Abstract
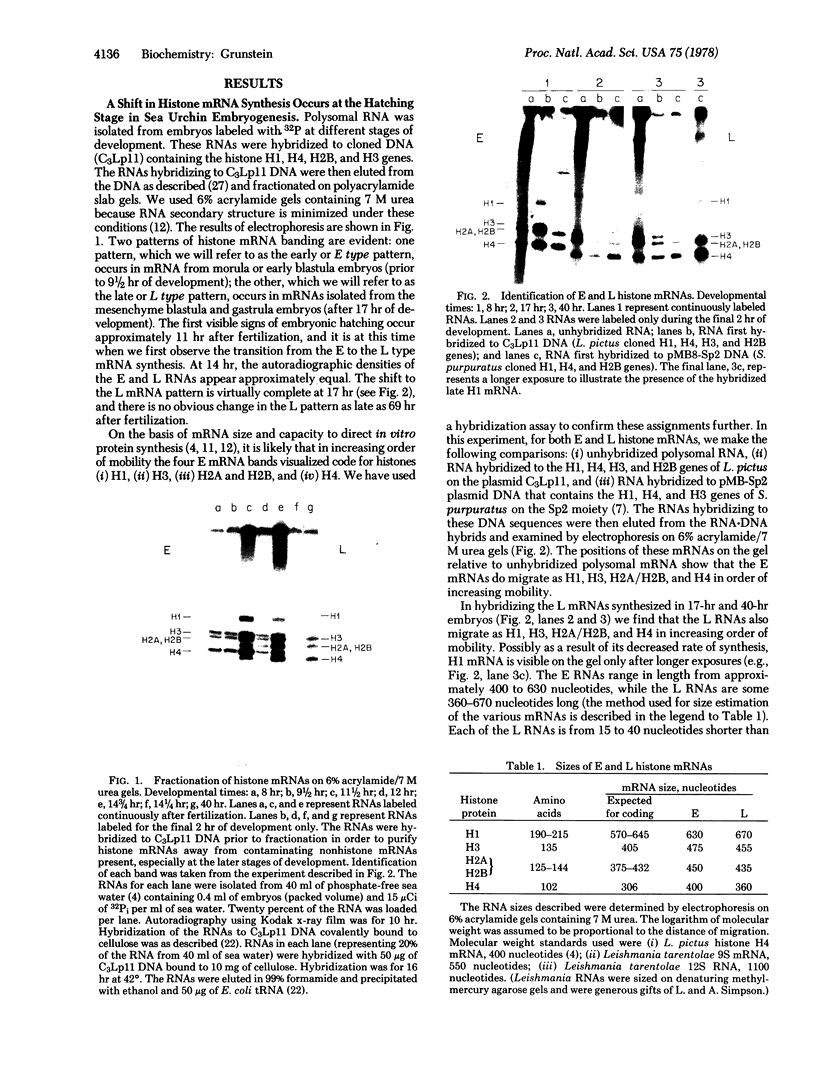
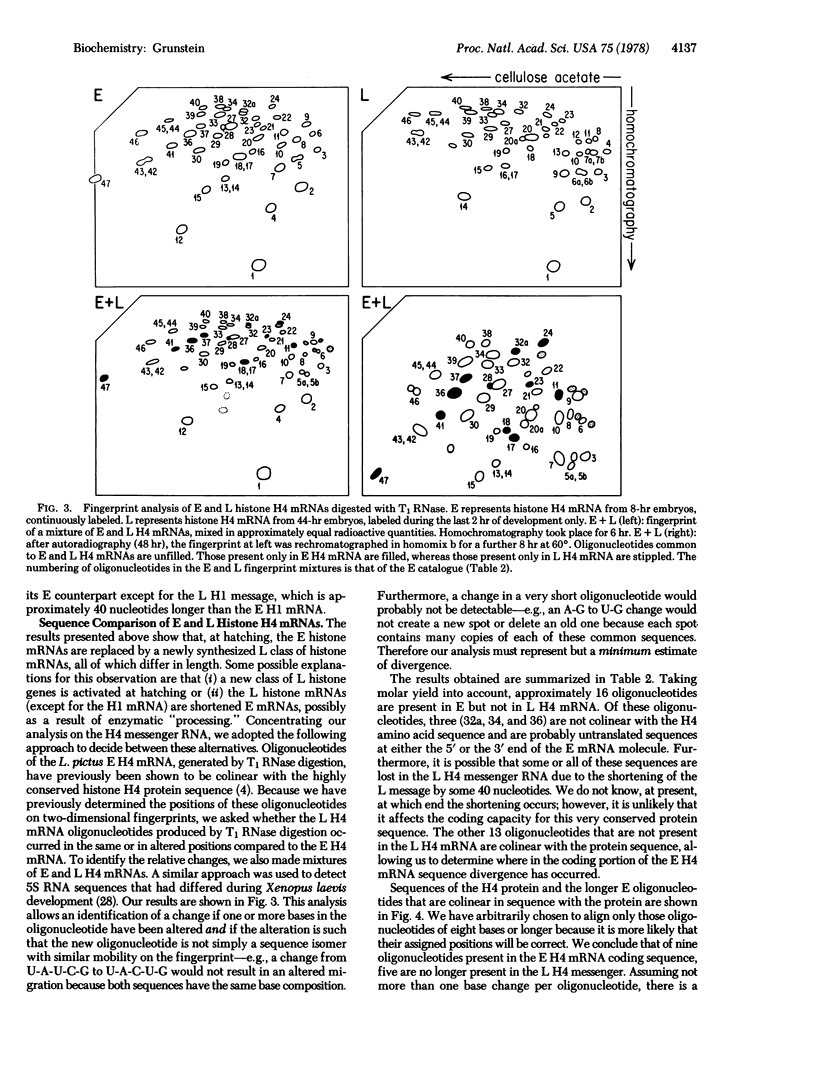
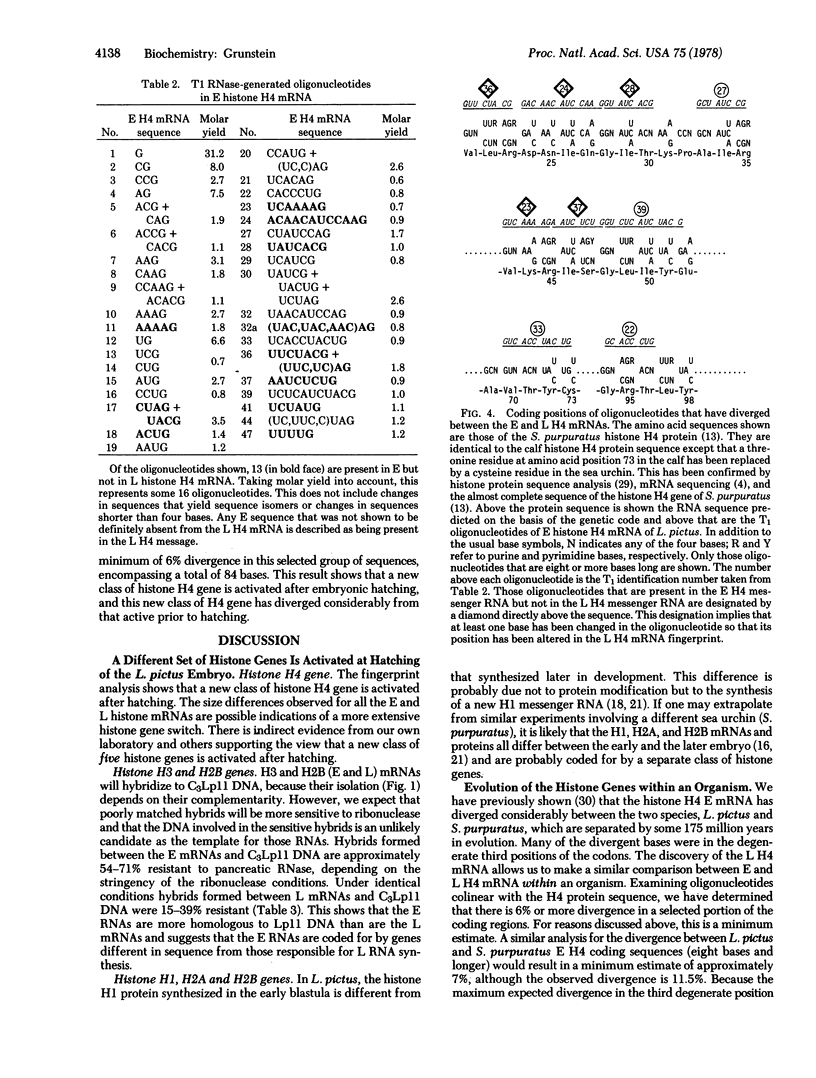
There is a distinct shift in histone mRNA synthesis at approximately 11--12 hr of sea urchin emhryogenesis, coincident with embryonic hatching. The synthesis of the blastula type (early) histone mRNAs gradually ceases at this stage and a new class of posthatching (late) histone mRNAs is produced. Briefly labeled early and late mRNAs were isolated and identified by means of RNA-DNA hybridization to different cloned histone genes. The late histone HI mRNA is approximately 40 nucleotides longer than the early HI mRNA. The H3, H2A, H2B, and H4 late mRNAs are 15--40 nucleotides shorter than their early counterparts. We present sequence evidence to show that the genes coding for the late H4 mRNA are a separate class from those that code for the early histone H4 message.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arceci R. J., Senger D. R., Gross P. R. The programmed switch in lysine-rich histone synthesis at gastrulation. Cell. 1976 Sep;9(1):171–178. doi: 10.1016/0092-8674(76)90062-3. [DOI] [PubMed] [Google Scholar]
- Birnstiel M., Telford J., Weinberg E., Stafford D. Isolation and some properties of the genes coding for histone proteins. Proc Natl Acad Sci U S A. 1974 Jul;71(7):2900–2904. doi: 10.1073/pnas.71.7.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Brown D. D., Wensink P. C., Jordan E. A comparison of the ribosomal DNA's of Xenopus laevis and Xenopus mulleri: the evolution of tandem genes. J Mol Biol. 1972 Jan 14;63(1):57–73. doi: 10.1016/0022-2836(72)90521-9. [DOI] [PubMed] [Google Scholar]
- Cohen L. H., Newrock K. M., Zweidler A. Stage-specific switches in histone synthesis during embryogenesis of the sea urchin. Science. 1975 Dec 5;190(4218):994–997. doi: 10.1126/science.1237932. [DOI] [PubMed] [Google Scholar]
- Cohn R. H., Lowry J. C., Kedes L. H. Histone genes of the sea urchin (S. purpuratus) cloned in E coli: order, polarity, and strandedness of the five histone-coding and spacer regions. Cell. 1976 Sep;9(1):147–161. doi: 10.1016/0092-8674(76)90060-x. [DOI] [PubMed] [Google Scholar]
