Abstract
Breast cancer is the most common cancer and the leading cause of cancer death in women. Although tamoxifen therapy is successful for some patients, it does not provide adequate benefit for those who have estrogen receptor (ER)-negative cancers. Therefore, we approached novel treatment strategies by combining two potential bioactive dietary supplements for the reactivation of ERα expression for effective treatment of ERα-negative breast cancer with tamoxifen. Bioactive dietary supplements such as green tea polyphenols (GTPs) and sulforaphane (SFN) inhibit DNA methyltransferases (DNMTs) and histone deacetylases (HDACs), respectively, which are of central importance to cancer prevention. In the present study, we have observed that treatment of ERα-negative breast cancer cells with GTPs and SFN alone or in combination leads to the reactivation of ERα expression. The combination of 20 µg/mL GTPs and 5 µM SFN was found to be the optimal dose of ERα-reactivation at 3 days in MDA-MB-231 cells. The reactivation of ERα expression was consistently correlated with ERα promoter hypomethylation and hyperacetylation. Chromatin immunoprecipitation (ChIP) analysis of the ERα promoter revealed that GTPs and SFN altered the binding of ERα-transcriptional co-repressor complex thereby contributing to ERα-reactivation. In addition, treatment with tamoxifen in combination with GTPs and SFN significantly increased both cell death and inhibition of cellular proliferation in MDA-MB-231 cells in comparison to treatment with tamoxifen alone. Collectively, our findings suggest that a novel combination of bioactive-HDAC inhibitors with bioactive-demethylating agents is a promising strategy for the effective treatment of hormonal refractory breast cancer with available anti-estrogens.
Introduction
Breast cancer is the most frequently diagnosed cancer and the leading cause of cancer death among women, accounting for 23% of the total cancer cases and 14% of the cancer deaths [1]. One of the important classifications of breast tumors is based on the presence or absence of the estrogen receptor (ER). While the majority of breast cancers are ER-positive, approximately 25–30% are ER-negative [2], [3]. Patients with ER-positive breast cancer receive hormonal therapy using either selective estrogen receptor modulators (SERMs) such as tamoxifen, raloxifene and lasofoxifene, or with aromatase inhibitors (AIs) such as anastrozole, letrozole, and exemestene, and have a better prognosis. However, treatment of patients with ER-negative tumor is challenging due to the poor response to hormonal therapies in the absence ER expression. Therefore alternative targeted therapies are aimed to prevent and treat hormonal refractory breast cancers.
Recently, many studies have addressed the possibilities of reactivation of ER expression in ER-negative breast cancer cells for the effective treatment with available SERMs. Further, the absence of ERα gene expression in ER-negative breast cancer is largely due to epigenetic silencing instead of DNA mutation or deletion of the ERα gene [4], [5]. Previous studies have shown that epigenetic silencing of ER is associated with DNA hypermethylation at the ER-promoter in ER-negative breast cancer cells [6], [7]. In addition, histone modifications, specifically histone acetylation/deactylations have also been implicated as common mechanisms underlying ER silencing in human malignant mammary cells [7], [8]. Hence, treatment of ER-negative breast cancer cells with DNA methyltransferase (DNMT) inhibitors such as 5-aza-2′-deoxycytidine and/or histone deacetylase (HDAC) inhibitors such as trichostatin A (TSA) leads to the reactivation of ER expression, underscoring the importance of DNMTs and HDACs in maintaining the repressive environment at the ERα gene [2], [9], [10]. However, the use of synthetic small molecules as the DNMT and HDAC inhibitors for the ER-reactivation in ER-negative breast tumor would be expected to result in too many adverse side-effects to warrant practical application to chemoprevention and therapy.
Many studies have demonstrated the chemopreventive properties of green tea polyphenols (GTPs) and sulforaphane (SFN) against various types of carcinoma through multiple mechanisms such as anti-oxidant, induction of apoptosis, cell cycle regulation, inhibition of angiogenesis and metastasis [11], [12]. Further, (-)-epigallocatechin-3-gallate (EGCG), a major constituent of GTPs, is known to complex with the DNA methyltransferases (DNMTs) which reduces methylating activity of many genes in cancer cells as well as in mouse models [13], [14]. The hypomethylation induced by EGCG has been shown to be associated with reactivation of methylation-silenced tumor suppressor genes such as p16INK4a, p21CIP/WAF and the DNA mismatch repair gene, human mutL homologue 1 (hMLH1), which eventually leads to tumor suppression [13], [15]. Further, SFN is a bioactive dietary supplement found in cruciferous vegetables, that has an established histone deactylation (HDAC) inhibition activity [16], [17]. The HDAC inhibition activity of SFN has been shown to lead to an increase in the global and local histone acetylation status of a number of genes including tumor promoter genes such as human telomerase reverse transcriptase (hTERT) in breast cancer [18]. Both DNA methylation and histone acetylations have been the focus of considerable attention in cancer prevention and therapy.
In addition to histone acetylation and promoter methylation, histone modifications-mediated transcriptional regulation of ERα expression has emerged. The ERα promoter is mostly hypermethylated in ER-negative breast cancer cells [6], [7]. Hypermethylation of CpG-islands may inhibit transcription by recruiting the methyl-CpG binding domain (MBD) proteins or by interfering with the recruitment and function of basal transcription factors or transcriptional coactivators [2], [7]. Similarly, ER-negative breast cancer cells also display a relative depletion of acetyl-H3 and acetyl-H4 which provide transcriptional repressive environment at the ERα gene [8] Therefore, in the present study, we tested our hypothesis that a combination of dietary DNMT and HDAC inhibitors may lead to transcriptional activation of ERα expression in ER-negative breast cancer cells. Our study demonstrates that treatment of ER-negative breast cancer cells with GTPs and SFN synergistically reactivates ERα expression through epigenetic alteration of CpG methylation and histone acetylation-mediated release of transcriptional inhibitor complex at the ERα promoter. Furthermore, our findings suggest a novel dietary combination of DNA methyltransferase and histone deacetylase inhibitors contribute to ER re-expression in ER-negative breast cancer for the effective treatment of hormonal refractory breast cancers (HRBCs) with available SERMs.
Materials and Methods
Materials
GTPs and R, S-sulforaphane were purchased from LKT laboratories (Minneapolis, MN). GTPs was freshly prepared at a stock concentration of 1 mg/mL in sterile PBS just before cellular treatment. SFN was prepared in DMSO and stored at a stock concentration of 10 mmol/L at −20°C.
Cell culture and cell proliferation assay
The human breast cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA). Breast cancer MCF-7 [ER (+)], MDA-MB-453 [ER (−)] and MDA-MB-231 [ER (−)] cells were cultured as a monolayer in phenol-red–free Dulbecco's modified Eagle's medium (DMEM) (Mediatech Inc, Manassas, VA) supplemented with 10% dextran-charcoal–stripped fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA) and 1% penicillin/streptomycin (Mediatech, Herndon, VA) as described previously [4], [18]. Control MCF10A cells were also procured from ATCC and maintained as described previously [18]. MCF10A is a non-tumorigenic human breast epithelial cell line and frequently used as a human breast cell control [19], [20], [21]. Cells were treated with GTPs or SFN and a combination of both at the indicated concentrations. The medium with GTPs and SFN was replaced every 24 h for the duration of the experiments. The maximum concentration of DMSO in the culture medium was 0.1% (v/v). MTT assay was performed for assessing cellular proliferation. Briefly, cells were plated at a density of 1×104 cells per well in 200 µL of complete medium containing different concentrations of GTPs or SFN and combination of both in a 96-well microtiter plate. Each treatment was repeated in 8 wells. The cells were incubated for 96 h at 37°C in a humidified chamber at the end of which MTT solution (50 µL, 5 mg/mL in media) was added to each well and incubated for 2 h. The microtiter plate containing the cells was centrifuged at 600 g for 5 min at 4°C. The MTT solution was removed from the wells by aspiration and the MTT-formazan crystals were dissolved in DMSO (150 µL). Absorbance was recorded at 540 nm wavelength. To observe the effects of 17β-estradiol (E2) (Sigma) and tamoxifen (Sigma) on cellular apoptosis, MDA-MB-231 cells were treated from the second day after treatments with GTPs and SFN.
