SUMMARY
Background
The centrosome has a well-established role as a microtubule organizer during mitosis and cytokinesis. In addition, it facilitates the union of parental haploid genomes following fertilization by nucleating a microtubule aster along which the female pronucleus migrates towards the male pronucleus. Stable associations between the sperm aster and the pronuclei are essential during this directed movement.
Results
Our studies reveal that the zebrafish gene futile cycle (fue) is required in the zygote for male pronucleus-centrosome attachment and female pronuclear migration. We show that fue encodes a novel, maternally-provided long form of lymphoid-restricted membrane protein (lrmp), a vertebrate-specific gene of unknown function. Both maternal lrmp mRNA and protein are highly localized in the zygote, in a largely overlapping pattern at nuclear membranes, centrosomes, and spindles. Truncated Lrmp-EGFP fusion proteins identified subcellular targeting signals in the C-terminus of Lrmp, however endogenous mRNA localization is likely important to ensure strict spatial expression of the protein. Localization of both Lrmp protein and lrmp RNA is defective in fue mutant embryos, indicating that correct targeting of lrmp gene products is dependent on Lrmp function.
Conclusions
Lrmp is a conserved vertebrate gene whose maternally-inherited products are essential for nucleus-centrosome attachment and pronuclear congression during fertilization. Precise subcellular localization of lrmp products also suggests a requirement for strict spatiotemporal regulation of their function in the early embryo.
Keywords: zebrafish, maternal-effect gene, futile cycle (fue), lymphoid-restricted membrane protein (lrmp), centrosome, sperm aster, fertilization, RNA localization
INTRODUCTION
In many species including zebrafish, pronuclear congression during fertilization depends on a microtubule aster that forms near the male pronucleus [1–3]. This sperm aster is nucleated by the zygotic centrosome, a structure built from paternally inherited centrioles and maternally provided pericentriolar material [2,4,5]. During pronuclear congression, the male pronucleus is associated with the center of this aster at the centrosome, while astral microtubules attach to the female pronucleus and facilitate its dynein/dynactin-dependent migration toward the centrosome and male pronucleus [3,6–9]. This movement results in the apposition and subsequent fusion of male and female pronuclei.
Zygotes from homozygous futile cycle zebrafish females (fue embryos) fail to undergo pronuclear congression and fusion [10]. Here we show that embryos lacking fue products exhibit defects in the ability of pronuclei to interact with the sperm aster. We determine that fue encodes a maternally expressed long isoform of Lymphoid-restricted membrane protein (Lrmp). Lrmp is conserved in vertebrate lineages and shares structural features with proteins that link the nuclear envelope to the cytoskeleton. We also find that fue/lrmp mRNA exhibits cell cycle-dependent subcellular localization patterns that likely facilitate Lrmp protein enrichment at the nuclear envelope. Our studies reveal an essential function for a conserved gene and offer novel insights into mechanisms of nuclear dynamics in early vertebrate embryos.
RESULTS
Pronuclei exhibit defects in their interactions with the sperm aster in fue zygotes
The fue mutation was previously shown to affect pronuclear migration and fusion ([10], Figure S1A,B). Formation of a sperm aster is critical for pronuclear migration in many species [2,5]. However, analysis of fue mutants revealed normal astral microtubule growth following fertilization (Figure S1, C–F).
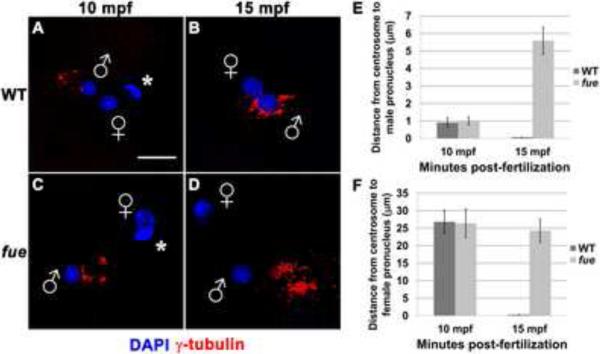
The zygotic centrosome resides at the core of the sperm aster, which is normally in close proximity to the male pronucleus during pronuclear congression [5]. To determine if the centrosome-male pronucleus association is affected in fue embryos, we conducted antibody labeling for the centrosomal component γ-tubulin. At 10 minutes post-fertilization (mpf) in both wild-type and fue embryos, centrosomal material lies within a few microns of the male pronucleus (Figure 1A,C,E). By 15 mpf in wild-type embryos, the centrosome is in complete juxtaposition with the male pronucleus (< 1 micron distance, Figure 1B,E, Movie S1). In fue embryos at the same time point, γ-tubulin appears separate from the male pronucleus, at a distance of 4 to 6 microns (Figure 1D,E, Movie S2). These data indicate a failure of the centrosome to attach to the male pronucleus in fue mutant embryos.
Figure 1. Centrosomes fail to attach to pronuclear envelopes in fue embryos.
In vitro fertilized embryos from wild-type and fue females fixed and labeled for centrosomes (γ-tubulin antibody, red) and DNA (DAPI, blue) at 10 (A,C) and 15 mpf (B,D). Asterisks indicate polar bodies, and male and female pronuclei are indicated with symbols. Scale bar represents 20 μm and applies to all panels. Images are projections from confocal z-stacks. (E,F) Distance between centrosomal γ-tubulin labeling and male or female pronuclear envelopes quantified at 10 and 15 mpf. Error bars indicate +/- 1 standard error. See also Figure S1, and Movies S1 and S2.
We next examined whether the female pronucleus still migrates toward the detached centrosome in mutant embryos. Immediately after fertilization in both wild-type and fue mutant zygotes, the female pronucleus is found at significance distance from the center of the sperm aster (average distance in both cases: approx. 25 μm; Figure 1F). By 15 mpf in wild-type zygotes, the female pronucleus is directly adjacent to the centrosome, indicating that female pronuclear migration has occurred, and pronuclear fusion is underway (Figure 1B,F, movie S1). At 15 mpf in fue mutants (Figure 1D,F, movie S2), the female pronucleus is at a distance from the centrosome similar to that at 10 mpf, suggesting that the female pronucleus does not undergo centrosome-directed migration along the sperm aster. Thus, fue function is required for pronuclei to interact successfully with the centrosome and the sperm aster.
