Abstract
The Polycomb repressive complex 2 is a multimeric aggregate that mediates silencing of a broad range of genes, and is associated with important biological contexts such as stem cell maintenance and cancer progression. PRC2 mainly trimethylates lysine 27 of histone H3 and is composed of three essential core subunits: EZH2, EED and SUZ12. The Xenopus orthologs of PRC2 subunits Ezh2 and Eed have been described but Suz12 remained unidentified. Here, we report the cloning of the Xenopus Suz12, and determine its spatiotemporal expression during development. Xsuz12 transcript is provided maternally and continues to be expressed throughout development, particularly in the anterior part of the developing central nervous system. Importantly, comparative analysis of the expression of the PRC2 subunits Xez, Xeed and Xrbbp4 indicates that their expression largely coincides with Xsuz12 in the nervous system, suggesting that PRC2 may have unexplored functions in the development of the frog central nervous system.
Keywords: Polycomb, PRC2, trimethylation, histone, SUZ12, Xenopus laevis, repression, neural development, RBBP4/7, XEZ, XEED
INTRODUCTION
The polycomb group (PcG) genes are highly conserved factors that were initially identified in Drosophila as repressors of Hox genes during developmental patterning (Sparmann and Lohuizen, 2006). Studies have demonstrated that PcG-mediated repression is not restricted to Hox genes, and has been implicated in the biology of embryonic stem cells and cancer (Lee et al, 2006; Schwartz et al, 2006; Boyer et al, 2006). How PcGs repress gene expression is not well understood but evidence suggests that they work in complexes that can directly interfere with transcription initiation or antagonize the function of the chromatin remodeling complex SWI/SNF (Francis and Kingston, 2001; Dellino et al, 2004). Three complexes with distinct biochemical and functional properties, termed Polycomb Repressive Complexes (PRCs), have been purified thus far: PRC1, PRC2 and PHORC (Sparmann and Lohuizen, 2006; Klymenko et al, 2006; Shao, et al, 1999; Saurin et al, 2001).
The polycomb repressive complex PRC2 consists of three core subunits: Suz12 (the mammalian ortholog of the Drosophila Suppressor of Zest Su(z)12), Ehz1/2 (the mammalian orthologs of the Drosophila Enhancer of zeste E(z)) and Eed (the mammalian ortholog of the Drosophila Extra Sex Combs Esc) (Kuzmichev et al, 2002). EZH2 bears a histone methyltransferase (HMTase) activity through its SET domain that catalyzes the trimethylation of H3K27 in vitro as well as in vivo (Kuzmichev et al, 2002; Muller et al, 2002; Su et al, 2003). H3K27 is a histone mark that is associated with the maintenance of gene repression in multiple developmental processes (Boyer et al, 2006; Schuettengruber et al, 2007; Barski et al, 2007). EZH2 can also interact with DNA methyltransferases thus directly controlling DNA methylation (Vire et al, 2006). The exact functions of SUZ12 and EED are largely unknown, but both proteins are required for the EZH1/2 methyltransferase activity (Pasini et al, 2004; Kuzmichev et al, 2005; Ketel et al, 2005; Margueron et al, 2008; Shen et al, 2008). EZH2, EED and SUZ12 constitute the minimal PRC2 subunits required for HMTase activity and subsequent initiation of gene repression (Sparmann and van Lohuizen, 2006; Ketel et al, 2005). PRC2 also contains other subunits that have been shown to be important for its function. For instance, the Drosophila NURF55 and its vertebrate ortholog RbAP48 (also named RBBP4) enhances the catalytic activity of PRC2 and is required for its nucleosome binding (Cao and Zhang, 2004; Nekrasov et al, 2005).
The roles of SUZ12 in development have been poorly explored in part because the mouse knockout of Suz12 suffers severe developmental abnormalities and dies shortly after gastrulation (Pasini et al, 2004). One of the emerging roles of Suz12 is its involvement in the mammalian embryonic stem (ES) cell pluripotency. In ES cells SUZ12 mediates repression of a large set of developmental genes that are implicated in differentiation and cell fate decisions (Lee et al, 2006, Boyer et al, 2006). In agreement with these findings, ES cells derived from mouse Suz12 knockout display a loss of H3K27me3 mark and upregulation of differentiation-specific genes, impairing ES cell differentiation in culture (Pasini et al, 2007).