Quantification of ERα expression by real-time PCR
Total RNA isolation and real-time quantification of ERα expression were followed as described previously [4]. Total RNA was extracted using the RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. Total RNA (2 µg) was reverse-transcribed into cDNA using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). The primers specific for ERα (Hs01046818_ml) and glyceraldehydes-3-phosphate dehydrogenase (GAPDH) (Hs99999905_ml) were obtained from Inventorial Gene Assay Products (Applied Biosystems, Foster City, CA). The reaction was performed in a Bio-Rad MyiQ thermocycler (Bio-rad, Hercules, CA) using platinum SYBR Green detection system (Invitrogen, Carlsbad, CA). Thermal cycling was initiated at 94°C for 4 min followed by 35 cycles of PCR (94°C, 15 s; 60°C, 30 s). The calculations for determining the relative level of gene expression were made using the cycle threshold (C t) method. The mean C t values from duplicate measurements were used to calculate the expression of the target gene using the formula: fold change in gene expression, 2−ΔΔCt = 2−{ΔCt (treated samples)−ΔCt (untreated control)}, where ΔCt = Ct (ERα)−Ct (GAPDH).
Western blot analysis
Protein was extracted from cultured cells using the RIPA-lysis buffer (Upstate Biotechnology, Lake Placid, NY) following the manufacturer's protocol. For immunoblot analysis, 100 µg of protein was resolved on a 10% SDS-PAGE and transferred onto nitrocellulose membrane. After incubation in blocking buffer for 1 h, the membranes were incubated with the primary antibodies specific for ERα (NeoMarkers, Fremont, CA), DNMT1, DNMT3a, DNMT3b, SUV39H1 (Santa Cruz Biotechnology, Santa Cruz, CA), HDAC antibody sampler kit (cat# 9928; Cell Signalling, Danvers, MA) and β-actin (Cell Signalling). The blot was then washed with TBS and 0.05% (v/v) Tween-20 and incubated with specific secondary antibody conjugated with horseradish peroxidase. Protein bands were then visualized using the ECL-detection system following the protocol of the manufacturer. The bands were analyzed by using Kodak 1D 3.6.1 image software for the intensity and normalized with respective β-actin.
5-methyl cytosine (5-mC) immunostaining
Cells were grown on the sterile cover slips and treated with GTPs and SFN for 3 days. After the treatment period, cells were fixed with cold-ethanol, permeabilized with 0.1% Triton- X100 in phosphate buffered saline (PBS), and washed with PBS for 10 min. The cells were then blocked with 5% goat serum in PBS for 30 min, followed by incubation with 3% H2O2 for 20 min to quench endogenous peroxidase. After washing the cells with PBS, cells were incubated with 5- mC specific antibody (1∶500, v/v, Calbiochem, Gibbstown, NJ) for 1 h, followed by sequential incubation of cells with biotinylated secondary antibody, and HRP-conjugated streptavidin, and finally with diaminobenzidine (DAB) substrate for 5-mC positive staining. Nuclei were counterstained with methyl green (Sigma).
South-western dot-blot analysis for 5-methyl cytosine (5-mC)
Cells were treated with GTP and SFN for 3 days as described above. Genomic DNA was isolated using the DNA Isolation Kit (Qiagen, Maryland, MD) according to the manufacturer's instructions, and dot-blot analysis was performed as described previously [22]. Briefly, 1 µg of genomic DNA was transferred onto Hybond-ECL nitrocellulose membranes (Amersham Biosciences, UK) using Bio-Dot Microfiltration Apparatus (Bio-Rad Laboratories, Inc. Hercules, CA), and fixed by baking the membrane for 30 min at 80°C. After blocking the non-specific-binding sites, the membrane was incubated with the antibody specific to 5-mC (1∶500, v/v) followed by incubation with a HRP-conjugated secondary antibody. The bands were then visualized using the ECL-detection system following the protocol of the manufacturer (Santa Cruz Biotechnology). The bands were analyzed by using Kodak 1D 3.6.1 image software for the intensity and equal DNA loading was verified by staining the membranes with 0.2% methylene blue.
DNMTs activity assay
DNMTs activity was determined using a colorimetric DNMTs activity assay kit (Epigentek, Brooklyn, NY) according to the manufacturer's instruction. The reaction was initiated by adding 20 µg of nuclear extracts, containing active DNMTs, to the unique cytosine-rich DNA substrate-coated ELISA plate and incubated for 60 min at 37°C. The methylated DNA can be recognized with anti-5-methylcytosine antibody. The amount of methylated DNA, which is proportional to enzyme activity, is calorimetrically quantified at 450 nm.
HDACs and HATs activity assays
Cultured MDA-MB-231 cells were harvested at the indicated time points and nuclear extracts were prepared using the nuclear extraction reagent (Pierce, Rockford, IL). The activities of HDACs (Active Motif, Carlsbad, CA) and HATs (Epigentek, Brooklyn, NY) were performed using the colorimetric kit according to the manufacturer's instruction as described previously [18]. The enzymatic activities of HDACs and HATs were detected by a microplate reader at 450 nm.
Chromatin immunoprecipitation (ChIP) analysis
Chromatin immunoprecipitation was performed using the EZ-ChIP kit according to the manufacturer's instructions (Upstate Biotechnology, Lake Placid, NY) as described previously [18]. MCF-7 cells were used as a positive control. Cells were cross-linked with 1% formaldehyde at 37°C for 10 min, washed twice with ice-cold PBS, re-suspended in SDS-lysis buffer (1% SDS, 50 mM Tris-HCL, pH 8.0, 10 mM EDTA and protease inhibitor cocktail), and then sonicated to an average length of sheered genomic DNA of approximately 400–1000 bp. The antibodies used in the ChIP assays were ChIP-validated acetyl-histone H3, acetyl-histone H3K9, acetyl-histone H4, trimethyl-histone H3K9 (Upstate Biotechnology), HDAC1, MeCP2, MBD1, SUV39H1 (Santa Cruz Biotechnology) and DNMT1 (Abcam, Cambridge, MA). A “no antibody” control was also used to evaluate-ChIP efficiency. ChIP-purified DNA was quantified by using quantitative-PCR (qPCR) using the Platinum SYBR Green detection system and q-PCR specific ERα primer (Invitrogen, Carlsbad, CA) as described earlier [18], [21]. The binding of various transcription factors to the ERα promoter was analyzed by standard PCR conditions as described previously [4]. Briefly, the ERα promoter primers were forward-5′-GAA CCG TCC GCA GCT CAA GAT C-3′, reverse-5′-GTC TGA CCG TAG ACC TGC GCG TTG-3′, with a total of 30 cycles at 94°C for 30 s, 56°C for 30 s, 72°C for 1 min and final extension was extended at 72°C for 5 min. After amplification, PCR products were separated on 1.5% agarose gels and visualized by ethidium bromide staining using Kodak 1D 3.6.1 image software and quantified. Quantitative data were analyzed by optical densitometry using ImageJ Software version 1.36b (http://rsb.info.nih.gov/ij/).
Bisulfite sequencing analysis
The DNA methylation status of the ERα promoter was assayed by sodium bisulfite methylation sequencing using the EpiTect-Bisulfite modification kit following the manufacture's protocol (Qiagen, Valencia, CA). Approximately 2 µg of genomic DNA was used for bisulfite modification and then amplified by PCR using Go Taq mix (Promega, Madison, WI). Primers and PCR-conditions were followed as described previously [4]. PCR amplified DNA was purified using the QIAquick PCR purification kit (Qiagen) and sequenced using the 3730 DNA Sequencer (Applied Biosystems, Foster City, CA). Percent methylation was calculated using the following formula: Number of methylated CpG×100/total number of CpGs assessed.
Small interfering RNA (siRNA) knockdown of ERα
Approximately 2.2×105 cells per well were placed in a 6-well plate and allowed to incubate overnight. The ERα siRNA (Santa Cruz Biotechnology) was made into 10 µM stock using nuclease free water and 9 nM siRNA was delivered to the cells using the Silencer siRNA Transfection kit (Ambion/Applied Biosystems, TX, USA) according to the manufacturer's instructions. siCONTROL Non-Targeting siRNA (Santa Cruz Biotechnology) was used as a negative control. Treatments incorporating GTPs and SFN with or without TAM were performed for an additional 72 h. Cells were harvested and checked for ERα knockdown after 3 days using western blot analysis.