- DeLange R. J., Fambrough D. M., Smith E. L., Bonner J. Calf and pea histone IV. II. The complete amino acid sequence of calf thymus histone IV; presence of epsilon-N-acetyllysine. J Biol Chem. 1969 Jan 25;244(2):319–334. [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford P. J., Southern E. M. Different sequences for 5S RNA in kidney cells and ovaries of Xenopus laevis. Nat New Biol. 1973 Jan 3;241(105):7–12. doi: 10.1038/newbio241007a0. [DOI] [PubMed] [Google Scholar]
- Glover D. M., White R. L., Finnegan D. J., Hogness D. S. Characterization of six cloned DNAs from Drosophila melanogaster, including one that contains the genes for rRNA. Cell. 1975 Jun;5(2):149–157. doi: 10.1016/0092-8674(75)90023-9. [DOI] [PubMed] [Google Scholar]
- Gross K., Probst E., Schaffner W., Birnstiel M. Molecular analysis of the histone gene cluster of Psammechinus miliaris: I. Fractionation and identification of five individual histone mRNAs. Cell. 1976 Aug;8(4):455–469. doi: 10.1016/0092-8674(76)90213-0. [DOI] [PubMed] [Google Scholar]
- Grunstein M., Grunstein J. E. The histone H4 gene of Strongylocentrotus purpuratus: DNA and mRNA sequences at the 5' end. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):1083–1092. doi: 10.1101/sqb.1978.042.01.109. [DOI] [PubMed] [Google Scholar]
- Grunstein M., Levy S., Schedl P., Kedes L. Messenger RNAs for individual histone proteins: fingerprint analysis and in vitro translation. Cold Spring Harb Symp Quant Biol. 1974;38:717–724. doi: 10.1101/sqb.1974.038.01.077. [DOI] [PubMed] [Google Scholar]
- Grunstein M., Schedl P. Isolation and sequence analysis of sea urchin (Lytechinus pictus) histone H4 messenger RNA. J Mol Biol. 1976 Jun 25;104(2):323–349. doi: 10.1016/0022-2836(76)90275-8. [DOI] [PubMed] [Google Scholar]
- Grunstein M., Schedl P., Kedes L. Sequence analysis and evolution of sea urchin (Lytechinus pictus and Strongylocentrotus purpuratus) histone H4 messenger RNAs. J Mol Biol. 1976 Jun 25;104(2):351–369. doi: 10.1016/0022-2836(76)90276-x. [DOI] [PubMed] [Google Scholar]
