karyopherins for Fus2p import, we examined localization in a collection of mutants, including temperature-sensitive srp1-31 and rsl1-4, affecting the yeast importin α and importin β, respectively. Nuclear accumulation of Fus2p54–99 was severely impaired at 37°C in both mutants and at 25°C in rsl1-4, relative to wild type (the protein was expressed normally in rsl1-4) (Fig. 1C). However, none of the other importin mutant strains showed defects in nuclear localization (Supplemental Fig. S1C), indicating that Fus2p import is mediated primarily by the classical importin α/β pathway.
We next investigated Fus2p export. The C-terminal region of Fus2p54–104 is enriched in hydrophobic residues (90IFQNVVRLNL99), and its deletion blocked export by pheromone (Fig. 1A; Supplemental Fig. S1A,B). This motif is an excellent match to the consensus NES recognized by the exportin Crm1p [Φ1-(X)2–3–Φ2-(X)2–3–Φ3-X–Φ4, where Φ is hydrophobic and X is any amino acid] (Kutay and Guttinger 2005). Mutations of pairs of hydrophobic residues in the context of full-length Fus2p partially blocked and mutation of all six (Fus2pNES6A) completely blocked Fus2p export (Fig. 1D). Moreover, Fus2p54–99 remained in the nucleus of pheromone-treated crm1-1 cells at the restrictive temperature (Fig. 1E). We conclude that the regulated nuclear export of Fus2p is mediated by a Crm1p-dependent NES.
Phosphorylation within and adjacent to the NLS and NES regulates the subcellular localization of Fus2p
A previous proteomic analysis identified three phosphorylation sites: 67S, 84S, and 100S (Li et al. 2007). Because they are within or adjacent to the NLS or NES (Fig. 1A), we asked whether phosphorylation regulates Fus2p localization. Fus2p54–104 was used because this shows regulated transport and contains all three relevant phosphorylation sites. Electrophoretic mobility shifts were analyzed using Phos-tag acrylamide, which specifically reduces the mobility of phosphorylated proteins (Kinoshita-Kikuta et al. 2007). Fus2p54–104 gave rise to multiple slower-migrating species (I, II, III, and IV) that were absent in AAA, in which all three putative phosphorylation sites were mutated to alanine (Fig. 2A). The fastest-migrating species (I) was present in mitotic cells (Fig. 2A, lane 1) and greatly reduced in pheromone-treated cells (Fig. 2A, lane 3). The upper bands (II, III, and IV) were prominent in pheromone-treated cells (Fig. 2A, lane 3), but essentially absent in mitotic cells (Fig. 2A, lane 1). In the pheromone-treated fus3Δ mutant, where Fus2p is nuclear (Fig. 2A; Ydenberg and Rose 2009), there was a strong reduction in the levels of bands II, III, and IV, as well as the reappearance of mitotic band I (Fig. 2A, lane 4). The fus3Δ mutant contained cln3Δ to suppress the cell cycle arrest defect of fus3Δ (Elion et al. 1991). It is likely that the partially redundant Kss1p is responsible for residual phosphorylation in fus3Δ (Supplemental Fig. S2). These results indicate a close relationship between phosphorylation and localization: Species I is correlated with nuclear localization, and species II, III, and IV are correlated with cytoplasmic localization (Fig. 2A).
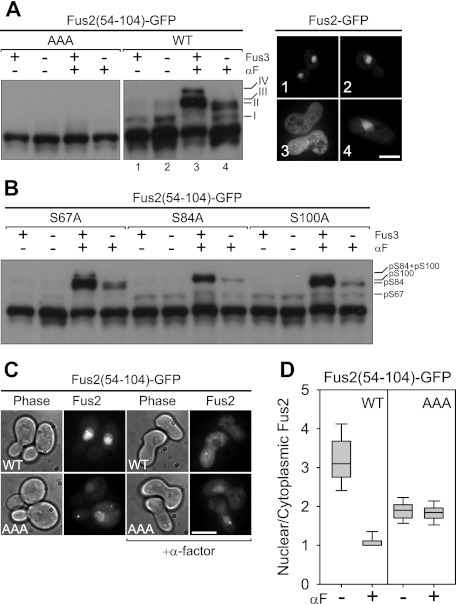
Figure 2.
The regulation of Fus2p nuclear localization by phosphorylations close to the NLS and NES. (A,B) Fus2p54–104 mutants (pMR6170, pMR6009, pMR6166, pMR6165, and pMR6169) were expressed in FUS3 (MY10174) or fus3Δ cln3Δ (MY10273) cells under the GAL1 promoter for 90 min during mitosis or after α-factor arrest for 2 h. GFP fluorescence was observed in living cells (A) and samples were run on 50 μM Phos-tag gels (A,B). AAA refers to residues 67, 84, and 100. (A) Each number of GFP fluorescence corresponds to that of lane in the immunoblot. Bar, 5 μm. (C,D) Fus2p54–104 (pMR6009) and AAA (pMR6170) mutants were expressed in fus2Δ (MY10174) under the GAL1 promoter for 90 min during mitosis or after α-factor arrest for 2 h. GFP fluorescence was observed in living cells (C), and its intensity of the nucleus relative to the cytoplasm was quantified (D) as described in the Materials and Methods. (C) Bar, 5 μm. (D) The lines inside the box and whisker plots indicate the median, the rectangles show the central 50%, and the error bars indicate the entire range.
To identify the bands, we individually mutated Ser 67, Ser 84, and Ser 100 to alanine. As shown in Figure 2B, S67A eliminated species I but not II, III, or IV. Species II and IV were lost only in S84A, and III and IV were lost only in the S100A (Fig. 2B). This clearly indicates that slower-migrating bands I, II, and III correspond to S67, S84, and S100 phosphorylated species, respectively. The absence of band IV in both the S84A and S100A mutants indicates that band IV corresponds to phosphorylation at both sites.
We tested whether phosphorylation affects the regulated transport of Fus2p. The ratio of nuclear to cytoplasmic fluorescence (N/C ratio) for Fus2p54–104 was 3.2 ± 0.7 (n = 63), which was reduced to 1.1 ± 0.7 (n = 33) in cells arrested with pheromone (Fig. 2C,D). In contrast, AAA showed constitutive weak nuclear localization in mitotic and pheromone-arrested cells (N/C = 1.9 ± 0.3, n = 40 and 1.8 ± 0.2, n = 28, respectively) (Fig. 2C,D). These results indicate that Fus2p localization is regulated primarily by phosphorylation and not by regulation of the transport machinery.
S67 phosphorylation promotes nuclear import of Fus2p
Because S67 is within the bipartite NLS and its phosphorylation is correlated with Fus2p nuclear localization, we asked whether it regulates Fus2p import. Fus2p54–83 carries only S67 and lacks the NES, allowing us to specifically examine the function of the NLS. Fus2p54–83 migrated as a doublet on Phos-tag gel (Fig. 3A) and accumulated in the nucleus of mitotic cells (N/C = 3.36 ± 0.74, n = 51) (Fig. 3B; Supplemental Fig. S3A). The S67A mutant protein ran as a single band and was largely cytoplasmic (N/C = 1.43 ± 0.18; n = 45) (Fig. 3A,B), demonstrating that S67 phosphorylation promotes nuclear localization. Furthermore, replacement of S67 with a phosphomimetic aspartate (S67E) resulted in a protein that was not phosphorylated, but that localized to the nucleus like wild type (N/C = 3.48 ± 0.83; n = 69) (Fig. 3A,B; Supplemental Fig. S3A).
