Abstract
Sepsis is an adverse systemic inflammatory response caused by microbial infection in blood. This paper reports a simple microfluidic approach for intrinsic, non-specific removal of both microbes and inflammatory cellular components (platelets and leukocytes) from whole blood, inspired by the invivo phenomenon of leukocyte margination. As blood flows through a narrow microchannel (20 × 20 µm), deformable red blood cells (RBCs) migrate axially to the channel centre, resulting in margination of other cell types (bacteria, platelets, and leukocytes) towards the channel sides. By using a simple cascaded channel design, the blood samples undergo a 2-stage bacteria removal in a single pass through the device, thereby allowing higher bacterial removal efficiency. As an application for sepsis treatment, we demonstrated separation of Escherichia coli and Saccharomyces cerevisiae spiked into whole blood, achieving high removal efficiencies of ∼80% and ∼90%, respectively. Inflammatory cellular components were also depleted by >80% in the filtered blood samples which could help to modulate the host inflammatory response and potentially serve as a blood cleansing method for sepsis treatment. The developed technique offers significant advantages including high throughput (∼1 ml/h per channel) and label-free separation which allows non-specific removal of any blood-borne pathogens (bacteria and fungi). The continuous processing and collection mode could potentially enable the return of filtered blood back to the patient directly, similar to a simple and complete dialysis circuit setup. Lastly, we designed and tested a larger filtration device consisting of 6 channels in parallel (∼6 ml/h) and obtained similar filtration performances. Further multiplexing is possible by increasing channel parallelization or device stacking to achieve higher throughput comparable to convectional blood dialysis systems used in clinical settings.
INTRODUCTION
Sepsis is a critical medical condition characterized by a systemic inflammatory response syndrome (SIRS) due to microbial infection and is the primary cause of death in intensive care unit (ICU) (∼751 000 cases in United States annually) with overall mortality between 28% and 50%.1 Upon infection, the innate immune system responds by initiating inflammatory pathways and leukocyte recruitment to the site of infection to combat the infectious agents. However, excess production of cytokines and inflammatory mediators can trigger a systemic inflammation, resulting in widespread activation of vascular endothelium, amplification of coagulation cascades through platelet activation, and increase of circulating peripheral leukocytes (mainly neutrophils and macrophages).2 These multi-factorial effects damage microvasculature and induce tissue hypoxia, leading to septic shocks or deaths in severe cases as a result of multiple organ dysfunctions.2, 3
Current sepsis treatments include antibiotics therapy with patients in critical conditions also receiving intravenous fluids to maintain blood pressure and prevent vital organs from failing. However, drug treatment is limited by dose toxicity and rapid emergence of drug-resistant pathogens.4, 5, 6 As inflammation continues to spread for a few hours before the drug effect takes place, infants with sepsis (neonatal sepsis) are especially at risk because their immune system are functionally immature and delay or inappropriate antibiotic treatment may cause fatality.7 Extracorporeal blood purification therapies have also been used for sepsis treatment and have shown to stabilize hemodynamics and improve clinical outcomes of patients.8, 9, 10 Similar to conventional dialysis system, semi-permeable membrane (diffusion/convection) or sorbent (adsorption) are used to non-selectively remove water-soluble inflammatory mediators and endotoxin from the plasma. However, drawbacks of these techniques include loss of beneficial molecules (nutrients and drugs) during dialysis and treatment is expensive due to large volume of replacement fluid post-filtration and high nursing workload.11, 12, 13 Currently, there is no general consensus on which components of the blood should be removed for better clinical outcome. In this regard, what is necessary is a hemofiltration technique that removes a broad array of targets (pathogens, immune cells, platelets, etc.) from blood, in order to help modulate (but not eliminate entirely) the overall inflammatory response.
Microfluidic technologies for blood separation applications are an attractive alternative due to numerous advantages such as reduced contamination issues, portability, and process automation.14, 15, 16 Typically, pathogenic microorganisms are very small, ranging from 1 to 3 µm in diameter and can be easily separated from the larger red blood cells (RBCs) and leukocytes. Several microfluidic approaches for microorganism separation from blood have been previously demonstrated based on either size differences17, 18 or affinity separation.19, 20 However, laborious sample preparations involving blood dilution and magnetic labeling of microorganisms limit their throughput. Large volume of buffer fluid is required for blood dilution or sheath flow in these methods, making them inconvenient to process large quantities of blood over prolonged period of time.
In this work, we introduce a novel microfluidic approach for intrinsic, non-specific removal of microorganisms and inflammatory cellular components from whole blood directly, inspired by the invivo phenomenon of leukocyte margination.21, 22 In blood vessels with diameter less than 300 µm, deformable RBCs migrate axially across streamlines to the channel centre due to Poiseuille flow profile. Mechanical collisions between the leukocytes and migrating RBCs result in the radial displacement of the larger leukocytes to the vessel wall, a phenomenon aptly termed as margination.23, 24 These effects have been demonstrated in microfluidic devices for leukocytes separation25, 26, 27 and recently by our group for deformability-based separation of malaria-infected RBCs.28 Besides leukocytes, platelet margination has also been observed in concentrated blood flow as excess platelets are found at the RBCs-depleted region next to the channel walls.29, 30 Although axial migration of RBCs to channel centre can partly lead to physical exclusion of platelets towards the wall, actual mechanisms responsible for platelet margination are known to be more complex and strongly dependent on hematocrit and shear rate.31, 32 For example, Zhao et al. highlighted the importance of RBCs influence by showing that platelet margination increases with hematocrit, most notably at hematocrit greater than 30%.32 Turitto and Weiss also proposed that collisions between RBCs and platelets can contribute to margination process due to enhanced platelet diffusion towards the vessel wall.33 Using computational fluid dynamics (CFD) modeling of platelet-RBCs interactions, Almonani et al. showed that RBCs motions (tumbling and translation) and cell-cell interactions cause localized flow fluctuation that leads to an increase in fluid shear forces acting on the platelets which results in their margination towards the channel walls.34 They also concluded that platelet margination is predominantly caused by the large size difference (not shape) between RBCs and platelets. Since microbes (∼1–3 µm) are similar in size as platelets, we hypothesize that microbes in blood flow can also undergo margination towards the wall region by similar mechanisms which can be used for bacteria separation applications. Here, we validated and applied the margination principle towards a high-throughput blood filtration technique for removal of pathogens from blood. The proposed device consists of a cascaded straight channel design which allows for 2-stage removal of microbes in a single processing step, thus achieving higher microbial removal efficiency.
