Abstract
Fluorescent probe pairs that can be selectively excited in the presence of Trp and Tyr are of great utility in studying conformational changes in proteins. However, the size of these probe pairs can restrict their incorporation to small portions of a protein sequence where their effects on secondary and tertiary structure can be tolerated. Our findings show that a thioamide bond — a single atom substitution of the peptide backbone — can quench fluorophores that are red shifted from intrinsic protein fluorescence, such acridone. Using steady state and fluorescence lifetime measurements, we further demonstrate that this quenching occurs through a dynamic electron transfer mechanism. In a proof-of-principle experiment, we apply this technique to monitor unfolding in a model peptide system, the villin headpiece HP35 fragment. Thioamide analogs of the natural amino acids can be placed in a variety of locations in a protein sequence, allowing one to make a large number of measurements to model protein folding.
Fluorescence spectroscopy is a powerful tool for studying dynamic processes in biological systems.1,2 In order to harness the full potential of this technique, red shifted fluorophores are often required that can be excited independently of the intrinsic fluorescence background of native proteins.3 When these experiments are used to monitor a distance-dependent event, such as a conformational rearrangement, a second probe is typically required that can act as an energy donor or acceptor.4,5 Because of their bulk, introducing such probe pairs at arbitrary positions in a protein sequence risks significantly perturbing native structure and function. If one of the partners in the probe pair were sufficiently non-perturbing to be tolerated at almost any position in a protein, much more detailed information could be obtained. Here, we show that a thioamide, a single-atom substitution of the peptide backbone, can serve as such a partner for the red-shifted fluorophores azaindole, coumarin, and acridone. Furthermore, we demonstrate that the acridone is quenched through dynamic electron transfer and that the acridone/thioamide probe pair can be used to monitor conformational changes in proteins.
Previously, we have shown that thioamides quench intrinsic protein fluorescence arising from excitation of tryptophan (Trp or W) or tyrosine (Tyr or Y), as well as extrinsic protein fluorescence arising from excitation of p-cyanophenylalanine (Cnf or F*).6,7 These fluorophores cannot be selectively excited in proteins containing other Trp or Tyr residues and are therefore limited to use in model proteins under highly purified conditions. However, the observation that Trp is quenched by thioamides without appreciable spectral suggested the possibility of finding a fluorophore that could be selectively excited in a protein and that could also be quenched by a thioamide. Mechanistically, such a process might involve a photo-induced electron transfer (PET) mechanism, rather than a Förster resonance energy transfer (FRET) or Dexter transfer mechanism, since FRET and Dexter require spectral overlap between the donor and acceptor chromophores.8 Note: Thioamide residues are represented by the one or three letter codes of the equivalent oxoamide amino acids with a prime symbol (e.g. A' or Ala' represent thioalanine).
