Abstract
Telomere serves two essential functions for the cell. It prevents the recognition of natural chromosome ends as DNA breaks (the end capping function). It counteracts incomplete end replication by adding DNA to the ends of chromosomes (the end elongation function). In most organisms studied, telomerase fulfills the end elongation function. In Drosophila, however, telomere specific retrotransposons have been coerced into performing this essential function for the host. In this review, we focus our discussion on transposition mechanisms and transcriptional regulation of these transposable elements, and present provocative models for the purpose of spurring new interests in the field.
1. Introduction
Transposable elements (TEs) are omnipresent in eukaryotes and can make up a significant portion of the genome. For example, the LINE-1 (L1) non-LTR retrotransposon alone has over half a million copies in the human genome, making up more than 17% of our haploid genome [1]. TEs generally impose detrimental effects on the host genome, for example, by altering the function of genes in their vicinity (e.g. [2]), or by inducing genome rearrangement via ectopic recombination between copies of related elements.
A general host response to a TE invasion, in particular one from retrotransposons that requires an RNA intermediate, is transcriptional silencing of the element to prevent its spreading. In addition, there are instances in which host-“parasite” conflicts resulted in domestication of a selfish element to fulfill cellular functions that are essential for host survival. One of the best examples is the non-LTR retrotransposons that populate the telomeric regions in Drosophila species. These elements transpose only to the ends of the chromosome and serve as the predominant if not the only mechanism to counter incomplete end replication, an essential function fulfilled by the telomerase reverse transcriptase and its RNA template in many other organisms.
For the general features of the Drosophila telomeric elements, the readers are referred to several recent review articles on the subject [3, 4]. In this article, we highlight areas in the study of these transposons that could yield rich mechanistic insights into retro-transposition in general, and basic principles governing the mutual dependence between the host and the selfish element.
2. Genomic organization of telomeric transposons in Drosophila
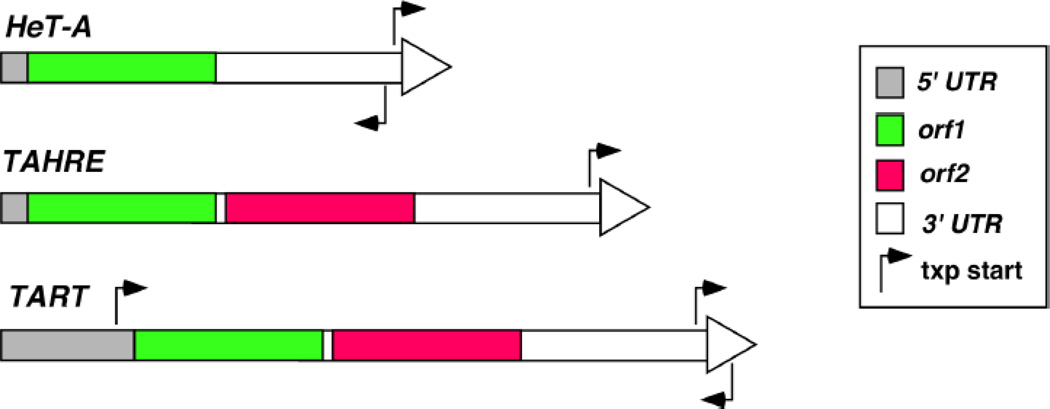
Telomeric DNA in D. melanogaster consists of ordered arrays of three retrotransposons, HeT-A, TART and TAHRE, in which the 5’ region of all elements is closer towards the telomere than the 3’. The HeT-A element is the most abundant. It contains a single open reading frame (orf) that encodes a protein with homology to Orf1 proteins (Orf1p) from other non-LTR elements (Figure 1, ref [5, 6]). HeT-A lacks a second orf that is present in other non-LTR elements, which would encode a protein with reverse transcriptase (RT) function and an endonuclease (EN) domain for target DNA incision [7]. Interestingly, the promoter for HeT-A transcription does not reside within the 5’ region, but at the most distal part of the 3’UTR (Figure 1, ref [8]). Therefore, transcription of a HeT-A element originates from the 3’UTR of its immediate upstream element (the one closer to the chromosome end). This represents an ingenious mechanism for preserving the 5’ region of a retrotransposon.
Figure 1. Molecular organization of telomeric transposons in Drosophila.
Each element is represented as a rectangular box with different parts of the element shaded differently according to their functions. The large arrowhead at the end of each element depicts the direction for the sense transcript. The approximate positions of transcriptional start sites (“txp start”) with their directions of transcription are represented by arrows. Elements are not drawn to scale.