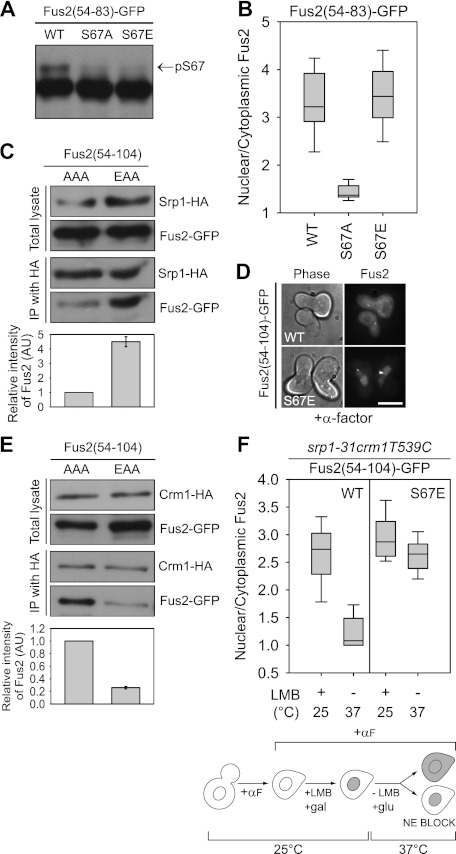
Figure 3.
The role of S67 phosphorylation in Fus2p transport. (A,B) Fus2p54–83 mutants (pMR6152, pMR6153, and pMR6154) were expressed in fus2Δ (MY10174) under the GAL1 promoter for 90 min. (A) Samples were run on 50 μM Phos-tag gels. The arrow indicates the S67 phosphorylated species. (B) GFP fluorescence was observed in living cells and quantified as described in Figure 2D. (C,E) Fus2p54–104 mutants (pMR6170 and pMR6173) were expressed in SRP1-3HA (MY12172) or CRM1-3HA (MY12170) cells under the GAL1 promoter for 90 min, and Srp1p or Crm1p was precipitated with anti-HA as described in the Materials and Methods. The intensity of copurified Fus2p was normalized by purified Srp1p or Crm1p and expressed relative to AAA. Error bars represent the means ± SEM of two independent experiments. AAA and EAA refer to residues 67, 84, and 100. (D) The indicated plasmids (pMR6009 and pMR6168) were expressed in fus2Δ (MY10174) under the GAL1 promoter for 90 min, after which glucose and α-factor were added for 2 h. GFP fluorescence was observed in living cells. Bar, 5 μm. (F) srp1-31 crm1T539C cells (MY12398) harboring the indicated plasmids (pMR6009 and pMR6168) were arrested with α-factor for 90 min, then galactose and 100 ng/mL LMB were added for 90 min at 25°C. After washing out the inhibitor, cells were incubated in glucose-containing and LMB-free medium for 90 min at 37°C. GFP fluorescence was observed in living cells and quantified as described in Figure 2D. (NE) Nuclear exit.
We examined whether S67 phosphorylation modulates interaction with the importin α Srp1p by coimmunoprecipitation. The S67 phosphorylation-defective Fus2p54–104-AAA and S67 phosphomimetic Fus2p54–104-EAA were used to exclude any effects of other phosphorylations. The AAA mutant showed weak binding to Srp1-HA3 compared with the almost 4.5-fold higher binding of the EAA mutant (Fig. 3C). Weak binding by the AAA mutant explains the residual nuclear localization of Fus2p54–83-S67A (Fig. 3B; Supplemental Fig. S3A). We conclude that the Fus2p NLS has a basal level of nuclear import activity and that the association with the importin Srp1p is strongly enhanced by S67 phosphorylation, stimulating nuclear import.
A significant amount of the S67 phosphomimetic mutant Fus2p54–104-S67E was retained in the nucleus of pheromone-treated cells (Fig. 3D) in spite of the presence of the NES in this protein. This could arise from re-entry into the nucleus, decreased nuclear export, or both. To determine whether S67 phosphorylation affects export, Fus2p's association with Crm1p was measured by coimmunoprecipitation. Crm1p binds to Fus2p54–104-AAA, but binding was greatly decreased in the EAA mutant (about fourfold reduction) (Fig. 3E), suggesting that S67 phosphorylation inhibits Fus2p export.
To verify this, we decoupled import and export using cells containing both a temperature-sensitive allele of srp1-31 and a leptomycin B (LMB)-sensitive allele of the exportin crm1T539C (Neville and Rosbash 1999). The srp1-31 crm1T539C cells were first arrested with α-factor at the permissive temperature, after which Fus2p54–104 and the Fus2p54–104-S67E were expressed by the GAL1 promoter. The proteins were trapped in the nucleus by treatment with LMB to inactivate Crm1p (N/C = 2.7 ± 0.5, n = 22 for wild type, and 2.9 ± 0.4, n = 24 for S67E) (Fig. 3F). After Fus2p expression was blocked by glucose, the temperature was raised to the nonpermissive temperature for srp1-31 to block re-entry into the nucleus, and the LMB was washed out to allow export. Almost all of the wild-type Fus2p was able to exit from the nucleus (N/C = 1.1 ± 0.3, n = 29) (Fig. 3F), whereas very little of the S67E mutant was able to exit (N/C = 2.6 ± 0.43, n = 31). We conclude that S67 phosphorylation strongly down-regulates Fus2p nuclear export, consistent with the decreased binding to Crm1p.
S84 and S100 phosphorylation promote Fus2p transport out of the nucleus
The phosphomimetic S84E mutant is significantly cytoplasmic (Ydenberg and Rose 2009). Two alternative mechanisms might explain this result: activation of nuclear export or inhibition of nuclear import. To examine whether S84 phosphorylation affects import, S84 mutations were introduced into Fus2p54–85-GFP3, which lacks the NES. The S84A accumulated in the nucleus like the wild-type protein (N/C = 4.4 ± 1.2, n = 77, compared with 4.4 ± 1.1, n = 64) (Fig. 4A; Supplemental Fig. S3B). However, the S84E mutation caused some accumulation in the cytoplasm of mitotic cells (N/C = 3.0 ± 0.7, n = 47) (Fig. 4A; Supplemental Fig. S3B), indicating that S84 phosphorylation partially down-regulates Fus2p import. Similar results were obtained for equivalent Fus2p54–85 S84 mutants in which S67 was also mutated to alanine (N/C = 2.3 ± 0.36, n = 57 for S67A; N/C = 2.3 ± 0.37, n = 54 for S67A S84A; N/C = 1.3 ± 0.2, n = 36 for S67A S84E) (Fig. 4A), excluding possible confounding effects of S67 phosphorylation.
