Abstract
The role of TMPRSS2–ERG gene fusion in prostate cancer prognostication remains controversial. We evaluated the prognostic role of TMPRSS2–ERG fusion using fluorescence in situ hybridization analysis in a case–control study nested in The Johns Hopkins retropubic radical prostatectomy cohort. In all, 10 tissue microarrays containing paired tumors and normal tissues obtained from 172 cases (recurrence) and 172 controls (non-recurrence) matched on pathological grade, stage, race/ethnicity, and age at the time of surgery were analyzed. All radical prostatectomies were performed at our institution between 1993 and 2004. Recurrence was defined as biochemical recurrence, development of clinical evidence of metastasis, or death from prostate carcinoma. Each tissue microarray spot was scored for the presence of TMPRSS2–ERG gene fusion and for ERG gene copy number gains. The odds ratio of recurrence and 95% confidence intervals were estimated from conditional logistic regression. Although the percentage of cases with fusion was slightly lower in cases than in controls (50 vs 57%), the difference was not statistically significant (P=0.20). The presence of fusion due to either deletion or split event was not associated with recurrence. Similarly, the presence of duplicated ERG deletion, duplicated ERG split, or ERG gene copy number gain with a single ERG fusion was not associated with recurrence. ERG gene polysomy without fusion was significantly associated with recurrence (odds ratio 2.0, 95% confidence interval 1.17–3.42). In summary, TMPRSS2–ERG fusion was not prognostic for recurrence after retropubic radical prostatectomy for clinically localized prostate cancer, although men with ERG gene copy number gain without fusion were twice more likely to recur.
Keywords: fluorescence in situ hybridization, nested case–control study, prognosis, prostate adenocarcinoma, TMPRSS2–ERG fusion
Prostate cancer remains a major health problem in the United States. At present, at the time of diagnosis, most cases present as localized disease and are treated by radical prostatectomy, radiation therapy, or active surveillance. Recently, calls for adjustment of our current approach to the diagnosis and management of prostate carcinoma have been voiced with concerns for ‘overtreatment’ being raised.1,2 A marker able to distinguish cases with the potential to progress would be of particular utility in helping to determine which individuals should pursue active surveillance and those who need more definitive or even adjuvant therapy. Described by Tomlins et al, the recurrent fusion between the androgen-regulated gene TMPRSS2 (21q22.3) and ETS transcription factor family member ERG (21q22.2) is a common occurrence in prostate carcinoma and has been reported in 15–80% of all cases.3–8 TMPRSS2–ERG fusion is an early event in prostate oncogenesis that results from either a small deletion on chromosome 21 (seen in approximately two-thirds of cases) or through a translocation.5 In either type of fusion, the ERG gene is brought under the control of an androgen-regulated promoter leading to overexpression of the ERG protein.
To date, the clinical significance of TMPRSS2–ERG fusion as a prognosticator for recurrence or progression remains controversial. Studies addressing the relationship of TMPRSS2–ERG fusion status to the natural history of the disease and to prostate cancer progression have so far led to conflicting results.9–14 Although earlier studies pointed to the presence of TMPRSS2–ERG fusion, or a particular subset of, as being a marker of aggressive outcome,9–11 more recent studies seem to downplay its role as a predictor of aggressive behavior.13–17 The aim of the current study was to evaluate TMPRSS2–ERG fusion status as a prognosticator for recurrence in a nested case–control study in a prostate-specific antigen (PSA) era cohort of men who underwent radical prostatectomy at our institution for clinically localized prostatic adenocarcinoma.
Materials and methods
The current study was approved by our Institutional Review Board.
Study Population and Nested Case–Control Design
We developed a case–control study nested in the cohort of 4860 men who underwent radical retropubic prostatectomy for clinically localized prostate cancer at The Johns Hopkins Medical Institutions between 1993 and 2004 and who had not had hormonal or radiation therapy before radical prostatectomy or as adjuvant therapy before recurrence. 18 The study was designed to efficiently evaluate prognostic and risk factors for recurrence after radical prostatectomy. Cases were 524 men who experienced biochemical recurrence (serum PSA ≥0.2 ng/ml), metastasis, or prostate cancer death after surgery. For each case, we used incidence density sampling to select a control who had not experienced recurrence by the date of the case’s recurrence and who was matched on age, race, pathological stage, and Gleason’s sum.19 In this nested design, a man could be initially sampled as a control and later be sampled as a case once he recurred. Controls who remained at risk for recurrence were eligible to be sampled more than once. The latter method of control sampling makes the odds ratio estimate an unbiased estimate of the hazard ratio that would have been obtained if the entire cohort had been studied. Sampling controls allowed us to test a smaller number of total men than if we had used the entire cohort making for a more time- and cost-efficient approach. Other clinicopathological data were available for these men, including preoperative PSA, clinical stage, and Gleason’s sum.
Tissues and Tissue Microarrays
A set of 16 tissue microarrays were constructed for the 524 matched cases and controls. Matched pairs were placed on the same tissue microarray, so that a subset of these could be used depending on sample size calculations. Paired prostate cancer and noncancer tissues were spotted (0.6mm) in triplicate from each radical prostatectomy specimen as described previously by Kononen et al.20 In specimens with multifocal tumors, only the dominant tumor (highest Gleason’s sum and usually the largest) was sampled. In all, 10 of the 16 available tissue microarrays were used for the current study based on a priori power calculations; 2521 observations for TMPRSS2–ERG fusions were available for a total of 631 cases and controls. There were 3470 fusion observations for 631 men, of which 1977 were for carcinoma tissues. Of these, 990 fusion observations were for 302 cases and 987 were for 191 controls. Upon excluding technically inadequate tissue microarray spots, 172 matched sets with complete data were available for the analysis.
