Abstract
Myosins have diverse mechanical properties reflecting a range of cellular roles. A major challenge is to understand the structural basis for generating novel functions from a common motor core. Myosin VI is specialized for processive motion toward the (-) end of actin filaments. We have used engineered myosin VI motors to test and refine the “redirected power stroke” model for (-) end directionality, and to explore poorly understood structural requirements for processive stepping. Guided by crystal structures and molecular modeling, we fused artificial lever arms to the catalytic head of myosin VI at several positions, retaining varying amounts of native structure. We found that an 18 residue alpha-helical insert is sufficient to reverse the directionality of the motor, with no requirement for any calmodulin light chains. Further, we observed robust processive stepping of motors with artificial lever arms, demonstrating that processivity can arise without optimizing lever arm composition or mechanics.
Members of the myosin superfamily have acquired specialized mechanical adaptations to perform cellular functions ranging from vesicle transport to muscle contraction. Myosin VI (M6) moves toward the (-) end of the actin filament, in the opposite direction from all other characterized myosins 1. Additionally, unlike muscle myosin II, individual dimers of M6 can travel processively for many steps before detaching from the filament 2. The determinants of M6 directionality and the mechanism of M6 processivity have recently been the subjects of considerable scrutiny 3-10.
Structural and functional studies have converged on a redirected power stroke model to explain directionality reversal in M6. According to current formulations of the power stroke or swinging cross-bridge model of myosin force generation 11, conformational changes at the nucleotide binding site are propagated through the catalytic head, driving a large rotation of the converter domain. This rotation is amplified by a rigid lever arm structure extending from the final helix of the converter. An x-ray crystal structure of the M6 post-stroke state showed that the lever arm emerges at an angle that differs from (+) end directed myosins by ∼120°, due to the presence of a unique insert in M6 following the converter (Figure 1A) 9. Engineered motors truncated before the unique insert show (+) end directed motion 3, as do chimeric motors in which M6 is fused immediately after the converter to the lever arm from myosin V 5. These studies demonstrate that redirection of the lever arm mediated by the unique insert is essential for (-) end directionality.
Figure 1.
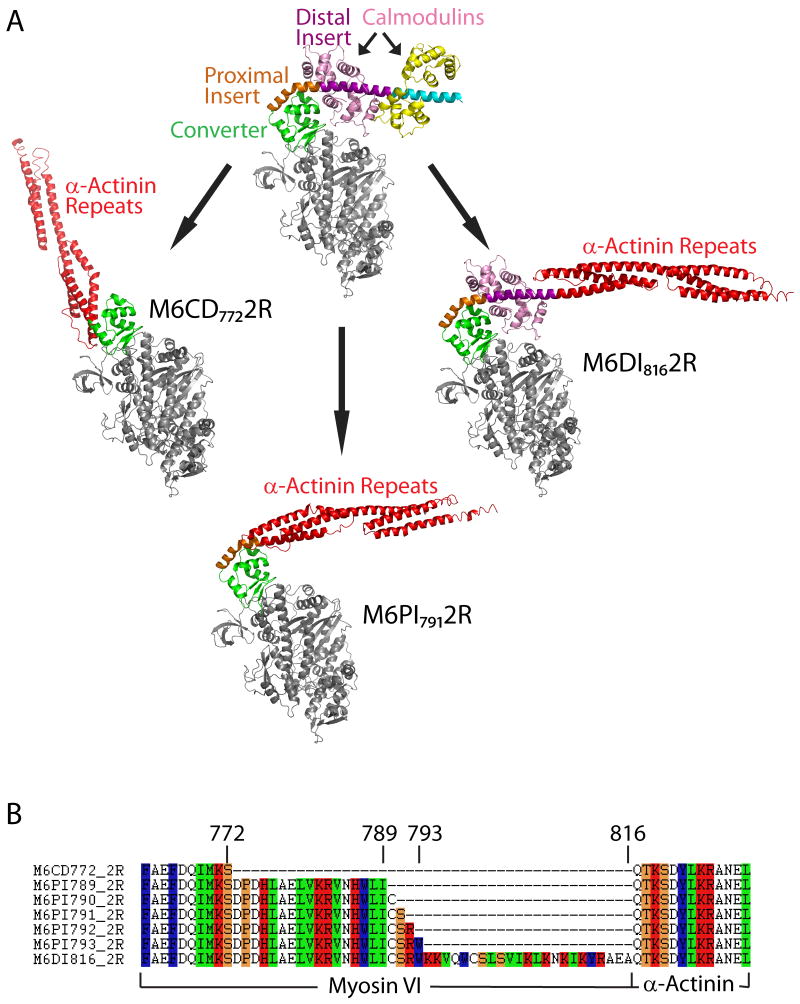
Design of engineered M6 motors. (A) Post-stroke structure of M6 (top) and model structures of chimeric constructs with artificial lever arms derived from α-actinin. In M6CD7722R, spectrin repeats from α-actinin are fused after the converter domain but prior to the unique insert; in M6PI7912R, the α-actinin repeats are fused immediately after the proximal part of the unique insert; in M6DI8162R, the α-actinin repeats are fused after the distal part of the insert. (B) Amino acid sequences of chimeric constructs at the site of fusion between myosin and α-actinin.