Apoptosis assay
Breast cancer cells transfected with ERα and control siRNA transfected cells were treated with the indicated concentrations of GTPs and SFN with or without TAM for 72 h. The cells were then lysed with nuclei lysis buffer provided for apoptosis assays using the Cell Death Detection ELISA Kit (Roche, Palo Alto, CA) as described previously [23]. Briefly, the cytoplasmic histone/DNA fragments were extracted and incubated in microtiter plate modules coated with anti-histone antibody. Subsequently, the peroxidase-conjugated anti-DNA antibody was used for the detection of immobilized histone/DNA fragments, followed by color development with 2,2′-azinobis(3-ethylbenzo-thiazoline-6-sulfonic acid) substrate for peroxidase. The spectrophotometric absorbance of the samples was recorded using Microplate Reader (Bio-Rad Model 680, Hercules, CA) at 405 nm. Percent apoptosis was calculated using the formula: (100×treatment cell absorbance/control cell absorbance)−100.
Statistical analysis
The statistical significance of differences between the values of treated samples and controls were determined with Kruskal-Wallis with Dunn's post test using GraphPad Prism version 4.00 for Windows, GraphPad Software, San Diego, California, USA (www.graphpad.com). In each case, P<0.05 was considered statistically significant.
Results
GTPs and SFN synergistically inhibits cellular proliferation of ERα-negative breast cancer cells
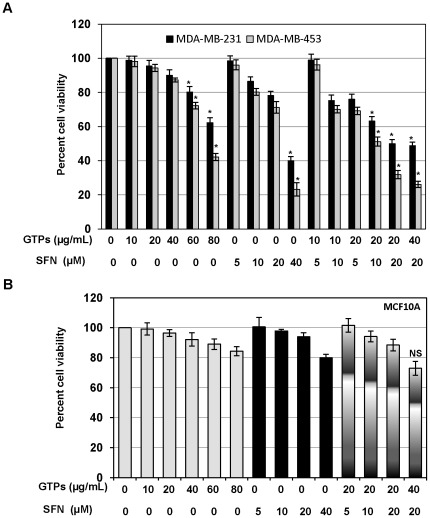
First to examine whether GTPs and SFN have any synergistic cellular proliferation inhibitory activity on human breast cancer cells, we performed cell viability assay with GTP and SFN alone or in combination treatments for 3 days. We intended to determine the optimal dose that will induce ERα transcriptional activation without causing cellular toxicity, thereby studying possible mechanisms involved in the ERα-reactivation in ERα-negative MDA-MB-231 cells. The MTT-cell viability assay was performed with the MDA-MB-231 and MDA-MB-453 cells treated with various concentrations of GTPs and SFN alone or in combinations as shown in Fig. 1A . We observed a dose-dependent cell growth inhibition with GTPs and SFN treatments in both ER-negative MDA-MB-453 and MDA-MB-231 cells, which became significant at 80 µg/ml and 40 µM, respectively. However, the combination of GTPs and SFN synergistically induced cell growth inhibition at 20 µg/ml GTPs and 10 µM SFN in these ERα-negative cells. The synergistic cell growth inhibitory effects were well pronounced at higher doses of GTPs and SFN. Further, MDA-MB-231 showed less cellular viability inhibitory effect than MDA-MB-453 at indicated GTPs and SFN doses, which might be due to the triple-negative in nature. Therefore, for further studies we chose MDA-MB-231 cells with minimum effect and triple-negative in nature; thereby we can study the mechanisms without toxicity. Control MCF10A cells were slightly inhibited in cell growth with combination of 40 µg/ml GTPs and 20 µM SFN after 3 days of treatment, indicating that higher combination doses might be toxic to the normal breast cells (Fig. 1B ). These results indicate that lower doses of combined GTPs and SFN selectively inhibit ERα-negative breast cancer cells; however, the optimal doses required for the transcriptional activation of ERα remained to be determined in our subsequent studies.
Figure 1. Combined GTPs and SFN synergistically inhibit cellular proliferation of ERα-negative breast cancer cells but has negligible effect on control MCF10A cells.
Human breast cancer MDA-MB-231 and MDA-MB-453 cells (panel A), and control MCF10A (panel B) cells were treated with varying concentrations of GTPs (0–80 µg/mL) and SFN (0–40 µM) as well as a combination of both the compounds for 3 days. Percent cell viability was obtained using MTT assay as described under Materials and Methods . Results were obtained from three independent experiments, mean ± SD. Statistical significance, *P<0.05.
GTPs and SFN activate ERα mRNA and protein expression in ERα-negative MDA-MB-231 breast cancer cells
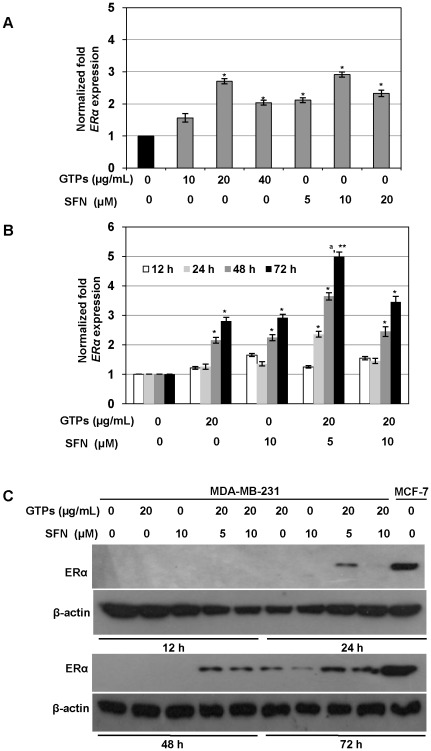
To determine the optimal dose of GTPs and SFN for the ERα reactivation, MDA-MB-231 cells were treated with increasing concentrations of GTPs and SFN for 3 days as shown in Fig. 2A . GTPs induced ERα mRNA expression at a concentration as low as 10 µg/ml, but the maximum significant effect was observed at 20 µg/ml. Similarly, SFN induced significant ERα expression starting from 5 µM and the maximum ERα reactivation was observed at 10 µM doses. Furthermore, in combination treatments, 20 µg/ml GTPs and 5 µM SFN were shown to induce optimal significant synergistic reactivation of ERα mRNA in ERα-negative MDA-MB-231 cells after 24 h of post-treatment compared to non-treated control (Fig. 2B ). However, although SFN at 10 µM alone achieved the maximum significant ERα reactivation in MDA-MB-231 cells, in combination with GTPs the optimal doses of SFN was found to be 5 µM (Fig. 2B ). Further, the combination of 20 µg/ml GTPs and 5 µM SFN at 72 h post-treatment with MDA-MB-231 cells induced a significant (P<0.001) ERα reactivation compared to the respective individual doses of GTPs and SFN at 72 h (Fig. 2B ). Western blot analysis showed that the combination of GTPs and SFN significantly reactivated ERα protein expression in ERα-negative MDA-MB-231 cells after 24 h of post-treatment (Fig. 2C ). The ERα reactivation was considerably higher at 72 h of post-combinational doses of 20 µg/ml GTPs and 5 µM SFN in MDA-MB-231 cells. These results indicated that the low concentrations of GTPs and SFN did not induce significant cellular toxicity, but induced transcriptional and translational reactivation of ERα expression in the ERα-negative human breast cancer cells. Based on these results, we therefore chose to use the concentrations of GTPs and SFN as optimized in Fig. 2 for subsequent experiments.
Figure 2. GTPs and SFN synergistically reactivated ERα expression in ERα-negative human breast cancer cells.