With an application for sepsis treatment in mind, we demonstrated separation of Escherichia coli (E. coli) and Saccharomyces cerevisiae (S. cerevisiae) spiked in whole blood, achieving removal efficiencies of ∼80% and ∼90%, respectively. Inflammatory cellular components (platelets and leukocytes) were also depleted by >80% in the filtered blood sample which could potentially help to modulate the excessive host inflammatory response. The technique offers significant advantages including high throughput (∼1 ml/h per channel) and label-free separation for non-specific removal of any blood-borne pathogens (bacteria and fungi). Unlike other techniques, no prior sample preparation is required as whole blood can be drawn from patients and pumped into the device directly, thus reducing processing time and cost. As the simplicity of the device architecture allows easy multiplexing, we designed and tested a larger filtration device consisting of 6 channels in parallel (∼6 ml/h) with similar filtration performance. We envisage further multiplexing (∼100 channels) to achieve higher throughput comparable to conventional blood dialysis techniques which can serve as an adjunctive therapy for more effective sepsis treatment.
MATERIALS AND METHODS
Fabrication
Channel layouts were designed using L-Edit (Tanner EDA, USA) and devices were fabricated in polydimethylsiloxane polymer (PDMS, Sylgard 184, Dow Corning, USA) using standard microfabrication soft-lithographic techniques.35 The channel designs were first patterned on polished silicon wafers and etched into silicon using deep reactive ion etching (DRIE). Following etching, the patterned silicon wafers were silanized with trichloro(1H, 1H, 2H, 2H-perfluorooctyl)silane (Sigma Aldrich, USA) for 1 h to facilitate PDMS mold release. PDMS prepolymer mixed in 10:1 (w/w) ratio with curing agent was poured onto the silicon wafer and cured at 70 °C for 2 h. The cured PDMS mold then acted as a template for subsequent PDMS casting (negative replica). The PDMS master template was silanized for 1 h before use to aid release of subsequent PDMS microchannels. Finally, holes (1.5 mm) for inlets and outlets were punched and the PDMS microchannels were irreversibly bonded to microscopic glass slides using an air plasma machine (Harrick Plasma Cleaner, USA) and left for 2 h at 70 °C to complete the bonding.
Cell culture
E. coli (HB101 K-12) bacteria expressing green fluorescent protein (GFP) was cultured overnight at 37 °C in LB Agar (Sigma Aldrich, USA) containing ampicillin (100 μg/ml) and arabinose (0.1%, inductor of GFP expression) then harvested and resuspended in 1 × phosphate buffer solution (PBS) for experimental use. Haploid S. cerevisiae (W303-1B strain) expressing yellow fluorescent protein (YFP) was a kind gift from Dr. Jacquin Niles from the Department of Biological engineering at MIT.
Sample preparation
For device characterization, human whole blood (Research Blood Components, Brighton, MA) was washed three times with washing buffer containing 1 × PBS, 0.5% v/v bovine serum albumin (BSA) (Miltenyi Biotec, USA) prior to running the experiment. Final RBC concentration was adjusted to physiological hematocrit (∼45%) in the sample buffer containing 1 × PBS, 0.5% v/v BSA, and 3% w/v Dextran 40 (Sigma Aldrich, USA) to prevent cell sedimentation. Alexa Fluor® 488 E. coli (K-12 strain) BioParticles® conjugates (Invitrogen, USA), cultured bacteria, and yeast were added to blood samples (106–7/ml) and vortexed for 30 s before use. For whole blood analysis, fresh human blood was incubated at 4 °C directly without washing for 40 min with fluorescein isothiocyanate (FITC) conjugated CD41a antibodies (1:50, BD Biosciences, USA) and allophycocyanin (APC) conjugated CD45 marker (1:100, BD Biosciences, USA) to identify platelets and leukocytes, respectively.
Device characterization and analysis
Whole blood samples were pumped through the microfluidic devices using a syringe pump (NE-1000, New Era Pump Systems Inc., USA). The microchannels were mounted on an inverted phase contrast microscope (Olympus IX71) equipped with a Hamamatsu Model C4742-80-12AG CCD camera (Hamamatsu Photonics, Japan). iplab (Scanalytics, Rockville, MD) software was used for video acquisition and captured videos were analyzed using imagej® software. For E. coli BioParticles® and whole blood analysis, separation efficiency was determined by performing flow cytometry (FACS) analysis on the sample and collected centre outlet using BD™ LSR II flow cytometer (BD Biosciences, USA). Different cell types were gated based on the forward and side scatters as well as the specific fluorescence intensity. For microbial experiments, bacteria concentrations at the inlet and filtered centre outlet were determined by plating 10 µl of solution on LB agar and colony-forming units (CFUs) were counted after 24 h of incubation at 37 °C. Bacteria and yeast concentrations were also diluted and evaluated using a haemocytometer separately.
Confocal imaging
A LSM 510 meta laser scanning confocal (Carl Zeiss, Jena, Germany) was mounted on an Axiovert 100 M Inverted Microscope (Carl Zeiss, MicroImaging) and images were acquired at 488 nm excitation. A 10× objective of the microscope was used with an additional digital zooming of 2.5× (from software) to prevent unnecessary photobleaching of adjacent areas. Image slices were acquired at an interval of 1µm in z-axis (vertical axis) with a resolution of at least 512 × 512 pixels at a minimum scanning speed of 0.96 µs per pixel. Captured images were then analyzed using imagej® software.
RESULTS
Microfluidic design and separation principle
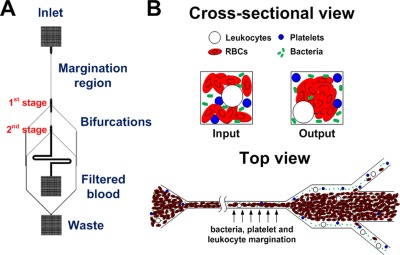
The microchannel design consists of two cascaded straight microchannels, 20 × 20 µm (W × H), with two bifurcations in series (1:8:1) (Fig. 1). As blood flows through the straight channel (margination region), deformable RBCs migrate to the axial centre of the channel to form a RBC-rich core and a cell-free layer adjacent to the channel walls. Mechanical collisions between the migrating RBCs and other cell types (bacteria, platelets and leukocytes) result in their margination towards the channel walls into the cell-free layer. The outlet of each margination channel expands into a 200 µm wide section to enhance the cell-free layer thereby allowing majority of the RBCs to enter the centre outlet, while the other cell types flowing at the sides are removed through the smaller side channels as waste.36 By cascading two margination channels in series, the bacteria-depleted blood at the first bifurcation undergo another round of margination, thus achieving a 2-stage separation and higher bacterial removal efficiency in a single pass through the device.