An examination of stroke sizes for a series of truncated M6 constructs led to a model in which the redirected lever arm rotates by ∼180° during the power stroke 3. This model was supported by a subsequent crystal structure of the putative pre-stroke state 10. However, the pre-stroke crystal structure was obtained using a fragment of M6 that lacks the distal part of the unique insert and its associated stabilizing light chain. The lever arm angle in this structure could only be deduced by modeling the missing distal insert. The functional relevance of the crystallized conformation, which contains large and surprising rearrangements of the converter domain, must remain tentative in the absence of experiments showing that the proximal part of the unique insert is sufficient to correctly position a lever arm for (-) end directed motion.
Functional replacement of the myosin II lever arm with rigid three-helix bundles derived from α-actinin provided critical tests of the swinging cross-bridge model for myosin II 12; 13. Here, we have used an extension of this strategy to challenge and refine the redirected power stroke model for M6 directionality. We have characterized M6 constructs in which α-actinin lever arms have been fused at several different locations following the converter domain. We include constructs in which the lever arm is fused immediately following the proximal part of the unique insert, probing whether this structure is sufficient for (-) end directionality.
Our strategy also provides a means to test models relating structure to processivity. Dimers of M6 have been shown to move processively with a hand-over-hand mechanism 8; 14 thought to depend on coordination mediated by strain in the lever arms 15. By replacing lever arms with alternative structural elements, we can directly test the effects on processivity of varying geometric and mechanical parameters in the dimer.
We designed chimeric constructs in which an artificial lever arm 12; 16 (two spectrin repeats from Dictyostelium α-actinin, designated 2R) was fused to M6 at one of three locations (Figure 1A and 1B): (1) after the converter domain but prior to the unique insert, M6CD7722R; (2) immediately after the proximal part of the unique insert, M6PI7912R; and (3) after the distal part of the insert, M6DI8162R. (Subscripts indicate the residue number of the last native M6 amino acid prior to the 2R junction.) M6CD7722R is predicted to have a lever arm orientation similar to myosin II. The longer constructs are predicted to yield lever arm orientations similar to native M6, but with differences in lever arm stability and mechanics due to the replacement of one or both native calmodulin-binding domains.
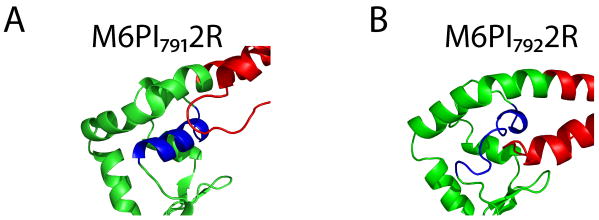
Precise fusion points at the level of amino acid residues were chosen with the aid of molecular modeling. For the constructs fused after the proximal part of the unique insert, five possible adjacent fusion points were modeled. Models of M6PI7892R and M6PI7932R showed steric collisions between the converter and the artificial lever arm. Initial models of M6PI7902R, M6PI7912R, and M6PI7922R were free of structural collisions, but molecular dynamics (MD) simulations for the three models predicted differing stabilities of local structures (Figure 2 and Supplemental Movies S1, S2). M6PI7902R and M6PI7912R remained stable for 5 ns of MD simulation. In contrast, a key α-helix in the converter domain of M6PI7922R became disordered during all-atom simulations. Thus, our models predicted that only two of these five fusions should retain functional lever arms: M6PI7902R and M6PI7912R.