To study the possible roles of PRC2 in X. laevis development it is necessary to characterize all subunits that are important for the complex to function properly. To date only two of the core subunits (Xez and Xeed) have been cloned in X. laevis and their expression patterns have been only partially explored. Thus, cloning the Suz12 gene and comparing its expression pattern to other PRC2 subunits in Xenopus is essential for understanding how this complex may function in proliferation, tissue specification and/or subsequent differentiation throughout the frog development. In this study we report the cloning of the X. laevis ortholog of Suz12 (named Xsuz12 hereafter) and characterize its spatiotemporal expression during development.
RESULTS AND DISCUSSION
Cloning Xsuz12
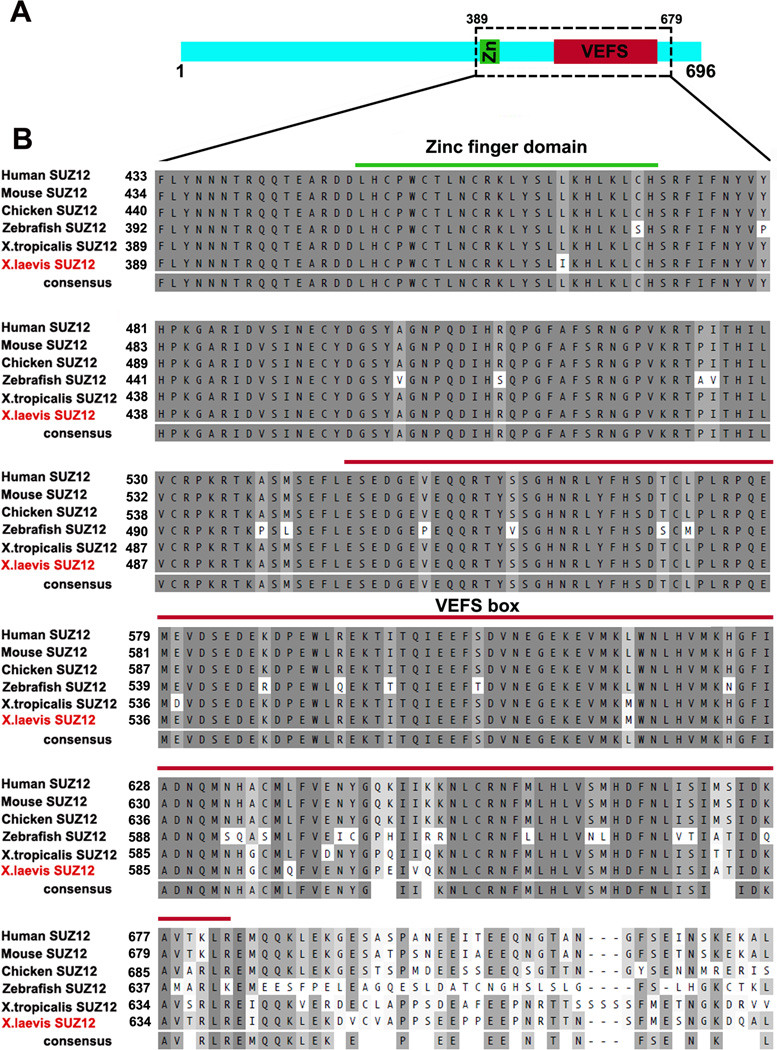
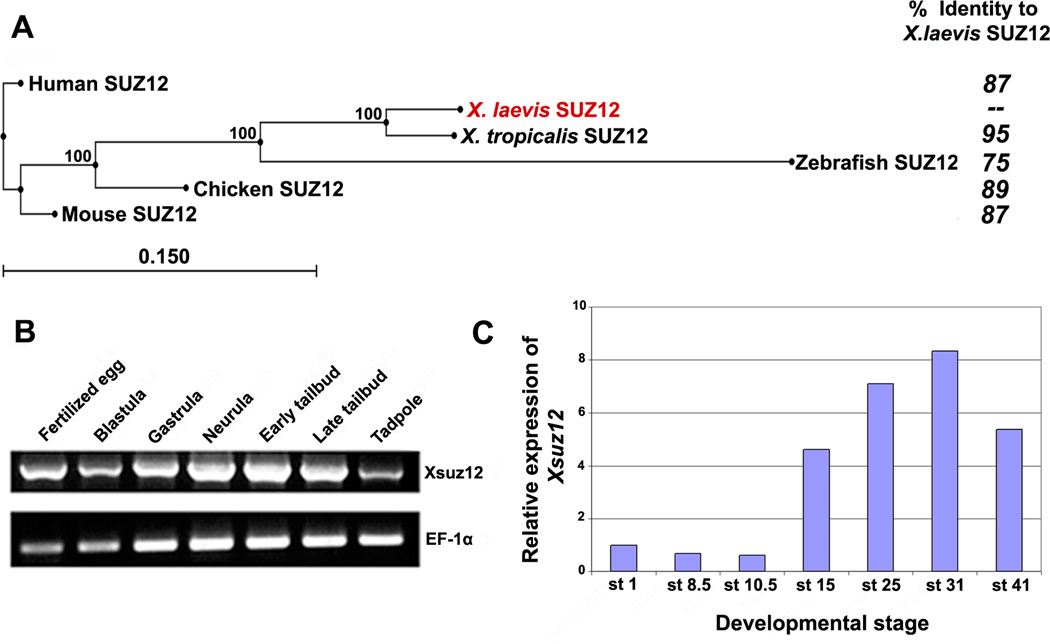
To isolate the full length cDNA of Xsuz12, we first identified Xenopus EST sequences in the NCBI database that showed high similarities to the mammalian Suz12. We designed forward and reverse primers based on two of these EST sequences, BU907606.1 and CA791061.1, respectively, and used them to amplify the full length Xsuz12 from a cDNA library prepared from whole embryos at stage 17/18 using PCR. We obtained a band of 2.1 kb, corresponding to the predicted molecular weight of Xsuz12 full length cDNA as compared to the molecular weight of its X. tropicalis ortholog. The band was sequenced and found to encode for a protein that is composed of 696 amino acids. Sequence alignment shows that the predicted protein is highly similar to its vertebrate orthologs, and contains two well conserved domains: a zinc finger motif and a VEFS box, which are characteristics of the SUZ12 protein, spanning amino acids 405–428 and 501–638, respectively (Fig.1A, B). Evidence suggests that both domains are important for SUZ12 function. For instance, the VEFS box of SUZ12 is required for SUZ12-EZH2 interaction (Yamamoto et al, 2003). Construction of a phylogenetic tree confirmed that Xsuz12 is closely related to its vertebrate orthologs (Fig. 2A).