Evaluation of TMPRSS2–ERG Fusion Status using Interphase ERG Break-Apart FISH Assay
Fluorescence in situ hybridization (FISH) analysis was performed using dual-color interphase breakapart probes for the 5′ and 3′ regions of the ERG gene as detailed previously.14,15 In brief, 4-μm paraffin-embedded tissue microarray sections were baked at 56°C for 2 h, and then deparaffinized and rehydrated using xylene and graded ethanol, respectively. Tissue microarray sections were pretreated using paraffin pretreatment reagent kit III (Abbott Molecular Inc., IL). BAC FISH probes used were SpectrumGreen d-UTP direct-labeled BAC RP11- 95I21 for 5′ERG, and SpectrumOrange d-UTP directlabeled BAC RP11-476D17 for 3′ERG (Nick transKit, Vysis, Abbott Park, IL). Tissue microarray and BAC FISH probes were co-denatured at 94°C for 5 min and hybridized overnight at 37°C in a humid chamber (StatSpin ThermoBrite, IRIS Inc., MA).
Detecting ERG gene rearrangement using breakapart probes provides indirect evidence for the occurrence of TMPRSS2–ERG fusions. FISH interpretation was performed by three urologic pathologists (AT, RA, and GJN). Tissue microarray sections were scored using a 100 × oil immersion lens on an Olympus BX-70 fluorescence microscope (Olympus, Center Valley, PA) equipped with appropriate filters. For photomicrographs, images were captured using a Nikon 50i epifluorescence microscope equipped with X-Cite series 120 illuminator (EXFO Photonics Solutions Inc., Ontario, Canada) and a 100 ×/1.4 NA oil immersion Neofluar lens. Fluorescence excitation/emission filters were as follows: Cy3 excitation, 546 nm/10nm BP; emission, 578nm LP (Carl Zeiss Inc.); DAPI excitation, 330 nm; emission, 400nm using an XF02 fluorescence set (Omega Optical, Brattleboro, VT); Alexa Fluor 488 excitation, 475 nm; emission, 535nm using a combination of 475RDF40 and 535RDF45 filters (Omega Optical). Gray-scale images were captured for presentation using Nikon NIS-Elements software and an attached Photometrics CoolsnapEZ digital camera, pseudocolored and merged.
In each case, a minimum of 50 cells were scored for the presence/absence of TMPRSS2–ERG gene fusion through deletion or split. Digitally scanned adjacent hematoxylin and eosin sections were available for side-by-side comparison with the FISH image to localize tumor cells. Gleason’s grade was confirmed in each TMA spot. Paired benign prostatic epithelium was also scored as a negative control.
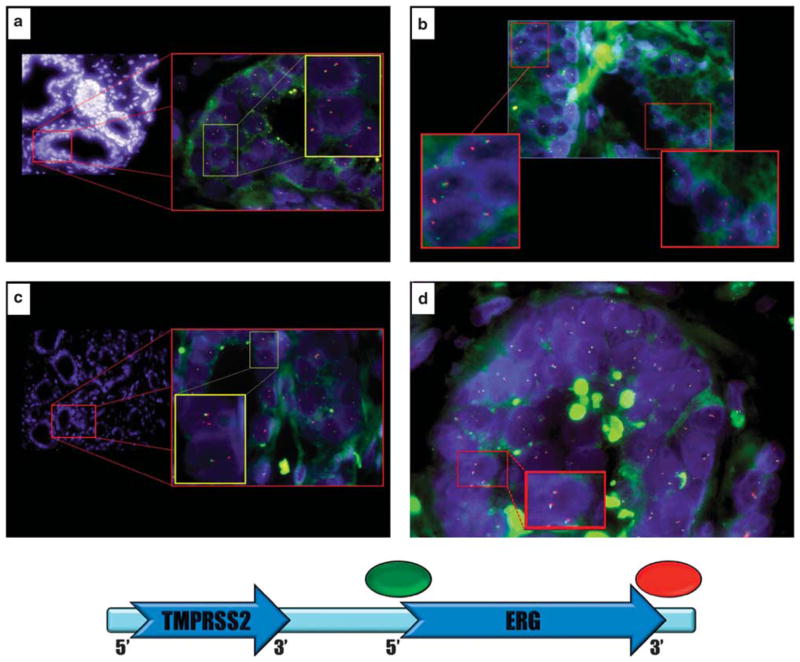
Each tissue microarray spot was assessed for TMPRSS2–ERG fusion as described previously by Attard et al11 with the following minor modifications: (1) class negative for fusion: a nucleus with two pairs of juxtaposed red and green signals forming yellow signals indicating the absence of ERG fusion (Figure 1a); (2) class ERG signal split: a nucleus with one juxtaposed red–green signal pair of the non-rearranged ERG allele and additional separate single red and single green signals of rearranged ERG allele (break-apart) reflecting a TMPRSS2–ERG fusion through split (Figure 1b); (3) class ERG deletion: a nucleus with one juxtaposed red–green signal pair for the non-rearranged allele and a single red signal of a rearranged allele indicating deletion of the telomeric (green) ERG probe region (Figure 1c).
Figure 1.
Detection of TMPRSS2–ERG fusion events by FISH. (a) No fusion events: Two intact ERG alleles are seen in the nuclei of benign prostatic epithelial cells. Two sets of juxtaposed red and green signals with occasional yellow overlap are noted in each nucleus (yellow box). (b) TMPRSS2–ERG fusion by split: One intact ERG allele and a second rearranged ERG allele in prostate adenocarcinoma (right box) and adjacent high-grade PIN epithelial cell (left box). The rearranged ERG alleles show split of the red and green signals indicative of an ERG fusion by translocation. (c) TMPRSS2–ERG fusion by deletion: One intact ERG allele and a second rearranged ERG allele in prostate adenocarcinoma. The rearranged ERG alleles show the absence of a green signal with a remaining red signal seen indicative of ERG fusion through deletion. (d) ERG gene copy number gain without fusion: increased ERG gene copy number in prostate adenocarcinoma (red box). Three sets of juxtaposed red–green signals are seen compared with background nuclei each showing only two sets of juxtaposed red–green signals.
