Abstract
Recent deep sequencing of cancer genomes has produced an explosion of new data implicating Notch signaling in several human cancers. Unlike most other pathways, these data indicate that Notch signaling can be either oncogenic or tumor suppressive, depending on the cellular context. In some instances, these relationships were predicted from mouse models or presaged by developmental roles for Notch, but in other cases were unanticipated. This review discusses the pathogenic and translational significance of these new findings.
Keywords: Notch signaling, tumor suppressor, oncogene
1. Introduction
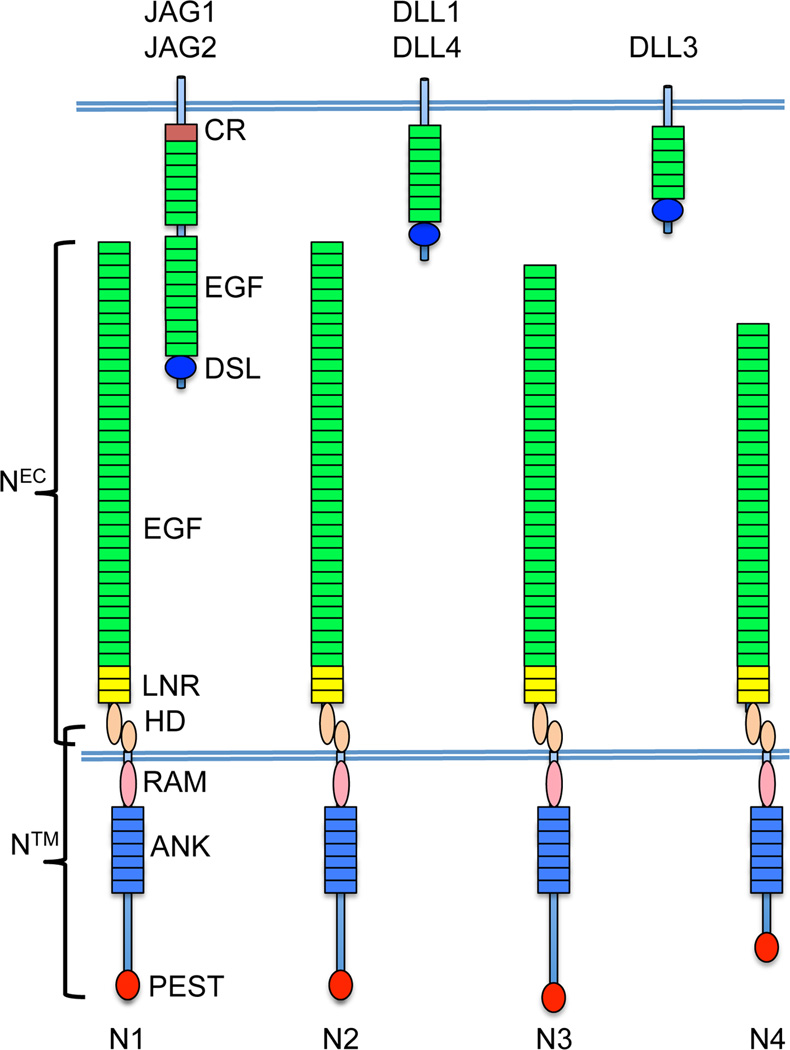
Notch receptors control many aspects of development and homeostasis in multicellular animals via a signal transduction pathway that relies on regulated intramembranous proteolysis (for recent review, see [1]). Mammals have four Notch receptors, Notch1–4 (Fig. 1). The extracellular domain of these proteins consists of variable numbers of epidermal growth factor(EGF)-like repeats that participate in ligand-binding, three Lin12/Notch repeats (LNRs), and a juxtamembrane heterodimerization domain. During maturation, Notch receptors are cleaved within the heterodimerization domain by furin-like proteases, producing two subunits that associate non-covalently through contacts in the heterodimerization domain and the LNRs, which together constitute a negative regulatory region (NRR) that is responsible for preventing ligand-independent receptor activation. The intracellular portions of Notch receptors include RAM and ankyrin repeat domains that are involved in protein:protein interactions and a C-terminal PEST degron domain.
Figure 1.
Structure of mammalian Notch receptors and ligands. Mammals express 5 Notch ligands, four of which activate Notch receptors (JAG1, JAG2, DLL1, and DLL4), and one of which (DLL3) may function as a decoy. Ligands have an N-terminal DSL domain, variable numbers of EGF repeats, and in the case of Serrate-like ligands (JAG1 and JAG2) a juxtamembrane cysteine-rich domain (CR). Mammals have four Notch receptors, Notch1–4, comprised of non-covalently associated extracellular (NEC) and transmembrane (NTM) subunits. NEC is comprised of 29–34 EGF repeats, 3 Lin12/Notch repeats (LNRs), and the N-terminal portion of the juxtamembrane heterodimerization domain (HD), while NTM is comprised of a RAM domain, 7 iterated ankyrin repeats (ANK) repeats, an structurally divergent unfolded region with variable transcriptional activation domain function (greatest in Notch1, least in Notch4), and a C-terminal PEST degron domain.
Notch signaling is normally initiated by ligands expressed on neighboring cells that belong to the Delta-Serrate-Lag2 (DSL) family, which are also transmembrane proteins (Fig. 1). Ligand binding first triggers cleavage by ADAM-10 or ADAM-17 metalloproteases at a site just external to the transmembrane domain, creating a short-lived membrane-bound intermediate lacking most of the Notch ectodomain that is a substrate for γ-secretase, a multisubunit intramembranous protease. Cleavage by g-secretase releases the intracellular domain of Notch (ICN), which translocates to the nucleus and forms a short-lived transcription activation complex with the DNA-binding factor RBPJ (also known as CSL) and coactivators of the MAML family. Rapid turnover of this complex is normally ensured in part by the C-terminal PEST degron in ICN.