Fig. 1.
Suz12 is highly conserved among vertebrates. A: schematic representation of XSUZ12 showing the relative positions of its zinc finger domain and VEFS box. B: Sequence alignment of Suz12 orthologs in vertebrates representing area boxed in A. The predicted amino acid sequence of X. laevis SUZ12 is aligned with human (Homo sapiens; accession no. NP_056170), mouse (Mus musculus; accession no. NP_954666.1), Chicken (Gallus gallus; accession no. XP_415658.), Zebrafish (Danio rerio; accession no. XP_694666) and X.tropicalis (accession no. NP_001072292). Green and red lines span the Zinc finger domain and the VEFS box, respectively.
Fig. 2.
A: Phylogenetic tree of SUZ12 proteins of different species created by neighbor-joining algorithm using CLC Sequence Viewer, and their respective amino acid identity compared to X. laevis Suz12. Percentage of branching point reproducibility is listed next to each node. Scale bar represents distance calculated on the basis of amino acid substitution rates. B: RT-PCR analysis of Xsuz12 expression during the frog development. cDNA pools were prepared from the following stages: Fertilized egg: stage 1, Blastula: stage 8–9, Gastrula: stage 10.5, Neurula: stage 14–18, Early tailbud: stage 25, Late tailbud; stage 35, Tadpole: stage 41. EF1-α was used as a loading control. C: Relative amounts of Xsuz12 transcripts during Xenopus development as revealed by qRT-PCR. The values of Xsuz12 was normalized to those of H4. Bars represent fold change in Xsuz12 expression at different stages as compared to stage 1.