TAHRE is the least abundant element. Although encoding one additional orf than HeT-A, TAHRE shares significant sequence homology with HeT-A. This led to the suggestion that HeT-A evolved from TAHRE by losing orf2 [9]. orf2 encodes an RT-like protein and with an EN domain. Promoter activities have also been detected at the 3’UTR of TAHRE (Figure 1, ref [10]), representing another similarity between HeT-A and TAHRE.
A full-length TART element has two orfs (Figure 1, ref [11]). Orf2p encodes an RT also with an EN domain. Both 5’ and 3’UTRs have promoter activities [12, 38], although evidence is lacking for the use of the promoter at 3’UTR in normal transposition. Recently, a novel template switching mechanism was proposed to explain how TART elements protect the integrity of the 5’ region. In this model, after one round of reverse transcription the RT molecule moves back to the 3’ region of the RNA template, adding DNA sequences from the 3’ region 5’ to the element [12].
3. The Biology of transposition
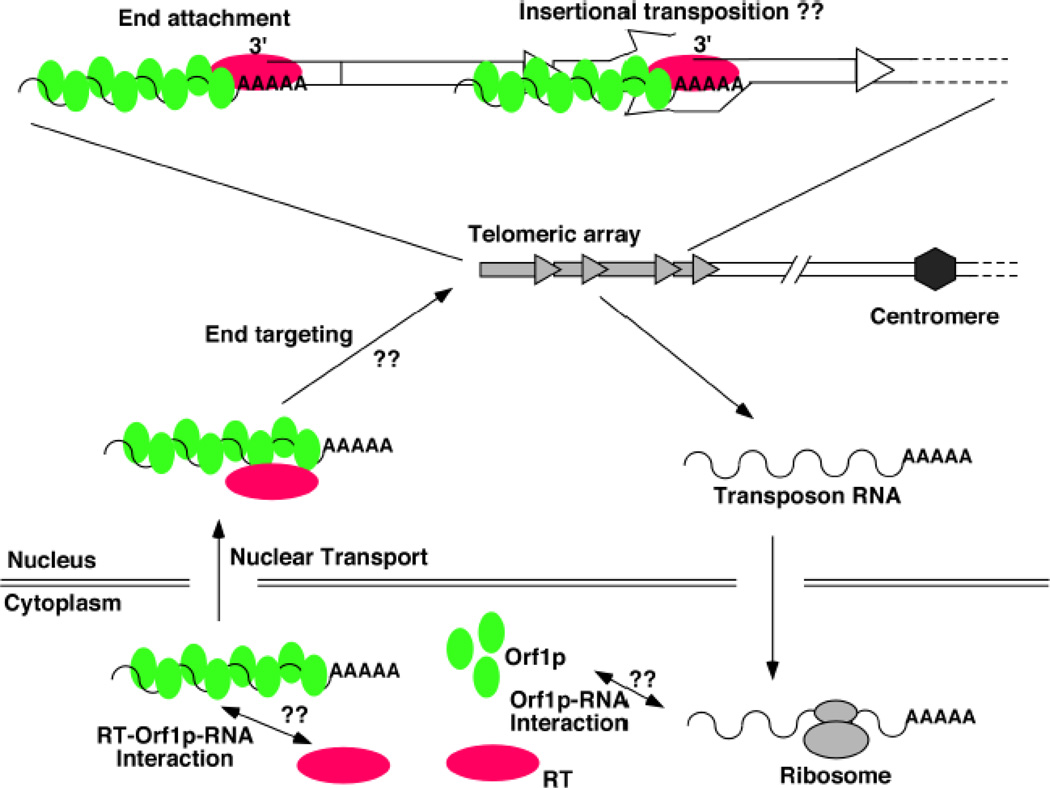
The exact mechanism of transposition remains obscure for the Drosophila telomeric elements. A generally agreed upon model has been presented multiple times before [3, 14], and our modified version is shown in Figure 2. Many aspects of the model were derived from our current knowledge on retro-transposition in general and from sequence analyses of cloned transposon arrays in Drosophila done by others. One essential aspect of the model, the mode of transposition, was deduced from studies on transposon attachment to DNA breaks during telomere healing of terminally deleted chromosomes (TD). We and others have shown that Drosophila cells can assemble a protective telomeric cap on chromosome ends with essentially any sequence so that TDs can be readily recovered by a variety of means [6, 15–18]. During the formation and/or propagation of these chromosomes, transposon attachment to the new end has been observed, suggesting that one mode of transposition is via reverse transcription primed with the 3’ end at the chromosome extremity.
Figure 2. A model for the life cycle of the Drosophila telomeric transposons.