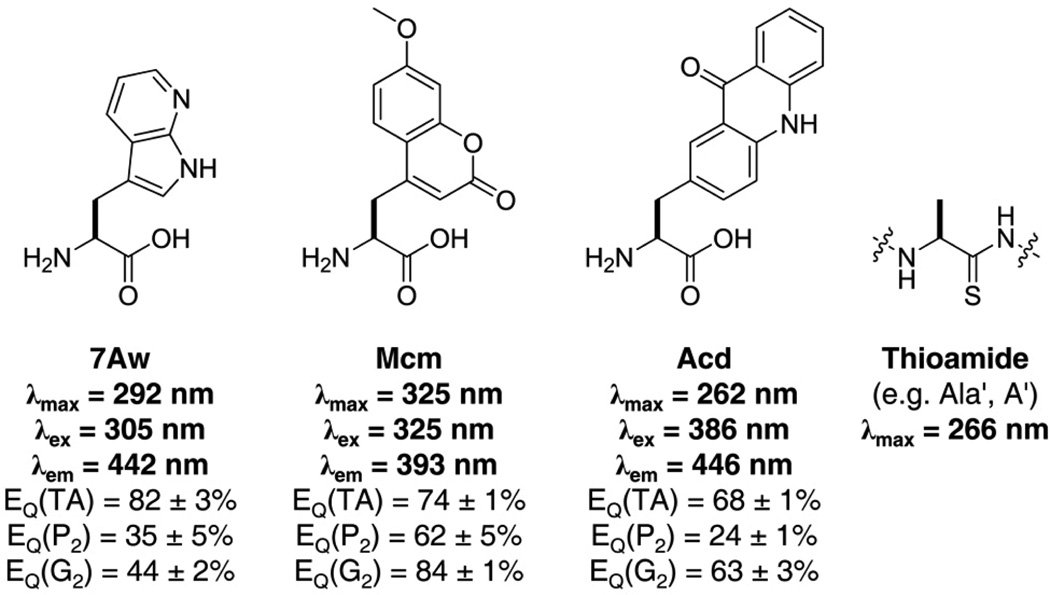
We sought a robust fluorophore with a large extinction coefficient and a high quantum yield that would be compact enough to be tolerated in several positions in a protein sequence and that could be incorporated genetically. We began by studying three fluorescent scaffolds that met these general requirements: 7-azaindole, 7-methoxycoumarin, and acridone, and their amino acid derivatives 7-azatryptophan (7Aw or ω), 7-methoxycoumarin-4-yl alanine (Mcm or μ), and acridon-2-yl alanine (Acd or δ), respectively (Fig. 1). In addition to their favorable optical properties, each of these amino acid scaffolds has been shown to be compatible with in vivo or in vitro ribosomal incorporation and has previously been used to probe biomolecular structure.9–13 We expect them to be minimally-perturbing as compared to other fluorophores since 7Aw is isosteric with Trp, Mcm is only 29% larger, and Acd 47% larger than Trp; whereas fluorescein is more than twice the size of Trp and Texas Red is more than three times larger.
Figure 1.
Chromophore Structures and Spectral Properties. Chemical structures of 7-azatryptophan (7Aw), 7-methoxycoumarin-4-yl alanine (Mcm), acridon-2-yl alanine (Acd), and a thioamide analog of alanine (A' or Ala') with corresponding wavelengths of maximum absorbance (λmax) and emission (λem). Wavelengths for selective excitation in a protein are listed as λex. Thioamide quenching efficiencies listed as EQ(TA) are for dilute solutions of the fluorophore in 50 mM thioacetamide and as EQ(P2) or as EQ(G2) are those in a short thioamide containing diproline or diglycine peptide, respectively, as described in the text.
Initially, we examined the fluorescence quenching of each chromophore by aqueous thioacetamide. We prepared dilute (< 50 µM) solutions of each fluorophore in 50 mM thioacetamide in phosphate buffer (100 mM, pH 7.00) and measured their fluorescence. As a control, we measured the fluorescence of equimolar solutions of the fluorophores in the absence of quencher (in phosphate buffer only) and in the presence of 50 mM acetamide. In all cases, we observed substantial quenching upon addition of thioacetamide and no quenching upon addition of acetamide. To quantify the extent of quenching, we calculated the quenching efficiency, EQ(TA), at the wavelength of maximum emission (Fig. 1; see Supporting Information for calculations). Since concentrated thioacetamide solutions absorb light very strongly below 350 nm, the observed quenching efficiencies of 7Aw and Mcm may be overestimates. Nonetheless, these results indicated that thioamides are capable of quenching each of these fluorophores.
