Abstract
Model organisms are vital to our understanding of human muscle biology and disease. The potential of the nematode Caenorhabditis elegans, the fruitfly, Drosophila melanogaster and the zebrafish, Danio rerio, as model genetic organisms for the study of human muscle disease is discussed by examining their muscle biology, muscle genetics and development. The powerful genetic tools available with each organism are outlined. It is concluded that these organisms have already demonstrated potential in facilitating the study of muscle disease and in screening for therapeutic agents.
Introduction
The major problems in investigating human muscle disease are that the organism concerned is long-lived, experimentally inaccessible during the important events of muscle development, and many essential experiments are impossible and/or unethical. Model genetic organisms, including mice (see Chapter by Hardeman) provide alternative approaches to understanding aspects of normal and diseased muscle. Genome sequencing of human and the major genetic model animals - mouse, zebrafish (Danio rerio), fruitfly (Drosophila melanogaster) and the nematode (Caenorhabditis elegans) has revealed close homologies in the genes and muscle proteins between these species and humans. Comparisons of muscle structure, function and development also show extensive homologies. However, their disadvantages are that although their muscles are similar to those from humans they do also differ to varying degrees from human muscle.
We aim to acquaint the reader with sufficient background to appreciate the opportunities that the non-mammalian model genetic organisms offer for study of human muscle genetic diseases and give examples to illustrate what these organisms have already contributed to disease investigations. We argue that there is enormous potential in using these non-mammalian genetic models to investigate human muscle disease that remains to be fully exploited
The major non-mammalian model genetic animals
The three major non-mammalian model organisms for genetic study of development, cell biology, physiology, neurobiology, ageing etc are the zebrafish, the fruitfly and the nematode (usually referred to as the ‘worm’). Comparisons of their genomes with humans, and their proven success as models with which to illuminate gene function have led to a wider acceptance, that they have important potential for the study of human disease.
Sequence analysis of these organisms’ genomes show that a large fraction of their genes have human homologues; 60-70% of human genes have counterparts in C. elegans and D. melanogaster. Having single genes where vertebrates have several paralogues these invertebrate organisms are well suited to identifying the core functions of gene families. The zebrafish has more orthologous genes and is particularly advantageous for dissecting the function of individual members of gene families.
Why or how were these organisms chosen and why are they such useful model genetic systems? In each case, the choice was made by an individual investigator. As the models proved amenable, each spawned very large international networks of researchers. It is the communal sharing and developing of genetic resources, tools, and information within these networks that make these organisms such powerful tools for genetics research.
Drosophila melanogaster, the fruitfly, was one of the first genetic model organisms. It contributed throughout the 20th Century to most major advances in genetics, especially pioneering genetic approaches to study animal development, cell biology, neurobiology etc. Thomas Hunt Morgan chose it as an experimental genetic organism around 1905 because it was inexpensive and easy to culture, had high female fecundity and flies could be kept in large numbers. Subsequently it transpired that it had only four chromosomes, a smallish genome, mutants were easy to obtain, and it was cytogenetically amenable. Its genome (180Mb) has been fully sequenced and annotated, along with those of 11 close relative species (see Flybase).
Sydney Brenner used similar arguments in the 1970s when initiating research on the nematode, Caenorhaditis elegans. This organism can be grown cheaply and easily on Escherichia coli lawns in Petri plates. Crosses are easily made and different mutant lines stored by freezing. Since the female worm is a self-fertilizing hermaphrodite, homozygotes are readily produced without the need to mate. Crippled animals survive as homozygotes, a great advantage when studying severe muscle mutations. The genome (97Mb) is sequenced1 and contains > 19,000 genes. It is unique in that a map of the invariant cell fate of every cell within the nematode is known.2
The zebrafish, Danio rerio was chosen as a suitable vertebrate model genetic organism by George Streisinger during the 1970s. It has excellent optical properties during early stages. Females have high fecundity and embryos develop rapidly, reaching a motile stage 24 hours after fertilisation. Eggs are fertilised outside the body which facilitates genetic and cellular manipulations. The zebrafish bridges the taxonomic gap between the worm and fly and the mouse. A major advantage of the fish as a genetic model for disease is that its organs and systems are more homologous to those of humans.3 Although the facilities required for culturing fish are more extensive than for either invertebrate model, sperm can be stored frozen which allows many different mutations and transgenic lines to be kept for long periods and transferred inexpensively.
Importantly, research communities associated with each of these organisms have developed and maintain publicly funded mutant stock collections, sequenced and annotated the genomes, and placed all this in open access databases, along with guides to aspects of the organisms’ genetics, biology and bibliography. These are accessible at Flybase (http://flybase.bio.indiana.edu/), Wormbase (http://www.wormbase.org/) and ZFIN, the Zebrafish Information Service (http://zfin.org/) and are linked to the major international bioinformatics resources. National and local mutant stock collections are maintained.
What makes these special as model genetic organisms for studies of muscle?
The simple answer is that compared to mammalian models all stages of muscle development are visible or easily accessible in vivo. In reality, it is a combination of our sophisticated knowledge of, and ability to manipulate, their genetics, their overall biology and specifically their muscle biology.
The generation of many separate populations of myogenic cells (myoblasts) is thought to underlie establishment of muscle fibre diversity in species from Drosophila to man.4,5 Four types of myoblast diversity are particularly significant. First, at, or before, terminal differentiation, myoblasts gain an ‘identity’ by expressing genes specifically required for the formation of particular muscles or muscle groups and display behaviours, such as adopting specific positions, orientations or morphologies within the tissue anlagen.6-10 ‘Identity’ ensures that muscle fibres interact correctly with connective tissues, become correctly positioned and oriented, and make correct paired attachments to skeletal and neural cells. Second, myoblasts that initiate new fibre formation may be distinct from those that contribute to growth. This is best understood in Drosophila, where founder myoblasts, in the embryo and at metamorphosis, express different genes and have different developmental capacities from so-called fusion-competent myoblasts.6,7 Tissue culture and zebrafish studies suggest a similar system operates in vertebrates.11,12 In fly embryos only founder cells appear to have muscle identity. 6,7 The situation in vertebrates is unresolved.4,13 Third, amniote myoblast clones have a cellular memory that determines the contractile protein genes they express, and hence the fibre type formed when they terminally differentiate.14-16 As fibres of similar fibre type exist in most muscles it is entirely unclear how this property relates to muscle identity. Is the slow fibre founder in slow soleus muscle distinct from a slow founder in the mainly fast plantaris? Fourth, myoblast clones, including those from human embryos, differ in their responsiveness to growth factors17 and capacity for proliferation 18 in ways reminiscent of the classical distinction between stem cells (capable of proliferating indefinitely) and progenitor, or transit amplifying, cells (capable of only a limited burst of proliferation followed by terminal differentiation). Stem cell-derived cells from non-muscle tissue participate in muscle repair and are particularly important in the context of muscle maintenance.19,20 Drosophila and zebrafish studies have begun to address these issues in the context of early muscle patterning.
Zebrafish (Danio rerio)
Zebrafish contain muscle types characteristic of all the vertebrates. As in mammals, skeletal muscles can be divided into somitic, somite-derived migratory hypaxial (e.g. fin) and head muscles.21-23 Skeletal muscle can be classified into slow and fast twitch fibres which, in embryos, are topologically separable. Mature fish have a subcutaneous layer of slow-twitch fibres at the dorsoventral midline; most of the rest of the skeletal musculature is fast-twitch. At least six fibre types have been identified in each somite as early as 24 hours post-fertilisation.24-27 Zebrafish cardiac myogenesis produces a heart with only two chambers and relies on many of the same genes as mammalian heart; mutations in genes that cause heart disease also affect fish hearts.28-29 Zebrafish also have vascular and visceral smooth muscle. Human muscle disease often preferentially affects specific muscle groups or fibre types, providing a major argument for investigating muscle disease in fish.
Importantly for studying human muscle disease, where muscle growth and repair can be major features, zebrafish have a dermomyotome,25 which contains muscle stem cells26 and may yield satellite cells. The phasic growth of fish muscle is similar to that in mammals.