futile cycle encodes a maternally-expressed Lrmp long isoform
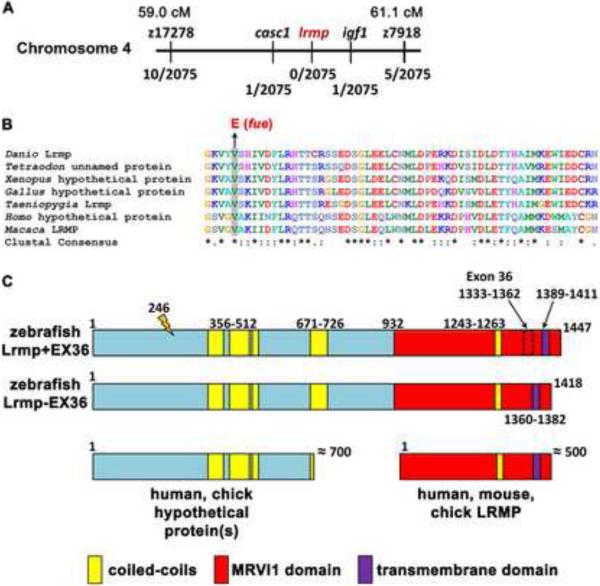
Positional cloning of fue traced the mutation to chromosome 4 in a region containing several transcripts including lymphoid-restricted membrane protein (lrmp) (Figure 2A). Further analysis uncovered a molecular lesion in exon 4 of lrmp consisting of a T-to-A transition predicted to cause a valine (V) – to – glutamate (E) substitution at residue 246 (Figure 2B,C). Sequence analysis of this portion of Lrmp revealed that the mutated residue is conserved across species (Figure 2B). Sequencing of maternally-derived cDNA products indicated the presence of two lrmp isoforms, which differ by 89 base pairs (bp). lrmp+EX36 (lrmp-001 in Vega) includes a penultimate exon (exon 36) not found in the shorter isoform (lrmp-EX36) (Figure 2C). The two isoforms behave similarly in expression studies (see below) and thus are referred to together as “Lrmp”.
Figure 2. The fue molecular lesion results in an amino acid substitution at a conserved residue of lymphoid-restricted membrane protein.
(A) Positional cloning indicated linkage of fue to chromosome 4 between markers z17278 and z7918. On the physical map, the mutation lies between markers in cancer susceptibility candidate 1 (casc1) and insulin-like growth factor 1 (igf1). Fractions below markers indicate the number of recombinant meiotic events at each position out of 2075 examined meioses. (B) A point mutation was discovered in the fourth exon of lrmp, predicted to cause a valine-to-glutamate amino acid substitution at a conserved residue. (C) Alternative splicing involving exon 36 results in proteins of 1447 (Lrmp+EX36, NCBI accession #CAI20727) and 1418 (Lrmp−EX36) residues. The N-terminal region of zebrafish Lrmp is homologous to hypothetical proteins in humans, chick, and several other species. The C-terminal 500 residues in zebrafish Lrmp correspond to Lrmp proteins described in mouse and human cells, and predicted in chick. Predicted domains and fue mutation site (orange marker at amino acid 246) are indicated. See also Figure S2 and Table S1.
The predicted open reading frame of zebrafish lrmp is consistent with the observed 5.1 kilobase (kb) transcript detected by Northern blot analysis (data not shown). The transcript encodes a protein significantly longer (1447 amino acids) than Lrmp isoforms previously described in mouse and humans (535 and 555 amino acids, respectively, [11]). The majority of annotated Lrmp genes in other vertebrates encompass only the C-terminal portion of zebrafish Lrmp (Figure 2C, Table S1). However, the N-terminal half of zebrafish Lrmp is homologous to a novel predicted protein directly upstream of the annotated lrmp gene in humans, chick and several other species (Figure 2C, Table S1). Moreover, species such as Taeniopygia guttata (zebra finch), Equus caballus (horse), and Macaca mulatta (rhesus macaque) have predicted long lrmp transcripts likely equivalent to zebrafish Lrmp (Table S1). While lrmp appears to have homologues in all vertebrate lineages, proteins with significant extended sequence similarity to zebrafish Lrmp are not found in most invertebrates including Drosophila and Caenorhabditis elegans, or in yeast and plants (Table S1, Figure S2A,B).
Quantitative RT-PCR analysis shows that zebrafish lrmp is most highly expressed in early cleavage embryos, decreases approximately 70% by 4 hours post-ferlization (hpf) (coincident with the midblastula transition [12–14], Figure S3), and cannot be detected at 24hpf (data not shown). Thus, zebrafish lrmp exhibits an expression pattern characteristic of a strict maternal gene.
Subcellular localization of lrmp mRNA
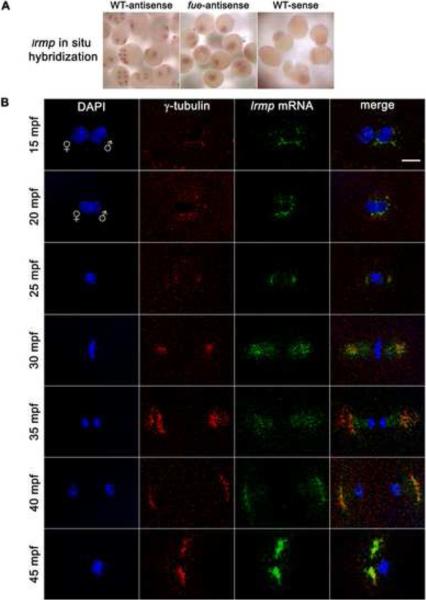
Whole-mount in situ hybridization showed that lrmp transcripts have an apparently perinuclear localization pattern in cleavage stage embryos (Figure 3A, left panel). Consistent with our qRT-PCR findings, lrmp transcript labeling decreased by the 512–1000 cell stage (3 hpf), although subcellular localization was still discernible (data not shown).
Figure 3. lrmp transcripts show dynamic localization patterns lost in fue mutants.
(A) Chromogenic in situ hybridization with lrmp antisense probes in wild-type (left) and fue mutants (center), and negative control sense probes (right). (B) Wild-type embryos fixed at 5-minute intervals and labeled with γ–tubulin antibody (red) and DAPI (blue), in combination with fluorescent in situ detection of lrmp mRNA (green). Scale bar represents 20 μm and applies to all panels in (B). See also Figure S3.
To examine the lrmp mRNA localization pattern in detail, we combined fluorescent in situ hybridization with γ-tubulin antibody labeling (Figure 3B). In 5 mpf wild-type embryos, γ-tubulin and lrmp mRNA localization could not be reliably detected, likely because maternally-derived centrosomal components have not yet robustly assembled around the paternally-derived centrioles. By 10 mpf, γ–tubulin and lrmp mRNA spatially overlap near the male pronucleus (data not shown, see Figure 1 for γ-tubulin labeling at 10 mpf). Just prior to pronuclear fusion (15 mpf), lrmp mRNA colocalizes with γ–tubulin, coating the male pronucleus (Figure 3B, top row). During pronuclear fusion, γ–tubulin and lrmp mRNA continue to colocalize as they spread around the fusing nuclei (20 mpf, Figure 3B, second row).