Figure 2.
Molecular dynamics simulations showing predicted differences in stability between alternative proximal insert fusion constructs that differ by one residue in the placement of the fusion point. (A) A snapshot from a simulation of M6PI7912R shows stable local structure in the converter domain. (B) A snapshot from a simulation of M6PI7922R shows a denatured α-helix (blue) in the converter domain.
A total of seven monomeric fusion constructs were expressed, purified, and assayed for motility, velocity, and directionality using dual-labeled gliding filament assays. In addition, the dimeric artificial lever arm construct M6DI8162RGCN4 was generated by fusion of M6DI8162R to the medial tail region of M6 followed by the leucine zipper GCN4 in order to ensure dimerization. This construct was expressed and assayed for processivity using single fluorophore tracking.
Constructs fused after the converter or after the unique insert are motile and have the expected directionalities
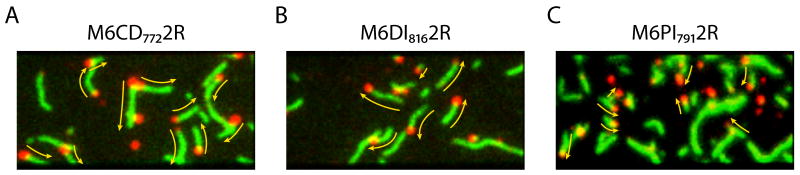
M6CD7722R shows (+) end directed motion in dual-labeled gliding filament assays (Figure 3A, Table 1, and Movie S3), consistent with its predicted structural similarity to myosin II. M6DI8162R retains the entire unique insert and has an extended lever arm, and was thus predicted to show rapid (-) end directed motion. Assays (Figure 3B and Movie S4) show the expected movement at an average velocity of -110 nm/s (Table 1), compared with -45 nm/s for a construct truncated after the unique insert alone 3.
Figure 3.
Images from dual-labeled gliding filament assays showing the directionality of engineered constructs (see also Supplemental Movies S3-S7). TMR-phalloidin labeled actin filaments are false-colored in green; Cy5-labeled (+) ends appear as red dots. The direction of gliding movement is indicated by yellow arrows. (A) M6CD7722R shows (+) end directed movement, while (B) M6DI8162R and (C) M6PI7912R show (-) end directed movement.
Table 1.
Gliding filament results for engineered M6 motors.
| M6CD7722R | M6PI7912R | M6DI8162R | |
|---|---|---|---|
| Directionality | (+) end (51/51 filaments) | (-) end (64/65 filaments) | (-) end (86/89 filaments) |
| Velocity | +40.64 ± 1.92 nm/s (n=44) | -29.03 ± 1.39 nm/s (n=32) | -110.66 ± 4.42 nm/s (n=51) |
All-atom modeling correctly predicts functional fusion sites
We tested all five constructs fused after the proximal part of the unique insert, and observed only stationary actin filaments in assays of the three constructs (M6PI7892R, M6PI7922R, and M6PI7932R) showing steric clashes or helix instability in MD simulations (Movie S5). Both of the constructs that remained stable in MD simulations (M6PI7902R and M6PI7912R) were motile (Movies S6, S7).
Constructs fused after the proximal part of the unique insert are (-) end directed, showing that the distal part of the insert is dispensable for directionality reversal
M6PI7902R and M6PI7912R show robust (-) end directed motion in gliding filament assays (Figure 3C and Table 1). These constructs move in the opposite direction from M6CD7722R due to the insertion of only 18-19 amino acids that form a proline kink and a short α-helical section, without any stabilizing light chains.