A fusion was considered to be present when a minimum of 10% of the cells counted contained a split or a minimum of 20% of the cells contained a deletion in a given spot. The latter stringent 20% cutoff point for deletion was based on evaluation of a set of 225 consecutive benign FISH-labeled nuclei from consecutive tissue microarray spots where a truncation rate of up to 15% was noted for either red or green signals. A tumor was considered fusion positive if any of its representative spots met the above cutoffs. Analyses were also repeated using the same cutoffs applied to the sum of positive nuclei in a given tumor combining all its representative tissue microarray spots. In addition, to evaluate the potential effect of ‘dosage’ of a given fusion type, analysis was also performed using the number of spots that were positive for fusion in each tumor (extent of fusion-positive tissue microarray spots) and using the calculated ratio of fusion-positive spots per total number of analyzed spots per each case (ratio of fusion-positive tissue microarray spots).
A spot with single fusion was classified as harboring a deletion or a split event. Spots with duplicated deletion or split-type fusions were classified as 2 + deletion or 2 + split, respectively. Tumors with two distinct sub-populations of cells with different types of fusion meeting the above cutoffs were assigned to more than one fusion class. In addition, the presence of a copy number gain of a non-rearranged ERG gene (>2 copies; presumably due to chromosome 21 polysomy without fusion) was simultaneously assessed in all evaluated nuclei in a given tissue microarray spot. Spots showing copy number gain of an intact ERG (>2 copies) were designated as ‘ERG gene copy number gain without fusion’ (Figure 1d). Tissue microarray spots with ERG gene copy number gain and a second allele showing fusion were designated as either ‘ERG gene copy number gain with single split event’ or ‘ERG gene copy number gain with single deletion event’. A tissue microarray spot was deemed technically inadequate for scoring if it lacked a diagnostic target tissue or was of weak non-interpretable probe signal. Spots with overlapping nuclei preventing accurate FISH assessment were also considered technically inadequate.
Statistical Analysis
Demographic, clinicopathological, and TMPRSS2– ERG data were analyzed using Wilcoxon’s sign rank test, paired t-test, and McNemar’s test. We calculated odds ratios of recurrence and 95% confidence intervals by TMPRSS2–ERG fusion status using conditional logistic regression taking into account matching factors of age, race, pathologic stage, and Gleason’s sum. We estimated the association between fusion status and recurrence for tissue microarray spots with cancer. We modeled TMPRSS2–ERG fusion status using the following eight categories: any fusion, fusion due to deletion event, fusion due to split event, duplicated fusion due to 2 + deletion events, duplicated fusion due to 2 + split events, ERG gene copy number gain with single deletion event, ERG gene copy number gain with single split event, and ERG gene copy number gain without fusion. Furthermore, we repeated the statistical analysis for all eight categories based on several permutations of classifying fusion status in a given radical prostatectomy case: each case classified based on fusion status of any individually assessed tumor tissue microarray spot; each case classified based on the combined assessment of all evaluated tumor cells in all represented tissue microarray spots; number of tissue microarray spots per case showing fusion and ratio of number of fusion-positive spot(s) divided per total number of tumor spots per case. All analyses were performed using SAS version 9.1 (SAS Institute Inc., Cary, NC). Statistical tests were two sided and P-values <0.05 were considered to be statistically significant.
Results
Patient Demographics and Clinicopathological Findings
Patient demographics and clinical data of recurrence cases and matched controls are summarized in Table 1. As expected, patients in the case group had higher mean preoperative PSA (P = 0.05) and were more likely to have positive surgical margins (P = 0.01). The median recurrence time was 2 years after surgery.
Table 1.
Demographic and clinicopathological characteristics of 172 prostate cancer recurrence cases and 172 matched controlsa nested in the Johns Hopkins radical prostatectomy cohort
| Case | Control | P-valueb | |
|---|---|---|---|
| Mean age at surgery (years)±s.d. | 59.5±6.8 | 59.7±6.3 | Matched |
| Race (%) | |||
| White | 79.7 | 83.7 | Matched |
| Black | 12.2 | 9.3 | |
| Other race/ethnicity | 8.1 | 7 | |
| Preoperative PSA concentration (ng/ml) | |||
| Mean±s.d. | 12.2±11.0 | 10.2±6.7 | 0.05 |
| Median (range) | 8.8 (0.10–79.1) | 8.5 (0.3–35.6) | 0.17 |
| <4 | 8.7 | 9.9 | |
| 4–10 | 48.8 | 54.7 | |
| >10 | 42.4 | 35.5 | |
| Pathological Gleason’s sum (%) | |||
| Mean±s.d. | 7.2±0.8 | 7.1±0.8 | Matched |
| ≤6 | 16.3 | 18.6 | |
| 3+4 | 38.4 | 45.9 | |
| 4+3 | 22.7 | 15.1 | |
| >7 | 22.7 | 20.4 | |
| Pathological stage (%) | |||
| pTNM | Matched | ||
| pT2N0 | 16.9 | 17.4 | |
| pT3aN0 | 12.2 | 18.6 | |
| pT3bN0 | 40.1 | 33.7 | |
| pT4N0 | 19.2 | 18.6 | |
| pT3bN+ | 5.8 | 4.7 | |
| pT4N+ | 5.2 | 5.8 | |
| Surgical margin positive (%) | 36.6 | 24.4 | 0.01 |
| Follow-up time (years) | |||
| Mean±s.d. | 2.5±2.0 | 5.4±2.5 | <0.0001 |
| Median (range) | 2 (1–9) | 5 (2–11) | <0.0001 |
Controls were matched to cases on age, race, pathological stage, and Gleason’s sum.
P-values were obtained from paired statistical tests.