Expression pattern of Xsuz12
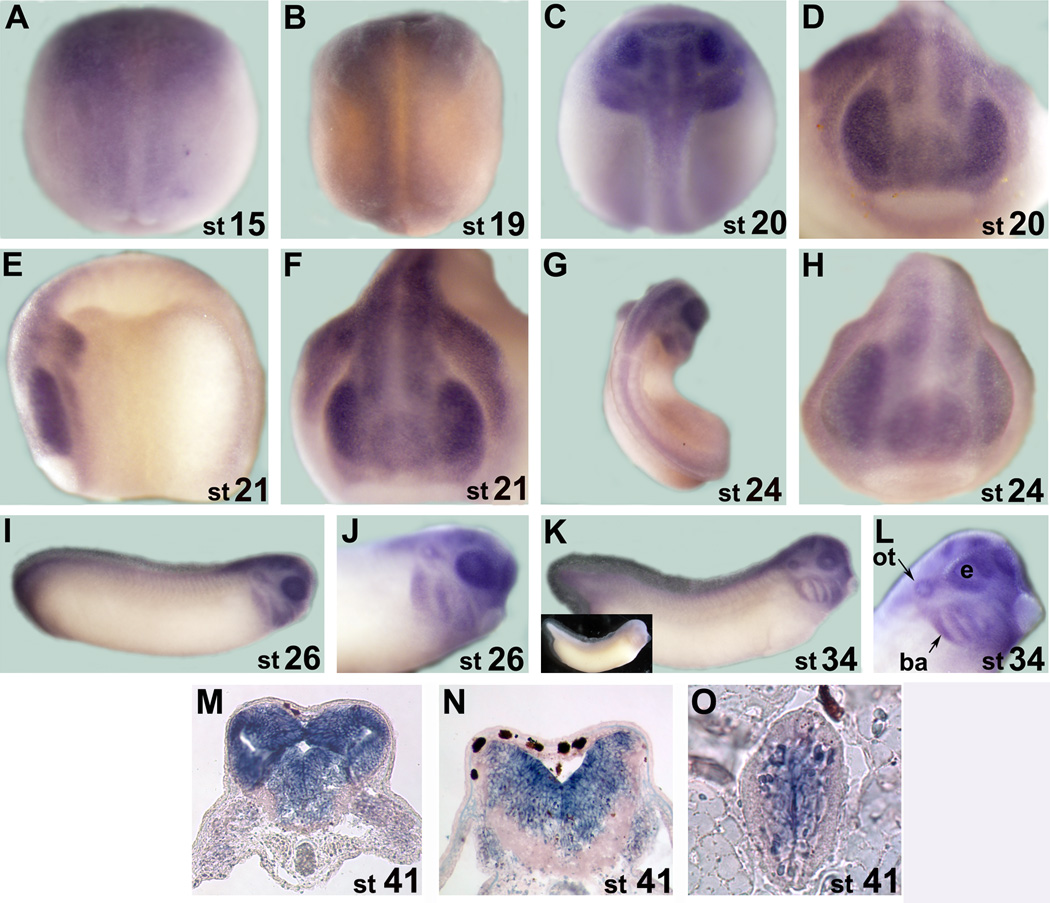
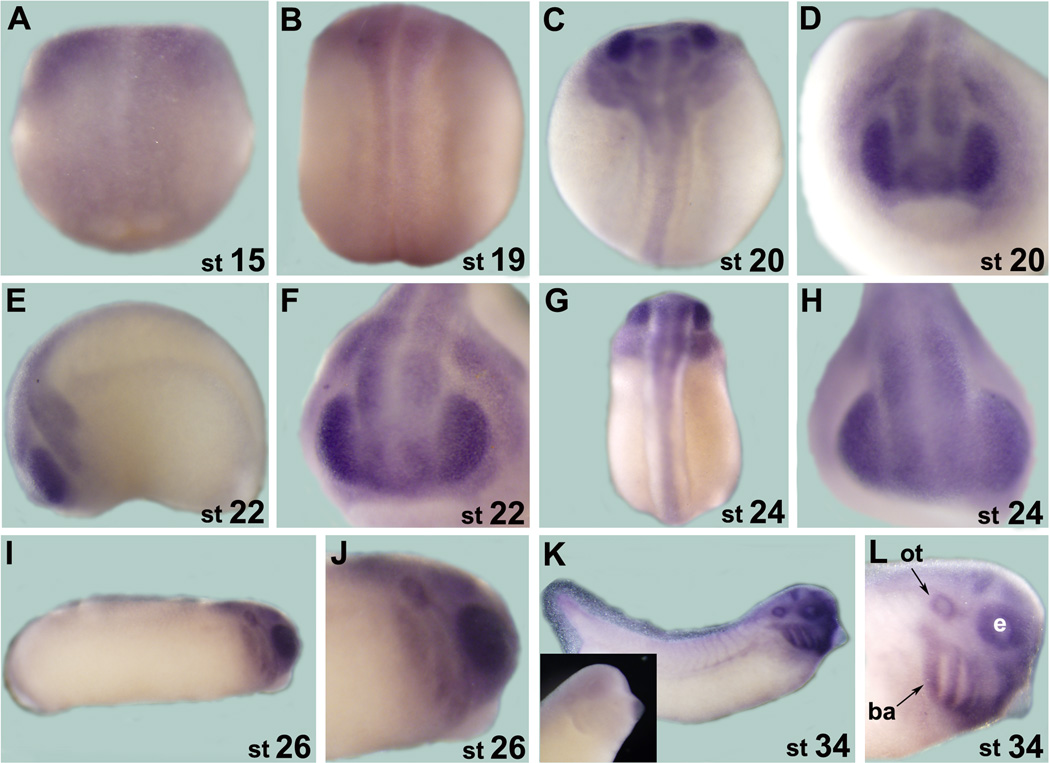
We first examined temporal expression of Xsuz12 by performing reverse transcriptase-polymerase chain reaction (RT-PCR) using RNA extracted from different developmental stages up to stage 41 (tadpole stage). We detected Xsuz12 expression at all stages examined, starting as early as fertilized egg (Fig. 2B). This indicates that Xsuz12 transcript is maternally supplied then zygotic expression is initiated later on, in agreement with the expression data of the PRC2 subunits, Xeed and Xez (Showell and Cunliffe, 2002; Satijn et al, 2001). We confirmed these results by real time quantitative RT-PCR (qRT-PCR), which also showed robust Xsuz12 expression after neural induction in neurula and tail bud stage embryos (Fig. 2C). To define tissues where Xsuz12 is expressed we performed whole-mount in situ hybridization analysis. Generally, Xsuz12 was mainly detectable in the developing central nervous system along the anterior-posterior axis with a particularly strong signal in the anterior part of the embryo (Fig. 3). Expression starts to become apparent in the open neural plate at around stage 15 (Fig. 3A, B). By stage 20–22 Xsuz12 transcript is seen in the developing spinal cord and in the head region of the embryo including the developing optic vesicles (Fig. 3C-F). From stage 24 onward, Xsuz12 expression is well defined in the head region, including developing