To ensure that these results would translate to a protein system, we synthesized several peptides bearing a fluorophore at the C-terminus and either leucine or thioleucine at the N-terminus, separated by two proline residues or by two glycine residues. Again, we calculated quenching efficiencies, EQ(P2) and EQ(G2), for each flourophore by comparing the fluorescence of the oxo- and thioamide containing peptides (Fig. 1). For 7Aw, EQ(G2) was much smaller than EQ(50 mM), likely due to the absence of the inner filter effect in the peptide system. In all cases, much greater quenching was observed in the glycine peptides than in the proline peptides. This is consistent with a collisional quenching mechanism that is enabled by a more flexible linker between fluorophore and thioamide.14 For further experiments, we chose to focus on the quenching of Acd rather than 7Aw or Mcm because of its high quantum yield (ΦAcd = 0.95;15 Φ7Aw = 0.01;16 ΦMcm = 0.18).17
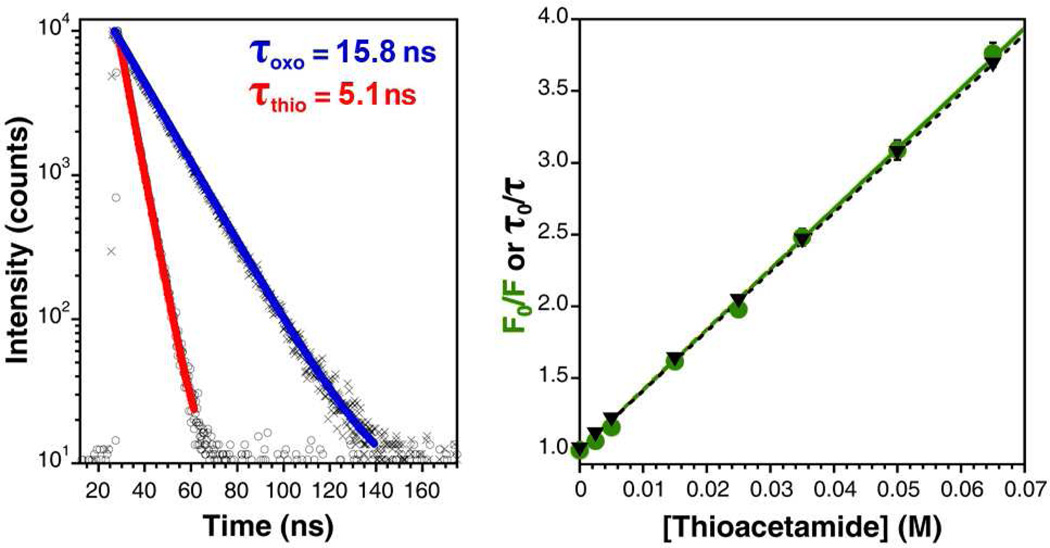
We began characterizing the mechanism of quenching by measuring the emission of Acd in varying concentrations of thioacetamide using variable-temperature fluorometry at 10, 25, 35, 45, and 65 °C. At each temperature, the fluorescence intensity varied inversely with thioacetamide concentration and appeared to follow Stern-Volmer quenching behavior (Fig. 2). The Stern-Volmer constant, KSV, increased with temperature from 34.16 ± 0.55 M−1 at 10 °C to 53.51 ± 0.40 M−1 at 65 °C, which suggested that thioamide quenching of Acd may arise from dynamic electron transfer.
Figure 2.
Stern-Volmer Quenching of Acridon-2-yl Alanine (Acd) by Thioacetamide. Left: Fluorescence lifetime measurements of 2 µM Acd in 100 mM phosphate buffer, pH 7.00 at 25 °C in the presence of 50 mM acetamide (×; blue fit) or 50 mM thioacetamide (○; red fit). Heavy lines are single exponential fits to the raw data. Right: Stern-Volmer plots of Acd fluorescence intensity (green circles) and lifetime (black triangles) as a function of thioacetamide concentration; error bars represent standard error. Linear fits to the Stern-Volmer equation are shown. (See Supporting Information).
Time-correlated single photon counting (TCSPC) spectroscopy on freshly prepared Acd solutions at 25 °C was employed to further characterize the mechanism of quenching. In 100 mM phosphate buffer, pH 7.00, we found the lifetime of Acd to be 15.73 ± 0.05 ns. The lifetime was unchanged upon addition of 50 mM acetamide (τ = 15.76 ± 0.05 ns), but decreased to 5.13 ± 0.02 ns upon addition of 50 mM thioacetamide. At intervening concentrations of thioacetamide, the lifetime varied with the concentration of quencher according to Stern-Volmer kinetics, with a Stern-Volmer constant KSV = 41.36 ± 0.05 M−1, which is nearly identical to that found from steady-state measurements at 25 °C, KSV = 41.96 ± 0.42 M−1 (Fig. 2). In all cases, lifetime data could be fit to single exponential functions as described in the Supporting Information.