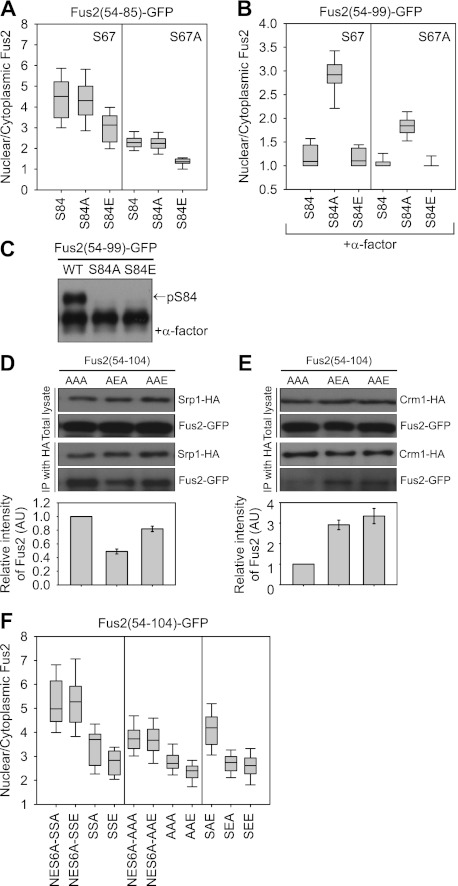
Figure 4.
The role of S67 and S100 phosphorylation in Fus2p transport. (A) Fus2p54–99 mutants (pMR6151, pMR6155, pMR6156, pMR6214, pMR6215, and pMR6157) were expressed in fus2Δ (MY10174) under the GAL1 promoter for 90 min. GFP fluorescence was observed in living cells and quantified as described in Figure 2D. (B,C) Fus2p54–85 mutants (pMR6159, pMR6218, pMR6160, pMR6161, pMR6162, and pMR6163) were expressed in fus2Δ (MY10174) under the GAL1 promoter for 90 min, after which glucose and α-factor were added for 2 h. (B) GFP fluorescence was observed in living cells and quantified as described in Figure 2D. (C) Samples were run on 50 μM Phos-tag gels. The arrow indicates the S84 phosphorylated species. (D,E) Fus2p54–104 mutants (pMR6170, pMR6178, and pMR6179) were expressed in SRP1-3HA (MY12172) or CRM1-3HA (MY12170) cells under the GAL1 promoter for 90 min and coprecipitated as described in Figure 3, C and E. The intensity of copurified Fus2p was normalized by purified Srp1p or Crm1p and expressed relative to AAA. Error bars represent the means ± SEM of two independent experiments. AAA, AEA, and AAE refer to residues 67, 84, and 100. (F) Fus2p54–104 mutants (pMR6184, pMR6185, pMR6169, pMR6177, pMR6182, pMR6183, pMR6170, pMR6179, pMR6175, pMR6176, and pMR6174) were expressed in fus2Δ (MY10174) under the GAL1 promoter for 90 min. GFP fluorescence was observed in living cells and quantified as described in Figure 2D. SSA, SSE, AAA, AAE, SAE, SEA, and SEE refer to residues 67, 84, and 100.
We next examined the effects of S84 mutations on Fus2p54–99-GFP3 export, which contains the NES. Both wild-type Fus2p and the S84E mutant exited the nucleus after α-factor treatment (N/C = 1.2 ± 0.2, n = 26 and 1.2 ± 0.2, n = 26, respectively) (Fig. 4B; Supplemental Fig. S3C). However, S84A remained in the nucleus of pheromone-treated cells (N/C = 2.9 ± 0.3, n = 18) (Fig. 4B; Supplemental Fig. S3C). Because S67 is not phosphorylated efficiently in pheromone-treated cells (Figs. 2A, 4C), it is unlikely that its nuclear localization results from reimport. To exclude any possible confounding effects of S67 phosphorylation, equivalent S84 mutations were constructed in Fus2p54–99-S67A, and similar results were obtained (N/C = 1.1 ± 0.17, n = 19 for S67A; N/C = 1.8 ± 0.21, n = 28 for S67A S84A; N/C = 1.0 ± 0.1, n = 28 for S67A S84E) (Fig. 4B). We conclude that S84 phosphorylation is required for Fus2p nuclear export.
To verify the effects of S84 phosphorylation on both nuclear import and export, the binding of Fus2p with Srp1p or Crm1p was examined by coimmunoprecipitation. Consistent with the localization analysis, binding of the S84 phosphomimetic mutant (AEA) to Srp1 was reduced ∼50% relative to the phosphorylation-defective AAA mutant (Fig. 4D), and binding of the AEA mutant to Crm1p was increased about threefold (Fig. 4E). Thus, we conclude that S84 phosphorylation weakly inhibits nuclear import and strongly stimulates nuclear export.
S100 is also phosphorylated by Fus3p, similar to S84 (Fig. 2A). To analyze a possible role in nuclear import, S100 was mutated in Fus2p54–104-NES6A, which lacks NES activity (Fig. 1D). Both NES6A-S100A (SSA) and NES6A-S100E (SSE) showed similar high levels of nuclear import (N/C = 5.0 ± 1.0, n = 29 and 5.3 ± 1.2, n = 33, respectively) (Fig. 4F; Supplemental Fig. S3D). Similarly, the S100E mutation (AAE) had little effect on coimmunoprecipitation with Srp1p (Fig. 4D). Next, a role in export was analyzed with Fus2p54–104 with an intact NES. Because S100 phosphorylation had no effect on import, any differences in nuclear accumulation must result from different export rates. S100E (SSE) caused a slight but significant reduction in nuclear localization relative to the S100A (SSA). N/C ratio is 2.8 ± 0.6, n = 40 versus 3.7 ± 0.9, n = 32, respectively (Fig. 4F; Supplemental Fig. S3D), indicating that S100 phosphorylation contributes to nuclear export. Similar results were obtained with Fus2p54–100-AAA and Fus2p54–100-AAE mutants, which exclude any possible confounding effects of other phosphorylations (Fig. 4F). Furthermore, coimmunoprecipitation with Crm1p showed increased binding of Fus2p54–100-AAE to Crm1p compared with Fus2p54–100-AAA (Fig. 4E). The residual binding of the Fus2p54–100-AAA mutant to Crm1p (Figs. 3E, 4E) suggests that the nonphosphorylated NES has a basal nuclear exit activity, consistent with the stronger nuclear localization when the NES6A mutation was introduced (Fig. 4F).
To ask whether S84 and S100 phosphorylation synergistically effect Fus2p export, we measured the nuclear localization of Fus2p54–104-SAE, Fus2p54–104-SEA, and Fus2p54–104-SEE mutants (Fig. 4F). The nuclear localization of SEA was similar to that of SEE (N/C = 2.8 ± 0.4, n = 35 and 2.6 ± 0.6, n = 36, respectively), whereas the SAE mutant was significantly more localized in the nucleus (N/C = 4.2 ± 0.8, n = 46). Thus, S84 phosphorylation is the major regulator, and S100 phosphorylation makes a minor contribution to Fus2p export.