As embryos enter mitosis and nuclear envelopes break down, some lrmp mRNA remains surrounding the condensing DNA while the majority localizes with γ-tubulin at the spindle poles (25 mpf, Figure 3B, third row). At metaphase, γ–tubulin and lrmp mRNA maintain colocalization at the spindle poles and also seem to extend onto spindle regions (Figure 3B, fourth row). As mitosis progresses, lrmp mRNA remains associated with centrosomes and appears to be present along the spindle (Figure 3B, fifth row). By late mitosis, localization of lrmp mRNA to the spindle is less extensive and the majority of lrmp transcript labeling again overlaps γ–tubulin labeling (Figure 3B, sixth row). As centrosomes separate, lrmp mRNA continues to colocalize with γ–tubulin near reforming nuclei (Figure 3B, bottom row). This pattern of localization is repeated throughout subsequent cell cycles (data not shown).
Parallel experiments using standard or fluorescent in situ hybridization showed that the lrmp subcellular localization pattern is entirely lost in fue embryos. Transcripts appear instead to be ubiquitously distributed throughout the blastodisc (Figure 3A, middle panel). Quantitative RT-PCR analysis revealed that lrmp mRNA levels do not differ significantly between wild-type and fue embryos (Figure S3), suggesting that localization but not stability of the lrmp transcript is affected in mutants. Activated, unfertilized wild-type eggs, which lack sperm-derived centrioles, also show ubiquitous distribution of lrmp transcripts (data not shown). These effects indicate a dependence of lrmp mRNA localization on fue function and possibly the presence of centrosomes (see Discussion).
Subcellular localization of Lrmp protein
Western blot analysis of cleavage-stage embryos with two antibodies against zebrafish Lrmp showed a common cross-reactive band at approximately 200 kDa (Figure S4A). In agreement with lrmp transcript abundance, the 200 kDa band was robust at cleavage stages but undetectable at 24 hpf. Notably, the 200 kDa band appeared reduced in fue embryo lysates compared to wild-type and only fue lysates showed faint 160 kDa bands, suggesting that Lrmp protein is either inefficiently translated or unstable in mutant embryos.
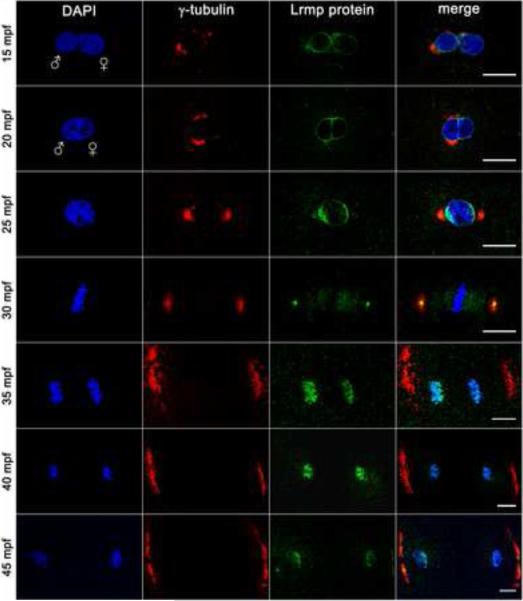
To determine the subcellular localization of Lrmp protein, in vitro fertilized wild-type embryos were fixed and labeled to detect γ-tubulin and Lrmp. In wild-type embryos at 10 mpf, Lrmp protein is present at both male and female pronuclear membranes (data not shown). By 15 mpf, when the pronuclei are in close proximity, Lrmp persists at the nuclear membranes with enrichment in regions adjacent to centrosomal γ–tubulin (Figure 4, top row). The membrane enrichment flanking centrosomes becomes more pronounced by 20 mpf during pronuclear fusion (Figure 4, second row).
Figure 4. Lrmp protein localizes to nuclear membranes and subregions of the mitotic apparatus.
Wild-type embryos fixed at 5-minute intervals and labeled with γ–tubulin antibody (red), anti-LrmpMD antiserum (green) and DAPI (blue). Scale bars represent 20 μm. See also Figure S4.
Beginning in prophase, a pool of centrosome-localized Lrmp protein can be detected on opposite sides of the zygotic nucleus (Figure 4, third row). At metaphase, Lrmp is present at the center of centrosomes and appears to extend along the mitotic spindle, but is absent from condensed chromosomes (Figure 4, fourth row). During chromosome segregation, Lrmp protein localizes with DNA again (Figure 4, fifth and sixth rows), likely as membrane begins to associate with chromatin to reform nuclear envelopes [15]. Centrosomal Lrmp is undetectable at the onset of anaphase, though it becomes apparent again by late mitosis (Figure 4, bottom row). Similar to the localization of lrmp transcript, this pattern of Lrmp protein localization is repeated in subsequent cell cycles (data not shown).
In parallel experiments, localized Lrmp protein was significantly reduced in mutant embryos during pronuclear migration and early cleavage stages (Figure S4B). Western blot analysis shows that Lrmp protein levels at 2hpf are only slightly lower in mutants compared to wild-type (Figure S4A), suggesting that the protein localization defect cannot be solely explained by a reduction in protein levels. In contrast to lrmp mRNA, Lrmp protein localizes to the pronuclear membrane even in unfertilized, activated wild-type eggs (Figure S4C), indicating that nuclear envelope targeting of Lrmp protein can occur independently of centrosomes.
Fluorescent in situ hybridization for lrmp transcript combined with antibody staining for Lrmp protein corroborated single-label experiments and provided additional insights. At prometaphase, when Lrmp protein resides adjacent to the condensed DNA, the majority of lrmp mRNA is concentrated just outside the zone of Lrmp protein, presumably closer to the centrosomes (Figure S4D, top row). In prometaphase and during early anaphase, there is significant overlap of lrmp mRNA and protein in regions corresponding to the spindle. Also during anaphase, Lrmp protein but not mRNA can been seen at the reforming nuclear membranes (Figure S4D, second row). By late mitosis (Figure S4D, bottom row), lrmp mRNA and protein again colocalize robustly to presumptive centrosomes. Our studies show that lrmp mRNA targets to centrosomes and spindles during mitosis, where it may provide a localized source of Lrmp protein at a time when nuclear membranes are undergoing dynamic assembly and disassembly (see Discussion).