Using these values, we calculated the quenching constant as kQ = 2.6 × 109 M−1 s−1, which is near the value predicted for a diffusion- limited, bimolecular quenching rate constant predicted by the Smoluchowski equation: k2 = 8.3 × 109 M−1 s−1.18 Although it is possible to assign the discrepancy in the values of kQ and k2 to a quenching efficiency factor fQ = 0.31, it is also possible that the theoretical model does not describe the system perfectly. Additionally, we examined the UV/Vis absorbance spectra of Acd solutions in the presence and absence of concentrated thioacetamide. We observed no difference in the near UV absorbance of these samples, which suggests that no static, ground-state complex is formed, even at high concentrations of thioacetamide (see Supporting Information). Taken together, these data point to an exclusively dynamic PET quenching mechanism.
To evaluate the energetic feasibility of a PET mechanism, we calculated the Gibbs energy for transferring an electron from thioacetamide to the excited fluorophore (ΔGET).19 We measured the electrochemical properties of acridone in anhydrous, deoxygenated DMF using cyclic voltammetry and observed irreversible oxidation (Epa = 1.4 V vs SCE) and reduction (Epc = −1.9 V vs SCE) events. Using these values, the reported oxidation potential of thioacetamide (0.97 V vs SCE),20 and the lowest energy, vibrational zero-zero electronic excitation of acridone (E0,0 = 3.0 eV), we calculated ΔGET = − 0.13 eV according to the Rehm-Weller Equation (see Supporting Information).21 A favorable ΔGET implies that transfer of an electron from a thioamide to excited Acd is possible. A similar analysis using previously reported values for the reduction potential and E0,0 of 7-methoxycoumarin provides a value of ΔGET = − 0.6 eV.22 We were unable to obtain satisfactory electrochemical data to perform a calculation of ΔGET for 7Aw.23 It is also possible that PET occurs in the other direction: the electron may be transferred from the fluorophore to the thioamide, but our current electrochemical data do not allow us to calculate ΔGET for this process.
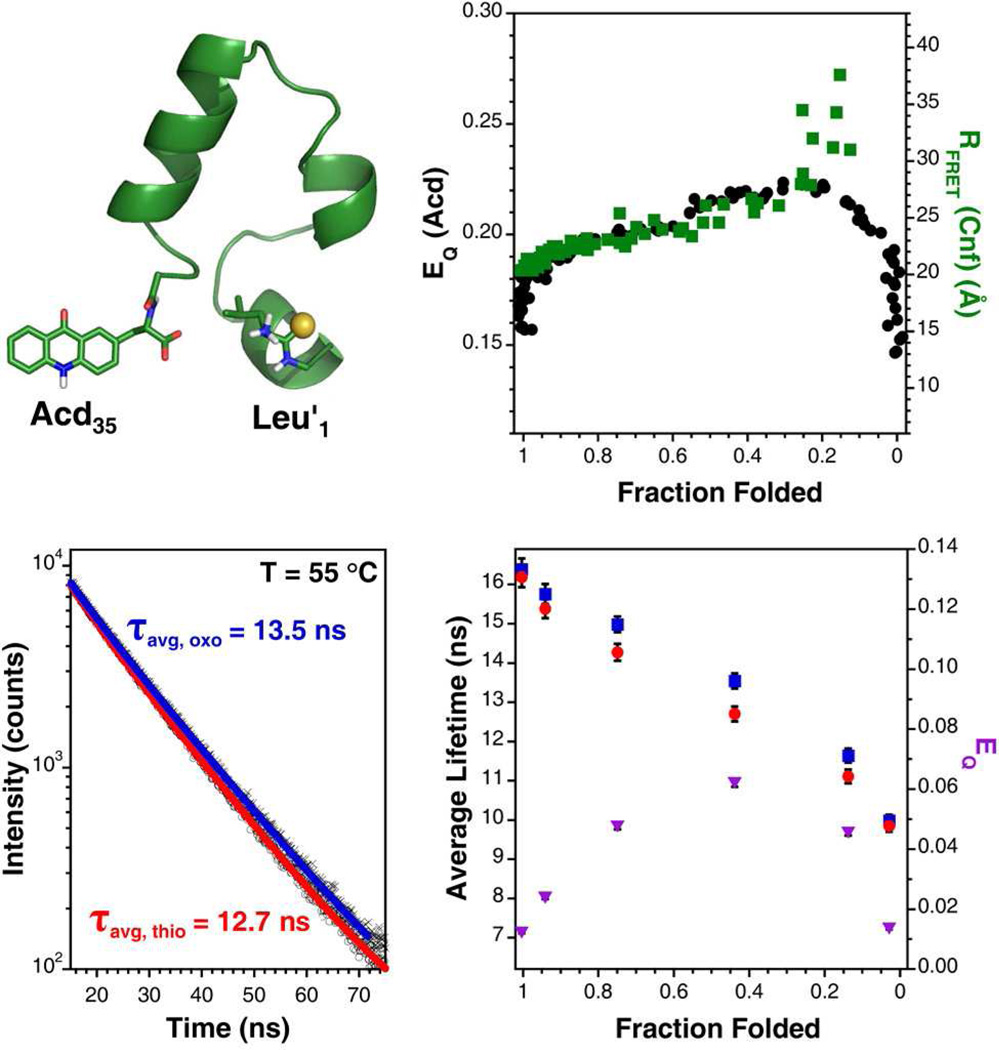
As a demonstration of the utility of the Acd/thioamide probe pair, we studied the thermal denaturation of a variant of the villin headpiece subdomain that has been well characterized, HP35.24,25 HP35 adopts a compact folded structure in which the N- and C-termini are within 20 Å.26 Previously, we used a FRET interaction between C-terminal Cnf35 and N-terminal Leu′1 in a double-labeled version of HP35 to monitor unfolding.6 We made this HP35-L′1F23F*35 construct without Trp (W23F mutation) because spectral overlap of Cnf with Trp makes deconvoluting the contributions of FRET to Trp and the thioamide difficult.7,27 The HP35 construct used in this study, HP35-L′1δ35 (containing a Trp at position 23), does not suffer this problem since Acd can be excited at wavelengths that do not excite Trp. Therefore, we can use PET quenching of Acd by the thioamide to track unfolding. In the following experiments, we compare the double-labeled HP35-L′1δ35 to the equivalent oxopeptide HP35-δ 35 using temperature-dependent circular dichroism (CD), steady state fluorescence, and TCSPC.