Once the early embryonic muscle pattern has been established, vertebrate muscles undergo huge growth. This entails the founding of the correct number of additional muscle fibres in each muscle, which occurs in the early phases of amniote life. Subsequently, the correct number of nuclei must be added to each founded syncitial fibre as the animal grows. In adulthood, this number must be dynamically maintained and adapted dependent on physiological demands. There is evidence from flies, birds and mammals that distinct myoblast populations may influence each of these growth processes.30-32 Experimental challenge can interconvert some populations.33 However, the normal embryological origin of vertebrate myoblast populations contributing to muscle growth are uncertain.
The role of myoblast diversity in adult muscle is obscure.34 Muscle maintenance and repair in the adult can be of several kinds. Mild muscle damage necessitates running repairs to existing fibres, while increased physiological demands enhance muscle mass or alter muscle character and inactivity leads to muscle atrophy. Serious muscle injury or muscle disease can necessitate de novo fibre reconstruction. There is evidence for stem cell populations in adult skeletal muscle.35 Current interest focuses on the nature, role, origin and capacities of multipotent mesenchymal stem cell populations found within muscle tissue and their relationship to satellite cells, the quiescent myoblasts of adult muscle.36,37 Change in myoblast diversity is observed in many pathological situations; the extent to which such changes contribute to altered muscle function and disease progression is unknown. Muscle pathology usually increases ‘fibroblast’ numbers leading to fibrosis. The origin and relationship of these cells to myogenic cells is unclear.
Amniotes exhibit a great diversity of myogenic routes. During fetal and postnatal myogenesis waves of distinct myoblasts appear and disappear, but their origin and fate are unknown. Molecules that indelibly mark particular cell lineages have not been found. Other myogenic populations exist within adult mammalian muscle. For example, so-called SP cells may be multipotent mesenchymal stem cells,19 but their normal role is unclear.
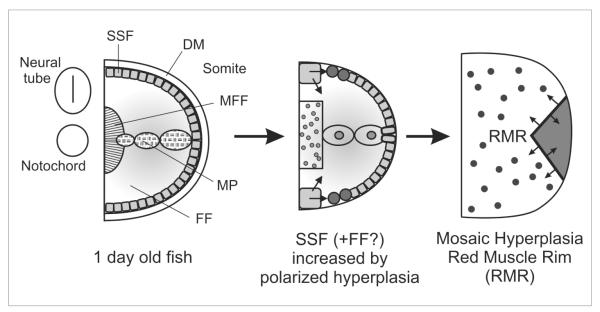
Fish myogenesis proceeds in waves similar to those in mammals. Several early myogenic cell populations give rise to diverse early muscle fibres (Figure 1). The muscle then grows by ‘polarized hyperplasia’, yielding extra fibres in specific somitic zones, the dorsal and ventral myotomal lips and the ‘red muscle rim’ between slow and fast muscle regions.26,38,39 Later ‘mosaic hyperplasia’ generates new fibres between existing early fibres similar to mammalian secondary fibre formation.38 Continuous growth of the fish requires fusion of myogenic cells to existing fibres. These may derive from satellite cells (SC) lying under the basal lamina of each fibre, as in mammals, and probably contribute to muscle repair. Zebrafish muscle growth and repair is essentially unstudied, although regeneration and growth is known to occur in teleosts.40
Figure 1.
Early myogenesis (left panel) generates four fibre types but leaves undifferentiated cells in the dermomyotome (DM), probably destined to support for later growth. Mononucleate superficial slow fibres (SSF) and muscle pioneer slow fibres (MP) form first, followed by multinucleate fast fibres (FF), a subset of which become specialised as medial fast fibres (MFF). Polarized hyperplasia (middle panel) generates further SSF and FF, probably from DM cells. Mosaic hyperplasia (right panel) arises from precursor cells scattered within the myotome and between the slow and fast regions at the so-called red muscle rim (RMR).
Zebrafish are readily mutagenized and a wide range of behavioural and developmental phenotypes visually screened for. Large scale screens41 for mutations affecting muscle development and function are a goldmine for understanding early muscle development. TILLING can be used to create allelic series of mutations in chosen genes. This involves random point (chemical) mutagenesis, followed by DNA sequencing of many fish to identify individuals carrying mutations in target genes. These approaches have tremendous potential to permit detailed analyses of gene function.
Fruitfly (Drosophila melanogaster)
All Drosophila muscles are striated. The organism contains many skeletal (attached to the exoskeleton) and visceral muscles, as well as a primitive heart. The musculature of Drosophila has been described in detail42 and images are available on Flybase.
The Drosophila life-cycle requires development of skeletal and visceral muscles twice; once for embryonic/larval development and another during pupal development prior to emergence of the adult (the heart undergoes some development during pupal metamorphosis). Myoblasts that form the embryonic muscles and those that will later differentiate into the adult muscles develop together during early embryogenesis, but while some differentiate immediately to form embryonic/larval muscles, others multiply during larval and early pupal development and then differentiate during metamorphosis to form the adult muscles. No satellite cells have been found in adult flies, though the myoblasts set aside during embryogenesis, only later to form the adult musculature could provide a model system for study of satellite cells.43 There is an extensive literature on Drosophila muscle development43,44 and an insightful detailed comparison of Drosophila and vertebrate muscle development.43 Drosophila embryonic myogenesis is rapid (<< 1 day) and easily visible. Mutant studies have identified many genes that are important during myogenesis and revealed cell interactions such as those between founder/pioneer myoblasts that inform studies in the vertebrate models.
Most studies of muscle disease in Drosophila have either focused on the later larval stages (instars) when the muscles are much larger or on the adult, especially the indirect flight muscles (IFM). IFM function can be easily assayed by measures of flight or an elevated wing position phenotype and have been used for fibre physiology/mechanics.45 The IFM are easily exposed by bisecting the thorax and readily removed for biophysical/biochemical experimentation.46 The effects of severe mutants can be studied, as flies, at least in the laboratory, do not need flight muscle function for survival and reproduction.
Flies are easily mutagenized by X- and γ-irradiation, various chemicals and, more recently, transposon mutagenesis. For embryonic muscle mutants, screens normally search for recessive lethal mutations which cause embryos to fail to hatch from the egg, followed by inspection to see if they have developed their cuticle and other organs, but remain immotile. Further investigation is then required to identify these mutants as muscle-defective. Muscle mutants in flies were recovered in the IFM by selecting for a dominant flightless phenotype.47,48 Many muscle protein genes were first identified in this way.
Nematode (Caenorhabditis elegans)
The nematode contains a number of muscle systems, but the major set is the bodywall muscles and these have been the target of most genetic studies.49 The other muscles include those of the pharynx, the 15 diagonal tail muscles with 26 other tail muscles involved in male copulation, 16 sex muscles (of uterus and vulva) in the hermaphrodite, two intestinal muscles and the anal muscles. The body-wall muscles are prominent and the organism small and translucent, so these muscles are readily visualized by polarised light or fluorescent microscopy of the whole organism. Details at the level of the sarcomere are visible. The bodywall muscles occur in 4 strips each consisting of 24 mononucleate, spindle-shaped cells. The first obvious difference is that these muscles contain an obliquely striated array of filaments that for those familiar with vertebrate or arthropod striated muscles is at first glance perplexing, but is wholly analogous to those in the cross-striated sarcomeres of vertebrates and arthropods. A detailed explanation is provided by Waterston49 in his general account of C. elegans muscle. The second difference is that the thick filaments are much thicker than in vertebrates, though less so than in most arthropods including Drosophila. Nematode muscle attachments are more complex. Though the ends of the muscle cells have dense plaques to which the thin filaments attach from only one direction, as in vertebrates and insects, nematode muscle is unusual in that most of the tension generated is transferred directly to the cuticle through a series of lateral attachments at the dense bodies, that are finger-like projections (analogous to Z-discs) into the muscle from the hypodermal side.49,50 There are similar dense body attachments between the neighbouring muscle cells. The M-lines are seen as finger-like projections that also originate within the muscle cell from the surface closest to the hypoderm.
Random genetic screens using a range of mutagens (X-/γ rays, chemical mutagens) are readily performed. Muscle mutants were recovered in the first screens for nematode uncoordinated (unc) mutants. Some of these mutants affect the nervous system, but many muscle genes were first identified on this basis49 and are still actively studied. More recently, severe muscle mutants have been recovered by arrested development with specific phenotypes e.g. the pat mutants.51 The ability to grow worms and flies in large numbers, quickly and cheaply not only facilitates the recovery of primary mutations, but also allows screening for secondary mutations in other genes (intergenic), that either reduce the severity of the primary mutation (suppressors), or enhance it (enhancers).
