Abstract
Sirt3 is a mitochondrial sirtuin, predominantly expressed in highly metabolic tissues. Germline ablation of Sirt3 has major metabolic consequences, including increased susceptibility to metabolic damage and oxidative stress after high fat feeding. In order to determine the contribution of liver and skeletal muscle to these phenotypes, we generated muscle-specific Sirt3 (Sirt3skm−/−) and liver-specific Sirt3 (Sirt3hep−/−) knock-out mice. Despite a marked global hyperacetylation of mitochondrial proteins, Sirt3skm−/− and Sirt3hep−/− mice did not manifest any overt metabolic phenotype under either chow or high fat diet conditions. Similarly, there was no evidence for increased oxidative stress in muscle or liver when Sirt3 was ablated in a tissue-specific manner. These observations suggest that the mitochondrial hyperacetylation induced by Sirt3-deletion in a tissue specific manner is not necessarily linked to mitochondrial dysfunction and does not recapitulate the metabolic abnormalities observed in the germline Sirt3 knock-out mice.
Sirt3 is one of the 7 members of the sirtuin family of NAD+-dependent deacetylases. Together with Sirt4 and Sirt5, Sirt3 localizes predominantly in the mitochondria1. Importantly, Sirt3 is a primary regulator of mitochondrial protein acetylation. The absence of Sirt3 enhances global acetylation of mitochondrial proteins, while no significant changes in protein acetylation were observed in Sirt4−/− and Sirt5−/− mitochondria2. Sirt3 expression is highly enriched in metabolic tissues like liver, brown adipose tissue (BAT), heart, skeletal muscle and kidney3. Sirt3 expression increases in muscle after exercise training, fasting and caloric restriction (CR), while it decreases upon long-term high fat feeding4,5. In liver and BAT Sirt3 expression also increases after CR and fasting, as well as upon cold exposure in BAT3,6.
Germline Sirt3−/− mice display increased levels of cellular reactive oxygen species (ROS)7,8,9,10,11,12 and impaired cellular respiration after prolonged fasting6 in different tissues, including the muscle, liver, brain or the inner ear. In contrast, Sirt3 overexpression in vitro enhances mitochondrial respiration13 and reduces ROS production3. Not surprisingly, whole body Sirt3 knock-out mice are sensitized to high fat diet (HFD)-induced obesity, insulin resistance, hyperlipidemia and steatohepatitis14. The etiology of such defects might be found in the ability of Sirt3 to enhance fat oxidation and improve anti-oxidant defences6,9,10,14,15,16. However, while recent reports show that Sirt3 suppresses oxidative stress under CR9,10,12, it still remains to be elucidated whether Sirt3 might influence HFD-induced oxidative stress.
All studies to date used germline Sirt3 deficient mice to study the role of Sirt3 on metabolism, making it impossible to distinguish the contribution of individual tissues to the phenotypic changes. Liver and muscle are two of the most important tissues determining whole body metabolism: skeletal muscle is the largest organ in mammals, contributing to ±40% of the body mass, and it plays a major role in whole body metabolism, as it is vital for insulin-mediated glucose disposal and lipid catabolism. In turn, the liver is central to regulate glucose, lipid and cholesterol homeostasis. Altered function of these tissues is, thus, likely to contribute to the systemic metabolic disturbances observed in the germline Sirt3 knock-out mice. Here, we report the generation of the first set of tissue-specific Sirt3 knockout mouse models in muscle and liver, and describe how Sirt3 deletion in these tissues, despite leading to mitochondrial protein hyperacetylation, has minor phenotypic consequences. This suggests that the metabolic abnormalities observed in the germline Sirt3-KO mice may not stem from the liver or muscle deficiencies.
Results
Generation of the Sirt3skm−/− and Sirt3hep−/− mice
To investigate the role of Sirt3 in muscle and liver, as well as its impact on whole body metabolism, we generated a mouse line in which exons 2 and 3 were flanked with LoxP sites, priming it for subsequent deletion using the Cre-LoxP system (Fig. 1A). Sirt3L2/L2 mice, bearing floxed Sirt3 L2 alleles, were bred with mice expressing the Cre recombinase under the control of the human α-skeletal actin promoter17, yielding HSA-CreTg/0/Sirt3L2/L2 mutants (Sirt3skm−/−), in which Sirt3 is selectively ablated in the skeletal muscles. A distinct mouse line, in which Cre expression was under the control of the albumin promoter18, was used to generate Alb-CreTg/0/Sirt3L2/L2 mice (Sirt3hep−/−), in which Sirt3 is selectively ablated in the liver.
