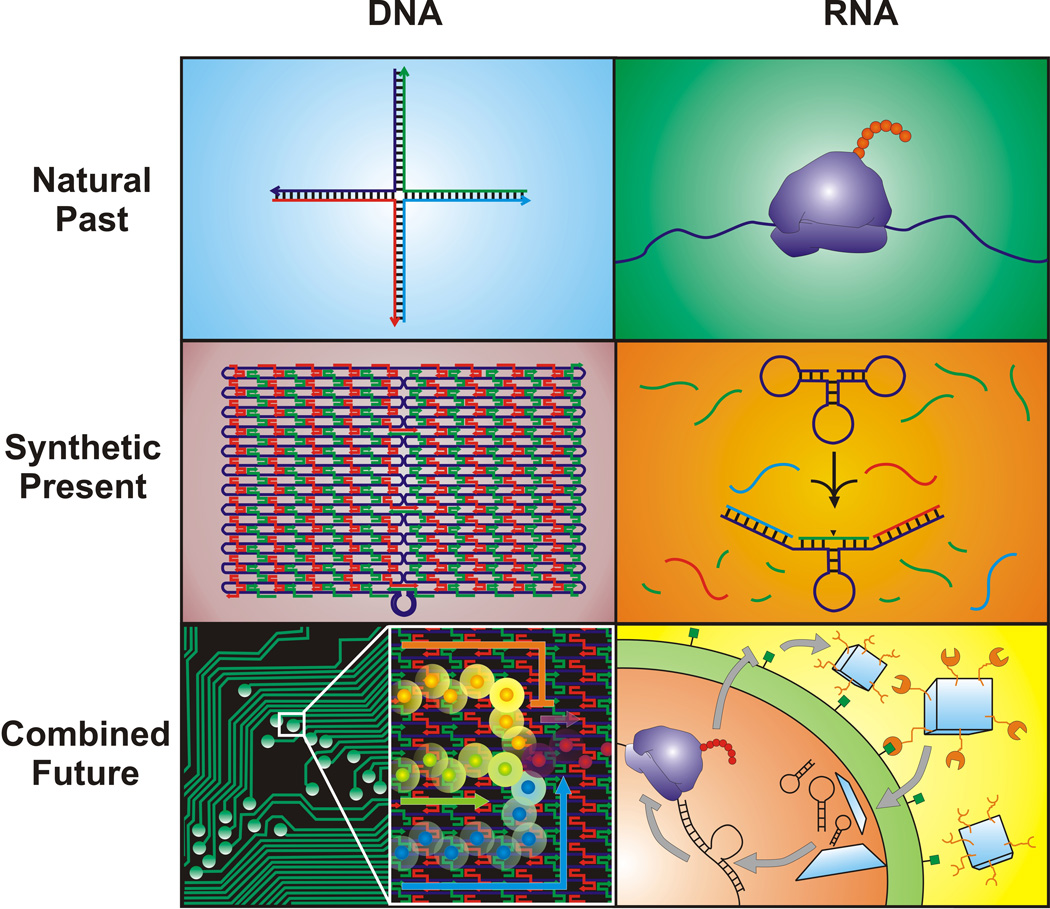
Abstract
Nucleic acid nanotechnology exploits the programmable molecular recognition properties of natural and synthetic nucleic acids to assemble structures with nanometer-scale precision. In 2006, DNA origami transformed the field by providing a versatile platform for self-assembly of arbitrary shapes from one long DNA strand held in place by hundreds of short, site-specific (spatially addressable) DNA ”staples”. This revolutionary approach has led to the creation of a multitude of 2D and 3D scaffolds that form the basis for functional nanodevices. Not limited to nucleic acids, these nanodevices can incorporate other structural and functional materials, such as proteins and nanoparticles, making them broadly useful for current and future applications in emerging fields such as nanomedicine, nanoelectronics, and alternative energy.
Introduction
Nucleic acid nanotechnology has been utilized by nature for billions of years1, 2. DNA in particular is chemically inert enough to reliably store genetic information over even millions of years3. Packaged in genomes, DNA is expressed in a regulated fashion with assistance from its more rapidly hydrolyzed structural analog, RNA, as well as protein factors2, 4, 5. Complementarily, the enhanced functionality of RNA is exploited in many biological nanomachines, such as the ribosome6 and the spliceosome7, 8, for the “data processing” that accompanies gene expression. Nature also uses the specificity of base pairing by nucleic acids for database maintenance and readout via the modulation of gene expression by non-coding RNAs4, 9.
Inspired by nature, researchers over the past four decades have explored nucleic acids as convenient building blocks to synthesize novel nanodevices1, 10. Because they are composed of only four different chemical building blocks and follow relatively simple, yet highly specific and thus predictable organizational base pairing rules at the molecular scale, nucleic acids are the preferred biological material for the design of structures with nanometer precision when compared with other candidates such as proteins. Short (typically < 50 nucleotides (nt) for RNA and < 100 nt for DNA) specific sequences of the building blocks can be chemically synthesized at low cost, whereas longer DNA strands of predefined sequence are provided by nature in the form of genomic DNA. A combination of these two sources led to the advent of the DNA origami method in 200611, which dramatically accelerated progress in nucleic acid nanotechnology by further increasing the simplicity, precision, and fidelity of the design principles available for generating spatially addressable nanoscale structures (see “DNA Origami” insert). Numerous DNA-origami-based nucleic acid nanodevices and nanomaterials have since been constructed12–16 with great potential for a multitude of useful applications, and with abundant prospects for innovation.
THE BUILDING MATERIALS: NATURAL NUCLEIC ACIDS
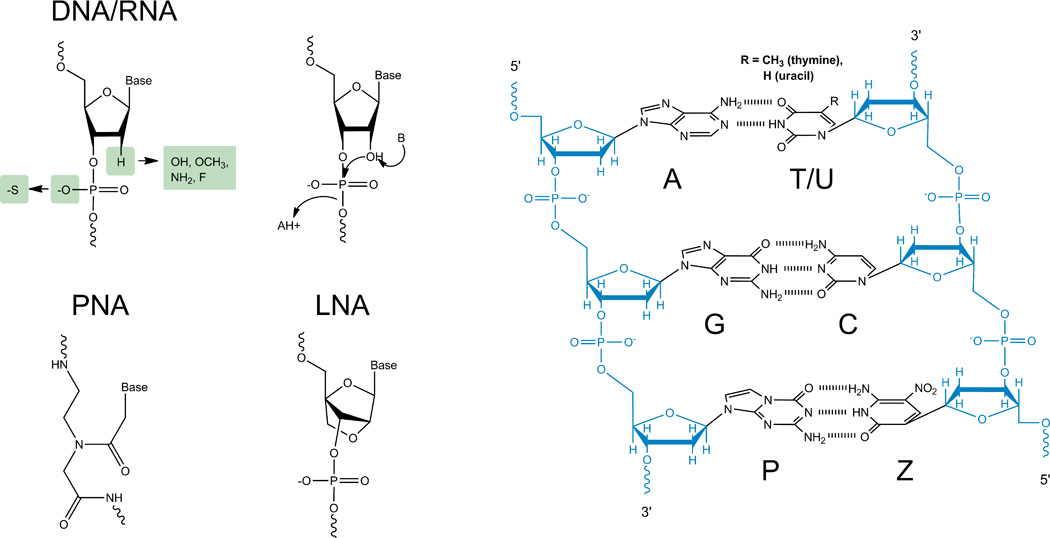
Nucleic acids are linear biopolymers found in all organisms, as well as many viruses, as they are the means by which genetic information is stored, transferred, and regulated. Their monomeric building blocks (nucleotides) each consist of three moieties (Figure 1a–c): a nucleobase, a five-carbon sugar, and a phosphate group. The phosphate group enables the formation of a phosphodiester bond between the sugars of adjacent nucleotides, creating a polymer known as a single-stranded nucleic acid. Complementary nucleobases can form hydrogen bonds which, along with stacking interactions between adjacent base pairs, results in two fully complementary polymers hybridizing to form a relatively rigid antiparallel double-stranded helix (Figure 1d)17. This famous Watson-Crick duplex has an average dimension (rise) of 0.34 nm (for canonical B-form DNA) between the base pairs along the helical axis and a diameter of 2 nm, ideally suited to engineer nanotechnological structures.
Figure 1.
Chemical structures of nucleic acids. Nucleotide backbone structures for (a) DNA (left), RNA (right), (b) PNA, and (c) LNA. The right side of panel a also indicates the backbone hydrolysis reaction of RNA, where “AH+” and “B” are an acid and base catalyst, respectively. (d) Structure of a double-stranded nucleic acid incorporating the four principal natural nucleobases A, T/U, G, and C, as well as artificial P:Z base pair.