Although some studies point to transcription-independent crosstalk between Notch and other signaling pathways such as Wnt and PI3K/Akt, genetic studies suggest that most Notch effects are mediated through transactivation of target genes by the RBPJ/ICN/MAML transcription complex. The outcome of signaling through this “canonical” pathway is strongly influenced by dose and cellular context; indeed, the lack of enzymatic amplification and putative transcription coupled degradation of ICN means that one activated Notch receptor probably transactivates at most one target gene prior to degradation, allowing for precise regulation of signal strength and duration.
Depending on the cellular context, Notch signaling can be an arbiter of survival versus death; proliferation versus growth arrest; or differentiation versus “stemness”. Given these varied effects, it is not surprising that widely divergent context-dependent roles for Notch have emerged in cancer. It is well established that Notch1 acts as an oncoprotein in T-cell acute lymphoblastic leukemia/lymphoma (T-ALL), an aggressive tumor that occurs mainly in children and adolescents. Recent application of deep sequencing approaches to hundreds of human cancer genomes and transcriptomes has detected somatic mutations in Notch receptor genes in an increasingly wide spectrum of tumors, expanding the breadth of Notch’s roles in cancer. Mouse modeling foretold some of these discoveries, but others were unexpected. This review mainly focuses on these new findings, which clearly document Notch’s ability to function as either an oncoprotein or a tumor suppressor depending on cellular context.
2. Notch as an oncoprotein: Notch1 in T-ALL as a paradigm
Before delving into recent discoveries (summarized in Table 1 and Table 2), brief review of oncogenic Notch mutations in T-ALL is in order as a point of reference (for detailed review, see [2]). To date somatic gain-of-function mutations in human T-ALL are confined to NOTCH1 and fall into two general classes. The most common class consists of point substitutions or small in-frame deletions, insertions or duplications involving the Notch1 NRR or transmembrane domain that allow ligand-independent proteolysis and activation. The second class consists of nonsense or frameshift mutations that result in the deletion of the C-terminal PEST degron domain. Some T-ALLs possess mutations of both classes in cis, an alignment that produces synergistic increases in signaling. Rarely in human T-ALL the NOTCH1 locus is broken by (7;9) translocations that fuse the 3’ end of NOTCH1 to TCRB promoter/enhancer elements. These rearranged NOTCH1 alleles express truncated transcripts encoding polypeptides that lack the NRR entirely. Similarly, in murine T-ALLs notch1 is commonly disrupted by RAG-mediated deletions that remove the 5’ end of the gene and activate a cryptic internal promoter that drives the expression of mRNAs encoding truncated polypeptides lacking the NRR. Thus, in T-ALL there is strong selection for somatic mutations that disrupt the Notch1 NRR and permit ligand-independent receptor activation. In the case of T-ALL, ligand-independent signaling may promote the spread of the tumor beyond the confines of the ligand-rich thymic microenvironment.
Table 1.
Activating Notch Mutations in Human Cancers
| NOTCH1 | Other Notch genes |
Somatic aberration | Comments | |
|---|---|---|---|---|
| T-ALL | ~60% in human T-ALL | ?NOTCH3 (rare) | In-frame NRR mutations and C-terminal PEST degron deletions | Mutations cause ligand-independent activation (NRR) or enhance protein half-life (PEST) |
| CLL | 5–12% | unknown | PEST degron deletions (>90% codon2514(del(CT)) | Associated with transformed and refractory CLL, absence of somatic hypermutation, trisomy 12 |
| MCL | ~10% | Confined to NOTCH1 | PEST degron deletions (>50% codon2514(del(CT)) | NOTCH1 locus also hypomethylated in MCL |
| Breast adenocarcinoma | <5% | NOTCH2 fusion genes also detected | Activating gene fusions | All rearrangements in ER- cancers, functionally validated |
Table 2.
Loss-of-function Notch mutations in human cancer.
| NOTCH1 | Other Notch genes |
Somatic aberration | Comments | |
|---|---|---|---|---|
| SCC (skin) | 60–70% | NOTCH2 > 25%, isolated NOTCH3 and NOTCH4 truncations | Frequent 5’ nonsense mutations, disruptive in-frame sequence variants | Functionally validated missense mutations, biallelic and heterozygous mutations identified, some predicted to have dominant negative activity |
| SCC (head and neck) | 15–20% | Confined to NOTCH1 | IRF6 and TP63 also recurrently mutated | |
| SCC (lung) | 5–10% | undefined | TCGA sequencing ongoing |
Another theme emerging from T-ALL is that the oncogenic role of Notch1 appears to be an exaggeration of its normal functions. Notch1 is essential at multiple junctures of T cell development, including T cell specification and thymocyte proliferation at β-selection (for recent review, see [3]). By analogy, selective pressures for gains or losses of Notch activity in other cancers may also reflect some normal aspect of Notch function in the cells of origin.
3. Evidence for oncogenic Notch signaling in chronic lymphocytic leukemia and other B-cell malignancies
Early work on Notch signaling in B lineage cells focused on its antagonism of B cell development. Strong gain-of-function NOTCH1 alleles skews the differentiation of hematopoietic progenitors towards T cell fate and away from B cell fate [4], and leads to growth arrest or apoptosis of several B cell neoplasms [5]. By contrast, notch2 is essential for murine splenic marginal zone B cell development [6], and notch1 has been implicated in B cell activation [7] and Ig secretion [8]. The possible ability of Notch to antagonize early steps in B cell development yet act positively at subsequent stages is reminiscent of the complex role of Notch in tissues such as the peripheral nervous system and the eye of Drosophila that arise through successive hierarchical cell fate decisions. It is thus possible that Notch may have additional uncharacterized roles in the development or function of various B cell subtypes.