Table 1 summarizes gliding filament results for monomeric engineered M6 motors. Directionalities agree with predictions and demonstrate that a short alpha-helical insert is sufficient to reverse the directionality of the motor. In these assays, polarity-labeled filaments occasionally become mislabeled, likely due to loss of the gelsolin cap on some Cy5-actin seeds 17. This explains results with M6PI7912R and M6DI8162R, in which fewer than 100% of filaments were scored as (-) end directed. As expected, the highest gliding filament velocity is seen for M6DI8162R, which has the longest lever arm length (Figure 1). M6CD7722R and M6PI7912R are expected to have relatively lower velocities due to shortened stroke sizes. Further reductions in velocity for the shortened constructs may occur due to increased actin attachment times, as seen previously for truncated M6 constructs lacking light chain binding regions 3.
Artificial lever arms can support processive movement
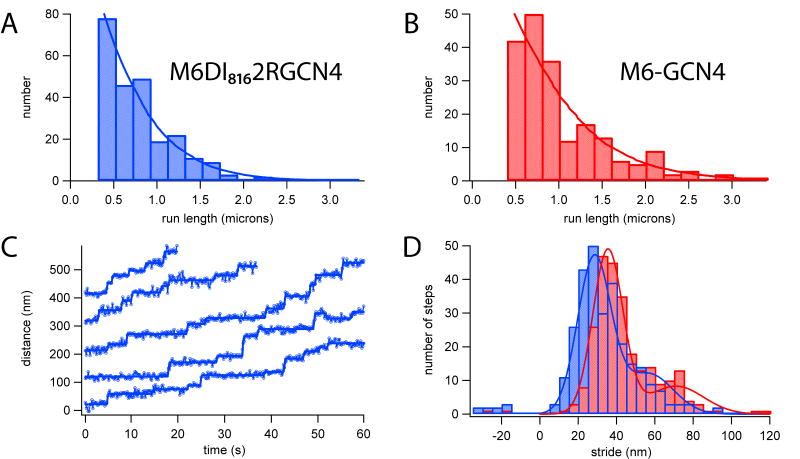
The dimeric construct M6DI8162RGCN4 was tested for processivity and compared to M6-GCN4 control (Figure 4A, B). We used total internal reflection fluorescence (TIRF) imaging to track the movement of individual tetramethylrhodamine (TMR)-labeled dimers on actin filaments (Movie S8). M6DI8162RGCN4 showed processive movement similar to control dimeric M6-GCN4, with run lengths approximately 70% as long as those of the control. Stepwise motion was detected during processive runs for both M6-GCN4 and M6DI8162RGCN4 (Figure 4C), allowing us to measure the distribution of stride sizes (Figure 4D). As expected from the extended lever arm design, M6DI8162RGCN4 stepped along the actin filament with long strides (28.4 +/- 0.5 nm). However, the peak stride size for M6DI8162RGCN4 was significantly smaller than control M6-GCN4 (35.5 +/- 0.5 nm), reflecting differences in lever arm geometry.
Figure 4.
Processive data from single-fluorophore tracking of TMR-labeled dimeric constructs. (A-B) Histograms of processive run lengths for a dimeric M6 with artificial lever arms, M6DI8162RGCN4 (mean run length 590 +/- 50 nm; N=243) and control M6-GCN4 (854 +/- 91 nm; N=199). (C) Example stepping traces for M6DI8162RGCN4. Blue circles, data from fluorophore localization; solid blue lines, step-finding fits. (D) Stride histograms show that M6DI8162RGCN4 (blue) has a slightly shorter stride (28.4 +/- 0.5 nm) than M6-GCN4 (red) (35.5 +/- 0.5 nm).
Directionality reversal in M6 was previously shown to depend on the presence of the 50 residue unique insert and its associated calmodulin light chain 3; 5. This light chain makes extensive contacts with the converter domain in the native M6 structure, suggesting that it might play an important structural role in redirection of the lever arm 9. However, we have shown here that the light chain and the distal part of the unique insert are dispensable for directionality reversal. Our results help validate the interpretation of a prestroke crystal structure for M6 in which the lever arm angle was assumed to be unperturbed by omission of the distal part of the unique insert and its associated light chain 10. This provides additional support for a model in which the M6 lever arm is nearly perpendicular to the actin filament and rotates close to 180° during the power stroke 3; 9; 10; 18.