A) GTPs and SFN at indicated doses induced ERα re-expression in ERα-negative MDA-MB-231 human breast cancer cells after 72 h of post-treatments. B) Treatment with GTPs or SFN and a combination of both of the compounds at indicated doses induced ERα re-expression in ERα-negative human breast cancer cells. Relative mRNA levels of ERα in GTPs and SFN treated cells were quantified at 12, 24, 48 and 72 h using real-time PCR. Data are in triplicates from three independent experiments and were normalized to GAPDH. The values were plotted against control as relative fold induction ± SD. Significance against nontreated control, *P<0.05, **P<0.001; aP<0.05 against individual GTPs and SFN doses at the same time interval. C) ERα protein expression with the treatment of GTPs or SFN alone or in combination at 12, 24, 48 and 72 h in MDA-MB-231 cells. ERα-positive MCF-7 cells served as a positive control. Actin was used as an equal loading control.
GTPs and SFN altered epigenetic enzymes expression and their activity
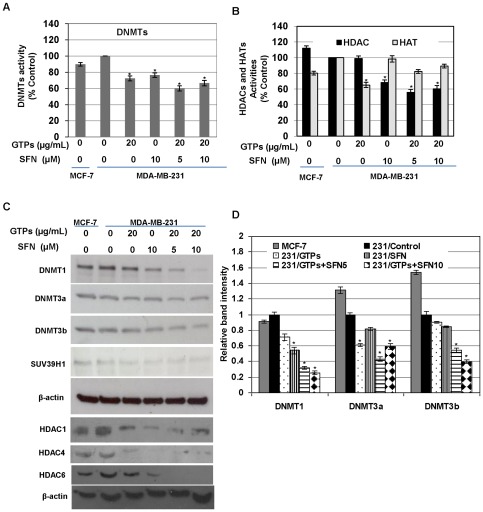
Previous studies have shown that ERα activation is associated with DNA methylation and histone modifications; we therefore assessed epigenetic-modulating enzymatic activity of the DNMTs (Fig. 3A ), HDACs (Fig. 3B ), HATs (Fig. 3B ) and DNMTs as well as HDACs expression (Fig. 3C ) in ERα-negative MDA-MB-231 breast cancer cells, using GTPs and SFN treatments. Interestingly, GTPs and SFN significantly reduced HATs and HDACs activities, respectively, at the optimal doses used as shown in Fig. 3B . This is in accordance with previous studies that EGCG has a HAT inhibitory activity, and SFN poses a HDACs inhibitory activity [16], [18], [24]. However, the combination of GTPs with SFN additively enhanced HDACs inhibitory activity of SFN in MDA-MB-231 cells but not HATs inhibitory activity. Further, HDAC expression analysis showed that the combination of GTPs with SFN considerably inhibits HDAC1, HDAC4 and HDAC6 expression in MDA-MB-231 cells (Fig. 3C ), in accordance with HDAC inhibitory activity observed in Fig. 3B . It is known that EGCG, an active compound present in GTPs, is a DNMTs inhibitor; similarly we have also observed that GTPs treatment considerably inhibited DNMTs activity and expression in MDA-MB-231 cells (Fig. 3A and 3D ). Interestingly, we found that SFN, a HDACs inhibitor, also inhibits DNMTs activity significantly in MDA-MB-231 cells (Fig. 3A ). The combinations of GTPs and SFN have more pronounced DNMTs inhibitory effects than when administrated separately. The combination doses mediated inhibition of DNMTs expression could be an important contributing factor in altering the binding of MBD-proteins at the ERα promoter. To our surprise, not only DNMTs but also histone methyltransferase, SUV39H1, is also inhibited by GTPs and SFN (Fig. 3C ). The GTPs- and SFN-mediated inhibition of DNMTs and SUV39H1 expressions could be an important contributing factor in facilitating demethylation of the ERα promoter, which leads to transcriptional activation of ERα expression [8], [25].
Figure 3. GTPs and SFN altered epigenetic enzymes expression and their activity in ERα-negative breast cancer cells.
Breast cancer MDA-MB-231 cells were treated with the indicated concentration of GTPs and SFN alone or in combinations of both for 3 days. Nuclear extracts were prepared and 20 µg of protein was used to estimate DNMTs (panel A), HDAC and HATs (panel B) activities using the colorimetric assay kit as described under Materials and Methods . Non-treated MCF-7 cells were used as a positive control. Values are representative of three independent experiments and represented as percent control ± SD; statistical significance against nontreated MDA-MB-231 control, *P<0.05. C) Effect of GTPs and SFN alone or in combinations of both on DNMTs, SUV39H1 and HDACs expression in ERα-negative human breast cancer MDA-MB-231 cells. Cell lysates were prepared at 3 days of post-treatments at the indicated doses followed by western blotting to analyze DNMTs (DNMT1, DNMT3a and DNMT3b), HDACs (HDAC1, HDAC4 and HDAC6) and SUV39H1 expression. Non-treated MCF-7 cells were used as a positive control. Actin was used as an equal loading control. D) Graphical representations are indicative of relative band intensity of DNMTs expression in MDA-MB-231 cells, normalized with β-actin. Values are mean band intensity of three independent blot ± SD.
GTPs and SFN altered histone modifications and DNA methylation of the ERα-promoter
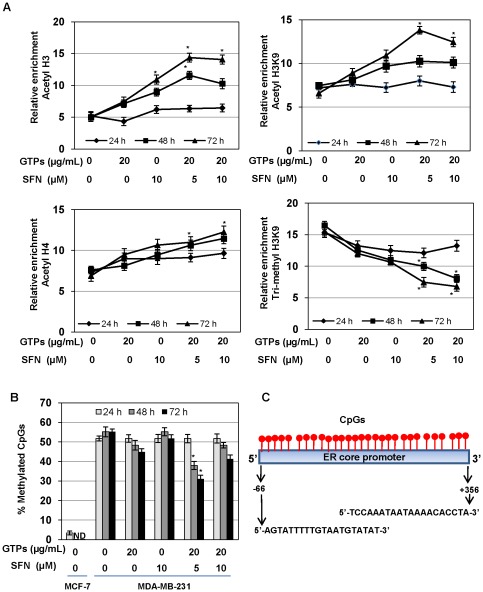
It is well known that histone modifications and DNA methylation play important roles in gene expression and regulation, especially in ERα activation in breast cancer cells [4], [25]. Our studies have shown that treatment with the GTPs and SFN significantly altered the activity as well as expression of epigenetic modulating enzymes in ERα-negative MDA-MB-231 cells, suggesting a potential role of histone modifications and DNA methylation in ERα regulations. Therefore, we sought to determine changes in histone modifications of the ERα promoter by GTPs and SFN treatment by different time intervals in MDA-MB-231 cells. We found that GTPs and SFN treatment can increase considerable enrichment of three histone acetylation chromatin markers, acetyl H3 (ac-H3), acetyl-H3 at lysine 9 (ac-H3K9) and ac-H4 in MDA-MB-231 cells after 48 h of post-treatment (Fig. 4A ). We also found a decrease in the methylation status of inactive histone markers such as trimethyl-H3 lysine 9 (tri-me-H3K9) in MDA-MB-231 cells with GTPs and SFN treatments (Fig. 4A ). Further, a significantly enriched level of histone acetylation and decreased tri-me-H3K9 was more pronounced in the combination treatments of GTPs and SFN at 48 h and 72 h, suggesting the importance of dietary combination-induced ERα-reactivation in ERα-negative breast cancer cells through histone modifications (Fig. 4A ). These changes of histone acetylation and deacetylation allow transcriptional factors binding into the ER regulatory region by maintaining a repressive environment [2], [7].
Figure 4. GTPs and SFN altered ERα promoter methylation and histone acetylations in MDA-MB-231 cells.
A) MDA-MB-231 cells were treated with GTPs and SFN alone, and in combinations as indicated for 24, 48 and 72 h. Histone modifications were analyzed by ChIP-qPCR using chromatin markers including acetyl-H3, trimethyl-H3K9, acetyl-H3K9 and acetyl-H4 in the promoter region of ERα. Mouse IgG antibody controls were assessed to verify the ChIP efficiency. The x-axis represents the different treatment groups, and the y-axis represents the relative enrichment of individual binding factors [the percentage of immunoprecipitates compared with the corresponding input samples (defined as 100)]. The experiment was repeated 3 times with triplicates in real-time PCR, and each point indicates the mean ± SD. Significance against nontreated MDA-MB-231 control, *P<0.05. B) GTPs and SFN altered ERα promoter DNA methylation in MDA-MB-231 breast cancer cells as assayed by bisulfite sequencing. MDA-MB-231 cells were treated with the indicated concentration of GTPs and SFN for 24 h, 48 h and 72 h. Percent methylation was obtained by dividing the number of methylated CpGs by the total number of CpGs (29) in the indicated ERα promoter region assessed. Values are representative of three independent experiments and are represented as percent control ± SD; statistical significance, *P<0.05. C) The ERα promoter region used for bisulfite sequencing is shown with PCR primer sequences, number of CpGs and the total amplification lengths.
