Abstract
Defining the mechanisms that control cell growth and division is crucial to understanding cell homeostasis, which impacts human diseases such as cancer and diabetes. IQGAP1, a widely conserved effector and/or regulator of the GTPase CDC42, is a putative oncoprotein that controls cell proliferation; however, its mechanism in tumorigenesis is unknown. The mechanistic target of rapamycin (mTOR) pathway, the center of cell growth control, is commonly activated in human cancers, but has proved to be an ineffective clinical target because of an incomplete understanding of its mechanisms in cell growth inhibition. Using complementary studies in yeast and mammalian cells, we examined a potential role for IQGAP1 in regulating the negative feedback loop (NFL) of mTOR complex 1 (mTORC1) that controls cell growth. Two-hybrid screens identified the yeast TORC1-specific subunit Tco89p as an Iqg1p-binding partner, sharing roles in rapamycin-sensitive growth, axial-bud-site selection and cytokinesis, thus coupling cell growth and division. Mammalian IQGAP1 binds mTORC1 and Akt1 and in response to epidermal growth factor (EGF), cells expressing the mTORC1–Akt1-binding region (IQGAP1IR-WW) contained attenuated phosphorylated ERK1/2 (ERK1/2-P) activity and inactive glycogen synthase kinase 3α/β (GSK3α/β), which control apoptosis. Interestingly, these cells displayed a high level of Akt1 S473-P, but an attenuated level of the mTORC1-dependent kinase S6K1 T389-P and induced mTORC1–Akt1- and EGF-dependent transformed phenotypes. Moreover, IQGAP1 appears to influence cell abscission and its activity is elevated in carcinoma cell lines. These findings support the hypothesis that IQGAP1 acts upstream on the mTORC1–S6K1→Akt1 NFL and downstream of it, to couple cell growth and division, and thus like a rheostat, regulates cell homeostasis, dysregulation of which leads to tumorigenesis or other diseases. These results could have implications for the development of the next generation of anticancer therapeutics.
Key words: IQGAP1, Proliferation, Secretion, mTOR
Introduction
The mechanisms that control cell proliferation continue to be central to cell biology research (Tapon et al., 2001; Sturgill and Hall, 2007; Moseley et al., 2009) and to understanding prevalent human diseases such as diabetes and cancer. The evolutionarily conserved serine/threonine protein kinase mechanistic target of rapamycin (mTOR), the center of cell growth control, interfaces nutrient and growth factor signals to regulate cell proliferation (Guertin and Sabatini, 2007, Sengupta et al., 2010). It is believed that mTOR couples cell growth and division by integrating the nutrient and growth factor signals through the phosphoinositide 3-kinase–RAC-α serine/threonine protein kinase–mTOR (PI3K–Akt1–mTOR) pathway to control cell size, a pre-requisite to entry into the cell cycle, but despite much progress, how the two activities are integrated remains unclear (Tapon et al., 2001; Fingar and Blenis, 2004; Sabatini, 2006; Wullschleger et al., 2006; Polak and Hall, 2006; Sturgill and Hall, 2007; Hall, 2008; Laplante and Sabatini, 2009; Huang and Manning, 2009; Sengupta et al., 2010).
Yeast and mammalian TORs form two distinct complexes, TORC1 and TORC2 (also known as CREB-regulated transcription coactivator 1 and 2), each containing shared and unique subunits (Fingar and Blenis, 2004; Sabatini, 2006; Wullschleger et al., 2006; Hall, 2008; Laplante and Sabatini, 2009). In addition to other subunits, yeast TORC1 contains the shared subunits Tor1p or Tor2p, Lst8p and the unique subunits Tco89p and Kog1p (Loewith et al., 2002; Reinke et al., 2004). In mammals, where mTOR is a shared subunit, regulatory-associated protein of mTOR (Raptor) defines mTORC1 and rapamycin-insensitive companion of mTOR (Rictor) defines mTORC2 (Sabatini, 2006). mTORC1 is a rapamycin-sensitive complex that regulates cell mass by activating mRNA translation by direct phosphorylation, thereby activating its effector ribosomal S6 kinase 1 (S6K1), therefore, S6K phosphorylation is a widely used marker of mTORC1 activation (Fingar and Blenis, 2004; Ruvinsky and Meyuhas, 2006; Guertin and Sabatini, 2007). By contrast, mTORC2, believed to control the actin cytoskeleton, exhibits rapamycin-insensitive properties, but responds to long-term-rapamycin treatment and acts upstream of mTORC1 by directly activating Akt1, the effector of PI3K (Sabatini, 2006). Akt1 in turn activates mTORC1 by inhibiting the GAP (Tsc1/2 complex) thus activating the GTPase Rheb, which activates mTOR and increases cell mass (Inoki et al., 2002; Inoki et al., 2003; Inoki et al., 2005; Manning, 2004; Laplante and Sabatini, 2009; Huang and Manning, 2009).
This activation process is regulated by a negative feedback loop (NFL) whereby activated S6K1 T389-P suppresses Akt1 S473-P to regulate the cell size. How this regulatory inhibitory mechanism is controlled remains unknown (Laplante and Sabatini, 2009; Huang and Manning, 2009; Dibble et al., 2009; Julien et al., 2010; Sengupta et al., 2010). It is important to define the mTORC1–S6K1 NFL regulation because although aberrant activation of mTOR and Akt1 is a common oncogenic and diabetic signal, the mTOR inhibitors have been ineffective in clinical trials or animal models because of their inhibition of the S6K NFL and activation of Akt (Manning, 2004; Guertin and Sabatini, 2005; Guertin and Sabatini, 2007; Huang and Manning, 2009; Hsieh et al., 2011). Therefore, understanding the regulations of the mTORC1–S6K1 NFL is crucial to developing the next generation of effective anticancer and anti-diabetic therapeutics.
This study reports a previously unknown role for IQGAP1 in integrating mTORC1 and Akt1 signaling by modulating the mTORC1–S6K1 NFL to control cell proliferation. IQGAP1 is a modular protein and a widely conserved effector and/or regulator of the putative oncogene CDC42 GTPase and has been implicated in regulating cell polarity, migration, actin cytoskeleton dynamics and epithelial cell organization (Osman and Cerione, 1998; Osman et al., 2002; Mateer et al., 2003; Noritake et al., 2004; Noritake et al., 2005; Bensenor et al., 2007; Le Clainche et al., 2007; Brandt and Grosse, 2007), and in integrating signaling networks (reviewed by Mateer et al., 2003; White et al., 2009, Osman, 2010). IQGAP1 has oncogenic activity; it induces transformed phenotypes in cell cultures and tumorigenesis in mice and its aberrant expression or mislocalization associates with a wide range of human carcinomas (Wang et al., 2009; White et al., 2009; Johnson et al., 2009; Osman, 2010; Chen et al., 2010). Despite substantial investigation, to date its molecular mechanism in oncogenesis remains unknown.
The yeast ortholog, Iqg1p, is similarly modular and promotes cytokinesis (Ko et al., 2007; Epp and Chant, 1997; Lippincott and Li, 1998; Osman and Cerione, 1998), cooperating with the mitotic exit network (Corbett et al., 2006). It regulates cytokinesis by serving as a positional marker for axial-bud-site selection in haploid cells, linking cytokinesis with bud-site selection and polarized growth (Osman and Cerione, 1998; Osman and Cerione, 2006; Osman et al., 2002), thus fulfilling the tenet of the ‘cytokinesis tag’ model, which predicts that proteins involved in bud-site selection early in the cell cycle, control cytokinesis at the end of the cycle (Madden and Snyder, 1998).
Together, these features support the concept that the essential role of IQGAP1 is to control cell homeostasis by coupling cell growth and division (Rittmeyer et al., 2008; Wang et al., 2009). It regulates insulin synthesis and secretion (Rittmeyer et al., 2008) and promotes cell size through its N-terminal domain, which binds mTOR (Wang et al., 2009), and it promotes cytokinesis and cell proliferation through its C-terminal domain, which binds and activates CDC42; however, it requires mTOR for this activity (Wang et al., 2009). The mechanism by which IQGAP1 regulates cell proliferation through the shared mTOR subunit remains to be defined. Because IQGAP1, CDC42 and mTORC2 are separately implicated in regulating the actin cytoskeleton it appeared that IQGAP1 would associate with mTORC2. Surprisingly, this appears to be not the case.
Using the conserved roles of yeast and mammalian IQGAPs, we investigated the involvement of IQGAP1 in modulating mTORC1–S6K1→Akt1 signaling to control cell proliferation. Screening for new binding partners of Iqg1p using a two-hybrid assay identified the TORC1-specific subunit Tco89p. Iqg1p and Tco89p bind through their N-terminal domains and share roles in coordinating rapamycin-sensitive cell growth and division. We further tested this in mammalian cells and demonstrated that human IQGAP1 regulates cell proliferation through mTORC1–Akt1 signaling. In both systems an association with TORC2 was not detected, supporting a conserved role of IQGAP1 through TORC1. Genetic and biochemical analyses of the effects of IQGAP1 on mTORC1–Akt1 and ERK1/2–GSK3α/β signaling support the model that IQGAP1 serves as an upstream regulator of the mTORC1/S6K1-dependent inhibitory mechanism that regulates Akt1 S473-P to modulate cell proliferation. Dysfunction of such a mechanism could explain the oncogenicity of IQGAP1 and its association with diverse carcinomas, making it a potential therapeutic target.