Figure 1.
Schematic illustration of the developed microfluidic device for pathogen removal from blood. (a) The design consists of two cascaded straight microchannels (20 × 20 µm (W × H), 5 mm long for first channel and 1 mm long for the cascaded channel) with two bifurcations (1:8:1) in series. The cascaded design allows removal of bacteria at each bifurcation, thereby achieving a 2-stage bacterial removal in a single step. (b) Cross-sectional and top view of the microchannel illustrating the separation principle. As blood flows through the channel (margination region), deformable RBCs migrate axially to the channel centre, resulting in margination of other cell types (bacteria, platelets, and leukocytes) towards the channel walls and subsequently removed from the side outlets while the centre outlet collects the bacteria-depleted blood.
Device optimization and confocal analysis
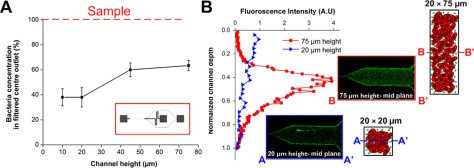
The microchannel design and testing conditions were first optimized by studying the effect of various parameters including microchannel aspect ratio, channel length, and flow rate on bacteria margination. The parameter “aspect ratio” in this work was studied by only varying the channel height and keeping the channel width constant as 20 µm. To study the effect of channel height/aspect ratio on bacteria margination, a single margination channel of 15 mm length, 20 µm width with varying heights of 10 µm, 20 µm, 45 µm, 75 µm were fabricated, yielding aspect ratios (height/width) of 0.5, 1, 2.25, and 3.75, respectively. The channel width was fixed at 20 µm due to enhanced Fahreaus effect in smaller channels as axial migration of RBCs becomes less efficient with increasing channel size.22, 37 Whole blood spiked with FITC-conjugated E. coli bioparticles was pumped into each device and the filtered blood at centre outlet was collected for FACS analysis. Bacteria concentration at the centre outlet was then normalized with the sample to determine the separation efficiency. Fig. 2a presents the normalized bacteria concentration in the filtered centre outlet at different channel heights. At low channel heights of 10 and 20 µm (aspect ratio ≤1), ∼40% of bacteria remained in the centre outlet after filtration. As channel height increased to 45 and 75 µm (aspect ratio ≥1), bacteria margination became less efficient with >60% of bacteria still remaining at the centre outlet. This is consistent with previous studies which reported that margination of leukocytes and malaria-infected RBCs was more prominent in low aspect ratio channels.25, 26, 28 To further understand bacteria margination in different aspect ratio channels, confocal microscopy was used to image the bacteria distribution at the outlet of 20 µm (aspect ratio 1) and 75 µm (aspect ratio 3.75) height channel. From the experimental results, confocal images and corresponding fluorescent intensity plots between the channel inlets and outlets illustrate a distinct difference in bacteria margination efficiency for 20 µm and 75 µm height channels (supplementary information, Fig. S1).38 As compared to the channel inlet, there was a ∼3× decrease in fluorescence signal at the mid plane of the channel outlet for 20 µm height channel, indicating lesser bacteria remaining at the centre region due to their efficient margination to the sides. Confocal images were also acquired along the channel height which showed that the vertical intensity profile of the outlet centre region for 20 µm height channel was almost uniform, indicative of efficient bacteria margination across the depth (Fig. 2b). Although bacteria undergo margination to the four channel walls, fluorescence intensity peaks were absent at the top and bottom channel wall proximities because of the sudden width expansion from 20 µm to 200 µm at the bifurcation which caused the bacteria to disperse across the channel width. However, for 75 µm height channel, there was uneven bacteria margination along the height, with a strong fluorescent intensity peak observed at the mid plane region. Confocal images of the channel outlet at the mid plane also showed negligible bacteria margination for 75 µm height channel as bacteria remained evenly distributed throughout the channel and fluorescence intensity was similar as compared to the channel inlet (supplementary information, Fig. S1).38 Several reasons can be accounted for the observed differences in margination efficiency. First, low aspect ratio channels (aspect ratio ≤1) closely resemble circular blood vessels in which the fluidic shear distribution is isotropic and uniform across the channel cross-section. As RBCs axial migration distances are the same in all directions, a well-defined RBC core can be easily formed at the centre region in circular and low aspect ratio channels, thereby allowing effective bacteria margination to the sidewalls. For high aspect ratio (ratio ≥ 1) channels, although there is a higher shear rate along the shorter channel width, shear rate is much lower along the height (z-direction) with more RBCs being packed at the cross-section centre region due to Fahreaus effect. This gives rise to a blunt velocity profile in the z-direction which can significantly influence the axial migration behaviour of RBCs at the mid plane region.37, 39 Moreover, bacteria margination at the mid plane region may be less effective as bacteria can be displaced vertically along the height in addition to axial margination towards to lateral walls, resulting in the bacteria being trapped at the channel centre. Similar observations were also made by Jain and Munn who showed that margination of leukocytes to the sidewalls decreased with channel height due to their lateral displacements towards top and bottom surfaces.26 Hence, high aspect ratio microchannels are unfavourable for separation applications and we limit the channel height to be 20 µm as channel aspect ratio of 1 gave the most effective bacteria margination with the highest throughput.
Figure 2.
Effect of microchannel height (aspect ratio) on bacteria margination. (a) Bacteria concentration in the filtered centre outlet with varying channel heights using a single margination channel of 15 mm length and 20 µm width (schematic single channel design in red box). Bacteria concentration was normalized with sample to determine the percentage of bacteria remaining after filtration. (b) Fluorescent intensity Z-profiles at the outlet centre region of 20 µm and 75 µm height channels and corresponding schematic illustration of their bacteria margination. Confocal images at the mid plane of 20 µm and 75 µm height channels indicate less efficient bacteria margination at the mid plane region of high aspect ratio channel.
After optimizing the channel height, the effect of channel length on bacteria margination was studied to determine the shortest channel length required for efficient bacteria margination. Experimental results indicate that bacteria margination occurred for all tested channel lengths but remained approximately constant for channel lengths between 5 mm and 20 mm (supplementary information, Fig. S2).38 Therefore, the channel length was fixed at 5 mm as this would minimize the pressure drop across the device and allow testing at higher working flow rate.