In the central region of the diagram is a single telomere with four transposon insertions (grey arrows). Transposon RNA (wavery line) with a poly-A tail is exported to cytoplasm for translation to produce Orf1p (green ovals) and RT (red oval). Orf1p and RT could come from different elements, e.g. Orf1p from a HeT-A and RT from a TAHRE. How Orf1p interacts with its RNA is unknown. How Orf1p-RNA interacts with RT is also unknown. Presumably, an RT-Orf1p-RNA complex is imported into the nucleus and targeted to the telomeric region of the chromosome. The mechanism for this “end targeting” process is unknown. A close-up view of the single representative telomere is given at the top. Two possible modes of transposition are given: end attachment, which has strong supporting evidence, and insertional transposition, which is speculative. During end attachment, the transposon machinery uses the 3’ end of the chromosome to prime reverse transcription. During insertional transposition, the EN activity of RT makes a nick near the junction between two arbitrarily chosen elements. The transpson machinery uses the 3’ end of this nick to prime reverse transcription.
In the model shown in Figure 2, the life cycle of an element starts as mRNA molecules generated from the telomeric regions. These transcripts are used for the production of the Orf1 and Orf2 proteins in the cytoplasm. Orf1p from other better-studied retro-elements forms RNP with transposon RNA [19, 20]. Therefore it is commonly believed that an Orf1p-RNA complex from the Drosophila elements is first assembled in the cytoplasm and then transported into the nucleus, and perhaps along with Orf2p. The transposition machinery, Orf1p-Orf2p-RNA, is then targeted to the ends of chromosomes, where transposition occurs. Here we highlight several interesting areas that remain poorly understood.
3.1. End targeting
The most fascinating aspect of the biology of these transposons is their specific and exclusive targeting to chromosome ends. Since Drosophila cells rely on these elements to elongate their chromosomes, it is important to have a sufficient rate of transposition to chromosome ends. It is equally important to prevent integration of these elements to other parts of the genome. Since a significant portion of Drosophila elements are full length and potentially transposition competent [21, 22], spurious attachment of these elements to spontaneous double strand breaks can have dire consequences for the host. How the retro-transposition machinery is recruited to telomeres, but not anywhere else is not known.
HeT-A Orf1p overproduced in Drosophila cultured cells form nuclear foci, some of which co-localizes with the HOAP protein specifically enriched at Drosophila telomeres [23]. Since overexpression was achieved with a heterologous expression system (not with the endogenous HeT-A regulatory elements), it is possible that Orf1p in these telomeric foci is not in complex with the endogenous HeT-A RNA. If so, it would suggest that Orf1p contains an intrinsic signal for the targeting to chromosome ends irrespective of the state of the RNP. If these foci indeed contained HeT-A RNA, it would suggest that the transposon RNA is an integral part of the end-targeting signal. Presumably, this targeting was accomplished by interaction between Orf1p-RNA and one or more of the resident telomeric proteins, such as HOAP or HipHop [15].
The most understood process of targeting molecular activities to chromosome ends has to do with the recruitment of the telomerase RNP in organisms with telomerase. Telomerase is an RT that adds short DNA repeats to chromosome ends using an RNA template. In yeast S. cerevisiae, the major player in telomerase recruitment is the Cdc13 protein, a component of the Cdc13-Stn1-Ten1 (CST) complex that binds the single strand overhangs of telomeres [24, 25]. CST has long been considered a yeast specific complex. Recently homologs of CST have been identified in both plants [26] and vertebrates [27] suggesting that the function of CST is evolutionarily conserved. Intriguingly, a recent study identified the Drosophila Verroccio capping protein that shares limited homolog to Stn1 proteins in other organisms [28], suggesting the tantalizing possibilities that Drosophila has its own CST and that it might participate in the recruitment of the retro-transposon machinery for the elongation of Drosophila telomeres.
3.2. Orf1p and RNA interaction
It is a foregone conclusion that Orf1p would interact with transposon RNA. Consistently, a recent structural analysis suggested that Orf1p from either HeT-A or TART contains two atypical RRM domains with RNA binding capability [29]. However, the temporal and spatial features of this interaction remain unknown.
Orf1p from L1 elements has a “cis-preference” for interacting with its RNA: it preferentially associates with the transcript that was once translated to make the protein itself [30]. The evolutionary implication here is that only elements with a functional coding capacity are allowed to transpose. In addition, since L1 Orf1p’s interaction with nucleic acid is sequence non-specific [31], this “cis-preference” suppresses spurious association of Orf1p with other RNA species in the cell. However, not all non-LTR retro-elements display a “cis-preference”. The telomeric transposons in Bombyx mori can act on heterologous RNA templates [32]. Results from HeT-A Orf1p overexpression experiments [23] suggest that Drosophila elements are unlikely to have a “cis-preference”. As discussed earlier, overproduced Orf1p can go to the telomeres either without forming a RNP or forming an RNP with endogenous transposon RNA molecules that did not encode the protein. However, it remains formally possible that the HeT-A Orf1p at the telomeres was in complex with the heterologous transcripts that encoded the protein. This is difficult to imagine since the only HeT-A homologous region on these transcripts is in orf1 coding sequences. Because HeT-A overexpression might not recapitulate the natural behavior of the transposon, further investigation is needed to definitively rule out “cis-preference” for the Drosophila elements.