Results
Yeast Iqg1p associates with TORC1
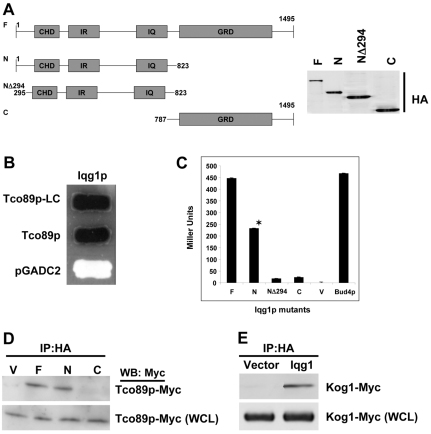
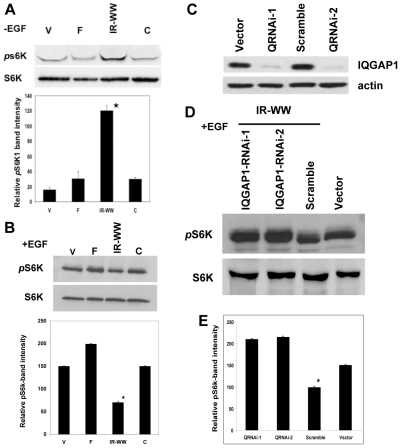
Previously we demonstrated a conserved role for IQGAP1/Iqg1p in coupling cell growth and division and promoting cell proliferation (Osman et al., 2002; Rittmeyer et al., 2008; Wang et al., 2009). To investigate the molecular basis of this role, we used Iqg1p as bait to screen three yeast two-hybrid libraries, and identified four positive library clones (LCs) encoding the first 300 amino acids of S. cerevisiae scYPL180w (Fig. 1A–D) that was identified as the TORC1 subunit Tco89p (Reinke et al., 2004). The specificity of the association was validated by testing the interaction with full-length Tco89p (Fig. 1B) and by mapping the binding region on Iqg1p using several mutants and quantitative two-hybrid assays, compared with the empty vector as control (Fig. 1C). Iqg1-N directly or indirectly mediated Iqg1p–Tco89p binding (Fig. 1C, second bar) and deletion of the 294 amino acids upstream of the CHD of Iqg1p significantly reduced this binding (Fig. 1C, third bar). These findings demonstrate that the interaction between Iqg1p and Tco89 is specific and mediated by the N-terminal region of each protein. Because Bud4p, another marker of axial bud site and cytokinesis, was previously identified from the same libraries as an Iqg1p-binding partner (Osman et al., 2002), we examined whether it would associate with Tco89p by using a two-hybrid quantitative assays. The two proteins strongly interacted (Fig. 1C, last bar).
Fig. 1.
Identification of yeast TORC1 as an Iqg1p partner. (A) Schematic representation of Iqg1p and the mutants used for two-hybrid or immunoprecipitation (IP) analyses: F, full length; N, N-terminus; NΔ294, lacking the N-terminus upstream sequences; CHD, calponin homology domain that binds actin; C, the C-terminus containing the Ras–Gap-related domain (GRD). (B) Tco89-N and full-length association with Iqg1p by two-hybrid plate assay. Yeast cells harboring β-Gal as a reporter gene and expressing HA–pGBDC2–Iqg1p full-length together with either the genomic library clone (LC) encoding the first 300 residues of Tco89p (black), the full-length pGADC2-Tco89p (black) or the empty vector pGADC2 as negative control (white), were patched on nutrient-dropout triple-selection plates containing β-Gal. (C) Mapping the domain of Iqg1p that interacts with Tco89p. Quantitative two-hybrid analysis of Tco89p association with Iqg1p mutants and with Bud4p. The β-Gal activity in Miller units, was calculated from five different clones and plotted as the means ± s.e.m., n=3. *P<0.005 compared with Iqg1-C or vector. (D) Iqg1p and Tco89p interact in vivo. Lysates from cells coexpressing the tagged chromosomal copies Tco89–Myc and Iqg1–HA mutants were immunoprecipitated with HA antibodies and blotted with Myc antibodies; 5% of the whole cell lysate (WCL) was blotted with Myc antibodies as a loading control. (E) Iqg1p co-precipitates with Kog1p, another TORC1-specific subunit. Cells coexpressing tagged chromosomal copies were processed as in D.
Next, we verified the two-hybrid results by in vivo biochemical assays. Tco89–Myc co-precipitated both ways with HA-tagged Iqg1p and Iqg1-N (Fig. 1D; data not shown), confirming the two-hybrid results. A mammalian counterpart of Tco89p has yet to be identified, but because Tco89p binds the TORC1-specific subunit Kog1p, the homologue of mammalian raptor, we were able to demonstrate that Iqg1–HA co-precipitates with Kog1–Myc (Fig. 1E) both ways (not shown), which affirms that Iqg1p interfaces TORC1. Furthermore, co-immunoprecipitation (co-IP) with Tor2 or TORC2-specific subunits was not detected (not shown). These results are the first reported link between TORC1 and Iqg1p, the effector of Cdc42p, a GTPase involved in bud-site selection and polarized growth, and support a potential role for Tco89p in bud-site selection and/or cytokinesis, which we investigated next.
Iqg1p and Tco89p confer rapamycin-sensitive growth and influence axial budding and cytokinesis
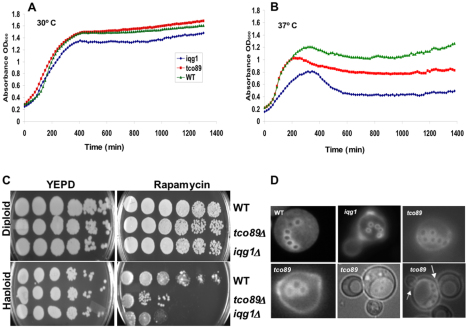
Genetic and cell biological analyses indicate that Tco89p, similar to Iqg1p, has a role in coupling cell growth and division by linking bud-site selection and cytokinesis (Fig. 2). Analysis of 22 tetrads from iqg1Δ tco89Δ heterozygous diploid cells did not yield double mutants (not shown), suggesting synthetic lethality and that the two genes affect the same essential function. Moreover, isogenic strains harboring iqg1Δ or tco89Δ grew at 30°C (Fig. 2A), but they were temperature sensitive at 37°C compared with the isogenic wild type (Fig. 2B), suggesting an effect on cell growth (proliferation). Because tco89Δ causes rapamycin sensitivity in a different genetic background (Reinke et al., 2004), we tested whether Tco89p and Iqg1p confer the same property in our strain background. We extended this analysis further by comparing the effects on diploid and haploid mutant strains growing on rapamycin-containing plates. Surprisingly, rapamycin caused cell cycle arrest in the haploid iqg1Δ and tco89Δ mutants (Fig. 2C, lower right) without affecting their isogenic homozygous diploids or the wild-type strains (Fig. 2C, upper right). Interestingly, the iqg1Δ haploid strain was at least twofold more sensitive to rapamycin than the tco89Δ strain (Fig. 2C, lower right). These findings demonstrate a previously unknown role for Iqg1p in rapamycin-sensitive growth, and uncover a role for TORC1 in haploid yeast cell growth, which we investigated further by testing its effect on bud-site selection, a well analyzed polarized event in the yeast cell cycle.
Fig. 2.
Iqg1p and Tco89p share functions in rapamycin-sensitive growth, axial-bud-site selection and cytokinesis. (A,B) IQG1 or TCO89 null strains are temperature sensitive. Haploid wild-type (WT) and mutant cells were grown in YEPD to OD600 0.2, serially diluted into 96-well plates and the optical density was determined every 10 minutes overnight with a Bioscreen C plate reader (A) at 30°C and (B) at 37°C. (C) Iqg1p confers rapamycin-sensitive growth. Isogenic wild-type, iqg1Δ, tco89Δ, diploid (upper) and haploid (lower) strains were grown in YEPD to mid-log phase (OD600=0.5), washed in sterile double-distilled water, serially diluted and plated on YEPD–agar for control (left) or on YEPD–agar containing 1.0 ng/ml rapamycin (right) and incubated at 30°C for 3 days then photographed with a Bio-Rad XRS imager. (D) Tco89p specifies axial budding. Isogenic haploid wild-type, iqg1Δ and tco89Δ strains were grown on YEPD, washed and stained with Calcofluor (which is a fluorescent dye specific for cell wall and bud-scar chitin), and photographed using a 100× oil immersion lens. The arrows denote additional buds from the same cell, indicating cytokinesis defects.
Haploid yeast cells bud according to an axial budding program and the diploid cells follow a bipolar program. Each program is controlled by a well-defined signaling cascade, sharing at the top the GTPases Bud1p and Cdc42p, which control general budding (Chant and Pringle, 1995; Madden and Snyder, 1998; Osman and Cerione, 2006). Components involved in the axial budding program directly control cytokinesis in the haploid cells and their dysfunction results in both cytokinesis and budding pattern defects (Madden and Snyder, 1998; Osman and Cerione, 2006). A comparison of the budding patterns of the isogenic haploid and homozygous diploid mutant strains with their respective wild types revealed that Tco89p impacts the axial budding program (Fig. 2D). Haploid iqg1Δ and tco89Δ cells, but not their diploid counterparts, both exhibited defects in budding pattern that was predominantly (~70%) semi-random with fewer (~10–20%) bipolar events (Fig. 2D), identical to the phenotype of iqg1Δ cells, reported previously (Osman et al., 2002). The random budding defect phenotype is a known marker of dysfunction of proteins upstream in the general budding control cascades, such as the GTPases Bud1p and Cdc42p (Chant and Pringle, 1995; Osman and Cerione, 2006). Thus this phenotype suggests that Tco89p serves upstream of the axial budding markers with Iqg1p–Cdc42p. Bipolar budding in haploid cells is a known marker of defects in the axial budding program (Chant and Pringle, 1995; Osman et al., 2002; Osman and Cerione, 2006) and supports the upstream role of Tco89p in the axial budding and growth cascade. Furthermore, the tco89Δ strains exhibited pronounced cell wall defects consistent with a previous report in a different strain background (Reinke et al., 2004). This defect in cell wall deposition was manifested as faint chitin staining (Fig. 2D, lower panels) and lyses and rupture in cell culture. These defects were rescued by re-expression of either Iqg1p or Tco89p in their respective mutant (Osman et al., 2002; and data not shown).
