Summary
Myogenic regulatory factors of the myod family (MRFs) are transcription factors essential for mammalian skeletal myogenesis. Here we show that a mutation in the zebrafish myod gene delays and reduces early somitic and pectoral fin myogenesis, reduces miR-206 expression, and leads to a persistent reduction in somite size until at least the independent feeding stage. A mutation in myog, encoding a second MRF, has little obvious phenotype at early stages, but exacerbates the loss of somitic muscle caused by lack of Myod. Mutation of both myod and myf5 ablates all skeletal muscle. Haploinsufficiency of myod leads to reduced embryonic somite muscle bulk. Lack of Myod causes a severe reduction in cranial musculature, ablating most muscles including the protractor pectoralis, a putative cucullaris homologue. This phenotype is accompanied by a severe dysmorphology of the cartilaginous skeleton and failure of maturation of several cranial bones, including the opercle. As myod expression is restricted to myogenic cells, the data show that myogenesis is essential for proper skeletogenesis in the head.
Keywords: muscle, zebrafish, myosin, slow, fiber, fast, myod, myogenin, myf5, miR-206, skeleton, bone, cartilage, head, fin, haploinsufficiency
Introduction
As its name implies, skeletal muscle develops and functions in intimate contact with the skeleton. It is clear that cartilage and bone can develop in the absence of skeletal muscle (Chevallier, 1979; Christ et al., 1977; Kardon, 1998). However, abundant evidence from sports science and exercise physiology shows that there is mutual dependency of muscular and skeletal maintenance and growth (Robling, 2009). Indeed, skeletal defects have been observed early in development of mice genetically modified to lack skeletal muscle. For example, mice lacking the key myogenic regulatory transcription factors (MRFs) not only lack certain early muscle fibres but also show skeletal defects (Hasty et al., 1993; Rawls et al., 1998; Rudnicki et al., 1992; Valdez et al., 2000; Venuti et al., 1995; Zhang et al., 1995). As these MRF genes are expressed only in the skeletal muscle lineage, it has been suggested that skeletal defects arise due to altered signalling between early muscle cells and skeletal precursors (Vinagre et al., 2010; Vivian et al., 2000). Additionally, alteration of expression of adjacent genes caused by the genomic manipulations may have a role, as new knockout alleles have less severe skeletal consequences (Kassar-Duchossoy et al., 2004; Kaul et al., 2000). To compare musculoskeletal developmental interactions and the roles of individual MRFs across the vertebrates, we turned to the zebrafish, in which point mutations and/or antisense morpholinos minimize genetic complications.
Like mice, zebrafish have four MRF family members: Myod, Myf5, Myogenin and Mrf4/Myf6. Previously, we analysed myf5 and myf6 mutants and found rather mild defects, less pronounced at early stages than those in mice lacking these MRFs individually (Hinits et al., 2009; Kassar-Duchossoy et al., 2004; Kaul et al., 2000; Zhang et al., 1995). Mice lacking Myod also have a surprisingly mild phenotype (Rudnicki et al., 1992). In contrast, we observed severe defects in somitic and cranial myogenesis in myod morphant embryos (Hinits et al., 2009). These differences raise the possibility that the functions of myf5 and myod have diverged during vertebrate evolution. On the other hand, the phenotype of double myf5;myod loss of function appears quite similar in mice and fish; most, but not all, early myogenesis is ablated (Hinits et al., 2009; Kassar-Duchossoy et al., 2004). Thus, the extent of divergence in role of MRF genes is unclear.
Here we report analysis of myod and myog mutant alleles that reveal that zebrafish Myod is essential for viability. Myod is required for most early cranial myogenesis and some early pectoral fin and somitic myogenesis. Mutants lack the ability to feed and die during the post-hatch larval period. Myotome size is reduced in myod mutants, prior to independent feeding. There is also a transient reduction in somite bulk in myod heterozygotes. Unlike mice, myf5;myod double mutant zebrafish lack all myogenesis at least until 5 days post fertilization (dpf). Most strikingly, the lack of cranial muscle in myod mutants leads to a severe deformation and failure of maturation of cranial skeletogenesis. The data indicate that early myogenesis is essential for normal head patterning.
Materials and methods
Zebrafish lines and maintenance
Wild type and transgenic lines Tg(acta1:GFP)zf13 (Higashijima et al., 1997), Tg(-2.2mylz2:GFP)i135 (Moore et al., 2007), Tg(βactin:HRAS-EGFP)vu119 (Cooper et al., 2005), myf5hu2022 (Hinits et al., 2009) were maintained on King’s wild type background and staging and husbandry were as described (Westerfield, 1995). Myodfh261 and myogfh265 mutant alleles were identified by TILLING (Draper et al., 2004) and were maintained on the *AB background and genotyped by sequencing of PCR products amplified from fin clip or embryo genomic DNA using primers 5′GGACCCCAGGCTTGTTC3′ and 5′GTTGGATCTCGGACTGGA3′ for myod, 5′AACCGGGCCATTGTCTCCA3′ and 5′CTCCATCGGCAGGCTGTAGTAGTAGTT3′ for myf5 and 5′TGACAGCTTTACCATCGCGCTTGA3′ and 5′GTCAGTTCACTCAACCAGCAGGAGCATGAC3′ for myog (the last contains a single base mutation for the dCAPS method, http://labs.fhcrc.org/moens/Tilling_Mutants/index.html).
In situ mRNA hybridisation, immunohistochemistry and histology
In situ mRNA hybridisation and immunohistochemistry were performed as described (Hinits and Hughes, 2007). Fluorescein- or digoxigenin-tagged probes used were myog (Weinberg et al., 1996), eng2a (Ekker et al., 1992), mylz2 and myhz1 (Xu et al., 2000), smyhc1 (Bryson-Richardson et al., 2005), actin (acta1b, IMAGE 7284336), dlx2a (Akimenko et al., 1994), klf2b (Oates et al., 2001). Dual-digoxigenin-labelled miR-206 specific LNA probes were obtained from Exiqon and hybridised and washed at 50 °C as described in (Sweetman et al., 2008). Antibodies were against Myod and Pax3/7 (Hammond et al., 2007), Myog (Hinits et al., 2009), striated muscle myosin heavy chain (MyHC; A4.1025 (Blagden et al., 1997) or MF20 (DHSB)), slow MyHC (F59; Devoto et al., 1996) and Smooth muscle Myosin (Myh11, Biomedical Technologies). Stains with alcian blue for cartilage and alizarin red for ossified bone was as described (Walker and Kimmel, 2007), followed by dissection to separate upper and lower head skeleton. 4,5-Diaminofluorescein diacetate (DAF-2DA, Santa Cruz) staining of bone was as described (Grimes et al., 2006).
Embryo manipulation
Myod MO (Gene-Tools, 5′-ATATCCGACAACTCCATCTTTTTTG-3′) was injected into 1-2 cell stage embryos at 4 ng/embryo. Cyclopamine (100 μM in fish medium) or vehicle control was added at 50% epiboly to embryos whose chorions had been punctured with a 30 G hypodermic needle.
Muscle size analysis
Tg(βactin:HRAS-EGFP)vu119 fish were bred onto myodfh261 and live 0.2 mM 1-phenyl-2-thiourea-treated embryos hemizygous for ßactin:GFP were embedded in 1.5% low melting point agarose in fish water containing MS222, and imaged by confocal microscopy on a Zeiss Exciter using a 20x1.0 W dipping objective on successive days. Fish were released from the agarose each day. A single lateral image and three equi-spaced transverse images within somite 17 were collected from each embryo at each stage. The somite was outlined in Zen software and an average transverse area calculated. This area was multiplied by the somite length, measured at the horizontal myoseptum, to yield an estimate of somite volume for each fish at each stage. After analysis, individual embryos were genotyped by sequencing of genomic PCR. Data are presented as mean ± s.e.m. with the number of embryos at each developmental stage indicated. Somite volume of fixed MF20-stained 72 hpf embryos was calculated similarly, and genotyped as mutants or sibs according to head muscle immunostaining. Data was analysed by two way ANOVA showing significant effect of both age and genotype using Graphpad Prism 4.0. Adult (15 month) fish from a single tank were anaesthetised, weighed and length measured, then tail-clipped for genotyping by sequencing of genomic PCR.
