Abstract
The lack of technologies that combine automated manipulation, sorting, as well as immobilization of single metazoan embryos remains the key obstacle to high-throughput organism-based ecotoxicological analysis and drug screening routines. Noticeably, the major obstacle hampering the automated trapping and arraying of millimetre-sized embryos on chip-based devices is their substantial size and mass, which lead to rapid gravitational-induced sedimentation and strong inertial forces. In this work, we present a comprehensive mechanistic and design rationale for manipulation and passive trapping of individual zebrafish embryos using only hydrodynamic forces. We provide evidence that by employing innovative design features, highly efficient hydrodynamic positioning of large embryos on a chip can be achieved. We also show how computational fluid dynamics-guided design and the Lagrangian particle tracking modeling can be used to optimize the chip performance. Importantly, we show that rapid prototyping and medium scale fabrication of miniaturized devices can be greatly accelerated by combining high-speed laser prototyping with replica moulding in poly(dimethylsiloxane) instead of conventional photolithography techniques. Our work establishes a new paradigm for chip-based manipulation of large multicellular organisms with diameters well above 1 mm and masses often exceeding 1 mg. Passive docking of large embryos is an attractive alternative to provide high level of automation while alleviating potentially deleterious effects associated with the use of active chip actuation. This greatly expands the capabilities of bioanalyses performed on small model organisms and offers numerous and currently inaccessible laboratory automation advantages.
INTRODUCTION
Advances in physics, electronics, and material sciences have recently led to the development of a plethora of miniaturized bioanalytical systems collectively known as Lab-on-a-Chip (LOC).1, 2, 3 Implementation of such miniaturized systems for in-situ analysis of small model organisms and embryos (both invertebrate and vertebrate) is attracting a mounting interest.2, 4 Despite spectacular progress in the development of LOC systems for the automated manipulation of micron-sized model organisms such as nematodes (Caenorhabditis elegans, Panagrolaimus davidi) and fruit fly (Drosophila melanogaster), there is, however, a noticeable lack of technologies capable of handling millimetre scale organisms such as fish and amphibian embryos/larvae.2, 5, 6, 7 Recent reports provided notable proof-of-concept that fish embryos can fully develop in a confined microfluidic environment and that LOC devices hold a substantial promise for miniaturized toxicological analysis.8, 9 These simple chip-based systems were, however, still based on a manual dispensing and loading of embryos on chip-based devices using conventional liquid handling. Such procedures are time consuming and error prone, limiting reproducibility and throughput of bioassays. There is, thus, a great need to develop innovative integrated manifolds to leverage automated loading, transport, positioning, and long-term immobilization of large millimetre scale embryos in ecotoxicology, drug discovery, and reproductive medicine.2, 10, 11
Millimetre-sized metazoan organisms like fish embryos are, however, not easily susceptible to laboratory automation.2, 10, 11, 12 The major obstacle hampering their manipulation on chip-based devices is their substantial diameter (above 1 mm) and mass (above 1 mg), which both lead to rapid gravitational-induced sedimentation and high inertial forces.11 Furthermore, the prototyping and fabrication of millimetre scale devices that can accommodate such large particles is very difficult and time consuming using the standard UV lithography due to the inherent processing time required to obtain pattern definition of the photoresist structures.13 As a result, only a handful of very thick channels with millimetre depth have been reliably fabricated. X-ray lithography and the LIGA (LIthographie, Galvanoformung, und Abformung) remain in this regard the preferred methods for fabricating ultra-thick structures.13, 14, 15 Both technologies are, however, very expensive and require extensive access to clean room environment.16 More recently, interesting alternatives to the lithographic technologies have emerged. These can accelerate the prototyping of microfluidic chip-based technologies and encompass the computer numeric controlled (CNC) mechanical micro-milling,17, 18 desktop digital craft cutting,19 water-jet machining,18 and a plethora of laser based direct micromachining techniques to directly ablate microfluidic channels in quartz, glass, or plastic substrates.15, 20, 21, 22, 23, 24 Conventional micro-milling has reportedly achieved minimum resolution of 200 μm.18 The problems associated with CNC milling involve, however, substantial roughness of the substrata following the processing, generation of significant amounts of debris, and lack of commonly available tools that could facilitate the fabrication of features of less than 100 μm in diameter. As such, mechanical micro-milling has mostly been restricted to direct write procedures and fabrication of simple channels. The mechanical micro-milling is also relatively complex, more expensive, and less user friendly as compared to the laser fabrication techniques. This stems from a great degree of automation incorporated in contemporary laser-based technologies.23 Laser fabrication systems range from simple CO2-based laser systems to sophisticated UV-lasers, such as excimer lasers and frequency-doubled solid-state laser that can achieve resolution down to 1 μm.23 As opposed to the UV lasers, CO2-lasers operate in the infrared range at 10.6 μm wavelength and ablate the material photo-thermally.15, 23, 24 The advantages of using CO2-laser micromachining include its sheer simplicity and common availability as desktop cutters/engravers. The latter feature fully integrated programmable CNC control and can rapidly fabricate complex features with minimal operator engagement.15, 22, 24 Regulating the laser power, velocity, and the number of laser beam pulses and passes can easily control the relative depth of microstructures.22 This provides a greater degree of control over fabrication process than mechanical milling. The tolerances achieved by laser-based methods are also superior to mechanical milling and even standard CO2 laser cutter can achieve accuracy of up to 3 μm and laser beam spot of 40 μm. Such features cannot be achieved on mechanical micro-milling systems. Recently, PMMA has become a material of choice for direct CO2-laser micromachining of single-use disposable prototypes. The CO2 laser machining in PMMA generates, however, some degree of surface roughness but usually less than 5–10 μm.22, 23 The latter can be reportedly eliminated by thermal annealing process.22
The direct write techniques described above such as micro-milling or direct laser ablation in polymeric substrata were so far limited to only very small channels and suffered from inapplicability to inexpensive replica molding techniques in soft, biocompatible polymers.22 In this work, we show that rapid prototyping and medium scale fabrication of ultra thick devices can be greatly accelerated by combining inexpensive high-speed laser prototyping with replica moulding in poly(dimethylsiloxane) (PDMS) instead of conventional photolithography techniques.25 This technique allows to replicate multiple PDMS prototypes from one laser-machined PMMA master.