Since the ERα promoter is mostly hypermethylated in ERα-negative breast cancer cells, we assessed the methylation status of the ERa promoter region from −66 to +356 covering 29 CpG dinucleotides and various overlapping transcription factor binding sites for 24 h, 48 h and 72 h (Fig. 4B ). We used bisulfite-sequencing to detect the ERa methylation patterns of GTPs and SFN treated MDA-MB-231 human breast cancer cells. Untreated MDA-MB-231 and MCF-7 cells served as controls. As shown in Fig. 4B , control MDA-MB-231 breast cancer cells maintain a high level of methylation at promoter sites at 54.02±2.36%. Although we found considerable inhibition in DNMTs expression levels with GTPs and SFN treatments, we did not find any significant changes in methylation status of the ERα promoter with GTPs and SFN treated MDA-MB-231 compared with untreated MDA-MB-231 cells (Fig. 4B ). This is in accordance with our previous study that EGCG treatment does not induce significant methylation changes in the CpG islands of the ERα promoter in MDA-MB-231 cells [4]. In contrast, combination treatments of GTPs with SFN significantly reduced ERα promoter methylation after 48 h post-treatment as we observed previously in combination of EGCG with Trichostatin A (TSA), a histone deacetylase inhibitor, in MDA-MB-231 cells [4]. Our results indicated that only combined treatments of GTPs and SFN can induce significant DNA hypomethylation at the ERα promoter. In summary, these results suggest that histone modifications and DNA methylation contribute a major role in GTPs and SFN induced ERα reactivation in ERα-negative human breast cancer cells.
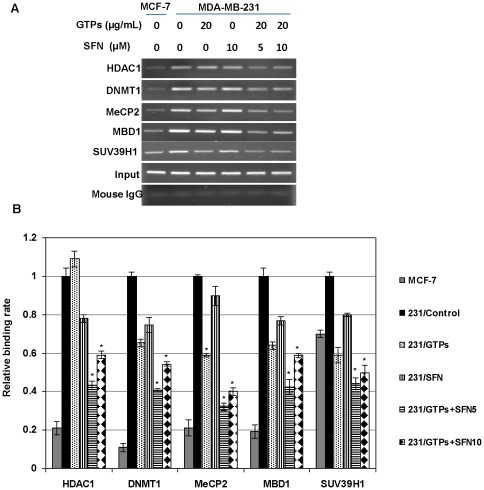
GTPs and SFN altered the binding of transcription complex to the ERα-promoter
Given the strong link between histone modification and DNA methylation, we asked whether GTP- and SFN-induced ERα re-expression is associated with reorganization of heterochromatin structure at the epigenetically regulated ERα promoter in MDA-MB-231 cells. Studies have shown that HDAC repressor complex, HDAC1 and DNMT1 involves gene silencing by recruiting co-repressor complexes to the methylated ERα promoter [26]. Studies have also shown that disruption of transcriptional repressor multi-molecular complex, HDAC1/DNMT1/SUV39H1, is associated with ERα transcriptional activation in ERα-negative breast cancer cells [8]. Therefore, ChIP-assays were performed to examine GTPs- and SFN-mediated changes in these transcriptional repressor complexes binding on the ERα promoter. As shown in Fig. 5A–B , GTPs and SFN can considerably lower the binding of these transcriptional repressor multi-molecular complexes to the ERα promoter and this effect was significant when treated with GTPs and SFN in combinations. Further, GTPs and SFN combination treatment also significantly disrupted binding of methyl-CpG binding domain (MBD) proteins, MeCP2 and MBD1, to the ERα promoter, might be due to the hypomethylation induced by GTPs and SFN at the ERα promoter (Fig. 5A–B ). Collectively, these data suggest that the binding alterations of transcriptional repressor complex to the ERα promoter contributed to the reactivation of ERα by the combination of dietary DNMT and HDAC inhibitors, GTPs and SFN, respectively.
Figure 5. GTPs and SFN altered binding of transcriptional factors to the ERα promoter in ERα-negative breast cancer cells.
A) MDA-MB-231 cells were treated with GTPs or SFN and a combination of both of the compounds as indicated for 3 days. Samples were prepared for ChIP-assay and analyzed for the binding of ERα transcription repressor proteins including HDAC1, DNMT1, MeCP2, MBD1 and SUV39H1 together with mouse IgG control. MCF-7 served as a positive control. PCR primers and conditions were used as described in Materials and Methods . Photographs are representative of an experiment that was repeated in triplicates. B) ChIP data were calculated from the corresponding DNA fragments amplified by PCR using Kodak 1D 3.6.1 image software; columns, mean; bars, SD; statistical significance, *P<0.05. The relative binding ratio was calculated as the ratio between the net intensity of each bound sample divided by the input and the untreated control sample divided by the input (bound/input)/(control/input).
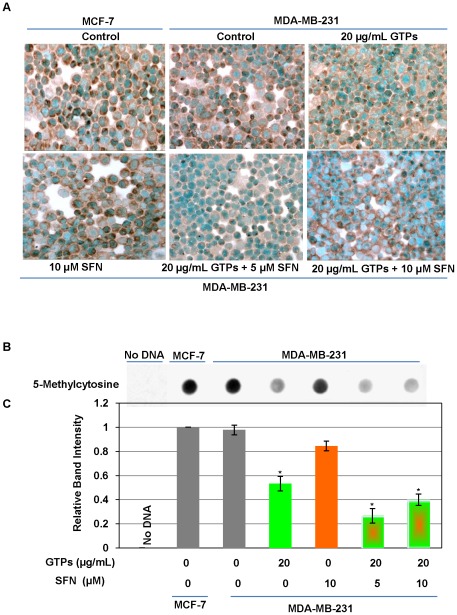
GTPs and SFN altered global DNA methylation in MDA-MB-231 cells
Studies have shown that EGCG can induce hypomethylation in various cell lines either by direct or indirect inhibition of DNA methyltransferases [13], [14], [27]. We previously discovered that SFN can also induce hypomethylation at the regulatory region of hTERT through inhibition of DNMTs expressions [18]. Therefore, we sought to determine the cause of GTPs and SFN treatment on global methylation status in ERα-negative MDA-MB-231 cells. We performed 5-methyl-cytosine (5-mC) immunostaining and dot-blot analysis to examine GTPs and SFN altered global methylation in MDA-MB-231 cells. As shown in Fig. 6A , GTPs-treatment considerably reduced 5-mC positive staining compared with control cells. The effect of demethylation was predominant in combination treatment of GTPs with SFN in MDA-MB-231 cells. The results were further semi-quantitatively analyzed by dot-blot analysis (Fig. 6B ). Treatment of GTPs resulted in a significant reduction in 5-mC levels in MDA-MB-231 cells compared with untreated MDA-MB-231 cells (Fig. 6C ). However, combined treatment of GTPs with SFN significantly reduced 5-mC expression in MDA-MB-231 cells, resulting in a synergistic global hypomethylation in CpG dinuclotides. These results suggest that GTPs may have a broad effect on DNA demethylation and this might be further accelerated by combination with SFN as observed in this study.
Figure 6. GTPs and SFN induced global hypomethylation in ERα-negative breast cancer cells.
MDA-MB-231 cells were treated with GTPs or SFN and combination of both the compounds as indicated for 3 days and analyzed for 5-methycytosine (5-mC). A) Immunocytochemical detection of DNA methylation using a 5-mC-specific antibody and counterstained with methyl green. 5-mC-positive staining is shown as dark brown. Magnification ×400. Photomicrographs are representative of three independent experiments. B) Cellular DNA was extracted and dot-blot analysis was performed for the presence of 5-mC. MCF-7 cells were used as a positive control and a no-DNA sample was used as a negative control. C) Graphical representations are indicative of relative band intensity of 5-mC expression in breast cancer cells as shown. Values are mean band intensity of three independent blot ± SD; statistical significance, *P<0.05.