Without a “cis-preference” for HeT-A Orf1p-RNA association, it would not be necessary to invoke a model in which the Orf1p-RNA complex forms only in the cytoplasm and that the RNA templates for transposition have to be imported from the cytoplasm. It is conceivable that Orf1p can interact with nascent RNA molecules from the transposons right off the chromosome and use those RNA species as transposition intermediates. In fact, Orf1p interaction of nascent transposon RNA could be an attractive mechanism for targeting the transposon machinery to chromosome ends. This cannot be the only mechanism, however, as it is inconsistent with the ability of these elements to attach to transposon-free ends. Nevertheless, our current knowledge does not rule out that the Orfp1-RNA interaction happens in the nucleus or even at the site of transposition.
3.3. RT interaction with Orf1p-RNA
It is interesting that HeT-A, the most abundant element at the telomeres, does not encode an RT. This implies that RT for HeT-A transposition has to be supplied in trans either from TARHE,TART, or an RT gene from other sources. This is reminiscent of the dichotomy of telomerase and its RNA template into two separate genes. How RT interacts with Orf1p-RNA complex remains obscure for the Drosophila elements. Are they packaged as a complex before entering the nucleus or does the incorporation of RT into the Orf1 RNP happen at the telomere? Since RT for HeT-A transposition must be supplied in trans, it is unlikely that RT and Orf1p-RNA are always present in the cytoplasm at the same time, unless there is coordinated protein translation from different elements. Therefore, it is possible that RT interacts with Orf1p-RNP inside the nucleus, perhaps at or around the site of genome integration. This implies that there is separate targeting of RT and Orf1p-RNA to chromosome ends. Alternatively, a functional end-targeting signal requires the participation of all three components. However, this is difficult to reconcile with results from HeT-A Orf1p overexpression, which likely resulted in vast overrepresentation of Orf1p in comparison with RT. An attractive model posits that Orf1p or RT proteins interact with nascent RNA in a chromatin context with high affinity resulting in the targeting of the transposon machinery to telomeres. How could we then explain targeting to transposon-free ends? It is possible that the transposition machinery is recruited to these de novo telomeres via telomere-telomere association, bringing these ends to transposon-capped ends. If true, it implies that DNA ends are receptive to transposon attachment only after it adopts a telomeric fate, e.g. occupied by telomere-specific proteins.
3.4. End attachment versus internal integration
It remains debatable whether end attachment is the predominant transposition mechanism operating at the natural environment of the telomere, i.e. in the presence of similar elements. It is formally possible that internal integration guided by primary DNA sequences at the existing elements represent another mode of transposition for Drosophila elements. We invoke this possibility because a peculiar feature of the RT proteins encoded by both TART and TAHRE: Orf2p in these two elements has a distinct endonuclease domain. In other retro-elements such as mammalian L1, EN nicks the target DNA, a function essential for transposition. Interestingly, EN-less L1 elements either naturally present or engineered in vitro can transpose to telomere ends [33, 34]. In addition, telomeric transposons from Bombyx mori and Tribolium castaneum use EN-mediated target incision for transposition to telomeres [35]. Therefore, it remains untested whether Drosophila element can integrate internally to transposon arrays at telomeres. Such a strategy may serve to protect functional elements from 5’ degradation due to incomplete end replication. Biochemical analyses using recombinant RT proteins would be needed to interrogate whether Drosophila telomeric elements encode EN activities.