We also present a comprehensive design rationale for manipulation and passive trapping of individual zebrafish embryos under hydrodynamic forces. Hydrodynamic techniques have been reportedly used to manipulate and trap single cells and microdroplets with diameters up to 50 μm.26, 27, 28, 29, 30 Recently, Tan and Takeuchi have reported an innovative principle of hydrodynamic immobilization of micrometer-sized polystyrene and alginate hydrogel beads using simple fluidic resistance method called μ-fluidic trap.29, 30 The μ-fluidic trap incorporated a unique geometry designed with lower flow resistance of the main channel as compared to the loop (trapping) channel. The differential resistance allowed immobilizing up to 100 beads of 15 μm in diameter under one-bead-to-one-trap principle where every trap was numbered for individual address ability.29, 30
Our work builds on the initial work by Tan and Takeuchi but establishes a new paradigm that the underlying hydrodynamic principles can be also employed for manipulation and docking of large metazoan organisms with diameters well above 1 mm and masses often exceeding 1 mg.29, 30 We also for the first time show that the hydrodynamic force can be effectively utilized to provide a higher density of trapping region creating an ordered microarray of traps with several traps per row without compromising the docking efficiency.
MATERIALS AND METHODS
Laser fabrication
Prototyping and fabrication was performed using a non-contact, 30 W CO2 laser cutting system equipped with high power density focusing optics (HPDFO)™ providing a 50 μm laser beam spot (Universal Laser Systems, Scottsdale, AZ, USA). The CO2 laser cutting system achieves an accuracy of up to 3 μm. The laser cut master shape was used for replica molding in PDMS (Sylgard 184; DowCorning Corp, Midland, MI, USA). The PDMS was mixed at a 10:1 (w/w) ratio of elastomer base to curing agent and degassed at 40 Torr to remove any residual air bubbles. The PDMS was then poured on a PMMA master to achieve approximately 5 mm thickness and cured thermally at 80 °C for up to 1 h. Cured PDMS devices were mechanically diced and attached to the glass microscope slides using oxygen plasma surface activation.
Chip setup, loading, and operation
Device was directly connected to the external high-precision Miniplus Evolution peristaltic pump (Gilson Inc, Middleton, WI, USA) using 1/16 in. polyurethane tubing (Cole-Parmer Instrument Company, Vernon Hills, Illinois, USA) with internal diameter (1.6 mm) allowing free passage of zebrafish embryos. The peristaltic pump was computer controlled and calibrated according to manufactures instructions. Changing the rotational speed of the pump rotor varied the flow rate.
The polyvinyl chloride (PVC) calibrated tubing (1.02 mm ID; Gilson, Inc.) was mounted inside the pump to provide flow rates over the desired range (0.1–2 ml/min). The chips were then positioned on the microscope stage, and embryos with intact chorions between 6 and 24 hours post-fertilization (hpf) were loaded by aspirating single embryos one by one at the flow rate of 1 ml/min. Embryo loading and trapping was confirmed microscopically.
Computational fluid dynamic (CFD) simulations
In order to estimate the variations of velocity and pressure throughout the flow domain and ultimately the hydrodynamic forces that guide the embryos towards the traps, three-dimensional (3D) models of the device were created with virtual embryos as spherical structures. The simulation was performed using Gambit 2.3 software (Fluent, Lebanon, NH, USA) to create the geometry and mesh generation. Finite-volume based Fluent 6.3 software (Fluent, Lebanon, NH, USA) was subsequently used to solve the associated differential equations as described in the supplementary material.31 The thermo-physical properties of the actual E3 medium used to culture the zebrafish embryos were comparable to that of the filtered tap water with only minor additional amounts of salts and an anti-fungal agent. Therefore, we applied the thermo-physical properties of water in our simulations and assumed them constant over the temperature range involved.
Computational “pressure balance” model was also developed to assess the hydrodynamic trapping efficiency. Accordingly, each trap was represented by four waypoints as shown in Supplementary Figure 1.31 The points were set to a height of 600 μm with respect to the bottom surface of the channel at the level of embryo geometrical center. The static pressure of each point was obtained using 3D CFD model, and the pressure gradients were calculated along and across the traps. A high pressure gradient along the trap indicated the embryo rolling in the main channel, while a higher pressure gradient across the trap predicted the embryo trapping and immobilization (Supplementary Figure 1).31
Figure 1.
Rapid prototyping of ultra thick devices. (a) Workflow of the technique: (1)—laser cutting of a PMMA negative relief pattern, (2)—dust particles removal, and (3)—PMMA thermal bonding. C-clamps are used to apply a uniform mechanical force during the bonding process, (4)—PDMS replica moulding. Note that PDMS does not bind electrostatically or covalently to PMMA allowing rapid removal without the need for releasing agents. (b) 1.5 mm thick negative relief pattern fabricated using laser cutting of PMMA. Note that small distortion of the pattern due to the laser cone geometry. Red arrow denotes the direction of the cutting laser beam, (c) Magnified view of the complex PMMA relief pattern for hydrodynamic trapping of single zebrafish embryos. Reflow of the polymer during the thermal bonding of PMMA layers removes the visible surface imperfections.