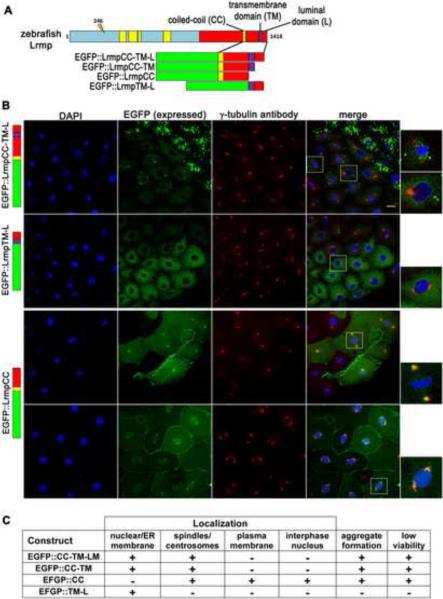
Lrmp C-terminal domains facilitate protein localization
Lrmp belongs to the class of tail-anchored or TA proteins, which are targeted to membrane compartments post-translationally [16,17]. All Lrmp proteins (long or short) contain a C-terminal MRVI1 domain [18,19] (Figure 2C, red segments) with a coiled-coil region (CC; Figure 2C, yellow bars) and a transmembrane segment (TM; Figure 2C, purple bar). In mouse and human Lrmp, the TM domain in combination with the C-terminal luminal (L) domain mediate localization to the ER membrane, where the C-terminus inserts into the ER lumen with the N-terminus exposed in the cytoplasm [11,16].
C. elegans ZYG–12 protein, like zebrafish Lrmp, is required for centrosome-nuclear attachment and pronuclear migration [20]. Lrmp and ZYG-12 do not show high levels of sequence homology but are structurally similar, with a CC domain followed by C-terminal TM and short L domains. We tested whether the zebrafish Lrmp C-terminal domains, as in C. elegans ZYG-12 and mammalian Lrmp [16,20,21], confer subcellular localization. Several EGFP-Lrmp fusion proteins were created with regions of the Lrmp C-terminus (Figure 6A): CC, TM and L domains and intervening regions (wild-type C-terminus, EGFP∷LrmpCC-TM-L), TM and L domains only (EGFP∷LrmpTM-L), CC and TM domains with the intervening region (EGFP∷LrmpCC-TM), and CC domain with the CC-to-TM intervening region (EGFP∷LrmpCC). Fusion constructs with and without exon 36 (Lrmp+EX36 or −EX36) behaved similarly. Fusion construct mRNAs were injected into 1-cell wild-type embryos and localized EGFP was normally detected by 2.5 hpf, when specimens were fixed and additionally labeled to detect γ-tubulin and DNA. In situ hybridization with EGFP antisense probe showed that exogenous mRNAs do not localize (data not shown), indicating that the observed protein localization patterns likely reflect direct protein targeting.
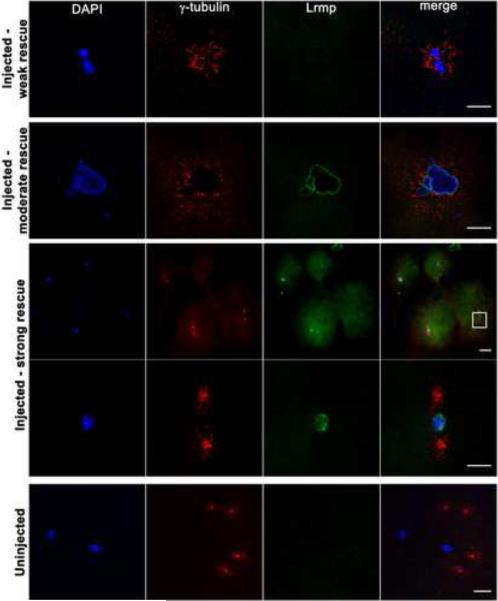
Figure 6. Mutant oocyte injection and rescue with wild-type lrmp mRNA.
Stage IV oocytes from fue mutant females were isolated and injected with wild-type lrmp mRNA. Following maturation and in vitro fertilization, embryos were fixed at 1 hpf and labeled for DNA, γ–tubulin, and Lrmp. Examples of weakly rescued embryos (top row), moderately rescued embryos (second row), and strongly rescued embryos (third and fourth rows) are shown. White box in the third row indicates the region shown at higher magnification in the fourth row. Embryos from uninjected fue oocytes, derived from the same set of mothers and treated in parallel with injected oocytes, showed the typical mutant phenotype (bottom row). Scale bars represent 20 μm. See also Table S2.
The fusion protein with the wild-type C-terminus (EGFP∷LrmpCC-TM-L) localized to nuclear membranes and regions corresponding to spindles, mirroring the endogenous Lrmp protein pattern (Figure 5B, top row, 5C). The construct with TM and L domains only (EGFP∷LrmpTM-L) localized to the nuclear membrane but not the spindle (Figure 5B second row, 5C). Conversely, the fusion protein containing the CC domain but lacking TM and L domains (EGFP∷LrmpCC) localized to the spindle and spindle poles but not to the nuclear envelope. (Figure 5B, third row, 5C). In addition, EGFP∷LrmpCC unexpectedly targeted to the plasma membrane and the interior of nuclei (Figure 5B, bottom row, 5C). The L domain was dispensable for all observed aspects of subcellular localization (EGFP∷LrmpCC-TM construct, Figures 5C and S5A).
Figure 5. C-terminal domains of Lrmp facilitate subcellular targeting.
(A) Diagram of EFGP-fusion constructs. (B) Fusion construct RNAs encoding EGFP::LrmpCC-TM-L (top row), EGFP::LrmpTM-L (second row), and EGFP::LrmpCC construct (bottom two rows), were injected into one-cell wild-type embryos. Embryos expressing EGFP were fixed and processed for DAPI and anti-γ–tubulin immunostaining between 2.5-3.5 hpf. White boxes indicate fields shown at higher magnification (right). Scale bar represents 20 μm in all lower magnification panels. (C) Summary of results from EGFP::Lrmp C-terminal protein expression. Low viability (far right column) manifested as cell division defects and failure to undergo gastrulation, which lead to embryo lysis. See also Figure S5.
Interestingly, fusion proteins containing the CC domain tended to aggregate, particularly in cells with high expression (e.g. Figure 5B, top row), and caused cell division defects and defective gastrulation (Figure 5C; data not shown), suggesting that CC domain-containing proteins or their aggregates interfere with normal cell functions. Expression of the various constructs in fue mutant embryos yielded localization patterns similar to those observed when expressed in wild-type, though mutants predictably lacked spindles and contained fewer nuclei (Figure S5B).