3.5. Genetically marked elements for the study of telomeric transposition
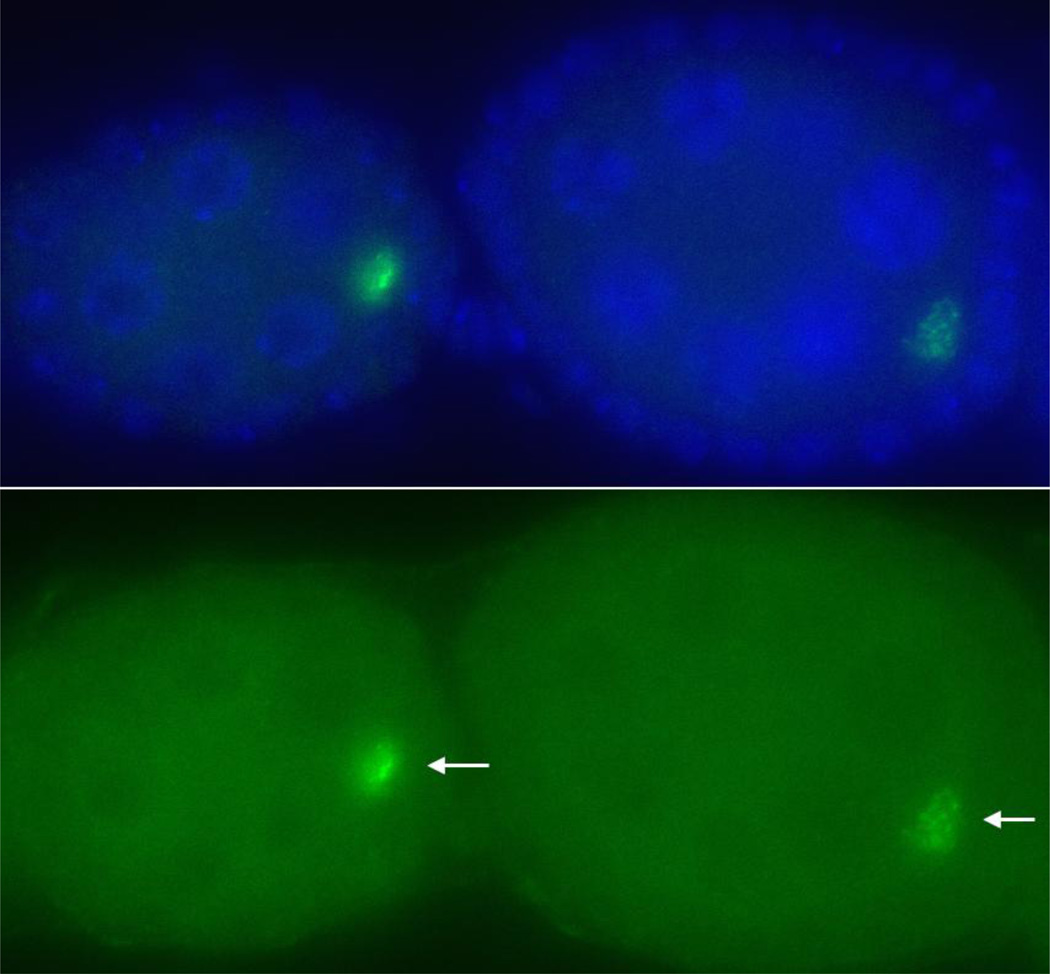
Genetically tractable transposons have greatly facilitated the study of TEs, especially in elucidating the mechanisms for transposition [36, 37]. Similar elements in Drosophila would no doubt help reveal rich mechanistic insights into transposon biogenesis and put our provocative models to test. Toward achieving that goal, we have constructed a genetically marked HeT-A element (unpublished results). Preliminary results suggest that the element likely encodes a functional Orf1p (Figure 3). Research is in progress to test whether this element can support transposition to telomeres.
Figure 3. HeT-A Orf1p is overproduced in defective piRNA germline.
Two mid-stage egg chambers are shown from a spnE-mutant female, who also had a transgene carrying a full length HeT-A element with a gfp-tagged orf1 gene. The top panel shows a merged image of DAPI staining for DNA (in purple) and anti-GFP signal (in green). The bottom panel shows GFP signals only with arrows pointing to the position of the oocyte.
4. The developmental regulation of transposon expression
Expression of Drosophila telomeric elements has been one of the most widely used readouts for defects in small RNA mediated transcriptional silencing. However, the normal transcriptional regulation of these elements is far from just silencing. In somatic tissues, all three elements are actively transcribed with full length sense transcripts detectable [10, 38–41]. In addition, TART elements produce a nearly full-length anti-sense transcript, whose function remains unknown. Transposon transcripts are generally limited to proliferating cells in the soma. Consistently, somatic transposition events can be readily detected as cell clones in which normal expression of visible markers (white or yellow genes) at the extremity of a terminally truncated chromosome is restored due to transposon attachment [42, 43]. These attachment events either relieve silencing on the visible marker or more likely provide promoter activities driving marker expression.
The conserved Heterochromatin Protein 1 (HP1) is a major player in somatic silencing of telomeric transposons. Even the loss of a single copy of the su(var)205 gene, which encoded HP1, leads to dramatic derepression of the transposons as assayed by Northern blots [42, 44]. This is consistent with ChIP results locating HP1 to the transposon arrays [15, 45]. Several other genes, when mutated, confer moderate effects on somatic silencing of transposons. These include the proliferation disrupter gene [46] and several genes with essential capping function [47].