Nature has optimized two different types of nucleic acids through evolution: DNA and RNA, which both consist of four nucleotide building blocks that tend to pair in a predictable Watson-Crick fashion (Figure 1d). The guanine-cytosine (G:C) base pair forms three hydrogen bonds, stronger than the two hydrogen bonds of the adenosine-thymine (A:T) pair, used in DNA, or the adenosine-uracil (A:U) pair, used in RNA. The primary chemical difference between DNA and RNA is their sugars (Figure 1a); ribose, the sugar found in RNA, contains an extra 2’-hydroxyl group not present in deoxyribose, the sugar found in DNA, causing the backbone of RNA to hydrolyze ~108- to 1010-fold more quickly than that of DNA (Figure 1a)18. The A-form RNA duplex, however, is more thermally stable than the B-form DNA duplex, even though RNA is predominantly found single-stranded in nature and forms mostly intramolecular helices19. These structural differences between DNA and RNA are functionally exploited by nature.
DNA
While the most notable purpose of DNA is storing and maintaining the genetic blueprint of an organism, DNA performs other structural roles in the cell as well. For instance, the guanine-rich single-stranded telomeres at the ends of chromosomes can form topologically more complex quadruplex structures built around a square arrangement of four stacked guanine-bases stabilized by a chelated metal ion (preferably K+) at their center. G-quadruplexes thus play important roles in the maintenance of linear eukaryotic genomes and possibly in gene expression regulation20. Similarly, so-called Holliday four-way junctions of two entwined DNA duplexes play critical roles in DNA recombination21. Forming and resolving such higher-order structures often requires the aid of external factors such as proteins. For example, complexes of histone proteins facilitate the compact folding of ~1 m of DNA into chromosomes that fit inside a ~6-µm-wide nucleus while still remaining accessible for gene expression22, 23. In cases where decreased stability would be most beneficial, a lower melting temperature may be achieved by increasing the ratio of A:T to G:C base pairs, as is desirable for promoter regions in DNA (regions that need to melt for an RNA polymerase to initiate gene expression24).
RNA
In the central dogma of molecular biology, the portrayed function of RNA is as the messenger between DNA and protein, carrying the blueprints for protein formation from the DNA genome so that they may be translated by the ribosome into proteins25. However, over the past few decades the pervasiveness and importance of RNA that does not code for proteins has been increasingly appreciated. Typically, these non-coding RNAs (ncRNAs) directly or indirectly regulate or mediate gene expression at the transcriptional or translational level, making them candidates for therapeutic applications4, 9, 26. Some ncRNAs simply make use of molecular recognition through base pairing, such as in the RNA interference pathway26. Others exploit conformational dynamics of RNA27. For example, riboswitches regulate gene expression via the conformational change induced by the RNA binding a small-molecule metabolite or second messenger28. Still other ncRNAs promote catalysis, such as in peptide bond formation within the ribosome6, 29 or cleavage of RNA via hydrolysis of the sugar-phosphate backbone30; the activity of each of these RNA enzymes (ribozymes) is dictated by its tertiary structure. While these activities are currently difficult to engineer from scratch due to the large number (hundreds to thousands) of nucleotides required and our limited understanding of the underlying mechanisms, these functional, structurally complex ncRNAs have already been optimized by nature as active components in cellular machines.
THE BUILDING MATERIALS: MODIFIED NUCLEIC ACIDS
Nucleic acids undergo chemical base modifications in vivo, impacting their cellular functions. For example, cytosine methylation prevents DNA from being digested by methylation-sensitive restriction enzymes in bacteria31, and in many eukaryotic organisms is involved in the epigenetic regulation of gene expression32. In an analogous fashion, researchers have synthetically modified nucleotides to increase thermodynamic and chemical stability as well as specificity of interactions. For instance, locked nucleic acids (LNA) are engineered with an extra methylene bridge between the 2′-oxygen and 4′-carbon of ribose, “locking” the ribose in the 3′-endo conformation (Figure 1c), which leads to a particularly stable A-form helix with enhanced base stacking and backbone pre-organization and significantly increased melting temperature, specificity of base pairing, and nuclease resistance33, 34. Other chemical modifications of the nucleic acid backbone commonly used for enhancing intracellular stability while still often supporting biological function of an ncRNA include 2’-O-methyl, 2’-amino, 2’-fluoro, and phosphorothioate substitutions (Figure 1a)35. As a more drastic modification, peptide nucleic acids (PNA) have an uncharged backbone of N-(2-aminoethyl)-glycine units joined by a peptide bond, resulting in higher thermal stability between PNA/DNA strands than corresponding DNA/DNA strands (Figure 1b)36. The uncharged nature and nuclease resistance of PNA make it an appealing option for nanodevices37 and 2D nucleic acid arrays38.
Not only can the nucleic acid backbone be modified, but also the nucleobases themselves. A particularly far-reaching example is the artificial expansion of the genetic alphabet by an additional, differently hydrogen bonded base pair such as Z or P (or 6-amino-5-nitro-3-(1’-β-D-2’-deoxyribofuranosyl)-2(1H)-pyridone and 2-amino-8-(1’-β-D-2’-deoxyribofuranosyl)-imidazo[1,2-a]-1,3,5-triazin-4(8H)-one, respectively, see Figure 1d), which increases hybridization specificity while still allowing for specific recognition by DNA-binding enzymes39.
Whereas the improved thermal and chemical stability often found in modified nucleic acids will prove useful for nanostructures, incorporation is nontrivial; nanostructure design is heavily dependent on helical twist, which is typically different between modified and unmodified nucleic acids10. This hurdle will, however, be overcome with improved and expanded design rules. In addition, these materials can already be used to site-specifically decorate DNA scaffolds since they form canonical Watson-Crick base pairs.
THE PRESENT STATE OF NUCLEIC ACID NANOTECHNOLOGY
By employing the knowledge acquired from observing nature and building upon it, nanostructures not found in nature have been engineered that encompass a broad range of design objectives. The following are examples of successful nanostructures grouped according to their purpose.
Scaffolding
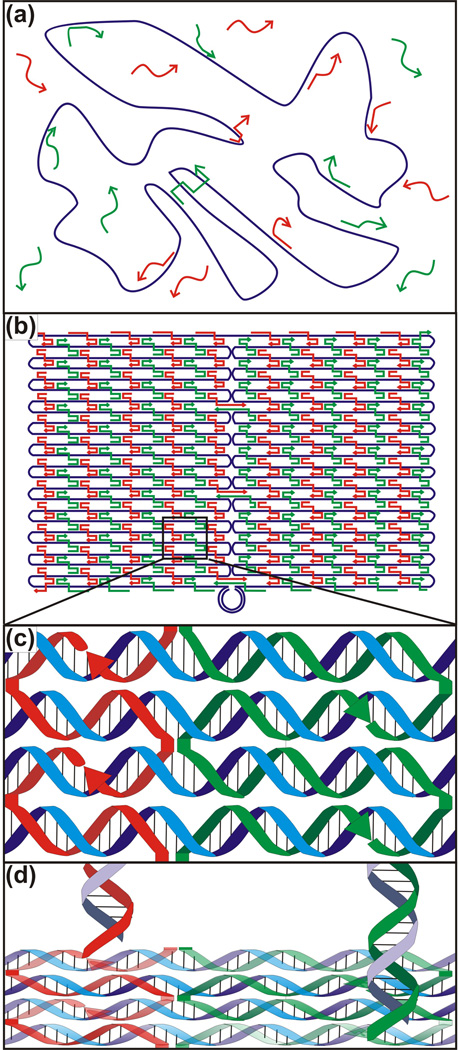
Nucleic acid scaffolds are nanostructures that may be functionalized for ordering and arraying materials with nanometer precision. Scaffolds were originally formed by combining different short double-stranded DNA (dsDNA) domains joined in a programmable fashion using single-stranded DNA (ssDNA) overhangs known as “sticky ends”40. While the stiffness of dsDNA makes it a suitable raw material for the edges of stable 2- and 3-dimensional structures, the vertices of such structures remain flexible, resulting in a range of angles between domains40. Inspired by naturally occurring Holliday junctions21, 41, more rigid structures were accomplished using reciprocal exchange to generate double42 and triple crossover motifs43 (Figure 2). As an alternative to combining multiple domains, which results in variable yields of assembled structures without defined boundaries, increased yield and stability are accomplished using Rothemund’s DNA origami method (see “Origami” insert and Figure 3), leading to more sophisticated 2D and 3D structures ranging from smiley faces11 to octahedra44 and nanoflasks45 that can be concatenated into larger structures11, 46.
Figure 2.