Moreover, the Lagrangian large particle tracking model was utilized to predict the trajectory of individual embryos, and consequently the trapping sequence of traps, as described in the supplementary material.31
Zebrafish husbandry and embryo culture
Wild-type zebrafish Danio rerio (AB line) were obtained from the Zebrafish International Resource Center (Oregon, Eugene, OR, USA). Embryos were then collected in embryo medium E3 and rinsed to remove any debris and dead embryos. Embryos were kept at 28.5 ± 0.5 °C in E3 medium, and for the mass transfer, experiments perfused and stained with 0.04% Trypan Blue dye (Life Technologies Corp, CA, USA). Animal research was conducted with approval from The University of Auckland Animal Ethics Committee (approval ID R661/1).
Imaging and data analysis
Stereoscopic images of embryos grown on chip-based devices were obtained using the Leica MZ7.5 stereomicroscope (Leica Microsystems, Wetzlar, Germany). Embryo trapping efficiency experiments were acquired using the Canon 600D Digital SLR camera (Canon Inc, Tokyo, Japan) equipped with a true 1:1 macro lens (Canon EF 100 mm f/2.8, Macro Lens; Canon, Inc.).
The data analysis and presentation were performed using the Leica Application Suite (LAS) (Leica Microsystems), ImageJ (Ref. 37, and Fluent 6.3 (Fluent) software. Student’s t-test was applied for comparison between groups with significance set at p < 0.05.
RESULTS AND DICUSSION
Rapid prototyping of ultra thick devices
The prototyping and fabrication of LOC devices that can accommodate millimeter scale particles such as metazoan embryos has not been extensively explored. Standard UV lithography methods are not particularly suitable for this purpose. The major problems associated with photoresist-based fabrication of structures with heights above 1 mm affect all processing steps such as the coating, baking, development, and residual stress effects during the curing of the photoresist resins.13
In this work, we show how rapid prototyping of ultra thick LOC devices can be greatly accelerated by combining high-speed laser prototyping with replica moulding in PDMS instead of conventional photolithography techniques. Laser cutter/engraver was used to cut out negative relief patterns in 1.5–2 mm thick poly-methyl methacrylate sheets in less than 3 min (Figures 1a, 1b, 1c). For the purposes of fabricating ultra think LOC devices, the feature sizes down to 0.35 mm were reliably obtained when using polymer sheets of 1.5 mm thickness (Figures 1a, 1b, 1c). Melting associated with polymer cutting and subsequent thermal bonding prevented, however, the fabrication of smaller features. The cutting laser beam created polished, high quality edges in the PMMA substrata (Figures 1b, 1c). The master shape could be subsequently thermally bonded to another PMMA sheet (110 °C for up to 2 h) providing that uniform mechanical force (0.05–0.1 kg/cm2) is applied for equal force distribution across both PMMA layers. Precise temperature control was a critical step needed to avoid PMMA deformation and excessive melting at temperatures higher than 130–140 °C. During the thermal bonding process, the limited reflow of the polymer reduced the surface imperfections resulting from the laser cutting (Figure 1c).
Interestingly, the PDMS did not bind electrostatically or covalently to PMMA, enabling its direct use for replica moulding without any need for releasing agents and/or distortion of the final patterns during the PDMS curing (Figure 2). As such, this rapid technique introduced a prototyping cycle that yields completed and ready to use LOC devices within less than 4 h of processing time and does not require any sophisticated clean room facilities. The process is easily applicable for both high-speed prototyping and medium scale fabrication of ultra thick LOC devices.
Figure 2.
Hydrodynamic embryo trapping array. (a) 2D computer-assisted design (CAD) drawing outlining the geometry of the device for trapping and immobilization of large metazoan embryos created using laser fabrication technique as described. Note the array of 48 traps interconnected with an array of 48 cross-flow suction channels. Blue arrows denote the direction of fluid flow. R—denotes the numbering of consecutive trapping rows; T—denotes the numbering of consecutive traps. (b) Magnified CAD drawing of the device section as shown by red circle in (a). Note the direction of the main flow and cross-flow across the traps thanks to the presence of interconnection suction channels. (c) 3D streamlines of fluid colored by flow velocity (m/s) obtained by computational fluid dynamic simulations for the initial two trapping rows (R1 and R2). Perfusion was simulated at a volumetric flow rate of 1 ml/min. Note a considerable flow passing through the suction channels. This phenomenon allows for robust immobilization of the embryos inside the traps and also efficient drug delivery and exchange. (d) Complete PDMS device for a one-step loading, trapping, and immobilization of large metazoan embryos (ca. 1.2 mm in diameter). Note large number of zebrafish embryos immobilized on a chip. (e) Magnified microphotograph of the section as shown in (d). Main channel, embryo trap, and interconnecting suction channels are clearly visible. Note the good representation of the features in PDMS following replica moulding on laser machined PMMA mold. Blue arrows denote the direction of fluid flow inside the device. (f) A 3D cartoon showing the embryo trapping principles: 1—embryo is aspirated from the storage vessel and injected into the main channel, 2-3—pressure difference across the trap guides the embryo into the trap, 4—next embryo is introduced and rolls on the previous one towards the next available trap, and 5-6—the process is repeated till all the traps are filled with embryos, while the hydrodynamic forces keep embryos securely docked for the duration of experiments. Blue arrows denote the direction of fluid flow and embryo movements.