Chronic lymphocytic leukemia (CLL) is an indolent but incurable neoplasm with a gene expression signature resembling that of normal memory B cells [9]. Several past reports have suggested a role for Notch signaling in CLL, but genetic evidence of Notch involvement was lacking. This changed in 2009, when focused resequencing of 43 cases of CLL by Falzetti’s group identified 2 cases with NOTCH1 exon 34 mutations leading to PEST degron deletions [10]. Subsequent study of 133 newly diagnosed CLL cases by the same group confirmed a low frequency of Notch1 PEST domain mutations (5.3%) [11]. In 2011, two unbiased whole exome studies of large European CLL cohorts also identified Notch1 gain-of-function mutations [12, 13]. In combination, these studies detected Notch1 mutations in 56 of 561 (10.0%) of newly diagnosed tumors. Notch1 mutations were associated with a worse outcome in all three of these series, and in the series from the Gaidano group [13] were associated with disease progression to large cell lymphoma and refractoriness to chemotherapy. Notch1 mutations were also frequently subclonal or undetectable at diagnosis and clonal in transformed tumors from the same patient, providing additional evidence of a link between Notch1 signaling and disease progression. Conversely, Notch1 mutations were identified in only 2 of 63 monoclonal CD5-positive B cell proliferations [14], an early stage of CLL development. Several groups have recently also reported that Notch1 mutations are enriched in CLLs associated with trisomy 12 [15–17], a common cytogenetic aberration in CLL.
Subsequently, Gascoyne’s group found NOTCH1 gain-of-function mutations in approximately 12% of mantle cell lymphoma (MCL) [18], an aggressive neoplasm usually derived from immunologically naïve mature B cells. As in CLL, most of these mutations lead to PEST degron deletions. Prior studies indicated that the NOTCH1 locus is frequently hypomethylated in MCL cells [19], but otherwise there was little reason to suspect a role for Notch signaling in this disease. Beyond MCL, Gaidano’s group also identified several diffuse large B cell lymphomas with Notch1 PEST domain mutations [13], and a recent report from Japan described a handful of diffuse large B cell lymphomas with Notch2 PEST domain mutations [20]. It is thus increasingly evident that Notch signaling has an oncogenic role in a number of B cell malignancies.
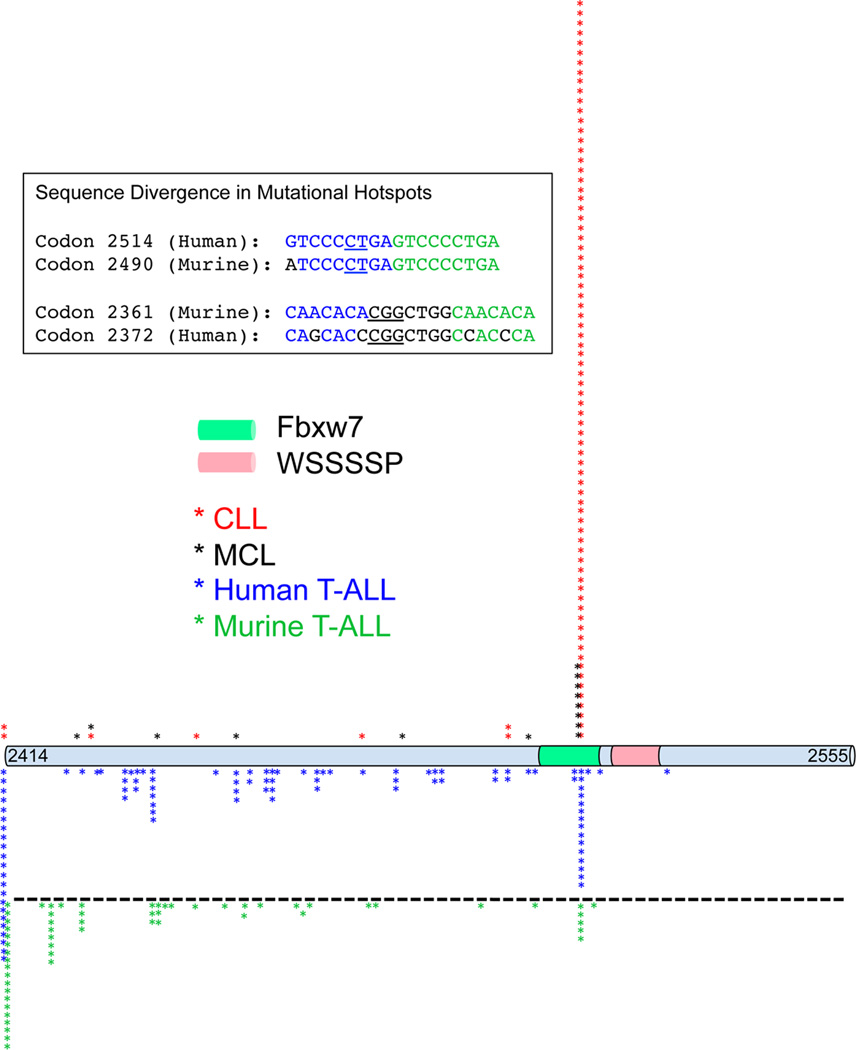
Several features of Notch1 mutations in CLL and MCL differ from those in T-ALL (Fig. 2). Virtually all of the mutations are PEST deletions, and of these roughly 90% in CLL and >50% in MCL consist of a del(CT) mutation involving codon 2514. This contrasts sharply with human and murine T-ALL, in which PEST mutations occur across a wide region (Fig. 2). Furthermore, while the NRR is the most common site of mutations in human T-ALL, and >95% of CLL and MCL have wild type NRR sequences. The Notch1 mutations in CLL and MCL thus raise several questions: 1) why are del(CT) mutations so prevalent (relative to T-ALL)?; 2) how is ICN1 generated in the absence of NRR mutations?; and 3) do Notch1 mutations reflect some normal role for Notch1 in mature human B cells?
Figure 2.