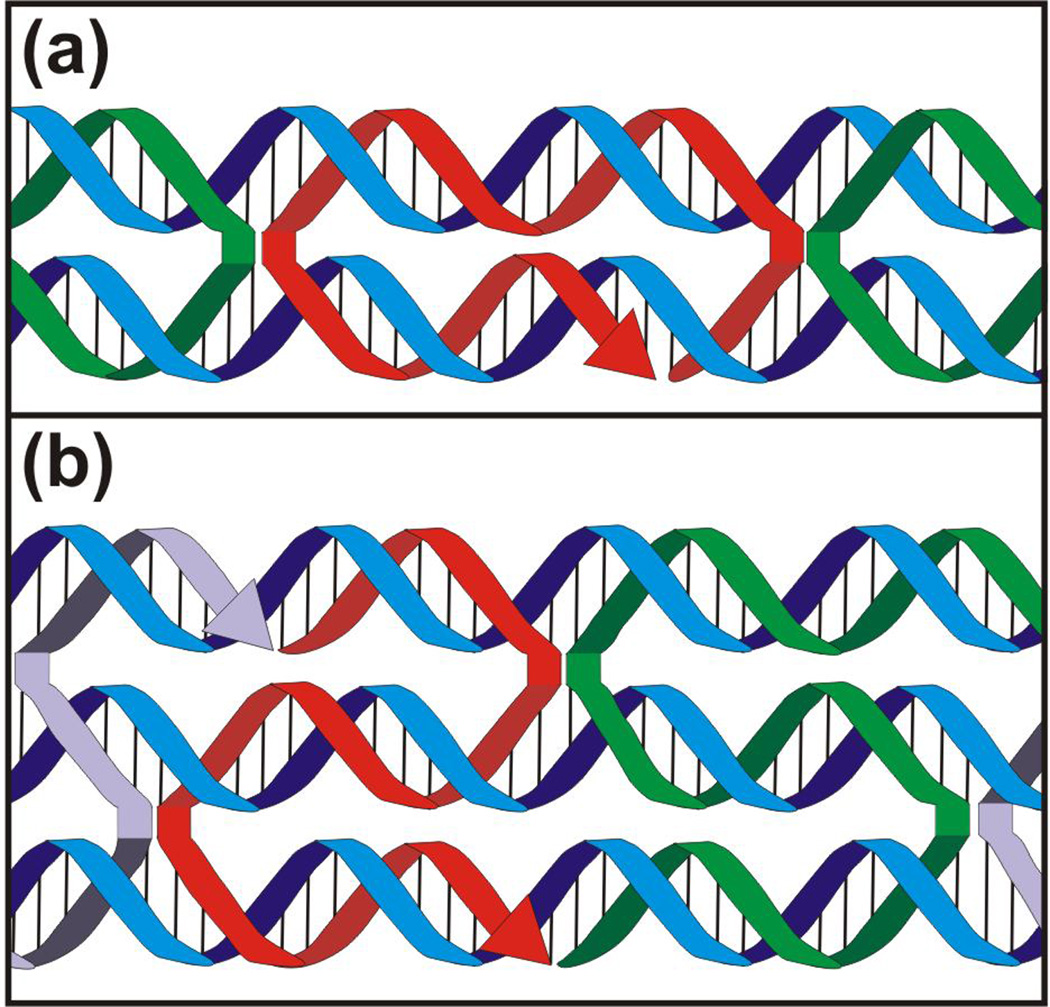
DNA crossover motifs. The (a) double and (b) triple crossover motifs are demonstrated by the red strand.
Figure 3.
The DNA origami method. (a) The single-stranded DNA “scaffold” strand (purple) is folded and held in place by specifically hybridizing “staple” strands (red and green). (b) Resultant rectangular origami tile once all the staples have bound to the scaffold. (c) Triple crossover motifs demonstrated by the staples interacting with the scaffold. (d) 5’ ends of the staples are extended to create overhangs, which then can be decorated with partially complementary oligonucleotides (purple).
Dynamic Devices Constructed on Nucleic Acid Scaffolds
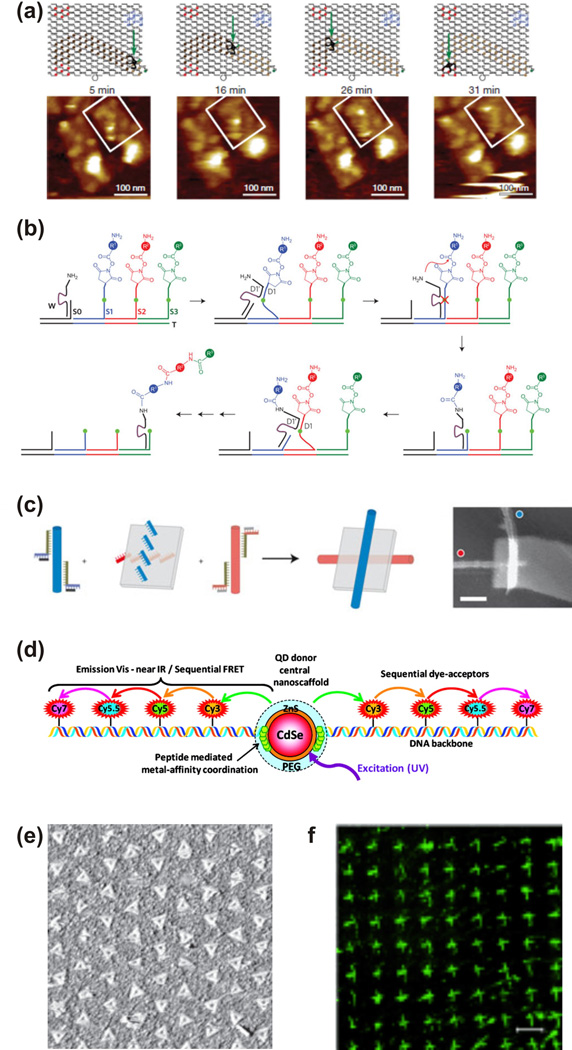
To add functionality to the scaffolds, DNA, RNA and protein have further been used to construct dynamic devices serving as proofs-of-principle for directed transport and nanoscale assembly factories. Molecular walkers analogous to the motor proteins myosin, kinesin, and dynein have been constructed completely or primarily from DNA, and have attained increasing levels of autonomy as well as responsiveness to external instruction (Figure 4a)52, 53. Some such transporters are capable of dozens to potentially thousands of directed steps and have been shown to walk processively along tracks on DNA origami47, 52. Directed walkers have also aided in the combinatorial assembly of different-sized gold nanoparticles48 or the multistep synthesis of a small organic compound49, in the former case aided by two-state DNA conformational switches (Figure 4b). Coupling such devices with DNA computing54, either on the scaffold or in solution, may further improve the complexity and range of responses to environmental stimuli or instructions.
Figure 4.
Current state-of-the-art of nucleic-acid-based nanotechnology. (a) Molecular nanorobot walking along a track generated by decorating a rectangular origami with leg footholds (substrates); below, tile (boxed) imaged using AFM over time, indicating spider movement47. (b) Multistep synthesis of an organic compound mediated by a deoxyribozyme49. (c) Junctions of carbon nanotubes and origami to create a field-effect transistor50. (d) Transfer of photonic energy along a DNA template using FRET56. (e) Triangular DNA origami arranged using electron beam lithography73. (f) DNA nanotubes arranged using soft lithography64. Each panel was reproduced with permission from the respective publisher.
In addition to the above devices made primarily of nucleic acids, several groups have sought to augment the functional repertoire of DNA and RNA nanotechnology through conjugation to other nanomaterials, using self-assembled DNA structures as a scaffold on which to position non-nucleic-acid components that impart some new catalytic, electronic, photonic, or structural functionality. For instance, placing the metabolically linked enzymes luciferase and NAD(P)H-dependent FMN oxidoreductase (NFOR) in close proximity on a DNA template led to more efficient recycling of their common FMN/FMNH2 cofactor and an overall enhancement of the catalyzed reaction rates, mimicking the strategy of substrate channeling often employed by nature55. DNA origami has been used to create junctions of carbon nanotubes with precisely defined geometry50, 51, in one case behaving like a nanoscale field-effect transistor (Figure 4c)50. Furthermore, DNA templates have directed the assembly of chromophores such as organic dyes and quantum dots for the directed transfer of photonic energy through FRET56–58, with the prospect of developing photonic circuits or artificial photosynthetic antenna complexes (Figure 4d). Conjugating DNA to small organic molecules or polymers has yielded materials with interesting thermal properties, such as solid DNA that only melts at 95°C59. Hybrids of nucleic acids and synthetic polymers show promise for drug delivery and diagnostics60. Finally, proteins may also be employed as integral structural components of nucleic acid nanoassemblies. For instance, assemblies shaped like equilateral triangles were formed from an RNA-binding protein and an RNA strand bearing the appropriate binding motif61.
Combining Bottom-Up with Top-Down Techniques
So far we have described nanoassemblies built using bottom-up techniques (i.e., the creation of more complicated structures self-assembled from fundamental units). While these techniques yield highly resolved structures that may be generated in parallel, they are so far still limited in their dimensions, making them unappealing for applications where truly large-scale patterning is required. By contrast, most current technology outside of nanotechnology uses top-down techniques, namely the ordered assembly of components by externally-controlled means. For example, many lithographic top-down fabrication methods are able to serially generate structures with features typically on the order of microns (e.g., for silicon-based computer chips), but at great capital expense for equipment. By combining top-down and bottom-up techniques, a more affordable large-scale (> 1 mm) patterned array with nanometer addressability may be accomplished62. Electron beam63 and soft64 lithography techniques have been used to pattern DNA nanostructures across a surface with spacings of ~300 nm and ~5 µm, respectively (Figure 4e,f).