Prevalence of TMPRSS2–ERG Fusion
TMPRSS2–ERG fusion and ERG gene copy number gain were not detected in paired benign prostate glandular tissue spots in either recurrence cases or controls. Although the overall frequency of TMPRSS2–ERG fusion assessed in cancer spots was slightly lower in cases than in controls (50 vs 57%), the difference was not statistically significant (P = 0.20; Table 2). We observed no statistically significant difference between prostate carcinoma cases and controls in the prevalence of split, combined deletion and split, or the presence of ERG gene copy number gain with ERG fusion. The prevalence of deletion events appeared to be lower in cases than in controls (P = 0.08). In recurrence cases who had TMPRSS2–ERG fusion, the prevalence of deletion and split fusion events were 35.5 and 25%, respectively; the latter was almost always of combined deletion and split classes due to the presence of two sub-populations of cells each showing one fusion class (see Table 2). The incidence of ERG gene copy number gain without fusion was statistically significantly higher in cases than in controls (28 vs 16%; P = 0.01).
Table 2.
TMPRSS2–ERG fusion status in prostate carcinoma tissues from 172 recurrence cases and 172 matched controlsa nested in the Johns Hopkins radical prostatectomy cohort
| Fusion status (%) | Case | Control | P-valueb |
|---|---|---|---|
| Any fusion event | 50.0 | 57.0 | 0.20 |
| Single fusion due to deletion event | 35.5 | 44.8 | 0.08 |
| Single fusion due to split event | 25.0 | 30.2 | 0.31 |
| Duplicated fusion events | 11.6 | 11.0 | 0.73 |
| ERG gene copy number gain with single fusion event | 20.4 | 19.2 | 0.48 |
| Any deletion event only | 18.0 | 19.8 | 0.77 |
| Any split event only | 0.6 | 0.6 | 1.00 |
| Deletion and split events | 25.6 | 30.8 | 0.31 |
| ERG gene copy number gain without fusion events | 27.9 | 16.3 | 0.01 |
Controls were matched to cases on age, race, pathological stage, and Gleason’s sum.
P-values were obtained from paired statistical tests.
Association between TMPRSS2–ERG Fusion Status and Prostate Carcinoma Recurrence
When classifying patients as fusion positive if the criteria for positivity (≥10% of cells with split or ≥20% with deletion events) were met in at least one tissue microarray spot, the presence of single TMPRSS2–ERG fusions or duplicated fusions with or without ERG gene copy number gain was not associated with risk of recurrence after radical prostatectomy, with the possible exception of an inverse association for single split fusion (Table 3). This inverse association was also statistically significant when classifying patients as fusion positive only when at least 10% of cancer cells across all tissue microarray spots contained a particular fusion (odds ratio 0.63, 95% confidence interval 0.40–0.99, P=0.04; Table 3).
Table 3.
Association between ERG fusion status and risk of prostate cancer recurrence following radical prostatectomy for clinically localized prostate cancer, 172 cases and 172 matched controls nested in the Johns Hopkins prostatectomy cohorta
| Fusion status | Criterion for positivityb |
|||
|---|---|---|---|---|
| In at least one tissue microarray spot
|
Across all tissue microarray spots assessed
|
|||
| Odds ratio (95% confidence intervals) | P-value | Odds ratio (95% confidence intervals) | P-value | |
| Any fusion event | 0.72 (0.45–1.14) | 0.16 | 0.73 (0.46–1.15) | 0.17 |
| Fusion due to deletion event | 0.75 (0.46–1.24) | 0.26 | 0.68 (0.37–1.21) | 0.18 |
| Fusion due to split event | 0.64 (0.40–1.03) | 0.07 | 0.63 (0.40–0.99) | 0.04 |
| Duplicated fusion due to 2+ split events | 1.00 (0.29–3.45) | 1.00 | 1.00 (0.14–7.10) | 1.00 |
| Duplicated fusion due to 2+ deletion events | 1.08 (0.51–2.29) | 0.85 | 1.67 (0.61–4.59) | 0.32 |
| ERG gene copy number gain with single split event | 1.06 (0.54–2.10) | 0.86 | 0.30 (0.08–1.09) | 0.07 |
| ERG gene copy number gain with single deletion event | 1.07 (0.53–2.16) | 0.86 | 1.25 (0.49–3.17) | 0.64 |
| ERG gene copy number without fusion events | 2.00 (1.17–3.42) | 0.01 | 2.70 (1.31–5.58) | 0.01 |
Estimated from conditional logistic regression taking into account the matching factors age, race, pathological stage, and Gleason’s sum.
In all, ≥10% cells were positive for a split or ≥20% for a deletion.
In contrast to the TMPRSS2–ERG fusion, the presence of ERG gene copy number gain without fusion was associated with an increased risk of recurrence (odds ratio 2.00, 95% confidence interval 1.17–3.47, P=0.01); further adjusting for pre-surgery PSA and calendar year of surgery slightly attenuated this association (odds ratio 1.81, 95% confidence interval 0.99–3.31). ERG gene copy number gain without fusion remained significantly associated with prostate cancer recurrence upon repeating the analyses based on the combined assessment of all evaluated tumor cells in all tissue microarray spots per radical prostatectomy cases (Table 3), the number of positive spots, and the ratio of number of positive spots to the number of spots assessed (Table 4).
Table 4.