Distribution of Notch1 PEST domain mutations in lymphoid cancers. Positions of nonsense and frameshift mutations in human chronic lymphocytic leukemia (CLL) [11–13], human mantle cell lymphoma (MCL)[18], and human [43–46] and murine T-cell acute lymphoblastic leukemia/lymphoma (T-ALL) [47, 48] are superimposed on the human C-terminal Notch1 amino acids 2414 and 2555, which includes a Fbxw7 E3-ligase target sequence and the sequence WSSSSP, both implicated in degradation of activated Notch1. Murine Notch1 mutations are aligned according to the homologous amino acid sequences of human Notch1. Disruptive mutations N-terminal of amino acid 2414 of human Notch1 are collected together and thus appear as a single peak. The inset box shows the murine and human genomic sequences around mutational hotspots in human lymphoid tumors (codon 2514) and in murine T-ALL (codon 2361).
Structure/function analyses suggest that the functional effect of the codon 2514del(CT) mutation is unlikely to differ from other PEST mutations, and indeed other Notch1 PEST mutations occur in CLL and MCL. A more likely possibility is that codon 2514 is embedded within a DNA context that is prone to spontaneous microdeletion. Of note, codon 2514 lies within the first of two 9 base pair direct tandem repeats (Fig. 2), which may make this sequence susceptible to “slippage” during DNA replication. If this is correct, codon 2514del(CT) mutations should be relatively common in all human cancers in which there is selective pressure for Notch1 gain-of-function. Consistent with this prediction, codon2514 del(CT) is the most common NOTCH1 mutation in human T-ALL as well. The idea that DNA sequence influences the distribution of PEST mutations is also supported by a comparison of human and murine T-ALL (Fig. 2). In contrast to human T-ALL, in murine T-ALL the most common PEST mutations are generated by insertions or deletions centered on codon 2361 (codon 2372 in human NOTCH1), which in the mouse (but not in the human) is framed by two direct 7 base pair tandem repeats that may promote illegitimate recombination events. Del(CT) mutations also occur in murine T-ALL, but at lower frequency than in human disease, possibly because of a single base substitution in the first of the two direct repeats (Fig. 2).
How ICN1 is generated in CLL and MCL is of biologic and therapeutic importance, since the mechanism will dictate the choice of targeting strategies. CLL cells proliferate in a specialized niche found mainly in lymph nodes that is defined by the presence of “nurse cells”, which are known to express membrane-bound factors such as BAFF that stimulate CLL growth and survival; one attractive possibility is that these cells also express Notch ligands. A second possibility is that the region encoding the Notch1 NRR may be disrupted by a mutation that might be missed by whole exome sequencing, such as a deletion or rearrangement; as mentioned, this type of mutation is common in murine T-ALL and also occurs in human breast cancer (described later). Finally, genetic studies in invertebrates have shown that aberrant Notch trafficking into late endosomes leads to ligand-independent receptor activation; the contribution of this mechanism to oncogenic Notch signaling in mammalian cancer is unknown.
It will also be of interest to compare and contrast the genes and pathways that are upregulated by Notch1 in B-cell tumors and T-ALL. Major oncogenic targets of Notch1 in T-ALL include MYC, the PI3K/Akt pathway, and possibly the NF-kB pathway as well. Of possible relevance, EBNA2, an Epstein-Barr virus (EBV) protein that binds CSL and functionally resembles ICN, directly upregulates MYC in EBV-transformed B cells [21]. Expression profiling suggests that MYC is also a direct target of Notch1 in MCL cell lines [18], supporting the idea that Notch signaling promotes the proliferation of transformed B cells. Proliferation index predicts outcome in MCL [22], bolstering the rationale for targeting Notch1 in patients with this disease.
4. Evidence for oncogenic Notch signaling in breast carcinoma
Interest in Notch signaling in breast carcinoma is longstanding. Notch4 was originally identified as the target of a retroviral insertion in a murine mammary tumor [23], and subsequent work confirmed the oncogenic activity of ICN1 and ICN4 in murine mammary epithelium. In models using ICN1, it appears that its oncogenic effects are again mediated through the upregulation of MYC [24]. Other work has suggested roles for Notch in maintenance of breast cancer cells with “stem-like” properties [25, 26] and in promotion of anchorage-independent growth [27]. However, a “smoking gun” in the way of activating mutations in human breast cancer has been lacking.
This changed recently when Chinnaiyan’s group identified abnormal Notch mRNAs in eight breast carcinoma cell lines and primary tumors [28]. All of these tumors were negative for expression of estrogen receptor (ER−), and 7 or 8 belonged to the so-called triple negative subgroup that is associated with a worse prognosis. The aberrant transcripts resulted from cytogenetically silent rearrangements of the NOTCH1 (6/8) or NOTCH2 (2/8) genes; although several of the rearrangements produce fusion genes, none appear to encode chimeric proteins. Instead, the resulting mRNAs are predicted to encode polypeptides that initiate from start codons within (Notch1) or just internal to (Notch2) the transmembrane domain, the familiar theme being the expression of Notch polypeptides lacking an NRR. MYC again appeared to be a downstream target of Notch in human breast cancers with Notch gene rearrangements.
Of therapeutic importance, the aberrant Notch polypeptides identified by Chinnaiyan’s group will not be inhibited by antibodies directed against Notch receptor ectodomains or Notch ligands, or (in the case of the Notch2 polypeptides) even GSIs, all of which are being tested or considered as therapies in breast cancer. By contrast, agents that directly target the Notch transcription complex should be effective; such inhibitors are also in preclinical development.