CD spectroscopy indicated that both proteins were primarily α-helical and underwent unfolding events that could be modeled as cooperative two-state transitions with comparable Tm values (53 °C for HP35-δ35 and 56 °C for HP35-L′1δ35). These thermal denaturation curves were used to determine the fraction of each peptide that was folded at any given temperature. Steady-state fluorescence measurements on equimolar solutions of the proteins in phosphate buffered saline, pH 7.00, showed that Acd in the thioamide-containing variant was quenched 16 % relative to the all oxo-amide containing protein at 1 °C. The quenching efficiency (EQ) increased gradually with temperature, reaching a maximum of 22 % at 66 °C, after which it decreased rapidly to 15 % at 95 °C.
The dependence of EQ on temperature can be displayed with respect to fraction folded as shown in Figure 3. Several observations are worth noting from this transformed data, which is plotted with data from our previous HP35-L′1F23F* 35 experiment for comparison. First, there is a sharp initial increase in quenching efficiency that occurs while the protein retains a mostly folded state. We attribute this to a slight increase in the flexibility of the N- and C-termini, favoring contact between the chromophores that leads to quenching. As the protein begins to unfold, EQ increases smoothly, presumably as a result of the increased rates of productive collisions that are accessible at higher temperatures. EQ drops dramatically when the protein globally unfolds at fraction folded of 0.2, which is consistent with the increase in separation between the ends of the protein seen in the HP35-L′1F23F* 35 FRET experiments. Previous experiments have shown that repacking of the HP35 core can occur well before global unfolding, consistent with our observations.28–30 Specifically, two other groups have recently observed separation of the villin N- and C-termini under partially-denaturing conditions using triplet-triplet energy transfer.31,32
Figure 3.
Thermal Denaturation of HP35. Upper Left: Structure of HP35-Leu′1Acd35 modified from PDB 1VII and rendered in PyMOL (Schrödinger; Portland, OR). Upper Right: Steady-state quenching efficiency (EQ(Acd); black circles) as a function of fraction of protein folded determined by CD spectroscopy and the separation of the N- and C-termini as a function of fraction folded determined previously by a FRET experiment with thioamide quenching of Cnf (RFRET(CNF); green squares). Lower Left: Normalized fluorescence lifetime measurements of Acd in HP35-L′1δ35 (○; red fit) and HP35-Acd35 (×; blue fit) at 55 °C. Average lifetimes are listed as determined by fits to a bi-exponential model as described in the Supporting Information. Lower Right: Average lifetimes of Acd in HP35-L′1δ35 (red circles) and HP35-δ35 (blue squares) as a function of fraction folded and determined from biexponential fits to the data. Quenching efficiency calculated by comparing lifetime measurements of the oxo- and thio-proteins as a function of fraction folded (purple triangles). Error bars are estimated from the fits.