Association between increasing ratio of the number of tissue microarray spots positive for ERG fusion with the number of spots assessed and risk of prostate cancer recurrence following radical prostatectomy for clinically localized prostate cancer, 172 cases and 172 matched controls nested in the Johns Hopkins prostatectomy cohort
| Fusion status | Odds ratio (95% confidence intervals)a | P-value |
|---|---|---|
| Any fusion event | 0.62 (0.39–1.00) | 0.05 |
| Fusion due to deletion event | 0.68 (0.38–1.20) | 0.18 |
| Fusion due to split event | 0.64 (0.39–1.03) | 0.07 |
| Duplicated fusion due to 2+ split events | 0.99 (0.21–4.61) | 0.99 |
| Duplicated fusion due to 2+ deletion events | 1.12 (0.49–2.56) | 0.80 |
| ERG gene copy number gain with single split event | 0.96 (0.39–2.38) | 0.93 |
| ERG gene copy number gain with single deletion event | 1.07 (0.44–2.62) | 0.88 |
| ERG gene copy number gain without fusion events | 2.34 (1.24–4.40) | 0.01 |
Per 1-Unit increase in the ratio of tissue spots positive for ERG fusion to number of spots assessed and estimated from conditional logistic regression taking into account the matching factors age, race, pathological stage, and Gleason’s sum.
We found no statistically significant associations between the presence of TMPRSS2–ERG fusion and the risk of recurrence in White men (137 pairs), younger (<60 years, 79 pairs) or older (≥60 years, 88 pairs) men, later-stage (N1 or T3b, 52 pairs) or early-stage (T2 or T3a and N0, 119 pairs) disease, Gleason’s sum disease ≥4 + 3 (42 pairs) vs ≤3 + 4 (75 pairs), men with poorer (N1 or T3b or ≥4 + 3, 80 pairs) and better (T2 or T3a and <4 + 3, 58 pairs) prognosis disease, men with negative surgical margins (89 pairs), and men who recurred <2 years (48 pairs) and ≥2 years (120 pairs) after radical prostatectomy (data not shown). ERG gene copy number gain without fusion was associated with increased risk for recurrence in White men, older but not younger men, men with early- but not laterstage disease, and men with negative surgical margins (Table 5).
Table 5.
Association of ERG gene copy number gain without fusion with prostatic carcinoma recurrence following radical prostatectomy in prognostic subgroups
| Odds ratio (95% confidence intervals) | |
|---|---|
| White men | 1.8 (1.02–3.43) |
| Older men | 2.4 (1.14–5.01) |
| Early stage prostate carcinoma | 2.9 (1.46–5.77) |
| Negative surgical margins | 3.4 (1.47–7.95) |
| Recurrence ≥2 years | 2.4 (1.21–4.94) |
Assessment of Chromosome 21 Numerical Alterations
Chromosome 21 copy number alterations were evaluated in a subset of two tissue microarrays using a second dual-color probe set targeting centromeric (RP11-22D1) and telomeric (RP11- 35C4) regions on the long arm of chromosome 21 separated by >27 megabases. Identical FISH processing and scoring parameters to those used with the first probe set for the 5′ and 3′ regions of ERG gene were used in the 60 evaluable tumors. Using the second probe set targeting centromeric and telomeric regions of the long arm of chromosome 21, we found evidence of chromosome 21 long-arm gains in 24 of 26 (93%) tumors that were originally classified as ERG gene copy number gain without fusion (Figure 2).
Figure 2.
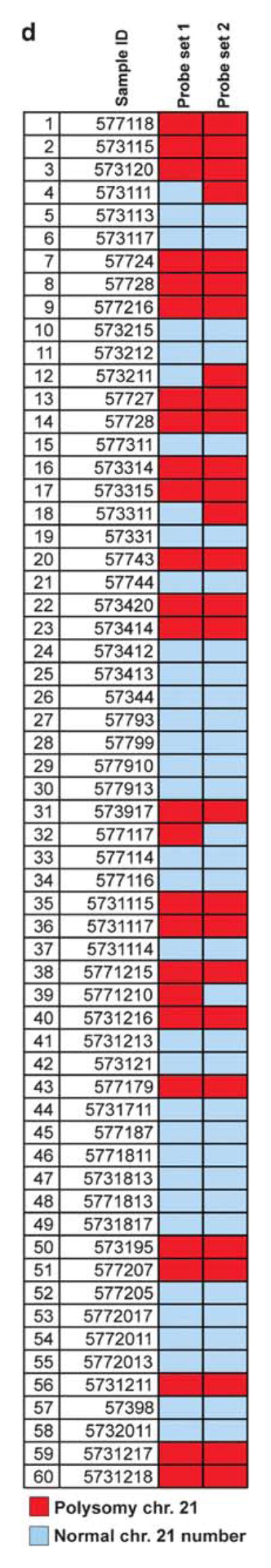
Evaluation of chromosome 21 numerical alterations. (a) Overview of BAC clones used in the study. Probe set 1 flanking the ERG genomic locus (RP11-476D17, RP11-95I21) is the set used to determine ERG gene rearrangement status in all cases in the current study. Probe set 2 was only used in a subset of cases to further assess chromosome 21 numerical alterations by targeting centromeric (RP11-22D1) and telomeric (RP11-35C4) regions on the long arm of chromosome 21. (b) Prostate adenocarcinoma showing no evidence of numerical chromosome 21 alteration using probe set 2. (c) Prostate adenocarcinoma showing chromosome 21 long-arm copy number gains as indicated by the presence of more than two red and/or more than two green signals per nucleus (white arrows). (d) Comparison of ERG gene copy number gain detected using probe set 1 and chromosome 21 numerical alteration assessed using probe set 2 in a subset of two tissue microarrays (60 cases) from our study. A high concordance rate (24/26; 93%) is found between the two sets of probes supporting that ERG gene copy number gain are a reflection of chromosome 21 copy number gains.
Discussion
Although earlier studies linked the presence of TMPRSS2–ERG fusions or a subset of fusion classes with a more aggressive biological behavior of prostate carcinomas,5–7,9–12,21,22 recent large cohort studies did not observe such a prognostic role.13,23–28 In this context, our recent report of a high incidence of TMPRSS2–ERG fusions in minute prostatic adenocarcinoma, comparable with that of nonminute prostatic adenocarcinoma, seems to lend support to the lack of prognostic role of fusion given the lack of clinical significance of minute tumors.15 In our current study conducted among men who underwent radical prostatectomy for clinically localized prostatic adenocarcinoma, TMPRSS2–ERG fusions were not associated with recurrence, a finding that contradicts previously reported associations between duplicated ERG fusions and aggressive outcome.11,13 We could not rule out a possible inverse association between TMPRSS2– ERG fusion by a single split event and risk of recurrence. Our results were consistent across methods of classifying the men as fusion positive.