ENVISIONING FUTURE APPLICATIONS
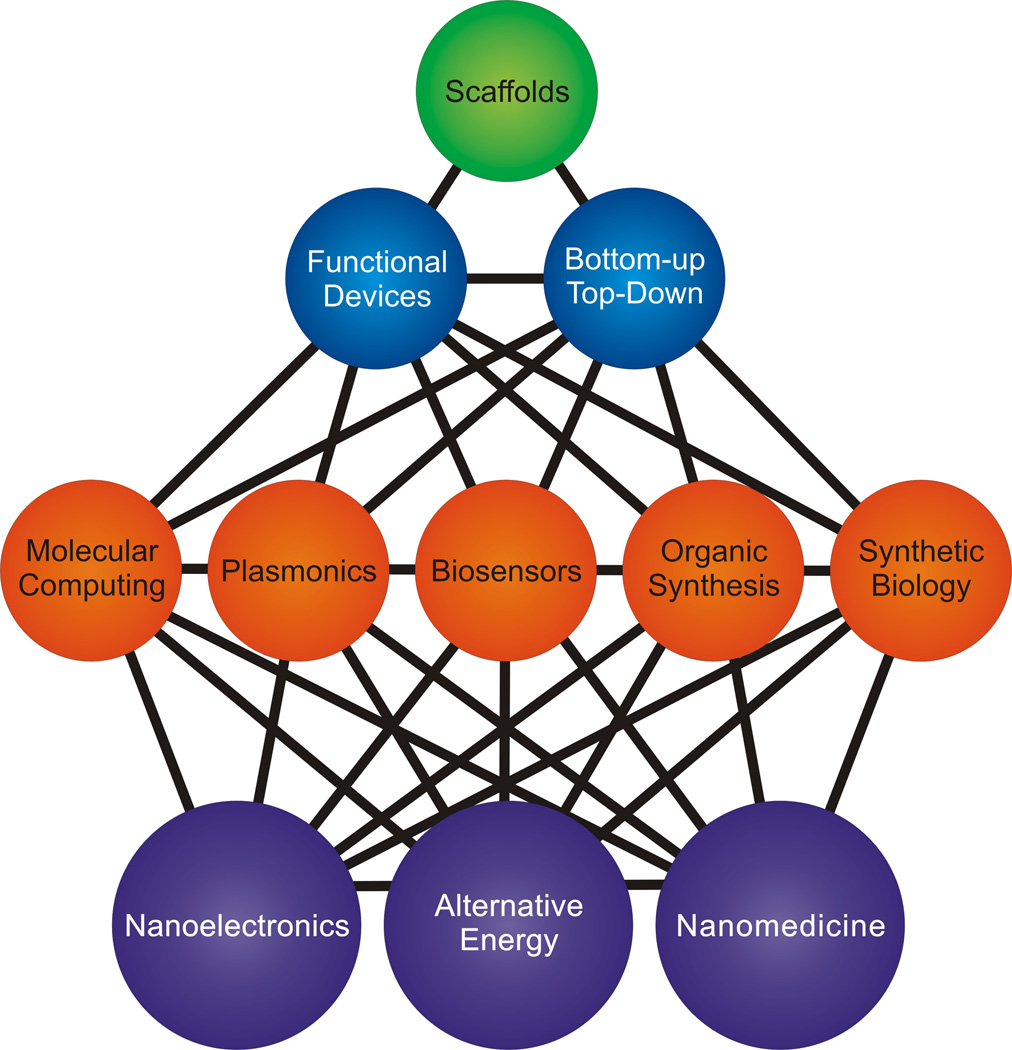
We envision that the basic materials and concepts of nucleic acid nanotechnology outlined so far will further be recombined to create devices capable of accomplishing an ever wider variety of tasks, particularly in the following fields (Figure 5).
Figure 5.
Hierarchy of areas impacted by DNA origami technology. The foundational DNA scaffolds (green) are used to create devices (blue) for a broad variety of applications (orange) that can be combined to enhance numerous emerging interdisciplinary fields (purple). Disclaimer: the nucleic acid nanotechnology field will not be limited by our current vision.
Molecular Computing
Modern day computers are capable of performing incredibly complex calculations with impressive speeds. At the computer’s core, these complex calculations are made possible by large series of binary digital circuits assembled in logic gates. Analogously, nucleic acids can behave as logic gates. For instance, the output “true” for an AND logic gate can be represented by a deoxyribozyme cleaving its substrate if and only if both DNA strand A and DNA strand B are present in solution65 (Figure 6). Using variations on this basic concept, nucleic acids have completed tasks with a range of complexities from competitively playing tic-tac-toe66 to mimicking field programmable gate arrays67 and neural networks68. Nucleic-acid-based logic gates inherently function based on thermodynamic mass action and chemical kinetics laws (i.e., probabilistic behavior), rather than the deterministic, digital on-off switches of silicon-based computers. Computing by nucleic acid nanotechnology therefore will more closely mimic the "fuzzy logic" and feedback loops of ant colonies and the human brain, with corresponding advantages in adaptability and complex pattern analysis69. As such, it will likely find ample application in both probing and interfacing with biology.
Figure 6.
Progression in DNA and RNA nanotechnology. . In nature, DNA forms structures such as the Holliday junction, which has inspired scientists to create more complex structures such as the rectangular DNA-origami tile. In the future, such tiles may be used in fields including nanoelectronics as a scaffold for plasmonic circuit components to generate circuits that mimic neuron behavior. As the simple example in the figure depicts, a new circuit connection (purple) may be strengthened by repeated cooperative stimuli from excitatory pathways (green and blue) and hindered by the stimuli from an inhibitory pathway (orange). In nature, RNA plays a catalytic role in peptide bond formation by the ribosome, arguably the most important enzyme on earth. The catalytic and exquisite molecular recognition activity of RNA is exploited in (deoxy)ribozyme computing, which may be used in the future for complex therapeutic nanomedicine applications. For instance, a drug carrier, specifically delivered to a diseased cell through endocytosis triggered by binding to a protein on the cell surface, opens after entering the cell to release the drug. The contents of the drug carrier include a miRNA mimic that causes repression of a specific protein that otherwise would inhibit the surface marker, resulting in a cell that becomes more receptive to the drug carriers.
Plasmonics
When metallic nanoparticles interact with visible electromagnetic waves, their conduction band electrons are excited, resulting in a phenomenon known as plasmon resonance70, 71. These nanoparticles placed in close proximity (<2.5 times the diameter of the nanoparticle) undergo strong near-field coupling with field enhancements72. However, the optimal field enhancement is dependent on the geometry of the nanoparticle array, requiring precise nanoparticle placement and spacing70, 72. To this end, gold nanoparticles have been assembled on DNA origami triangles73, 74. While DNA origami tiles are advantageous in their rigidity and addressability, they are limited in length (~100 nm) unless concatenated, in turn limiting the propagation of the plasmon resonance signal. In the future, the longer scaffolds, such as the nanopeapod75 or DNA origami nanotube76 with lengths on the order of tens of microns, will be paired with top-down microfabrication techniques to construct larger nanoparticle arrays with precision and spacing on the order of tens of nanometers. Applications will likely include communication systems that merge electronics and photonics (optics) at the nanoscale70.
Biosensors
Biosensors are small, self-contained devices that detect and report the concentration of an analyte through physical interaction77. Several types of biosensors have been constructed from nucleic acids (e.g., aptazyme-based78 or pH-sensitive79), the most common being optical biosensors in which the hybridization between the analyte and its complementary capture strand immobilized on the surface results in a change in fluorescent signal77. To be effective, such a technique requires a highly specific interaction between the capture strand and the analyte, and low yield of false positive results. Increased specificity has already been accomplished by incorporating synthetic nucleic acids80, 81, although there is still room for improvement, for example, with the further development of modified nucleotides. One of the causes of false positive signals is crosstalk between neighboring capture strands82. The spacing between neighboring biosensor units may be determined with nanometer precision by using a DNA origami scaffold, enabling a high density of capture strands on the surface while limiting their interactions. Such scaffold techniques have recently been utilized to detect analyte concentrations as low as 200 pM, and it was found that the hybridization sensitivity was dependent on probe position83. In addition, precisely arranged biosensor arrays with enhanced and possibly synergistic detection properties for complex samples are now within grasp.