In the female germline, low levels of HeT-A and TAHRE transcripts can be detected in early-staged egg chambers, while TART sense transcripts are found in nurse cells of late-staged egg chambers [10, 48]. This low level of expression is achieved by PIWI-interacting RNA (piRNA) mediated silencing as germlines mutant for the pathway have highly elevated transposon transcripts [13, 49, 50]. Interesting, a 434 bp fragment from HeT-A 3’UTR is sufficient to confer piRNA-mediated silencing of a lacZ gene placed 3’ to the HeT-A fragment, but inserted at non-telomeric positions [13]. If piRNA silencing of HeT-A were mainly executed through post-transcriptional degradation of transposon RNAs, one would have to conclude from these experiments that only about 30 bp, spanning from the HeT-A transcriptional start site to the start codon of lacZ, would be sufficient to induce instability of lacZ RNA in the female germline. There might be multiple elements within HeT-A that bring about piRNA-mediated silencing as it has been shown that HeT-A orf1 cDNA overexpressed in the female germline was subjected to piRNA mediated silencing [50]. We constructed a full length HeT-A element that encodes a GFP-tagged Orf1p (unpublished results). Consistent with results from Shpiz et al. [13], this element is subject to piRNA mediated silencing even at non-telomeric positions. Mutations in the piRNA pathway resulted in strong signals of GFP-Orf1p (Figure 3), whereas it is undetectable under normal condition (not shown). This suggests that full length HeT-A transcripts are stabilized in the mutants, and these transcripts have full coding capacity. Interestingly, piRNA mutants are female sterile. It remains undetermined whether highly abundant transposon-encoded proteins as a result of the derepression of HeT-A and other retrotransposons contribute to germline dysfunction.
5. Concluding remarks
Transposons generally affect host fitness in negative ways. However, conflicts between the host and the selfish elements can sometimes result in co-dependence of the two parties as host “tames” the transposons to fulfill essential function. The study on the regulation of Drosophila telomeric retrotransposons promises to be a fertile ground for the discovery of fundamental principles on host parasite interaction.
Acknowledgement
Research in the lab is supported by the Intramural program of the National Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Kazazian HH, Jr, Wong C, Youssoufian H, Scott AF, Phillips DG, Antonarakis SE. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature. 1988;332:164–166. doi: 10.1038/332164a0. [DOI] [PubMed] [Google Scholar]
- 3.Mason JM, Frydrychova RC, Biessmann H. Drosophila telomeres: an exception providing new insights. Bioessays. 2008;30:25–37. doi: 10.1002/bies.20688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pardue ML, Debaryshe PG. Retrotransposons that maintain chromosome ends. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1100278108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pardue ML, Danilevskaya ON, Lowenhaupt K, Wong J, Erby K. The gag coding region of the Drosophila telomeric retrotransposon, HeT-A, has an internal frame shift and a length polymorphic region. J Mol Evol. 1996;43:572–583. doi: 10.1007/BF02202105. [DOI] [PubMed] [Google Scholar]
- 6.Biessmann H, Champion LE, O'Hair M, Ikenaga K, Kasravi B, Mason JM. Frequent transpositions of Drosophila melanogaster HeT-A transposable elements to receding chromosome ends. EMBO J. 1992;11:4459–4469. doi: 10.1002/j.1460-2075.1992.tb05547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 8.Danilevskaya ON, Arkhipova IR, Traverse KL, Pardue ML. Promoting in tandem: the promoter for telomere transposon HeT-A and implications for the evolution of retroviral LTRs. Cell. 1997;88:647–655. doi: 10.1016/s0092-8674(00)81907-8. [DOI] [PubMed] [Google Scholar]
- 9.Abad JP, De Pablos B, Osoegawa K, De Jong PJ, Martin-Gallardo A, Villasante A. TAHRE, a novel telomeric retrotransposon from Drosophila melanogaster, reveals the origin of Drosophila telomeres. Mol Biol Evol. 2004;21:1620–1624. doi: 10.1093/molbev/msh180. [DOI] [PubMed] [Google Scholar]
- 10.Shpiz S, Kwon D, Uneva A, Kim M, Klenov M, Rozovsky Y, Georgiev P, Savitsky M, Kalmykova A. Characterization of Drosophila telomeric retroelement TAHRE: transcription, transpositions, and RNAi-based regulation of expression. Mol Biol Evol. 2007;24:2535–2545. doi: 10.1093/molbev/msm205. [DOI] [PubMed] [Google Scholar]