Additionally, tco89 mutant strains exhibited cytokinesis defects, evident as more than one bud per cell (Fig. 2D, lower panels), reminiscent of iqg1sec3 double mutants (Osman et al., 2002). The significance of these findings is that the axial bud-site-selection program in yeast links cell growth to cytokinesis directly (Madden and Snyder, 1998; Osman et al., 2002; Osman and Cerione, 2006) and mimics directed cell division in mammals required for pattern formation and cell fate determination, dysfunction of which leads to cancer, diabetes and a myriad of developmental diseases (Gladfelter et al., 2001; Nelson, 2003). Accordingly, these data raised the possibility that mammalian IQGAP1 controls cell proliferation by coupling cell growth and division through the mTOR pathway, a hypothesis that we investigated next.
Mammalian IQGAP1 interfaces mTORC1 and modulates mTORC1→Akt signaling
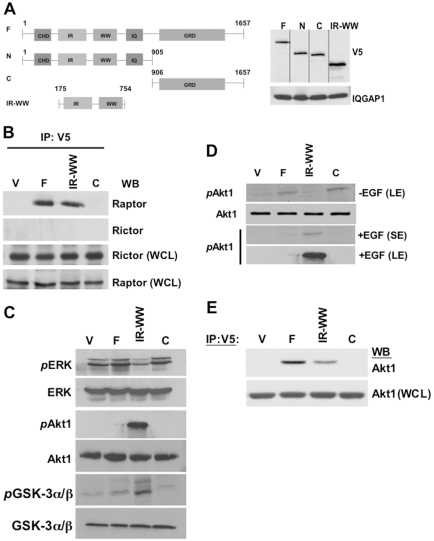
Previous evidence revealed that IQGAP1 regulates cell size and binds, and requires, mTOR for promoting cell proliferation (Wang et al., 2009). However, mTOR is a shared subunit of mTORC1 and mTORC2, and the results obtained above from yeast suggest that the conserved IQGAP1 interfaces mTORC1. Thus we examined the association of IQGAP1 with both complexes (Fig. 3), using the constructs shown in Fig. 3A. Both ways, V5-IQGAP1-F and V5-QGAP1IR-WW but not V5-IQGAP1-C, co-precipitated with Raptor, the mTORC1-specific subunit, but not with Rictor, the mTORC2-specific subunit (Fig. 3B; data not shown). This indicates that IQGAP1 acts through mTORC1 and explains previous reports of its roles in modulating protein synthesis (Rittmeyer et al., 2008) and cell size (Wang et al., 2009). Accordingly, we examined the probable effects of IQGAP1 on mTORC1 signaling.
Fig. 3.
IQGAP1 associates with mTORC1 and modulates mTOR–Akt1 signaling. (A) Schematic representation and expression level of the V5-IQGAP1 and mutants in stable HeLa or NIH3T3 cells (see Rittmeyer et al., 2008; Wang et al., 2009) used for signaling or IP; IR, IQGAP-specific repeats; WW, a tryptophan-rich domain resembling the SH3 protein interacting domains; IQ, 4 IQ motifs that bind calmodulin; GRD, Ras–Gap-related domain containing sequences that bind activated CDC42. The V5-IQGAP1 constructs: Full-length, IQGAP1-F; N-terminal domain, IQGAP1-N; C-terminal domain, IQGAP1-C; IR-WW domain, IQGAP1IR-WW. (B) IQGAP1 associates with Raptor (mTORC1) and not with Rictor (mTORC2). IQGAP1 or mutants were precipitated with V5 antibodies from HeLa cell lysate, separated on SDS-PAGE and blotted with antibodies for Raptor or Rictor. Rictor and Raptor in the whole cell lysate (WCL) were blotted as a loading controls. (C) IQGAP1 modulates mTOR–Akt-signaling. 80 μg of total proteins from HeLa cells stably expressing the indicated IQGAP1 constructs, serum-starved and treated with EGF, were blotted with total or phospho-specific antibodies for ERK1/244/42 [Thr202 and Tyr204 of Erk1 (Thr185 and Tyr187 of Erk2], Akt1 Ser473 or GSK3 Ser21/9. (D) Signal-dependent differential activation of Akt1 by IQGAP1. Cells were serum-starved and either left untreated or treated with EGF and blots obtained as in C. Blots were processed at both 30 seconds (SE; short exposure) and 2 minutes (LE, long exposure). The −EGF (control) and +EGF (from C) blots were processed on the same gel. (E) Akt1 co-precipitates with IQGAP1. IQGAP1 and mutants were precipitated with V5 antibodies and blotted with Akt1 antibodies. Akt in the cell lysates (WCL) used for IP was used as a loading control. The results are representative of different stable cell types: NIH3T3, β-cell lines, MDCK or kidney.
Both IQGAP1 and CDC42 are activated downstream of the epidermal growth factor receptor (EGFR) (Tu et al., 2003; Rittmeyer et al., 2008). EGF induces phosphorylation of serine in IQGAP1 (IQGAP Ser-P) leading to binding and activation of CDC42 (Rittmeyer et al., 2008; Wang et al., 2009). Binding of active CDC42–GTP to IQGAP1 inhibits insulin secretion (Rittmeyer et al., 2008) and reduces the cell size, but enhances IQGAP1 S1443-P-dependent cell proliferation (Wang et al., 2009). Thus we examined EGF-mediated effects of IQGAP1 on mTOR signaling. HeLa cells stably expressing wild-type or mutant V5-IQGAP1 (Fig. 3A) as reported previously (Rittmeyer et al., 2008; Wang et al., 2009) showed a fast response to EGF that was maximal at 5 minutes, and remained constant for 15 or 30 minutes (not shown); therefore treatment for 5 minutes was selected for measuring signaling. Interestingly, phosphorylation of the MAPK extra cellular signal-regulated kinase ERK1/2 (also known as MAPK3/1 and p44/42) was attenuated in IQGAP1IR-WW cells (Fig. 3C, upper two panels) whereas phosphorylation of the serine/threonine protein kinase Akt1 at Ser473 (S473-P; middle two panels), and its substrate the glycogen-synthase kinase-3 (GSK3α/β) at S21 and S9, respectively (bottom two panels) were augmented. These results demonstrate that expression of IQGAP1IR-WW downregulates ERK1/2 and GSK3α/β signaling and augments Akt1 signaling in response to EGF and they are consistent with the known phosphorylated Akt1 inactivation of GSK3, whose activity alters glucose and fat metabolism and induces cell apoptosis (Jope and Johnson, 2004; Manning, 2004).
Cells that were not treated with EGF as a control had no detectable level of phosphorylation of any of the tested kinases except for Akt1 S473-P, which was slightly increased in IQGAP1-F and -C cells (−EGF; Fig. 3D). This finding explains why these cells were previously shown to induce transformed phenotypes that were sensitive to rapamycin (Wang et al., 2009) and to the PI3K and Akt1 inhibitor LY294002 (Fig. 5) irrespective of EGF. This confirms that IQGAP1-F or IQGAP1-C expression bypasses EGF stimulation and activates Akt1.
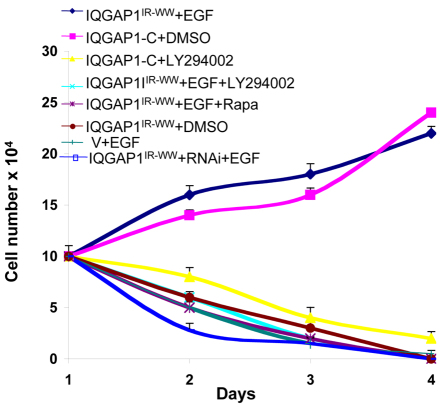
Fig. 5.
IQGAP1 induces an EGF-stimulated transformed phenotype. (A) IQGAP1IR-WW induces EGF-stimulated and mTORC1–PI3K–Akt1-dependent cell proliferation. NIH3T3 cells stably expressing IQGAP1IR-WW, IQGAP1-C (positive control), vector (V; as negative control) or IQGAP1 siRNA, were serum-starved then left untreated or treated with EGF, DMSO (as drug-vehicle control), 10 μM LY294002 (PI3K and Akt1 inhibitor) or 100 nM rapamycin (mTOR inhibitor), and evaluated by growth in low-serum medium. Cell number was determined in triplicate every other day for 6 days and is presented as the means ± s.e.m., n=3. EGF significantly (P<0.001) induced the transformed phenotype, and the drug inhibitors significantly (P<0.001) reversed this phenotype in treated versus untreated IQGAP1IR-WW cells.
Because mTOR binds IQGAP1 (Wang et al., 2009) and Akt1 is an mTOR substrate (Sarbassov et al., 2005), these data raised the possibility of an association between IQGAP1 and Akt1, which we detected by co-immunoprecipitation (Fig. 3E), indicating that they directly or indirectly form a complex in vivo. Although these data explain how phosphorylated IQGAP1 induces cell proliferation through Akt1, they do not explain how it augments Akt1 S473-P (Fig. 3) and reduces the cell size (Wang et al., 2009). To investigate the molecular basis of these observations, we examined mTOR downstream signaling.