Organic Synthesis
Unlike conventional organic synthesis, which is performed one step at a time with pure reagents, the cell is capable of catalyzing thousands of reactions simultaneously in the same vessel with extremely high specificity and efficiency. DNA scaffolds and devices may provide a way to emulate this approach by virtue of their programmable nanoscale structure and ability to spatially organize catalysts and reagents. DNA origami, in combination with single-stranded overhangs, conformational DNA switches, and/or DNA walkers, have been used for multi-step organic synthesis49, programmable assembly of dendrimeric oligomers84, and dynamically controlled combinatorial assembly of gold nanoparticles48. Since DNA origami are currently expensive to produce in large quantities, practical applications may be restricted in the short term to small-scale syntheses, those that must be performed under mild conditions, or those that are not feasible using conventional synthetic chemistry – for example, when high-resolution control of reactant positioning (different than that afforded by surface or metal ion coordination chemistry)84 is required. Such syntheses are abundant in nature and exemplified by the assembly lines of multi-domain polyketide synthases that generate a large number of bioactive compounds85. Another exciting prospect enabled by coupling DNA origami to other dynamic devices is the ability to make chemical reactions or noncovalent molecular assembly responsive to environmental cues48. For instance, signals from a host organism might provide cues for a smart molecular “assembly line” to synthesize one of many possible products depending intracellular conditions at a particular time.
Synthetic Biology
Synthetic biology is concerned with engineering natural or artificial biological systems to better understand biological phenomena or for practical uses such as food, drug, or fuel production, or for novel therapeutic approaches86, 87. While most synthetic biology has made use of pre-existing pathways by altering them for a particular purpose or inserting them into a different organism (top-down approach)86, it will be desirable to construct entirely novel artificial organisms88 and pathways to allow for more predictable and tractable performance (bottom-up approach)86. DNA origami and similar scaffolds could contribute to such systems by allowing for spatial and chemical coupling of related enzymes in a manner analogous to natural multi-enzyme complexes, a concept that has recently been demonstrated on simpler DNA templates89 in vitro and in highly concatenated RNA scaffolds in bacteria. (The latter demonstrates one clear advantage of RNA for many synthetic-biology applications: it is readily transcribed from a plasmid inserted into the host organism.) Three-dimensional nucleic-acid scaffolds could also serve to physically compartmentalize artificial biochemical reaction networks like natural organelles, since they can be engineered to assemble with high efficiency into vessels of arbitrary shape with well-defined dimensions45, 90, 91. Furthermore, due to the relative ease of engineering nucleic-acid-based reaction networks54 and the existence of numerous elementary components such as aptamers, ribozymes, and riboswitches, some of the first artificial biological systems or organisms may have nucleic acids as integral components of their information networks. Although true bottom-up synthetic biology remains an elusive prospect for now, nucleic acid scaffolds and devices are beginning to serve as a useful means to perturb biochemical networks92–94. Since DNA origami are stable in cells93, 94 and cell lysate95, ever more complex structures may soon find similar uses in interfacing with and programming of natural biological pathways.
EMERGENT INTERDISCIPLINARY FIELDS
Beyond the horizon of advantageous applications in more or less well-established fields, it is illuminating to explore the current state of some emerging interdisciplinary fields (Figure 5) to better understand how they are expected to benefit from future advancements in nucleic acid nanotechnology.
Nanoelectronics
There exists a strong effort to make more highly sophisticated, conveniently sized electronic devices, requiring the further minimization of electronic components. Currently, top-down-produced assemblies are limited by the size of nanowires and use of lithographic techniques to tens of nanometers in spatial resolution. The wires are also limited in the flux of digital information they are able to transport when compared to optical digitization techniques such as optical fibers96. Exploiting plasmonic properties between precisely-placed nanoparticles is a plausible alternative. Positioned using nucleic acid scaffolds, such an architecture would not only allow for signal enhancement of the propagated information, but theoretically, the nanoparticles could behave as various, reconfigurable circuit components based on their materials and geometries97 (Figure 6). Integrating this approach with established lithographic techniques will lead to synergy. Scaffolded components could also serve as templates for metal deposition to generate solid-state devices with nanometer precision, although it will be necessary to develop methods for sustaining the template in the adverse conditions required for deposition. Arraying various bottom-up circuit elements using top-down assembling techniques may generate larger, more complex circuits.
Alternative Energy
Rising global demand for energy and concerns about the environmental impact of fossil fuel use motivate the search for viable alternative energy sources 98, 99. While progress has been made toward improving alternative energy production and storage using nanomaterials, emphasis has been placed on inorganic materials rather than biomolecules. Owing to their complexity, stand-alone nucleic acid nanodevices are not likely to meet large-scale energy needs anytime soon, but become more formidable when combined with living systems or inorganic materials. For example, genetically encoded nucleic acid scaffolds can be used to increase the flux through engineered biochemical pathways by colocalizing functionally related enzymes, including in the production of fuels such as hydrogen100. Nanoelectronic and photonic devices constructed using nucleic acid scaffolds and suitable chromophores could, with sufficient improvements in efficiency, mimic photosynthetic antenna complexes for nanoscale solar energy harvesting.
Nanomedicine
Nanomedicine has been defined as the utilization of nanotechnology for medicinal purposes including diagnostic assays, therapeutic agents, and monitoring devices101. One of the current main objectives in the field is targeted drug delivery in which a drug carrier is able to selectively deliver a drug only to diseased cells, resulting in lower drug dosages and reduced harm to healthy cells101. Three-dimensional DNA smart materials have several features that make them appealing candidates for nanocarriers: they are biocompatible; they can selectively contain particles, fully encapsulate them, and release them when presented with a trigger; the exteriors may be decorated with targeting signals; and differing functional components may be incorporated into their design with defined spatial positioning. Current designs initiate cargo release by introducing a nucleic acid strand to the carrier’s environment90. By alternatively incorporating functional strands that are sensitive to natural changes in cellular environments, such as potassium-sensitive deoxyribozymes or DNA strands that undergo conformational changes in response to pH93 or an intrinsic cellular component, cargo release may be controlled intracellularly by natural means (much like intracellular genome release from a viral capsid102). Computational nucleic acid devices that mimic ncRNAs are being created to implement this goal by using self-cleaving deoxyribozyme hairpins that contain within their sequence a short ssDNA strand that is complementary to the portion of mRNA needing to be suppressed, but is not active unless triggered to be cleaved by disease-indicators in the cellular environment54, 65, 103. These devices could be integrated with the nanocarrier design to be transported only to the diseased cells. Beyond that, complex molecular “communities” could integrate biosensors, molecular computation, and chemical synthesis to diagnose and treat illnesses at the level of individual cells. For instance, one such system might enter a cell through a carrier-mediated interaction, detect abnormal levels of a certain subset of messenger RNAs, compute the probability that a given cell is cancerous, and then decide whether to initiate apoptosis through an intrinsic cellular pathway (Figure 6). While such an application remains technically very challenging and expensive, most of the required device classes have been constructed.
DNA does have the disadvantage that it is susceptible to nucleases, potentially leading to release of cargo before triggered to do so. To overcome this challenge, modified nucleic acids may be incorporated104, although introducing foreign materials will need to be rigorously tested for cytotoxicity and unexpected side effects. Before these devices can be used as therapeutic agents, failsafe features (that lead, for example, to instability of the device outside of its target area) will need to be included in the design to prevent any undesired side effects.
Another primary objective of nanomedicine is the creation of point-of-care devices, which provide a convenient way of testing and diagnosing a patient immediately. Biosensors constructed with top-down techniques hold much promise for this goal. However, the currently limited sensitivity of biosensors generally requires the amplification of the analyte, typically accomplished using PCR, which limits the detection efficiency82. Plasmonics may help amplify fluorescent signals, enabling a binding event to be detected even with low concentrations of the analyte71. Integration of biosensors with nanoelectronic devices could potentially accomplish the types of analysis required to accurately diagnose a patient.
Conclusion
Nucleic acid nanotechnology has come a long way since its inception a quarter century ago. It has advanced from purely static scaffolds to dynamic functional devices that include other natural and synthetic materials. The currently existing nanodevices show great promise for useful future applications in a broad variety of fields as sampled here. It is even more exciting to contemplate that the precise structures and functional versatility of devices such as these will spawn new applications and perhaps entire new fields that lie beyond the current bounds of our insight. Buttressing these advances will be the further development and exploitation of single molecule tools that can monitor the dynamic (bio)chemical reactions, actuation, movement (diffusion), and integrity of individual nanodevices, for example, super-resolution single molecule fluorescence microscopy47, 52, 105, 106.
DNA Origami.
The invention of scaffolded DNA origami was a milestone in the advancement of nucleic acid nanotechnology. Developed by Paul Rothemund, DNA origami is the assisted folding of one long single-stranded DNA “scaffold” strand from a bacterial phage genome consisting of 7,249 nucleotides into a predetermined shape by ~200 typically 32-nt-long single-stranded DNA “staple” strands containing sequences that are complementary to specific regions of the scaffold strand. Two or more nonadjacent segments of the scaffold strand are brought together and held in place by hybridization to different portions of the same staple oligonucleotide in aggregate (Figure 3a), enabling the creation of arbitrary shapes based solely on the staple sequences (Figure 3b,c). The staples may be extended at their 5’ ends to make them addressable for patterning with a resolution of 6 nm on the assembled origami, imposed by the inter-staple distance on the assembled DNA duplexes of the origami11 (Figure 3d).