Fus3p down-regulates S67 phosphorylation independently of S84 phosphorylation
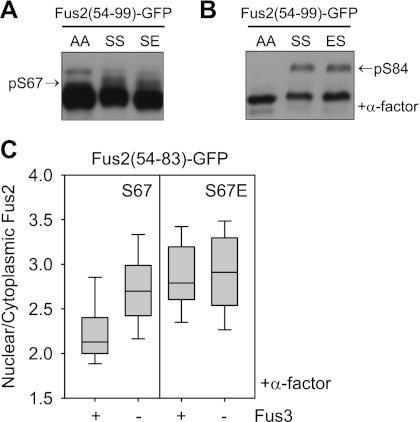
S67 and S84 phosphorylation act antagonistically on Fus2p transport. To test whether phosphorylation at one site might block phosphorylation at the other site, we examined phosphorylation of one site when the other was mutated to mimic phosphorylation in Fus2p54–99. In the S84E, S67 phosphorylation was equal to that of the wild type (Fig. 5A). Likewise, in the S67E, S84 phosphorylation by pheromone was equal to that of the wild type (Fig. 5B). Thus, each phosphorylation appears to be independent of the other phosphorylation.
Figure 5.
Regulation of S67 and S84 phosphorylation. (A,B) Fus2p54–99 mutants (pMR6162, pMR6159, and pMR6160 for A; pMR6162, pMR6159, and pMR6219 for B) were expressed in fus2Δ (MY10174) under the GAL1 promoter for 90 min during mitosis (A) or after α-factor arrest for 2 h (B). Samples were run on 50 μM Phos-tag gels. AA, SS, SE, and ES refer to residues 67 and 84. (A) The arrow indicates the S67 phosphorylated species. (B) The arrow indicates the S84 phosphorylated species. (C) Fus2p54–83 mutants (pMR6152 and pMR6154) were expressed in FUS3 (MY10174) or fus3Δcln3Δ (MY10273) from the GAL1 promoter for 90 min after α-factor arrest for 2 h. GFP fluorescence was observed in living cells and quantified as described in Figure 2D.
S67 phosphorylation was strongly reduced in pheromone-arrested wild-type cells but not in fus3Δ mutant cells (Fig. 2A), suggesting down-regulation by Fus3p. If so, in pheromone-arrested fus3Δ, Fus2p import may be facilitated. To verify an influence of Fus3p on S67 phosphorylation, the nuclear localization of Fus2p54–83 and S67E mutant was measured in pheromone-arrested FUS3 or fus3Δ cells. These proteins lack the NES and Fus3p-dependent phosphorylation sites; nuclear import is dependent solely on the activity of the NLS. Wild-type Fus2p54–83 was partially nuclear in the FUS3 cells (N/C = 2.1 ± 0.3, n = 34), which increased to 2.7 ± 0.4, n = 64 in the fus3Δ mutant (Fig. 5C). The nuclear localization of S67E in FUS3 cells (N/C = 2.8 ± 0.4, n = 37) was comparable with that of wild type in fus3Δ cells and was not affected by fus3Δ (2.9 ± 0.5, n = 54) (Fig. 5C). Thus, Fus3p down-regulates S67 phosphorylation and Fus2p import independent of S84 phosphorylation. Fus3p might regulate S67 phosphorylation by inhibiting the S67 kinase activity or interfering with substrate accessibility.
Mitotic Cdk facilities Fus2p nuclear import via S67 phosphorylation
Cells in the G2/M phase of the cell cycle exposed to pheromone do activate mating signaling, but Fus2p is retained in the nucleus, dependent on the Cdk Cdc28p (Ydenberg and Rose 2009). Cdc28p might regulate localization by activating import (via S67 phosphorylation or by regulating the nuclear import machinery) or inhibiting export (by regulating S84 phosphorylation or the nuclear export machinery.
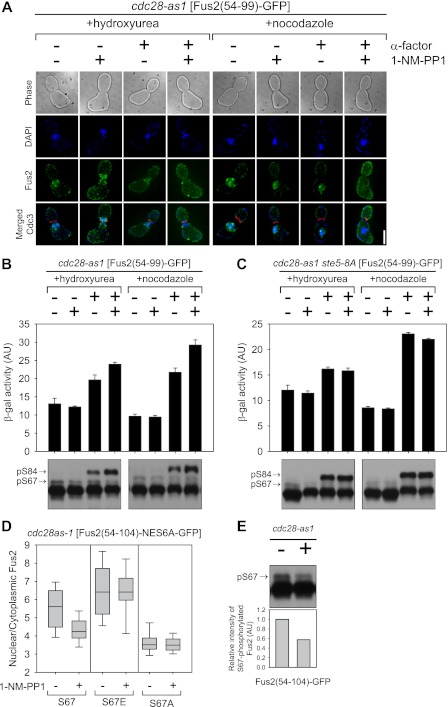
To assess the role of Cdk, its activity was modulated using the cdc28-as1 mutation, which is specifically sensitive to the inhibitor 1-NM-PP1 (Bishop et al. 2000). Cells were arrested in S phase with hydroxyurea or in G2/M with nocodazole and then treated with α-factor and/or 1-NM-PP1, after which Fus2p54–99 was induced by the GAL1 promoter. Mitotic arrest was verified by Cdc3p, a septin that splits into two rings at cytokinesis (Longtine and Bi 2003). As expected, Fus2p was nuclear in cells arrested in S and G2/M but cytoplasmic in cells treated simultaneously with both α-factor and 1-NM-PP1 (Fig. 6A). In both S- and G2/M-arrested cells, addition of α-factor induced expression of FUS1-lacZ, a reporter for pheromone signaling (Fig. 6B), showing that pheromone signaling can occur in both S and G2/M. Moreover, S84 phosphorylation was evident in cells treated with α-factor, showing that pheromone signaling is not restricted to transcriptional activation (Fig. 6B). The minor reduction in signaling and phosphorylation relative to cells treated with both α-factor and 1-NM-PP1 was due to residual G1-phase Cln/Cdk activity. Cln/Cdk blocks pheromone signaling by phosphorylation of Ste5p (Strickfaden et al. 2007); 1-NM-PP1 addition had no effect in a ste5-8A mutant (Fig. 6C), which is refractory to Cln/Cdk inhibition (Strickfaden et al. 2007). Taken together, these results indicate that Clb/Cdk activity does not inhibit pheromone signaling or Fus3p phosphorylation of Fus2p in S or G2/M.
Figure 6.
Cdk involvement in Fus2p nuclear import. (A–C) cdc28-as1 (MY10323) or cdc28as-1 ste5-8A (MY10469) harboring fus1-LacZ (pMR0727) and PGAL-Fus2p54–99-GFP3 (pMR6254) were arrested in S phase with 0.2 M hydroxyurea or in G2/M phase with 15 μg/mL nocodazole, then treated with α-factor and 500 nM 1-NM-PP1 in galactose medium. (A) Cells were fixed and stained with DAPI. Bar, 5 μm. (B,C) Pheromone signaling was measured by induction of pheromone-responsive genes using fus1-LacZ expression as described in Ydenberg and Rose (2009). Data are mean ± SD from three independent experiments. For S84 phosphorylation, samples were run on 50 μM Phos-tag gels. Arrows indicates the S67 or S84 phosphorylated species. (D) cdc28-as1 (MY10323) harboring Fus2p54–104 mutants (pMR6193, pMR6389, and pMR6388) were treated with 500 nM 1-NM-PP1 for 30 min, after which galactose was added for 90 min. GFP fluorescence was observed in living cells and quantified as described in Figure 2D. (E) cdc28-as1 (MY10323) harboring Fus2p54–104 (pMR6009) were treated with 500 nM 1-NM-PP1 for 30 min, after which galactose was added for 90 min. Samples were run on 50 μM Phos-tag gels. The arrow indicates the S67 phosphorylated species. Intensity of phosphorylated S67 was normalized by that of total Fus2p and expressed relative to an untreated sample.