Our findings suggest that different regions of the Lrmp C-terminus mediate different aspects of Lrmp targeting (Figure S5C): the CC domain and/or CC-to-TM regions confer localization to spindles and centrosomes, as well as the ability to form aggregates, while the TM domain mediates localization to the nuclear membrane and perinuclear ER. Targeting by the TM domain also appears to prevent mislocalization of Lrmp to the plasma membrane and the interior of the nucleus. We conclude that the overall localization pattern of endogenous Lrmp protein can be recapitulated by the combined action of CC and TM domains. These results are generally concordant with previous analyses of ZYG-12 protein domains in C. elegans [20,21].
Genetic rescue by injection of wild-type lrmp mRNA into fue oocytes
To test for functional rescue of the fue maternal-effect embryonic phenotype by exogenous gene expression, mRNAs for wild-type lrmp and other constructs were injected into oocytes dissected from fue mutant females. Oocytes were then matured in vitro and fertilized with wild-type sperm [22]. Embryos were fixed after approximately 1 hour of development (4-cell stage in wild-type) and labeled for γ–tubulin, DNA, and Lrmp protein. Varying degrees of rescue were observed in the resulting embryos (Figure 6; Table S2). Embryos with weak rescue showed no detectable localized Lrmp protein, but contained a single DNA mass with γ–tubulin in close proximity (Figure 6, top row), indicating that pronuclear fusion and centrosome-nuclear attachment had occurred. Embryos with moderate rescue exhibited these traits as well as robust nuclear membrane localization of Lrmp protein (Figure 6, second row). Embryos exhibiting the greatest degree of rescue showed normal chromosome segregation, as reflected by the presence of a nucleus in each blastomere, and wild-type Lrmp and γ–tubulin localization (Figure 6, third and fourth rows). Sibling in vitro matured uninjected oocytes gave rise to zygotes with the expected nucleus-centrosome detachment and pronuclear congression defects (Figure 6, bottom row). In preliminary experiments, oocytes injected with RNAs encoding fue mutant Lrmp or wild-type C. elegans ZYG-12 did not show evidence of rescue (data not shown). Rescue of the fue phenotype by wild-type lrmp mRNA corroborates that lrmp is the gene affected by the fue mutation.
DISCUSSION
Function of Lrmp in the early zygote
Our studies indicate that Lrmp function is required for nuclear-cytoskeletal interactions that facilitate pronuclear congression after fertilization. In general, proteins involved in nuclear-cytoskeletal attachment belong to LInker of the Nucleoskeleton and Cytoskeleton (LINC) complexes [23], which bridge inner and outer nuclear membranes (INM and ONM). LINC complexes include two types of membrane proteins: SUN (Sad1/UNC-84) domain-containing proteins in the INM, and KASH (Klarsicht/Anc-1/Syne homology) domain proteins in the ONM [24]. LINC function is essential for nuclear migration in Drosophila and mouse neuronal cells [25–27], as well as during pronuclear migration in C. elegans [20,21].
The domain structures of Fue/Lrmp and KASH proteins are similar, containing coiled-coil regions adjacent to tail-anchored membrane insertion motifs at the C-terminus. In addition, the transmembrane and luminal domains of zebrafish Fue/Lrmp align partially with the KASH domain consensus sequence (Figure S5D). Moreover, the C-terminal coiled-coil and transmembrane domains of both the C. elegans KASH domain protein ZYG-12 and zebrafish Fue/Lrmp confer comparable subcellular targeting [20,21]. These similarities suggest that Fue/Lrmp may represent a LINC complex component.
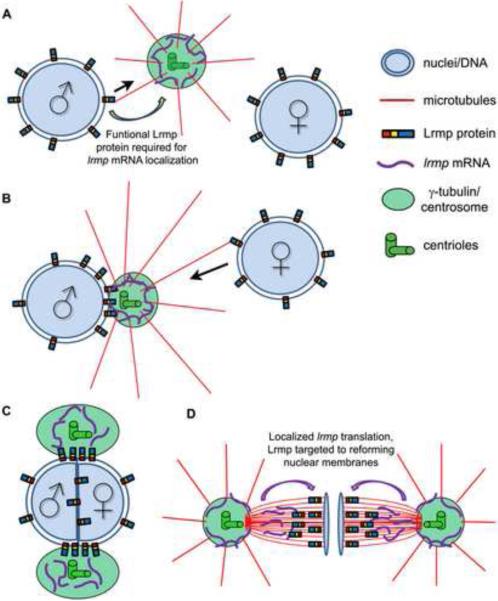
C. elegans ZYG-12 is required to localize the minus end-directed motor protein dynein to the nuclear envelope [20] where it facilitates centrosome-male pronucleus attachment as well as pronuclear migration [8,28]. A unifying hypothesis, based on our work and previous studies [20,21,25,29], proposes a link between Fue/Lrmp function and the known role of dynein-dependent transport along sperm aster microtubules during pronuclear congression (Figure 7). When astral microtubules nucleate from the centrosome after fertilization, they first encounter the male pronucleus where Lrmp at the nuclear membrane may mediate centrosomal attachment through minus end-directed movement (Figure 7A). As sperm aster microtubules lengthen, they contact the female pronucleus where Lrmp may again facilitate dynein-driven nuclear migration towards the centrosome (Figure 7B). These movements lead to pronuclear apposition and subsequent fusion (Figure 7C).
Figure 7. Model for Lrmp function in early zebrafish development.
(A, B) At fertilization, Lrmp protein localizes to pronuclear membranes and lrmp mRNA accumulates at centrosomes. Lrmp facilitates the association of pronuclei to the sperm aster (black arrows indicate direction of hypothesized minus-oriented motor movement), promoting pronuclear congression and fusion (C). (D) Spindle-associated lrmp mRNA may provide a localized source of newly synthesized Lrmp protein to the reforming nuclear membranes in late mitosis.
Multiple mechanisms ensure the dynamic subcellular localization of Fue/Lrmp
lrmp mRNA localizes to precise subcellular regions, suggesting a requirement for strict spatial and/or temporal regulation of Lrmp protein translation. Lrmp contains a tail-anchor and such domains are thought to bind with high affinity to lipid bilayers/membrane compartments [17]. Localizing lrmp mRNA just proximal to nuclei may prevent Lrmp protein insertion into non-nuclear membranes.
During mitosis, both fue/lrmp mRNA and protein products display dynamic subcellular localization to centrosomes, spindles, and nuclear membranes (Figure 7D). fue/lrmp mRNA and protein localization patterns largely overlap with two exceptions, both occurring during anaphase when 1) Fue/Lrmp protein is largely absent from centrosomes and 2) Fue/Lrmp protein, but not its transcript, becomes highly localized to reforming nuclear membranes. These observations are consistent with a model wherein Lrmp protein and lrmp mRNA are co-translationally transported from the centrosome along spindles towards the assembling nuclear membranes. Such a mechanism would allow completion of Lrmp protein, in particular its membrane-anchoring domain, at the right time and place for correct targeting to the reforming nuclear envelope. Future studies will address this possibility.