The conflicting evidence on the prognostic significance of TMPRSS2–ERG fusion is potentially due to methodological differences and/or differences in examined cohorts. Some of the studies pointing to the association with aggressive behavior were performed on conservatively managed, population-based, watchful waiting cohorts,9,11 in contrast to the more recent studies assessing large, PSA-screened populations treated by radical prostatectomy for clinically localized prostate carcinoma. 13 Watchful waiting cohorts were often diagnosed by transurethral resection with predominantly transition-zone cancers9 as opposed to the more recent studies of primarily peripheral-zone cancer diagnosed on needle biopsy. Recent series have shown that in transition-zone prostate cancers, ERG fusions are less prevalent (12–13% of all cases) than in tumors originating in the peripheral zone,29–31 suggesting that the underlying molecular abnormalities might be different between these subsets of prostatic adenocarcinomas. Nevertheless, in our study, TMRSS2–ERG fusions were present in 50 and 57% of recurrence cases and controls, respectively, which is consistent with the previous percentages (40–60%) reported in surgical cohort studies assessing ERG alterations3,5,7,10,13,32 using similar FISH break-apart methodology.5
In our study population, ERG fusion was more likely to be the result of deletion, either homogeneously throughout a given tumor or in association with a sub-population of tumor cells harboring a split event. The preponderance of deletion events is in line with previous observations.5,24,32 Intronic loss of genomic DNA between ERG and TMPRSS2 on chromosome 21q22.2–3 appears to be a main mechanism of TMPRSS2–ERG fusion. Previous studies have shown the presence of TMPRSS2– ERG fusion to be homogeneous in a given tumor focus5 but heterogeneous in the context of multiple cancer foci.33 Although the identification in the present series of combined deletion and split fusions events within the same focus contradicts these earlier results, our findings are in agreement with those published by Clark et al34,35 who have demonstrated different categories of ERG gene alteration to be present either together in a single cancerous region or within separate foci of cancer in the same prostate slice. These results further support the notion that TMPRSS2–ERG gene fusions may arise independently in different regions of a single prostate or even within the same tumor focus.
Although TMPRSS2–ERG fusion was not associated with recurrence in this study, ERG gene copy number gain without fusion was associated with twice the risk of recurrence. Our findings are consistent with a recent report by Gopalan et al.13 This low-level increase of ERG gene copy number is presumably the result of tumor aneuploidy status, potentially leading to chromosome 21 numerical gains. The latter is further supported by our reassessment of chromosome 21 numerical alterations using a second probe set targeting telomeric and centromeric regions at the long arm of chromosome 21 ,suggesting that chromosome 21 polysomy might be responsible for the detected ERG gene copy number gain in this group. ERG gene copy number gain was associated with the presence of chromosome 21 long-arm gains in 93% of analyzed cases. Seen in such context, the association of ERG gene copy number gain without fusion with higher likelihood of cancer progression is not surprising given the previous evidence supporting aneuploidy as a negative prognosticator in prostatic adenocarcinomas. 36–38 The lack of association between ERG gene copy number gain in the presence of a second allele with ERG fusion and cancer progression is counterintuitive and requires further investigation.
In addition to the strong study design, which takes into account clinical features, pathological stage, and Gleason’s sum, the detailed analysis of the different fusion classes with multiple approaches to assigning positive fusion status for a given man, represent some of the strengths of our current study. On the other hand, the lack of confirmatory RT-PCR-based analysis or additional molecular methods to further characterize the fusion variants at the exonic level could be viewed as a potential weakness. In this regard, given the previously cited potential prognostic role for specific exonic variant of TMRSS2–ERG deletions such as the T1/E4 variant,27 additional molecular analysis could be warranted in our group of prostate cancer patients. Another plausible limitation of the present series is the effect that ERG fusion status heterogeneity might have on the results considering that, when facing multicentric prostate carcinomas, we only sampled the dominant tumor showing the highest Gleason’s sum. Indeed, several studies have shown that up to 41% of multicentric TMPRSS2–ERG fusion-positive prostatic adenocarcinomas exhibit heterogeneity in different foci of the same gland.33,39,40 However, this study was not designed to evaluate the heterogeneity of TMPRSS2–ERG fusion status within the same tumor but to determine the association between its presence and outcome. Moreover, the rationale for sampling only the dominant tumor is supported by the argument that these high-grade areas are the most likely to dictate prognosis in cases with multicentric lesions.
In summary, our study is the first to evaluate the prognostic role of TMPRSS2–ERG fusion in a large nested case–control study of men who underwent radical prostatectomy for clinically localized prostatic adenocarcinoma and that took into account clinicopathological prognostic parameters. We found a comparable prevalence of TMPRSS2–ERG fusion in men who did and did not recur, further supporting recent mounting evidence for a lack of prognostic significance of the ERG gene fusion by FISH. Copy number increase of ERG gene, likely as the result of aneuploidy, was strongly predictive of prostate cancer recurrence in our study.
Acknowledgments
We acknowledge the contributions of Helen L Fedor and Marcella Southerland from the Brady Urological Research Institute Prostate Specimen Repository at the Johns Hopkins School of Medicine for the generation of the tissue microarrays for this nested case–control study. This study was supported in part by grants from the Patrick C Walsh of the Brady Urological Institute, David H Koch Prostate Cancer Fund, the Patana Fund of the Brady Urological Institute, the Prostate SPORE Pathology Core (P50 CA58236), and a DOD grant (DAMD 17-03-0273).