While building on previous approaches to self-assembly of DNA nanostructures, the DNA origami technique has multiple advantages: (i) It accomplishes highly specific topologies with a spatially addressable resolution comparable to that accomplished by AFM or STM surface manipulation. (ii) The well-formed, redundant self-assembly is relatively insensitive to varying stoichiometric ratios of the staples, eliminating the need for intermittent purification steps and resulting in higher yields. (iii) Multiple nanostructures may be obtained simultaneously and possibly further assembled into larger structures with high fidelity. (iv) Creating arbitrary shapes that are largely unrestricted by symmetry considerations is relatively straightforward11.
The DNA origami method has so far provided a scaffold for many applications including forming the track for molecular nanorobots47, constructing ordered molecular assembly lines48, 49, and assembling components of putative nanoelectronic circuits50, 51. The scope and limitations of these applications are enumerated throughout.
Footnotes
Further Reading/Resources
Nadrian Seeman, DNA in a material world. Nature 2003, 421:427–431.
Nadrian Seeman, Nanomaterials based on DNA. Annu Rev Biochem 2010, 79:65–87.
Rothemund, Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440:297–302.
Faisal Aldaye, Alison Palmer and Hanadi Sleiman, Assembling materials with DNA as the guide. Science 2008, 321:1795–1799
Max von Delius and David Leigh, Walking molecules. Chem Soc Rev 2011, 40:3656–3676.
http://en.wikipedia.org/wiki/DNA_nanotechnology
http://en.wikipedia.org/wiki/Nanomedicine
Contributor Information
Nicole Michelotti, Department of Physics, University of Michigan.
Alexander Johnson-Buck, Department of Chemistry, University of Michigan.
Anthony J. Manzo, Department of Chemistry, University of Michigan
Nils G. Walter, Email: nwalter@umich.edu, Department of Chemistry, University of Michigan.
References
- 1.Seeman NC. DNA in a material world. Nature. 2003;421:427–431. doi: 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]
- 2.Voet D, Voet JG. Biochemistry. 4th ed. New York: J. Wiley & Sons; 2011. [Google Scholar]
- 3.Paabo S, Poinar H, Serre D, Jaenicke-Despres V, Hebler J, Rohland N, Kuch M, Krause J, Vigilant L, Hofreiter M. Genetic analyses from ancient DNA. Annu Rev Genet. 2004;38:645–679. doi: 10.1146/annurev.genet.37.110801.143214. [DOI] [PubMed] [Google Scholar]
- 4.Mattick JS, Taft RJ, Faulkner GJ. A global view of genomic information--moving beyond the gene and the master regulator. Trends Genet. 2010;26:21–28. doi: 10.1016/j.tig.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 5.Marek MS, Johnson-Buck A, Walter NG. The shape-shifting quasispecies of RNA: one sequence, many functional folds. Phys Chem Chem Phys. 2011;13:11524–11537. doi: 10.1039/c1cp20576e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frank J, Gonzalez RL., Jr Structure and dynamics of a processive Brownian motor: the translating ribosome. Annu Rev Biochem. 2010;79:381–412. doi: 10.1146/annurev-biochem-060408-173330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Will CL, Luhrmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2011:3. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newman AJ, Nagai K. Structural studies of the spliceosome: blind men and an elephant. Curr Opin Struct Biol. 2010;20:82–89. doi: 10.1016/j.sbi.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 9.Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5:e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seeman NC. Nanomaterials based on DNA. Annu Rev Biochem. 2010;79:65–87. doi: 10.1146/annurev-biochem-060308-102244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rothemund PW. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 12.Seeman NC. An overview of structural DNA nanotechnology. Mol Biotechnol. 2007;37:246–257. doi: 10.1007/s12033-007-0059-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aldaye FA, Palmer AL, Sleiman HF. Assembling materials with DNA as the guide. Science. 2008;321:1795–1799. doi: 10.1126/science.1154533. [DOI] [PubMed] [Google Scholar]
- 14.Nangreave J, Han D, Liu Y, Yan H. DNA origami: a history and current perspective. Curr Opin Chem Biol. 2010;14:608–615. doi: 10.1016/j.cbpa.2010.06.182. [DOI] [PubMed] [Google Scholar]
- 15.Shih WM, Lin C. Knitting complex weaves with DNA origami. Curr Opin Struct Biol. 2010;20:276–282. doi: 10.1016/j.sbi.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torring T, Voigt NV, Nangreave J, Yan H, Gothelf KV. DNA origami: a quantum leap for self-assembly of complex structures. Chem Soc Rev. 2011 doi: 10.1039/c1cs15057j. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 18.Williams NH, Takasaki B, Wall M, Chin J. Structure and nuclease activity of simple dinuclear metal complexes: Quantitative dissection of the role of metal ions. Acc Chem Res. 1999;32:485–493. [Google Scholar]
- 19.Wang S, Kool ET. Origins of the large differences in stability of DNA and RNA helices: C-5 methyl and 2'-hydroxyl effects. Biochemistry. 1995;34:4125–4132. doi: 10.1021/bi00012a031. [DOI] [PubMed] [Google Scholar]
- 20.Lipps HJ, Rhodes D. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol. 2009;19:414–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 21.Grindley ND, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu Rev Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 22.Schones DE, Zhao K. Genome-wide approaches to studying chromatin modifications. Nat Rev Genet. 2008;9:179–191. doi: 10.1038/nrg2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 24.Chen J, Darst SA, Thirumalai D. Promoter melting triggered by bacterial RNA polymerase occurs in three steps. Proc Natl Acad Sci USA. 2010;107:12523–12528. doi: 10.1073/pnas.1003533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crick F. Central dogma of molecular biology. Nature. 1970;227:561–563. doi: 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- 26.Davidson BL, McCray PB., Jr Current prospects for RNA interference-based therapies. Nat Rev Genet. 2011;12:329–340. doi: 10.1038/nrg2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Al-Hashimi HM, Walter NG. RNA dynamics: it is about time. Curr Opin Struct Biol. 2008;18:321–329. doi: 10.1016/j.sbi.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dambach MD, Winkler WC. Expanding roles for metabolite-sensing regulatory RNAs. Curr Opin Microbiol. 2009;12:161–169. doi: 10.1016/j.mib.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 30.Ferre-D'Amare AR, Scott WG. Small self-cleaving ribozymes. Cold Spring Harb Perspect Biol. 2010;2:a003574. doi: 10.1101/cshperspect.a003574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ehrlich M, Gama-Sosa MA, Carreira LH, Ljungdahl LG, Kuo KC, Gehrke CW. DNA methylation in thermophilic bacteria: N4-methylcytosine, 5-methylcytosine, and N6-methyladenine. Nucleic Acids Res. 1985;13:1399–1412. doi: 10.1093/nar/13.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 33.Campbell MA, Wengel J. Locked vs. unlocked nucleic acids (LNA vs. UNA): contrasting structures work towards common therapeutic goals. Chem Soc Rev. 2011 doi: 10.1039/c1cs15048k. [DOI] [PubMed] [Google Scholar]
- 34.Braasch DA, Corey DR. Locked nucleic acid (LNA): fine-tuning the recognition of DNA and RNA. Chem Biol. 2001;8:1–7. doi: 10.1016/s1074-5521(00)00058-2. [DOI] [PubMed] [Google Scholar]
- 35.Rana TM. Illuminating the silence: understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol. 2007;8:23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- 36.Egholm M, Buchardt O, Christensen L, Behrens C, Freier SM, Driver DA, Berg RH, Kim SK, Norden B, Nielsen PE. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules. Nature. 1993;365:566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 37.Williams KA, Veenhuizen PT, de la Torre BG, Eritja R, Dekker C. Nanotechnology: carbon nanotubes with DNA recognition. Nature. 2002;420:761. doi: 10.1038/420761a. [DOI] [PubMed] [Google Scholar]