Embryo array device design
The major obstacle hampering on-chip manipulations of millimetre sized embryos is their large diameter and mass which are not easily transferable to LOC devices as it results in rapid gravitational-induced sedimentation and strong inertial forces affecting their translational and rotational movements. Using the rapid prototyping technique described above, we fabricated a proof-of-concept polymeric device that was employed for one step automated loading and passive, hydrodynamic trapping of large numbers of zebrafish embryos with a diameter of 1.2 ± 0.1 mm and a mass of 1 ± 0.25 mg. The chip featured three integrated modules: (1) the serpentined shaped main channel (width = 1.7 mm and height = 1.5 mm) for embryo loading and medium perfusion, (2) an array of embryo traps in consecutive rows, and (3) an array of small suction channels (width = 0.5 mm and height = 1.5 mm), which interconnected embryo traps with the main channel (Figures 2a, 2b. The latter design feature provided a direct hydrodynamic force to drag the embryos into the traps (Figure 2c). The volume of the device was approximately 825.9 μl, while the volume of a single trap was 2.77 μl. This innovative design was based on the principle that the fluid stream in the main serpentine channel generated suction through an array of smaller cross (suction) channels (Figures 2c, 2d, 2e). The resulting suction force changed the embryo trajectory allowing their docking and long-term immobilization inside an array of traps interconnected with the suction channels (Figures 2e, 2f). Our design builds on the initial work by Tan and Takeuchi but establishes a new paradigm that the underlying hydrodynamic principles of can be also employed for manipulation and docking of large metazoan organisms in contrast to micrometer sized polystyrene beads or human cells.29, 30 We also for the first time show that the hydrodynamic force can be effectively utilized to provide a higher density of trapping region creating an ordered microarray of traps with several traps per row without compromising the docking efficiency. In our design, multiple embryos are deflected perpidicularly to the main flow by a suction force that drags them from the main channel towards the traps. Trapped embryos act as plugs that increase the flow resistance across the occupied trap and redirect more flow to the subsequent traps in each row.29, 30 Our design proved capable of docking and immobilizing large metazoan embryos with diameters well above 1 mm and masses in excess of 1 mg. Interstingly, all trapped embryos could also be recovered and collected simply by reversing the flow direction.
Hydrodynamic trapping of millimetre-sized embryos
To validate the trapping performance of the embryo array, the chip was connected to a computer-controlled stepper motor-driven peristaltic pump. The calibrated pump was characterized by 0.01 rpm readability below 10 rpm that translated into the variation ≤0.1% at the flow rate of up to 2 ml/min. The embryo array design achieved 80% ± 3% trapping efficiency when positioned on a horizontal microscope stage. Our observations indicated that the device exhibited similar trapping characteristics in the flow rates ranging from 0.4–2 ml/min. However, both numerical and experimental results are shown and discussed at the flow rate of 1 ml/min. This flow rate was selected as it provides the optimal trapping of embryos while minimizing the shear stress on developing embryo. Loading of the 48 traps took approximately 3–4 min at the flow rate of 1 ml/min. Importantly, the previously described surface roughness of less than 10 μm generated during laser cutting of the PMMA master did not affect the performance of the device. This might be due to several factors including: the smoothing of the walls during the thermal annealing process,22 the very low Reynolds number (Re ∼ 21 at the inlet of the serpentine channel at a flow rate of 1 ml/min), negligible effect owing to the overall size of the device (depth = 1.5 mm and width = 1.7 mm), precluding occurrence of any substantial eddies in the main channel, and also the strong inertial forces and elastic surface of embryos which dampened any undesired disturbances.
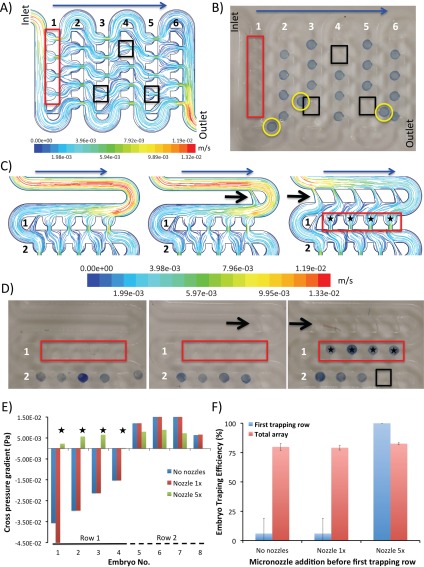
We observed, however, that irrespective of the applied flow rate, two discrete sections of the chip exhibited a suboptimal trapping leading to over 20% loss of embryos across the total of 48 traps. The principal problematic region was the first trapping row comprising of traps T1-4 close to the inlet and averaging only 6% ± 13% trapping efficiencies (Figures 3a, 3b). Lack of docking in this region translated to up to 8% ± 3% loss in overall trapping efficiency per entire embryo array. The second region of suboptimal docking was the traps located at the end of each row (T8, T12, T16,… T48) where the flow velocity was low and the lateral force generated by the suction channel was inadequate to deflect the embryos towards the traps (Figure 3b).
Figure 3.