Footnotes
This study was presented in part at the 2009 United States and Canadian Academy of Pathology (USCAP) annual meeting, Boston, USA.
Disclosure/conflict of interest
The authors declare no conflict of interest.
References
- 1.Schroder FH, Hugosson J, Roobol MJ, et al. Screening and prostate-cancer mortality in a randomized European study. N Engl J Med. 2009;360:1320–1328. doi: 10.1056/NEJMoa0810084. [DOI] [PubMed] [Google Scholar]
- 2.Andriole GL, Crawford ED, Grubb RL, III, et al. Mortality results from a randomized prostate-cancer screening trial. N Engl J Med. 2009;360:1310–1319. doi: 10.1056/NEJMoa0810696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang J, Cai Y, Yu W, et al. Pleiotropic biological activities of alternatively spliced TMPRSS2/ERG fusion gene transcripts. Cancer Res. 2008;68:8516–8524. doi: 10.1158/0008-5472.CAN-08-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cerveira N, Ribeiro FR, Peixoto A, et al. TMPRSS2- ERG gene fusion causing ERG overexpression precedes chromosome copy number changes in prostate carcinomas and paired HGPIN lesions. Neoplasia. 2006;8:826–832. doi: 10.1593/neo.06427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perner S, Mosquera JM, Demichelis F, et al. TMPRSS2- ERG fusion prostate cancer: an early molecular event associated with invasion. Am J Surg Pathol. 2007;31:882–888. doi: 10.1097/01.pas.0000213424.38503.aa. [DOI] [PubMed] [Google Scholar]
- 6.Rajput AB, Miller MA, De Luca A, et al. Frequency of the TMPRSS2:ERG gene fusion is increased in moderate to poorly differentiated prostate cancers. J Clin Pathol. 2007;60:1238–1243. doi: 10.1136/jcp.2006.043810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mehra R, Tomlins SA, Shen R, et al. Comprehensive assessment of TMPRSS2 and ETS family gene aberrations in clinically localized prostate cancer. Mod Pathol. 2007;20:538–544. doi: 10.1038/modpathol.3800769. [DOI] [PubMed] [Google Scholar]
- 8.Lapointe J, Kim YH, Miller MA, et al. A variant TMPRSS2 isoform and ERG fusion product in prostate cancer with implications for molecular diagnosis. Mod Pathol. 2007;20:467–473. doi: 10.1038/modpathol.3800759. [DOI] [PubMed] [Google Scholar]
- 9.Demichelis F, Fall K, Perner S, et al. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. 2007;26:4596–4599. doi: 10.1038/sj.onc.1210237. [DOI] [PubMed] [Google Scholar]
- 10.Nam RK, Sugar L, Wang Z, et al. Expression of TMPRSS2:ERG gene fusion in prostate cancer cells is an important prognostic factor for cancer progression. Cancer Biol Ther. 2007;6:40–45. doi: 10.4161/cbt.6.1.3489. [DOI] [PubMed] [Google Scholar]
- 11.Attard G, Clark J, Ambroisine L, et al. Duplication of the fusion of TMPRSS2 to ERG sequences identifies fatal human prostate cancer. Oncogene. 2008;27:253–263. doi: 10.1038/sj.onc.1210640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheville JC, Karnes RJ, Therneau TM, et al. Gene panel model predictive of outcome in men at high-risk of systemic progression and death from prostate cancer after radical retropubic prostatectomy. J Clin Oncol. 2008;26:3930–3936. doi: 10.1200/JCO.2007.15.6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gopalan A, Leversha MA, Satagopan JM, et al. TMPRSS2-ERG gene fusion is not associated with outcome in patients treated by prostatectomy. Cancer Res. 2009;69:1400–1406. doi: 10.1158/0008-5472.CAN-08-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lotan TL, Toubaji A, Albadine R, et al. TMPRSS2-ERG gene fusions are infrequent in prostatic ductal adenocarcinomas. Mod Pathol. 2009;22:359–365. doi: 10.1038/modpathol.2008.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Albadine R, Latour M, Toubaji A, et al. TMPRSS2-ERG gene fusion status in minute (minimal) prostatic adenocarcinoma. Mod Pathol. 2009;22:1415–1422. doi: 10.1038/modpathol.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hermans KG, van Marion R, van Dekken H, et al. TMPRSS2:ERG fusion by translocation or interstitial deletion is highly relevant in androgen-dependent prostate cancer, but is bypassed in late-stage androgen receptor-negative prostate cancer. Cancer Res. 2006;66:10658–10663. doi: 10.1158/0008-5472.CAN-06-1871. [DOI] [PubMed] [Google Scholar]
- 17.Rubio-Briones J, Fernandez-Serra A, Calatrava A, et al. Clinical implications of TMPRSS2-ERG gene fusion expression in patients with prostate cancer treated with radical prostatectomy. J Urol. 2010;183:2054–2061. doi: 10.1016/j.juro.2009.12.096. [DOI] [PubMed] [Google Scholar]
- 18.Han M, Partin AW, Pound CR, et al. Long-term biochemical disease-free and cancer-specific survival following anatomic radical retropubic prostatectomy. The 15-year Johns Hopkins experience. Urol Clin North Am. 2001;28:555–565. doi: 10.1016/s0094-0143(05)70163-4. [DOI] [PubMed] [Google Scholar]
- 19.Wang MH, Shugart YY, Cole SR, et al. A simulation study of control sampling methods for nested casecontrol studies of genetic and molecular biomarkers and prostate cancer progression. Cancer Epidemiol Biomarkers Prev. 2009;18:706–711. doi: 10.1158/1055-9965.EPI-08-0839. [DOI] [PubMed] [Google Scholar]