- 38.Lukeman PS, Mittal AC, Seeman NC. Two dimensional PNA/DNA arrays: estimating the helicity of unusual nucleic acid polymers. Chem Commun. 2004:1694–1695. doi: 10.1039/b401103a. [DOI] [PubMed] [Google Scholar]
- 39.Chen F, Yang Z, Yan M, Alvarado JB, Wang G, Benner SA. Recognition of an expanded genetic alphabet by type-II restriction endonucleases and their application to analyze polymerase fidelity. Nucleic Acids Res. 2011;39:3949–3961. doi: 10.1093/nar/gkq1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petrillo ML, Newton CJ, Cunningham RP, Ma RI, Kallenbach NR, Seeman NC. The Ligation and Flexibility of 4-Arm DNA Junctions. Biopolymers. 1988;27:1337–1352. doi: 10.1002/bip.360270902. [DOI] [PubMed] [Google Scholar]
- 41.Holliday R. Mechanism for Gene Conversion in Fungi. Genet Res. 1964;5:282–304. doi: 10.1017/S0016672308009476. [DOI] [PubMed] [Google Scholar]
- 42.Fu TJ, Seeman NC. DNA double-crossover molecules. Biochemistry. 1993;32:3211–3220. doi: 10.1021/bi00064a003. [DOI] [PubMed] [Google Scholar]
- 43.LaBean TH, Yan H, Kopatsch J, Liu FR, Winfree E, Reif JH, Seeman NC. Construction, analysis, ligation, and self-assembly of DNA triple crossover complexes. J Am Chem Soc. 2000;122:1848–1860. [Google Scholar]
- 44.Shih WM, Quispe JD, Joyce GF. A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nature. 2004;427:618–621. doi: 10.1038/nature02307. [DOI] [PubMed] [Google Scholar]
- 45.Han D, Pal S, Nangreave J, Deng Z, Liu Y, Yan H. DNA origami with complex curvatures in three-dimensional space. Science. 2011;332:342–346. doi: 10.1126/science.1202998. [DOI] [PubMed] [Google Scholar]
- 46.Woo S, Rothemund PW. Programmable molecular recognition based on the geometry of DNA nanostructures. Nat Chem. 2011;3:620–627. doi: 10.1038/nchem.1070. [DOI] [PubMed] [Google Scholar]
- 47.Lund K, Manzo AJ, Dabby N, Michelotti N, Johnson-Buck A, Nangreave J, Taylor S, Pei R, Stojanovic MN, Walter NG, et al. Molecular robots guided by prescriptive landscapes. Nature. 2010;465:206–210. doi: 10.1038/nature09012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gu HZ, Chao J, Xiao SJ, Seeman NC. A proximity-based programmable DNA nanoscale assembly line. Nature. 2010;465:U202–U286. doi: 10.1038/nature09026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He Y, Liu DR. Autonomous multistep organic synthesis in a single isothermal solution mediated by a DNA walker. Nat Nanotechnol. 2010;5:778–782. doi: 10.1038/nnano.2010.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maune HT, Han SP, Barish RD, Bockrath M, Goddard WA, Rothemund PWK, Winfree E. Self-assembly of carbon nanotubes into two-dimensional geometries using DNA origami templates. Nat Nanotechnol. 2010;5:61–66. doi: 10.1038/nnano.2009.311. [DOI] [PubMed] [Google Scholar]
- 51.Eskelinen AP, Kuzyk A, Kaltiaisenaho TK, Timmermans MY, Nasibulin AG, Kauppinen EI, Torma P. Assembly of Single-Walled Carbon Nanotubes on DNA-Origami Templates through Streptavidin-Biotin Interaction. Small. 2011;7:746–750. doi: 10.1002/smll.201001750. [DOI] [PubMed] [Google Scholar]
- 52.Michelotti N, de Silva C, Johnson-Buck AE, Manzo AJ, Walter NG. A bird's eye view tracking slow nanometer-scale movements of single molecular nano-assemblies. Methods Enzymol. 2010;475:121–148. doi: 10.1016/S0076-6879(10)75006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.von Delius M, Leigh DA. Walking molecules. Chem Soc Rev. 2011;40:3656–3676. doi: 10.1039/c1cs15005g. [DOI] [PubMed] [Google Scholar]
- 54.Qian L, Winfree E. Scaling up digital circuit computation with DNA strand displacement cascades. Science. 2011;332:1196–1201. doi: 10.1126/science.1200520. [DOI] [PubMed] [Google Scholar]
- 55.Niemeyer CM, Koehler J, Wuerdemann C. DNA-directed assembly of bienzymic complexes from in vivo biotinylated NAD(P)H : FMN oxidoreductase and luciferase. Chembiochem. 2002;3:242–245. doi: 10.1002/1439-7633(20020301)3:2/3<242::AID-CBIC242>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 56.Boeneman K, Prasuhn DE, Blanco-Canosa JB, Dawson PE, Melinger JS, Ancona M, Stewart MH, Susumu K, Huston A, Medintz IL. Self-Assembled Quantum Dot-Sensitized Multivalent DNA Photonic Wires. J Am Chem Soc. 2010;132:18177–18190. doi: 10.1021/ja106465x. [DOI] [PubMed] [Google Scholar]
- 57.Ruiz-Carretero A, Janssen PGA, Stevens AL, Surin M, Herz LM, Schenning APHJ. Directing energy transfer in discrete one-dimensional oligonucleotide-templated assemblies. Chem Commun. 2011;47:884–886. doi: 10.1039/c0cc04128a. [DOI] [PubMed] [Google Scholar]
- 58.Stein IH, Steinhauer C, Tinnefeld P. Single-molecule four-color FRET visualizes energy-transfer paths on DNA origami. J Am Chem Soc. 2011;133:4193–4195. doi: 10.1021/ja1105464. [DOI] [PubMed] [Google Scholar]
- 59.Singh A, Tolev M, Meng M, Klenin K, Plietzsch O, Schilling CI, Muller T, Nieger M, Brase S, Wenzel W, et al. Branched DNA that forms a solid at 95 degrees C. Angew Chem Int Ed Engl. 2011;50:3227–3231. doi: 10.1002/anie.201006992. [DOI] [PubMed] [Google Scholar]
- 60.Kwak M, Herrmann A. Nucleic acid/organic polymer hybrid materials: synthesis, superstructures, and applications. Angew Chem Int Ed Engl. 2010;49:8574–8587. doi: 10.1002/anie.200906820. [DOI] [PubMed] [Google Scholar]
- 61.Ohno H, Kobayashi T, Kabata R, Endo K, Iwasa T, Yoshimura SH, Takeyasu K, Inoue T, Saito H. Synthetic RNA-protein complex shaped like an equilateral triangle. Nat Nanotechnol. 2011;6:115–119. doi: 10.1038/nnano.2010.268. [DOI] [PubMed] [Google Scholar]
- 62.Hung AM, Noh H, Cha JN. Recent advances in DNA-based directed assembly on surfaces. Nanoscale. 2010;2:2530–2537. doi: 10.1039/c0nr00430h. [DOI] [PubMed] [Google Scholar]
- 63.Kershner RJ, Bozano LD, Micheel CM, Hung AM, Fornof AR, Cha JN, Rettner CT, Bersani M, Frommer J, Rothemund PW, et al. Placement and orientation of individual DNA shapes on lithographically patterned surfaces. Nat Nanotechnol. 2009;4:557–561. doi: 10.1038/nnano.2009.220. [DOI] [PubMed] [Google Scholar]
- 64.Lin CX, Ke YG, Liu Y, Mertig M, Gu J, Yan H. Functional DNA nanotube arrays: Bottom-up meets top-down. Angew Chem Int Ed Engl. 2007;46:6089–6092. doi: 10.1002/anie.200701767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stojanovic MN, Mitchell TE, Stefanovic D. Deoxyribozyme-based logic gates. J Am Chem Soc. 2002;124:3555–3561. doi: 10.1021/ja016756v. [DOI] [PubMed] [Google Scholar]
- 66.Stojanovic MN, Stefanovic D. A deoxyribozyme-based molecular automaton. Nat Biotechnol. 2003;21:1069–1074. doi: 10.1038/nbt862. [DOI] [PubMed] [Google Scholar]
- 67.Pei R, Matamoros E, Liu M, Stefanovic D, Stojanovic MN. Training a molecular automaton to play a game. Nat Nanotechnol. 2010;5:773–777. doi: 10.1038/nnano.2010.194. [DOI] [PubMed] [Google Scholar]
- 68.Qian L, Winfree E, Bruck J. Neural network computation with DNA strand displacement cascades. Nature. 2011;475:368–372. doi: 10.1038/nature10262. [DOI] [PubMed] [Google Scholar]
- 69.Morris MK, Saez-Rodriguez J, Sorger PK, Lauffenburger DA. Logic-based models for the analysis of cell signaling networks. Biochemistry. 2010;49:3216–3224. doi: 10.1021/bi902202q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ozbay E. Plasmonics: merging photonics and electronics at nanoscale dimensions. Science. 2006;311:189–193. doi: 10.1126/science.1114849. [DOI] [PubMed] [Google Scholar]
- 71.Anker JN, Hall WP, Lyandres O, Shah NC, Zhao J, Van Duyne RP. Biosensing with plasmonic nanosensors. Nature Materials. 2008;7:442–453. doi: 10.1038/nmat2162. [DOI] [PubMed] [Google Scholar]