Implementation of diverging channels to improve the trapping efficiency. (a) Streamlines of fluid colored by flow velocity (m/s) across the original prototype at the vertical middle plane when perfused at a flow rate of 1 ml/min. Red and black rectangles highlight the regions of suboptimal trapping efficiency due to the high velocity of fluid within the main channel of the first row and the inadequate suction within the last traps of each row. (b) Experimental validation of conditions shown in (a). Note that a good agreement of experimental embryo docking as compared to the CFD simulations. Red and black rectangles highlight the regions of suboptimal trapping efficiency. Yellow circles highlight the un-trapped embryos travelling in the main channel. (c) Streamlines of fluid colored by flow velocity (m/s) for modified design implementing diverging channels (black arrows). The data are shown for the initial two trapping rows only. Note that the addition of the five diverging channels decreases the flow velocity within the main channel considerably while increases the cross flow towards the first four traps. Blue arrows denote the direction of flow, black arrows denote the implementation of diverging channels and black stars refer to the regions with improved trapping efficiency. Note that the implementation of diverging channels in the first channel help slow the flow velocity and redirect more flow towards the traps. (d) Experimental validation of improved trapping due to diverging channels. Note that 100% trapping achieved in the first row following the implementation of five diverging channels. Black arrows denote the implementation of diverging channels and black stars refer to the regions with improved trapping efficiency. (e) Variations of cross pressure gradient at different traps of the designs with 0, 1×, and 5× diverging channels, as obtained by the pressure balance model. Note the excellent agreement between CFD simulations and experimental data as shown in (d). Improvement (traps 1–4) and deterioration (trap 8) of the embryo docking following the implementation of diverging channels is clearly visible. (f) Comparative representation of results from embryo docking experiments performed at a volumetric flow rate of 1 ml/min. Note that localized improvement in trapping efficiency following the implementation of new geometry does not necessarily translate into the improvements across the whole array.
As there are virtually no reports on CFD-guided modeling of chip-based technologies for manipulation of large particles such as embryos, we set out to validate how CFD simulation-guided optimization of the device geometry can lead to enhanced device performance needed for high-throughput laboratory automation. Two main design criteria were then experimentally evaluated to ensure: (1) complete, 100% trapping and long-term immobilisation of large particles; (2) ability for post-analysis recovery of the embryos when the device was actuated under a reversed flow regime from the outlet port. The previosly descibed surface roughness was ignored in numerical simulations. However, this simplification did not affect the accuracy of the model in terms of predicting the trapping efficiency of the system. Initially, the addition of miniaturized diverging channels that increase the lateral force in the first trapping row was evaluated (Figure 3c). We found that the addition of five diverging channels to the first row slowed down the flow velocity at the entrance of the main channel from 1.19 E-02 m/s to 3.98 E-03 m/s (Figure 3c). At the same time, more flow was streamlined into the four traps of the first row, obtaining 100% trapping efficency in this region (Figure 3d). The outcomes were consistent with the pressure balance model results, indicating a negative cross pressure gradient for the first four traps (T1-T4) for no-diverging channel and 1×-diverging channel designs, while a positive cross pressure gradient for 5×-diverging channel design (Figure 3e). However, this local improvement did not translate into the increased total trapping efficiency (Figure 3f, Supplementary Figure 2).31 The observed trapping losses were due to the increased suboptimal embryo trapping among the traps located at the end of each row. This constituted up to 20% of the docking loss and correlated well with the pressure balance model results predicting the deteriorated cross pressure gradient for the 5×-diverging channel design across the trap T8 (Figure 3e). Moreover, the diverging channels severely hampered the post-analysis recovery of the embryos from the device when a reversed flow was applied at the outlet port. We found that during the reversed flow regime, the embryos were pushed towards the outlet of diverging channels leading to the main channel clogging.
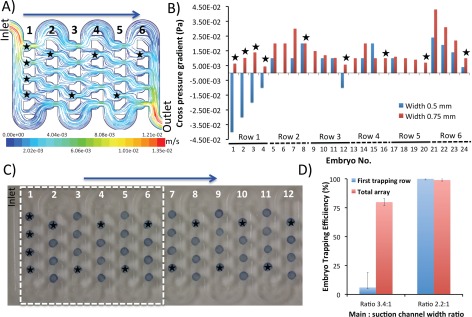
We next validated whether the increase of the suction channel width by 50% (from 0.5 to 0.75 mm) will lead to a substantially improved embryo trapping efficiency. The CFD simulations indicated that implementation of this modification in all 48 traps across the embryo array led to a better streamline profile (Figure 4a)) and a considerably improved cross pressure gradient compared to the original design, as obtained by the pressure balance model (Figure 4b). Experimental validation proved that the CFD simulations translated well into the overall 100% trapping efficiency (Figures 4c, 4d). Although the CFD-guided design favoured the rapid and efficient embryo docking/immobilization, it proved at the same time difficult to perform the post-analysis recovery of the embryos from the device. The increased width of the suction channels led to the trapping of embryos at the oulets of the suction channels during the reverse flow regime. This led to the clogging of the main channel and damage to the embryos during the recovery process, precluding the fullfilment of the second design criterion.
Figure 4.
Varying the suction channel width to improve the embryo trapping efficiency. (a) Streamlines of fluid colored by flow velocity (m/s) at the vertical middle plane when perfused at a flow rate of 1 ml/min. Note a considerable flow passing through the suction channels with the increased width. Red and black rectangles highlight the regions of suboptimal trapping efficiency. Black stars refer to the regions with improved trapping efficiency. Due to the computational limitations only first six rows were simulated. (b) Variations of cross pressure gradient at different traps of the designs with suction channel widths of 0.5 and 0.75 mm, as obtained by the pressure balance model. Note that considerable improvement in suction across all traps as compared to the original design. (c) Experimental validation of improved trapping due to suction channel widening. Note that 100% trapping efficiency is achieved across the whole array following the implementation of new design geometry. Also note the excellent agreement between CFD simulations and experimental validation data as shown in (b); (d) Comparative representation of results from embryo docking experiments performed at a volumetric flow rate of 1 ml/min. The increase of the suction channel width by 50% allows for robust immobilization of the embryos inside the traps.