eye, branchial arches, otic vesicles and in the forebrain (Fig. 3G-L). We also noticed the presence of a recognizable signal in the tail region that is obvious in the tail bud stages (Fig. 3I, K). Further, we performed in situ hybridization analysis on several cross sections taken along the anterior posterior axis of embryos at stage 41 (tadpole stage). We found that Xsuz12 expression was particularly robust in areas surrounding the ventricular zones in the brain (Fig. 3M, N) and in the spinal cord (Fig. 3O). Brain and spinal cord expression of Xsuz12 is also detected in cross sections made at tailbud stages (data not shown). In situ hybridization signal cannot be detected when a sense probe is used (Fig. 3K inset).
Fig. 3.
A-L: Whole mount in situ hybridization analysis of Xsuz12 expression starting from stage 15 (neurula). A, B, C and G are dorsal views. D, F and H are anterior views. E, I, G, K and L are lateral views. G and L are magnifications of the head region as viewed laterally at stage 26 and 34, respectively. Inset in K shows an example of a whole mount in situ hybridization that was performed with Xsuz12 sense probe. M-O: in situ hybridization analysis performed on traverse sections from the head region at stage 41. M and N sections are at the levels of forebrain and hindbrain, respectively. O is a traverse section in the spinal cord. Stages are indicated at the lower right corner of each image. ot, otic vesicle; ba, branchial arches; e, eye.