Results
Myod mutation delays slow and reduces fast myogenesis
TILLING (Draper et al., 2004) was used to isolate a zebrafish mutant in myod with a stop codon at residue 126 early in the second helix, thereby generating a predicted null mutation (Fig. 1A; (Davis et al., 1990; Ma et al., 1994)). One quarter of embryos from a carrier in-cross of myodfh261/+ lacked Myod immunoreactivity. DNA sequencing of individual stained embryos confirmed that those lacking Myod were mutants and that heterozygotes had less staining than wild types (Fig. 1B; quantification of all experiments is presented in Table S1). Myod mRNA was reduced in heterozygotes and greatly diminished in mutants at 10 somite stage (10s), suggesting nonsense-mediated decay of the mutant mRNA (Fig. 1C). Consistent with the result of morpholino (MO) knockdown of Myod, slow myogenesis was delayed in mutants (Fig. 1B)(Hinits et al., 2009). In adaxial precursors of superficial slow fibres, myog mRNA and other muscle differentiation markers examined were expressed, although delayed (Fig. 1D). However, eng2a, a marker of the specialised muscle pioneer cells, was diminished (Fig. 1E). Thus, this mutant confirms that Myod contributes to, but is not essential for, early slow myogenesis (Hinits et al., 2009).
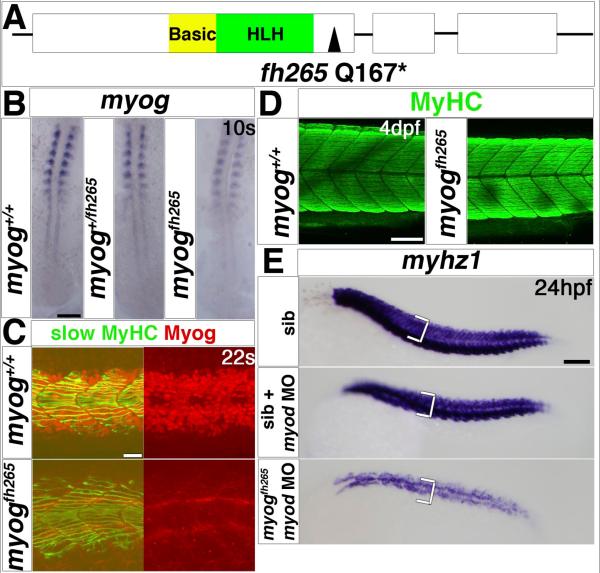
Fig. 1. Myodfh261 mutants have delayed muscle differentiation.
A. Schematic of Myod gene and protein showing the fh261 arginine to stop mutation within the helix-loop-helix domain, which is essential for dimerisation and DNA binding. B. Dual immunodetection of Myod and slow myosin in embryos from a myodfh261/+ incross, genotyped by sequencing of genomic PCR. Dorsal view, anterior to top. C. In situ mRNA hybridization reveals reduction in myod mRNA in embryos from a myodfh261/+ incross. Wholemount, anterior to top. D-F. In situ mRNA hybridization for indicated probes on myodfh261 mutants and their siblings. D. Adaxial slow precursors show reduced myog and smyhc1 at 10s and miR-206 at 15s, but actin mRNA appeared less affected (white arrowheads). Fast muscle precursors had no myog or miR-206 expression at this stage (black arrowheads). Dorsal flatmount, anterior to top. E. eng2a expression is reduced in myodfh261 mutant in regions of muscle pioneers (black arrowhead) and medial fast fibres (red arrowhead). Lateral wholemount, anterior to left. F. Embryos labelled for fast mylz2 with Fast Red were cryo-sectioned at the region indicated (arrow, left panels). Immunofluorescent detection of Pax3/7 protein revealed increased nuclei in the lateral somite in myodfh261 mutants (yellow arrowheads). Dorsoventral extent of fast myhz1 mRNA (bracket, right panels) is reduced in myodfh261 mutants at 24 hpf. Lateral wholemounts, anterior to left. Bars = 100 μm.
Loss of Myod caused more dramatic defects in fast myogenesis in the somite. Early myog and later mylz2 and myhz1 myosin mRNAs were diminished in fast muscle (Fig. 1D,F). Conversely, Pax3/7, a marker of dermomyotome was up-regulated in the lateral somite at 24 hpf, with more Pax3/7-immunoreactive nuclei in sections of mutants (Fig. 1F). In summary, fast myogenesis was reduced in myodfh261 mutants, extending results from MO knockdown studies (Hammond et al., 2007; Hinits et al., 2009; Maves et al., 2007).
Myogenin cooperates with Myod in fast myogenesis
Myogenin is a Myod target gene. We previously suggested that Myogenin cooperates with Myod in somitic fast myogenesis (Hinits et al., 2009). We isolated a myogfh265 mutant by TILLING, which has a stop codon in place of residue 167, just downstream of the helix-loop-helix domain (Fig. 2A). As with myod, myog mRNA was reduced in heterozygotes and greatly diminished in mutants at 10s, suggesting nonsense-mediated decay of the mutant mRNA (Fig. 2B). Although Myog immunoreactivity was lost in mutants, no defect was observed either in in-crosses from heterozygous myogfh265/+ carrier fish or in genotyped myogfh265/+ mutants: pectoral fin, jaw and somite morphology and movement appeared normal, at least until 5 dpf (Fig. 2C,D and data not shown). However, to date no myogfh265/+ mutants were obtained among in-cross progeny genotyped at 10 weeks of age.
Fig. 2. Myogenin cooperates with Myod in fast myogenesis.
A. Schematic of Myog gene and protein showing the fh265 glutamine to stop mutation following the helix-loop-helix domain. B-E. In situ mRNA hybridization for myog (B) or myhz1 (E) or immunodetection of Myog and slow MyHC (C) or MyHC (D) in myog+/fh265 incrosses, injected with myod morpholino as indicated (E). B. Expression of myog mRNA is little affected in myogfh265 mutants. Dorsal flatmounts, anterior to top. C. Nuclear immunoreactivity with a polyclonal anti-rat Myog antibody is lost in myogfh265 mutants at 22s. D. All embryos from a myog+/fh265 incross show normal levels of MyHC at 4 dpf. E. No change in myhz1 mRNA is detected in myogfh265 mutants at 24 hpf (top panel). Injection of myod MO into myogfh265 (bottom panel) leads to a greater loss of fast muscle than when injected into their siblings (middle panel), which are comparable to myodfh261 mutants (see Fig. 1F). Lateral views, anterior to left (C-E). Bars = 100 μm (except C = 20 μm).
To test the hypothesis that Myod and Myog cooperate to drive fast myogenesis, myod MO was injected into a myogfh265/+ in-cross. In siblings, myod MO alone reduced myhz1 expression to a similar extent to that observed in myodfh261 mutants (Fig. 2E, compare to Fig. 1F). In 25% of embryos from a myogfh265/+ incross, myod MO elicited an exacerbated loss of fast myhz1 mRNA (Fig. 2E). We conclude that although full length Myog is dispensable for embryonic development, it functions to promote early fast myogenesis in the somite in co-operation with Myod.
Myod promotes miR-206 expression
Myod drives normal fast muscle differentiation in zebrafish acting, in part, through induction myog (Hinits et al., 2009). By 72 hpf, myog mRNA has declined in differentiated muscle, although its expression in dorsal and ventral somite extremes is still Myod-dependent, suggesting that Myod is required for ongoing myogenesis (Fig. 3D). miR-206 is a muscle-specific microRNA that, like Myog, has been suggested to promote myoblast differentiation (Hirai et al., 2010). In zebrafish, miR-206 expression was similar to that of myog in early stage somites, accumulating weakly in adaxial slow fibres as they terminally differentiate, and more strongly in medial fast muscle as it formed (Figs 1D, S1A). At 24 hpf, miR-206 was abundant in fast muscle of the somite (Fig. S1) and at later stages also in head and pectoral fin muscle, persisting beyond myog in differentiated fast fibres (Fig. S1B-D). Ablation of Myod function, either in myodfh261 or morphants, led to down-regulation of miR-206 in both slow and fast cells at 15s, although faint signal in the PSM was unchanged (Fig. 1D). Lateral miR-206 is also reduced by lack of Hh signalling, suggesting that either expression persists in slow fibres at 24 hpf or Hh signalling is required for miR-206 expression in some fast muscle (Fig. S1E). At this stage, miR-206 is clearly reduced in fast muscle of the myodfh261 mutant (Fig. S1E). Reduction in miR-206 persisted during the pre-hatching period after loss of myod function (Fig. 3A,B), and was comparable in myod morphants and myodfh261 mutant (Fig. 3C,D). By contrast, no alteration in miR-206 was detected at 15s, 24 or 48 hpf embryos from a myf5hu2022/+ in-cross (data not shown). These findings suggest that miR-206 accumulates in differentiating muscle fibres and that its loss could contribute to the reduction in muscle differentiation observed at 24 hpf in myodfh261 mutants.
Fig. 3. Muscle growth is impaired in myodfh261 mutants.