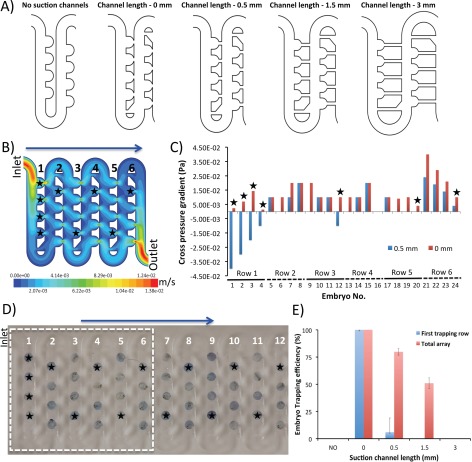
As a result, we next validated the effects of suction channel length on the trapping efficiency of the device. Five distinctive designs were simulated and consisted of prototypes featuring: (1) no interconnecting suction channels; (2) suction channel length of 0 mm, (3) suction channel length of 0.5 mm (original design); (4) suction channel length of 1.5 mm, and (5) suction channel length of 3 mm (Figure 5a, Supplementary Figure 3).31 The CFD simulations of the flow velocity profile indicated that to generate the maximum suction force the length of the suction channels should be minimised (Figures 5b, 5c). Accordingly, only the design featuring suction channel length of 0 mm considerably improved the cross pressure gradient, as obtained by the pressure balance model (Figure 5c). Experimental validation has confirmed that this translated well into the trapping efficiency reaching 99% ± 1 (Figures 5d, 5e). Moreover, this design allowed the trapped embryos to retain their position during the course of long-term experiments (ca. 72 h). Importantly, the one-step loading and trapping process was straightforward and did not require any active on-chip or off-chip actuators apart from a single pump. The modified geometry of suction channels did not hamper the post-analysis recovery of the embryos from the device. We found that during the reversed flow regime both embryos and eletheuro-embryos could be rapidly recovered without the main channel clogging.
Figure 5.
Varying the suction channel length to improve the trapping efficiency. (a) 2D CAD drawings outlining the evaluated designs. (b) Velocity contour (m/s) across the design featuring suction channels length reduced to 0 mm. Simulation was performed at the vertical middle plane when perfused at a flow rate of 1 ml/min. Note a considerable flow passing through the suction channels with the decreased length. Due to the computational limitations, only first six rows were simulated. (c) Variations of cross pressure gradient at different traps of the designs with suction channel lengths of 0 and 0.5 mm, as obtained by the pressure balance model. Note that considerable improvement in suction across all traps as compared to the original design featuring suction channels of 0.5 mm long. Black stars refer to the regions with improvement in the simulated trapping efficiency. (d) Experimental validation of improved trapping due to suction channel shortening. Note that 100% trapping efficiency is achieved across the whole array following the implementation of new design geometry. Also note the excellent agreement between CFD simulations and experimental validation data as shown in (b) and (c). (e) Comparative representation of results from embryo docking experiments performed at a volumetric flow rate of 1 ml/min.
Interstingly, the lack of interconnecting suction channels as well as their extension above 1.5 mm resulted in a greatly deteriorated hydrodynamic docking of the embryos. This was due to the lack of the sufficient suction force capable of changing the trajectory of embryos travelling in the main channel (Figure 5e, Supplementary Figure 3).31 Positioning the devices with suction channel lengths of 1.5 and 3 mm on an elevated stage with a tilt angle of 45° could, however, provide up to 50% and 75% improvement in trapping efficency, for both designs, respectively (Supplementary Figure 3).31 The experimental data indicated, however, that gravitational pull alone was not a leading factor that can support efficient embryo docking and immobilization as the design with removed cross-flow suction channels did not deliver any trapping even at the highest degrees of elevation. Our data support the notion that hydrodynamic trapping and immoblization of large metazoan embryos can be rapidly implemented on meso-scale chip-based devices. Moreover, the CFD-guided prototyping and rapid fabrication can greatly accelerate the optimization process leading to the implementation of new on-chip functionalities.
Implementation of Lagrangian particle tracking model
Although the pressure balance model can predict both the local and overall trapping efficiencies of different design modifications, they cannot be used to predict the dynamic interaction between the traps to understand how the trap occupancy influences the downstream performance of the array. In order to overcome these limitations, we for the first time applied the Lagrangian particle-tracking model to perform a real-time prediction of simulated large embryo behaviour under the effect of hydrodynamic and sedimentation forces.31, 32, 33 The model takes into account the variations of velocity, pressure, and shear stress within the system due to either the geometry of the design or the occupancy of the existing traps. It also takes into the account the dimensions and density of the embryos immobilized across the whole chip-based device.
Figure 6 depicts the simulated patterns of embryo trapping for the design featuring suction channel width of 0.75 mm at four distinctive time steps while the complete numerical filling process. Interestingly, the model recapitulated the deflection of embryos and docking in empty traps provided that they generated sufficient suction as stated above (Figure 6a). The experimental validation indicated a good agreement with the computational assumption of the Lagrangian particle-tracking model (Figure 6b). The simulation was capable of predicting the inability of the original design (featuring suction channel width of 0.5 mm) to trap the embryos at the first row and the last traps of each row (Figures 7a, 7b), which is in line with our earlier observations. Moreover, the model showed the inability of the design featuring suction channel length of 3 mm to trap even one single embryo (Figures 8a, 8b). We conclude that the application of innovative particle tracking and simulation algorithms can considerably supplement the conventional CFD modeling and lead to the accelerated development of chip-based devices for manipulation of millimetre sized metazoan small model organisms.