Expression of Xeed, Xez and Xrbbp4
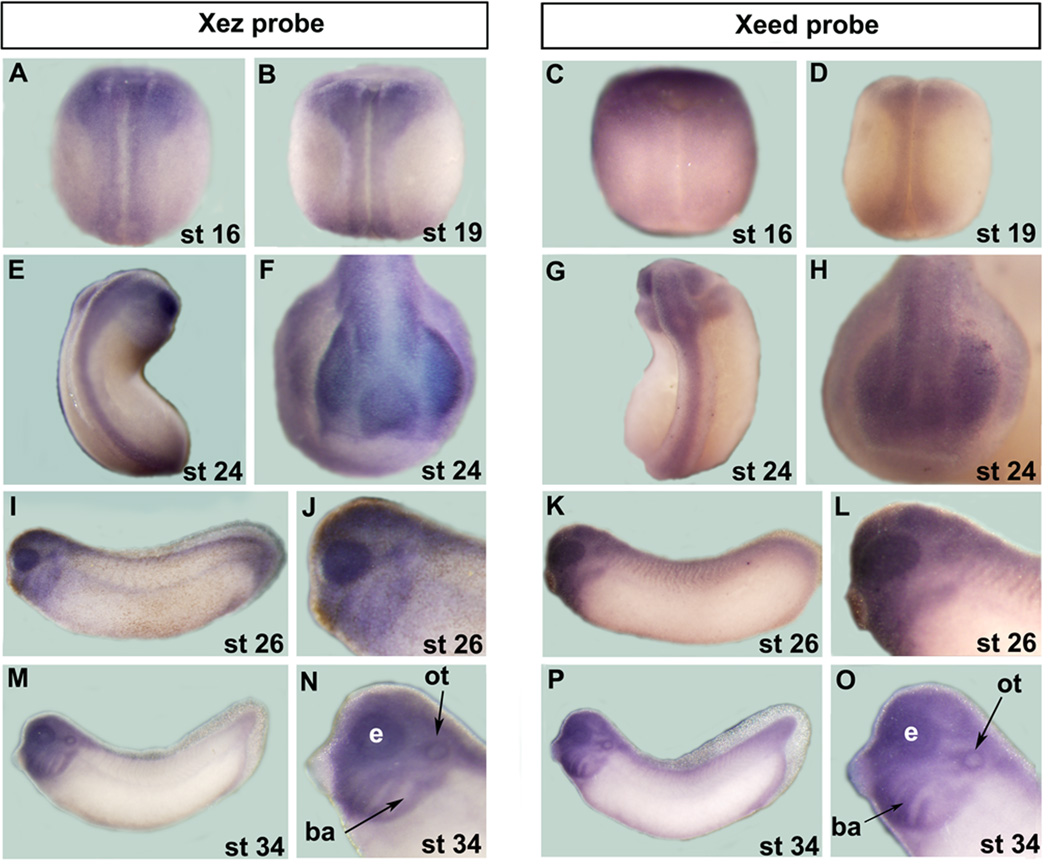
If the PRC2 complex is active and plays important roles in the developing nervous system in Xenopus, then we should expect that the expression pattern of its subunits largely, if not fully, coincide. Accordingly, we first sought to compare the spatial expression of Xsuz12 to those of Xez and Xeed, as both genes have been previously cloned but their expression was not fully characterized (Showell and Cunliffe, 2002; Barnett et al, 2001; Satijn et al, 2001). Xez and Xeed expression can be clearly seen at stage 16 in the anterior open neural plate (Fig. 4A, C). We found that Xez is expressed along the anterior-posterior axis and is not restricted to the anterior embryo as was previously reported (Fig. 4A, B, E; Barnett et al, 2001). At stage 19, Xez and Xeed transcripts are obvious in the presumptive spinal cord and by stage 24 both transcripts are expressed in the head region, including the emerging optic vesicle, as well as in the developing spinal cord (Fig. 4B-H). In the tail bud stages, Xeed and Xez expression persists anteriorly in the branchial arches, the otic vesicle, the forebrain and the developing eyes (Fig. 4I-Q). As the case of Xsuz12 a weak but detectable staining can be seen in the tail region (Fig. 4I, K, M and P).
Fig. 4.
Comparison of Xez and Xeed spatial expression during frog development as assessed by whole mount in situ hybridization. A B, C, D, E and G are dorsal views. F and H are anterior views. I, K, M and P are lateral views. J, L, N and O are magnifications of the head region in I, K, M and P, respectively. ot, otic vesicle; ba, branchial arches; e, eye.
Finally, we sought to characterize the expression of Xrbbp4, a vertebrate ortholog of the Drosophila Nurf55. This subunit has been purified from the PRC2 complex in Drosophila embryos and from HeLa cells, and is required for H3 binding (Cao et al, 2002, Czremin et al, 2002; Nekrasov et al, 2005). RBBP4 is also known to interact with other protein complexes as well, such as the N-CoR complex (Jones et al, 2001). Xrbbp4 has been cloned but its expression pattern in the frog was not determined (Vermaak et al, 1999). Therefore, we performed a whole mount in situ hybridization analysis on different embryonic stages of Xenopus and found that Xrbbp4 expression in the developing nervous system is virtually identical to that of Xsuz12, Xeed and Xez. Xrbbp4 is expressed in the developing eyes, branchial arches, otic vesicles and in the brain (Fig. 5A-L). Taken together we conclude that PRC2 principle components are expressed in the developing nervous system, suggesting an important role in neural tissue proliferation and/or differentiation.