- Kedes L. H., Birnstiel M. L. Reiteration and clustering of DNA sequences complementary to histone messenger RNA. Nat New Biol. 1971 Apr 7;230(14):165–169. doi: 10.1038/newbio230165a0. [DOI] [PubMed] [Google Scholar]
- Kedes L. H., Chang A. C., Houseman D., Cohen S. N. Isolation of histone genes from unfractionated sea urchin DNA by subculture cloning in E. coli. Nature. 1975 Jun 12;255(5509):533–538. doi: 10.1038/255533a0. [DOI] [PubMed] [Google Scholar]
- Kedes L. H., Gross P. R. Identification in cleaving embryos of three RNA species serving as templates for the synthesis of nuclear proteins. Nature. 1969 Sep 27;223(5213):1335–1339. doi: 10.1038/2231335a0. [DOI] [PubMed] [Google Scholar]
- Kimura M. Genetic variability maintained in a finite population due to mutational production of neutral and nearly neutral isoalleles. Genet Res. 1968 Jun;11(3):247–269. doi: 10.1017/s0016672300011459. [DOI] [PubMed] [Google Scholar]
- Levy S., Wood P., Grunstein M., Kedes L. Individual histone messenger RNAs: identification by template activity. Cell. 1975 Mar;4(3):239–248. doi: 10.1016/0092-8674(75)90171-3. [DOI] [PubMed] [Google Scholar]
- Newrock K. M., Alfageme C. R., Nardi R. V., Cohen L. H. Histone changes during chromatin remodeling in embryogenesis. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):421–431. doi: 10.1101/sqb.1978.042.01.045. [DOI] [PubMed] [Google Scholar]
- Noyes B. E., Stark G. R. Nucleic acid hybridization using DNA covalently coupled to cellulose. Cell. 1975 Jul;5(3):301–310. doi: 10.1016/0092-8674(75)90105-1. [DOI] [PubMed] [Google Scholar]
- Ruderman J. V., Baglioni C., Gross P. R. Histone mRNA and histone synthesis during embryogenesis. Nature. 1974 Jan 4;247(5435):36–38. doi: 10.1038/247036a0. [DOI] [PubMed] [Google Scholar]
- Ruderman J. V., Gross P. R. Histones and histone synthesis in sea urchin development. Dev Biol. 1974 Feb;36(2):286–298. doi: 10.1016/0012-1606(74)90052-9. [DOI] [PubMed] [Google Scholar]