To examine how Cdk regulates Fus2p import, we used the NES-defective Fus2p54–104-NES6A. This protein showed very high levels of nuclear localization (N/C = 5.6 ± 1.3, n = 30), which was reduced by 1-NM-PP1 (N/C = 4.0 ± 0.7, n = 32), demonstrating that Cdk activity simulates Fus2p nuclear import (Fig. 6D). If Cdk activity regulated import via S67 phosphorylation, then we expect that the S67 phosphomimetic mutation will be epistatic to varying Cdk activity. Indeed, inhibition of Cdc28p-as1 had no effect on the S67E-NES6A nuclear localization (N/C = 6.4 ± 1.6, n = 41 versus mock-treated cells 6.41 ± 1.4, n = 33) (Fig. 6D). Moreover, the equivalent S67A mutant showed lower levels of nuclear localization, which were unaffected by Cdk inhibition (N/C = 3.5 ± 0.6, n = 33 and 3.5 ± 0.4, n = 26 in mock-treated and 1-NM-PP1-treated cells, respectively) (Fig. 6D). These results demonstrate that the Cdk regulates Fus2p localization via S67 phosphorylation and not by regulation of the import machinery, consistent with Figure 2D.
Finally, we examined whether S67 phosphorylation was dependent on Cdk. S67 phosphorylation was partially decreased in 1-NM-PP1-treated cells (Fig. 6E), consistent with the effects on nuclear import. However, the high residual level phosphorylation suggests that S67 is a not a direct substrate of Cdk, but must be phosphorylated by a kinase that is regulated by Cdk.
The PAK kinase Cla4p phosphorylates S67 and promotes Fus2p nuclear import
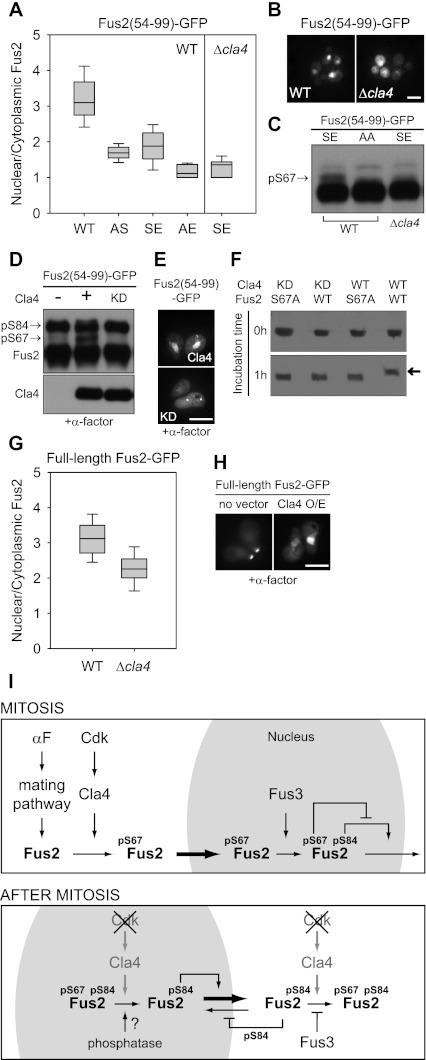
To identify potential S67 kinases, mutations affecting 113 of the 126 predicted yeast kinases were tested for their effects on Fus2p54–99-S84E (SE) import. The S84E phosphomimetic mutant is exquisitely sensitive to the level of S67 phosphorylation in mitotic cells; the AE is essentially cytoplasmic (N/C = 3.1 ± 0.7, n = 63 for wild type and SS and 1.7 ± 0.2, n = 87 for AS, compared with 1.9 ± 0.5, n = 97 for SE and 1.1 ± 0.2, n = 24 for AE) (Fig. 7A). A strong defect was observed in cla4Δ cells lacking the PAK kinase Cla4p (N/C = 1.4 ± 0.2, n = 43) (Fig. 7A,B). We next examined S67 phosphorylation in the cla4Δ cells. In wild-type cells, Fus2p54–99-SE migrated as a doublet due to S67 phosphorylation, but in cla4Δ cells, it migrated identically to the unphosphorylated AA mutant (Fig. 7C). Thus, Cla4p is required for normal Fus2p nuclear import and for S67 phosphorylation.
Figure 7.
S67 phosphorylation and Fus2p import by a PAK kinase, Cla4p. (A–C) Wild type (BY4741) harboring Fus2p54–99 mutants (pMR6159, pMR6161, pMR6160, pMR6163, and pMR6162) or cla4Δ (MY12156) harboring Fus2p54–99-S84E (SE, pMR6160) was expressed from the GAL1 promoter for 90 min. SS, AS, SE, AE, and AA refer to residues 67 and 84. (A) GFP fluorescence was observed in living cells and quantified as described in Figure 2D. (B) Bar, 5 μm. (C) Samples were run on 50 μM Phos-tag gels. The arrow indicates the S67 phosphorylated species. (D,E) cla4Δ cells (MY12156) harboring Fus2p54–99 (pMR6254) were transformed with PGAL-CLA4 (pMR6194) or PGAL-CLA4KD (pMR6195). Cells were arrested with α-factor for 2 h, and then galactose was added for 90 min. (D) Samples were run on 50 μM Phos-tag gels. The arrows indicate the S67 or S84 phosphorylated species. (E) GFP fluorescence was observed in living cells. Bar, 5 μm. (F) Fus2p1–328-His6 (pMR5630), Fus2p1–328-S67A-His6 (pMR6181), GST-Cla4p (pMR6196), and GST-Cla4pKD (pMR6197) were purified in Escherichia coli as described in the Materials and Methods. Purified Fus2p and Fus2p-S67A mutants were treated with equivalent amounts of either purified GST-Cla4p or GST-Cla4pKD in the presence of ATP for 1 h at 30°C as described in the Materials and Methods. Phosphorylation was detected by anti-6XHis immunoblot after running on 50 μM Phos-tag gels. The arrow indicates the S67 phosphorylated Fus2p. (G) Full-length Fus2p (pMR5738) was expressed in wild-type (BY4741) or cla4Δ (MY12156) cells under GAL1 promoter for 3 h. GFP fluorescence was observed in living cells and quantified as described in Figure 2D. (H) Wild-type cells harboring PGAL-full-length Fus2p (pMR5738) or wild-type cells harboring both PGAL-full-length Fus2p (pMR5738) and PGAL-Cla4p (pMR6194) were arrested with α-factor for 2 h, and then galactose was added for 90 min. GFP fluorescence was observed in living cells. Bar, 5 μm. (I) A model for the regulated Fus2p transport during mitosis and after mitosis in pheromone-treated cells. See the Discussion for details.
To verify a role in Fus2p import, Cla4p and a kinase-dead mutant (Cla4pKD and Cla4pK594A) were ectopically expressed in α-factor-arrested cells, where S67 phosphorylation is not observed and Fus2p is normally cytoplasmic. Ectopic expression of Cla4p resulted in S67 phosphorylation (Fig. 7D) and caused relocalization of Fus2p to the nucleus (Fig. 7E). Expression of the Cla4pKD was ineffective for either S67 phosphorylation or nuclear localization. Therefore, Cla4p kinase activity is both required and sufficient for S67 phosphorylation and Fus2p import.