Genomes/Genetic technologies of the model organisms
The combination of recombinant DNA technology with transgenesis techniques has transformed our ability to manipulate the genomes of model organisms. This has led to the generation and application of an almost bewildering array of powerful molecular tools.
Stable genomic transgenesis is an important process required of model genetic organisms in the current era. Unlike the mouse, in none of these three model genetic organisms is it yet possible to target specific genes for replacement by homologous recombination. Gene replacement thus requires transgenesis of mutant gene copies into genomes containing ‘null’ mutations for the gene of interest. In C. elegans for genes already containing a transposon (‘tagged’) it is possible to copy a modified version of the gene directly into the native chromosome; similarly in Drosophila an inserted transposon (P-element, see below) can be replaced at a high frequency with another P-element, that could contain a novel version of the same gene.
The nematode genome contains a number of transposable elements including Tc1 and Tc3, members of the Tc1/mariner transposon superfamily.52 Mutations due to transposon insertion have been developed into ‘gene-tagging’ (also used in Drosophila and zebrafish) whereby mutations with specific phenotypes are recovered and the genes identified using PCR techniques to obtain the insert flanking sequences. These mutations may be unstable or have weak phenotypes. Stable mutations can be recovered by remobilising the original transposon by imprecise excision that removes flanking genomic DNA creating deletions detectable using PCR methods. More recently, a significant number of C. elegans genes have been tagged using the Drosophila Mos1 transposon, which is more stable than Tc elements.53 Nematode transgenesis is routinely achieved by non-integration of plasmid vectors by either DNA injection or sometimes particle bombardment54 and allows a wide variety of gene manipulations. to look at the in vivo effects. These include gene ‘rescue’ (with wild type gene copies) of mutants, insertion of dominant negative mutations made in vitro, expressing gene and protein fusions, to investigate gene promoters using promoter-reporter constructs, and ectopic expression of genes. Not only do these allow genetic investigations, but also provide a wide variety of powerful microscopical tools for investigating the organismal cell biology through the expression of protein reporters such as fluorescent tags (e.g. GFP, YFP etc.), enzymes and epitope tags etc.
In Drosophila a stable transgenesis system has been developed using the P-element, a native transposon, though other native transposons have also been developed as vectors. P-element vector DNA is injected into the posterior part of preblastoderm eggs along with a source of transposase (either the enzyme or a defective P-element able to express functional transposase). A variety of P-element vectors are available and whole genes can be inserted for e.g. ‘rescue’ experiments, to make promoter-reporter constructs for in vivo gene expression analysis, insert in vitro mutagenized genes, or other DNA sequences. Specific P-elements (and some other transposons e.g. pigyBac) have been made for a range of application. For instance, they are used for insertinal mutagenesis55 Berkely Drosophila Genome Project http://www.fruitly.org) and to generate deletions by imprecise excision or by recombination between two inserts. A novel approach is to ‘enhancer trap’ different genes (using reporter sequences – GFP, lacZ etc.) by relying on insertion near a gene enhancer to overcome the weak endogenous expression of the reporter gene in the P-element. Protein trapping canbe achieved by random insertion of a P-element containing GFP as an exon into a genomic intron that leads to the appearance of green fluorescent embryos or tissues. This allows for rapid screening for useful protein-GFP fusions.56 These activities have produced massive collections of gene knockouts, deletions,57 enhancer traps, expression reporters etc.
Arguably, the most powerful application of P-element transgenesis has been the development and use of the GAL4-UAS system.58 In this system transgenes expressing the yeast GAL4 transcription factor ‘drive’ expression of any other transgenic sequence inserted downstream of the yeast UAS sequence, the normal GAL4 binding target. Large collections of GAL4 lines expressing GAL4 from gene-specific promoter sequences, or from ‘enhancer traps’ now exist. Simply by crossing an appropriate (by expression level/pattern) GAL4 line with any of the different UAS construct lines expressing a vast array of heterologous proteins (GFP, GFP-protein fusions, dominant negative alleles, human disease proteins etc.) one can control the ectopic expression of the protein of choice to interfere with the system, to follow gene expression patterns or to determine the cellular locations of proteins of interest.58
Zebrafish transgenesis is based on either retroviral infection or on the ubiquitous Tc1/mariner transposons. Retroviral systems are used for efficient insertional mutagenesis,59 enhancer trapping60 and for a variety of other transgenic purposes. Retroviral systems are relatively laborious. Use of the Tc1/mariner transposon to achieve transgenesis has great potential to exploit the breadth of applications demonstrated by the Drosophila transposon systems.
The discovery that short double-stranded RNA (dsRNA) molecules can cause specific degradation of the corresponding mRNA was made in the nematode.61 This technique, known as RNAi, is now used widely to ‘knockdown’ mRNA levels in many different systems. Its use has been developed further in C. elegans, in Drosophila and more recently in the zebrafish. Originally applied to worms by injection, 61 it is now routinely achievable by soaking worms in dsRNA, or by feeding dsRNA in liquid culture, but most easily by feeding worms on E. coli containing plasmids with the specific dsRNA sequence.62 A large library of E. coli strains containing different dsRNAs covering the nematode genome is available; almost any gene may be screened using the appropriate clones.63 In flies the major technique is to make transgenic lines with specific dsRNA sequences downstream of the UAS sequence. This allows one to drive RNAi expression in different tissues, dependent only on the availability of a transgenic GAL4 driver. This is a very effective and flexible approach. A large available collection of transgenic UAS-RNAi strains has recently been made.64
In the zebrafish, morpholinos, short chemically modified RNA molecules that bind to mRNA provide an effective ‘knock down’ approach.65 Recently, RNAi technology has become available through injection of dsRNA.66
Model organism studies of human muscular disease
There is already considerable model organism muscle research that is of relevance to the study of human muscle disease, including both heart and skeletal muscle. The use of zebrafish genetics to study congenital heart disease has been reviewed,29 as has the potential of the Drosophila heart as a model system for human heart disease.67
Mutations of the myosin heavy chain can cause skeletal muscle myopathy and hypertrophic cardiomyopathy. Many myosin mutations are known in nematodes and flies. Trangenesis of a mutated copy of the nematode unc-54 (MyHC-B) gene into an unc54 null strain to study the effects of the human (skeletal) myosin heavy chain IIa E706R mutation that leads to contractions of the joints, and a progressive muscle weakness was able to show that the mutant affects myosin function rather than causing a structural effect.68 At least one Drosophila mutant myosin suppressor of hypercontraction occurs in the myosin ‘hypertrophic cardiomyopathy loop’.69
Arthrogryposis is a complex, rare human congenital disorder exhibiting multiple joint contractures, usually of the distal limb. It is often associated with congenital muscular dystrophy, congenital myopathies (central core, nemaline), myasthenic syndromes, intrauterine viral myositis, and mitochondrial disorders. Two types of arthrogryposis are caused by mutations of the fast twitch muscle-specific TnI (TNNI2) or the TPM2 tropomyosin gene.70 Tropomyosin/troponin complex mutations are known in nematodes, flies and zebrafish and are usually associated with hypercontraction, a phenotype where normally developed muscles undergo severe contractions causing muscle damage and dysfunction. Sarcomeric troponin mutations in C. elegans e.g. in the TnT (CeTnT-1) gene can cause detachment of body wall muscles.71,72 Mutants of the single Drosophila TnI and TnT genes cause IFM hypercontraction and muscle detachment.72,73 IFMs appear especially sensitive to hypercontraction. The held-up2 troponin I mutation at a highly conserved alanine residue (from humans to flies) causes IFM hypercontraction before adult emergence. Adult flies are initially normal, but flightless; then hypercontraction produces progressive crippling due to leg muscle detachment.74
Drosophila single myosin heavy chain Mhc gene mutations can cause hypercontraction.75,76 The Mhc6 and Mhc13 myosin rod mutations do so by reducing thick filament mechanical stability.75. In zebra fish silent heart mutants of the TNNT2 troponin T, gene affect the heart.28
Suppressor mutations of the Drosophila hdp2 (TnI) mutation have been used to investigate hypercontraction and the function of the troponin/tropomyosin complex in the activation of muscle contraction.69,74,77,78 Suppressors were found intragenically (a second TnI mutation that ‘corrects’ the effects of the first),77 in the Tm2 tropomyosin gene74 and in the Mhc motor domain.69,78 Suppression by Mhc mutations is probably due to reduced contraction force.69 Certain mutations in the IFM-specific Act88F actin gene suppress hdp2 hypercontraction (Haigh, Cammarato and Sparrow, unpublished). Suppression studies are a powerful way to study the functional relationships of interacting muscle proteins. They may be invaluable in exploring how genetic variance impacts the severity of specific human muscle disease mutations.