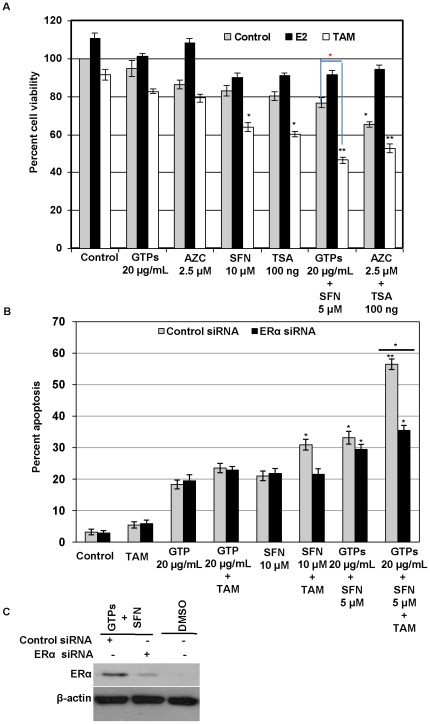
GTPs and SFN sensitized ERα-negative breast cancer cell to SERM through epigenetic reactivation of ERα
Collectively our aforementioned observations conclude that the combination of dietary DNMT and HDAC inhibitors, GTPs and SFN, respectively, epigenetically reactivates ERα expression in ERα-negative MDA-MB-231 cells. Furthermore, we sought to determine whether the ERα-reactivation could be used along with available SERMs such as tamoxifen therapy in hormonal refractory breast cancer. We therefore investigated the changes in cellular viability and apoptosis in ERα-negative MDA-MB-231 cells treated with GTPs and SFN alone or in combinations along with tamoxifen (TAM). As shown in Fig. 7A , untreated MDA-MB-231 cells showed an increased cellular proliferation with 17β-estradiol (E2), an ERα-ligand activator. Treatments with GTPs and 5-aza-2′-deoxycytidine (AZC), a DNMT inhibitor, alone or in combinations with TAM did not inhibit significant cellular proliferation in MDA-MB-231 cells, which is likely due to the limited ERα reactivations. However, MDA-MB-231 cells treated with SFN and TSA combined with TAM had significantly reduced cellular proliferation, likely due to the pronounced effect of histone modifications as well as DNA demethylation-mediated ERα activation in MDA-MB-231 cells. Furthermore, combined treatment of GTP and SFN with TAM showed a significantly greater effect in reducing cellular proliferation in ERα-negative MDA-MB-231 cells. GTPs and SFN combined with TAM had pronounced cellular proliferation inhibitory effect than the combination of synthetic DNMT inhibitor, AZC, and HDAC inhibitor, TSA, along with TAM (Fig. 7A ). It was found that MDA-MB-231 cells treated with 20 µg/mL GTPs and 10 µM SFN induced a considerable level of cellular apoptosis in both control as well as ERα-knockdown cells. Furthermore, combined treatment with GTP and SFN showed a significantly higher apoptosis in both control siRNA as well as ERα-knockdown siRNA cells (P<0.05) (Fig. 7B ). MDA-MB-231 cells treated with GTPs and SFN combined with TAM had significantly higher cellular apoptosis (P<0.01). Conversely, ERα knockdown MDA-MB-231 cells treated with GTPs and SFN combined with TAM induced a significantly lesser apoptosis than control siRNA treated MDA-MB-231 cells (P<0.05). This might be the fact that GTPs and SFN induced cellular apoptosis in MDA-MB-231 cells in both ERα-dependent as well as ERα-independent mechanisms. However, TAM required ERα-reactivation to induce significant level of apoptosis in ERα-negative MDA-MB-231 cells at the low dose used. Collectively, these results indicated that the combination of GTPs and SFN can induce functional ERα re-expression and re-sensitize ERα-negative breast cancer cells to available SERM, TAM, which could provide an extremely important clinical implication in potential application of combination of bioactive dietary supplements as a therapeutic strategy for hormonal refractory breast cancer.
Figure 7. Combined treatments of GTPs and SFN sensitize ERα-negative breast cancer cells for tamoxifen therapy.
MDA-MB-231 cells treated with GTPs and SFN together with tamoxifen induced cellular apoptosis and inhibited cellular proliferation. A) ERα-negative MDA-MB-231 cellular viability in response to estradiol (10 nM) or tamoxifen (1 µM) alone, or in combination with GTP and SFN for 3 days. For a comparison, DNMTs inhibitor, AZC (2.5 µM), and HDAC inhibitor, TSA (100 ng), also administrated to MDA-MB-231 cells. Cell viability was determined and plotted against percent control. Data were obtained from three independent experiments, mean ± SD. Statistical significance, *P<0.05; **P<0.01. B) Knockdown of ERα decreases GTPs- and SFN-sensitized, TAM-induced apoptosis in ERα-negative MDA-MB-231 cells. MDA-MB-231 cells were subjected to treatment with 9 nM ERα-siRNA or control-siRNA. Cells were further treated with GTPs and SFN in combinations with or without TAM as indicated for 72 h. The cells were lysed with nuclear lysis buffer and analyzed for apoptosis as described in Materials and Methods . Values are representative of three independent experiments. Significance, *P<0.05; **P<0.01. C) Effect of siRNA interference with ERα gene expression was assayed after 72 h using specific antibodies to ERα and β-actin by western blot analysis. For ERα reactivation, 20 µg/mL GTPs and 5 µM SFN were used for 72 hrs. Data shown are representative of the three separate experiments.
Discussion
Epigenetic regulation has attracted considerable interest as a molecular target for cancer prevention and therapy as well as a target of many bioactive dietary components. Growing evidence suggests that bioactive dietary components impact epigenetic processes often involved with silencing of tumor suppressor genes, activation of cell survival proteins and induction of cellular apoptosis in many types of cancer [4], [15], [28], [29]. GTPs and SFN have been found to have anti-cancer properties in various cancers through genetic and epigenetic mechanisms [11], [12], [14], [16]. The most abundant bioactive compound present in GTPs is catechins, which include (–)-epicatechin (EC), (–)-epicatechin-3-gallate (ECG), (–)-epigallocatechin (EGC) and (–)-epigallocatechin-3-gallate (EGCG) [12], [30]. Of these, EGCG accounts for more than 50% of the total polyphenol and effective content in green tea [31]. EGCG, a well studied green tea polyphenol, has DNMTs inhibitory activity, however, other catechins in GTPs such as (–)-epicatechin (EC), (–)-epicatechin-3-gallate (ECG) and (–)-epigallocatechin (EGC) have also been found to share similar properties although they are less efficient than EGCG [13], [32]. Therefore, the use of GTPs as a whole not only mimics the natural environment but also enhances its synergistic epigenetic activity against various cancers including breast cancer. Another bioactive dietary supplement used in this study is sulforaphane (SFN), an isothiocyanate naturally abundant in widely consumed cruciferous vegetables, found to have HDACs inhibitory activity [16], [17]. Therefore the focus of the current study is the use of combined dietary DNMT and HDAC inhibitors for the prevention and therapeutics of hormonal refractory breast cancer.
The epigenetics of ERα re-expression in ERα-negative breast cancer cells has been studied in many laboratories, including our laboratory, and has been of intense interest as a novel strategy for the treatment of hormonal refractory breast cancer [4], [7], [25]. Since ERα-negative tumors are difficult to treat with available SERMs due to the lack of hormonal receptor, it is very crucial to formulate a new treatment strategy for this type of hormonal refractory breast cancer. Many studies have advanced our knowledge that treatment with AZC, a DNMT inhibitor, and TSA, a HDAC inhibitor, can reactivate ER expression in ER-negative breast cancer cells, suggesting that epigenetic mechanisms play an important role in ERα transcriptional regulations [7], [9], [25]. However, the use of synthetic molecules might induce potential adverse side effects and higher cost. Therefore, the use of bioactive dietary supplements as DNMT and HDAC inhibitors for the reactivation of ER-expression in ER-negative breast cancer could greatly mimic more natural dietary milieu, reduced treatment cost and most importantly minimum adverse effects.