Figure 6.
Implementation of Lagrangian particle tracking model to predict the trapping characteristics of the design featuring suction channel width of 0.75 mm. (a) Simulated particle tracking model just after an embryo has filled the traps T2, T9, T22, and T23, increasing the total number of trapped embryos to 7, 8, 22, and 23, respectively. (b) Experimental validation of the particle tracking model as shown in (a). Note a high degree of agreement between the numerical and experimental results. White circles denote embryos travelling in the main channel towards the available trapping region (enhanced online).
Figure 7.
Lagrangian particle tracking model for the original design featuring suction channel width of 0.5 mm. (a) Simulation of embryo docking process across six consecutive trapping rows. Perfusion was simulated at a volumetric flow rate of 1 ml/min. Note the predicted inability to trap the embryos at the first row and the last traps of each row. (b) Experimental validation of the particle-tracking model that shows the embryo docking process across six consecutive trapping rows. Embryos were counter stained with 0.04% Trypan Blue to improve their visibility during the videomicroscopy. Perfusion was conducted at a volumetric flow rate of 1 ml/min. Note that the predicted by the Lagrangian particle tracking model inability to trap the embryos at the first row and the last traps of each row is confirmed during the experimental validation of the prototype. Red arrows highlight the small differences between the CFD simulation and the actual experimental validation of trapping efficiency. The overall accuracy of the CFD simulations varied between 92% and 98%. Yellow circles highlight the un-trapped embryos travelling in the main channel (enhanced online) .
Figure 8.
Implementation of Lagrangian particle tracking model to predict the trapping characteristics for the prototype featuring suction channel length of 3 mm. (a) Simulated particle tracking model predicted no trapping across the length of the device due to greatly deteriorated hydrodynamic conditions. (b) Experimental validation of the particle tracking model as shown in (a). Note a high degree of agreement between the numerical and experimental results. Yellow circles highlight the un-trapped embryos travelling in the main channel (enhanced online) .
CONCLUSIONS
High throughput and automated analysis of small model organisms is still a challenging task.2, 34, 35, 36 Manual handling procedures are not only time consuming but also error prone, limiting the reproducibility and introduction of laboratory standards. The present study was designed to provide a comprehensive mechanistic and design rationale for accelerated handling of millimetre sized metazoan embryos on LOC devices. It provides a new foundation for rapid prototyping and automated manipulation of large developing embryos on miniaturized polymeric device. Our results support the notion that by employing innovative CFD-guided design features and accelerated fabrication methods, hydrodynamic positioning of large particles such as embryos on LOC devices can be an attractive alternative to provide high levels of laboratory automation.
ACKNOWLEDGMENTS
Grant sponsor: Supported by Faculty Research and Development Fund, University of Auckland, New Zealand (K.K., J.A., D.W.); The Australia Endeavour Awards, Department of Education, Employment and Workplace Relations, Australia (K.K.); The Australian Research Council’s Discovery Early Career Researcher Award funding scheme (ARC DE120101402) (K.K.); Ministry of Science & Innovation, New Zealand (C.J.H., K.E.C., P.S.C.); and Biotechnology and Biological Sciences Research Council (BBSRC), Engineering and Physical Sciences Research Council (EPSRC) and Scottish Funding Council, UK, funded under the RASOR Program (Radical Solutions for Researching the proteome) (J.M.C., D.W.).
The authors are grateful to A. Mahagaonkar for his expert management of the zebrafish facility.
References
- Whitesides G. M., Nature 442(7101), 368–373 (2006). 10.1038/nature05058 [DOI] [PubMed] [Google Scholar]
- Wlodkowic D., Khoshmanesh K., Akagi J., Williams D. E., and Cooper J. M., Cytometry. Part A 79(10), 799–813 (2011). 10.1002/cyto.a.21070 [DOI] [PubMed] [Google Scholar]
- Becker H., Lab Chip 9(15), 2119–2122 (2009). 10.1039/b911553f [DOI] [PubMed] [Google Scholar]
- Hulme S. E., Shevkoplyas S. S., and Samuel A., Nat. Methods 5(7), 589–590 (2008). 10.1038/nmeth0708-589 [DOI] [PubMed] [Google Scholar]
- Hulme S. E., Shevkoplyas S. S., McGuigan A. P., Apfeld J., Fontana W., and Whitesides G. M., Lab Chip 10(5), 589–597 (2010). 10.1039/b919265d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoshmanesh K., Kiss N., Nahavandi S., Evans C. W., Cooper J. M., Williams D. E., and Wlodkowic D., Electrophoresis 32(22), 3129–3132 (2011). 10.1002/elps.201100160 [DOI] [PubMed] [Google Scholar]