5. The Flipside of the Coin: Notch as a Tumor Suppressor
Radtke and co-workers were the first to describe an increased incidence in skin cancers in conditional notch1 knockout mice [29]. Deletion of Notch1 in the skin of one-week old mice resulted in spontaneous basal cell carcinoma (BCC)-like lesions with increased Gli2 expression in 95% of mice by 12 months of age. The incidence of papilloma formation in notch1 null skin was also markedly increased by exposure to chemical carcinogens plus phorbol ester, with a small proportion progressing to either BCC or squamous cell carcinoma (SCC). Proweller et al. next noted that expression of a specific inhibitor of the Notch transcription complex, dominant negative MAML1 (DN-MAML), in murine skin led to the development of cutaneous SCC but not BCC [30]. Differences in tumor phenotypes observed in these two models may have been related to the degree of Notch loss-of-function, as multiple Notch receptors are expressed in keratinocytes and DN-MAML is a pan-Notch inhibitor that produced a much more severe preneoplastic phenotype consisting of cutaneous hyperplasia and alopecia. Subsequent work showed that human keratinocytes expressing oncogenic RAS and deficient in Notch signaling form SCC in immunocompromised mice [31], setting the stage for the genomic discoveries described below.
A number of tumor suppressive mechanisms have been proposed for Notch in the skin. Dotto’s group identified several Notch targets that may mediate pro-differentiation and anti-growth effects, such as the cell cycle regulator p21 [32] and the transcription factor Irf6 [33]. Crosstalk between Notch and other pathways linked to skin carcinogenesis, including Ras, NF-kB, Wnt, and Hedgehog, have also been described in various contexts, each of which could contribute to a cell autonomous tumor suppressive activity. By contrast, Kopan and co-workers have postulated that epidermal barrier defects caused by Notch loss-of-function produce a chronic cutaneous inflammatory state that promotes cancer development through factors released from dermal stromal and inflammatory cells [34]. Thus, it is possible loss of Notch function contributes to skin carcinogenesis through both cell autonomous and non-cell autonomous effects.
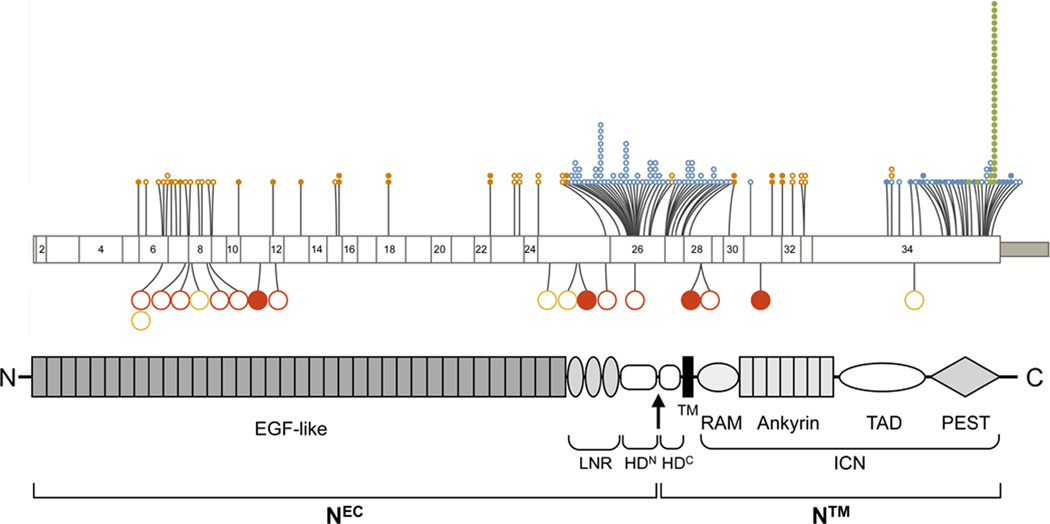
While the mechanism(s) remain to be defined, abundant genetic evidence emerging in the past year is consistent with Notch having a tumor suppressive role in multiple types of human SCC (Table 2). Whole exome deep sequencing by two groups independently identified likely loss-of-function mutations in a least one Notch signaling component in roughly 15% to 20% of head and neck SCC (Fig. 3) [35, 36]. In parallel, in studies also using whole exome deep sequencing, we noted the presence of at least one putative loss-of-function mutation involving either NOTCH1 or NOTCH2 in 19 of 26 primary cutaneous SCC or derived cell lines [37]. Most of these mutations were truncating or likely to be structurally disruptive, and one mutation predicted to be “benign” based on polyphen-2 analysis, a R1594Q substitution in the Notch1 NRR, interfered with ligand-mediated receptor activation in cell-based assays [37]. Several other mutations identified in our series were also confirmed to produce loss-of-function, including a point substitution in intracellular Notch1 that abrogates binding of ICN1 to RBPJ, and a point substitution in EGF repeat 12 that disrupts ligand-binding [37, 38]. Despite the preceding murine data, no NOTCH1 or NOTCH2 mutations were seen in 8 BCC [37]; sequencing of additional tumors will be needed to see if this difference is real or merely a product of small sample size.
Figure 3.
Distribution of cancer-associated missense (open circle) and nonsense (closed circle) mutations in NOTCH1, organized by exon and protein domain. Selected activating mutations in T-ALL (blue) and CLL (green) are compared with putative loss-of-function mutations for head and neck SCC (orange), cutaneous SCC (red) and lung SCC (yellow).
These results raise the question of whether Notch signaling is disrupted in all SCCs in one way or another. Of note in this regard, MAML1 was originally identified as a human papilloma virus 16 (HPV16) E6-binding protein. Targeting of the Notch signaling pathway by high-risk human papilloma viruses such as HPV16 (a possibility yet to be thoroughly explored) would further solidify the fundamental role of the Notch signaling pathway as a tumor suppressor in human SCC. However, Agrawal et al. [36] noted several HPV-positive tumors in their series of head and neck cancers that nevertheless exhibit Notch gene mutations, and the contribution of HPV interference with Notch to squamous cell carcinogenesis requires further investigation.
A smaller fraction of lung SCC have acquired Notch receptor mutations [37], and the importance of Notch as a tumor suppressor in this other type of SCC is unsettled at this time. Conversely, a paper relying on focused Sanger resequencing reported that gain-of-function Notch1 PEST domain mutations were found in roughly 10% of human lung carcinomas [39], but deep sequencing has yet to confirm this association.