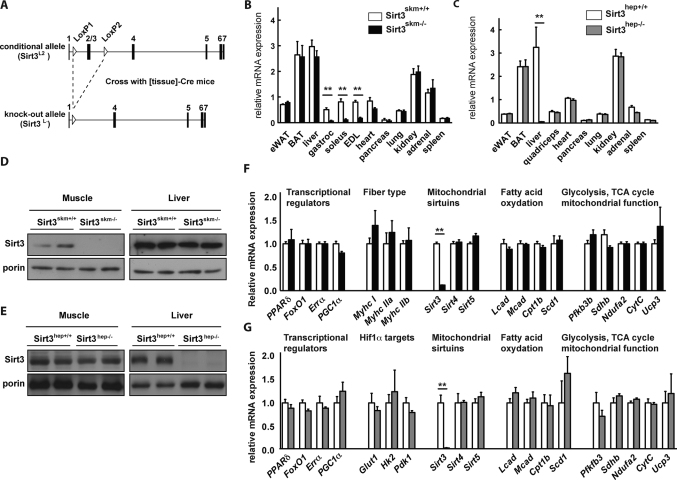
Figure 1. Generation, validation and expression of metabolic genes for the Sirt3hep−/− and Sirt3skm−/− mice.
(A) Targeting strategy used to generate the tissue-specific Sirt3-deficient mice. Maps of the Sirt3 genomic locus showing the conditional allele (upper panel) and the KO allele (lower panel). The white arrowheads indicate the LoxP sites and the black vertical bars represent the respective exons. (B–C) Sirt3 mRNA expression levels in different tissues of Sirt3skm+/+ and Sirt3skm−/− mice (B) and of Sirt3hep+/+ and Sirt3hep−/− mice (C); n = 4 per group. (D–E) Sirt3 protein expression as assessed by western blotting in muscle (D) and liver (E) of the Sirt3-deficient mouse models. Porin is used as a loading control. (F–G) Gene expression in gastrocnemius muscle of Sirt3skm+/+ and Sirt3skm−/− mice (F) and in liver of Sirt3hep+/+ and Sirt3hep−/− mice (G) after 18 weeks of HFD. mRNA levels were normalized to 36B4. Data represent the mean ± SEM for at least four animals per group.
Sirt3skm−/− and Sirt3hep−/− mice were born at a normal Mendelian ratio and had a normal appearance. Only residual Sirt3 mRNA levels were detected in skeletal muscle (gastrocnemius, soleus and EDL) of 8-week-old Sirt3skm−/− mice (Fig. 1B), demonstrating that Sirt3 was efficiently ablated in Sirt3skm−/− muscles. In addition to the skeletal muscle, a 35% reduction in Sirt3 mRNA levels was observed in the heart of Sirt3skm−/− mice, whereas no significant reduction was seen in other tissues (Fig. 1B). Sirt3 mRNA levels were also blunted in livers from Sirt3hep−/− mice of 8 weeks of age, but normal in other tissues (Fig. 1C). In line with the above results, the Sirt3 protein was undetectable in the skeletal muscle of Sirt3skm−/− mice, but remained unaffected in the liver (Fig. 1D); conversely the Sirt3 protein was only absent in the liver in the Sirt3hep−/− mice (Fig. 1E), confirming that the Sirt3 deletion is efficient and restricted to the targeted tissue.
The absence of Sirt3 had no impact on the expression of a vast set of metabolic genes in either muscle (Fig. 1F) or liver (Fig. 1G). Of note, we did not detect any compensatory increase in the expression of the other mitochondrial sirtuins (Sirt4 and Sirt5) in the two Sirt3-deficient tissues. We also determined Pgc1α and Scd1 expression, two genes whose levels were reported to be altered in germline Sirt3-KO tissues5,14; however, we could not detect any significant change in their expression in our mutant mice. These results suggest that muscle or liver-specific deletion of Sirt3 does not have a major impact on the expression of genes involved in metabolic control. We only performed this gene expression analysis in mice subjected to HFD, since young, unchallenged mice show no phenotype whatsoever, both in our hands and in previous reports2.