IQGAP1 modulates S6K1 activity
As mentioned earlier, it is well established that Akt1 S473-P activates mTORC1 leading to S6K1 activation by phosphorylation on Thr389. Subsequently, S6K1 T389-P suppresses Akt1 S473-P activity to control the cell size, thus the level of S6K1 T389-P is widely used as a marker of mTORC1 activity and cell growth. Evaluation of the S6K1 T389-P level in IQGAP1 mutant cell lines revealed that under nutrient conditions (full serum), S6K1 T389-P activity increased in IQGAP1IR-WW cells (Fig. 4A) and decreased (30–50%, P>0.001) when EGF was applied (Fig. 4B, third lane; lower Fig. 4B, third bar). This attenuation of S6K1 T389-P was reproducible and specific to IQGAP1 because knockdown of endogenous IQGAP1 by RNAi in the IQGAP1IR-WW cells (Fig. 4C) restored the S6K1 T389-P level (Fig. 4D, first two lanes; Fig. 4E, first two bars). Notably, EGF increased (P<0.05) the S6K1 T389-P level in cells expressing full-length IQGAP-F compared with control cells expressing the vector (V; Fig. 4B). These data explain our previous finding that expression of IQGAP1IR-WW enhanced nutrient-induced protein synthesis (Rittmeyer et al., 2008) and cell size (Wang et al., 2009) by binding to mTORC1 (Fig. 3) and increasing S6K1 T389-P. They also demonstrate that IQGAP1IR-WW mediates EGF-responsive augmentation of Akt1 S473-P by attenuating S6K1 T389-P, thus suppressing its known inhibitory effects on Akt1 S473-P. Collectively, our data support the concept that although IQGAP1-C serves as a dominant-active (DA) gain-of-function mutant in cell proliferation, and IQGAP1IR-WW serves as a dominant-negative (DN) mutant (Rittmeyer et al., 2008; Wang et al., 2009), it can serve as a DA mutant in response to EGF. If so, then expression of IQGAP1IR-WW, which arrests cytokinesis in the presence of nutrients (Wang et al., 2009), would induce an EGF-mediated cell proliferation, a hypothesis that we tested next.
Fig. 4.
IQGAP1 modulates S6K1 T389 activity. (A) IQGAP1 stimulates S6K T389-P (pS6KT389) activity in high-serum (-EGF). Upper panel: a representative blot. Lysates from HeLa cells expressing the V5-IQGAP1 constructs and growing in full serum were blotted with total and phosphorylated Thr389 S6K1 antibodies. Lower panel: bands intensities were quantified by densitometry, normalized against total S6K and presented as the means ± s.e.m., n=3 independent experiments. *Significantly (P<0.001) lower than IQGAP1-F cells. (B) IQGAP1 attenuates S6K T389-P activity in response to EGF (+EGF): stable HeLa cell were serum-starved and treated with EGF and equal amount of lysates were processed and the band intensities quantified as in A. (C) Lysates from cells expressing two IQGAP1 siRNAs, which down regulate IQGAP1 by >90%. (D) Knockdown of endogenous IQGAP1 in cells expressing IQGAP1IR-WW and treated with EGF, restores the S6K T389-P level. Control and stable IQGAP1IR-WW HeLa cells expressing the IQGAP1 siRNAs were serum-starved and treated with EGF and their lysates blotted with antibodies for total and phosphorylated Thr389 S6K1. (E) Quantification by densitometry of the bands in D, presented as the means ± s.e.m., n=3 independent experiments. *Significantly (P<0.001) lower than the siRNA-treated cells. The results are representative of several cell lines including pancreatic β-cell and kidney cell lines.
IQGAP1 induces EGF-stimulated Akt-mediated proliferation and localizes to the midbody during cell abscission
We demonstrated the capacity of IQGAP1IR-WW to induce EGF-stimulated and Akt1–mTORC1-dependent transformed phenotypes, using RNAi, pharmacology and growth in low serum (Fig. 5). The IQGAP1IR-WW-mediated, EGF-stimulated transformed phenotype in fibroblasts relied on mTORC1–Akt1, because it was abolished by applying rapamycin or the PI3K and Akt1 inhibitor LY294002. It was specific to IQGAP1 because it was abolished by knockdown of IQGAP1, indicating requirement of the endogenous protein (Fig. 5), consistent with the behavior of these dominant mutants (Rittmeyer et al., 2008; Wang et al, 2009). Similarly, the positive control transformed IQGAP1-C cells, which are rapamycin sensitive (Wang et al., 2009) and contain EGF-independent Akt1 S473-P (Fig. 3D), are sensitive to LY294002, whereas IQGAP1IR-WW cells that were not treated with EGF (as a negative control) whether they express IQGAP1 siRNA or not, failed to proliferate in low serum (Fig. 5), as previously reported (Wang et al., 2009). Collectively, these results demonstrate that IQGAP1 differentially responds to nutrient and mitogenic signals and accordingly modulates ERK1/2–GSK3α/β and mTORC1–S6k1→Akt1 signaling to regulate cell proliferation by coupling cell growth and division, thus raising the possibility that IQGAP1 differentially localizes during the cell cycle.
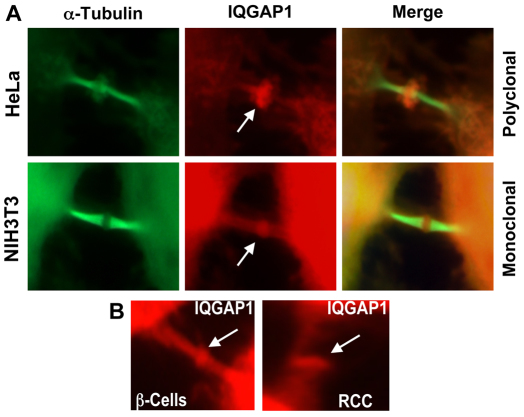
We tested this possibility by determining IQGAP1 localization during the cell cycle in synchronized HeLa and NIH3T3 cells, using α-tubulin as a cytokinesis marker. IQGAP1 and α-tubulin distributed diffusely in the cytoplasm throughout the cell cycle then colocalized on the midzone mitotic spindles (supplementary material Fig. S1) during telophase, as revealed previously by proteomics (Skop et al., 2004; Morita et al., 2007). However, in late cytokinesis, during cell abscission, α-tubulin remained on the mitotic spindles whereas IQGAP1 mostly concentrated in the middle of the midbody (Fig. 6, arrows; supplementary material Fig. S2), resembling the centrosome localization (Gromley et al., 2005). This localization pattern was consistent in several cell lines that we screened with IQGAP1 antibodies, including pancreatic β-cells and renal cell carcinoma (RCC; Fig. 6B). This finding implies that IQGAP1 has a role in midbody abscission, which is the distinct final step in animal cytokinesis (Gromley et al., 2005). This result explains the previous finding that although IQGAP1-F and IQGAP1-C served as DA mutants and accelerated the cell cycle and induced transformed phenotypes, IQGAP1IR-WW acted as a DN mutant and arrested cytokinesis (Wang et al., 2009). Together, these findings support an additional potential role for IQGAP1 in cell abscission downstream of the mTORC1–S6K–Akt1 pathway.
Fig. 6.
IQGAP1 localizes in the midbody during cell abscission. (A) Synchronously growing cells were fixed every 15 minutes for 16 hours and stained with the indicated rabbit polyclonal or rabbit monoclonal IQGAP1 antibodies (red, arrows) and FITC–α-tubulin (green), and photographed under a confocal microscope. Enlarged midbody regions are shown. For full views of representative cells and throughout the cell cycle see supplementary material Figs S1 and S2. (B) Different cell lines were screened for midbody localization with antibodies for IQGAP1. Two examples: the RCC cell line (SW839) and β-cells (βTC-6 insulin-secreting insulinoma) are shown.
IQGAP1 activity is deregulated in carcinoma cell lines
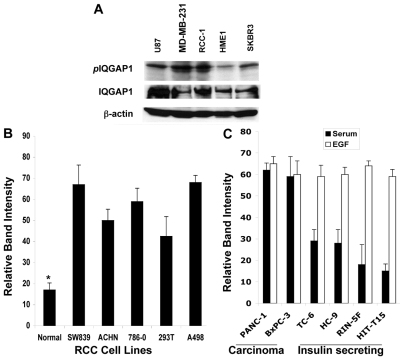
EGF or overexpression of IQGAP1 activates both IQGAP1 and CDC42, resulting in their binding and induction of cell proliferation (Rittmeyer et al., 2008; Wang et al., 2009). Thus we hypothesized that unregulated phosphorylated IQGAP1 signaling resulting from overexpression (Rittmeyer et al., 2008) but not the mere overexpression, underlies tumorigenesis. To begin testing this hypothesis we evaluated the level of IQGAP1 Ser-P in a variety of carcinoma cell lines, including brain, breast RCC and pancreatic carcinomas. We detected a significant increase in the level of IQGAP1 Ser-P in carcinomas compared with normal human mammary epithelia (HME1; Fig. 7A) and normal kidney epithelia (Fig. 7B). Interestingly, when comparing pancreatic β-cell lines, we found higher levels of IQGAP1 Ser-P in carcinoma than in insulin-secreting insulinoma cell lines (compare black bars). Furthermore, EGF treatment evoked higher levels of IQGAP1 Ser-P in the insulin-secreting cells but not in the pancreatic carcinoma cell lines (compare white bars), perhaps because the pancreatic carcinomas had maximal starting levels. These findings substantiate our hypothesis and will be tested by further studies. The collective data discussed in this study support the model that IQGAP1, regulated by phosphorylation in response to nutrient or mitogenic signals, has a role in modulating mTORC1–S6K→Akt1 NFL and serves downstream of it to control cell homeostasis. Dysfunction of this process can underlie tumorigenesis caused by IQGAP1 and explain its association with human carcinomas (Fig. 8).