To determine whether Cla4p directly phosphorylates S67, we performed an in vitro kinase assay using GST-tagged Cla4p and His6-tagged Fus2p fragments (Fus2p1–328) purified from Escherichia coli. Phosphorylation was determined by the electrophoretic mobility shift on Phos-tag gels. Incubation with Cla4p led to the complete conversion of Fus2p to the phosphorylated species (Fig. 7F), but not the Fus2p-S67A, indicating S67 phosphorylation. Moreover, phosphorylation was not observed for the Cla4pKD, showing that Cla4p was responsible for the phosphorylation. We conclude that Fus2p-S67 is a direct substrate for the Cla4p kinase.
To determine whether Cla4p has the same effect on full-length Fus2p as the N-terminal fragments, the cellular distribution of full-length Fus2p was examined in mitotic cla4Δ cells (Fig. 7G) and pheromone-treated cells in which Cla4p was ectopically expressed (Fig. 7H). Nuclear localization of full-length Fus2p in mitotic cells was reduced in cla4Δ relative to the wild-type cells (N/C = 2.3 ± 0.42, n = 33 for cla4Δ and 3.1 ± 0.5, n = 22 for wild type) (Fig. 7G). Moreover, Cla4p overexpression resulted in nuclear accumulation of full-length Fus2p in pheromone-treated cells (Fig. 7H). Thus, Cla4p regulates the transport of full-length Fus2p.
Discussion
In this study, we show how budding yeast spatially regulates a key mating regulator, Fus2p, to temporally coordinate cell proliferation and mating differentiation. Fus2p is largely nuclear in mitotic cells at steady state. However, nuclear accumulation became even more pronounced in crm1-1 cells at 37°C or when the NES was mutated or deleted (Fig. 4F; Supplemental Fig. S4), showing that even in mitotic cells, Fus2p shuttles between the nucleus and cytoplasm. Therefore, shifts in nuclear/cytoplasmic localization must result from regulated changes in the rates of import and/or export.
We found that Fus2p import and export are regulated by three phosphorylation sites (S67, S84, and S100) adjacent to or within the NLS and NES. S67 phosphorylation dictates nuclear localization by promoting the association with the import receptor and down-regulating association with the export receptor (Fig. 3). Dual regulation of import and export was also characteristic of S84 phosphorylation, which functioned in the opposite way to facilitate nuclear export (Fig. 4). However, phosphomimetic mutations at S67 and S84 did not inhibit phosphorylation at the other site (Fig. 5A,B). This is somewhat surprising for S67, which is located within the S84 MAP kinase docking site described below. It is possible that the phosphomimetic glutamate residue may be insufficient to inhibit Fus3p docking where the presence of a phosphate at that site may sterically occlude binding. However, we observed that pheromone treatment caused S84 phosphorylation, but not nuclear export, in cells arrested in mitosis when S67 is also phosphorylated (Fig. 6B,C). These results indicate that S84 phosphorylation at least does not block S67 phosphorylation, suggesting that the two sites do not function as a bistable switch. Nevertheless, the two phosphorylation sites act antagonistically toward each other's functions; S84 phosphorylation suppressed the ability of S67 phosphorylation to promote interaction with Srp1p and nuclear import (Supplemental Figs. S5, S6A). Similarly, S67 phosphorylation blocked the effect of S84 phosphorylation on Crm1p binding and export (Figs. 3F, 4E; Supplemental Fig. S6B).
Interestingly, mutation of 63KR64 to 63AA64 in the NLS reduced both S67 phosphorylation (Supplemental Fig. S7A) and S84/S100 phosphorylation (Supplemental Fig. S7B). This was not due to decreased nuclear localization; mutation of 80KK81 to 80AA81, also in the NLS, had no effect (Supplemental Fig. S7). 63KR64 may be part of the S67 kinase recognition motif, whereas 63KRPASLDL70 fits the consensus for a MAP kinase docking site, (R/K)1–2X4–6LXL (where X is any amino acid). Kinase docking sites are thought to regulate the enzymatic efficiency and specificity in the MAP kinase pathway (Remenyi et al. 2005). Thus, residues surrounding S67 are required for efficient phosphorylation of both S67 and S84. Fus3p might down-regulate S67 phosphorylation by interfering with substrate accessibility, or alternatively, Fus3p may inhibit, directly or indirectly, the S67 kinase Cla4p. In the KR63AA mutant treated with pheromone, weak S67 phosphorylation was observed (Supplemental Fig. S7B), consistent with the former model.
Structural studies of yeast importin α revealed that the NLS-binding domain is surrounded by negatively charged amino acids, which provide electrostatic interactions with positively charged residues in the NLS in addition to hydrophobic interactions (Conti et al. 1998). S84 phosphorylation adds a negatively charged phosphate group proximal to the Fus2p NLS, which is likely to disrupt electrostatic interactions and decrease the binding of the NLS to its import receptor, Srp1p. Remarkably, in contrast to S84, both Fus2p nuclear import and binding to importin α appear to be dependent on S67 phosphorylation, which is located within the bipartite NLS. Because mutation of 63KR64 to alanines also disrupted S67 phosphorylation (Supplemental Fig. S7B), we considered the possibility that the basic cluster 63KR64 may be important for S67 phosphorylation by Cla4p, but does not act as part of the NLS. In an analogous system, in SV40 large T-antigen, phosphorylation upstream of the NLS enhances nuclear import by modulating recognition by importin α1 (Nardozzi et al. 2010). However, this is not the case for Fus2p. The S67 phosphomimetic mutation does not override the Fus2p-KR63AA defect in nuclear import (Supplemental Fig. S8). These results indicate that 63KR64 plays an important role in both regulatory phosphorylation events as well as being part of the NLS. Because S67 phosphorylation, although far from the NES, was able to regulate the NES by decreasing the interaction with Crm1p (Fig. 3E), we propose that S67 phosphorylation might induce a conformational change through which Fus2p interaction with its transport receptors is modulated. Other proteins are known to up-regulate nuclear import by phosphorylation, such as EBNA-1, HBV core-antigen, and STAT1, which enhance the binding affinity for the importin or induces a conformational change (Nardozzi et al. 2010).
S67 is phosphorylated by Cla4p, a member of the PAK family kinase of serine/threonine protein kinases. Phosphorylation of 63KRPAS67 is consistent with the site selectivity of Cla4p for arginine at the −2 or −3 position (Mok et al. 2010). The best-characterized function of Cla4p is in septin ring assembly, and recent studies have shown that it is regulated during mitotic entry and exit (Hofmann et al. 2004). Cla4p is hyperphosphorylated in a Cdk-dependent fashion and is activated during mitosis; its activity drops as cells complete cytokinesis and enter the G1 phase (Benton et al. 1997; Tjandra et al. 1998). Although it is not known whether Cla4p kinase activity requires phosphorylation, its activity in vivo coincides with Clb2p/Cdk-dependent phosphorylation, consistent with cell cycle-dependent activation by Cdk. We propose that the mitotic Cdk facilitates Fus2p nuclear import by activating Cla4p, which in turn phosphorylates S67. Supporting this hypothesis, the S67 phosphomimetic mutant overrode the defect in nuclear import of Fus2p when Cdk activity was blocked by 1-NM-PP1 (Fig. 6D).