- 72.Tan SJ, Campolongo MJ, Luo D, Cheng W. Building plasmonic nanostructures with DNA. Nat Nanotechnol. 2011;6:268–276. doi: 10.1038/nnano.2011.49. [DOI] [PubMed] [Google Scholar]
- 73.Hung AM, Micheel CM, Bozano LD, Osterbur LW, Wallraff GM, Cha JN. Large-area spatially ordered arrays of gold nanoparticles directed by lithographically confined DNA origami. Nat Nanotechnol. 2010;5:121–126. doi: 10.1038/nnano.2009.450. [DOI] [PubMed] [Google Scholar]
- 74.Ding B, Deng Z, Yan H, Cabrini S, Zuckermann RN, Bokor J. Gold nanoparticle self-similar chain structure organized by DNA origami. J Am Chem Soc. 2010;132:3248–3249. doi: 10.1021/ja9101198. [DOI] [PubMed] [Google Scholar]
- 75.Lo PK, Karam P, Aldaye FA, McLaughlin CK, Hamblin GD, Cosa G, Sleiman HF. Loading and selective release of cargo in DNA nanotubes with longitudinal variation. Nat Chem. 2010;2:319–328. doi: 10.1038/nchem.575. [DOI] [PubMed] [Google Scholar]
- 76.Rothemund PW, Ekani-Nkodo A, Papadakis N, Kumar A, Fygenson DK, Winfree E. Design and characterization of programmable DNA nanotubes. J Am Chem Soc. 2004;126:16344–16352. doi: 10.1021/ja044319l. [DOI] [PubMed] [Google Scholar]
- 77.Palchetti I, Mascini M. Nucleic acid biosensors for environmental pollution monitoring. Analyst. 2008;133:846–854. doi: 10.1039/b802920m. [DOI] [PubMed] [Google Scholar]
- 78.Sekella PT, Rueda D, Walter NG. A biosensor for theophylline based on fluorescence detection of ligand-induced hammerhead ribozyme cleavage. RNA. 2002;8:1242–1252. doi: 10.1017/s1355838202028066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Modi S, Swetha MG, Goswami D, Gupta GD, Mayor S, Krishnan Y. A DNA nanomachine that maps spatial and temporal pH changes inside living cells. Nat Nanotechnol. 2009;4:325–330. doi: 10.1038/nnano.2009.83. [DOI] [PubMed] [Google Scholar]
- 80.Wang J. DNA biosensors based on peptide nucleic acid (PNA) recognition layers. A review. Biosens Bioelectron. 1998;13:757–762. doi: 10.1016/s0956-5663(98)00039-6. [DOI] [PubMed] [Google Scholar]
- 81.Martinez K, Estevez MC, Wu Y, Phillips JA, Medley CD, Tan W. Locked nucleic acid based beacons for surface interaction studies and biosensor development. Anal Chem. 2009;81:3448–3454. doi: 10.1021/ac8027239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Piunno PA, Krull UJ. Trends in the development of nucleic acid biosensors for medical diagnostics. Anal Bioanal Chem. 2005;381:1004–1011. doi: 10.1007/s00216-004-3024-0. [DOI] [PubMed] [Google Scholar]
- 83.Ke Y, Lindsay S, Chang Y, Liu Y, Yan H. Self-assembled water-soluble nucleic acid probe tiles for label-free RNA hybridization assays. Science. 2008;319:180–183. doi: 10.1126/science.1150082. [DOI] [PubMed] [Google Scholar]
- 84.Liu HJ, Torring T, Dong MD, Rosen CB, Besenbacher F, Gothelf KV. DNA-Templated Covalent Coupling of G4 PAMAM Dendrimers. J Am Chem Soc. 2010;132:18054–18056. doi: 10.1021/ja109677n. [DOI] [PubMed] [Google Scholar]
- 85.Crawford JM, Townsend CA. New insights into the formation of fungal aromatic polyketides. Nat Rev Microbiol. 2010;8:879–889. doi: 10.1038/nrmicro2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Andrianantoandro E, Basu S, Karig DK, Weiss R. Synthetic biology: new engineering rules for an emerging discipline. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100073. 2006 0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee SK, Chou H, Ham TS, Lee TS, Keasling JD. Metabolic engineering of microorganisms for biofuels production: from bugs to synthetic biology to fuels. Curr Opin Biotechnol. 2008;19:556–563. doi: 10.1016/j.copbio.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 88.Gibson DG, Glass JI, Lartigue C, Noskov VN, Chuang RY, Algire MA, Benders GA, Montague MG, Ma L, Moodie MM, et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 89.Wilner OI, Shimron S, Weizmann Y, Wang ZG, Willner I. Self-assembly of enzymes on DNA scaffolds: en route to biocatalytic cascades and the synthesis of metallic nanowires. Nano Letters. 2009;9:2040–2043. doi: 10.1021/nl900302z. [DOI] [PubMed] [Google Scholar]
- 90.Andersen ES, Dong M, Nielsen MM, Jahn K, Subramani R, Mamdouh W, Golas MM, Sander B, Stark H, Oliveira CL, et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature. 2009;459:73–76. doi: 10.1038/nature07971. [DOI] [PubMed] [Google Scholar]
- 91.Endo M, Hidaka K, Kato T, Namba K, Sugiyama H. DNA prism structures constructed by folding of multiple rectangular arms. J Am Chem Soc. 2009;131:15570–15571. doi: 10.1021/ja904252e. [DOI] [PubMed] [Google Scholar]
- 92.Venkataraman S, Dirks RM, Ueda CT, Pierce NA. Selective cell death mediated by small conditional RNAs. Proc Natl Acad Sci USA. 2010;107:16777–16782. doi: 10.1073/pnas.1006377107. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 93.Surana S, Bhat JM, Koushika SP, Krishnan Y. An autonomous DNA nanomachine maps spatiotemporal pH changes in a multicellular living organism. Nat Commun. 2011;2:340. doi: 10.1038/ncomms1340. [DOI] [PubMed] [Google Scholar]
- 94.Bhatia D, Surana S, Chakraborty S, Koushika SP, Krishnan Y. A synthetic icosahedral DNA-based host-cargo complex for functional in vivo imaging. Nat Commun. 2011;2:339. doi: 10.1038/ncomms1337. [DOI] [PubMed] [Google Scholar]
- 95.Mei Q, Wei X, Su F, Liu Y, Youngbull C, Johnson R, Lindsay S, Yan H, Meldrum D. Stability of DNA origami nanoarrays in cell lysate. Nano Letters. 2011;11:1477–1482. doi: 10.1021/nl1040836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lu W, Lieber CM. Nanoelectronics from the bottom up. Nat Mater. 2007;6:841–850. doi: 10.1038/nmat2028. [DOI] [PubMed] [Google Scholar]
- 97.Engheta N. Circuits with light at nanoscales: Optical nanocircuits inspired by metamaterials. Science. 2007;317:1698–1702. doi: 10.1126/science.1133268. [DOI] [PubMed] [Google Scholar]
- 98.Tour JM, Kittrell C, Colvin VL. Green carbon as a bridge to renewable energy. Nat Mater. 2010;9:871–874. doi: 10.1038/nmat2887. [DOI] [PubMed] [Google Scholar]
- 99.Solomon S, Plattner GK, Knutti R, Friedlingstein P. Irreversible climate change due to carbon dioxide emissions. Proc Natl Acad Sci U S A. 2009;106:1704–1709. doi: 10.1073/pnas.0812721106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Delebecque CJ, Lindner AB, Silver PA, Aldaye FA. Organization of intracellular reactions with rationally designed RNA assemblies. Science. 2011;333:470–474. doi: 10.1126/science.1206938. [DOI] [PubMed] [Google Scholar]
- 101.Moghimi SM, Hunter AC, Murray JC. Nanomedicine: current status and future prospects. FASEB J. 2005;19:311–330. doi: 10.1096/fj.04-2747rev. [DOI] [PubMed] [Google Scholar]
- 102.Brandenburg B, Lee LY, Lakadamyali M, Rust MJ, Zhuang X, Hogle JM. Imaging poliovirus entry in live cells. PLoS Biol. 2007;5:e183. doi: 10.1371/journal.pbio.0050183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Benenson Y, Gil B, Ben-Dor U, Adar R, Shapiro E. An autonomous molecular computer for logical control of gene expression. Nature. 2004;429:423–429. doi: 10.1038/nature02551. [DOI] [PubMed] [Google Scholar]
- 104.Krishna H, Caruthers MH. Solid-Phase Synthesis, Thermal Denaturation Studies, Nuclease Resistance, and Cellular Uptake of (Oligodeoxyribonucleoside)methylborane Phosphine-DNA Chimeras. J Am Chem Soc. 2011;133:9844–9854. doi: 10.1021/ja201314q. [DOI] [PubMed] [Google Scholar]
- 105.Rueda D, Walter NG. Single molecule fluorescence control for nanotechnology. J Nanosci Nanotechnol. 2005;5:1990–2000. doi: 10.1166/jnn.2005.505. [DOI] [PubMed] [Google Scholar]
- 106.Walter NG, Huang CY, Manzo AJ, Sobhy MA. Do-it-yourself guide: how to use the modern single-molecule toolkit. Nat Methods. 2008;5:475–489. doi: 10.1038/nmeth.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]