Fluorescence lifetime measurements of the oxo- and thioversions of HP35 at several temperatures are generally consistent with the steady state observations (Fig. 3). The magnitude of EQ as determined by comparing the average lifetimes of HP35-δ35 and HP35-L′1δ35 is less than that obtained by comparing steady-state measurements, but the overall trend as a function of fraction folded is the same. The change in magnitude of EQ might indicate that there is a static quenching component in the protein context that we did not observe in bimolecular thioacetamide quenching experiments.
Although we were able to fit the lifetime data collected for the free amino acid in the presence and absence of thioacetamide to a single exponential, we were not able to model the lifetime of Acd in HP35 with this function. Instead, we found a bi-exponential fit described the data well, giving average lifetimes of 15.76 ± 0.25 ns for HP35-δ35 and 15.38 ± 0.24 ns for HP35-L′1δ35 at 25 °C. We used this function as a simple model of a system that placed Acd in at least two environments with distinct quenching. Gaussian fits to the data produced nearly identical results to those found with the bi-exponential model, in which 5 % quenching was observed in the thioamide-containing peptide.
Since the data from HP35-δ35 could not be fit to a single exponential, we measured the fluorescence lifetime of Acd in other all oxo-amide systems in order to determine whether this resulted from incorporation of the amino acid into a peptide backbone. In the cases of both the LP2δ (τ = 16.68 ± 0.05 ns at 25 °C) and LG2δ (τ = 16.35 ± 0.04 ns at 25 °C) peptides described above, we found that the Acd lifetime data for the oxo-peptides could be fit with a simple single exponential function, but a satisfactory fit to the corresponding data for the thiopeptides could be achieved with a bi-exponential model. We observed a substantially shorter average lifetime for the L′G2δ (τavg = 12.92 ± 0.16 ns at 25 °C) than for L′P2δ (τavg = 16.08 ± 0.28 ns at 25 °C), which is consistent with steady state measurements and the idea that access to favorable conformations is important for quenching. In a separate experiment, we found that the presence or absence of C-terminal amidation also does not seem to have an effect on the oxopeptide: both LP2δ-OH (τ = 16.68 ± 0.05 ns) and LP2δ-NH2 (τ = 16.67 ± 0.05 ns) could be fit to a single exponential model.
The difference between the lifetime of Acd in HP35-δ35 and LP2δ or LG2δ shows that there is some quenching of Acd in HP35 independent of the thioamide. This is likely to be true in any real protein, where almost every fluorophore experiences some quenching when located near redox-active amino acids such as Trp, Tyr, His, Met, and Cys.33,34 Through protein electron transfer can be very efficient, and may further complicate fluorescence quenching.35 Even so, since the HP35 proteins studied here differ only by the single O to S substitution in the Leu′ amide bond, we can assign quenching to be specifically due to the thioamide. It is crucial that measurements on thioamide-containing proteins be compared to the all oxo-amide version to control for the effects of temperature, solvent accessibility, and neighboring residues on fluorescence that may arise during a conformational change. With these controls, one can follow the unfolding of events specific to the thioamide site with fluorescence spectroscopy. At this point, we do not attempt to assign interchromophore distances based on the efficiency of quenching, rather we merely interpret quenching as resulting from transient molecular contact so that fluorophores are less than 20 Å apart if quenching is observed. Further mechanistic study may allow us to assign distances in a more precise manner.