6. Other Possible Tumor Suppressive Roles for Notch
Aifantis’ group reported recently that knockout of nicastrin, a component of g-secretase, produces a myeloproliferative disorder in mice [40]. This same report identified several mutations involving Notch pathway components in human chronic myelomonocytic leukemia, including a truncation mutation in MAML1 predicted to yield a dominant negative polypeptide. Loss-of-function mutations in Notch receptors were not detected, however, and since γ-secretase cleaves many substrates besides Notch and MAMLs interact with multiple signaling pathways, these phenotype could stem from “off-Notch” effects. Sequencing of the genomes of human myeloproliferative disorders will soon reveal the extent of Notch’s involvement as a tumor suppressor in this type of human malignancy.
Other work has raised the specter of a tumor suppressive role for Notch signaling in vascular tumors. Notch and VEGF co-regulate many facets of vasculogenesis, angiogenesis, and endothelial cell homeostasis. Rats treated chronically with DLL4 blocking antibody develop subcutaneous vascular tumors and hepatic vascular abnormalities associated with endothelial cell activation [41]. Furthermore, conditional notch1 deficiency in mice is also associated with vascular proliferations, most commonly in the liver [42]. It will thus be of interest to determine if deep sequencing of human angiosarcomas or other human vascular neoplasms reveals Notch loss-of-function mutations.
7. Concluding Remarks
Recent discoveries emphasize the multifaceted role of Notch signaling in cancer. Notch is an increasingly attractive therapeutic target in multiple human malignancies, including some not discussed here, such as melanoma and ovarian cancer. Notch signaling also has roles in the immune system that suggests opportunities for therapeutic intervention in other cancer-relevant conditions, such as graft versus host disease. Conversely, given the frequency of SCC in light-skinned sun-exposed populations, it is likely that the tumor suppressive role of Notch signaling supersedes its oncogenic role, a relationship that will further complicate attempts to treat patients with drugs or antibodies that chronically inactivate Notch. This includes the use of GSIs in patients with Alzheimer disease, which has been attributed to pathogenic peptides generated by cleavage of amyloid precursor protein by γ-secretase. Notably, a phase III trial of the GSI semagacestat was stopped in 2010, mainly due to worsening of cognitive function, but also because the treatment group had an increased incidence of skin cancers. Going forward, this risk will need to be calculated into any attempt to target oncogenic Notch signaling with broad-spectrum inhibitors such as GSIs.
On a basic level, the divergent context-dependent effects of Notch signaling in cancer cells speaks to a fundamental question: why are certain genes commonly mutated in some cancers but not others? Notch1 takes this question to a new level by being the most frequently mutated oncoprotein in one human cancer (T-ALL) and one of the most frequently mutated tumor suppressors in a second (SCC). Recently developed whole genomic approaches for understanding how factors regulate transcriptomes globally, such as ChIP-Seq, now provide a means for elucidating the basis for the double-edged effect of Notch signaling in various cancers genome-wide. Together with detailed study of Notch/genome interactions during normal development, these studies should lead to a fuller understanding of Notch’s cell autonomous roles in cancer.
Highlights.
We present new findings linking activating mutations in Notch1 to B cell neoplasms
We discuss the discovery of Notch gene rearrangements in breast cancer
We review the discovery of loss-of-function mutations in Notch receptors in squamous cell carcinoma
Acknowledgements
We thank Zack Sanborn for assistance with illustrations. We apologize for the need to overlook the contributions of many workers in the field due to space constraints. Supported by grants from the National Institutes of Health and the Leukemia and Lymphoma Society (J.C.A.), the Dermatology Foundation (R.J.C.), and DebRA International and the British Skin Foundation (A.P.S).
Abbreviations
- GSI
γ-secretase inhibitor
- NRR
negative regulatory region
- EGF
epidermal growth factor
- LNR
Lin12/Notch repeat
- ADAM
a disintegrin and metalloprotease
- DSL
Delta-Serrate-Lag2
- ICN
intracellular domain of Notch
- MAML
Mastermind-like
- RBPJ
recombining signal binding protein for immunoglobulin kappa J region
- CLL
chronic lymphocytic leukemia
- T-ALL
T-cell acute lymphoblastic leukemia/lymphoma
- MCL
mantle cell lymphoma
- SCC
squamous cell carcinoma
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009 Apr 17;137(2):216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aster JC, Pear WS, Blacklow SC. Notch signaling in leukemia. Annu Rev Pathol. 2008;3:587–613. doi: 10.1146/annurev.pathmechdis.3.121806.154300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Radtke F, Fasnacht N, Macdonald HR. Notch signaling in the immune system. Immunity. 2010 Jan 29;32(1):14–27. doi: 10.1016/j.immuni.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 4.Pui JC, Allman D, Xu L, DeRocco S, Karnell FG, Bakkour S, et al. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 1999 Sep;11(3):299–308. doi: 10.1016/s1074-7613(00)80105-3. [DOI] [PubMed] [Google Scholar]