Fus3p is both nuclear and cytoplasmic, but is predominantly in the nucleus (van Drogen et al. 2001; Blackwell et al. 2003). After pheromone stimulation, Fus3p nuclear localization increases (Blackwell et al. 2003), and a fraction localizes to the tip of the mating projection (van Drogen et al. 2001). In contrast, Cla4p is cytoplasmic and is mainly localized to the bud cortex (Holly and Blumer 1999). We hypothesize that the spatial separation of these two proteins facilitates Fus2p's sequestration in the nucleus in mitotic cells exposed to pheromone (Fig. 7I). When Fus2p is initially expressed in mitotic cells, cytoplasmic Cla4p's phosphorylation of S67 would stimulate binding to Srp1p, such that the rate of import becomes greater than the rate of export. In the nucleus, S84 becomes phosphorylated by activated Fus3p. However, Fus2p export would be suppressed by the inhibitory effect of S67 phosphorylation decreasing association with Crm1p (Fig. 4E; Supplemental Fig. S6B). Therefore, Cla4p in mitotic cells overrides activated Fus3p by the pheromone signaling and keeps Fus2p inactive in the nucleus. As cells exit the cell cycle, Cdk activity declines, which leads to reduced Cla4p activity, allowing for S67 dephosphorylation. Alternatively, S67 may be actively dephosphorylated by an as yet unidentified phosphatase because S67 was phosphorylated in pheromone-arrested fus3Δ cells (Fig. 2A). To prevent S67 phosphorylation, Fus3p could positively regulate the S67 phosphatase or negatively regulate the S67 kinase Cla4p. In the absence of S67 phosphorylation, the Fus3p-dependent phosphorylation of S84 and S100 stimulates binding to Crm1p and nuclear export. S84 phosphorylation also reduces binding to importin Srp1p. Independent of S84 phosphorylation, Fus3p suppresses S67 phosphorylation by an as yet unidentified mechanism. Increased nuclear export activity and decreased reimport synergize to assure that Fus2p is retained in the cytoplasm during mating differentiation. Thus, although Fus3p is activated by pheromone early in mitosis, Cdk and Cla4p act as a mitotic brake to prevent premature nuclear export of Fus2p. Only when the brake is released is Fus3p able to accelerate the export of Fus2p and allow progression into the cell fusion pathway.
Materials and methods
Yeast strains and general methods
All yeast culture and genetic techniques were performed as described by Ydenberg and Rose (2009). The S. cerevisiae strains used in this study are listed in Supplemental Table S1. SRP1 and CRM1 were tagged with HA3 by PCR-based methods. The crm1T1539C mutant was integrated into the CRM1 locus of srp1-31 (MY12594) using XbaI-cut pMR6338. Cells were synchronized in S phase with 0.2 M hydroxyurea for 2–3 h or in G2/M in 15 μg/mL nocodazole for 2 h. To activate pheromone signaling, cells were treated with 10 μg/mL α-factor. To inhibit Cdk activity, cdc28-as1 cells were treated with 500 nM 1-NM-PP1 (Bishop et al. 2000). The GAL1 promoter was induced for full-length Fus2p and Cla4p expression using 2% galactose and for Fus2p truncations with 2% galactose + 0.2% glucose.
Plasmids used in this study are described in Supplemental Table S2. Point mutations of FUS2 were generated by PCR-based site-directed mutagenesis. To generate pMR6337, a fragment of CRM1-HA3 comprising codons 329–1085 was amplified and cloned into integrative pRS405 (pMR1863). This plasmid was used as a template to generate pMR6338 by site-directed mutagenesis.
Protein analysis and coimmunoprecipitation
Proteins were prepared by TCA precipitation, resolved on 7.5% polyacrylamide gel with or without 50 μM Phos-tag (FMS Laboratory). For immunoprecipitation of HA-tagged Srp1p and Crm1p, total cellular lysate (1 mg in 700 μL of lysis buffer) was incubated for 2 h at 4°C with Dynabeads (Invitrogen) and conjugated with anti-HA for 2 h at 4°C. Lysis buffer contained 50 mM Tris-Cl (pH 7.4), 150 mM NaCl, 1 mM DTT, 0.1 mM NaVO3, 60 mM β-glycerophosphate, 0.1% NP-40, and protein inhibitor cocktail (Roche).
Microscopy and imaging analysis
Fluorescence microscopy and quantification of GFP florescence intensity were performed essentially as described by Ydenberg and Rose (2009). Cellular labeling was visualized on a microscopy system (DeltaVision, Applied Precision, LLC) using an inverted microscope (TE200, Nikon), a charge-coupled device camera (CoolSNAP HQ, Roper Scientific), and a 100× objective with a 1.4 NA. The fluorescence intensity of GFP-fused proteins was quantitatively analyzed by softWoRx and ImageJ (National Institutes of Health). After fluorescence intensity of selected regions of the nucleus or cytoplasm was quantified and the background fluorescence was subtracted by placing the same measurement circle in nearby regions without a GFP signal, the N/C ratio was calculated for each cell. No further manipulations other than adjustments in brightness and contrast were made.
Screening for the kinase responsible for S67 phosphorylation
One-hundred-thirteen kinase mutant cells were transformed by PGAL-Fus2p54–99-S84E-GFP3 and grown to mid-logarithmic phase. The Fus2p mutant protein was ectopically expressed under control of the GAL1 promoter for 90 min, and its nuclear localization was observed by fluorescence microscopy and quantified as described above.
In vitro kinase assay
For the in vitro kinase assay, the fusion proteins 6×HN-Fus2p1–328, Fus2p1–328-S67A, GST-Cla4p, and GST-Cla4pKD were expressed in BL21PRO cells and purified as described by Ydenberg and Rose (2009) and Versele and Thorner (2004). Approximately 200 ng of Fus2p fragments was mixed with ∼400 ng of either GST-Cla4p or GST-Cla4pKD in 15 μL of kinase buffer (50 mM Tris-Cl at pH 7.5, 10 mM MgCl2, 1 mM DTT) with 200 mM ATP. After incubation for 1 h at 30°C, Laemmli SDS sample buffer was added to terminate the phosphorylation reaction. Proteins were separated by 7.5% SDS-PAGE containing 50 μM Phos-tag and detected by anti-His immunoblot.
Acknowledgments
We thank Jeremy Thorner, Pamela Silver, Eduard Hurt, John Aitchison, Anita Corbett, and Juan Carlos Igual for providing yeast strains and plasmids. We thank May Husseini and Rebecca Davis for excellent help with media and materials. We acknowledge Casey Ydenberg and members of the laboratory for helpful discussion on this work. This work was supported by NIH grant number GM37739 to M.D.R.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.187260.112.