In situ mRNA hybridization for miR-206, myog and myhz1 and immunodetection of MyHC. Lateral (A,C,D) views, anterior to left or transverse cryosections (B). A. miR-206 in sequence-genotyped 48 hpf embryos from a myod+/fh261 incross is primarily in fast fibres and is strongly reduced. B. Embryos from A were sectioned and stained with MyHC. miR-206 expression is strongly reduced in myodfh261 mutants compared with their siblings, while MyHC immunodetection is only mildly reduced. C. At 72hpf, miR-206 has increased in myod morphants, but is still less than in siblings. D. Compared to sibs, myodfh261 mutants (identified by head muscle defects) had less myog mRNA (arrowheads) and reduction in dorsoventral extent of somitic miR-206 and myhz1 mRNA (brackets). E. Somite volume increases with age, but is reduced in myodfh261 mutant. These differences were consistent in live and fixed embryos. Number of embryos per condition are shown on columns and t-test statistics are indicated above columns. F. Adult fish mass and length. Bars = 100 μm.
The reduction in miR-206 accumulation appeared to be greater than the loss of overall muscle mass in myodfh261 mutants at 48 hpf (Fig. 3B). Congruently, myodfh261/+ heterozygotes showed a reduction in miR-206 compared to wild types (Fig. 3A). These data suggest that Myod function up-regulates miR-206 in differentiated muscle fibres.
Myod requirement for somitic myogenesis appears transient
The reduction in miR-206 suggested that muscle growth might be decreased in myodfh261 mutants. Myod morphants have reduced muscle differentiation at 24 hpf, but growth did occur between 24 and 72 hpf ((Hinits et al., 2009); Fig. 3C). We used the myodfh261 mutant to test whether this growth was truly Myod-independent or reflected loss of MO effectiveness. At 24 hpf, myodfh261 mutants had approximately half the somite volume of wild type siblings. Strikingly, even heterozygous myodfh261/+ embryos had significantly reduced somite volume, intermediate between wild type and mutant siblings (Fig. 3E). These differences appear to be maintained at 48 hpf (Fig. 3A). At 72 hpf, both myhz1- and miR-206-expressing tissue was reduced, as in myod morphants (Fig. 3C,D). In parallel, myodfh261 mutants had reduced myotome volume compared to siblings (Fig. 3E). Significant somite growth was, nevertheless, observed in myodfh261 mutants between 24, 72 and 120 hpf (Fig. 3A-E; P<0.001). At 120 hpf, myodfh261 mutants were slightly smaller than wild type siblings, but increase in somite volume since 24 hpf seemed to have been normal (Fig. 3E). Studies of growth after 5 dpf were not possible in mutants owing to cranial defects (see below). We conclude that Myod is required for normal somitic myogenesis and its lack leads initially to smaller muscle bulk.
At 24 hpf, heterozygotes had significantly less somitic muscle than wild type siblings, but this defect became less significant in larvae (Fig. 3E). We compared the growth into adulthood of fish with one or two functional copies of the myod gene. Heterozygous myodfh261/+ fish survived well to adulthood, and were not significantly lighter than their wild type siblings (P = 0.27; Fig 3F). Fish growth is difficult to measure because any slight size advantage leads to success in competition for food and consequent faster growth. We used fish length to control for overall growth differences. Heterozygotes were not significantly different in length from wild type (P = 0.36; Fig. 3F). These data show that loss of one myod allele has little effect on growth of zebrafish.
Myod drives cranial and pectoral fin myogenesis
Like myod morphants, myodfh261 mutants lacked miR-206 in most cranial muscle at 72 hpf, with the exception of the sternohyoideus muscle (Fig. 4A). Other cranial muscle markers, myog, myhz1 and myosin accumulation, were also greatly reduced or entirely absent (Fig. 4B-D). Fin muscle was also diminished (Fig. 4B-E), but to a variable extent amongst mutant embryos, with 68% showing bilaterally symmetrical reduction (28/41) and 32% (13/41) having asymmetry between fins of the same embryo, with greater reduction on the right more common (11/13 P = 0.013 X2; Fig. 4E). Similarly, the levator and adductor operculi muscles were often present but reduced in fibre number. In contrast, the adjacent dilatator operculi and adductor hyomandibulae and most other cranial muscles were missing (Fig. 4F). These data confirm our morphant finding that Myod is the primary MRF driving early myogenesis in cranial and pectoral fin muscles (Hinits et al., 2009).
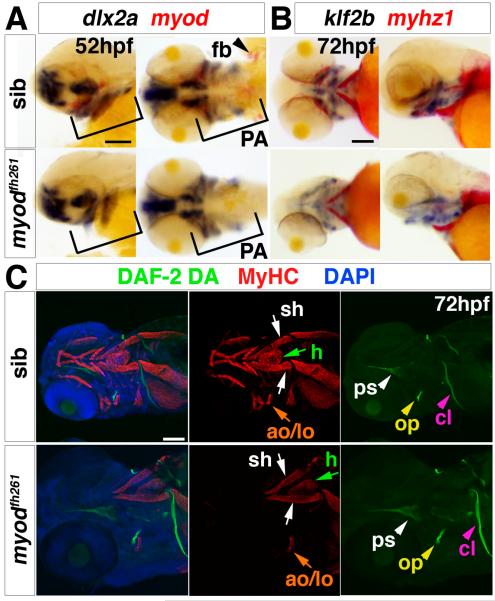
Fig. 4. Myodfh261 mutants lack head muscles and have defects in cartilage morphogenesis.
In situ RNA hybridisation for miR-206 (A), myog (B), myhz1 (C, E) and klf2b (E) or immunodetection of MyHC (MF20, D; A4.1025, F). Ventral (A, D and left panels of B and C), lateral (F, right panels of B and C) or dorsal (E) views, anterior to left. A. Myod morphant and myodfh261 mutant have similar loss of most cranial muscles except sternohyoideus (yellow arrows). B-D. In myodfh261 mutants, myog and myhz1 mRNAs and MyHC are lost from many cranial muscles (blue arrows). Expression remains in sternohyoideus (yellow arrows), Weak myog mRNA is detected in adductor mandibulae (B, black arrows) and occasional fibres differentiate in adductor mandibulae. Hypaxial muscles (C, green arrows) seem unaffected and pectoral fin muscles (B-F, white arrows, dotted area in F) are variably reduced. E. In myodfh261 mutants, most head muscles are consistently missing, even though sternohyoid (yellow arrows) and cartilage precursors expressing klf2b are present, Pectoral fin muscle is variably reduced (white arrows). F. Higher magnification shows that levator and adductor operculi are partially spared in myodfh261mutants, whereas pectoral fin and other muscles are missing, revealing the underlying somitic muscle (s). do dilatator operculi, ah adductor hyomandibulae, lo levator operculi, ao adductor operculi, l-5 levator 5, pp protractor pectoralis, Bars = 100 μm.
We examined the effect of loss of Myod on various other muscles linking the hypobranchial and pectoral regions. Whereas sternohyoideus was substantially preserved, most other muscles were reduced or missing entirely in myodfh261 mutants. However, small groups of myhz1-expressing cells are observed in the pectoral zone (Fig. 4C,E). We analysed the protractor pectoralis, in particular, because this is thought to be the teleost homologue of the cucullaris muscle group (Diogo et al., 2008; Greenwood and Lauder, 1981). Looking anterior to the cleithrum, no protractor pectoralis or levator 5 muscles were detected in myodfh261 mutants (Fig. 4F).
Lack of Myod is larval lethal. At embryonic stages, myodfh261 mutants were identified by sequence genotyping at the expected frequencies. In breeding programmes, we have not found an homozygous mutant at 13 dpf or beyond. We also separated myodfh261 mutant embryos from siblings at 5 dpf, when cranial defects become apparent (Fig. 5A) and reared them with feeding. Whereas all siblings survived (36/36), all myodfh261 mutant embryos died (9/9) in the period 6-12 dpf, presumably due to failure to feed.
Fig. 5. Myodfh261 mutants have cranial bone defects.
A. Bright field image of a live 6 dpf myodfh261 mutant (retrospectively sequence genotyped) and a 5 dpf myod morphant showing craniofacial defects of lowered jaw and open mouth compared with sibling and control, respectively. B,C. Alcian blue and alizarin red double staining for cartilage and bone in myodfh261 mutants and siblings. At 5 dpf (B), cartilage components in mutants have different morphology with the whole lower jaw dropping, forming a gaping mouth. Both first and second arch derivatives, such as Meckel’s (mk) and ceratohyal (ch) cartilage respectively, are bent ventrally. The ethmoid plate (eth) is only mildly affected. Little bone staining is detected in the mutant. Anterior to left, ventral (left panel) and lateral (right panel) views. At 6 dpf (C), myodfh261 mutants show various cartilage and bone defects; the opercle (arrowheads) is missing, ceratobranchial 5 and the parasphenoid (white arrows) less ossified bone and the cleithrum is reduced. Cartilage cells in the mutant remain rounded, failing to mature into flattened columns. Anterior to left. Wholemounts in lateral view (left panel), or flatmounts of the dissected pharyngeal skeleton (ventral view, second from left) or neurocranium (dorsal view, central panel). Large boxed areas are magnified in the fourth panels, showing reduced ossification of ceratobranchial 5 and cleithrum. Small boxed areas are magnified in righthandmost panels to reveal rounded morphology of cartilage cells in the joint between Meckel’s and palatoquadrate (pq) cartilages in the myodfh261 mutant. cl, cleithrum; nc, notochord; op, opercle; ps, parasphenoid; cb5 ceratobranchial 5. Bars = 100 μm.