In the present study, we provided evidences that the combination of GTPs and SFN can induce re-expression of endogenous ERα in ERα-negative MDA-MB-231 human breast cancer cells. For the first time, our results demonstrate that the reactivation of ERα by GTPs and SFN is mediated, at least partly, through the epigenetic alterations in DNA methylation and chromatin remodeling in ERα gene promoter. Recent evidences suggest that epigenetic regulation is one of the most important molecular events associated with ERα silencing in ERα-negative breast cancers [2], [7], [9]. Recently, extensive studies have focused on EGCG, a major component in GTPs, mediated DNMTs and HAT inhibitory activity in various cancer cells, including breast cancer cells [13], [21], [24]. Besides direct inhibition of DNMT by EGCG, it was also reported that consumption of GTPs could lead to a decrease in available S-adenosyl-L-methionine (SAM) and an increase in S-adenosyl-L-homocysteine (SAH) and homocysteine levels, thereby providing evidence of an indirect inhibition of DNA methylation by EGCG/GTPs [27]. This conjecture is supported by animal studies demonstrating that GTPs consumption through drinking water can moderately decrease the level of SAM in the intestine [14]. Beside HDAC inhibitory activity of SFN, we also observed DNMTs inhibitory activity in human breast cancer cells. This is in accordance with earlier findings that SFN-treatment significantly inhibited HDAC activity and DNMTs expression in breast cancer cells; however, we did not find any significant alteration in HAT activity [18].
Several studies have reported that DNA methylation and histone acetylation play important roles in ER transcriptional regulation [7]–[9], [25]. Together, our results suggest that GTPs and SFN-induced down-regulation of DNMTs expression and histone modifications is not only causing a repressive environment at the ERα promoter but also altered the binding of transcriptional repressor complex at the ERα promoter. This is confirmed further with our ChIP-analysis that GTPs- and SFN-induced enrichment of transcriptional active chromatin markers such as acetylated histone H3, H3K9 and acetyl-H4 in ER-negative MDA-MB-231 cells, whereas chromatin inactive markers such as trimethyl-H3K9 was decreased. Importantly, we found that the histone H3K9 methyltransferase, SUV39H1, was released from the ER-promoter since presence of SUV39H1 has been shown to be crucial for maintenance of the H3-methylation and epigenetic control of heterochromatin assembly in cancer cells [33], [34]. In accordance, we found that GTPs- and SFN-mediated release of SUV39H1 protein from ER promoter leads to suppression of trimethyl-H3K9 methylation in ER-negative MDA-MB-231 cells.
Studies have shown that CpG methylation of the ERα promoter results in transcriptional ER silencing [26]. We found that bioactive dietary DNA demethylating and histone deacetylating agents such as GTPs and SFN can alter the binding of methyl-CpG binding proteins, DNMT and HDAC, which are actively involved in ERα transcriptional regulations [2], [7]. Further our results demonstrate that combinations of GTPs and SFN induced the release of co-repressor complexes to the demethylated ERα promoter and the disruption of transcriptional repressor multi-molecular complex, HDAC1/DNMT1/SUV39H1, is actively associated with ERα transcriptional activation in ERα-negative breast cancer cells [8]. Further, it is also reported that release of co-repressor complex leads to concomitant enrichment of ac-H3, ac-H3K9 and ac-H4 [7]. Besides gene specific DNA demethylations, we also observed a global DNA hypomethylation by GTPs and SFN in MDA-MB-231 cells. This might be due to the GTPs- and SFN-mediated DNMTs inhibition in these human breast cancer cells [15], [18], [21]. Taken together, it is apparent that DNMTs-induced promoter demethylation and HDAC-associated chromatin remodelling altered binding of transcriptional repressor multi-molecular complex, which is closely, linked to the ERα re-activation by GTPs and SFN in ERα-negative human breast cancer cells.
In our potential application study, we have clearly demonstrated that GTPs and SFN-mediated ERα-reactivation can be utilized for the treatment with available SERMs, tamoxifen in ERα-negative breast cancer cells. For the first time we demonstrated that the combination of bioactive dietary supplements, GTPs and SFN can reactivate ERα-expression in ERα-negative breast cancer cells through DNA demethylation and histone modifications associated epigenetic alterations. These findings are of importance not only for understanding epigenetic regulation of the ERα gene but also to provide evidence for the combined anticancer mechanism of bioactive dietary DNMT and HDAC inhibitors in cancer prevention and therapy.
In conclusion, our results suggest that the combination of dietary bioactive supplements GTPs and SFN could enhance the possible novel treatment strategy for hormonal refractory breast tumors. Further, epigenetic regulation of ERα re-activation by combination of GTPs and SFN could help in designing novel therapeutic strategies. However, further studies with in vivo transgenic models such as C3(1)/SV40 and Her2/neu are necessary to validate our observations during different stages of breast cancer progression. These in vivo mouse models can produce ER-negative breast tumors which closely resemble the development, progression and morphology of human breast tumors [35], [36]. These in vivo models can be manipulated to use for ER-reactivation studies by potential bioactive dietary supplements with more close resemblance to humans for the treatment of hormonal refractory breast cancer in combination with available SERMs.
Acknowledgments
We thank Mr. Amiya Ahmed and Ms. Isha Soni for their technical assistance. CSIR-CDRI communication Number-8240.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported in part by grants to TOT from the National Cancer Institute, National Institutes of Health (R01 CA129415) and the American Institute for Cancer Research. Part of the work was also supported by the Ministry of Health and Family Welfare, Government of India, India (SMM). Junior Research Fellowship grant from CSIR, Government of India, is also acknowledged (SS). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jemal A, Bray F Center MM, Ferlay J, Ward E, et al. CA Cancer J Clin. United States; 2011. Global cancer statistics. pp. 69–90. [DOI] [PubMed] [Google Scholar]
- 2.Saxena NK, Sharma D. Epigenetic Reactivation of Estrogen Receptor: Promising Tools for Restoring Response to Endocrine Therapy. Mol Cell Pharmacol. 2010;2:191–202. [PMC free article] [PubMed] [Google Scholar]
- 3.Ni M, Chen Y, Lim E, Wimberly H, Bailey ST, et al. Targeting androgen receptor in estrogen receptor-negative breast cancer. Cancer Cell. 2011;20:119–131. doi: 10.1016/j.ccr.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y, Yuan YY, Meeran SM, Tollefsbol TO. Synergistic epigenetic reactivation of estrogen receptor-alpha (ERalpha) by combined green tea polyphenol and histone deacetylase inhibitor in ERalpha-negative breast cancer cells. Mol Cancer. 2010;9:274. doi: 10.1186/1476-4598-9-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roodi N, Bailey LR, Kao WY, Verrier CS, Yee CJ, et al. Estrogen receptor gene analysis in estrogen receptor-positive and receptor-negative primary breast cancer. J Natl Cancer Inst. 1995;87:446–451. doi: 10.1093/jnci/87.6.446. [DOI] [PubMed] [Google Scholar]