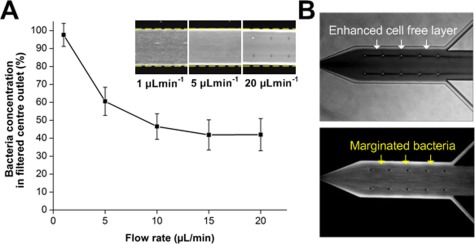
Lastly, experiments were done to characterize the effect of flow rate on bacteria margination. Fig. 3a presents the normalized bacteria concentration at the filtered centre outlet for increasing flow rates. At low flow rate of 1 µl min−1, bacteria concentration at the centre outlet was comparable to the initial sample, indicating negligible bacteria margination. As flow rate increases, bacteria separation efficiency improved evident by the lower bacteria concentrations at the collected outlet but remained approximately constant beyond 10 µl min−1. These results were expected because at such high flow rates (Reynold’s number ∼4–5), RBCs experience higher shear and inertia in the margination channel which expedite their axial migration to the channel centre to form a well-defined RBC core, resulting in margination of small bacteria towards the channel walls. However, inertia migration would have negligible effects on RBCs axial migration as the flow rates tested were still far from inertia-dominated flow conditions (Reynold’s number > 50),40, 41 and inertial focusing is greatly affected in whole blood due to significant cell-cell interactions.42 A flow rate of 15 µl min−1 was chosen for subsequent experiments as the PDMS device and connection setup used was not rigid enough to withstand the pressure drop at higher flow rates for long period of device operation. In addition, it was also observed that the cell free layer was enhanced at the expanded bifurcation with most of the marginated bacteria residing in the cell free region (Fig. 3b). This is useful for filtration purposes as the large centre outlet of the bifurcation (1:8:1) was able to collect most of the RBCs, while the marginated bacteria were efficiently removed via the side channels.
Figure 3.
Effect of flow rate on bacteria margination. (a) Normalized bacteria concentration in the filtered centre outlet for increasing flow rate. Bacteria separation efficiency improved as flow rate increases but remained approximately constant beyond 10 µl min−1. Averaged composite images of the bifurcation indicate increase in concentration of FITC-conjugated bacteria at the channel sides with increasing flow rates. (b) Optical images illustrating the enhanced cell free layer at the expanded bifurcation. Most of the marginated bacteria reside within the cell free layer next to the channel wall which allows their efficient removal from the side channels, while the densely packed RBCs are filtered into the larger centre outlet with minimal loss.
Device characterization of the cascaded design
Fig. 4a presents the averaged fluorescence composite images at the margination channel and their corresponding intensity linescans across the channel width. At the inlet of the channel, FITC-conjugated bacteria were evenly distributed and the fluorescence intensity profile was uniform across the channel although stronger intensities were observed at the channel sides due to slower flow at the corners. At the outlet of the margination channel, fluorescence intensity at the channel centre decreased with stronger fluorescence intensity peaks at the sides, evident of the higher number of bacteria present near the channel walls after margination. Margination efficiency was determined based on the assumptions that (1) complete bacteria margination to the four channel walls occurred and (2) they would occupy the entire cross section perimeter uniformly. The theoretical separation efficiency was calculated by taking the ratio of the wetted perimeter of the centre region (blue region in schematic) to the total cross section perimeter. As shown in Fig. 4b, the separation efficiencies obtained from the experimental results at each stage were in good agreement with the theoretical values. We also characterized the hematocrit of filtered blood from centre outlet and found that it increased by ∼10% and ∼25% after first and second stage, respectively, as compared to inlet sample (supplementary information, Fig. S3).38 Recent simulation work by Zhao et al. has shown that increase in hematocrit enhances platelet migration towards channel sides due to (1) greater platelet diffusivity at the cell-laden region (RBC core) and (2) higher frequency of platelet-RBCs collisions at the peripheral of RBC core which result in higher platelet lateral drift velocity towards wall.43 Hence, by removing a fraction of the plasma and marginated bacteria at the first stage bifurcation, the filtered blood (with higher hematocrit and less bacteria) would experience additional RBCs axial migration (Fahreaus effect) and RBCs-bacteria hydrodynamic interactions due to higher hematocrit in the second margination channel which lead to further bacteria margination towards the channel periphery. This allows the cascaded design to remove marginated bacteria (via the side channels) at the first and second stage bifurcation successively, thus achieving an overall bacteria separation efficiency of >80%.
Figure 4.
Device characterization of the cascaded design. (a) Averaged fluorescence composite images at the margination channel (20 µm × 20 µm) and corresponding intensity linescans illustrating the larger number of FITC-conjugated bacteria found at the channel sides after margination. Dotted lines indicate the approximate position of channel walls. (b) Optical images and plot of bacteria filtration efficiency at different stages. Experimental results were similar to the theoretical separation efficiencies calculated based on the bifurcation ratio (blue region in schematic) and the complete bacteria margination to the four channel walls. A high bacteria separation efficiency of >80% was achieved at the collected centre outlet after two stages of filtration (enhanced online).
Whole blood analysis
Following the characterization of device dimensions and operating conditions, experiments using human whole blood were carried out to quantify the separation efficiency of other cellular components such as platelets and leukocytes. The cells were stained with specific fluorophore-conjugated monoclonal antibodies (see Methods section) and identified using FACS analysis. As shown in Fig. 5a, >80% of leukocytes and platelets were removed in the collected centre outlet, consistent with previous work which reported on the phenomenon of platelets32, 44 and leukocytes margination24, 25 in microchannels. RBCs concentration at the centre outlet increased by ∼30% as the expanded channel bifurcation (1:8:1) enabled efficient filtration of the densely packed RBCs into the larger centre outlet. Negligible haemolysis was observed in the filtered blood based on optical absorption at 414 nm due to the short RBCs transit time (<0.01 s) in the margination channel where the shear rate is the highest (data not shown). As an application for sepsis treatment, we tested blood sample spiked with two different kinds of microbes, GFP-transfected E. coli bacteria and YFP-transfected S. cerevisiae yeast, at high microbial concentrations (∼106–7/ml) typically found in sepsis patients. As expected, our results indicate successful removal of ∼80% of bacteria and ∼90% of yeast at the filtered centre outlet, indicating effective margination for both kinds of microbes in our device. Due to the larger yeast size dimension (∼5 µm), most of them occupied the four corners of the margination channel instead of the entire channel peripheral, resulting in complete margination to the sidewalls and thus better separation efficiency (Fig. 5b). This clearly indicates the versatility of the device as it can be used for non-specific removal of pathogens present in blood without any labeling or prior information on the infecting species.