Fig. 5.
Expression of Xrbbp4 by whole mount in situ hybridization analysis. A B, C and G are dorsal views. D, F and H are anterior views. I, K, M and P are lateral views. Whole mount in situ hybridization performed with a sense probe is shown as inset in 3K. ot, otic vesicle; ba, branchial arches; e, eye.
Because polycomb genes are highly conserved among species, we speculate that some of the physiological roles of PRC2, at least in certain developmental processes, may also be conserved. To date, the developmental expression pattern of mammalian PRC2 genes has not been thoroughly investigated but available data suggest that they may be expressed in neural tissues in a pattern similar to that of the frog Xenopus. For instance, mouse Ezh2 is expressed during embryogenesis and is detected in the neural tube, optic vesicle, branchial arches and in the developing brain in general (Laible et al, 1997; Caretti et al, 2008). This may indicate that at least some of the biological functions of PRC2 in neural tissues are evolutionarily conserved.
EXPERIMENTAL PROCEDURES
Cloning of Xsuz12
Primers were designed based on available X. laevis Suz12 ESTs found in the NCBI database (BU907606.1 and CA791061.1). The following primers were used to amplify Suz12 from a cDNA library prepared from whole embryos at stage 17–18: forward, 5’-TAATTACCCCGTATGGCC- CCTCAGAAGCAC-3’, reverse, 3’-ACACAGCAAAAAGCAGAAGCCCTGAAGG-5’. PCR was performed using Pfu turbo polymerase (Stratagene), and a band of 2.1 kb, corresponding to the predicted molecular weight of XSuz12 full length cDNA (as compared to the molecular weight of its X. tropicalis orthologue) was isolated and sequenced by the University of Utah Sequencing Core Facility. Final Xsuz12 sequence was submitted to GenBank (accession number FJ905047). qRT-PCR and data analyses were performed as was previously described (Logan et al, 2005).
Alignment and phylogenetic analyses were performed using MacVector and CLC Sequence Viewer software (http://www.clcbio.com/index.php?id=28), respectively. Neighbor-joining algorithm was used to construct SUZ12 phylogenetic tree, and reproducibility of branching points was determined by performing 100 bootstrap reiterations. EF1α was used as a loading control for PCR, using the following primers, forward, 5’-CAGATTGGTGCTGGATA- TGC-3’; reverse, 5’-ACTGCCTTGATGACTCCTAG-3’.
In Situ Hybridization
Digoxigenin-labeled antisense probes that span the first 700 bp and 744 bp of Xsuz12 and Xrbbp4 cDNA, respectively, were transcribed using T7 RNA polymerase after plasmid linearization. Xrbbp4 probe was synthesized from a full length cDNA clone (accession number BC077257) that was purchased from Open Biosystems. Xez and Xeed probes were generous gifts from Dr Elizabeth A. Jones and Dr Vincent T. Cunliffe, respectively. Embryos were fixed with 4% paraformaldehyde (PFA) in phosphate buffer solution (PBS). In situ hybridization was performed as previously described (Hutcheson and Vetter, 2001).
Acknowledgements
We would like to thank Dr. Elizabeth A. Jones and Dr Vincent T. Cunliffe for providing Xez and Xeed cDNA as well as their respective probes. We appreciate the help of Yangsook Green Song and Michael Steele with the qRT-PCR analysis. This work was funded by an NIH grant (EY012274) to MLV.