- 11.Sheen FM, Levis RW. Transposition of the LINE-like retrotransposon TART to Drosophila chromosome termini. Proc Natl Acad Sci U S A. 1994;91:12510–12514. doi: 10.1073/pnas.91.26.12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.George JA, Traverse KL, DeBaryshe PG, Kelley KJ, Pardue ML. Evolution of diverse mechanisms for protecting chromosome ends by Drosophila TART telomere retrotransposons. Proc Natl Acad Sci U S A. 2010;107:21052–21057. doi: 10.1073/pnas.1015926107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shpiz S, Olovnikov I, Sergeeva A, Lavrov S, Abramov Y, Savitsky M, Kalmykova A. Mechanism of the piRNA-mediated silencing of Drosophila telomeric retrotransposons. Nucleic Acids Res. 2011;39:8703–8711. doi: 10.1093/nar/gkr552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pardue ML, DeBaryshe PG. Drosophila telomeres: A variation on the telomerase theme. Fly (Austin) 2008;2:101–110. doi: 10.4161/fly.6393. [DOI] [PubMed] [Google Scholar]
- 15.Gao G, Walser JC, Beaucher ML, Morciano P, Wesolowska N, Chen J, Rong YS. HipHop interacts with HOAP and HP1 to protect Drosophila telomeres in a sequence-independent manner. EMBO J. 2010;29:819–829. doi: 10.1038/emboj.2009.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahmad K, Golic KG. The transmission of fragmented chromosomes in Drosophila melanogaster. Genetics. 1998;148:775–792. doi: 10.1093/genetics/148.2.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levis RW. Viable deletions of a telomere from a Drosophila chromosome. Cell. 1989;58:791–801. doi: 10.1016/0092-8674(89)90112-8. [DOI] [PubMed] [Google Scholar]
- 18.Donaldson KM, Karpen GH. Trans-suppression of terminal deficiency-associated position effect variegation in a Drosophila minichromosome. Genetics. 1997;145:325–337. doi: 10.1093/genetics/145.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin SL. Ribonucleoprotein particles with LINE-1 RNA in mouse embryonal carcinoma cells. Mol Cell Biol. 1991;11:4804–4807. doi: 10.1128/mcb.11.9.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hohjoh H, Singer MF. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 1996;15:630–639. [PMC free article] [PubMed] [Google Scholar]
- 21.George JA, DeBaryshe PG, Traverse KL, Celniker SE, Pardue ML. Genomic organization of the Drosophila telomere retrotransposable elements. Genome Res. 2006;16:1231–1240. doi: 10.1101/gr.5348806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abad JP, De Pablos B, Osoegawa K, De Jong PJ, Martin-Gallardo A, Villasante A. Genomic analysis of Drosophila melanogaster telomeres: full-length copies of HeT-A and TART elements at telomeres. Mol Biol Evol. 2004;21:1613–1619. doi: 10.1093/molbev/msh174. [DOI] [PubMed] [Google Scholar]
- 23.Rashkova S, Karam SE, Kellum R, Pardue ML. Gag proteins of the two Drosophila telomeric retrotransposons are targeted to chromosome ends. J Cell Biol. 2002;159:397–402. doi: 10.1083/jcb.200205039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Evans SK, Lundblad V. Est1 and Cdc13 as comediators of telomerase access. Science. 1999;286:117–120. doi: 10.1126/science.286.5437.117. [DOI] [PubMed] [Google Scholar]
- 25.Nugent CI, Hughes TR, Lue NF, Lundblad V. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- 26.Surovtseva YV, Churikov D, Boltz KA, Song X, Lamb JC, Warrington R, Leehy K, Heacock M, Price CM, Shippen DE. Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol Cell. 2009;36:207–218. doi: 10.1016/j.molcel.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyake Y, Nakamura M, Nabetani A, Shimamura S, Tamura M, Yonehara S, Saito M, Ishikawa F. RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol Cell. 2009;36:193–206. doi: 10.1016/j.molcel.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 28.Raffa GD, Raimondo D, Sorino C, Cugusi S, Cenci G, Cacchione S, Gatti M, Ciapponi L. Verrocchio, a Drosophila OB fold-containing protein, is a component of the terminin telomere-capping complex. Genes Dev. 2010;24:1596–1601. doi: 10.1101/gad.574810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khazina E, Weichenrieder O. Non-LTR retrotransposons encode noncanonical RRM domains in their first open reading frame. Proc Natl Acad Sci U S A. 2009;106:731–736. doi: 10.1073/pnas.0809964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV. Human L1 retrotransposition: cis preference versus trans complementation. Mol Cell Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Callahan KE, Hickman AB, Jones CE, Ghirlando R, Furano AV. Polymerization and nucleic acid-binding properties of human L1 ORF1 protein. Nucleic Acids Res. doi: 10.1093/nar/gkr728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osanai M, Takahashi H, Kojima KK, Hamada M, Fujiwara H. Essential motifs in the 3' untranslated region required for retrotransposition and the precise start of reverse transcription in non-long-terminal-repeat retrotransposon SART1. Mol Cell Biol. 2004;24:7902–7913. doi: 10.1128/MCB.24.18.7902-7913.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morrish TA, Garcia-Perez JL, Stamato TD, Taccioli GE, Sekiguchi J, Moran JV. Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres. Nature. 2007;446:208–212. doi: 10.1038/nature05560. [DOI] [PubMed] [Google Scholar]