- 6.Issa JP, Zehnbauer BA, Civin CI, Collector MI, Sharkis SJ, et al. The estrogen receptor CpG island is methylated in most hematopoietic neoplasms. Cancer Res. 1996;56:973–977. [PubMed] [Google Scholar]
- 7.Sharma D, Blum J, Yang X, Beaulieu N, Macleod AR, et al. Release of methyl CpG binding proteins and histone deacetylase 1 from the Estrogen receptor alpha (ER) promoter upon reactivation in ER-negative human breast cancer cells. Mol Endocrinol. 2005;19:1740–1751. doi: 10.1210/me.2004-0011. [DOI] [PubMed] [Google Scholar]
- 8.Macaluso M, Cinti C, Russo G, Russo A, Giordano A. pRb2/p130-E2F4/5-HDAC1-SUV39H1-p300 and pRb2/p130-E2F4/5-HDAC1-SUV39H1-DNMT1 multimolecular complexes mediate the transcription of estrogen receptor-alpha in breast cancer. Oncogene. 2003;22:3511–3517. doi: 10.1038/sj.onc.1206578. [DOI] [PubMed] [Google Scholar]
- 9.Yang X, Phillips DL, Ferguson AT, Nelson WG, Herman JG, et al. Synergistic activation of functional estrogen receptor (ER)-alpha by DNA methyltransferase and histone deacetylase inhibition in human ER-alpha-negative breast cancer cells. Cancer Res. 2001;61:7025–7029. [PubMed] [Google Scholar]
- 10.Jang ER, Lim SJ, Lee ES, Jeong G, Kim TY, et al. The histone deacetylase inhibitor trichostatin A sensitizes estrogen receptor alpha-negative breast cancer cells to tamoxifen. Oncogene. 2004;23:1724–1736. doi: 10.1038/sj.onc.1207315. [DOI] [PubMed] [Google Scholar]
- 11.Cheung KL, Kong AN. Molecular targets of dietary phenethyl isothiocyanate and sulforaphane for cancer chemoprevention. AAPS J. 2010;12:87–97. doi: 10.1208/s12248-009-9162-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mukhtar H, Ahmad N. Tea polyphenols: prevention of cancer and optimizing health. Am J Clin Nutr 71: 1698S–1702S; discussion. 2000;1703S–1694S doi: 10.1093/ajcn/71.6.1698S. [DOI] [PubMed] [Google Scholar]
- 13.Fang M, Wang Y, Ai N, Hou Z, Sun Y, et al. Tea polyphenol (-)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 2003;63:7563–7570. [PubMed] [Google Scholar]
- 14.Fang M, Chen D, Yang C. Dietary polyphenols may affect DNA methylation. J Nutr. 2007;137:223S–228S. doi: 10.1093/jn/137.1.223S. [DOI] [PubMed] [Google Scholar]
- 15.Nandakumar V, Vaid M, Katiyar SK. (-)-Epigallocatechin-3-gallate reactivates silenced tumor suppressor genes, Cip1/p21 and p16INK4a, by reducing DNA methylation and increasing histones acetylation in human skin cancer cells. Carcinogenesis. 2011;32:537–544. doi: 10.1093/carcin/bgq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Myzak MC, Karplus PA, Chung FL, Dashwood RH. A novel mechanism of chemoprotection by sulforaphane: inhibition of histone deacetylase. Cancer Res. 2004;64:5767–5774. doi: 10.1158/0008-5472.CAN-04-1326. [DOI] [PubMed] [Google Scholar]
- 17.Nian H, Delage B, Ho E, Dashwood R. Modulation of histone deacetylase activity by dietary isothiocyanates and allyl sulfides: studies with sulforaphane and garlic organosulfur compounds. Environ Mol Mutagen. 2009;50:213–221. doi: 10.1002/em.20454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meeran SM, Patel SN, Tollefsbol TO. Sulforaphane causes epigenetic repression of hTERT expression in human breast cancer cell lines. PLoS One. 2010;5:e11457. doi: 10.1371/journal.pone.0011457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Golubovskaya V, Virnig C, Cance W. TAE226-induced apoptosis in breast cancer cells with overexpressed Src or EGFR. Mol Carcinog. 2008;47:222–234. doi: 10.1002/mc.20380. [DOI] [PubMed] [Google Scholar]
- 20.Ciftci K, Su J, Trovitch P. Growth factors and chemotherapeutic modulation of breast cancer cells. J Pharm Pharmacol. 2003;55:1135–1141. doi: 10.1211/002235703322277177. [DOI] [PubMed] [Google Scholar]
- 21.Meeran SM, Patel SN, Chan TH, Tollefsbol TO. A novel prodrug of epigallocatechin-3-gallate: differential epigenetic hTERT repression in human breast cancer cells. Cancer Prev Res (Phila) 2011;4:1243–1254. doi: 10.1158/1940-6207.CAPR-11-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meeran SM, Mantena SK, Meleth S, Elmets CA, Katiyar SK. Interleukin-12-deficient mice are at greater risk of UV radiation-induced skin tumors and malignant transformation of papillomas to carcinomas. Mol Cancer Ther. 2006;5:825–832. doi: 10.1158/1535-7163.MCT-06-0003. [DOI] [PubMed] [Google Scholar]
- 23.Meeran SM, Katiyar S, Katiyar SK. Berberine-induced apoptosis in human prostate cancer cells is initiated by reactive oxygen species generation. Toxicol Appl Pharmacol. 2008;229:33–43. doi: 10.1016/j.taap.2007.12.027. [DOI] [PubMed] [Google Scholar]
- 24.Choi KC, Jung MG, Lee YH, Yoon JC, Kwon SH, et al. Epigallocatechin-3-gallate, a histone acetyltransferase inhibitor, inhibits EBV-induced B lymphocyte transformation via suppression of RelA acetylation. Cancer Res. 2009;69:583–592. doi: 10.1158/0008-5472.CAN-08-2442. [DOI] [PubMed] [Google Scholar]
- 25.Zhou Q, Atadja P, Davidson NE. Histone deacetylase inhibitor LBH589 reactivates silenced estrogen receptor alpha (ER) gene expression without loss of DNA hypermethylation. Cancer Biol Ther. 2007;6:64–69. doi: 10.4161/cbt.6.1.3549. [DOI] [PubMed] [Google Scholar]
- 26.Vaute O, Nicolas E, Vandel L, Trouche D. Functional and physical interaction between the histone methyl transferase Suv39H1 and histone deacetylases. Nucleic Acids Res. 2002;30:475–481. doi: 10.1093/nar/30.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee W, Zhu B. Inhibition of DNA methylation by caffeic acid and chlorogenic acid, two common catechol-containing coffee polyphenols. Carcinogenesis. 2006;27:269–277. doi: 10.1093/carcin/bgi206. [DOI] [PubMed] [Google Scholar]
- 28.Landis-Piwowar KR, Milacic V, Dou QP. Relationship between the methylation status of dietary flavonoids and their growth-inhibitory and apoptosis-inducing activities in human cancer cells. J Cell Biochem. 2008;105:514–523. doi: 10.1002/jcb.21853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Majid S, Kikuno N, Nelles J, Noonan E, Tanaka Y, et al. Genistein induces the p21WAF1/CIP1 and p16INK4a tumor suppressor genes in prostate cancer cells by epigenetic mechanisms involving active chromatin modification. Cancer Res. 2008;68:2736–2744. doi: 10.1158/0008-5472.CAN-07-2290. [DOI] [PubMed] [Google Scholar]
- 30.Graham H. Green tea composition, consumption, and polyphenol chemistry. Prev Med. 1992;21:334–350. doi: 10.1016/0091-7435(92)90041-f. [DOI] [PubMed] [Google Scholar]
- 31.Lin J, Liang Y. Cancer chemoprevention by tea polyphenols. Proc Natl Sci Counc Repub China B. 2000;24:1–13. [PubMed] [Google Scholar]
- 32.Lee W, Shim J, Zhu B. Mechanisms for the inhibition of DNA methyltransferases by tea catechins and bioflavonoids. Mol Pharmacol. 2005;68:1018–1030. doi: 10.1124/mol.104.008367. [DOI] [PubMed] [Google Scholar]
- 33.Shi L, Sun L, Li Q, Liang J, Yu W, et al. Histone demethylase JMJD2B coordinates H3K4/H3K9 methylation and promotes hormonally responsive breast carcinogenesis. Proc Natl Acad Sci U S A. 2011;108:7541–7546. doi: 10.1073/pnas.1017374108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 35.Green JE, Shibata MA, Yoshidome K, Liu ML, Jorcyk C, et al. The C3(1)/SV40 T-antigen transgenic mouse model of mammary cancer: ductal epithelial cell targeting with multistage progression to carcinoma. Oncogene. 2000;19:1020–1027. doi: 10.1038/sj.onc.1203280. [DOI] [PubMed] [Google Scholar]
- 36.Rossi C, Di Lena A, La Sorda R, Lattanzio R, Antolini L, et al. Intestinal tumour chemoprevention with the antioxidant lipoic acid stimulates the growth of breast cancer. Eur J Cancer. 2008;44:2696–2704. doi: 10.1016/j.ejca.2008.08.021. [DOI] [PubMed] [Google Scholar]