Figure 5.
Spiked whole blood analysis. (a) Normalized concentration of different cellular components at the filtered centre outlet using human whole blood spiked with E. coli and S. cerevisiae separately. RBCs concentration increased by ∼30% as more RBCs were packed at the centre due to Fahreaus effect. Inflammatory cellular components (platelets and leukocytes) and microbes undergo margination to the channel sides, resulting in >80% decrease in concentration at the centre outlet. Optical images (100× magnification) of each component are indicated at the top of their corresponding histogram bar. (b) Optical images illustrating yeast filtration at different stages. The larger yeast (∼5 µm) undergoes margination to the four corners in the straight channel (red arrows), resulting in complete margination to the sides and thus higher separation efficiency (enhanced online).
High throughput multiplexing using parallel design
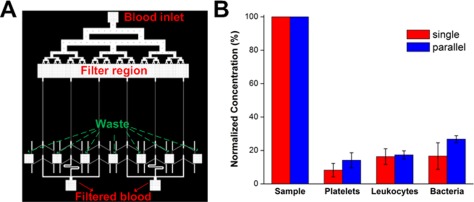
In terms of throughput, our single channel design (whole blood processing at ∼1 ml/h) is higher than other reported microfluidic approaches used for blood separation applications, yet significantly slower than conventional marcoscale blood purification methods (∼1–2 l/h). To demonstrate the scalability of our device for higher throughput, we designed a parallel system consisting of 6 channels to filter blood simultaneously at ∼6 ml/h (Fig. 6a). Similar filtration performances were obtained as compared to the single channel device (Fig. 6b) and the simple experimental setup enables easy and continuous collection of the filtered blood from the two centre outlets (supplementary information, Fig. S4).38 This parallelization scheme can further be extended to larger devices by stacking layers of devices to achieve even higher throughputs comparable to conventional dialysis systems (supplementary information, Fig. S5).38
Figure 6.
High throughput blood filtration using a parallel system. (a) Schematic layout of the device consisting of an additional filter region to remove clogs and debris and 6 channels with cascaded design in parallel to achieve higher flow rates. (b) Experimental results indicating similar device performances as compared to single channel in removal of different blood components and bacteria using the parallel system at 100 µl/min.
DISCUSSION
Blood flow in microvessels displays complex rheological behaviours due to the high volume fraction of RBCs (∼45%) in blood and their interactions with the vessel wall.22 As blood flows through vessels with dimensions less than 300 µm, RBCs undergo lateral migration to the axial centre of the channel due to Poiseuille flow profile to form a RBC-rich core with a thin layer of cell-free region adjacent to the vessel wall. This gives rise to a decrease in tube hematocrit (Fahreaus effect) as RBCs at the channel centre attain higher flow velocities than the overall blood flow. Interestingly, this effect is also known to limit the wetting capability of whole blood in capillary tubes (<50 µm diameter) as the faster flowing RBCs tend to form a concentrated plug at the meniscus, which would arrest the flow due to high hydrodynamic resistance.45, 46 The thickness of the cell-free layer also becomes significant with smaller tube diameter which serves as a lubrication layer and helps to reduce the effective blood viscosity (Fahreaus-Linquvist effect).21, 47 As RBCs migrate towards the centre, mechanical interactions between the deformable RBCs and the larger leukocytes lead to the physical displacement of leukocytes to the vessel wall, a phenomenon termed as leukocyte margination.23, 24 This allows subsequent leukocyte rolling and adherence on the endothelium which can affect microvasculature blood flow significantly due to the viscoelastic properties of leukocytes and their biochemical interactions with the endothelial cells when activated.48, 49 Besides leukocytes, the margination effect is also observed for platelets in concentrated blood flow29, 30 and applied for separation of malaria-infected RBCs.28 In these applications, the separated cells are either similar or larger in size as compared to RBCs thereby allowing efficient margination to the channel walls. Here, we show for the first time that microbes such as bacteria and yeast can also undergo margination in microchannels and be used as a high throughput blood filtration technique for removal of pathogenic microbes from blood. It has been previously demonstrated both invitro and invivo that RBCs tend to adhere to each other to form rouleaux (or aggregates) at low shear rates (50–600 s−1) which not only increase the blood viscosity but also help to exclude the larger and stiffer leukocytes from the axial core of RBCs, resulting in efficient leukocyte margination to the vessel walls.24, 26, 50, 51 In this work however, the small channel geometry and high flow conditions employed result in significantly higher wall shear rate (∼6 × 104 s−1) whereby RBCs aggregation will be minimal,52 and we postulate that margination of microbes occurs mainly due to Fahreaus effect by the formation of a well-defined RBCs core at the channel centre and hydrodynamic forces arising from intercellular microbe-RBCs interactions. This hypothesis has been demonstrated by us and others who showed that margination of platelets and malaria-infected RBCs can still take place in the absence of RBCs aggregation (by using washed blood and high working flow rates).28, 32 Furthermore, Freund also reported that RBCs aggregation is not necessary for leukocyte margination using fluid mechanics simulations and attributed the distinct physical characteristics and hydrodynamics interactions between leukocytes and RBCs as the more important criteria for margination process.53 Hence, unlike leukocyte margination at low shear rates, RBCs aggregation is not critical at high flow conditions in our device for microbe margination, but further studies are warranted to better understand the actual mechanism responsible so as to improve the channel design for better separation performance.
To demonstrate the viability for sepsis treatment, E. coli and S. cerevisiae were spiked in whole blood at clinically relevant concentrations and separated using the developed microfluidic device. E. coli is a Gram-negative, rod-shaped bacterium typically about 2.0 μm long and 0.5 μm in diameter. Almomani et al. have previously shown that platelets migration occurs due to the large size difference between platelets and RBCs which leads to an increase in lateral fluid and shear forces acting on the platelets.34 Hence, although not specifically tested, we postulate that the large size difference between bacteria and RBCs might also contribute to the bacteria margination process. Secondly, as the bacterial cell wall consists of a rigid layer of peptidoglycan network (Young’s Modulus ∼107 Pa) to help maintain the cell shape by resisting the turgor pressure,54, 55 it is suggestive that the distinct difference in stiffness between E. coli and RBCs (membrane Young’s Modulus ∼104 Pa)56 can aid in the margination process. Another possible reason is due to the non-spherical shape of the bacteria because as compared to spherical particles which are only subjected to translational motion, rod-shaped particles are also subjected to torques which result in additional tumbling and rotational motions.57 This enables the rod-shaped bacteria to attain higher lateral drift velocities which may enhance their margination capability across streamlines towards the channel sides.58 To illustrate the non-specific microbial removal of the margination technique, we spiked another type of microbe, S. cerevisiae, which is a widely studied model yeast type in cell biology. Its morphology (∼5–7 μm in diameter, ovoid shape) is similar to pathogenic fungus species such as Candida albicans which is the most common and life-threatening fungus infection in humans.59 Similarly for S. cerevisiae, the cell wall elastic modulus (∼108 Pa) is much higher than RBCs membrane60 and as they are more comparable in size with the RBCs, yeast/fungi undergo better margination in microchannel than bacteria which results in a higher removal efficiency (Fig. 5).