Fig. 7.
Increased level of IQGAP1 Ser-P in carcinoma cell lines. (A) 80 μg of total proteins from glioblastoma (U87), breast carcinoma (MD-MB-231 and SKBR3) and renal cell carcinomas (RCC-1) and normal (as control) human mammary epithelia HME1, cell lines were immunoprecipitated with antibodies for IQGAP1 and blotted with PKC substrate phosphorylated-Ser-specific antibodies (upper lanes). Equal amounts of cell lysate were blotted with antibodies for IQGAP1 (middle panel) or actin, as loading control. (B) The IQGAP1 Ser-P level is elevated in several clear cell renal cell carcinoma (RCC) cell lines. Blots were obtained as described in A and the level of IQGAP1 Ser-P bands was quantified by band densitometry, normalized against total IQGAP1 and expressed as the means ± s.e.m., n=3. *IQGAP1 Ser-P in normal epithelia is significantly (P<0.05) lower than any of the RCC cell lines. (C) Effects of EGF on IQGAP1 Ser-P levels in differentiated and undifferentiated carcinoma cell lines. EGF evoked higher IQGAP1 Ser-P levels in insulin-secreting insulinoma cell lines compared with undifferentiated pancreatic carcinomas, PANC-1, which already contain high levels in serum alone. Cells were starved for 12 hours and were either left untreated (black bars) or treated (white bars) with EGF and equal amount of lysates immunoprecipitated with IQGAP1 antibodies and processed as in A.
Fig. 8.
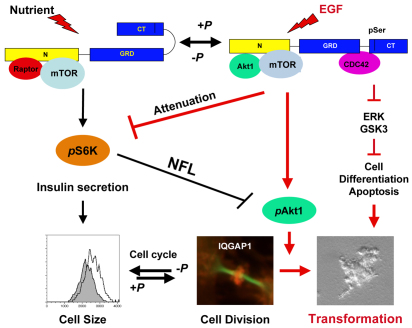
IQGAP1 regulates cell homeostasis through mTORC1. A model depicting the action of IQGAP1 in coupling cell growth and division through a previously unidentifed CDC42-mTORC1–S6K1–Akt1 pathway in response to nutrient or mitogenic signal. Black arrows and lines denote previously identified regulation of cell size by IQGAP1 and a known mTORC1–S6K1 negative feedback loop (NFL), and red arrows and lines denote new finings in this study. In closed conformation, IQGAP1 enhances insulin secretion, protein synthesis and cell growth (Rittmeyer et al., 2008; Wang et al., 2009) through mTORC1–S6K1. In open conformation IQGAP1-P promotes cell division and proliferation (Wang et al., 2009) through IQGAP1 S1443-P↑, mTORC1 and S6K1↓, and Akt1 S473-P↑ and suppresses differentiation and/or apoptosis by attenuating ERK1/2 and GSK3α/β. IQGAP1 attenuates S6K1 T389-P activity to enhance Akt1 S473-P activity, which in turn activates mitosis where IQGAP1 has a role in cell abscission. Thus IQGAP1 may act like a rheostat to modulate the NFL of S6K1 T389-P→Akt1 S473-P and maintain cell homeostasis, dysregulation of which could underlie tumorigenesis and/or aberrant insulin signaling in diabetes.
Discussion
This study presents a number of findings relevant to the central question of how cells coordinate their growth and division to maintain homeostasis, which is crucial to understanding the mechanism of tumorigenesis as well as diabetes in humans. It demonstrates for the first time, involvement of yeast TORC1 in bud-site selection and cytokinesis in baker's yeast through the actin cytoskeleton regulators Iqg1p and Cdc42p. Thus, despite the prevailing model, which assigns cytoskeleton regulation to TORC2, our results indicate that TORC1 also impacts the actin cytoskeleton, an idea that requires further investigation. Moreover, this study reveals for the first time, a role for yeast and mammalian IQGAPs in rapamycin-sensitive growth. Interestingly, yeast Iqg1p was more sensitive to temperature and rapamycin-mediated growth inhibition than the TORC1 subunit Tco89p (Fig. 2B,C), which could potentially be used to predict rapamycin sensitivity in human cancers. Together these findings support the concept that IQGAP1 serves as a TORC1-scaffolding regulator. Furthermore, they suggest a potentially conserved role for IQGAP1 in coupling cell growth and division through a potentially conserved CDC42–IQGAP1–TORC1–S6K1–Akt1 pathway. Because we did not detect an interaction with mTORC2, our findings raise the possibility that mTOR and IQGAP1 form a distinct mTOR sub-complex that impacts Akt1 signal specificity towards mitosis and cell proliferation; experiments are underway to test this hypothesis.
Previously we reported that yeast Iqg1p specifies axial bud site and links budding to cytokinesis (Osman et al., 2002; Osman and Cerione, 2006). In yeast, budding and cytokinesis both require directed deposition of new cell wall material to the growing bud site as well as to the cytokinesis plate to bisect the daughter cells (Chuang and Schekman, 1996; Hales et al., 1999; Wolfe and Gould, 2005; Zhang et al., 2006; Osman and Cerione, 2006). Cell wall synthesis is part of the cellular growth programs and is co-regulated by the action of positional cues with the cell polarity machinery and the secretory pathway (Lesage and Bussey, 2006) and requires coordination with the Tor pathway (Fuchs and Mylonakis, 2009). However, the molecular basis of the proposed crosstalk between Tor and the polarity machinery has not been determined (Fuchs and Mylonakis, 2009). Our finding that Iqg1p, the effector of Cdc42p GTPase, implicated as a positional marker in cell polarity, actin cytoskeleton and cytokinesis (Ziman et al., 1991; Osman and Cerione, 1998; Johnson, 1999; Osman et al., 2002), interacts with TORC1 subunit Tco89p (Fig. 1), which is implicated in cell wall synthesis (Loewith et al., 2002; Reinke et al., 2004), represents the first molecular evidence for such crosstalk. Moreover, our results indicate, for the first time, that TORC1 is involved in budding yeast cytokinesis, which appears to be a conserved function, as recent evidence has also implicated TORC1 in fission yeast cytokinesis (Hartmuth and Petersen, 2009). However, a prevailing concept in the field is that TORC1 controls cell growth whereas TORC2 controls the actin cytoskeleton, our results now invite a revision of this model.
For relevance to human health, we examined dominant mutants of IQGAP1 and translated our findings into mammalian cells, as they permit study of growth factors signals. Biochemical and yeast two-hybrid assays suggest that IQGAP1 undergoes intramolecular interaction where the C-terminus folds on itself and the N-terminus folds on the C-terminus (Grohmanova et al., 2004; Rittmeyer et al., 2008). These findings led to the adoption of autoinhibition-folding as a mechanism, which has general support from structural evidence from similar proteins (Agazadeh et al., 2000). On this basis, we demonstrated previously that IQGAP1IR-WW is DA in secretion and its effect on cell size and is DN in cell proliferation and migration, with the converse being true for IQGAP1-C, which binds and activates CDC42 (Rittmeyer et al., 2008; Wang et al., 2009). However, these two activities are interdependent because the phenotype of these mutants requires the endogenous protein as substantiated by RNAi (Rittmeyer et al., 2008; Wang et al., 2009). Although this finding supports the autoinhibition-folding model, whereby these mutants probably heterodimerize with the endogenous protein to cause dominant effects revealing cryptic functions of IQGAP1, it also supports the concept that cell growth is a prerequisite to cell division (Tapon et al., 2001); explaining why the endogenous protein is required. Overall, we propose that these features underlie the mechanism of actions of these mutants reported in this study.
On the basis of the above concept, we propose a potential mechanism by which IQGAP1 inhibits mTORC1 to augment Akt1 signaling and promote cell proliferation and survival, while attenuating ERK1/2–GSK3α/β signaling (Figs 3, 4 and 5) implicated in cell differentiation and apoptosis. This finding is consistent with the opposing signals of ERK1/2–GSK3α/β and Akt1 (Ding et al., 2005). GSK3α/β regulates apoptosis, lipid and fat metabolism and is a crucial PI3K–Akt1 substrate where active Akt1 S473-P inhibits GSK3 by phosphorylating Ser21 on GSK3α and Ser9 on GSK3β (Jope and Johnson, 2004; Manning, 2004). Furthermore, the finding that IQGAP1IR-WW attenuates ERK1/2 agrees with and further extends with new insights, previous reports that IQGAP1 modulates ERK2 (Roy et al., 2004) and serves as a scaffold for Raf-MEK1/2–ERK1/2 and Akt1 and controls stress-mediated cardiomyocyte apoptotic death (Sbroggiò et al., 2011). ERK1 and 2 are effectors of Ras, Raf and MEK1, and they regulate cell differentiation (Minden and Karin, 1997), therefore their attenuated activity is consistent with the increased Akt1 S473-P signal, which promotes cell survival and mitosis, eliciting transformed phenotypes (Figs 3, 5). Thus, our results demonstrate, for the first time, the capacity of IQGAP1 to modulate ERK1/2 and Akt1 signaling to promote cell proliferation and potentially suppress apoptosis; a concept that is currently under investigation.