We have also shown that processive motion can be achieved in a dimeric myosin using artificial lever arms and without extensive optimization of lever arm geometry or mechanics. (Lack of geometric optimization in our constructs is underscored by the significantly shorter stride of the chimera, which fails to match the actin pseudo-repeat.) In most models of hand-over-hand motion, coordination of the two myosin heads is hypothesized to depend on internal strain generated when both heads are bound to the actin filament 15; 19. The mechanical properties of the lever arms might therefore be tuned for strain-mediated communication between the heads, and the lever arms might also be optimized for thermally sampling conformations during the binding site search that precedes front head reattachment. Computational models have led researchers to propose stringent mechanical requirements for lever arms: Lan et al. found that bending anisotropy was a key feature of computational models for myosin V stepping 20. Our findings imply that either coordination is robust to changes in lever arm composition, or else coordination is relatively unimportant for processive motion. Indeed, as previously suggested 19, dimers with independently cycling heads may theoretically be expected to show substantial processive motion, as long as the duty ratio of each monomer is high.
Finally, we note that while chimeric proteins can be powerful tools for structure/function studies, they must fold correctly in order to provide useful information. Here, we used all-atom molecular modeling to predict misfolding at junctions due to steric clashes or other unfavorable interactions. We found that relatively simple atomistic computations helped discriminate between functional and non-functional chimeras. For chimeras involving extensive new domain interfaces, it may be necessary to apply sequence selection by structure-based protein design, as demonstrated earlier for a homing endonuclease 21.
Supplementary Material
Acknowledgments
We thank D. Parker, B. Spink, S. Sivaramakrishnan, S. Sutton, M. Footer, and H. Flyvbjerg for technical assistance; and A. Dunn and R. Altman for helpful discussions. This work was supported by the National Institutes of Health through the NIH Roadmap for Medical Research Grant U54 GM072970 (to J.-C.L., S.L.D., and J.A.S.). J.A.S. was supported by National Institutes of Health Grant GM33289. Z.B. was supported by a Helen Hay Whitney Postdoctoral Fellowship and by NIH Grant DP2 OD004690. M.W.E. was supported by a National Science Foundation Graduate Research Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wells AL, Lin AW, Chen LQ, Safer D, Cain SM, Hasson T, Carragher BO, Milligan RA, Sweeney HL. Myosin VI is an actin-based motor that moves backwards. Nature. 1999;401:505–8. doi: 10.1038/46835. [DOI] [PubMed] [Google Scholar]
- 2.Rock RS, Rice SE, Wells AL, Purcell TJ, Spudich JA, Sweeney HL. Myosin VI is a processive motor with a large step size. Proc Natl Acad Sci U S A. 2001;98:13655–9. doi: 10.1073/pnas.191512398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryant Z, Altman D, Spudich JA. The power stroke of myosin VI and the basis of reverse directionality. Proc Natl Acad Sci U S A. 2007;104:772–7. doi: 10.1073/pnas.0610144104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iwaki M, Tanaka H, Iwane AH, Katayama E, Ikebe M, Yanagida T. Cargo-binding makes a wild-type single-headed myosin-VI move processively. Biophys J. 2006;90:3643–52. doi: 10.1529/biophysj.105.075721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park H, Li A, Chen LQ, Houdusse A, Selvin PR, Sweeney HL. The unique insert at the end of the myosin VI motor is the sole determinant of directionality. Proc Natl Acad Sci U S A. 2007;104:778–83. doi: 10.1073/pnas.0610066104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park H, Ramamurthy B, Travaglia M, Safer D, Chen LQ, Franzini-Armstrong C, Selvin PR, Sweeney HL. Full-length myosin VI dimerizes and moves processively along actin filaments upon monomer clustering. Mol Cell. 2006;21:331–6. doi: 10.1016/j.molcel.