Many, mostly dominant, congenital myopathy mutants are known in the human ACTA1 gene.79 A unique feature of the Drosophila IFM for the study of these diseases is that they express all their sarcomeric actin from a single gene, Act88F, that is expressed (almost) solely in these muscles.80 Act88F null mutations are viable, so mutated Act88F transgenic copies can be used to study effects on muscle structure and function, and the mutant actin can be purified for biophysical and biochemical experiments.81 Since the Act88F and ACTA1 proteins show 94% residue identity one can study the congenital actin myopathies using the Drosophila IFMs.82 Biopsies from human disease patients show a range of subcellular effects – nemaline rods, intranuclear rods, homogenously staining regions containing large concentrations of thin filaments, zebra bodies83 and congenital fibre type proportion (CFTD).84 Transgenesis of Act88F genes containing ACTA1 nemaline mutations all affect muscle structure and function (Sevdali and Sparrow, unpublished). Effects range from mild disturbances of sarcomeric structure to complete absence of thin filaments, ‘zebra bodies’ (comparatively rare in human biopsies) (see Figure 2), some nemaline-rod like structures (smaller than usually seen in humans) and novel Z-disc structures enclosing other sarcomeric proteins. Some mutations reduce fibre diameter but since all IFM fibres are equivalent whether these mimic CFTD effects is unclear.
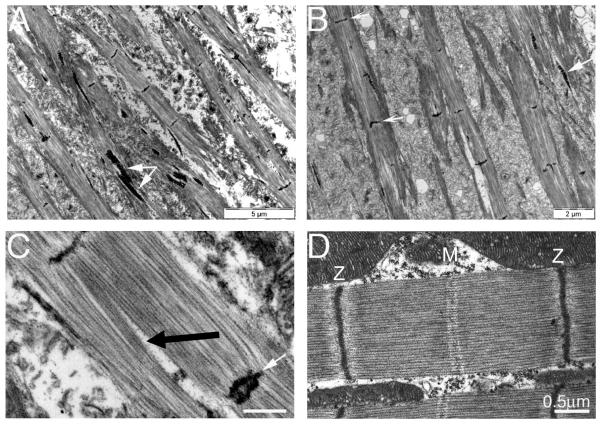
Figure 2.
Electron micrographs of flight muscle sarcomeres from Act88F-G15R homozygous mutant (A-C), and wild type flies (D). In panel A and B aberrant myofibrils are seen; there is a lack of regular structure; Z-discs never extend across the myofibril width and large arrays of ‘zebra-bodies’ comprising stacks of short sections of Z-disc-like material, spacing by about 200nm, are seen mostly within the muscles. In this and other mutants they are also found separated from the myofibrillar materials. Panel C highlights other aspects of these mutant sarcomeres including longitudinal gaps (black arrow) and dark staining material which appears to be split or duplicated Z-discs (white arrow). Panel D, wild sarcomere shows the very regular structure and much reduced I-band width, typical of this muscle type. EMs courtesy of Vikash Kumar.
Studies of early IFM myogenesis reveal at least two pathways to the fibre defects; most of the actin mutants cannot assemble normal sarcomeres during development; others form sarcomeres normally, but cannot maintain structural integrity during use (Sevdali and Sparrow, unpublished). Nemaline mutations occur throughout the actin molecule with little evidence of clustering of mutant residues with respect to phenotypic effects.79 The IFM system allows exploration of the relationships between mutant residues, the biochemical interactions of actin and effects on sarcomerogenesis and structural integrity. ‘Zebra bodies’ also occur in the IFM of Drosophila troponin T mutants. 73
This example highlights an issue of working with a model system for human muscle disease. The Drosophila IFM express a single sarcomeric actin from the outset; during early human skeletal myogenesis the ACTC cardiac isoform is expressed, switching to the ACTA1 skeletal isoform during late foetal stages. One can analyse the basic molecular lesion in Drosophila without complications from other isoforms.
Only one well-known human muscle disease (but not a sarcomeric muscle disease) has been focused on in all three model genetic organisms - muscular dystrophy. We describe these studies to illustrate the benefits in applying these approaches to the investigation of human muscle diseases, including the sarcomeric diseases.
Muscular dystrophy
Duchenne muscular dystrophy (DMD) in humans is due to mutations in the gene that encodes dystrophin. Dystrophins and dystrophin-like proteins are widely distributed in animals.85 The dystrophin-glycoprotein complex (DGC) includes dystrophin, as well as transmembrane and cytoplasmic proteins. Homologues of this complex are known in all three model organisms.
C. elegans possesses single conserved dystrophin-like (dys-1) and dystrobrevin-like (dyb-1) genes.86 Conserved homologues have been found also for dystroglycan, delta/gamma-sarcoglycan, syntrophin and divergent, but related, proteins to α- and β-sarcoglycans.87 Mutations inactivating the dys-1 gene cause only mild muscle degeneration, but cause behavioural phenotypes of hyperactivity, unusual movements of the head and a tendency to hypercontract88,89 that are also seen in mutants of other genes encoding DGC components.86,89 In combination with a weak hlh-1 gene mutation, the dys-1 mutation produces a time-dependent muscular dystrophy. The hlh-1 gene encodes a MyoD homologue. In mdx mice MyoD mutations also enhance the severity of dystrophy.90 Dysfunction of the dys-1 mutants is partly rescued transgenically by human dystrophin88 revealing a functional homology. This C. elegans genetic combination can thus be used as a model system for human muscular dystrophy.
Microarray studies of two dystrophin mutants, dys-1(cx35) and dys-1(cx18) revealed expression changes for many genes.91 Comparison with microarray analyses of human dystrophic patients showed many similarities but some significant differences probably because of important differences in DGC composition in the two organisms.91
Characterisation of the C. elegans syntrophin gene89 illustrates where the relative genetic simplicity and the tools available underpinned substantial progress in understanding a specific gene/protein of the DGC. Syntrophin is a DGC component and searching across the C. elegans genome revealed two nematode genes with homology to a human syntrophin sequence. A transgenic syntrophin-1 (stn-1) promoter-GFP reporter construct was made and revealed a nerve/muscle-specific expression pattern, consistent with syntrophin localisation in humans. An stn-1 intragenic deletion mutation, ordered from the Gene Knockout Consortium, showed phenotypic effects typical of dys-1 gene mutants. Using transgenesis expression of a wild-type stn-1 gene controlled by a muscle-specific promoter in a stn-1 knockout mutant fully rescued the phenotype, indicating that the mutant effects were caused by an absence of protein in the muscles, rather then the nerves. Although the stn-1 knockout behaves like the dys-1 and dyb-1 mutants, its mild effects on muscle degeneration were not potentiated by hlh-1.
Proposals that elevated Ca2+ levels increased the progression of the disease in mdx mice have not been directly tested in the mdx mouse but were addressed in C. elegans by using a gain of function mutation of the egl-19 calcium channel or knocking it down with RNAi-mediated inhibition.92 These studies showed that increased calcium is bad for dystrophic muscle.
C. elegans can be used, quickly and inexpensively, to screen for unknown genetic/biological interactions or as an early stage screen for chemicals that may lead to therapy in humans. This was demonstrated in successful screens for molecules, including serotonin and prednisone, that alleviate the symptoms of the dystrophin mutations.88,93,94 Identifying potential drugs for therapeutic development also highlights cellular processes for further investigation. A screen for mutations with a phenotype similar to dys-1 recovered a number of mutations including dyb-1 encoding a homologue of dystrobrevin,95 and the SLO-1 gene.96 SLO-1 regulates neurotransmitter release in nematode motorneurons and had not previously been implicated in dystrophic effects. The double dys-1; SLO-1 mutant worm does not have an additive phenotypic effect, suggesting that this channel is involved in the same pathway as the nematode dystrophin homologue. Random mutagenic screens have successfully identified mutations that suppress the phenotypic effects of dys-1 mutants, thereby uncovering new genes likely to be involved in dystrophy,95 including the dyc-1 gene.