Our data reveal, for the first time, a potential mechanism by which IQGAP1 augments Akt1 S473-P and promotes cell proliferation by inhibiting mTORC1–S6K1 signaling. IQGAP1 mediated an EGF-stimulated attenuation of S6K1 activity thereby directly or indirectly activating Akt1 S473 (Figs 3, 4); however, binding to Akt1 (Fig. 3E) suggests a direct role, which requires further investigation. It has been demonstrated that mTORC2 (mTOR-Rictor) activates Akt1 by phosphorylation on Ser473, subsequently, S6K1 T389-P phosphorylates Rictor on T1135 (Dibble et al., 2009; Julien et al., 2010) to suppresses mTORC2 and inhibit Akt1 S473 activity through a negative feedback loop. Expression of the mutant Rictor T1135A resistant to phosphorylation by S6K1T389-P increases Akt1 S473-P activity and enhances mitosis by activating FoxO proteins in response to EGF (Julien et al., 2010). This finding agrees with our results that attenuated S6K1 T389-P (Fig. 4) led to enhanced Akt1 S473-P activity (Fig. 3C) and increased EGF-stimulated cell proliferation (Fig. 5). Moreover, several lines of evidence support the proposal that inhibition of mTORC1 advances mitosis (Hartmuth and Petersen, 2009; Gwinn et al., 2010; Julien et al., 2010) through Akt1 and its downstream effectors, the forkhead (FKH-Tfs) FoxO transcription factors; however, regulation of this signaling axis remains incompletely defined (Alvarez et al., 2001; Shiota et al., 2006; Frias et al, 2006; Jacinto et al., 2006; Dibble et al., 2009; Julien et al., 2010). Our results present a physiologically plausible mechanism whereby IQGAP1 inhibits mTORC1–S6K1 and prevents its inhibitory feedback mechanism on Akt1 S473-P, thus advancing mitosis and cell proliferation. The role of IQGAP1 in cytokinesis lends additional credence to this hypothesis.
As in yeast, animal cytokinesis requires membrane expansion and directed secretion in the cleavage furrow to bisect the daughter cells (Gromley et al., 2005). Our data support a role for IQGAP1 downstream of mTORC1–Akt1 in promoting mitosis and executing cytokinesis, which is consistent with our previous observation that IQGAP1-F and IQGAP1-C were DA and accelerated the cell cycle, whereas IQGAP1IR-WW was DN and arrested cytokinesis (Wang et al., 2009). The spatial distribution of IQGAP1 as a ring in the midbody during cytokinesis (Fig. 6) implies a direct role in cell abscission (Fig. 8). In support of this concept, IQGAP1 was identified as a partner of midbody proteins, both by proteomics and two-hybrid assay (Skop et al., 2004; Morita et al., 2007). This localization pattern is identical to that of the centrosome and Exo70, an IQGAP1-binding partner in secretion (Rittmeyer et al., 2008), found to cooperate in midbody abscission by promoting asymmetric secretion (Gromley et al., 2005). Thus, IQGAP1 midbody localization, binding to the exocyst, regulation of exocytosis (Rittmeyer et al., 2008), acceleration of the cell cycle and promotion of cell proliferation (Fig. 5) (Wang et al., 2009), predict a key role in mammals in bisecting the daughter cells, which is currently being characterized. This is the first report of localization of a member of the IQGAP family to the cleavage furrow and may not apply to all three members of the family, which may regulate cytokinesis by different mechanisms, as yet, to be determined. Collectively, our data suggest that IQGAP1 has a conserved role in cell homeostasis by coupling cell growth and division, serving both upstream and downstream of mTORC1–S6k1–Akt1 NFL (Fig. 8).
This mechanism would also explain the role of IQGAP1 in tumorigenesis. Several studies reported that IQGAP1 expression or mislocalization is associated with a diversity of human carcinomas (reviewed by Johnson et al., 2009). However, our data suggest that it is not mere overexpression, but unregulated IQGAP1 signaling that leads to tumorigenesis (Figs 7, 8), marked by increased IQGAP1 S1443-P and possibly aberrant subcellular redistribution (Figs 6, 7 and 8; supplementary material Fig. S1). Thus IQGAP1 could serve as a marker in tumors and as a potential therapeutic target. Because of the roles of IQGAP1 and mTOR in insulin secretion, it is appealing to suggest that the same mechanism applies to controlling insulin homeostasis and that its deregulation underlies diabetes.
Materials and Methods
Yeast cell culture, genetic manipulations and two-hybrid analyses
Yeast strains, culture and two-hybrid screens and analyses were used as described previously (Osman et al., 2002). Briefly, IQG1 was cloned in frame into HA-pGBD-C2 vector, verified by sequencing and western blotting of the right sized HA-tagged protein with HA antibody and used as bait for screening the three genomic libraries, YL2H-C1 to 3 in the yeast strain PJ694A (James et al., 1996) as described previously (Osman et al., 2002). The putative interacting library clones (LCs) were identified by sequencing and the relevant gene was cloned by high fidelity PCR into the two hybrid vectors, verified by sequencing, and the specificity of the interaction retested by qualitative (plate) and quantitative (liquid) two-hybrid assays. The chromosomal copy of the genes was epitope-tagged and the interaction confirmed by immunoprecipitation. Gene cloning, C-terminal tagging or deletions were performed as described previously (Osman et al., 2002) using the pFA6a plasmid cassettes and procedures of Longtine et al. (Longtine et al., 1998) and Baudin et al. (Baudin et al., 1993). For quantitative two-hybrid analyses, β-galactosidase activity (Miller units) was calculated from at least five independent clones as previously described (Osman et al., 2002). Growth curves were determined by using a Bioscreen C Machine (Growth Curves, Piscataway, NJ). The cells were grown to OD600 0.2, serially diluted with yeast extract peptone dextrose (YEPD) in 96-well sterile plates in duplicate and incubated overnight (16 hours) with shaking at 30 or 37°C. The optical densities were automatically determined every 10 minutes and the average of two independent experiments was used to generate the growth curves with the algorisms in Microsoft Excel software.
Mammalian cell culture
All cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% calf serum (NIH3T3) or 10% fetal bovine serum (FBS) and 100 units/ml penicillin, 100 μg/ml streptomycin (Invitrogen) in a humidified 5% CO2 incubator at 37°C. HeLa cells (obtained from ATCC, Manassas, VA) were cultured in MEM plus FBS under the same conditions. NIH3T3 and HeLa cell lines stably expressing the V5-IQGAP1 constructs were described previously (Rittmeyer et al., 2008; Wang et al., 2009). Glioblastoma U87, breast carcinomas, kidney RCC and normal HME1 and normal kidney cell lines as well as the human pancreatic carcinoma PANC-1, the insulin-secreting insulinomas βHC-9 and βTC-6 and the insulin-secreting RIN-5F and HIT-T15 cell lines were obtained from ATCC.
Cell synchronization
Mammalian cells were cultured under the conditions described above and synchronized at the G1–S transition with a double aphidicolin block (1.25 μg/ml) in growth medium overnight, followed by release for 9 hours and treatment again overnight, as described previously (Surka et al., 2002). Sample cells growing on coverslips were fixed every 15 minutes over a 16-hour period and used for immunofluorescence.
Fluorescence microscopy
Calcofluor staining of chitin in cell walls and bud scars in yeast was performed as described previously (Osman et al., 2002) following the method of Pringle (Pringle, 1991). Briefly, cells at log phase were collected and stained with Calcofluor (Fluorescent Brightener; Sigma-Aldrich) and cells with more than three bud scars were scored for budding pattern. Using a 100× oil immersion lens on an Olympus fluorescence microscope fitted with a Hamamatsu ORCAER monochrome CCD camera. Synchronized cells were fixed as described previously (Rittmeyer et al., 2008; Wang et al., 2009) and stained with IQGAP1 primary antibodies [the polyclonal was from Santa Cruz (H-109) or Abcam (ab86064) and the monoclonal was from Epitomocs (RabMAb cat. no. 3548-1) or Novus Biologicals (2C5)] followed by secondary antibodies (Texas Red and Alexa Fluor 488; Molecular Probes) and α-tubulin–FITC monoclonal antibody (Sigma) for 1 hour each, at room temperature. The nuclei were stained with DAPI (Sigma) and the images were captured with a Leica confocal microscope.
Immunoprecipitation (IP) and western blotting
The stable cells were washed with ice-cold phosphate-buffered saline (PBS), scraped into 500 μl lysis buffer (25 mM HEPES, pH 7.4, 15 mM MgCl2, 150 mM NaCl, 1% NP40, 10 μg/ml each of leupeptin and aprotinin and 0.2 mg/ml phenylmethylsulfonic chloride) and incubated on ice prior to centrifugation at 13,000 g for 30 minutes. Yeast cells were disrupted with acid-washed glass beads and the lysate collected by centrifugation. As controls, lysates were incubated with beads alone or with an unrelated antibody. Protein concentrations were determined with the Bio-Rad Dc kit and equal amounts were precleared with beads (15 μl) for 1 hour at 4°C and used for IP. Briefly, the antibody was added to the precleared lysate and incubated on ice for 1 hour and 40 μl of PBS-equilibrated protein-A or -G beads were added and gently rocked for 3 hours, or overnight, at 4°C. The beads were washed 4× with 1 ml lysis buffer, boiled for 10 minutes in 40 μl 2× SDS sample buffer and resolved with SDS-PAGE. For IP with TOR, the detergent in the buffer was replaced with 3% CHAPS to preserve the TOR complexes (Sorbassov et al., 2005). Immunoblotting was performed with the antibodies indicated in the figures. IQGAP1 and yeast Tor1 and Tor2 antibodies were from Santa Cruz and the antibodies for mTOR, S6K, PCKC substrate pan Ser-P and Akt1 were from Cell Signaling Technology. The antibodies were used in combination with Signal Enhancer Hikari (Nacalai USA, San Diego, CA). Band signal was detected and quantified as below.