In conclusion, we have demonstrated that thioamides are capable of quenching a variety of fluorophores, including some that can be selectively excited in the context of a protein. Our findings from steady-state and lifetime measurements of Acd fluorescence indicate that this quenching arises through a dynamic photo-induced electron transfer mechanism. Furthermore, we have shown that the Acd-thioamide fluorophore-quencher probe pair can be used to study conformational changes in proteins. Although previous experiments have gained valuable information using visible wavelength fluorescent donors and Trp quenchers, we believe that our reporter system will allow us to study protein dynamics in far greater detail because the thioamide will be tolerated at more positions in the protein.19,36 Other recent work in our laboratory has demonstrated that thioamides can be incorporated into full-sized proteins through native chemical ligation, so that studies of the type conducted on HP35 could be carried out on larger proteins.37 Taken together, these studies lay the groundwork for the application of thioamide quenching to the study of protein folding and the design of conformational biosensors.38
Supplementary Material
ACKNOWLEDGMENT
This work was supported by funding from the University of Pennsylvania, the National Science Foundation (CHE-1020205 to EJP), and the Searle Scholars Program (10-SSP-214 to EJP). We thank Jerome Robinson and Eric Schelter for assistance with the electrochemical measurements, Jeff Saven for use of the fluorometer, Feng Gai for assistance with the CD spectrometer (supported by NSF DMR05-20020), and Rakesh Kohli for assistance with MALDI-MS (supported by NSF MRI-0820996). We are grateful to Tom Troxler for assistance with the TCSPC measurements collected at the Regional Laser and Biomedical Technology Laboratories at the University of Pennsylvania (supported through NIH/NCRR P41RR001348).
Footnotes
SUPPORTING INFORMATION AVAILABLE. Descriptions of peptide and small molecule synthesis, purification, and characterization; CD, fluorescence, and UV/Vis spectroscopy; electrochemical measurements; and calculations are provided in Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Giepmans BNG, Adams SR, Ellisman MH, Tsien RY. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 2.Royer CA. Chem. Rev. 2006;106:1769–1784. doi: 10.1021/cr0404390. [DOI] [PubMed] [Google Scholar]
- 3.Ross JB, Szabo AG, Hogue CWV. Methods Enzymol. 1997;278:151–190. doi: 10.1016/s0076-6879(97)78010-8. [DOI] [PubMed] [Google Scholar]
- 4.Selvin PR. Nat. Struct. Biol. 2000;7:730–734. doi: 10.1038/78948. [DOI] [PubMed] [Google Scholar]
- 5.Weiss S. Nat. Struct. Biol. 2000;7:724–729. doi: 10.1038/78941. [DOI] [PubMed] [Google Scholar]
- 6.Goldberg JM, Batjargal S, Petersson EJ. J. Am. Chem. Soc. 2010;132:14718–14720. doi: 10.1021/ja1044924. [DOI] [PubMed] [Google Scholar]
- 7.Goldberg JM, Wissner RF, Klein AM, Petersson EJ. Chem. Commun. 2012;48:1550–1552. doi: 10.1039/c1cc14708k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Speiser S. Chem. Rev. 1996;96:1953–1976. doi: 10.1021/cr941193+. [DOI] [PubMed] [Google Scholar]
- 9.Budisa N, Pal PP. Biol. Chem. 2004;385:893–904. doi: 10.1515/BC.2004.117. [DOI] [PubMed] [Google Scholar]
- 10.Hamada H, Kameshima N, Szymanska A, Wegner K, Lankiewicz L, Shinohara H, Taki M, Sisido M. Bioorg. Med. Chem. 2005;13:3379–3384. doi: 10.1016/j.bmc.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 11.Taki M, Yamazaki Y, Suzuki Y, Sisido M. Chem. Lett. 2010;39:818–819. [Google Scholar]