Besides the presence of pathogenic microbes in the blood of sepsis patients, the widespread activation of circulating leukocytes (neutrophils and monocytes) also contributes to the pathogenesis of sepsis.61, 62 Impaired deformability of leukocytes during sepsis63, 64 and coagulation in small blood vessels through platelet activation65 may alter microvascular flow and lead to inadequate tissue perfusion and organ failure. Our device not only removes infecting microbial agents from blood but also allows simultaneous margination and removal of platelets and leukocytes. The rapid removal of both infecting species and immune cells help reduce the concentration of bacterial endotoxin (the main component responsible for inducing inflammatory responses) and production of inflammatory molecules from the activated leukocytes. This may prevent excessive vascular endothelial damage and potentially help to modulate the host immune response for sepsis treatment. However, the current lack of understanding of the pathophysiology of sepsis makes it difficult to judge how much of the immune cells should be removed. In the current device, the outlet bifurcation was designed such that the side channels (20 µm in width) could skim off marginated leukocytes (8–15 µm) with high efficiency. Yang et al. have previously demonstrated the principle of plasma skimming and Zweifach–Fung effect in microchannel to isolate plasma with high purity.66 They reported a critical flow rate ratio of 1:6 between the plasma (side) outlet and main outlet to prevent RBCs from entering the plasma channel. Similarly, other groups have also reported the importance of flow rate ratio for optimizing plasma collection efficiency using plasma skimming.67, 68 As flow rate ratio between side and center channels in our device was ∼1:4, we were able to remove >80% of leukocytes and platelets, but some RBCs were inevitably lost through the side outlets (Fig. 3b), which might affect the final hematocrit and volume of the filtered blood. In future, proper design of the outlets by changing the flow rate ratio or varying the outlet channel lengths to adjust relative resistance between filtered (centre) and waste (side) channels27 would allow control over the RBCs concentration in the filtered blood as well as optimizing the removal efficiency of leukocyte, bacteria, and platelets for sepsis treatment. The clinical efficacy of this idea should be verified in a future animal/clinical testing.
Major limitations for current microfluidics approaches for microorganism removal from blood are laborious sample preparation and the requirement of large volume of buffer solution either as sheath flow or for blood sample dilution.17, 18, 19, 20 These make them inconvenient for blood dialysis applications which require processing large quantity of blood over long period of time. Moreover, the “cleaned” blood has to undergo post-processing such as reconstitution back to physiological hematocrit level or removal of excess, unbound magnetic beads before returning to the bloodstream of patients. Here, we applied invivo microcirculatory phenomenon including Fahreaus effect and leukocyte margination in a simple microfluidic device for intrinsic, non-specific removal of microorganisms and inflammatory cellular components from whole blood. The technique allows whole blood processing at ∼1 ml/h per channel with high microbial separation efficiency (>80%). The label-free separation also enables non-specific pathogen removal which is useful for separating unknown infecting species in sepsis patients. No sample preparation is necessary and the continuous processing and collection mode enable the return of the filtered blood back to the patient, thereby achieving a simple and complete dialysis circuit setup. Finally, we also tested a larger scale device consisting of 6 channels in parallel (∼6 ml/h) and obtained similar filtration performance. Further multiplexing is possible by increasing the number of parallel channels or stacking layers of parallel devices to achieve higher throughput comparable to convectional blood dialysis techniques used in clinical settings.
CONCLUSIONS
In this work, a high-throughput microfluidic blood filtration technique for microbial removal from whole blood, inspired by the invivo microcirculatory phenomenon of leukocyte margination was introduced. We demonstrated filtration of E. coli and S. cerevisiae spiked in whole blood, achieving a high removal efficiency of ∼80% and ∼90%, respectively. Key advantages in the developed technique include high throughput (∼1 ml/h per channel) and label-free separation which allows non-specific removal of any blood-borne pathogens. Inflammatory cellular components (platelets and leukocytes), which contribute to the pathogenesis of sepsis, were also depleted by >80% in the filtered blood and could potentially help to modulate the host inflammatory response. Lastly, the device simplicity allows easy multiplexing by channel parallelization or device stacking to further improve the throughput for clinical applications.
ACKNOWLEDGMENTS
Confocal microscopy was performed at the W. M. Keck Biological Imaging Facility in Whitehead Institute. Financial support by the Singapore-MIT Alliance for Research and Technology (SMART) Centre (BioSyM IRG) is gratefully acknowledged. This work is also supported by the use of MIT’s Microsystems Technology Laboratories as well as the financial support by DARPA DLT (Dialysis-Like Therapeutics) program, under SSC Pacific grant N66001-11-1-4182. Any opinions, findings, and conclusions or recommendations expressed in this publication are those of the authors and do not necessarily reflect the views of the DARPA. H.Y.G. acknowledges Agency for Science, Technology and Research (A*STAR) Singapore for the post-doctoral fellowship support.