- 5.Zweidler-McKay PA, He Y, Xu L, Rodriguez CG, Karnell FG, Carpenter AC, et al. Notch signaling is a potent inducer of growth arrest and apoptosis in a wide range of B-cell malignancies. Blood. 2005 Dec 1;106(12):3898–3906. doi: 10.1182/blood-2005-01-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saito T, Chiba S, Ichikawa M, Kunisato A, Asai T, Shimizu K, et al. Notch2 is preferentially expressed in mature B cells and indispensable for marginal zone B lineage development. Immunity. 2003 May;18(5):675–685. doi: 10.1016/s1074-7613(03)00111-0. [DOI] [PubMed] [Google Scholar]
- 7.Thomas M, Calamito M, Srivastava B, Maillard I, Pear WS, Allman D. Notch activity synergizes with B-cell-receptor and CD40 signaling to enhance B-cell activation. Blood. 2007 Apr 15;109(8):3342–3350. doi: 10.1182/blood-2006-09-046698. [DOI] [PubMed] [Google Scholar]
- 8.Santos MA, Sarmento LM, Rebelo M, Doce AA, Maillard I, Dumortier A, et al. Notch1 engagement by Delta-like-1 promotes differentiation of B lymphocytes to antibody-secreting cells. Proc Natl Acad Sci U S A. 2007 Sep 25;104(39):15454–15459. doi: 10.1073/pnas.0702891104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klein U, Tu Y, Stolovitzky GA, Mattioli M, Cattoretti G, Husson H, et al. Gene expression profiling of B cell chronic lymphocytic leukemia reveals a homogeneous phenotype related to memory B cells. J Exp Med. 2001 Dec 3;194(11):1625–1638. doi: 10.1084/jem.194.11.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Ianni M, Baldoni S, Rosati E, Ciurnelli R, Cavalli L, Martelli MF, et al. A new genetic lesion in B-CLL: a NOTCH1 PEST domain mutation. Br J Haematol. 2009 Sep;146(6):689–691. doi: 10.1111/j.1365-2141.2009.07816.x. [DOI] [PubMed] [Google Scholar]
- 11.Sportoletti P, Baldoni S, Cavalli L, Del Papa B, Bonifacio E, Ciurnelli R, et al. NOTCH1 PEST domain mutation is an adverse prognostic factor in B-CLL. Br J Haematol. 2010 Nov;151(4):404–406. doi: 10.1111/j.1365-2141.2010.08368.x. [DOI] [PubMed] [Google Scholar]
- 12.Puente XS, Pinyol M, Quesada V, Conde L, Ordonez GR, Villamor N, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011 Jul 7;475(7354):101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fabbri G, Rasi S, Rossi D, Trifonov V, Khiabanian H, Ma J, et al. Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation. J Exp Med. 2011 Jul 4;208(7):1389–1401. doi: 10.1084/jem.20110921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rasi S, Monti S, Spina V, Foa R, Gaidano G, Rossi D. Analysis of NOTCH1 mutations in monoclonal B cell lymphocytosis. Haematologica. 2011 Oct 11; doi: 10.3324/haematol.2011.053090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Del Giudice I, Rossi D, Chiaretti S, Marinelli M, Tavolaro S, Gabrielli S, et al. NOTCH1 mutations in +12 chronic lymphocytic leukemia (CLL) confer an unfavorable prognosis, induce a distinctive transcriptional profiling and refine the intermediate prognosis of +12 CLL. Haematologica. 2011 Dec 29; doi: 10.3324/haematol.2011.060129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balatti V, Bottoni A, Palamarchuk A, Alder H, Rassenti LZ, Kipps TJ, et al. NOTCH1 mutations in CLL associated with trisomy 12. Blood. 2011 Nov 15; doi: 10.1182/blood-2011-10-386144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med. 2011 Dec 29;365(26):2497–2506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kridel R, Meissner B, Rogic S, Boyle M, Telenius A, Woolcock B, et al. Whole transcriptome sequencing reveals recurrent NOTCH1 mutations in mantle cell lymphoma. Blood. 2011 doi: 10.1182/blood-2011-11-391474. epublished December 30, 2011. [DOI] [PubMed] [Google Scholar]
- 19.Leshchenko VV, Kuo PY, Shaknovich R, Yang DT, Gellen T, Petrich A, et al. Genomewide DNA methylation analysis reveals novel targets for drug development in mantle cell lymphoma. Blood. 2010 Aug 19;116(7):1025–1034. doi: 10.1182/blood-2009-12-257485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee SY, Kumano K, Nakazaki K, Sanada M, Matsumoto A, Yamamoto G, et al. Gain-of-function mutations and copy number increases of Notch2 in diffuse large B-cell lymphoma. Cancer Sci. 2009 May;100(5):920–926. doi: 10.1111/j.1349-7006.2009.01130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao B, Zou JY, Wang H, Johannsen E, Peng C-W, Quackenbush J, et al. Epstein-Barr virus exploits intrinsic B-lymphocyte transcription programs to achieve immortal cell growth. Proc Natl Acad Sci U S A. 2011;108:14902–14907. doi: 10.1073/pnas.1108892108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenwald A, Wright G, Wiestner A, Chan WC, Connors JM, Campo E, et al. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell. 2003 Feb;3(2):185–197. doi: 10.1016/s1535-6108(03)00028-x. [DOI] [PubMed] [Google Scholar]
- 23.Robbins J, Blondel BJ, Gallahan D, Callahan R. Mouse mammary tumor gene int-3: a member of the notch gene family transforms mammary epithelial cells. J Virol. 1992 Apr;66(4):2594–2599. doi: 10.1128/jvi.66.4.2594-2599.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klinakis A, Szabolcs M, Politi K, Kiaris H, Artavanis-Tsakonas S, Efstratiadis A. Myc is a Notch1 transcriptional target and a requisite for Notch1-induced mammary tumorigenesis in mice. Proc Natl Acad Sci U S A. 2006 Jun 13;103(24):9262–9267. doi: 10.1073/pnas.0603371103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kakarala M, Wicha MS. Implications of the cancer stem-cell hypothesis for breast cancer prevention and therapy. J Clin Oncol. 2008 Jun 10;26(17):2813–2820. doi: 10.1200/JCO.2008.16.