The D. melanogaster genome contains one or two genes for most of the essential components of the DGC, including dystrophin, dystrobrevin, dystroglycan, syntrophin and sarcoglycans, but no sarcospan homologue.97 This is in contrast to the genetic complexity and potential redundancy found with the multiple isoforms of vertebrate DGC components. The conservation of the DGC genes together with their smaller number suggests that the fly system, like that of the nematode, will be a relatively simple, but good model for its human counterpart. Detailed examination of the single dystrophin/utrophin gene homologue shows substantial sequence and predicted structural homology to the human products.98 However, this Drosophila gene shows a highly complex structure expressing at least six dystrophin-like isoforms, DLP1-3 (long forms), Dp186,97-99 Dp205 and Dp117,100 with different expression patterns.98,101 Only the DLP2 isoform in known to be expressed in muscle fibres and at muscle attachment sites.98,101 As in vertebrates, the dystrophin gene products are expressed in nerves and muscles and recent genetic studies have begun to illuminate the gene complexity and the functions of its products.100,102 Transposon mutagenesis reveals that absent or reduced expression of the large gene products expressed in the musculature does not cause degeneration of larval body wall muscles. However, neurophysiological studies showed that the DLP2 dystrophin isoform is required postsynaptically to maintain normal levels of neurotransmitter release.102 This indicates a novel role for dystrophin in neuromuscular function, though its relevance to human disease is unknown. Using the Drosophila GAL4–UAS system to drive expression of RNAi constructs specifically directed against all dystrophins in the muscles and tendon cells did not compromise myogenesis or muscle attachment to tendon cells, but caused a progressive degeneration of larval and adult muscles. Reduction of the Dp117 isoform alone induced this degeneration. Clearly the ability to manipulate gene expression of a complex gene in Drosophila has revealed at least two roles for dystrophin gene products – maintaining synaptic homeostasis and preserving the structural stability of the muscle.100
The zebrafish genome contains 29 genes orthologous to those in humans known to cause dystrophy and databases show transcripts for 28 of these genes.103 There is a high level of conservation, enhancing the potential of zebrafish for studies of muscular dystrophies. RNAi against the Duchenne/dystrophin gene (dmd) produces muscle disease phenotypes in embryos showing one can target the range of different transcripts produced by the gene.66
Zebrafish mutants are known which exhibit various phenotypes of dystrophy, one of which is a premature termination mutation in the dystrophin gene,104 where muscle degeneration becomes apparent as soon as three days after egg fertilization.105 The sapje mutation in the fish orthologue of the human DMD gene causes a degeneration phenotype similar to human muscular dystrophies.106 The mutation reveals that degeneration effects are due to failure of the muscle end attachments in embryonic muscle, indicating that dystrophin is required for the formation of stable muscle attachments.106 Such an involvement of dystrophin in vertebrate myotendinous junctions was proposed earlier from studies of mdx mice.107 This zebrafish data is the first direct evidence for this dystrophin function and highlights the importance of studying human orthologues in different models.
Human mutations of DGC components have effects on other tissues,108,109 including the nervous system. The effects of mutations in DGC complexes are being explored in other tissues within model genetic organisms. Gain-of-function mutants in Drosophila dystroglycan, laminin A and dystrophin genes affect epithelial cells and oocytes and have been characterised using genetic mosaic analysis and RNAi methods.110 Using Drosophila dystrophin mutants, dsRNA constructs and a range of GAL4/UAS lines, it has been shown that dystroglycan and dystrophin are required in the brain for targeting neurons and glial cells for correct neuronal path-finding.111
Conclusions
As genetic systems for studying muscle development and functions in their own right, all three of these organisms have made fundamental contributions to muscle biology. Studies of disease-causing genes and their human mutations in these model genetic organisms are being increasingly used to address questions specifically related to understanding human muscle disease. This suggests that muscle biologists working with these organisms now have the genetic tools to do so, as detailed above, but also that clinically focused muscle scientists are beginning to see the potential of such studies.
An issue of working with a model system for human muscle disease is how well the model system recapitulates the human disease. Neither nematode nor Drosophila muscle has any regeneration capacity in response to dysfunction and neither organism has an adaptive immune response. These can be considered advantages and disadvantages. The model system will not recapitulate the whole disease, and in the examples studied to date do not. The defects seen are related, but different. However, this is an advantage, allowing investigation of the basic mutant molecular lesions without complications from repair or immune responses.
In the context of sarcomeric muscle disease, the value of these model organisms will be judged by contributions they make. In this chapter we have sought to illuminate what these model organisms have to offer. The choice of a model organism for studies of human muscle disease must be a balance of different advantages and disadvantages. Thus while zebrafish muscle has much closer homologies to humans it does not yet have the various sophisticated transgenic techniques and resources available in Drosophila, or, as especially in the nematode, the rapidity of screening large numbers of organisms for muscle mutations, RNAi effects across the whole transcriptome, or for chemicals that suppress the phenotype in disease models.
Mammalian genomes contain only slightly larger numbers of genes than these model organisms, yet mammalian genomes produce very many different proteins from them. Comparing homologous muscle protein genes shows that those of nematode and Drosophila, while also complex, are less so than those found in fish or mammals. The fish genes are only slightly less complex than those in mammals. This makes detailed genetic analysis much easier in the invertebrate models.
In conclusion, these three genetic models have enormous potential in the study of human sarcomeric muscle disease. The literature increasingly contains reports of human muscle disease mutations introduced into these model genetic organisms. This approach promises to be especially informative. The European Union recently funded a large research network Myores on ‘Multi-organismic approaches to muscle development and disease’ (www.myores.org).
Acknowledgements
The contributing authors are all members of the EU-funded FP6 Network of Excellence, Myores.
Contributor Information
Prof John Sparrow, Department of Biology, University of York, York, YO1 5DD, UK. Tel: 44-1904-328675; Fax: 44-1904-328825; jcs1@york.ac.uk.
Dr Simon M. Hughes, Randall Division of Cell and Molecular Biophysics and MRC Centre for Developmental Neurobiology, New Hunt’s House, King’s College London, Guy’s Campus, London, SE1 1UL. Tel: 44-20 7848 6445; Fax: 44-7848 6435; simon.hughes@kcl.ac.uk.
Dr Laurent Segalat, CNRS-CGMC, Universite Lyon-1 Claude Bernard, Batiment Mendel, 43 bld du 11 Novembre, 69622 Villeurbanne Cedex, France. Tel: 33-4-72-43-29-51; Fax: 33-4-72-43-29-51; segalat@cgmc.univlyon1.fr.