Our analyses indicate that the C-terminal Fue/Lrmp protein domains mediate protein localization to centrosomes, spindles, and the nuclear membrane, and that the disruption of these domains results in protein mistargeting. Notably, these domains are capable of mediating protein targeting independent of mRNA localization. Together, these data suggest that multiple mechanisms, including localized Lrmp translation and protein-protein interactions, ensure specific enrichment of Lrmp protein at the nuclear envelope and prevent its ectopic localization.
Role for centrosomes and Fue/Lrmp function in fue/lrmp mRNA localization
In unfertilized eggs, fue/lrmp mRNA fails to exhibit any subcellular localization, suggesting that targeting of lrmp mRNA is dependent on entry of sperm-derived centrioles. It is possible that the zygotic centrosome acts as an anchoring site for maternally provided fue/lrmp mRNA. In contrast, unfertilized eggs show nuclear envelope localization of maternal Fue/Lrmp protein, indicating that it can readily insert into membranes independently of lrmp mRNA localization. This is also indicated by the targeting of overexpressed C-terminal protein fusions without localization of the injected mRNAs. By providing a dock for fue/lrmp mRNA localization, the centriole and/or centrosome may allow for a more robust and selective pattern of Fue/Lrmp protein enrichment.
Unexpectedly, lrmp transcripts fail to localize in fue mutant zygotes. It is possible that the mutation interferes with a key interaction between Fue/Lrmp protein and fue/lrmp mRNA, although Fue/Lrmp does not contain recognizable RNA binding motifs and preliminary RNA-immunoprecipitation experiments did not reveal differential abilities of mutant and wild-type Lrmp protein to associate with lrmp RNA (R.L., unpublished observations). We favor an alternative scenario wherein localized Fue/Lrmp activity is required for the formation of a protein complex or subcellular domain critical for fue/lrmp mRNA recruitment. Such a structure could be a LINC-associated complex, the nuclear envelope-centrosome interface, or a specialized domain of the nucleoplasmic membrane.
A defect in this proposed structure may also contribute to the reduction in mutant Fue/Lrmp protein localization in fue zygotes, given that the N-terminal molecular lesion in this protein would not be expected to interfere directly with the identified C-terminal protein targeting domains. Indeed, we find that Fue mutant protein localizes largely normally when overexpressed in wild-type embryos (R.L. unpublished observations), suggesting that it may be the cellular environment in fue/lrmp mutants, and not the Fue/Lrmp mutant protein itself, that is inconducive to proper Fue/Lrmp protein targeting.
Long Lrmp may have evolved in an ancestral vertebrate lineage
Long Lrmp, which we show is maternally provided and involved in nucleus-centrosome attachment at fertilization in zebrafish, is present in most analyzed vertebrate species, though it is often predicted as two or three adjacent transcripts (Table S1). The presence of Lrmp homologues in vertebrate lineages mirrors the occurrence of lymphocytes as a vertebrate-specific cell type [30], suggesting a scenario where a gene encoding long Lrmp arose in a basal vertebrate lineage and acquired functions in both fertilization and lymphocytes. Rodent lineages, which lack paternally-inherited centrioles [4], may have maintained only the shorter, lymphocyte-specific C-terminal Lrmp.
Conclusions
We identify a new form of Lrmp affected in the zebrafish maternal-effect mutant futile cycle, which is required for centrosome-pronucleus attachment, and describe a novel pattern of subcellular targeting for its gene products in the zygote. In addition to their implications for vertebrate reproductive biology, these studies may offer insights into the role of lrmp in lymphoid cells [11], lung cancer and diabetes susceptibility, [31,32], as well as in muscular dystrophy and premature aging [33,34].
EXPERIMENTAL PROCEDURES
Fish maintenance
Wild-type AB, WIK, and fue mutant fish were maintained at 28° C under standard conditions [35]. Embryos were cultured in E3 medium [36] and staged by time after fertilization/activation or with standardized morphological markers [37]. Genotyping of fue mutants was accomplished using flanking SSLP markers or with lrmp-specific RFLP markers.
Positional cloning of fue
Analysis with genome-spanning DNA markers established linkage of fue to chromosome 4 between 59.0 and 61.1 cM on the zebrafish MGH recombinant map. Candidate genes within this region were sequenced, revealing additional polymorphic markers and uncovering the molecular lesion in transcript lrmp-001 (gene designation from Vega database, also listed as NCBI accession # CAI20727).
Immunofluorescence labeling
Embryos from natural matings or in vitro fertilized eggs [36] were dechorionated and fixed in 4% paraformaldehyde or microtubule fix (for α-tubulin labeling) as previously described [38]. Anti-LrmpMD serum was used at 1:1000 and anti-LrmpCT serum at 1:750. Commercial primary and secondary antibodies were as follows: anti-α-tubulin (Sigma, mouse monoclonal B5-1-2, 1:2500), anti-γ-tubulin (Sigma, mouse monoclonal GTU-88, 1:2000; Sigma, rabbit polyclonal, 1:2000); goat anti-mouse-Cy3 (Jackson ImmunoResearch, 1:100), goat anti-rabbit-Alexa488 (Molecular Probes, 1:100), goat anti-rabbit-Cy3 (Jackson ImmunoResearch 1:100). DNA was detected by incubation in 0.5 μg/ml DAPI for 10 min or by mounting in ProLong Gold antifade reagent with DAPI (Invitrogen). Male/female pronuclear identities after fertilization were inferred based on the relative proximity of nuclei to centrosomes (male) and polar bodies (female).
In situ hybridization
In situ hybridizations using chromogenic substrates were performed as in [39]. Fluorescent in situ hybridization for lrmp used anti-DIG-POD antibody (1:1000, Roche) and an Alexa Fluor 488 tyramide signal amplification kit (Invitrogen) as in [38]. All probes were made by in vitro transcription with T7 or SP6 RNA polymerase (Fermentas) from DNA fragments cloned into the pGEM-T Easy vector (Promega). Three different lrmp RNA probes were used, corresponding to exons 2–4, exons 12–17, and exons 27–32 of zebrafish lrmp and all showed similar results.