- Lucchetta E. M., Munson M. S., and Ismagilov R. F., Lab Chip 6(2), 185–190 (2006). 10.1039/b516119c [DOI] [PubMed] [Google Scholar]
- Wielhouwer E. M., Ali S., Al-Afandi A., Blom M. T., Riekerink M. B., Poelma C., Westerweel J., Oonk J., Vrouwe E. X., Buesink W., vanMil H. G., Chicken J., van’t Oever R., and Richardson M. K., Lab Chip 11(10), 1815–1824 (2011). 10.1039/c0lc00443j [DOI] [PubMed] [Google Scholar]
- Yang F., Chen Z. G., Pan J. B., Li X. C., Feng J., and Yang H., Biomicrofluidics 5(2), 024115 (2011). 10.1063/1.3605509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammer E., Kamp H. G., Hisgen V., Koch M., Reinhard D., Salinas E. R., Wendler K., Zok S., and Braunbeck T., Toxicol. In Vitro 23(7), 1436–1442 (2009). 10.1016/j.tiv.2009.05.014 [DOI] [PubMed] [Google Scholar]
- Graf S. F., Hotzel S., Liebel U., Stemmer A., and Knapp H. F., J. Assoc. Lab. Autom. 16 (2), 105–111 (2011). 10.1016/j.jala.2010.11.002 [DOI] [PubMed] [Google Scholar]
- Giacomotto J. and Segalat L., Br. J. Pharmacol. 160(2), 204–216 (2010). 10.1111/j.1476-5381.2010.00725.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. H., Lee G. B., Chang B. W., and Chang G. L., J Micromech. Microeng. 12(5), 590–597 (2002). 10.1088/0960-1317/12/5/312 [DOI] [Google Scholar]
- Mohr J., Proc. Int. School Phys. 128, 181–207 (1996). [Google Scholar]
- Khan Malek C. G., Anal. Bioanal. Chem. 385(8), 1362–1369 (2006). 10.1007/s00216-006-0517-z [DOI] [PubMed] [Google Scholar]
- Hruby J., AIP Conf. Proc. 625, 55–61 (2002). 10.1063/1.1498183 [DOI] [Google Scholar]
- Ogilvie I. R. G., Sieben V. J., Floquet C. F. A., Zmijan R., Mowlem M. C., and Morgan H., J. Micromech. Microeng. 20(6), 065016 (2010). 10.1088/0960-1317/20/6/065016 [DOI] [Google Scholar]
- Islam M. Z., McMullin J. N., and Tsui Y. Y., Cytometry, Part A 79(5), 361–367 (2011). 10.1002/cyto.a.21063 [DOI] [PubMed] [Google Scholar]
- Yuen P. K. and Goral V. N., Lab Chip 10(3), 384–387 (2010). 10.1039/b918089c [DOI] [PubMed] [Google Scholar]
- Suriano R., Kuznetsov A., Eaton S. M., Kiyan R., Cerullo G., Osellame R., Chichkov B. N., Levi M., and Turri S., Appl. Surf. Sci. 257(14), 6243–6250 (2011). 10.1016/j.apsusc.2011.02.053 [DOI] [Google Scholar]
- Waddell E. A., Barker S. L. R., Rose D. J., Locascio L. E., and Kramer G. W., Abstr. Pap. - Am. Chem. Soc. 220, U91–U92 (2000). [Google Scholar]
- Hong T. F., Ju W. J., Wu M. C., Tai C. H., Tsai C. H., and Fu L. M., Microfluid. Nanofluid. 9(6), 1125–1133 (2010). 10.1007/s10404-010-0633-0 [DOI] [Google Scholar]
- Klank H., Kutter J. P., and Geschke O., Lab Chip 2(4), 242–246 (2002). 10.1039/b206409j [DOI] [PubMed] [Google Scholar]
- Khan Malek C. G., Anal. Bioanal. Chem. 385(8), 1351–1361 (2006). 10.1007/s00216-006-0514-2 [DOI] [PubMed] [Google Scholar]
- Sia S. K. and Whitesides G. M., Electrophoresis 24(21), 3563–3576 (2003). 10.1002/elps.200305584 [DOI] [PubMed] [Google Scholar]
- Wlodkowic D., Faley S., Zagnoni M., Wikswo J. P., and Cooper J. M., Anal. Chem. 81(13), 5517–5523 (2009). 10.1021/ac9008463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobel S., Valero A., Latt J., Renaud P., and Lutolf M., Lab Chip 10(7), 857–863 (2010). 10.1039/b918055a [DOI] [PubMed] [Google Scholar]
- Sun M., Bithi S. S., and Vanapalli S. A., Lab Chip (23), 3949–3952 (2011). 10.1039/c1lc20709a [DOI] [PubMed] [Google Scholar]
- Tan W. H. and Takeuchi S., Proc. Natl. Acad. Sci. U.S.A. 104(4), 1146–1151 (2007). 10.1073/pnas.0606625104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W. H. and Takeuchi S., Lab Chip 8(2), 259–266 (2008). 10.1039/b714573j [DOI] [PubMed] [Google Scholar]
- See supplementary material at http://dx.doi.org/10.1063/1.3699971 for materials and methods, mathematical equations, and data.
- Berlemont A., Chang Z. Z., and Gouesbet G., Flow, Turbul. Combust. 60(1), 1–18 (1998). 10.1023/A:1009963215802 [DOI] [Google Scholar]
- Kim M. C., Wang Z. H., Lam R. H. W., and Thorsen T., J. Appl. Phys. 103(4), 044701 (2008). 10.1063/1.2840059 [DOI] [Google Scholar]
- Vogt A., Codore H., Day B. W., Hukriede N. A., and Tsang M., J. Vis. Exp. 40, 1900 (2010). 10.3791/1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-Martin C., Chang T. Y., Koo B. K., Gilleland C. L., Wasserman S. C., and Yanik M. F., Nat Methods 7(8), 634–636 (2010). 10.1038/nmeth.1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler G. N. and Brandli A. W., Dev. Dyn. 238(6), 1287–1308 (2009). 10.1002/dvdy.21967 [DOI] [PubMed] [Google Scholar]
- See http://rsb.info.nih.gov/ij/ for more details about ImageJ.