Myod is essential for normal cranial skeletogenesis
By 5 dpf, myodfh261 mutants lacking cranial muscle have a striking morphological defect of the mouth (Fig. 5A). The defect was phenocopied in myod morphants, eliminating the possibility that a second mutation linked to the myodfh261 allele accounts for the skeletal defects (Fig. 5A). At 5 dpf, Meckel’s and ceratohyal cartilages are down-turned, giving a permanently open protruding mouth (Fig. 5B). Dissection and flatmounting of cranial cartilages revealed that all major cartilage elements are present at 6 dpf (Fig. 5C). However, the morphology of chondrocytes in the head of the palatoquadrate was rounded in myodfh261 mutants and they failed to form the stacks observed in siblings, correlating with failure of morphogenesis of the joint articulation between Meckel’s and palatoquadrate cartilages (Fig. 5C). We conclude that Myod function is required for aspects of cranial cartilage morphogenesis.
Some dermal bones, such as the opercle, cleithrum, and parasphenoid, which form directly from mesenchyme, were reduced or missing at 5 dpf in myodfh261 mutants (Fig. 5B). A day later, ossified cleithrum and parasphenoid were apparent, but the opercle was still missing. Moreover, ossification of cartilaginous bones such as the ceratobranchial 5 was reduced (Fig. 5C). Myod is therefore required for normal formation of bone.
No skeletal defects were observed in myodfh261 mutants prior to, or at, the normal time of cranial muscle differentiation at 3 dpf (Fig. 6A-C). We examined dlx2a mRNA, which is a transcription factor expressed in branchial arch skeletal precursors, klf2b mRNA, another transcription factor which prefigures cartilage formation, and the fluorescent nitric oxide indicator 4,5-diaminofluorescein diacetate (DAF-2DA), which strongly labels dermal bones (Lepiller et al., 2007). None was disrupted in myodfh261 mutants at 72 hpf (Fig. 6A-C). Thus, the striking skeletal defects in myodfh261 mutants only arise after the failure of muscle formation, consistent with the restriction of myod mRNA to myogenic zones.
Fig. 6. Early cranial skeletal development is normal in myodfh261 mutants.
A,B. In situ mRNA hybridisation for dlx2a and myod (A), klf2b and myhz1 (B) in 52 hpf (A) and 72 hpf (B) myodfh261 mutant and sibling embryos, shown in wholemount, anterior to left. Outer panels, lateral view, central panels, ventral view. Myod mRNA is absent but dlx2a unaffected in the pharyngeal arches (PA, brackets). Myhz1 mRNA is detected in sternohyoideus (sh, arrows) but not in other head muscles in the mutant. klf2b mRNA in skeletal parts is normal. C. Confocal stacks of heads of myodfh261 mutant and sibling stained with DAF-2DA (green), MyHC (MF20, red) and DAPI (blue). Anterior to left, ventrolateral view. Muscle in the mutant is missing except for the sternohyoideus (sh, white arrows), adductor operculi and levator operculi (ao and lo, orange arrows). Bones such as the opercle (op, yellow arrowhead), cleithrum (cl, pink arrowheads) and parasphenoid (ps, white arrowheads) are normal at this stage. Bar = 100 μm.
Myf5 and Myod are essential for embryonic myogenesis
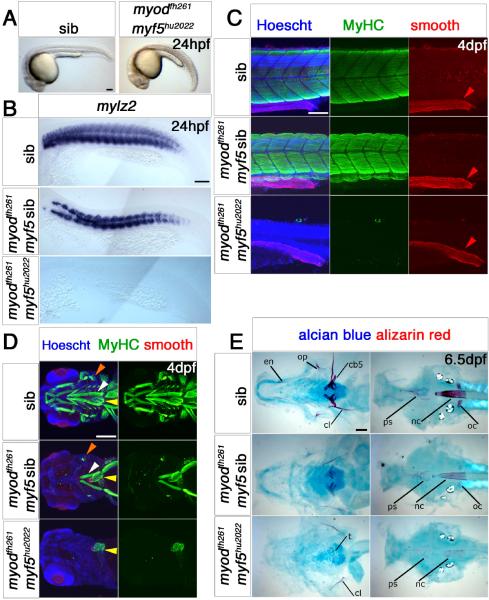
Myf5 and myod double morphants lack muscle at 15s (Hammond et al., 2007; Maves et al., 2007), but by 24 hpf small amounts of somitic muscle form even in myf5hu2022 mutants treated with myod morpholino (Hinits et al., 2009). To test whether this muscle is formed independent of both Myf5 and Myod activities, we bred double mutant myf5hu2022;myodfh261 fish. Such embryos lacked all muscle differentiation at 15s (Fig. S2A). Even a single functional myf5 allele was sufficient for partial differentiation at this stage. At 24 hpf, neither slow nor fast muscle was found in the double mutants (Fig. 7A,B and Fig. S2B), suggesting that the residual muscle we observed in myf5hu2022mutant+myod MO (see (Hinits et al., 2009)) differentiated due to inefficiency of the myod MO. All cranial, pectoral fin and somitic muscle was entirely lacking at 72 and 120 hpf (Fig. 7C,D and Fig. S2C-E), indicating that either Myf5 or Myod is essential for early myogenesis in the zebrafish.
Fig. 7. Myf5 or Myod is required for myogenesis.
Wholemounts (A), flatmounts (B) and confocal stacks (C,D) in lateral (A-C) or ventral (D) view of zebrafish embryos from myf5hu2022/+;myodfh261/+ in-cross analysed at the indicated stage by in situ mRNA hybridization for mylz2 (B) or immunohistochemistry for all-MyHC (green), smooth muscle myosin heavy chain 11 (red) and Hoechst (blue) in C,D, anterior to left. A. By 24 hpf, double mutant embryos are curved and have misshapen somites with signs of cell death. B. mRNA encoding fast myosin is reduced in putative myodfh261 mutants and absent in doubles. C. Confocal stacks of somite-16 region at 4 dpf embryos show that no skeletal muscle MyHC is detected in genotyped double mutants, in which embryos are severely reduced in size. Smooth muscle myosin in the gut (red arrowheads) are unaffected. D. Confocal stacks of the head region of 4 dpf embryos show that all skeletal muscle is missing in myf5hu2022;myodfh261 double mutant, while heart (yellow arrows) and smooth muscle are intact. myodfh261 mutant is missing all muscle except for the sternohyoideus (white arrows), adductor operculi and levator operculi (orange arrows). E. Alcian blue and alizarin red double staining for cartilage and bone in 6.5 dpf myf5hu2022;myodfh26 1 shows a more severe phenotype than in myodfh261 mutants and siblings. Flatmounts of the dissected pharyngeal skeleton (ventral view, left panels) or neurocranium (dorsal view, right panels). Cartilage components in double mutants stain poorly. Bone is only detected in the double mutant in cleithrum (cl) and teeth (left panel) and faintly in the notochord (nc) and parasphenoid (ps) (right panel). oc, occipitals; op, opercle; en, enteropterygoid; cb5, ceratobranchial 5. Bars = 100 μm.
Double mutant myf5hu2022;myodfh261 fish show cranial skeletal defects more severe than those of myodfh261 single mutants (Fig. 7E). As both myodfh261 and myf5hu2022;myodfh261 mutant fish die prior to trunk skeletogenesis, the effect of muscle loss on vertebral and rib development could not be assessed. However, the skeletons of adult myodfh261/+ and 13 dpf myodfh261/+ and myf5hu2022/+;myodfh261/+ dual heterozygous fish showed no consistent defects (data not shown). Among the cohort of fish analysed at 13 dpf no myf5hu2022;myodfh261/+ or myf5hu2022 mutants were found among 30 larvae analysed, showing that these genotypes fail to survive.
Discussion
The findings described here provide genetic demonstration of three major points. First, that Myod is essential for viability, normal somite and limb myogenesis and most of cranial myogenesis in zebrafish. Second, that either myf5 or myod is essential for all embryonic muscle differentiation. Third, that formation of a normal cranial skeleton is dependent upon the proper generation of musculature.