References
- 1.C. elegans sequencing consortium Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 2.Sulston JE, Schierenberg E, White JG, et al. The embryonic lineage of the nematode C. elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- 3.Lieschke GJ, Currie PD. Animal models of human disease: zebrafish swim into view. Nature Rev Genet. 2007;8:353–367. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- 4.Hughes SM, Salinas PC. Control of muscle fibre and motoneuron diversification. Curr Opin Neurobiol. 1999;9:54–64. doi: 10.1016/s0959-4388(99)80007-5. [DOI] [PubMed] [Google Scholar]
- 5.Pownall ME, Gustafsson MK, Emerson CP. Myogenic regulatory factors and the specification of muscle progenitors in vertebrate embryos. Ann Rev Cell Dev Biol. 2002;18:747–783. doi: 10.1146/annurev.cellbio.18.012502.105758. [DOI] [PubMed] [Google Scholar]
- 6.Bate M. The embryonic development of larval muscles in Drosophila. Development. 1990;110:791–804. doi: 10.1242/dev.110.3.791. [DOI] [PubMed] [Google Scholar]
- 7.Dietrich S, Schubert FR, Healy C. Specification of the hypaxial musculature. Development. 1998;125:2235–2249. doi: 10.1242/dev.125.12.2235. [DOI] [PubMed] [Google Scholar]
- 8.Dohrmann C, Azpiazu N, Frasch M. A new Drosophila homeo box gene is expressed in mesodermal precursor cells of distinct muscles during embryogenesis. Genes Dev. 1990;4:2098–2111. doi: 10.1101/gad.4.12a.2098. [DOI] [PubMed] [Google Scholar]
- 9.Kardon G. Muscle and tendon morphogenesis in the avian hind limb. Development. 1998;125:4019–4032. doi: 10.1242/dev.125.20.4019. [DOI] [PubMed] [Google Scholar]
- 10.Mankoo BS, Collins NS, Ashby P, et al. Mox2 is a component of the genetic hierarchy controlling limb muscle development. Nature. 1999;400:69–73. doi: 10.1038/21892. [DOI] [PubMed] [Google Scholar]
- 11.Horsley V, Jansen KM, Mills ST, et al. IL-4 acts as a myoblast recruitment factor during mammalian muscle growth. Cell. 2003;113:483–494. doi: 10.1016/s0092-8674(03)00319-2. [DOI] [PubMed] [Google Scholar]
- 12.Srinivas BP, Woo J, Leong WY, et al. A conserved molecular pathway mediates myoblast fusion in insects and vertebrates. Nat Genet. 2007;39:781–786. doi: 10.1038/ng2055. [DOI] [PubMed] [Google Scholar]
- 13.Hughes SM, Blau HM. Muscle fibre pattern is independent of cell lineage in postnatal rodent development. Cell. 1992;68:659–671. doi: 10.1016/0092-8674(92)90142-y. [DOI] [PubMed] [Google Scholar]
- 14.DiMario JX, Fernyak SE, Stockdale FE. Myoblasts transferred to the limbs of embryos are committed to specific fiber fates. Nature. 1993;362:165–167. doi: 10.1038/362165a0. [DOI] [PubMed] [Google Scholar]
- 15.Hoh JFY, Hughes S. Basal lamina and superfast myosin expression in regenerating cat jaw muscle. Muscle Nerve. 1991;14:316. doi: 10.1002/mus.880140503. [DOI] [PubMed] [Google Scholar]
- 16.Robson LG, Hughes SM. Local signals in the chick limb bud can override myoblast lineage commitment: induction of slow myosin heavy chain in fast myoblasts. Mech Dev. 1999;85:59–71. doi: 10.1016/s0925-4773(99)00060-x. [DOI] [PubMed] [Google Scholar]
- 17.Seed J, Hauschka SD. Clonal analysis of vertebrate myogenesis. 8. Fibroblast growth factor (FGF) dependent and FGF-independent muscle colony types during chick wing development. Dev Biol. 1988;128:40–49. doi: 10.1016/0012-1606(88)90264-3. [DOI] [PubMed] [Google Scholar]
- 18.Quinn LS, Holtzer H, Nameroff M. Generation of chick skeletal muscle cells in groups of 16 from stem cells. Nature. 1985;313:692–694. doi: 10.1038/313692a0. [DOI] [PubMed] [Google Scholar]
- 19.Gussoni E, Soneoka Y, Strickland CD, et al. Dystrophin expression in the mdx mouse restored by stem cell transplantation. Nature. 1999;401:390–394. doi: 10.1038/43919. [DOI] [PubMed] [Google Scholar]
- 20.LaBarge MA, Blau HM. Biological progression from adult bone marrow to mononucleate muscle stem cell to multinucleate muscle fibre in response to injury. Cell. 2002;111:589–601. doi: 10.1016/s0092-8674(02)01078-4. [DOI] [PubMed] [Google Scholar]
- 21.Felsenfeld AL, Curry M, Kimmel CB. The FUB-1 mutation blocks initial myofibril formation in zebrafish muscle pioneer cells. Dev Biol. 1991;148:23–30. doi: 10.1016/0012-1606(91)90314-s. [DOI] [PubMed] [Google Scholar]
- 22.Schilling TF, Kimmel CB. Musculoskeletal patterning in the pharyngeal segments of the zebrafish embryo. Development. 1997;124:2945–2960. doi: 10.1242/dev.124.15.2945. [DOI] [PubMed] [Google Scholar]
- 23.Neyt C, Jagla K, Thisse C, et al. Evolutionary origins of vertebrate appendicular muscle. Nature. 2000;408:82–86. doi: 10.1038/35040549. [DOI] [PubMed] [Google Scholar]
- 24.Holloway G, Currie P. Vertebrate myotome development. Birth Defects Res C Embryo Today. 2005;75:172–9. doi: 10.1002/bdrc.20046. [DOI] [PubMed] [Google Scholar]
- 25.Devoto SH, Stoiber W, Hammond CL, et al. Generality of vertebrate developmental patterns: evidence for a dermomyotome in fish. Evol Dev. 2006;8:101–110. doi: 10.1111/j.1525-142X.2006.05079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stellabotte F, Devoto SH. The teleost dermomyotome. Dev Dyn. 2007;236:2432–2443. doi: 10.1002/dvdy.21253. [DOI] [PubMed] [Google Scholar]
- 27.Stickney HL, Barresi MJ, Devoto SH. Somite development in zebrafish. Dev Dyn. 2000;219:287–303. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1065>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 28.Sehnert JA, Huq A, Weinstein BM, et al. Cardiac troponin T is essential in sarcomere assembly and cardiac contractility. Nat Genet. 2002;31:106–110. doi: 10.1038/ng875. [DOI] [PubMed] [Google Scholar]
- 29.Lambrechts D, Carmeliet P. Genetics in zebrafish, mice, and humans to dissect congenital heart disease: insights in the role of VEGF. Curr Top Dev Biol. 2004;62:189–224. doi: 10.1016/S0070-2153(04)62007-2. [DOI] [PubMed] [Google Scholar]
- 30.Pavlath GK, Thaloor D, Rando TA, et al. Heterogeneity among muscle precursor cells in adult skeletal muscles with differing regenerative capacities. Dev Dyn. 1998;212:495–508. doi: 10.1002/(SICI)1097-0177(199808)212:4<495::AID-AJA3>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 31.Roy S, VijayRaghavan K. Patterning muscles using organizers: larval muscle templates and adult myoblasts actively interact to pattern the dorsal longitudinal flight muscles of Drosophila. J Cell Biol. 1998;141:1135–1145. doi: 10.1083/jcb.141.5.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang JW, Kelly R, Daood M, et al. Alteration in myosatellite cell commitment with muscle cell maturation. Dev Dyn. 1998;211:141–152. doi: 10.1002/(SICI)1097-0177(199802)211:2<141::AID-AJA3>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 33.Heslop L, Beauchamp JR, Tajbakhsh S, et al. Transplanted primary neonatal myoblasts can give rise to functional satellite cells as identified using the MYf5(nlacZ/+) mouse. Gen Ther. 2001;8:778–783. doi: 10.1038/sj.gt.3301463. [DOI] [PubMed] [Google Scholar]
- 34.Rosenblatt JD, Parry DJ, Partridge TA. Phenotype of adult mouse muscle myoblasts reflects their fibre type of origin. Differentiation. 1996;60:39–45. doi: 10.1046/j.1432-0436.1996.6010039.x. [DOI] [PubMed] [Google Scholar]
- 35.Beauchamp JR, Morgan JE, Pagel CN. Dynamics of myoblast transplantation reveal a discrete minority of precursors with stem cell-like properties as the myogenic source. J Cell Biol. 1999;144:1113–1121. doi: 10.1083/jcb.144.6.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee JY, Qu-Petersen Z, Cao BH, et al. Clonal isolation of muscle-derived cells capable of enhancing muscle regeneration and bone healing. J Cell Biol. 2000;150:1085–1099. doi: 10.1083/jcb.150.5.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Asakura A, Seale P, Girgis-Gabardo A, et al. Myogenic specification of side population cells in skeletal muscle. J Cell Biol. 2002;159:123–134. doi: 10.1083/jcb.200202092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rowlerson A, Mascarello F, Radaelli G, et al. Differentiation and growth of muscle in the fish Sparus aurata (L). 2. Hyperplastic and hypertrophic growth of lateral muscle from hatching to adult. J Muscle Res Cell Motil. 1995;16:223–236. doi: 10.1007/BF00121131. [DOI] [PubMed] [Google Scholar]