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrison H, Farnie G, Howell SJ, Rock RE, Stylianou S, Brennan KR, et al. Regulation of breast cancer stem cell activity by signaling through the Notch4 receptor. Cancer Res. 2010 Jan 15;70(2):709–718. doi: 10.1158/0008-5472.CAN-09-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mazzone M, Selfors LM, Albeck J, Overholtzer M, Sale S, Carroll DL, et al. Dose-dependent induction of distinct phenotypic responses to Notch pathway activation in mammary epithelial cells. Proc Natl Acad Sci U S A. 2010 Mar 16;107(11):5012–5017. doi: 10.1073/pnas.1000896107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson DR, Kalyana-Sundaram S, Wu YM, Shankar S, Cao X, Ateeq B, et al. Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nat Med. 2011;17(12):1646–1651. doi: 10.1038/nm.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicolas M, Wolfer A, Raj K, Kummer JA, Mill P, van Noort M, et al. Notch1 functions as a tumor suppressor in mouse skin. Nat Genet. 2003 Mar;33(3):416–421. doi: 10.1038/ng1099. [DOI] [PubMed] [Google Scholar]
- 30.Proweller A, Tu L, Lepore JJ, Cheng L, Lu MM, Seykora J, et al. Impaired notch signaling promotes de novo squamous cell carcinoma formation. Cancer Res. 2006 Aug 1;66(15):7438–7444. doi: 10.1158/0008-5472.CAN-06-0793. [DOI] [PubMed] [Google Scholar]
- 31.Lefort K, Mandinova A, Ostano P, Kolev V, Calpini V, Kolfschoten I, et al. Notch1 is a p53 target gene involved in human keratinocyte tumor suppression through negative regulation of ROCK1/2 and MRCKalpha kinases. Genes Dev. 2007 Mar 1;21(5):562–577. doi: 10.1101/gad.1484707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rangarajan A, Talora C, Okuyama R, Nicolas M, Mammucari C, Oh H, et al. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 2001 Jul 2;20(13):3427–3436. doi: 10.1093/emboj/20.13.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Restivo G, Nguyen BC, Dziunycz P, Ristorcelli E, Ryan RJ, Ozuysal OY, et al. IRF6 is a mediator of Notch pro-differentiation and tumour suppressive function in keratinocytes. EMBO J. 2011 Sep 9; doi: 10.1038/emboj.2011.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Demehri S, Turkoz A, Kopan R. Epidermal Notch1 loss promotes skin tumorigenesis by impacting the stromal microenvironment. Cancer Cell. 2009 Jul 7;16(1):55–66. doi: 10.1016/j.ccr.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011 Aug 26;333(6046):1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011 Aug 26;333(6046):1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang NJ, Sanborn Z, Arnett KL, Bayston LJ, Liao W, Proby CM, et al. Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma. Proc Natl Acad Sci U S A. 2011 Oct 25;108(43):17761–17766. doi: 10.1073/pnas.1114669108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cordle J, Redfieldz C, Stacey M, van der Merwe PA, Willis AC, Champion BR, et al. Localization of the delta-like-1-binding site in human Notch-1 and its modulation by calcium affinity. J Biol Chem. 2008 Apr 25;283(17):11785–11793. doi: 10.1074/jbc.M708424200. [DOI] [PubMed] [Google Scholar]
- 39.Westhoff B, Colaluca IN, D'Ario G, Donzelli M, Tosoni D, Volorio S, et al. Alterations of the Notch pathway in lung cancer. Proc Natl Acad Sci U S A. 2009 Dec 29;106(52):22293–22298. doi: 10.1073/pnas.0907781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klinakis A, Lobry C, Abdel-Wahab O, Oh P, Haeno H, Buonamici S, et al. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature. 2011 May 12;473(7346):230–233. doi: 10.1038/nature09999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan M, Callahan CA, Beyer JC, Allamneni KP, Zhang G, Ridgway JB, et al. Chronic DLL4 blockade induces vascular neoplasms. Nature. 2010 Feb 11;463(7282):E6–E7. doi: 10.1038/nature08751. [DOI] [PubMed] [Google Scholar]
- 42.Liu Z, Turkoz A, Jackson EN, Corbo JC, Engelbach JA, Garbow JR, et al. Notch1 loss of heterozygosity causes vascular tumors and lethal hemorrhage in mice. J Clin Invest. 2011 Feb 1;121(2):800–808. doi: 10.1172/JCI43114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weng AP, Ferrando AA, Lee W, Morris JP, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004 Oct 8;306(5694):269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 44.Breit S, Stanulla M, Flohr T, Schrappe M, Ludwig WD, Tolle G, et al. Activating NOTCH1 mutations predict favorable early treatment response and long-term outcome in childhood precursor T-cell lymphoblastic leukemia. Blood. 2006 Aug 15;108(4):1151–1157. doi: 10.1182/blood-2005-12-4956. [DOI] [PubMed] [Google Scholar]
- 45.van Grotel M, Meijerink JP, van Wering ER, Langerak AW, Beverloo HB, Buijs-Gladdines JG, et al. Prognostic significance of molecular-cytogenetic abnormalities in pediatric T-ALL is not explained by immunophenotypic differences. Leukemia. 2008 Jan;22(1):124–131. doi: 10.1038/sj.leu.2404957. [DOI] [PubMed] [Google Scholar]
- 46.Asnafi V, Buzyn A, Le Noir S, Baleydier F, Simon A, Beldjord K, et al. NOTCH1/FBXW7 mutation identifies a large subgroup with favorable outcome in adult T-cell acute lymphoblastic leukemia (T-ALL): a Group for Research on Adult Acute Lymphoblastic Leukemia (GRAALL) study. Blood. 2009 Apr 23;113(17):3918–3924. doi: 10.1182/blood-2008-10-184069. [DOI] [PubMed] [Google Scholar]
- 47.Lin YW, Nichols RA, Letterio JJ, Aplan PD. Notch1 mutations are important for leukemic transformation in murine models of precursor-T leukemia/lymphoma. Blood. 2006 Mar 15;107(6):2540–2543. doi: 10.1182/blood-2005-07-3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Neil J, Calvo J, McKenna K, Krishnamoorthy V, Aster JC, Bassing CH, et al. Activating Notch1 mutations in mouse models of T-ALL. Blood. 2006 Jan 15;107(2):781–785. doi: 10.1182/blood-2005-06-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]