MRF roles in muscle development
We previously suggested, based on morpholino knockdown experiments, that myod functions to drive differentiation of a specific subset of somitic muscle fibres (Hinits et al., 2009). The results presented here provide independent genetic confirmation that Myod is essential for both cranial and somitic myogenesis. We show here that, in zebrafish, Myod is essential for viability, in sharp contrast to mice in which Myod1 null mutants are viable and fertile (Rudnicki et al., 1992). The molecular nature of myodfh261 and the severity of the phenotype suggest that the allele is a functional null. As a truncated N-terminal peptide could theoretically be produced in myodfh261, either the truncated protein is not significantly accumulated or the in vitro finding that a Myod protein lacking the second helix loses its ability to bind DNA, dimerise and function in myogenesis (Davis et al., 1990)) is also true for Myod function in vivo. In this regard, we note that the mutant myod mRNA is less abundant, probably due to nonsense-mediated decay, indicating a low translation rate of the truncated N-terminal peptide.
We describe a persistent somitic muscle defect in myodfh261 mutants, at least until 5 dpf, when lack of ability to feed complicates further analysis (see below). Nevertheless, significant somitic muscle growth does occur during the larval period. Myf5 is required for this growth, as myf5;myod double mutants fail to make any skeletal muscle. The myf5-dependent cells do not fully compensate for lack of Myod. Yet Myf5 can support significant pre-hatching growth in myodfh261 mutants, despite the low levels of myf5 mRNA detected at these stages. Numerous minor defects are present in Myod mutant mice, even though they are viable (Brack et al., 2005; Chargé et al., 2008; Hughes et al., 1997; Megeney et al., 1996; Tiidus et al., 1996; Wang et al., 2003; Yablonka-Reuveni et al., 1999). MyoDm1 mutant mice show poor postnatal survival when in competition with littermates carrying a wild type MyoD allele, but survive well without such competition (Rudnicki et al., 1992). In the case of myodfh261 mutant zebrafish, by contrast, all mutants fail to survive in bulk rearing, in which competition for resources is less intense than in mouse pups, and even when reared alone. It seems that despite elements of redundancy between Myf5 and Myod, a significant role for each explains their conservation across the vertebrates.
The identification of haploinsufficiency in myodfh261/+ embryos demanded detailed study. By comparing fish of molecularly verified genotype, we describe both a significant reduction in Myod protein immunoreactivity and miR-206 at early stages and a reduction of embryonic muscle bulk in myodfh261/+ heterozygotes. Haploinsufficiency in MyoD and Mrf4 has been observed at the molecular level in mice (Patapoutian et al., 1995; Rudnicki et al., 1992). In humans, haploinsufficiency in Mrf4 has been suggested to exacerbate muscular dystrophy (Kerst et al., 2000). With growth during the larval period, the muscle defect in myodfh261/+ heterozygotes appears to diminish, at least as a proportion of total muscle bulk. Adult myodfh261/+ heterozygous fish had no significant growth defect.
Pectoral fin muscle derives from cells of somitic origin (Neyt et al., 2000). Fin muscle was delayed and diminished in myodfh261 mutants, although some fin movement was observed by 5 dpf. This confirms our finding of variable defects in fin musculature in myod morphants and is consistent with the delayed limb myogenesis in MyoD1 mutant mice (Chen and Goldhamer, 2004; Hinits et al., 2009; Kablar et al., 1997). Recent analyses have highlighted the importance in amniotes and basal chordates of the cucullaris muscle group that links forelimb and neck movement and stabilizes the shoulder girdle (Kusakabe et al., 2011; Theis et al., 2011). The development and even the existence of cucullaris muscles in teleosts has been unclear, as the group is reduced and appears to develop late, being known as the protractor pectoralis (Diogo et al., 2008; Greenwood and Lauder, 1981). The zebrafish protractor pectoralis depends on Myod for its formation, like many other head muscles. However, as some somitic myogenesis is also Myod-dependent, our finding does not address the developmental origin of the teleost protractor pectoralis.
Although most intrinsic head muscle are consistently missing in myodfh261 mutants, remnants of certain head muscles are observed. Variable loss of each individual branchiomeric muscle occurs in Tbx1 mutant mice and only posterior arch muscles are lost in tbx1 mutant zebrafish (Kelly et al., 2004; Piotrowski and Nusslein-Volhard, 2000). In contrast, we observe a consistent loss of some muscles in all arches, and a size reduction of other muscles in the first two arches. Vestiges of both the adductor mandibulae, the putative masseter homologue, and two specific hyoid arch muscles, adductor operculi and levator operculi are identifiable based on their location and orientation in myodfh261 mutants. Both adductor muscles appear to be common osteichthyan plesiomorphies (Diogo et al., 2008). The greater role of Myf5 in these muscles is reminiscent of the situation in mice, in which either Myf5 or MyoD is sufficient for head myogenesis (reviewed in (Sambasivan et al., 2011). Most strikingly, it is precisely the two hyoid opercular muscles that remain in myodfh261 mutants that are partially lost in Ret signalling mutants, raising the possibility of complementary mechanisms of myogenesis (Knight et al., 2011).
The myogfh265 mutant had little phenotype in both somitic and cranial muscles, and was thus much milder than we previously reported in myog morphants (Hinits et al., 2009). However, on a myodfh261 mutant background, myogfh265 caused a loss of fast muscle similar to, but less extensive than, that caused by myog MO. These findings raise the possibility that the myogfh265 allele is hypomorphic. Using an antiserum raised to the entire Myogenin protein, but for which the major epitopes are unknown, we could not detect protein in myogfh265 mutants. Nevertheless, an in vitro study in which the introduction of a termination codon 3′ of the HLH motif of mouse Myogenin (a similar change to the myogfh265 allele) resulted in a partial, rather than a complete, loss in myogenic activity (Schwarz et al., 1992). Clarification of the role of Myog in fish awaits isolation of an unambiguous null allele.
The lack of skeletal muscle in myf5;myod double mutants shows that zebrafish Mrf4 alone cannot drive early somitic myogenesis, unlike the situation in the mouse (Kassar-Duchossoy et al., 2004). Our data raise the possibility that, early in vertebrate evolution, Myf5 and Myod were the primary MRF drivers of myogenesis and that Mrf4 secondarily gained a myogenic role, somewhere in the amniote lineage. Alternatively, zebrafish could have lost an early role for Mrf4 present in the common ancestor. In either case, our data predict that, although Mrf4 and Myf5 have retained close genetic linkage for at least 400 Myr, the regulation in the locus has diverged significantly. Mrf4 clearly evolved prior to the divergence of sarcopterygian and actinopterygian fish (Atchley et al., 1994). The major conserved site of expression of Mrf4 is in differentiated muscle fibres, so it is likely that its ancestral role was in these cells and that elements controlling fibre expression may be conserved across the vertebrates (Hinits et al., 2007).
Myod regulation of miR-206 in vivo
miR-206 is expressed primarily in zebrafish differentiated muscle of both slow and fast lineages in somitic, limb and cranial muscles. Like mice, zebrafish lacking Myod express miR-206 in their muscle (Sweetman et al., 2008). The loss of miR-206 in myodfh261 mutant cranial regions is most simply explained by the loss of cranial muscles in the myodfh261 mutant. However, the loss of miR-206 in the trunk musculature is less readily explained by loss of muscle cells, because miR-206 appears more highly reduced than myhz1 mRNA. In contrast, the apparently normal accumulation of miR-206 in myf5hu2022 mutant fish, is quite different from the lack of miR-206 in somitic muscle of Myf5 mutant mice until at least E14, when muscle is already differentiated (Sweetman et al., 2008). miR-206 can drive muscle differentiation and promote cell survival in mammals by regulating Pax3/7 genes (Dey et al., 2011; Hirai et al., 2010; Rao et al., 2006). Lack of miR-206 accumulation may contribute to the maintenance of Pax3/7 in extra cells we observe in the somite of myodfh261 mutant fish. However, based on its abundant expression, the major function of miR-206 in fish appears to be in muscle fibres themselves, consistent with its reported role in neuromuscular interaction (Williams et al., 2009).
Effect of muscle development on skeletogenesis
The data presented show that Myod, and presumably muscle, are essential for proper development of the ventral head, jaw and pectoral girdle. Not only muscle, but also skeletal and, presumably, connective tissue elements are affected. These cranial defects prevent survival of myod mutant fish beyond larval development. This finding is phenocopied in myod morphants. Therefore, effects on adjacent genes or other genetic changes arising during the tilling screen can be eliminated as contributing to the phenotype.