- 39.Barresi MJF, D’Angelo JA, Hernandez LP, et al. Distinct mechanisms regulate slow muscle development. Curr Biol. 2001;11:1432–1438. doi: 10.1016/s0960-9822(01)00428-6. [DOI] [PubMed] [Google Scholar]
- 40.Rowlerson A, Radaelli G, Mascarello F, et al. Regeneration of skeletal muscle in two teleost fish: Spartus aurata and Brachydanio rerio. Cell Tiss Res. 1997;289:311–322. doi: 10.1007/s004410050878. [DOI] [PubMed] [Google Scholar]
- 41.Haffter P, Granato M, Brand M, et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- 42.Bate M. ‘The mesoderm and its derivatives. In: Bate M, Martinez Arias A, editors. The Development of Drosophila melanogaster. Cold Spring Harbor Press; 1993. [Google Scholar]
- 43.Taylor MV. Comparison of muscle development in Drosophila and vertebrates. In: Sink H, editor. Muscle Development in Drosophila. Landes Bioscience USA; 2006. [Google Scholar]
- 44.Sink H. Muscle Development in Drosophila. Landes Bioscience USA; 2006. [Google Scholar]
- 45.Peckham M, Molloy JE, Sparrow JC, et al. Physiological properties of the dorsal longitudinal flight muscle and the tergal depressor of the trochanter muscle of Drosophila melanogaster. J Muscle Res Cell Motil. 1990;11:203–215. doi: 10.1007/BF01843574. [DOI] [PubMed] [Google Scholar]
- 46.Sparrow JC, Geeves MA. Molecular assays for acto-myosin interactions. In: Vigoreaux JO, editor. Nature’s Versatile Engine: Insect Flight Muscle Inside and Out. Landes Bioscience, USA; 2004. [Google Scholar]
- 47.Mogami K, Hotta Y. Isolation of Drosophila flightless mutants which affect myofibrillar proteins of indirect flight muscle. Mol Gen Genet. 1981;183:409–417. doi: 10.1007/BF00268758. [DOI] [PubMed] [Google Scholar]
- 48.Cripps RM, Ball E, Stark M, et al. Dominant flightless mutants of Drosophila melanogaster and identification of a new gene required for normal muscle structure and function. Genetics. 1994;137:151–164. doi: 10.1093/genetics/137.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waterston RH. Muscle. In: Wood WB, editor. The Nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory; 1988. pp. 281–336. [Google Scholar]
- 50.Lecroisey C, Segalat L, Gieseler K. The C. elegans dense body: a dynamic anchoring point of the muscle. J Muscle Res Cell Motil. 2007;28:79–87. doi: 10.1007/s10974-007-9104-y. [DOI] [PubMed] [Google Scholar]
- 51.McArdle K, Allen TS, Bucher EA. Ca2+-dependent muscle dysfunction caused by mutation of the Caenorhabditis elegans troponin T-1 gene. J Cell Biol. 1998;143:1201–1213. doi: 10.1083/jcb.143.5.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bessereau J-L. Transposons in C. elegans. The C. elegans Research Community , WormBook. 2006 doi/10.1895/wormbook.1.70.1, http://www.wormbook.org.
- 53.Granger L, Martin E, Ségalat L. Mos as a tool for genome-wide insertional mutagenesis in Caenorhabditis elegans: results of a pilot study. Nucleic Acids Res. 2004;32:e117. doi: 10.1093/nar/gnh111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Evans TC. The C. elegans. Research Community WormBook; 2006. Transformation and microinjection. doi/10.1895/wormbook.1.108.1, http://www.wormbook.org. [Google Scholar]
- 55.Spradling AC, Stern D, Beaton A, et al. The Berkeley Drosophila Genome Project Gene Disruption Project: single P-element insertions mutating 25% of vital Drosophila genes. Genetics. 1999;153:135–177. doi: 10.1093/genetics/153.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clyne PJ, Brotman JS, Sweeney ST, et al. Green fluorescent protein tagging Drosophila proteins at their native genomic loci with small P elements. Genetics. 2003;165:1433–1441. doi: 10.1093/genetics/165.3.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parks AL, Cook KR, Belvin M, et al. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet. 2004;36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- 58.Duffy JB. GAL4 system in Drosophila: a fly geneticist’s Swiss army knife. Genesis. 2002;34:1–15. doi: 10.1002/gene.10150. [DOI] [PubMed] [Google Scholar]
- 59.Amsterdam A, Burgess S, Golling G, et al. A large scale insertional mutagenesis screen in zebrafish. Genes and Development. 1999;13:2713–2724. doi: 10.1101/gad.13.20.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ellingspen S, Laplante MA, Konig M, et al. Large scale enhancer detection in the zebrafish genome. Development. 2005;132:3799–3811. doi: 10.1242/dev.01951. [DOI] [PubMed] [Google Scholar]
- 61.Fire A, Xu S, Montgomery MK, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 62.Ahringer J. Reverse Genetics. The C. elegans Research Community Wormbook. 2006 doi/10.1895/wormbook.1.47.1, http://www.wormbook.org.
- 63.Kamath RS, Fraser AG, Dong Y, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 64.Dietz G, Chen D, Schnorrer F, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:51–56. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 65.Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- 66.Dodd A, Chambers SP, Love DR. Short interfering RNA-mediated gene targeting in the zebrafish. FEBS Lett. 2004;561:89–93. doi: 10.1016/S0014-5793(04)00129-2. [DOI] [PubMed] [Google Scholar]
- 67.Wolf MJ, Amrein H, Izatt JA, et al. Drosophila as a model for the identification of genes causing adult human heart disease. Proc Natl Acad Sci USA. 2006;103:1394–1399. doi: 10.1073/pnas.0507359103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tajsharghi H, Pilon M, Oldfors A. A Caenorhabditis elegans model of the myosin heavy chain IIa E706R mutation. Ann Neurol. 2005;58:442–448. doi: 10.1002/ana.20594. [DOI] [PubMed] [Google Scholar]
- 69.Nongthomba U, Cummins M, Clark S, et al. Suppression of muscle hypercontraction by mutations in the myosin heavy chain gene of Drosophila melanogaster. Genetics. 2003;164:209–222. doi: 10.1093/genetics/164.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sung SS, Brassington A-M, Grannatt K. Mutations in genes encoding fast-twitch contractile proteins cause distal arthrogryposis syndromes. Am J Human Genet. 2003;72:681–690. doi: 10.1086/368294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Myers CD, Goh PY, Allen TS, et al. Developmental genetic analysis of troponin T mutations in striated and nonstriated muscle cells of Caenorhabditis elegans. J. Cell Biol. 1996;132:1061–1077. doi: 10.1083/jcb.132.6.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nongthomba U, Clark S, Cummins M, et al. Troponin I is required for myofibrillogenesis and sarcomere formation in Drosophila flight muscle. J Cell Sci. 2004;117:1795–1805. doi: 10.1242/jcs.01024. [DOI] [PubMed] [Google Scholar]
- 73.Nongthomba U, Ansari MA, Stark M, et al. Aberrant splicing of a jump and flight muscle-specific exon in the Drosophila troponin-T gene. Genetics. 2007;177:295–306. doi: 10.1534/genetics.106.056812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Naimi B, Harrison A, Cummins M, et al. A tropomyosin-2 mutation suppresses a troponin-I myopathy in Drosophila. Mol Biol Cell. 2001;12:1529–1539. doi: 10.1091/mbc.12.5.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Montana ES, Littleton JT. Characterization of a hypercontraction induced myopathy in Drosophila caused by mutants in Mhc. J Cell Biol. 2004;164:1045–1054. doi: 10.1083/jcb.200308158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kronert WA, O’Donnell PT, Fieck A, et al. Defects in the Drosophila myosin rod permit sarcomere assembly but cause flight muscle degeneration. J Mol Biol. 1995;248:111–125. doi: 10.1006/jmbi.1995.0283. [DOI] [PubMed] [Google Scholar]