For co-labeling of lrmp mRNA and either γ-tubulin or Lrmp protein, dechorionated embryos were fixed in 4% paraformaldehyde overnight and the standard in situ protocol [39] was conducted through probe hybridization. Embryos were deyolked and blocked, then anti-DIG-POD antibody and primary γ-tubulin or Lrmp antibodies were simultaneously incubated with embryos. The Alexa 488 tyramide kit was used for RNA probe detection followed without a fixation step by blocking and an anti-rabbit-Cy3 secondary antibody incubation for γ-tubulin or Lrmp antibody detection.
RNA injections
mRNA for injection was synthesized using either an mMessage mMachineSP6 kit (Ambion, for all EGFP fusions) as in [39], or the mMessage mMachine T7 Ultra kit and manufacturers protocols (Ambion, for full-length lrmp constucts). All constructs included the full 3' UTR of lrmp. For injection, RNAs were diluted to 250 ng/μl in 0.1 M nuclease-free KCl. Matured oocytes and 7–25 mpf embryos were injected with approximately 0.5–1 nl mRNA solution.
Oocyte Culture and Injection
Dissection of ovaries, in vitro culture of oocytes, and fertilization of matured eggs was carried out as described in [22], with an injection step during the maturation period (manuscript in preparation). After fertilization, embryos developed for 1–2 hours and were then fixed for immunofluorescence labeling.
Imaging
Fluorescently labeled embryos were imaged with a Zeiss LSM 510 confocal microscope. Images were processed using ImageJ software and the Sync Measure 3D plug-in was employed for distance measurements within z-stacks. Embryos labeled by whole-mount in situ hybridization were imaged using a Leica FLII microscope, color camera (Diagnostic Instruments Spot Insight), and Spot imaging software.
Supplementary Material
HIGHLIGHTS
futile cycle is required for centrosome-nucleus attachment in early embryos
fue encodes lymphoid-restricted membrane protein, a vertebrate-specific gene
lrmp mRNA and protein show dynamic localization patterns during the cell cycle
Targeting of lrmp mRNA may facilitate spatiotemporal control of Lrmp translation
ACKNOWLEDGEMENTS
We are grateful to Dr. Nüsslein-Volhard (MPI, Tübingen), for generously providing the futile cycle mutation, laboratory members for discussions and advice, Dr. Amy Cooke from Dr. Marvin Wickens' laboratory (UW-Madison) for assistance with antibody production, Dr. Taijiro Yabe for identifying fue-linked markers, and Dr. Michael Sheets (UW-Madison) for critical reading of the manuscript. Funding was provided by NIH grant RO1 GM065303.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Chambers EL. The movement of the egg nucleus in relation to the sperm aster in the echinoderm egg. J. Exp. Biol. 1939;16:409–424. [Google Scholar]
- 2.Navara CS, First NL, Schatten G. Microtubule organization in the cow during fertilization, polyspermy, parthenogenesis, and nuclear transfer: the role of the sperm aster. Dev. Biol. 1994;162:29–40. doi: 10.1006/dbio.1994.1064. [DOI] [PubMed] [Google Scholar]
- 3.Reinsch S, Gonczy P. Mechanisms of nuclear positioning. J. Cell. Sci. 1998;111:2283–2295. doi: 10.1242/jcs.111.16.2283. [DOI] [PubMed] [Google Scholar]
- 4.Delattre M, Gonczy P. The arithmetic of centrosome biogenesis. J. Cell. Sci. 2004;117:1619–1630. doi: 10.1242/jcs.01128. [DOI] [PubMed] [Google Scholar]
- 5.Schatten G. The centrosome and its mode of inheritance: the reduction of the centrosome during gametogenesis and its restoration during fertilization. Dev. Biol. 1994;165:299–335. doi: 10.1006/dbio.1994.1256. [DOI] [PubMed] [Google Scholar]
- 6.Bestor TH, Schatten G. Anti-tubulin immunofluorescence microscopy of microtubules present during the pronuclear movement of sea urchin fertilization. Dev. Biol. 1981;88:80–91. doi: 10.1016/0012-1606(81)90220-7. [DOI] [PubMed] [Google Scholar]
- 7.Reinsch S, Karsenti E. Movement of nuclei along microtubules in Xenopus egg extracts. Curr. Biol. 1997;7:211–214. doi: 10.1016/s0960-9822(97)70092-7. [DOI] [PubMed] [Google Scholar]
- 8.Gonczy P, Pichler S, Kirkham M, Hyman AA. Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J. Cell Biol. 1999;147:135–150. doi: 10.1083/jcb.147.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Payne C, Rawe V, Ramalho-Santos J, Simerly C, Schatten G. Preferentially localized dynein and perinuclear dynactin associate with nuclear pore complex proteins to mediate genomic union during mammalian fertilization. J. Cell. Sci. 2003;116:4727–4738. doi: 10.1242/jcs.00784. [DOI] [PubMed] [Google Scholar]
- 10.Dekens MPS, Pelegri FJ, Maischein HM, Nüsslein-Volhard C. The maternal-effect gene futile cycle is essential for pronuclear congression and mitotic spindle assembly in the zebrafish zygote. Development. 2003;130:3907–3916. doi: 10.1242/dev.00606. [DOI] [PubMed] [Google Scholar]
- 11.Behrens TW, Jagadeesh J, Scherle P, Kearns G, Yewdell J, Staudt LM. Jaw1, A lymphoid-restricted membrane protein localized to the endoplasmic reticulum. J. Immunol. 1994;153:682–690. [PubMed] [Google Scholar]
- 12.Kane DA. Cell cycles and development in the embryonic zebrafish. In: Detrich HW, Zon LI, Westerfield M, editors. The Zebrafish: Biology. 1999. pp. 11–26. [DOI] [PubMed] [Google Scholar]
- 13.Mathavan S, Lee SGP, Mak A, Miller LD, Murthy KRK, Govindarajan KR, Tong Y, Wu YL, Lam SH, Yang H, et al. Transcriptome analysis of zebrafish embryogenesis using microarrays. PLOS Genet. 2005;1:260–276. doi: 10.1371/journal.pgen.0010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zamir E, Kam Z, Yarden A. Transcription-dependent induction of G1 phase during the zebra fish midblastula transition. Mol. Cell. Biol. 1997;17:529–536. doi: 10.1128/mcb.17.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schoft VK, Beauvais AJ, Lang C, Gajewski A, Prüfert K, Winkler C, Akimenko MA, Paulin-Levasseur M, Krohne G. The lamina-associated polypeptide 2 (LAP2) isoforms ß, g and w of zebrafish: developmental expression and behavious during the cell cycle. J. Cell Sci. 2003;116:2505–2517. doi: 10.1242/jcs.00450. [DOI] [PubMed] [Google Scholar]