- 12.Katritzky AR, Narindoshvili T. Org. Biomol. Chem. 2009;7:627–634. doi: 10.1039/b818908k. [DOI] [PubMed] [Google Scholar]
- 13.Wang JY, Xie JM, Schultz PG. J. Am. Chem. Soc. 2006;128:8738–8739. doi: 10.1021/ja062666k. [DOI] [PubMed] [Google Scholar]
- 14.Doose S, Neuweiler H, Barsch H, Sauer M. Proc. Natl. Acad. Sci. U. S. A. 2007;104:17400–17405. doi: 10.1073/pnas.0705605104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szymanska A, Wegner K, Lankiewicz L. Helv. Chim. Acta. 2003;86:3326–3331. [Google Scholar]
- 16.Lotte K, Plessow R, Brockhinke A. Photochem. Photobiol. Sci. 2004;3:348–359. doi: 10.1039/b312436c. [DOI] [PubMed] [Google Scholar]
- 17.Brun MP, Bischoff L, Garbay C. Angew. Chem. Int. Ed. 2004;43:3432–3436. doi: 10.1002/anie.200454116. [DOI] [PubMed] [Google Scholar]
- 18.Lakowicz JR. Principles of fluorescence spectroscopy. Springer; 2006. [Google Scholar]
- 19.Doose S, Neuweiler H, Sauer M. ChemPhysChem. 2009;10:1389–1398. doi: 10.1002/cphc.200900238. [DOI] [PubMed] [Google Scholar]
- 20.Bordwell FG, Algrim DJ, Harrelson JA. J. Am. Chem. Soc. 1988;110:5903–5904. [Google Scholar]
- 21.Rehm D, Weller A. Israel J. Chem. 1970;8:259–271. [Google Scholar]
- 22.Seidel CAM, Schulz A, Sauer MHM. J. Phys. Chem. 1996;100:5541–5553. [Google Scholar]
- 23.Mukherjee M, Karmakar S, Chakraborty T. J. Phys. Chem. A. 2011;115:1830–1836. doi: 10.1021/jp1084422. [DOI] [PubMed] [Google Scholar]
- 24.McKnight CJ, Doering DS, Matsudaira PT, Kim PS. J. Mol. Biol. 1996;260:126–134. doi: 10.1006/jmbi.1996.0387. [DOI] [PubMed] [Google Scholar]
- 25.Cellmer T, Buscaglia M, Henry ER, Hofrichter J, Eaton WA. Proc. Natl. Acad. Sci. U. S. A. 2011;108:6103–6108. doi: 10.1073/pnas.1019552108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKnight CJ, Matsudaira PT, Kim PS. Nat. Struct. Biol. 1997;4:180–184. doi: 10.1038/nsb0397-180. [DOI] [PubMed] [Google Scholar]
- 27.Tucker MJ, Oyola R, Gai F. Biopolymers. 2006;83:571–576. doi: 10.1002/bip.20587. [DOI] [PubMed] [Google Scholar]
- 28.Brewer SH, Song BB, Raleigh DP, Dyer RB. Biochemistry. 2007;46:3279–3285. doi: 10.1021/bi602372y. [DOI] [PubMed] [Google Scholar]
- 29.Glasscock JM, Zhu YJ, Chowdhury P, Tang J, Gai F. Biochemistry. 2008;47:11070–11076. doi: 10.1021/bi8012406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu L, Ghosh K, King M, Cellmer T, Bakajin O, Lapidus LJ. Journal of Physical Chemistry B. 2011;115:12632–12637. doi: 10.1021/jp206238y. [DOI] [PubMed] [Google Scholar]
- 31.Beauchamp KA, Ensign DL, Das R, Pande VS. Proc. Natl. Acad. Sci. U. S. A. 2011;108:12734–12739. doi: 10.1073/pnas.1010880108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reiner A, Henklein P, Kiefhaber T. Proc. Natl. Acad. Sci. U. S. A. 2010;107:4955–4960. doi: 10.1073/pnas.0910001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen H, Ahsan SS, Santiago-Berrios MEB, Abruna HD, Webb WW. J. Am. Chem. Soc. 2010;132:7244–7245. doi: 10.1021/ja100500k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y, Barkley MD. Biochemistry. 1998;37:9976–9982. doi: 10.1021/bi980274n. [DOI] [PubMed] [Google Scholar]
- 35.Gray HB, Winkler JR. Ann. Rev. Biochem. 1996;65:537–561. doi: 10.1146/annurev.bi.65.070196.002541. [DOI] [PubMed] [Google Scholar]
- 36.Mansoor SE, DeWitt MA, Farrens DL. Biochemistry. 2011;49:9722–9731. doi: 10.1021/bi100907m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Batjargal SB, Wang YJ, Goldberg JM, Wissner RF, Petersson EJ. J. Am. Chem. Soc. doi: 10.1021/ja2113245. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vallee-Belisle A, Plaxco KW. Curr. Opin. Struct. Biol. 2010;20:518–526. doi: 10.1016/j.sbi.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.