- 20.Kononen J, Bubendorf L, Kallioniemi A, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 21.Nam RK, Sugar L, Yang W, et al. Expression of the TMPRSS2:ERG fusion gene predicts cancer recurrence after surgery for localised prostate cancer. Br J Cancer. 2007;97:1690–1695. doi: 10.1038/sj.bjc.6604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mehra R, Tomlins SA, Yu J, et al. Characterization of TMPRSS2-ETS gene aberrations in androgenindependent metastatic prostate cancer. Cancer Res. 2008;68:3584–3590. doi: 10.1158/0008-5472.CAN-07-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hermans KG, Boormans JL, Gasi D, et al. Overexpression of prostate-specific TMPRSS2(exon 0)-ERG fusion transcripts corresponds with favorable prognosis of prostate cancer. Clin Cancer Res. 2009;15:6398–6403. doi: 10.1158/1078-0432.CCR-09-1176. [DOI] [PubMed] [Google Scholar]
- 24.Yoshimoto M, Joshua AM, Chilton-Macneill S, et al. Three-color FISH analysis of TMPRSS2/ERG fusions in prostate cancer indicates that genomic microdeletion of chromosome 21 is associated with rearrangement. Neoplasia. 2006;8:465–469. doi: 10.1593/neo.06283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lapointe J, Li C, Higgins JP, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci USA. 2004;101:811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rouzier C, Haudebourg J, Carpentier X, et al. Detection of the TMPRSS2-ETS fusion gene in prostate carcinomas: retrospective analysis of 55 formalin-fixed and paraffin-embedded samples with clinical data. Cancer Genet Cytogenet. 2008;183:21–27. doi: 10.1016/j.cancergencyto.2008.01.021. [DOI] [PubMed] [Google Scholar]
- 27.Barwick BG, Abramovitz M, Kodani M, et al. Prostate cancer genes associated with TMPRSS2-ERG gene fusion and prognostic of biochemical recurrence in multiple cohorts. Br J Cancer. 2010;102:570–576. doi: 10.1038/sj.bjc.6605519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saramaki OR, Harjula AE, Martikainen PM, et al. TMPRSS2:ERG fusion identifies a subgroup of prostate cancers with a favorable prognosis. Clin Cancer Res. 2008;14:3395–3400. doi: 10.1158/1078-0432.CCR-07-2051. [DOI] [PubMed] [Google Scholar]
- 29.Bismar TA, Trpkov K. TMPRSS2-ERG gene fusion in transition zone prostate cancer. Mod Pathol. 2010;23:1040–1041. doi: 10.1038/modpathol.2010.89. [DOI] [PubMed] [Google Scholar]
- 30.Liu S, Yoshimoto M, Trpkov K, et al. Detection of ERG gene rearrangements and PTEN deletions in unsuspected prostate cancer of the transition zone. Cancer Biol Ther. 2011;11:562–566. doi: 10.4161/cbt.11.6.14376. [DOI] [PubMed] [Google Scholar]
- 31.Falzarano SM, Navas M, Simmerman K, et al. ERG rearrangement is present in a subset of transition zone prostatic tumors. Mod Pathol. 2010;23:1499–1506. doi: 10.1038/modpathol.2010.150. [DOI] [PubMed] [Google Scholar]
- 32.Tu JJ, Rohan S, Kao J, et al. Gene fusions between TMPRSS2 and ETS family genes in prostate cancer: frequency and transcript variant analysis by RT-PCR and FISH on paraffin-embedded tissues. Mod Pathol. 2007;20:921–928. doi: 10.1038/modpathol.3800903. [DOI] [PubMed] [Google Scholar]
- 33.Furusato B, Gao CL, Ravindranath L, et al. Mapping of TMPRSS2-ERG fusions in the context of multi-focal prostate cancer. Mod Pathol. 2008;21:67–75. doi: 10.1038/modpathol.3800981. [DOI] [PubMed] [Google Scholar]
- 34.Clark J, Attard G, Jhavar S, et al. Complex patterns of ETS gene alteration arise during cancer development in the human prostate. Oncogene. 2008;27:1993–2003. doi: 10.1038/sj.onc.1210843. [DOI] [PubMed] [Google Scholar]
- 35.Clark J, Merson S, Jhavar S, et al. Diversity of TMPRSS2-ERG fusion transcripts in the human prostate. Oncogene. 2007;26:2667–2673. doi: 10.1038/sj.onc.1210070. [DOI] [PubMed] [Google Scholar]
- 36.Epstein JI, Amin M, Boccon-Gibod L, et al. Prognostic factors and reporting of prostate carcinoma in radical prostatectomy and pelvic lymphadenectomy specimens. Scand J Urol Nephrol Suppl. 2005;216:34–63. doi: 10.1080/03008880510030932. [DOI] [PubMed] [Google Scholar]
- 37.Brown JA, Slezak JM, Lieber MM, et al. Fluorescence in situ hybridization aneuploidy as a predictor of clinical disease recurrence and prostate-specific antigen level 3 years after radical prostatectomy. Mayo Clin Proc. 1999;74:1214–1220. doi: 10.4065/74.12.1214. [DOI] [PubMed] [Google Scholar]
- 38.Lau WK, Blute ML, Bostwick DG, et al. Prognostic factors for survival of patients with pathological Gleason score 7 prostate cancer: differences in outcome between primary Gleason grades 3 and 4. J Urol. 2001;166:1692–1697. [PubMed] [Google Scholar]
- 39.Barry M, Perner S, Demichelis F, et al. TMPRSS2-ERG fusion heterogeneity in multifocal prostate cancer: clinical and biologic implications. Urology. 2007;70:630–633. doi: 10.1016/j.urology.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mehra R, Han B, Tomlins SA, et al. Heterogeneity of TMPRSS2 gene rearrangements in multifocal prostate adenocarcinoma: molecular evidence for an independent group of diseases. Cancer Res. 2007;67:7991–7995. doi: 10.1158/0008-5472.CAN-07-2043. [DOI] [PubMed] [Google Scholar]