Detection of kinase activity
Confluent cells stably expressing the IQGAP1 constructs and vector control (neomycin-resistant cells) were serum-starved for 12 hours and stimulated with epidermal growth factor (EGF; 100 ng/ml) in serum-free medium for 5 minutes or were left untreated for the control. After washing with ice-cold PBS, the cells were lysed and ~80 μg proteins were resolved in a 7–20% gradient SDS-PAGE, transferred to PVD membranes and immunoblotted with total and phospho-specific (active) antibodies (Cell Signaling Technology) for ERK (p44/42), Akt1 (Ser473), GSK3α/β (Ser21/Ser9) and S6K (Thr389). The signal was developed using Enhanced Chemiluminescence (Amersham) and band intensities detected and quantified from three experiments using ChemiDocXRS and Quantity One, 4.4.1 (Bio-Rad). Statistical analysis was performed using the algorithms in the Microsoft Excel software.
Transformation assays
Ability to proliferate in low serum is a phenotype acquired by transformed cells. A transformation assay in low serum was used to compare NIH3T3 stable cells with neomycin-resistant control cells expressing the empty vector, as described previously (Wang et al., 2009). Briefly, to measure serum-independent growth of the transformed IQGAP1-C and the EGF-treated IQGAP1IR-WW cells, confluent cells were trypsinized and seeded in triplicate at 10×104 in 12-well plates in DMEM plus 10% calf serum and incubated for 5 hours to allow cell adherence. Thereafter, the cells were washed with low-serum DMEM containing 1% CS and grown in the same medium that was changed every other day for 6 days. At 2, 4 and 6 days, the cells were washed thoroughly with PBS, trypsinized and counted with a hemocytometer. To measure EGF-induced transformation, 100 ng/ml EGF was added to the growth medium. Experiments were performed in triplicate from three clones with similar expression levels of IQGAP1 constructs. Data are presented as the means of three independent experiments and statistical analyses were performed using the algorithms in the Microsoft Excel software.
RNA interference
RNAi was performed as described previously (Rittmeyer et al., 2008; Wang et al., 2009). Two human IQGAP1 siRNAs, scramble and the siCONTROL oligomers that do not target IQGAP1-C or other IQGAP family members as demonstrated previously (Rittmeyer et al., 2008; Wang et al., 2009), were obtained from Dharmacon.
Supplementary Material
Acknowledgements
We thank Richard A. Cerione (Cornell University, Ithaca, NY) for his help, John Helmann (Cornell University) for the Bioscreen machine, Ted Powers (UC, Davis, CA) for reagents, Kun-Liang Guan (UC, San Diego, CA) and Brendan Manning (Harvard University, Cambridge, MA) for insightful discussion and suggestions. Stacey Tiam Fook (currently at Novartis-North Western, Boston, MA) and Melissa Williams (currently at NIH, Washington DC) both contributed to the initial yeast studies.
Footnotes
Funding
This work was supported by the National Cancer Institute [grant number CA104285 to M.O.]. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.098947/-/DC1
References
- Aghazadeh B., Lowry W. E., Xin Y. H., Rosen M. K. (2000). Structural basis for relief of autoinhibition of the Dbl homology domain of proto-oncogene Vav by tyrosine phosphorylation. Cell 102, 625-633 [DOI] [PubMed] [Google Scholar]
- Alvarez B. C., Martinez B., Burgering M., Carrera A. C. (2001). Forkhead transcription factors contribute to execution of the mitotic programme in mammals. Nature 413, 744-747 [DOI] [PubMed] [Google Scholar]
- Baudin A., Ozier-Kalogeropoulos O., Denouel A., Lacroute F., Cullin C. (1993). A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21, 3329-3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensenor L., Kan H.-M., Wang N., Wallrabe H., Davidson L. A., Cai Y., Schafer D. A., Bloom G. S. (2007). IQGAP1 regulates cell motility by linking growth factor signaling to actin assembly. J. Cell Sci. 120, 658-669 [DOI] [PubMed] [Google Scholar]
- Brandt D. T., Grosse R. (2007). Get to grips: steering local actin dynamics with IQGAPs. EMBO Rep. 8, 1019-1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chant J., Pringle J. R. (1995). Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. J. Cell Biol. 129, 751-765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Zhu H. H., Zhou L. F., Wu S. S., Wang J., Chen Z. (2010). IQGAP1 is overexpressed in hepatocellular carcinoma and promotes cell proliferation by Akt activation. Exp. Mol. Med. 42, 477-483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang J. S., Schekman R. W. (1996). Differential trafficking and timed localization of two chitin synthase proteins, Chs2p and Chs3p. J. Cell Biol. 135, 597-610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett M., Xiong Y., Boyne J. R., Wright D. J., Munro E., Price C. (2006). IQGAP and mitotic exit network (MEN) proteins are required for cytokinesis and re-polarization of the actin cytoskeleton in the budding yeast, Saccharomyces cerevisiae. Eur. J. Cell Biol. 85, 1201-1215 [DOI] [PubMed] [Google Scholar]
- Dibble C. C., Asara J. M., Manning B. D. (2009). Characterization of Rictor phosphorylation sites reveals direct regulation of mTOR complex 2 by S6K1. Mol. Cell. Biol. 29, 5657-5670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q., Xia W., Liu J. C., Yang J. Y., Lee D. F., Xia J., Bartholomeusz G., Li Y., Pan Y., Li Z., et al. (2005). Erk associates with and primes GSK-3 >for its inactivation resulting in upregulation of β-catenin. Mol. Cell 19, 159-170 [DOI] [PubMed] [Google Scholar]
- Epp J. A., Chant J. (1997). An IQGAP-related protein controls actin-ring formation and cytokinesis in yeast. Curr. Biol. 7, 921-929 [DOI] [PubMed] [Google Scholar]
- Fingar D. C., Blenis J. (2004). Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene 23, 3151-3171 [DOI] [PubMed] [Google Scholar]
- Frias M. A., Thoreen C. C., Jaffe J. D., Schroder W., Sculley T., Carr S. A., Sabatini D. M. (2006). mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr. Biol. 16,1865-1870 [DOI] [PubMed] [Google Scholar]
- Fuchs B. B., Mylonakis E. (2009). Our paths might cross: the role of the fungal cell wall integrity pathway in stress response and cross talk with other stress response pathways. Eukaryot. Cell 8, 1616-1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter A., Pringle S. J., Lew D. J. (2001). The septin cortex at the yeast mother-bud neck. Curr. Opin. Microbiol. 4, 681-689 [DOI] [PubMed] [Google Scholar]
- Grohmanova K., Schlaepfer D., Hess D., Gutierrez P., Beck M., Kroschewski R. (2004). Phosphorylation of IQGAP1 modulates its binding to Cdc42, revealing a new type of Rho-GTPase regulator. J. Biol. Chem. 279, 48495-48504 [DOI] [PubMed] [Google Scholar]
- Gromley A., Yeaman C., Rosa J., Redick S., Chen C.-T., Mirabelle S., Guha M., Sillibourne J., Doxsey S. J. (2005). Centriolin anchoring of exocyst and SNARE complexes at midbody is required for secretory vesicle-mediated abscission. Cell 123, 75-87 [DOI] [PubMed] [Google Scholar]
- Guertin D. A., Sabatini D. M. (2005). An expanding role for mTOR in cancer. Trends Mol. Med. 11, 353-361 [DOI] [PubMed] [Google Scholar]
- Guertin D. A., Sabatini D. M. (2007). Defining the role of mTOR in cancer. Cancer Cell 12, 9-22 [DOI] [PubMed] [Google Scholar]
- Gwinn D. M., Asara J. M., Shaw R. J. (2010). Raptor is phosphorylated by cdc2 during mitosis. PLoS ONE 5, e9197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales K. G., Bi E., Wu J. Q., Adam J. C., Yu I. C., Pringle J R. (1999). Cytokinesis: an emerging unified theory for eukaryotes? Curr. Opin. Cell Biol. 11, 717-725 [DOI] [PubMed] [Google Scholar]
- Hall M. N. (2008). mTOR-what does it do? Transplant Proc. 40, S5-S8 [DOI] [PubMed] [Google Scholar]
- Hartmuth S., Petersen J. (2009). Fission yeast Tor1 functions as part of TORC1 to control mitotic entry through the stress MAPK pathway following nutrient stress. J. Cell Sci. 122, 1737-1746 [DOI] [PubMed] [Google Scholar]
- Hsieh A. C., Truitt M. L., Ruggero D. (2011). Oncogenic AKTivation of translation as a therapeutic target. Br. J. Cancer 105, 329-336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Manning B. D. (2009). A complex interplay between Akt, TSC2 and the two mTOR complexes. Biochem. Soc. Trans. 37, 217-222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K., Li Y., Zhu T., Wu J., Guan K. L. (2002). TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 4, 648-657 [DOI] [PubMed] [Google Scholar]
- Inoki K., Zhu T., Guan K. L. (2003). TSC2 mediates cellular energy response to control cell growth and survival. Cell 115, 577-590 [DOI] [PubMed] [Google Scholar]
- Inoki K., Corradetti M. N., Guan K. L. (2005). Dysregulation of the TSC-mTOR pathway in human disease. Nat. Genet. 37, 19-24 [DOI] [PubMed] [Google Scholar]