- 77.Prado A, Canal I, Barbas JA, et al. Functional recovery of troponin-I in a Drosophila heldup mutant after a 2nd site mutation. Mol Biol Cell. 1995;6:1433–1441. doi: 10.1091/mbc.6.11.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kronert WA, Acebes A, Ferrus A, et al. Specific myosin heavy chain mutations suppress TnI effects in Drosophila muscles. J Cell Biol. 1999;144:989–1000. doi: 10.1083/jcb.144.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sparrow JC, Nowak K, Durling HJ, et al. Muscle disease caused by mutations in the skeletal muscle alpha-actin gene, ACTA1. Neuromusc Disorders. 2003;13:519–531. doi: 10.1016/s0960-8966(03)00101-9. [DOI] [PubMed] [Google Scholar]
- 80.Nongthomba U, Pasalodos-Sanchez S, Clark S, et al. Expression and function of the Drosophila ACT88F actin isoform is not restricted to the indirect flight muscles. J Muscle Res Cell Motil. 2001;22:111–119. doi: 10.1023/a:1010308326890. [DOI] [PubMed] [Google Scholar]
- 81.Sparrow JC. Actin and arthrin’. In: Vigoreaux JO, editor. ‘Nature’s Versatile Engine: Insect Flight Muscle Inside and Out. Landes Bioscience, USA; 2004. [Google Scholar]
- 82.Domazetovska A, Ilkovski B, Kumar V, et al. Intranuclear rod myopathy: molecular pathogenesis and mechanisms of weakness. Ann Neurol. 2007;62:597–608. doi: 10.1002/ana.21200. [DOI] [PubMed] [Google Scholar]
- 83.Nowak KJ, Sewry CA, Navarro C, et al. Nemaline myopathy caused by absence of alpha-skeletal muscle actin. Ann Neurol. 2007;61:175–184. doi: 10.1002/ana.21035. [DOI] [PubMed] [Google Scholar]
- 84.Laing NG, Clarke NF, Dye DE, et al. Actin mutations are one cause of congenital fibre type disproportion. Ann Neurol. 2004;56:689–694. doi: 10.1002/ana.20260. [DOI] [PubMed] [Google Scholar]
- 85.Roberts RG, Bobrow M. Dystrophins in vertebrates and invertebrates. Human Mol Genet. 1998;7:589–595. doi: 10.1093/hmg/7.4.589. [DOI] [PubMed] [Google Scholar]
- 86.Gieseler K, Bessou C, Segalat L. Dystrobrevin- and dystrophin-like mutants display similar phenotypes in the nematode Caenorhabditis elegans. Neurogenet. 1999;2:87–90. doi: 10.1007/s100480050057. [DOI] [PubMed] [Google Scholar]
- 87.Grisoni K, Martin E, Gieseler K, et al. Genetic evidence for a dystrophin-glycoprotein complex (DGC) in Caenorhabditis elegans. Gene. 2002;294:77–86. doi: 10.1016/s0378-1119(02)00762-x. [DOI] [PubMed] [Google Scholar]
- 88.Bessou C, Giugia JB, Franks CJ, et al. Mutations in the Caenorhabditis elegans dystrophin-like gene dys-1 lead to hyperactivity and suggest a link with cholinergic transmission. Neurogenetics. 1998;2:61–72. doi: 10.1007/s100480050053. [DOI] [PubMed] [Google Scholar]
- 89.Grisoni K, Gieseler K, Mariol M, et al. The stn-1 gene of C. elegans is functionally related to dystrophin and dystrobrevin. J Mol Biol. 2003;332:1037–1046. doi: 10.1016/j.jmb.2003.08.021. [DOI] [PubMed] [Google Scholar]
- 90.Megeney L, Kablar B, Garrett K, et al. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Develop. 1996;10:1173–1183. doi: 10.1101/gad.10.10.1173. [DOI] [PubMed] [Google Scholar]
- 91.Towers PR, Lescure P, Baban D, et al. Gene expression profiling studies on Caenorhabditis elegans dystrophin mutants dys-1(cx-35) and dys-1(cx18) Genomics. 2006;88:642–649. doi: 10.1016/j.ygeno.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 92.Mariol MC, Segalat L. Muscular degeneration in the absence of dystrophin is a calcium-dependent process. Curr Biol. 2001;11:1691–1694. doi: 10.1016/s0960-9822(01)00528-0. [DOI] [PubMed] [Google Scholar]
- 93.Gaud A, Simon JT, Witzel T, et al. Prednisone reduces muscle degeneration in dystrophin-deficient Caenorhabditis elegans. Neuromusc Disorders. 2004;427:451–457. doi: 10.1016/j.nmd.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 94.Carre-Pierrat M, Mariol M-C, et al. Blocking of striated muscle degeneration by serotonin in C. elegans. J Muscle Res Cell Motil. 2006;27:253–258. doi: 10.1007/s10974-006-9070-9. [DOI] [PubMed] [Google Scholar]
- 95.Gieseler K, Grisoni K, Segalat L. Genetic suppression of phenotypes arising from mutations in dystrophin-related genes in Caenorhabditis elegans. Curr Biol. 2000;10:1092–1097. doi: 10.1016/s0960-9822(00)00691-6. [DOI] [PubMed] [Google Scholar]
- 96.Carre-Pierrat M, Grisoni K, Giesler K, et al. The SLO-1 BK channel of Caenorhabditis elegans is critical for muscle function and is involved in dystrophin dependent muscle dystrophy. J Mol Biol. 2006;358:387–395. doi: 10.1016/j.jmb.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 97.Greener MJ, Roberts RG. Conservation of components of the dystrophin complex in Drosophila. FEBS lett. 2000;482:13–18. doi: 10.1016/s0014-5793(00)02018-4. [DOI] [PubMed] [Google Scholar]
- 98.Neuman S, Kaban A, Volk T, et al. The dystrophin/ utrophin homologues in Drosophila and in sea urchin. Gene. 2001;263:17–29. doi: 10.1016/s0378-1119(00)00584-9. [DOI] [PubMed] [Google Scholar]
- 99.Neuman S, Kovalio M, Yaffe D, et al. The Drosophila homologue of the dystrophin gene - Introns containing promoters are the major contributors to the large size of the gene. FEBS Letts. 2005;579:5365–537. doi: 10.1016/j.febslet.2005.08.073. [DOI] [PubMed] [Google Scholar]
- 100.Van der Plas MC, Pilgram GSK, de Jon AWM, et al. Drosophila dystrophin is required for integrity of the musculature. Mech Dev. 2007;124:617–630. doi: 10.1016/j.mod.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 101.Dekkers LC, van der Plas MC, van Loenen PB, et al. Embryonic expression patterns of the Drosophila dystrophin-associated glycoprotein complex orthologs. Gene Expr Patterns. 2004;4:153–159. doi: 10.1016/j.modgep.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 102.Van der Plas MC, Pilgram GSK, Plomp JJ, et al. Dystrophin is required for appropriate retrograde control of neurotransmitter release at the Drosophila neuromuscular junction. J Neurosci. 2006;26:333–344. doi: 10.1523/JNEUROSCI.4069-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Steffen LS, Guyon JR, Vogel ED, et al. Zebrafish orthologs of human muscular dystrophy genes. BMC Genomics. 2007;8 doi: 10.1186/1471-2164-8-79. Art 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kunkel LM, Bachrach E, Bennett RR, et al. Diagnosis and cell-based therapy for Duchenne muscular dystrophy in humans, mice and zebrafish. J. Human Genet. 2007;51:397–406. doi: 10.1007/s10038-006-0374-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Guyon JR, Steffen LS, Howell MH, et al. Modeling human muscle disease in zebrafish. Bioch Biophys Acta. 2007;1772:205–215. doi: 10.1016/j.bbadis.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 106.Bassett DI, Bryson-Richardson RJ, Daggett DF, et al. Dystrophin is required for the formation of stable muscle attachments in the zebrafish embryo. Development. 2003;130:5851–5860. doi: 10.1242/dev.00799. [DOI] [PubMed] [Google Scholar]
- 107.Law DJ, Tidball JG. Dystrophin deficiency is associated with myotendinous junction defects in prenecrotic and fully regenerated skeletal muscle. Am J Pathol. 1993;142:1513–1523. [PMC free article] [PubMed] [Google Scholar]
- 108.Durbeej M, Campbell KP. Muscular dystrophies involving the dystroglycan complex. An overview of current mouse models. Curr Opin Gen Dev. 2002;12:349–361. doi: 10.1016/s0959-437x(02)00309-x. [DOI] [PubMed] [Google Scholar]
- 109.Cohn RD. Dystroglycan: important player in skeletal muscle and beyond. Neuromusc Disorders. 2005;15:827–841. doi: 10.1016/j.nmd.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 110.Deng W-M, Schneider M, Frock R, et al. Dystroglycan is required for polarizing the epithelial cells and oocyte in Drosophila. Development. 2003;130:173–184. doi: 10.1242/dev.00199. [DOI] [PubMed] [Google Scholar]
- 111.Shcherbata HR, Yatsenko AS, Patterson L, et al. Dissecting muscle and neuronal disorders in a Drosophila model of muscular dystrophy. EMBO J. 2007;26:481–493. doi: 10.1038/sj.emboj.7601503. [DOI] [PMC free article] [PubMed] [Google Scholar]