References
- Angus D. C., Linde-Zwirble W. T., Lidicker J., Clermont G., Carcillo J., and Pinsky M. R., Crit. Care Med. 29(7), 1303–1310 (2001). 10.1097/00003246-200107000-00002 [DOI] [PubMed] [Google Scholar]
- Cohen J., Nature 420(6917), 885–891 (2002). 10.1038/nature01326 [DOI] [PubMed] [Google Scholar]
- Legrand M., Klijn E., Payen D., and Ince C., J. Mol. Med. 88(2), 127–133 (2010). 10.1007/s00109-009-0585-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbarth S., Garbino J., Pugin J., Romand J. A., Lew D., and Pittet D., Am. J. Med. 115(7), 529–535 (2003). 10.1016/j.amjmed.2003.07.005 [DOI] [PubMed] [Google Scholar]
- Valles J., Rello J., Ochagavia A., Garnacho J., Alcala M. A., and Spanish Collaborative Grp I., Chest 123(5), 1615–1624 (2003). 10.1378/chest.123.5.1615 [DOI] [PubMed] [Google Scholar]
- Neu H. C., Science 257(5073), 1064–1073 (1992). 10.1126/science.257.5073.1064 [DOI] [PubMed] [Google Scholar]
- Edmond K. and Zaidi A., PLoS Med. 7(3), e1000213 (2010). 10.1371/journal.pmed.1000213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimmelé T. and Kellum J., Crit. Care 15(1), 1–10 (2011). 10.1186/cc10118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann J. N., Hartl W. H., Deppisch R., Faist E., Jochum M., and Inthorn D., Kidney Int. 48(5), 1563–1570 (1995). 10.1038/ki.1995.448 [DOI] [PubMed] [Google Scholar]
- Cole L., Bellomo R., Journois D., Davenport P., Baldwin I. and Tipping P., Intensive Care Med. 27(6), 978–986 (2001). 10.1007/s001340100963 [DOI] [PubMed] [Google Scholar]
- Boldt J., Lenz M., Kumle B., and Papsdorf M., Intensive Care Med. 24(2), 147–151 (1998). 10.1007/s001340050536 [DOI] [PubMed] [Google Scholar]
- Druml W., Kidney Int. 56(S72), S56–S61 (1999). 10.1046/j.1523-1755.56.s72.1.x [DOI] [PubMed] [Google Scholar]
- Moerer O., Schmid A., Hofmann M., Herklotz A., Reinhart K., Werdan K., Schneider H., and Burchardi H., Intensive Care Med. 28(10), 1440–1446 (2002). 10.1007/s00134-002-1429-9 [DOI] [PubMed] [Google Scholar]
- Pratt E. D., Huang C., Hawkins B. G., Gleghorn J. P., and Kirby B. J., Chem. Eng. Sci. 66(7), 1508–1522 (2010). 10.1016/j.ces.2010.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagat A., Bow H., Hou H., Tan S., Han J., and Lim C., Med. Biol. Eng. Comput. 48(10), 999–1014 (2010). 10.1007/s11517-010-0611-4 [DOI] [PubMed] [Google Scholar]
- Hou H. W., Bhagat A. A. S., Lee W. C., Huang S., Han J., and Lim C. T., Micromachines 2(3), 319–343 (2011). 10.3390/mi2030319 [DOI] [Google Scholar]
- Wu Z. G., Willing B., Bjerketorp J., Jansson J. K., and Hjort K., LabChip 9(9), 1193–1199 (2009). 10.1039/B817611F [DOI] [PubMed] [Google Scholar]
- Mach A. J. and Di Carlo D., Biotechnol. Bioeng. 107(2), 302–311 (2010). 10.1002/bit.22833 [DOI] [PubMed] [Google Scholar]
- Xia N., Hunt T., Mayers B., Alsberg E., Whitesides G., Westervelt R., and Ingber D., Biomed. Microdevices 8(4), 299–308 (2006). 10.1007/s10544-006-0033-0 [DOI] [PubMed] [Google Scholar]
- Yung C. W., Fiering J., Mueller A. J., and Ingber D. E., LabChip 9(9), 1171–1177 (2009). 10.1039/B816986A [DOI] [PubMed] [Google Scholar]
- Goldsmith H. L., Cokelet G. R., and Gaehtgens P., Am. J. Physiol. 257(3), H1005–H1015 (1989). [DOI] [PubMed] [Google Scholar]
- Pries A. R., Secomb T. W., and Gaehtgens P., Cardiovasc. Res. 32(4), 654–667 (1996). 10.1016/S0008-6363(96)00065-X [DOI] [PubMed] [Google Scholar]
- Fiebig E., Ley K., and Arfors K. E., Int. J. Microcirc.: Clin. Exp. 10(2), 127–144 (1991). [PubMed] [Google Scholar]
- Goldsmith H. L. and Spain S., Microvasc. Res. 27(2), 204–222 (1984). 10.1016/0026-2862(84)90054-2 [DOI] [PubMed] [Google Scholar]
- Shevkoplyas S. S., Yoshida T., Munn L. L., and Bitensky M. W., Anal. Chem. 77(3), 933–937 (2005). 10.1021/ac049037i [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A. and Munn L. L., PLoS ONE 4(9), e7104 (2009). 10.1371/journal.pone.0007104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A. and Munn L. L., Lab Chip 11(17), 2941–2947 (2011). 10.1039/c1lc20401g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H. W., Bhagat A. A. S., Chong A. G. L., Mao P., Tan K. S. W., Han J. Y., and Lim C. T., LabChip 10(19), 2605–2613 (2010). 10.1039/C003873C [DOI] [PubMed] [Google Scholar]
- Aarts P., van den Broek S., Prins G., Kuiken G., Sixma J., and Heethaar R., Arterioscler., Thromb.,Vasc. Biol. 8(6), 819–824 (1988). 10.1161/01.ATV.8.6.819 [DOI] [PubMed] [Google Scholar]
- Zhao R., Kameneva M. V., and Antaki J. F., Biorheology 44(3), 161–177 (2007). [PubMed] [Google Scholar]
- Yeh C. and Eckstein E. C., Biophys. J. 66(5), 1706–1716 (1994). 10.1016/S0006-3495(94)80962-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R., Marhefka J. N., Shu F. J., Hund S. J., Kameneva M. V., and Antaki J. F., Ann. Biomed. Eng. 36(7), 1130–1141 (2008). 10.1007/s10439-008-9494-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turitto V. and Weiss H., Science 207(4430), 541–543 (1980). 10.1126/science.7352265 [DOI] [PubMed] [Google Scholar]
- AlMomani T., Udaykumar H. S., Marshall J. S., and Chandran K. B., Ann. Biomed. Eng. 36(6), 905–920 (2008). 10.1007/s10439-008-9478-z [DOI] [PubMed] [Google Scholar]
- McDonald J. C. and Whitesides G. M., Acc. Chem. Res. 35(7), 491–499 (2002). 10.1021/ar010110q [DOI] [PubMed] [Google Scholar]
- Faivre M., Abkarian M., Bickraj K., and Stone H. A., Biorheology 43(2), 147–159 (2006). [PubMed] [Google Scholar]
- Lima R., Ishikawa T., Imai Y., Takeda M., Wada S., and Yamaguchi T., J. Biomech. 41(10), 2188–2196 (2008). 10.1016/j.jbiomech.2008.04.033 [DOI] [PubMed] [Google Scholar]
- See supplementary material at http://dx.doi.org/10.1063/1.4710992 for additional figures on device characterization and multiplexing system through channel parallelisation and stacking.