REFRENCES
- Barnett MW, Seville RA, Nijjar S, Old RW, Jones EA. Xenopus Enhancer of Zeste (XEZ); an anteriorly restricted polycomb gene with a role in neural patterning. Mech Dev. 2001;102:157–167. doi: 10.1016/s0925-4773(01)00304-5. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED—EZH2 complex. Mol Cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila Enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- Dellino GI, Schwartz YB, Farkas G, McCabe D, Elgin SC, Pirrotta V. Polycomb silencing blocks transcription initiation. Mol. Cell. 2004;13:887–893. doi: 10.1016/s1097-2765(04)00128-5. [DOI] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE. Mechanisms of transcriptional memory. Nature Rev. Mol. Cell Biol. 2001;2:409–421. doi: 10.1038/35073039. [DOI] [PubMed] [Google Scholar]
- Hutcheson DA, Vetter ML. The bHLH factors Xath5 and XNeuroD can upregulate the expression of XBrn3d, a POU-homeodomain transcription factor. Dev. Biol. 2001;232:327–338. doi: 10.1006/dbio.2001.0178. [DOI] [PubMed] [Google Scholar]
- Jones PL, Sachs LM, Rouse N, Wade PA, Shi YB. Multiple N-CoR complexes contain distinct histone deacetylases. J. Biol. Chem. 2001;276:8807–8811. doi: 10.1074/jbc.C000879200. [DOI] [PubMed] [Google Scholar]
- Ketel CS, Andersen EF, Vargas ML, Suh J, Strome S, Simon JA. Subunit contributions to histone methyltransferase activities of fly and worm polycomb group complexes. Mol Cell Biol. 2005;16:6857–6868. doi: 10.1128/MCB.25.16.6857-6868.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymenko T, Papp B, Fischle W, Köcher T, Schelder M, Fritsch C, Wild B, Wilm M, Müller J. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 2006;20:1110–1122. doi: 10.1101/gad.377406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laible G, Wolf A, Dorn R, Reuter G, Nislow C, Lebersorger A, Popkin D, Pillus L, Jenuwein T. Mammalian homologues of the Polycomb-group gene Enhancer of zeste mediate gene silencing in Drosophila heterochromatin and at S. cerevisiae telomeres. EMBO J. 1997;16:3219–3232. doi: 10.1093/emboj/16.11.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K, Odom DT, Otte AP, Volkert TL, Bartel DP, Melton DA, Gifford DK, Jaenisch R, Young RA. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan MA, Steele MR, Vetter ML. Expression of synaptic vesicle two-related protein SVOP in the developing nervous system of Xenopus laevis. Dev Dyn. 2005;234:802–807. doi: 10.1002/dvdy.20618. [DOI] [PubMed] [Google Scholar]
- Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32:503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O'Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- Nekrasov M, Wild B, Müller J. Nucleosome binding and histone methyltransferase activity of Drosophila PRC2. EMBO Rep. 2005;6:348–353. doi: 10.1038/sj.embor.7400376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satijn DP, Hamer KM, den Blaauwen J, Otte AP. The polycomb group protein EED interacts with YY1, and both proteins induce neural tissue in Xenopus embryos. Mol Cell Biol. 2001;21:1360–1369. doi: 10.1128/MCB.21.4.1360-1369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin AJ, Shao Z, Erdjument-Bromage H, Tempst P, Kingston RE. A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature. 2001;412:655–660. doi: 10.1038/35088096. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nature Genet. 2006;38:700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu CT, Bender W, Kingston RE. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showell C, Cunliffe VT. Identification of putative interaction partners for the Xenopus Polycomb-group protein Xeed. Gene. 2002;291:95–104. doi: 10.1016/s0378-1119(02)00588-7. [DOI] [PubMed] [Google Scholar]
- Sparmann A, Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nature Rev. Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- Su IH, Dobenecker MW, Dickinson E, Oser M, Basavaraj A, Marqueron R, Viale A, Reinberg D, Wülfing C, Tarakhovsky A. Polycomb group protein ezh2 controls actin polymerization and cell signaling. Cell. 2005;121:425–436. doi: 10.1016/j.cell.2005.02.029. [DOI] [PubMed] [Google Scholar]
- Vermaak D, Wade PA, Jones PL, Shi YB, Wolffe AP. Functional analysis of the SIN3-histone deacetylase RPD3-RbAp48-histone H4 connection in the Xenopus oocyte. Mol. Cell. Biol. 1999;19:5847–5860. doi: 10.1128/mcb.19.9.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viré E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, Bollen M, Esteller M, Di Croce L, de Launoit Y, Fuks F. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Sonoda M, Inokuchi J, Shirasawa S, Sasazuki T. Polycomb group suppressor of zeste 12 links heterochromatin protein 1alpha and enhancer of zeste 2. J Biol Chem. 2004;279:401–406. doi: 10.1074/jbc.M307344200. [DOI] [PubMed] [Google Scholar]