- Schaffner W., Gross K., Telford J., Birnstiel M. Molecular analysis of the histone gene cluster of psammechinus miliaris: II. The arrangement of the five histone-coding and spacer sequences. Cell. 1976 Aug;8(4):471–478. doi: 10.1016/0092-8674(76)90214-2. [DOI] [PubMed] [Google Scholar]
- Smith G. P. Unequal crossover and the evolution of multigene families. Cold Spring Harb Symp Quant Biol. 1974;38:507–513. doi: 10.1101/sqb.1974.038.01.055. [DOI] [PubMed] [Google Scholar]
- Strickland M., Strickland W. N., Brandt W. F., Von Holt C. Sequence of the cysteine-containing portion of histone F2al from the sea urchin Parechinus angulosus. FEBS Lett. 1974 Apr 1;40(2):346–348. doi: 10.1016/0014-5793(74)80260-7. [DOI] [PubMed] [Google Scholar]
- Strickland M., Strickland W. N., Brandt W. F., Von Holt C. The complete amino-acid sequence of histone H2B(1) from sperm of the sea urchin Parechinus angulosus. Eur J Biochem. 1977 Jul 15;77(2):263–275. doi: 10.1111/j.1432-1033.1977.tb11665.x. [DOI] [PubMed] [Google Scholar]
- Strickland W. N., Strickland M., Brandt W. F., Von Holt C. The complete amino-acid sequence of histone H2B(2) from sperm of the sea urchin Parechinus angulosus. Eur J Biochem. 1977 Jul 15;77(2):277–286. doi: 10.1111/j.1432-1033.1977.tb11666.x. [DOI] [PubMed] [Google Scholar]
- Weinberg E. S., Birnstiel M. L., Purdom I. F., Williamson R. Genes coding for polysomal 9S RNA of sea urchins: conservation and divergence. Nature. 1972 Nov 24;240(5378):225–228. doi: 10.1038/240225a0. [DOI] [PubMed] [Google Scholar]
- Weinberg E. S., Overton G. C., Hendricks M. B., Newrock K. M., Cohen L. H. Histone gene heterogeneity in the sea urchin Strongylocentrotus purpuratus. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):1093–1100. doi: 10.1101/sqb.1978.042.01.110. [DOI] [PubMed] [Google Scholar]
- Wu M., Holmes D. S., Davidson N., Cohn R. H., Kedes L. H. The relative positions of sea urchin histone genes on the chimeric plasmids pSp2 and pSp17 as studied by electronmicroscopy. Cell. 1976 Sep;9(1):163–169. doi: 10.1016/0092-8674(76)90061-1. [DOI] [PubMed] [Google Scholar]