- 34.Gladyshev EA, Arkhipova IR. Telomere-associated endonuclease-deficient Penelope-like retroelements in diverse eukaryotes. Proc Natl Acad Sci U S A. 2007;104:9352–9357. doi: 10.1073/pnas.0702741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Osanai-Futahashi M, Fujiwara H. Coevolution of Telomeric Repeats and Telomeric Repeat-Specific Non-LTR Retrotransposons in Insects. Mol Biol Evol. 2011 doi: 10.1093/molbev/msr135. [DOI] [PubMed] [Google Scholar]
- 36.Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 37.Curcio MJ, Garfinkel DJ. Single-step selection for Ty1 element retrotransposition. Proc Natl Acad Sci U S A. 1991;88:936–940. doi: 10.1073/pnas.88.3.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maxwell PH, Belote JM, Levis RW. Identification of multiple transcription initiation, polyadenylation, and splice sites in the Drosophila melanogaster TART family of telomeric retrotransposons. Nucleic Acids Res. 2006;34:5498–5507. doi: 10.1093/nar/gkl709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walter MF, Biessmann H. Expression of the telomeric retrotransposon HeT-A in Drosophila melanogaster is correlated with cell proliferation. Dev Genes Evol. 2004;214:211–219. doi: 10.1007/s00427-004-0400-x. [DOI] [PubMed] [Google Scholar]
- 40.George JA, Pardue ML. The promoter of the heterochromatic Drosophila telomeric retrotransposon, HeT-A, is active when moved into euchromatic locations. Genetics. 2003;163:625–635. doi: 10.1093/genetics/163.2.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Danilevskaya ON, Traverse KL, Hogan NC, DeBaryshe PG, Pardue ML. The two Drosophila telomeric transposable elements have very different patterns of transcription. Mol Cell Biol. 1999;19:873–881. doi: 10.1128/mcb.19.1.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Savitsky M, Kravchuk O, Melnikova L, Georgiev P. Heterochromatin protein 1 is involved in control of telomere elongation in Drosophila melanogaster. Mol Cell Biol. 2002;22:3204–3218. doi: 10.1128/MCB.22.9.3204-3218.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Golubovsky MD, Konev AY, Walter MF, Biessmann H, Mason JM. Terminal retrotransposons activate a subtelomeric white transgene at the 2L telomere in Drosophila. Genetics. 2001;158:1111–1123. doi: 10.1093/genetics/158.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perrini B, Piacentini L, Fanti L, Altieri F, Chichiarelli S, Berloco M, Turano C, Ferraro A, Pimpinelli S. HP1 controls telomere capping, telomere elongation, and telomere silencing by two different mechanisms in Drosophila. Mol Cell. 2004;15:467–476. doi: 10.1016/j.molcel.2004.06.036. [DOI] [PubMed] [Google Scholar]
- 45.Frydrychova RC, Mason JM, Archer TK. HP1 is distributed within distinct chromatin domains at Drosophila telomeres. Genetics. 2008;180:121–131. doi: 10.1534/genetics.108.090647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Torok T, Benitez C, Takacs S, Biessmann H. The protein encoded by the gene proliferation disrupter (prod) is associated with the telomeric retrotransposon array in Drosophila melanogaster. Chromosoma. 2007;116:185–195. doi: 10.1007/s00412-006-0090-4. [DOI] [PubMed] [Google Scholar]
- 47.Bi X, Srikanta D, Fanti L, Pimpinelli S, Badugu R, Kellum R, Rong YS. Drosophila ATM and ATR checkpoint kinases control partially redundant pathways for telomere maintenance. Proc Natl Acad Sci U S A. 2005;102:15167–15172. doi: 10.1073/pnas.0504981102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Savitsky M, Kwon D, Georgiev P, Kalmykova A, Gvozdev V. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes Dev. 2006;20:345–354. doi: 10.1101/gad.370206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klenov MS, Lavrov SA, Stolyarenko AD, Ryazansky SS, Aravin AA, Tuschl T, Gvozdev VA. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res. 2007;35:5430–5438. doi: 10.1093/nar/gkm576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lim AK, Tao L, Kai T. piRNAs mediate posttranscriptional retroelement silencing and localization to pi-bodies in the Drosophila germline. J Cell Biol. 2009;186:333–342. doi: 10.1083/jcb.200904063. [DOI] [PMC free article] [PubMed] [Google Scholar]