References
- Benton BK, Tinkelenberg A, Gonzalez I, Cross FR 1997. Cla4p, a Saccharomyces cerevisiae Cdc42p-activated kinase involved in cytokinesis, is activated at mitosis. Mol Cell Biol 17: 5067–5076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, et al. 2000. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407: 395–401 [DOI] [PubMed] [Google Scholar]
- Blackwell E, Halatek IM, Kim HJ, Ellicott AT, Obukhov AA, Stone DE 2003. Effect of the pheromone-responsive G(α) and phosphatase proteins of Saccharomyces cerevisiae on the subcellular localization of the Fus3 mitogen-activated protein kinase. Mol Cell Biol 23: 1135–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brizzio V, Gammie AE, Rose MD 1998. Rvs161p interacts with Fus2p to promote cell fusion in Saccharomyces cerevisiae. J Cell Biol 141: 567–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F, Herskowitz I 1992. Phosphorylation of FAR1 in response to α-factor: A possible requirement for cell-cycle arrest. Mol Biol Cell 3: 445–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti E, Uy M, Leighton L, Blobel G, Kuriyan J 1998. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin α. Cell 94: 193–204 [DOI] [PubMed] [Google Scholar]
- Davis LI 1995. The nuclear pore complex. Annu Rev Biochem 64: 865–896 [DOI] [PubMed] [Google Scholar]
- Dawson JC, Legg JA, Machesky LM 2006. Bar domain proteins: A role in tubulation, scission and actin assembly in clathrin-mediated endocytosis. Trends Cell Biol 16: 493–498 [DOI] [PubMed] [Google Scholar]
- Elion EA 2000. Pheromone response, mating and cell biology. Curr Opin Microbiol 3: 573–581 [DOI] [PubMed] [Google Scholar]
- Elion EA, Brill JA, Fink GR 1991. FUS3 represses CLN1 and CLN2 and in concert with KSS1 promotes signal transduction. Proc Natl Acad Sci 88: 9392–9396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen H, Humphries C, Ho Y, Schub O, Colwill K, Andrews B 2006. Characterization of the yeast amphiphysins Rvs161p and Rvs167p reveals roles for the Rvs heterodimer in vivo. Mol Biol Cell 17: 1306–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galderisi U, Jori FP, Giordano A 2003. Cell cycle regulation and neural differentiation. Oncogene 22: 5208–5219 [DOI] [PubMed] [Google Scholar]
- Gammie AE, Brizzio V, Rose MD 1998. Distinct morphological phenotypes of cell fusion mutants. Mol Biol Cell 9: 1395–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrenton LS, Braunwarth A, Irniger S, Hurt E, Künzler M, Thorner J 2009. Nucleus-specific and cell cycle-regulated degradation of mitogen-activated protein kinase scaffold protein Ste5 contributes to the control of signaling competence. Mol Cell Biol 29: 582–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henchoz S, Chi Y, Catarin B, Herskowitz I, Deshaies RJ, Peter M 1997. Phosphorylation- and ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor Far1p in budding yeast. Genes Dev 11: 3046–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann C, Shepelev M, Chernoff J 2004. The genetics of Pak. J Cell Sci 117: 4343–4354 [DOI] [PubMed] [Google Scholar]
- Holly SP, Blumer KJ 1999. PAK-family kinases regulate cell and actin polarization throughout the cell cycle of Saccharomyces cerevisiae. J Cell Biol 147: 845–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita-Kikuta E, Aoki Y, Kinoshita E, Koike T 2007. Label-free kinase profiling using phosphate affinity polyacrylamide gel electrophoresis. Mol Cell Proteomics 6: 356–366 [DOI] [PubMed] [Google Scholar]
- Kurjan J 1993. The pheromone response pathway in Saccharomyces cerevisiae. Annu Rev Genet 27: 147–179 [DOI] [PubMed] [Google Scholar]
- Kutay U, Guttinger S 2005. Leucine-rich nuclear-export signals: Born to be weak. Trends Cell Biol 15: 121–124 [DOI] [PubMed] [Google Scholar]
- Li X, Gerber SA, Rudner AD, Beausoleil SA, Haas W, Villen J, Elias JE, Gygi SP 2007. Large-scale phosphorylation analysis of α-factor-arrested Saccharomyces cerevisiae. J Proteome Res 6: 1190–1197 [DOI] [PubMed] [Google Scholar]
- Longtine MS, Bi E 2003. Regulation of septin organization and function in yeast. Trends Cell Biol 13: 403–409 [DOI] [PubMed] [Google Scholar]
- Madhani HD, Styles CA, Fink GR 1997. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell 91: 673–684 [DOI] [PubMed] [Google Scholar]
- McKinney JD, Chang F, Heintz N, Cross FR 1993. Negative regulation of FAR1 at the Start of the yeast cell cycle. Genes Dev 7: 833–843 [DOI] [PubMed] [Google Scholar]
- Mok J, Kim PM, Lam HY, Piccirillo S, Zhou X, Jeschke GR, Sheridan DL, Parker SA, Desai V, Jwa M, et al. 2010. Deciphering protein kinase specificity through large-scale analysis of yeast phosphorylation site motifs. Sci Signal 3: ra12 doi: 10.1126/scisignal2000482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardozzi JD, Lott K, Cingolani G 2010. Phosphorylation meets nuclear import: A review. Cell Commun Signal 8: 32 doi: 10.1186/1478-811X-8-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neville M, Rosbash M 1999. The NES–Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae. EMBO J 18: 3746–3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson JM, Ydenberg CA, Rose MD 2008. Dynamic localization of yeast Fus2p to an expanding ring at the cell fusion junction during mating. J Cell Biol 181: 697–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter M, Herskowitz I 1994. Direct inhibition of the yeast cyclin-dependent kinase Cdc28-Cln by Far1. Science 265: 1228–1231 [DOI] [PubMed] [Google Scholar]
- Remenyi A, Good MC, Bhattacharyya RP, Lim WA 2005. The role of docking interactions in mediating signaling input, output, and discrimination in the yeast MAPK network. Mol Cell 20: 951–962 [DOI] [PubMed] [Google Scholar]
- Sheltzer JM, Rose MD 2009. The class V myosin Myo2p is required for Fus2p transport and actin polarization during the yeast mating response. Mol Biol Cell 20: 2909–2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickfaden SC, Winters MJ, Ben-Ari G, Lamson RE, Tyers M, Pryciak PM 2007. A mechanism for cell-cycle regulation of MAP kinase signaling in a yeast differentiation pathway. Cell 128: 519–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjandra H, Compton J, Kellogg D 1998. Control of mitotic events by the Cdc42 GTPase, the Clb2 cyclin and a member of the PAK kinase family. Curr Biol 8: 991–1000 [DOI] [PubMed] [Google Scholar]
- van Drogen F, Stucke VM, Jorritsma G, Peter M 2001. MAP kinase dynamics in response to pheromones in budding yeast. Nat Cell Biol 3: 1051–1059 [DOI] [PubMed] [Google Scholar]
- Versele M, Thorner J 2004. Septin collar formation in budding yeast requires GTP binding and direct phosphorylation by the PAK, Cla4. J Cell Biol 164: 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JM, Rose MD 2001. Yeast mating: Getting close to membrane merger. Curr Biol 11: R16–R20 doi: 10.1016/S0960-9822(00)00036-1 [DOI] [PubMed] [Google Scholar]
- Ydenberg CA, Rose MD 2009. Antagonistic regulation of Fus2p nuclear localization by pheromone signaling and the cell cycle. J Cell Biol 184: 409–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Skoultchi AI 2001. Coordinating cell proliferation and differentiation. Curr Opin Genet Dev 11: 91–97 [DOI] [PubMed] [Google Scholar]