- Jacinto E., Facchinetti V., Liu D., Soto N., Wei S., Jung S., Huang Q., Qin J., Su B. (2006). SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell 127, 125-137 [DOI] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E. A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425-1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D. I. (1999). Cdc42: An essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol. Mol. Biol. Rev. 63, 54-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M., Sharma M., Henderson B. R. (2009). IQGAP1 regulation and roles in cancer. Cell. Signal. 21, 1471-1478 [DOI] [PubMed] [Google Scholar]
- Jope R. S., Johnson G. V. (2004). The glamour and gloom of glycogen synthase kinase-3. Trends Biochem. Sci. 29, 95-102 [DOI] [PubMed] [Google Scholar]
- Julien L.-A., Carriere A., Moreau J., Roux P. P. (2010). mTORC1-activated S6K1 phosphorylates Rictor on threonine 1135 and regulates mTORC2 signaling. Mol. Cell. Biol. 30, 908-921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko N., Nishihama R., Tully G. H., Ostapenko D., Solomon M. J., Morgan D. O., Pringle J. R. (2007). Identification of yeast IQGAP (Iqg1p) as an anaphase-promoting-complex substrate and its role in actomyosin-ring-independent cytokinesis. Mol. Biol. Cell 18, 5139-5153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M., Sabatini D. M. (2009). mTOR signaling at a glance. J. Cell Sci. 122, 3589-3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Clainche C., Schlaepfer D., Ferrari A., Klingauf M., Grohmanova K., Veligodskiy A., Dirdy D., Le D., Egile C., Carlier M.-F., et al. (2007). IQGAP1 stimulates actin assembly through the N-Wasp-Arp2/3 pathway. J. Biol. Chem. 282, 426-435 [DOI] [PubMed] [Google Scholar]
- Lesage G., Bussey H. (2006). Cell wall assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70, 317-343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J., Li R. (1998). Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis. J. Cell Biol. 140, 355-366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J. L., Bonenfant D., Oppliger W., Jenoe P., Hall M. N. (2002). Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10, 457-468 [DOI] [PubMed] [Google Scholar]
- Longtine M. S., Mckenzie A., Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953-961 [DOI] [PubMed] [Google Scholar]
- Madden K., Snyder M. (1998). Cell polarity and morphogenesis in budding yeast. Annu. Rev. Microbiol. 52, 687-744 [DOI] [PubMed] [Google Scholar]
- Manning B. D. (2004). Balancing Akt with S6K: implications for both metabolic diseases and tumorigenesis. J. Cell Biol. 167, 399-403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateer S. C., Wang N., Bloom G. S. (2003). IQGAPs: integrators of the cytoskeleton, cell adhesion machinery, and signaling networks. Cell Motil. Cytoskeleton 55, 147-155 [DOI] [PubMed] [Google Scholar]
- Minden A., Karin M. (1997). Regulation and function of the JNK subgroup of MAP kinases. Biochim. Biophys. Acta 1333, F85-F104 [DOI] [PubMed] [Google Scholar]
- Morita E., Sandrin V., Chung H.-Y., Morham S. G., Gygi S. P., Rodesch C. K., Sundquist W. I. (2007). Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 26, 4215-4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley J. B., Mayeux A., Paoletti A., Nurse P. (2009). A spatial gradient coordinates cell size and mitotic entry in fission yeast. Nature 459, 857-860 [DOI] [PubMed] [Google Scholar]
- Nelson W. J. (2003). Adaptation of core mechanisms to generate cell polarity. Nature 422, 766-774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noritake J., Fukata M., Sato K., Nakagawa M., Watanabe T., Izumi N., Wang S., Fukata Y., Kaibuchi K. (2004). Positive role of IQGAP1, an effector of Rac1, in actin-meshwork formation at sites of cell-cell contact. Mol. Biol. Cell 15, 1065-1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noritake J., Watanabe T., Sato K., Wang S., Kaibuchi K. (2005). IQGAP1: a key regulator of adhesion and migration. J. Cell Sci. 118, 2085-2092 [DOI] [PubMed] [Google Scholar]
- Osman M. (2010). An emerging role for IQGAP1 in regulating protein traffic. ScientificWorldJournal 10, 944-953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman M. A., Cerione R. A. (1998). Iqg1p, a yeast homologue of the mammalian IQGAPs, mediates Cdc42p effects on the actin cytoskeleton. J. Cell Biol. 142, 443-455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman M. A., Cerione R. A. (2006). Actin doesn't do the locomotion. Secretion drives cell polarization. In Protein Trafficking: Mechanisms and Regulation (ed. Segev N.), pp. 388-399 Georgetown, Texas: Landes Bioscience/Eurekah; [Google Scholar]
- Osman M. A., Konopka J. B., Cerione R. A. (2002). Iqg1p links spatial and secretion landmarks to polarity and cytokinesis. J. Cell Biol. 159, 601-611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak P., Hall M. N. (2006). mTORC2 caught in SINful Akt. Dev. Cell 11, 433-434 [DOI] [PubMed] [Google Scholar]
- Pringle J. R. (1991). Staining of bud scars and mother cell wall chitin with calcofluor. Methods Enzymol. 194, 732-735 [DOI] [PubMed] [Google Scholar]
- Reinke A., Anderson S., McCaffery J. M., Yates J., Aronova S., Chu S., Fairclough S., Iverson C., Wedaman K. P., Powers T. (2004). TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J. Biol. Chem. 279, 14752-14762 [DOI] [PubMed] [Google Scholar]
- Rittmeyer E. N., Daniel S., Hsu S.-C., Osman M. A. (2008). A dual role for IQGAP1 in regulating exocytosis. J. Cell Sci. 121, 391-408 [DOI] [PubMed] [Google Scholar]
- Roy M., Li Z., Sacks D. B. (2004). IQGAP1 binds ERK2 and modulates its activity. J. Biol. Chem. 279, 17329-17337 [DOI] [PubMed] [Google Scholar]
- Ruvinsky I., Meyuhas O. (2006). Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem. Sci. 31, 342-348 [DOI] [PubMed] [Google Scholar]
- Sabatini D. M. (2006). mTOR and cancer: insights into a complex relationship. Nat. Rev. Cancer 6, 729-734 [DOI] [PubMed] [Google Scholar]
- Sarbassov D. D., Guertin D. A., Siraj M. A., Sabatini D. M. (2005). Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098-1101 [DOI] [PubMed] [Google Scholar]
- Sbroggiò M., Carnevaleb D., Berteroa A., Cifelli G., Blasioa E. D., Mascio G., Hirsch E., Bahou W. F., Turco E., Silengo L., et al. (2011). IQGAP1 regulates ERK1/2 and AKT signaling in the heart and sustains functional remodeling upon pressure overload. Cardiovasc Res. 91, 456-464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S., Peterson T. R., Sabatini D. M. (2010). Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol. Cell 40, 310-322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiota C., Woo J., Lindner J., Shelton K., Magnuson M. (2006). Multiallelic disruption of the rictor gene in mice reveals that mTOR complex 2 is essential for fetal growth and viability. Dev. Cell 11, 583-589 [DOI] [PubMed] [Google Scholar]
- Skop A. R., Liu H., Yates J., Jr, Meyer B. J., Heald R. (2004). Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms. Science 305, 61-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill T. W., Hall M. N. (2007). Holding back TOR advances mitosis. Nat. Cell Biol. 9, 1221-1222 [DOI] [PubMed] [Google Scholar]
- Surka M. C., Tsang C. W., Trimble W. S. (2002). The mammalian septin MSF localizes with microtubules and is required for completion of cytokinesis. Mol. Biol. Cell 13, 3532-3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapon N., Moberg K. H., Hariharan I. (2001). The coupling of cell growth to the cell cycle. Curr. Opin. Cell Biol. 13, 731-737 [DOI] [PubMed] [Google Scholar]
- Tu S., Wu W. J., Wang J., Cerione R. A. (2003). Epidermal growth factor-dependent regulation of Cdc42 is mediated by the Src tyrosine kinase. J. Biol. Chem. 278, 49293-49300 [DOI] [PubMed] [Google Scholar]
- Wang J.-B., Sonn R., Tekletsadik Y. K., Samorodnitsky D., Osman M. A. (2009). IQGAP1 regulates cell proliferation through a novel CDC42-mTOR/PI3K pathway. J. Cell Sci. 122, 2024-2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White C. D., Brown M. D., Sacks D. B. (2009). IQGAPs in cancer: a family of scaffold proteins underlying tumorigenesis. FEBS Lett. 18, 1817-1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe B. A., Gould K. L. (2005). Split decisions: coordinating cytokinesis in yeast. Trends Cell Biol. 15, 10-18 [DOI] [PubMed] [Google Scholar]
- Wullschleger S., Loewith R., Hall M. N. (2006). TOR signaling in growth and metabolism. Cell 124, 471-484 [DOI] [PubMed] [Google Scholar]
- Zhang G., Kashimshetty R., Ng K. E., Tan H. B., Yeong F. M. (2006). Exit from mitosis triggers Chs2p transport from the endoplasmic reticulum to mother-daughter neck via the secretory pathway in budding yeast. J. Cell Biol. 174, 207-220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziman M., O'Brien J. M., Ouellette L. A., Church W. R., Johnson D. I. (1991). Mutational analysis of CDC42Sc, a Saccharomyces cerevisiae gene that encodes a putative GTP-binding protein involved in the control of cell polarity. Mol. Cell. Biol. 11, 3537-3544 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.