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 7.Rock RS, Ramamurthy B, Dunn AR, Beccafico S, Rami BR, Morris C, Spink BJ, Franzini-Armstrong C, Spudich JA, Sweeney HL. A flexible domain is essential for the large step size and processivity of myosin VI. Mol Cell. 2005;17:603–9. doi: 10.1016/j.molcel.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 8.Yildiz A, Park H, Safer D, Yang Z, Chen LQ, Selvin PR, Sweeney HL. Myosin VI steps via a hand-over-hand mechanism with its lever arm undergoing fluctuations when attached to actin. J Biol Chem. 2004;279:37223–6. doi: 10.1074/jbc.C400252200. [DOI] [PubMed] [Google Scholar]
- 9.Menetrey J, Bahloul A, Wells AL, Yengo CM, Morris CA, Sweeney HL, Houdusse A. The structure of the myosin VI motor reveals the mechanism of directionality reversal. Nature. 2005;435:779–85. doi: 10.1038/nature03592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Menetrey J, Llinas P, Mukherjea M, Sweeney HL, Houdusse A. The structural basis for the large powerstroke of myosin VI. Cell. 2007;131:300–8. doi: 10.1016/j.cell.2007.08.027. [DOI] [PubMed] [Google Scholar]
- 11.Holmes KC, Schroder RR, Sweeney HL, Houdusse A. The structure of the rigor complex and its implications for the power stroke. Philos Trans R Soc Lond B Biol Sci. 2004;359:1819–28. doi: 10.1098/rstb.2004.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anson M, Geeves MA, Kurzawa SE, Manstein DJ. Myosin motors with artificial lever arms. EMBO J. 1996;15:6069–74. [PMC free article] [PubMed] [Google Scholar]
- 13.Ruff C, Furch M, Brenner B, Manstein DJ, Meyhofer E. Single-molecule tracking of myosins with genetically engineered amplifier domains. Nat Struct Biol. 2001;8:226–9. doi: 10.1038/84962. [DOI] [PubMed] [Google Scholar]
- 14.Okten Z, Churchman LS, Rock RS, Spudich JA. Myosin VI walks hand-over-hand along actin. Nat Struct Mol Biol. 2004;11:884–7. doi: 10.1038/nsmb815. [DOI] [PubMed] [Google Scholar]
- 15.Sweeney HL, Park H, Zong AB, Yang Z, Selvin PR, Rosenfeld SS. How myosin VI coordinates its heads during processive movement. EMBO J. 2007;26:2682–92. doi: 10.1038/sj.emboj.7601720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kliche W, Fujita-Becker S, Kollmar M, Manstein DJ, Kull FJ. Structure of a genetically engineered molecular motor. EMBO J. 2001;20:40–6. doi: 10.1093/emboj/20.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Connell CB, Mooseker MS. Native Myosin-IXb is a plus-, not a minus-end-directed motor. Nat Cell Biol. 2003;5:171–2. doi: 10.1038/ncb924. [DOI] [PubMed] [Google Scholar]
- 18.Sun Y, Schroeder HW, 3rd, Beausang JF, Homma K, Ikebe M, Goldman YE. Myosin VI walks “wiggly” on actin with large and variable tilting. Mol Cell. 2007;28:954–64. doi: 10.1016/j.molcel.2007.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Veigel C, Wang F, Bartoo ML, Sellers JR, Molloy JE. The gated gait of the processive molecular motor, myosin V. Nat Cell Biol. 2002;4:59–65. doi: 10.1038/ncb732. [DOI] [PubMed] [Google Scholar]
- 20.Lan G, Sun SX. Flexible light-chain and helical structure of F-actin explain the movement and step size of myosin-VI. Biophys J. 2006;91:4002–13. doi: 10.1529/biophysj.106.089888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chevalier BS, Kortemme T, Chadsey MS, Baker D, Monnat RJ, Stoddard BL. Design, activity, and structure of a highly specific artificial endonuclease. Mol Cell. 2002;10:895–905. doi: 10.1016/s1097-2765(02)00690-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.