- Lima R., Wada S., Tanaka S., Takeda M., Ishikawa T., Tsubota K.-i., Imai Y., and Yamaguchi T., Biomed. Microdevices 10(2), 153–167 (2008). 10.1007/s10544-007-9121-z [DOI] [PubMed] [Google Scholar]
- Di Carlo D., Irimia D., Tompkins R. G., and Toner M., Proc. Natl. Acad. Sci. U.S.A. 104(48), 18892–18897 (2007). 10.1073/pnas.0704958104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagat A. A. S., Hou H. W., Li L. D., Han J. Y., and Lim C. T., LabChip 11(11), 1870–1878 (2011). 10.1039/C0LC00633E [DOI] [PubMed] [Google Scholar]
- Lim E. J., Ober T. J., Edd J. F., McKinley G. H., and Toner M., LabChip (2012). 10.1039/C2LC21100A [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Shaqfeh E. S. G., and Narsimhan V., Phys. Fluids 24(1), 011902–011921 (2012). 10.1063/1.3677935 [DOI] [Google Scholar]
- Yeh C. and Eckstein E. C., Biophys. J. 66(5), 1706–1716 (1994). 10.1016/S0006-3495(94)80962-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R. and Chang H.-C., J. Colloid Interface Sci. 287(2), 647–656 (2005). 10.1016/j.jcis.2005.02.023 [DOI] [PubMed] [Google Scholar]
- Zhou R., Gordon J., Palmer A. F., and Chang H.-C., Biotechnol. Bioeng. 93(2), 201–211 (2006). 10.1002/bit.20672 [DOI] [PubMed] [Google Scholar]
- Fedosov D. A., Caswell B., Popel A. S., and Karniadakis G. E., Microcirculation 17(8), 615–628 (2010). 10.1111/j.1549-8719.2010.00056.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid-Schönbein G. W., Sung K. L., Tözeren H., Skalak R., and Chien S., Biophys. J. 36(1), 243–256 (1981). 10.1016/S0006-3495(81)84726-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamm R. D., Annu. Rev. Fluid Mech. 34(1), 211–232 (2002). 10.1146/annurev.fluid.34.082401.165302 [DOI] [PubMed] [Google Scholar]
- Sun C. and Munn L. L., Physica A 362(1), 191–196 (2006). 10.1016/j.physa.2005.09.027 [DOI] [Google Scholar]
- Pearson M. J. and Lipowsky H. H., Microcirculation 11(3), 295–306 (2004). 10.1080/10739680490425994 [DOI] [PubMed] [Google Scholar]
- Baskurt O. K. and Meiselman H. J., Semin. Thromb. Hemost. 29(05), 435–450 (2003). 10.1055/s-2003-44551 [DOI] [PubMed] [Google Scholar]
- Freund J. B., Phys. Fluids 19(2), 13 (2007). 10.1063/1.2472479 [DOI] [Google Scholar]
- Boulbitch A., Quinn B., and Pink D., Phys. Rev. Lett. 85(24), 5246 (2000). 10.1103/PhysRevLett.85.5246 [DOI] [PubMed] [Google Scholar]
- Huang K. C., Mukhopadhyay R., Wen B., Gitai Z., and Wingreen N. S., Proc. Natl. Acad. Sci. 105(49), 19282–19287 (2008). 10.1073/pnas.0805309105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulinska I., Targosz M., Strojny W., Lekka M., Czuba P., Balwierz W., and Szymonski M., J. Biochem. Biophys. Methods 66(1–3), 1–11. (2006). 10.1016/j.jbbm.2005.11.003 [DOI] [PubMed] [Google Scholar]
- Randall T. et al. , Nanotechnology 22(11), 115101 (2011). 10.1088/0957-4484/22/11/115101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sei-Young L. et al. , Nanotechnology 20(49), 495101 (2009). 10.1088/0957-4484/20/49/495101 [DOI] [PubMed] [Google Scholar]
- Pfaller M. A. and Diekema D. J., Clin. Microbiol. Rev. 20(1), 133–163 (2007). 10.1128/CMR.00029-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. E., Zhang Z., Thomas C. R., Moxham K. E., and Middelberg A. P. J., Proc. Natl. Acad.Sci. 97(18), 9871–9874 (2000). 10.1073/pnas.97.18.9871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K. A., Brain S. D., Pearson J. D., Edgeworth J. D., Lewis S. M., and Treacher D. F., Lancet 368(9530), 157–169 (2006). 10.1016/S0140-6736(06)69005-3 [DOI] [PubMed] [Google Scholar]
- Muller Kobold A. C., Tulleken J. E., Zijlstra J. G., Sluiter W., Hermans J., Kallenberg C. G. M., and Cohen Tervaert J. W., Intensive Care Med. 26(7), 883–892 (2000). 10.1007/s001340051277 [DOI] [PubMed] [Google Scholar]
- Rosenbluth M. J., Lam W. A., and Fletcher D. A., Lab Chip 8(7), 1062–1070 (2008). 10.1039/b802931h [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Backer D., Creteur J., Preiser J.-C., Dubois M.-J., and Vincent J.-L., Am. J. Respir. Crit. Care Med. 166(1), 98–104 (2002). 10.1164/rccm.200109-016OC [DOI] [PubMed] [Google Scholar]
- Levi M., Hematology 10(1), 129–131 (2005). 10.1080/10245330512331390177 [DOI] [PubMed] [Google Scholar]
- Yang S., Undar A., and Zahn J. D., Lab Chip 6(7), 871–880 (2006). 10.1039/b516401j [DOI] [PubMed] [Google Scholar]
- Blattert C., Jurischka R., Tahhan I., Schoth A., Kerth P., and Menz W., presented at the Engineering in Medicine and Biology Society, 2004. IEMBS ‘04. 26th Annual International Conference of the IEEE, 2004. [DOI] [PubMed]
- Jaggi R. D., Sandoz R., and Effenhauser C. S., Microfluid. Nanofluid. 3(1), 47–53 (2007). 10.1007/s10404-006-0104-9 [DOI] [Google Scholar]