Myod mutant mice show no defects in skeletogenesis, presumably reflecting the ability of Myf5 to compensate for loss of Myod in mouse cranial myogenesis (Sambasivan et al., 2009). However, mice with no muscles or with reduced muscle mass, such as mutants affecting the Mrf4/Myf5 locus, Myf5;Myod double mutants and Pax3 (Splotch) mutants have dramatic skeletal defects, the origin of which is complex (Gensch et al., 2008; Haldar et al., 2008; Kassar-Duchossoy et al., 2004; Kaul et al., 2000; Nowlan et al., 2010; Vinagre et al., 2010). Unlike myf5, myod lacks synteny between fish and mammals (www.metazome.org) and, more importantly, is not significantly expressed in presomitic mesoderm. So a cell autonomous function of Myod in skeletogenic cells is unlikely, as suggested in mice (Vinagre et al., 2010). We conclude that lack of myogenesis per se leads to skeletal defects.
The head phenotype of myodfh261 mutant fish bears some similarity to the open mouth and cranial cartilage defects resulting from mutation of mef2ca (hoover) (Miller et al., 2007). Elegant transplantation experiments in fish and lineage-specific deletion of Mef2c in mouse have shown a cell autonomous requirement for Mef2ca in neural crest-derived cartilage cells (Miller et al., 2007; Verzi et al., 2007). Mef2ca is expressed in both cranial neural crest and muscle (Hinits and Hughes, 2007; Miller et al., 2007). In contrast, myod expression is only detected in myogenic cells (Hinits et al., 2009; Weinberg et al., 1996), showing that it acts on cartilage and bone non-cell autonomously. As mef2ca is expressed in muscle cells after terminal differentiation (Hinits and Hughes, 2007), our data raise the possibility that defects in muscle fibre function may also contribute to the mef2ca mutant skeletal phenotype.
The effects of the lack of head muscle differentiation on the cranial bone and cartilage elements, make the zebrafish myod mutant an excellent model to study the signalling events and interactions between these tissues.
Supplementary Material
Fig. S1. Expression of miR-206 in zebrafish development.
In situ RNA hybridization for miR-206. Dorsal (A) or lateral (E) flatmounts, lateral (B,C,D,D”), dorsal (C”) or ventral (D’) wholemounts, anterior to top (A) or left (B-E). A. Expression is detected late in adaxial slow fibre differentiation and early in fast myogenesis. B. At 24 hpf, expression is restricted to the differentiated somitic muscle. C. By 48 hpf, expression is abundant in somitic fast muscle (arrowheads). A transverse section at the level indicated by the line in C, shows signal in somitic fast muscle (C’, black arrowheads). Notochord signal (red arrow) is detected in some embryos. Pectoral fin muscle expression is also detected (C", white arrowheads). D. At 72 hpf, the forming cranial muscles also contain miR-206. E. miR-206 expression is reduced in somitic muscle of myodfh261 mutant and further reduced after cyclopamine treatment, indicating that miR-206 is expressed in slow muscle fibres. Bars = 100 μm.
Fig. S2: Myf5 or Myod is required for myogenesis.
Dorsal flatmounts (A,D-bottom panel), ventral (D-top panel) and lateral (B,C) wholemounts or transverse cryosections (E) of zebrafish embryos from myf5hu2022/+;myodfh261/+ in-cross analysed at the indicated stage by immunohistochemistry for slow MyHC (A) or all MyHC (E) or in situ mRNA hybridization for smyhc1 (B) or mylz2 (C,D), anterior to top (A -D), dorsal to top (E). A. At 15s, most embryos show strong slow muscle differentiation, but approximately 3/16ths (genotyped as myodfh261/fh261;myf5hu2022/+ or myodfh261/fh261;myf5+/+) have less muscle and 1/16th (genotyped as myodfh261/fh261;myf5hu2022/hu2022) have no detectable muscle. B. mRNAs encoding slow myosin is reduced in putative myodfh261 mutants and absent in doubles. C,D. At 72 hpf, no mylz2 mRNA is detected in genotyped double mutants, in which the trunk and tail is severely reduced in size. E. At 120 hpf, double mutant lacks MyHC in the gut extension region and has a reduced somite area. Note the similar sizes of non-muscle tissues. Bars = 100 μm.
Table S1 Quantification of effects of mutations, morpholinos and drug treatments.
Acknowledgments
SMH is a member of MRC Scientific Staff with Programme Grant support. TILLING of myod and myog was supported by NIH HG002995 to CBM. CBM is an Investigator with the Howard Hughes Medical Institute. DS was supported by grants to Andrea Münsterberg and by a University of Nottingham Early Career Research and Knowledge Transfer award.
References
- Akimenko MA, Ekker M, Wegner J, Lin W, Westerfield M. Combinatorial expression of three zebrafish genes related to distal-less: part of a homeobox gene code for the head. J. Neurosci. 1994;14:3475–3486. doi: 10.1523/JNEUROSCI.14-06-03475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchley WR, Fitch WM, Bronner-Fraser M. Molecular evolution of the MyoD family of transcription factors. Proc. Natl Acad. Sci. USA. 1994;91:11522–11526. doi: 10.1073/pnas.91.24.11522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagden CS, Currie PD, Ingham PW, Hughes SM. Notochord induction of zebrafish slow muscle mediated by Sonic Hedgehog. Genes Dev. 1997;11:2163–2175. doi: 10.1101/gad.11.17.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brack AS, Bildsoe H, Hughes SM. Evidence that satellite cell decrement contributes to preferential decline in nuclear number from large fibres during murine age-related muscle atrophy. J Cell Sci. 2005;118:4813–4821. doi: 10.1242/jcs.02602. [DOI] [PubMed] [Google Scholar]
- Bryson-Richardson RJ, Daggett DF, Cortes F, Neyt C, Keenan DG, Currie PD. Myosin heavy chain expression in zebrafish and slow muscle composition. Dev. Dyn. 2005;233:1018–1022. doi: 10.1002/dvdy.20380. [DOI] [PubMed] [Google Scholar]
- Chargé SB, Brack AS, Bayol SA, Hughes SM. MyoD- and nerve-dependent maintenance of MyoD expression in mature muscle fibres acts through the DRR/PRR element. BMC Dev. Biol. 2008;8:5. doi: 10.1186/1471-213X-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JC, Goldhamer DJ. The core enhancer is essential for proper timing of MyoD activation in limb buds and branchial arches. Dev. Biol. 2004;265:502–512. doi: 10.1016/j.ydbio.2003.09.018. [DOI] [PubMed] [Google Scholar]
- Chevallier A. Role of the somitic mesoderm in the development of the thorax in bird embryos. II. Origin of thoracic and appendicular musculature. J. Embryol. Exp. Morphol. 1979;49:73–88. [PubMed] [Google Scholar]
- Christ B, Jacob HJ, Jacob M. Experimental analysis of the origin of the wing musculature in avian embryos. Anat. Embryol. (Berl) 1977;150:171–186. doi: 10.1007/BF00316649. [DOI] [PubMed] [Google Scholar]
- Cooper MS, Szeto DP, Sommers-Herivel G, Topczewski J, Solnica-Krezel L, Kang HC, Johnson I, Kimelman D. Visualizing morphogenesis in transgenic zebrafish embryos using BODIPY TR methyl ester dye as a vital counterstain for GFP. Dev. Dyn. 2005;232:359–368. doi: 10.1002/dvdy.20252. [DOI] [PubMed] [Google Scholar]
- Davis RL, Cheng PF, Lassar AB, Weintraub H. The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell. 1990;60:733–746. doi: 10.1016/0092-8674(90)90088-v. [DOI] [PubMed] [Google Scholar]
- Devoto SH, Melancon E, Eisen JS, Westerfield M. Identification of separate slow and fast muscle precursor cells in vivo, prior to somite formation. Development. 1996;122:3371–3380. doi: 10.1242/dev.122.11.3371. [DOI] [PubMed] [Google Scholar]
- Dey BK, Gagan J, Dutta A. miR-206 and -486 induce myoblast differentiation by downregulating Pax7. Mol. Cell. Biol. 2011;31:203–214. doi: 10.1128/MCB.01009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diogo R, Hinits Y, Hughes SM. Development of mandibular, hyoid and hypobranchial muscles in the zebrafish: homologies and evolution of these muscles within bony fishes and tetrapods. BMC Dev. Biol. 2008;8:24. doi: 10.1186/1471-213X-8-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper BW, McCallum CM, Stout JL, Slade AJ, Moens CB. A high-throughput method for identifying N-ethyl-N-nitrosourea (ENU)-induced point mutations in zebrafish. Methods Cell Biol. 2004;77:91–112. doi: 10.1016/s0091-679x(04)77005-3. [DOI] [PubMed] [Google Scholar]