- 16.Behrens TW, Kearns GM, Rivard JJ, Bernstein HD, Yewdell JW, Staudt LM. Carboxyl-terminal targeting and novel post-translational processing of JAW1, a lymphoid protein of the endoplasmic reticulum. J. Biol. Chem. 1996;271:23528–23534. doi: 10.1074/jbc.271.38.23528. [DOI] [PubMed] [Google Scholar]
- 17.Rabu C, Schmid V, Schwappach B, High S. Biogenesis of tail-anchored proteins: the beginning for the end? J. Cell. Sci. 2009;122:3605–3612. doi: 10.1242/jcs.041210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schlossmann J, Ammendola A, Ashman K, Zong X, Huber A, Neubauer G, Wang GX, Allescher HD, Korth M, Wilm M, et al. Regulation of intracellular calcium by a signalling complex of IRAG, IP3 receptor and cGMP kinase Ibeta. Nature. 2000;404:197–201. doi: 10.1038/35004606. [DOI] [PubMed] [Google Scholar]
- 19.Shaughnessy JD, Jr, Largaespada DA, Tian E, Fletcher CF, Cho BC, Vyas P, Jenkins NA, Copeland NG. Mrvi1, a common MRV integration site in BXH2 myeloid leukemias, encodes a protein with homology to a lymphoid-restricted membrane protein Jaw1. Oncogene. 1999;18:2069–2084. doi: 10.1038/sj.onc.1202419. [DOI] [PubMed] [Google Scholar]
- 20.Malone CJ, Misner L, Le Bot N, Tsai MC, Campbell JM, Ahringer J, White JG. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 2003;115:825–836. doi: 10.1016/s0092-8674(03)00985-1. [DOI] [PubMed] [Google Scholar]
- 21.Minn IL, Rolls MM, Hanna-Rose W, Malone CJ. SUN-1 and ZYG-12, mediators of centrosome-nucleus attachment, are a functional SUN/KASH pair in Caenorhabditis elegans. Mol. Biol. Cell. 2009;20:4586–4595. doi: 10.1091/mbc.E08-10-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seki S, Kouya T, Tsuchiya R, Valdez DM, Jin B, Hara T, Saida N, Kasai M, Edashige K. Development of a reliable in vitro maturation system for zebrafish oocytes. Reproduction. 2008;135:285–292. doi: 10.1530/REP-07-0416. [DOI] [PubMed] [Google Scholar]
- 23.Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. Coupling of the nucleus and cytoplasm: role of the LINC complex. J. Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Starr DA, Fridolfsson HN. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu. Rev. Cell Dev. Biol. 2010;26:421–444. doi: 10.1146/annurev-cellbio-100109-104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mosley-Bishop KL, Li Q, Patterson L, Fischer JA. Molecular analysis of the klarsicht gene and its role in nuclear migration within differentiating cells of the Drosophila eye. Curr. Biol. 1999;9:1211–1220. doi: 10.1016/s0960-9822(99)80501-6. [DOI] [PubMed] [Google Scholar]
- 26.Kracklauer MP, Banks SM, Xie X, Wu Y, Fischer JA. Drosophila klaroid encodes a SUN domain protein required for Klarsicht localization to the nuclear envelope and nuclear migration in the eye. Fly (Austin) 2007;1:75–85. doi: 10.4161/fly.4254. [DOI] [PubMed] [Google Scholar]
- 27.Zhang X, Lei K, Yuan X, Wu X, Zhuang Y, Xu T, Xu R, Han M. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron. 2009;64:173–187. doi: 10.1016/j.neuron.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoder JH, Han M. Cytoplasmic dynein light intermediate chain is required for discrete aspects of mitosis in Caenorhabditis elegans. Mol. Biol. Cell. 2001;12:2921–2933. doi: 10.1091/mbc.12.10.2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malone CJ, Fixsen WD, Horvitz HR, Han M. UNC-84 localizes to the nuclear envelope and is required for nuclear migration and anchoring during C. elegans development. Development. 1999;126:3171–3181. doi: 10.1242/dev.126.14.3171. [DOI] [PubMed] [Google Scholar]
- 30.Litman GW, Cannon JP, Dishaw LJ. Reconstructing immune phylogeny: new perspectives. Nat. Rev. Immunol. 2005;5:866–879. doi: 10.1038/nri1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manenti G, Galbiati F, Pettinicchio A, Spinola M, Piconese S, Leoni VP, Conti B, Ravagnani F, Incarbone M, Pastorino U, et al. A V141L polymorphism of the human LRMP gene is associated with survival of lung cancer patients. Carcinogenesis. 2006;27:1386–1390. doi: 10.1093/carcin/bgi332. [DOI] [PubMed] [Google Scholar]
- 32.Duarte N, Lundholm M, Holmberg D. The Idd6.2 diabetes susceptibility region controls defective expression of the Lrmp gene in nonobese diabetic (NOD) mice. Immunogenetics. 2007;59:407–416. doi: 10.1007/s00251-007-0194-x. [DOI] [PubMed] [Google Scholar]
- 33.Salpingidou G, Smertenko A, Hausmanowa-Petrucewicz I, Hussey PJ, Hutchison CJ. A novel role for the nuclear membrane protein emerin in association of the centrosome to the outer nuclear membrane. J. Cell Biol. 2007;178:897–904. doi: 10.1083/jcb.200702026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Broers JL, Ramaekers FC, Bonne G, Yaou RB, Hutchison CJ. Nuclear lamins: laminopathies and their role in premature ageing. Physiol. Rev. 2006;86:967–1008. doi: 10.1152/physrev.00047.2005. [DOI] [PubMed] [Google Scholar]
- 35.Brand M, Granato M, Nüsslein-Volhard C. Keeping and raising zebrafish. In: Nüsslein-Volhard C, Dahm R, editors. Zebrafish. Oxford University Press; Oxford: 2002. pp. 7–37. [Google Scholar]
- 36.Pelegri F, Schulte-Merker S. A gynogenesis-based screen for maternal-effect genes in the zebrafish, Danio rerio. Methods Cell Biol. 1999;60:1–20. doi: 10.1016/s0091-679x(08)61891-9. [DOI] [PubMed] [Google Scholar]
- 37.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 38.Theusch EV, Brown KJ, Pelegri F. Separate pathways of RNA recruitment lead to the compartmentalization of the zebrafish germ plasm. Dev. Biol. 2006;292:129–141. doi: 10.1016/j.ydbio.2005.12.045. [DOI] [PubMed] [Google Scholar]
- 39.Pelegri F, Maischein HM. Function of zebrafish ß-catenin and TCF-3 in dorsoventral patterning. Mech. Dev. 1998;77:63–74. doi: 10.1016/s0925-4773(98)00132-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.