- Ekker M, Wegner J, Akimenko MA, Westerfield M. Coordinate embryonic expression of three zebrafish engrailed genes. Development. 1992;116:1001–1010. doi: 10.1242/dev.116.4.1001. [DOI] [PubMed] [Google Scholar]
- Gensch N, Borchardt T, Schneider A, Riethmacher D, Braun T. Different autonomous myogenic cell populations revealed by ablation of Myf5-expressing cells during mouse embryogenesis. Development. 2008;135:1597–1604. doi: 10.1242/dev.019331. [DOI] [PubMed] [Google Scholar]
- Greenwood PH, Lauder GV. The protractor pectoralis muscle and the classification of teleost fishes. Bulletin of the British Museum (Natural History) Zoology. 1981;41:213–234. [Google Scholar]
- Grimes AC, Stadt HA, Shepherd IT, Kirby ML. Solving an enigma: arterial pole development in the zebrafish heart. Dev. Biol. 2006;290:265–276. doi: 10.1016/j.ydbio.2005.11.042. [DOI] [PubMed] [Google Scholar]
- Haldar M, Karan G, Tvrdik P, Capecchi MR. Two Cell Lineages, myf5-Dependent and myf5-Independent, Participate in Mouse Skeletal Myogenesis. Dev. Cell. 2008;14:437–445. doi: 10.1016/j.devcel.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond CL, Hinits Y, Osborn DP, Minchin JE, Tettamanti G, Hughes SM. Signals and myogenic regulatory factors restrict pax3 and pax7 expression to dermomyotome-like tissue in zebrafish. Dev. Biol. 2007;302:504–521. doi: 10.1016/j.ydbio.2006.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasty P, Bradley A, Morris JH, Edmondson DG, Venuti JM, Olson EN, Klein WH. Muscle deficiency and neonatal death in mice with targeted mutation in the myogenin gene. Nature. 1993;364:501–506. doi: 10.1038/364501a0. [DOI] [PubMed] [Google Scholar]
- Higashijima S, Nose A, Eguchi G, Hotta Y, Okamoto H. Mindin/F-spondin family: novel ECM proteins expressed in the zebrafish embryonic axis. Dev. Biol. 1997;192:211–227. doi: 10.1006/dbio.1997.8760. [DOI] [PubMed] [Google Scholar]
- Hinits Y, Hughes SM. Mef2s are required for thick filament formation in nascent muscle fibres. Development. 2007;134:2511–2519. doi: 10.1242/dev.007088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinits Y, Osborn DP, Carvajal JJ, Rigby PW, Hughes SM. Mrf4 (myf6) is dynamically expressed in differentiated zebrafish skeletal muscle. Gene Expr. Patterns. 2007;7:738–745. doi: 10.1016/j.modgep.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinits Y, Osborn DP, Hughes SM. Differential requirements for myogenic regulatory factors distinguish medial and lateral somitic, cranial and fin muscle fibre populations. Development. 2009;136:403–414. doi: 10.1242/dev.028019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai H, Verma M, Watanabe S, Tastad C, Asakura Y, Asakura A. MyoD regulates apoptosis of myoblasts through microRNA-mediated down-regulation of Pax3. J. Cell Biol. 2010;191:347–365. doi: 10.1083/jcb.201006025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes SM, Koishi K, Rudnicki M, Maggs AM. MyoD protein is differentially accumulated in fast and slow skeletal muscle fibres and required for normal fibre type balance in rodents. Mech. Dev. 1997;61:151–163. doi: 10.1016/s0925-4773(96)00631-4. [DOI] [PubMed] [Google Scholar]
- Kablar B, Krastel K, Ying C, Asakura A, Tapscott SJ, Rudnicki MA. MyoD and Myf-5 differentially regulate the development of limb versus trunk skeletal muscle. Development. 1997;124:4729–4738. doi: 10.1242/dev.124.23.4729. [DOI] [PubMed] [Google Scholar]
- Kardon G. Muscle and tendon morphogenesis in the avian hind limb. Development. 1998;125:4019–4032. doi: 10.1242/dev.125.20.4019. [DOI] [PubMed] [Google Scholar]
- Kassar-Duchossoy L, Gayraud-Morel B, Gomes D, Rocancourt D, Buckingham M, Shinin V, Tajbakhsh S. Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature. 2004;431:466–471. doi: 10.1038/nature02876. [DOI] [PubMed] [Google Scholar]
- Kaul A, Koster M, Neuhaus H, Braun T. Myf-5 revisited: loss of early myotome formation does not lead to a rib phenotype in homozygous Myf-5 mutant mice. Cell. 2000;102:17–19. doi: 10.1016/s0092-8674(00)00006-4. [DOI] [PubMed] [Google Scholar]
- Kelly RG, Jerome-Majewska LA, Papaioannou VE. The del22q11.2 candidate gene Tbx1 regulates branchiomeric myogenesis. Hum. Mol. Genet. 2004;13:2829–2840. doi: 10.1093/hmg/ddh304. [DOI] [PubMed] [Google Scholar]
- Kerst B, Mennerich D, Schuelke M, Stoltenburg-Didinger G, von Moers A, Gossrau R, van Landeghem FK, Speer A, Braun T, Hubner C. Heterozygous myogenic factor 6 mutation associated with myopathy and severe course of Becker muscular dystrophy. Neuromuscul. Disord. 2000;10:572–577. doi: 10.1016/s0960-8966(00)00150-4. [DOI] [PubMed] [Google Scholar]
- Knight RD, Mebus K, d’Angelo A, Yokoya K, Heanue T, Roehl H. Ret signalling integrates a craniofacial muscle module during development. Development. 2011;138:2015–2024. doi: 10.1242/dev.061002. [DOI] [PubMed] [Google Scholar]
- Kusakabe R, Kuraku S, Kuratani S. Expression and interaction of muscle-related genes in the lamprey imply the evolutionary scenario for vertebrate skeletal muscle, in association with the acquisition of the neck and fins. Dev. Biol. 2011;350:217–227. doi: 10.1016/j.ydbio.2010.10.029. [DOI] [PubMed] [Google Scholar]
- Lepiller S, Laurens V, Bouchot A, Herbomel P, Solary E, Chluba J. Imaging of nitric oxide in a living vertebrate using a diamino-fluorescein probe. Free Radic. Biol. Med. 2007;43:619–627. doi: 10.1016/j.freeradbiomed.2007.05.025. [DOI] [PubMed] [Google Scholar]
- Ma PC, Rould MA, Weintraub H, Pabo CO. Crystal structure of MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation. Cell. 1994;77:451–459. doi: 10.1016/0092-8674(94)90159-7. [DOI] [PubMed] [Google Scholar]
- Maves L, Waskiewicz AJ, Paul B, Cao Y, Tyler A, Moens CB, Tapscott SJ. Pbx homeodomain proteins direct Myod activity to promote fast-muscle differentiation. Development. 2007;134:3371–3382. doi: 10.1242/dev.003905. [DOI] [PubMed] [Google Scholar]
- Megeney LA, Kablar B, Garrett K, Anderson JE, Rudnicki MA. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev. 1996;10:1173–1183. doi: 10.1101/gad.10.10.1173. [DOI] [PubMed] [Google Scholar]
- Miller CT, Swartz ME, Khuu PA, Walker MB, Eberhart JK, Kimmel CB. mef2ca is required in cranial neural crest to effect Endothelin1 signaling in zebrafish. Dev. Biol. 2007;308:144–157. doi: 10.1016/j.ydbio.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore CA, Parkin CA, Bidet Y, Ingham PW. A role for the Myoblast city homologues Dock1 and Dock5 and the adaptor proteins Crk and Crk-like in zebrafish myoblast fusion. Development. 2007;134:3145–3153. doi: 10.1242/dev.001214. [DOI] [PubMed] [Google Scholar]
- Neyt C, Jagla K, Thisse C, Thisse B, Haines L, Currie PD. Evolutionary origins of vertebrate appendicular muscle. Nature. 2000;408:82–86. doi: 10.1038/35040549. [DOI] [PubMed] [Google Scholar]
- Nowlan NC, Bourdon C, Dumas G, Tajbakhsh S, Prendergast PJ, Murphy P. Developing bones are differentially affected by compromised skeletal muscle formation. Bone. 2010;46:1275–1285. doi: 10.1016/j.bone.2009.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oates AC, Pratt SJ, Vail B, Yan Y, Ho RK, Johnson SL, Postlethwait JH, Zon LI. The zebrafish klf gene family. Blood. 2001;98:1792–1801. doi: 10.1182/blood.v98.6.1792. [DOI] [PubMed] [Google Scholar]
- Patapoutian A, Yoon JK, Miner JH, Wang S, Stark K, Wold B. Disruption of the mouse MRF4 gene identifies multiple waves of myogenesis in the myotome. Development. 1995;121:3347–3358. doi: 10.1242/dev.121.10.3347. [DOI] [PubMed] [Google Scholar]
- Piotrowski T, Nüsslein-Volhard C. The endoderm plays an important role in patterning the segmented pharyngeal region in zebrafish (Danio rerio) Dev. Biol. 2000;225:339–356. doi: 10.1006/dbio.2000.9842. [DOI] [PubMed] [Google Scholar]
- Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc. Natl. Acad. Sci. USA. 2006;103:8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls A, Valdez MR, Zhang W, Richardson J, Klein WH, Olson EN. Overlapping functions of the myogenic bHLH genes MRF4 and MyoD revealed in double mutant mice. Development. 1998;125:2349–2358. doi: 10.1242/dev.125.13.2349. [DOI] [PubMed] [Google Scholar]
- Robling AG. Is bone’s response to mechanical signals dominated by muscle forces? Med. Sci. Sports Exerc. 2009;41:2044–2049. doi: 10.1249/MSS.0b013e3181a8c702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnicki MA, Braun T, Hinuma S, Jaenisch R. Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell. 1992;71:383–390. doi: 10.1016/0092-8674(92)90508-a. [DOI] [PubMed] [Google Scholar]
- Sambasivan R, Gayraud-Morel B, Dumas G, Cimper C, Paisant S, Kelly RG, Tajbakhsh S. Distinct regulatory cascades govern extraocular and pharyngeal arch muscle progenitor cell fates. Dev. Cell. 2009;16:810–821. doi: 10.1016/j.devcel.2009.05.008. [DOI] [PubMed] [Google Scholar]
- Sambasivan R, Kuratani S, Tajbakhsh S. An eye on the head: the development and evolution of craniofacial muscles. Development. 2011;138:2401–2415. doi: 10.1242/dev.040972. [DOI] [PubMed] [Google Scholar]
- Schwarz JJ, Chakraborty T, Martin J, Zhou J, Olson EN. The basic region of myogenin cooperates with two transcription activation domains to induce muscle-specific transcription. Mol. Cell. Biol. 1992;12:266–275. doi: 10.1128/mcb.12.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetman D, Goljanek K, Rathjen T, Oustanina S, Braun T, Dalmay T, Munsterberg A. Specific requirements of MRFs for the expression of muscle specific microRNAs, miR-1, miR-206 and miR-133. Dev. Biol. 2008;321:491–499. doi: 10.1016/j.ydbio.2008.06.019. [DOI] [PubMed] [Google Scholar]
- Theis S, Patel K, Valasek P, Otto A, Pu Q, Harel I, Tzahor E, Tajbakhsh S, Christ B, Huang R. The occipital lateral plate mesoderm is a novel source for vertebrate neck musculature. Development. 2011;137:2961–2971. doi: 10.1242/dev.049726. [DOI] [PubMed] [Google Scholar]
- Tiidus PM, Bombardier E, Xeni J, Bestic NM, Vandenboom R, Rudnicki MA, Houston ME. Elevated catalase activity in red and white muscles of MyoD gene-inactivated mice. Biochem. Mol. Biol. Int. 1996;39:1029–1035. doi: 10.1080/15216549600201192. [DOI] [PubMed] [Google Scholar]
- Valdez MR, Richardson JA, Klein WH, Olson EN. Failure of Myf5 to support myogenic differentiation without myogenin, MyoD, and MRF4. Dev. Biol. 2000;219:287–298. doi: 10.1006/dbio.2000.9621. [DOI] [PubMed] [Google Scholar]
- Venuti JM, Morris JH, Vivian JL, Olson EN, Klein WH. Myogenin is required for late but not early aspects of myogenesis during mouse development. J. Cell Biol. 1995;128:563–576. doi: 10.1083/jcb.128.4.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verzi MP, Agarwal P, Brown C, McCulley DJ, Schwarz JJ, Black BL. The Transcription Factor MEF2C Is Required for Craniofacial Development. Dev. Cell. 2007;12:645–652. doi: 10.1016/j.devcel.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinagre T, Moncaut N, Carapuco M, Novoa A, Bom J, Mallo M. Evidence for a myotomal Hox/Myf cascade governing nonautonomous control of rib specification within global vertebral domains. Dev. Cell. 2010;18:655–661. doi: 10.1016/j.devcel.2010.02.011. [DOI] [PubMed] [Google Scholar]
- Vivian JL, Olson EN, Klein WH. Thoracic skeletal defects in myogenin- and MRF4-deficient mice correlate with early defects in myotome and intercostal musculature. Dev. Biol. 2000;224:29–41. doi: 10.1006/dbio.2000.9788. [DOI] [PubMed] [Google Scholar]
- Walker MB, Kimmel CB. A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech. Histochem. 2007;82:23–28. doi: 10.1080/10520290701333558. [DOI] [PubMed] [Google Scholar]
- Wang ZZ, Washabaugh CH, Yao Y, Wang JM, Zhang L, Ontell MP, Watkins SC, Rudnicki MA, Ontell M. Aberrant development of motor axons and neuromuscular synapses in MyoD-null mice. J. Neurosci. 2003;23:5161–5169. doi: 10.1523/JNEUROSCI.23-12-05161.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg ES, Allende ML, Kelly CS, Abdelhamid A, Murakami T, Andermann P, Doerre OG, Grunwald DJ, Riggleman B. Developmental regulation of zebrafish MyoD in wild-type, no tail and spadetail embryos. Development. 1996;122:271–280. doi: 10.1242/dev.122.1.271. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book - a guide for the laboratory use of zebrafish (Danio rerio) 3 ed University of Oregon Press; 1995. [Google Scholar]
- Williams AH, Valdez G, Moresi V, Qi X, McAnally J, Elliott JL, Bassel-Duby R, Sanes JR, Olson EN. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 2009;326:1549–1554. doi: 10.1126/science.1181046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, He J, Wang X, Lim TM, Gong Z. Asynchronous activation of 10 muscle-specific protein (MSP) genes during zebrafish somitogenesis. Dev. Dyn. 2000;219:201–215. doi: 10.1002/1097-0177(2000)9999:9999<::aid-dvdy1043>3.3.co;2-9. [DOI] [PubMed] [Google Scholar]
- Yablonka-Reuveni Z, Rudnicki MA, Rivera AJ, Primig M, Anderson JE, Natanson P. The transition from proliferation to differentiation is delayed in satellite cells from mice lacking MyoD. Dev. Biol. 1999;210:440–455. doi: 10.1006/dbio.1999.9284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Behringer RR, Olson EN. Inactivation of the myogenic bHLH gene MRF4 results in up-regulation of myogenin and rib anomalies. Genes Dev. 1995;9:1388–1399. doi: 10.1101/gad.9.11.1388. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Expression of miR-206 in zebrafish development.
In situ RNA hybridization for miR-206. Dorsal (A) or lateral (E) flatmounts, lateral (B,C,D,D”), dorsal (C”) or ventral (D’) wholemounts, anterior to top (A) or left (B-E). A. Expression is detected late in adaxial slow fibre differentiation and early in fast myogenesis. B. At 24 hpf, expression is restricted to the differentiated somitic muscle. C. By 48 hpf, expression is abundant in somitic fast muscle (arrowheads). A transverse section at the level indicated by the line in C, shows signal in somitic fast muscle (C’, black arrowheads). Notochord signal (red arrow) is detected in some embryos. Pectoral fin muscle expression is also detected (C", white arrowheads). D. At 72 hpf, the forming cranial muscles also contain miR-206. E. miR-206 expression is reduced in somitic muscle of myodfh261 mutant and further reduced after cyclopamine treatment, indicating that miR-206 is expressed in slow muscle fibres. Bars = 100 μm.
Fig. S2: Myf5 or Myod is required for myogenesis.
Dorsal flatmounts (A,D-bottom panel), ventral (D-top panel) and lateral (B,C) wholemounts or transverse cryosections (E) of zebrafish embryos from myf5hu2022/+;myodfh261/+ in-cross analysed at the indicated stage by immunohistochemistry for slow MyHC (A) or all MyHC (E) or in situ mRNA hybridization for smyhc1 (B) or mylz2 (C,D), anterior to top (A -D), dorsal to top (E). A. At 15s, most embryos show strong slow muscle differentiation, but approximately 3/16ths (genotyped as myodfh261/fh261;myf5hu2022/+ or myodfh261/fh261;myf5+/+) have less muscle and 1/16th (genotyped as myodfh261/fh261;myf5hu2022/hu2022) have no detectable muscle. B. mRNAs encoding slow myosin is reduced in putative myodfh261 mutants and absent in doubles. C,D. At 72 hpf, no mylz2 mRNA is detected in genotyped double mutants, in which the trunk and tail is severely reduced in size. E. At 120 hpf, double mutant lacks MyHC in the gut extension region and has a reduced somite area. Note the similar sizes of non-muscle tissues. Bars = 100 μm.
Table S1 Quantification of effects of mutations, morpholinos and drug treatments.