Normal metabolic phenotype in Sirt3hep−/− mice fed chow or high fat diet
We next subjected Sirt3hep+/+ and Sirt3hep−/− mice to a standardized phenotyping protocol before and after high fat diet (HFD) feeding (Fig. 2A). Body weight and body composition were similar in both genotypes before and after HFD (Fig. 2B and 2C). The weight gain curves of the Sirt3hep−/− mice were indistinguishable from those of the Sirt3hep+/+ mice (data not shown). During indirect calorimetry using the comprehensive lab animal monitoring system (CLAMS), no difference was observed in food intake (Fig. 2D), mean VO2 (Fig. 2E), mean respiratory exchange ratio (RER) (Fig. 2F) or spontaneous locomotor activity (Fig. 2G), before and after HFD. At sacrifice, the weight of the liver, epididymal white fat or interscapular brown fat depots were not different between the genotypes (Fig. 2H). To specifically test the contribution of Sirt3 deletion in the liver to glucose homeostasis we performed an intraperitoneal glucose tolerance test (ipGTT) and an insulin tolerance test (ipITT) both before (data not shown) and after HFD. Again no significant differences between Sirt3hep+/+ and Sirt3hep−/− animals were detected in these experiments (Fig. 2I, 2J and data not shown). All these data contrast with the results obtained in germline Sirt3-deficient mice, where Sirt3-deficiency was shown to sensitize the liver to HFD-induced damage14.
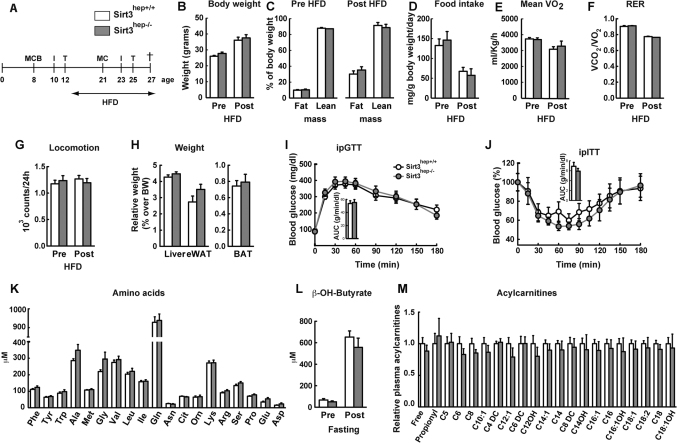
Figure 2. Metabolic phenotyping of Sirt3hep−/− mice.
(A) Experimental schedule of the clinical phenotyping protocol. MCB: non-invasive monitoring of body fat and lean mass by EchoMRI, energy expenditure by indirect calorimetry (CLAMS system), and blood sampling for metabolite analysis; I: intra-peritoneal glucose tolerance test (ipGTT); T: intra-peritoneal insulin tolerance test (ipITT); †: sacrifice and collection of blood and organs for further analysis. (B) Body weight and (C) body composition of Sirt3hep+/+ and Sirt3hep−/− mice before and after 8 weeks of HFD. (D–G) Daily food intake (D), mean O2 consumption (VO2; E), mean respiratory exchange ratio (RER; F) and total ambulatory locomotor activity (G) as measured over a 24 h period before and after HFD. (H) Relative weight of tissues normalized by body weight in Sirt3hep+/+ and Sirt3hep−/− mice. (I–J) Intra-peritoneal glucose tolerance test (ipGTT; I), and intra-peritoneal insulin tolerance test (ipITT; J) in mice fed a HFD for 15-17 weeks. The bar graphs represent the average area under the curve (AUC). (K) Levels of the indicated aminoacids were determined in plasma from 8 weeks old male mice. (L) β-hydroxy-butyrate was analysed in plasma from 8 weeks old male mice before and after a 24h fasting period. (M) Acylcarnitine levels in plasma from 8 weeks old male mice. Results are represented as mean values (n = 8−10 mice for each group) ± standard error of the mean (SEM).
Germline Sirt3-KO mice have also been reported to display altered aminoacid profiles in blood and liver15. We therefore also analyzed the blood aminoacid profiles, but could not detect differences between the Sirt3hep−/− and Sirt3hep+/+ samples, both in the fed state (data not shown) and after a 24 h fast (Fig. 2K).
Sirt3 has also been suggested to act as a key modulator of ketone body production, a glucose-sparing energy substrate in the fasting state mostly synthesized by hepatocytes19. However, blood levels of β-hydroxybutyrate, a surrogate marker of the activity of this pathway, were similar in Sirt3hep+/+ and Sirt3hep−/− mice both before and after a 24 h fast at 8-weeks of age (Fig. 2L). This contrasts with the germline Sirt3 knock-out mice, which displayed a maximal decrease in β-hydroxybutyrate after a 24 h fast19.
Finally, since germline Sirt3-KO mice display abnormal fatty acid metabolism and aberrant blood acylcarnitine profiles14,15, we analyzed the blood acylcarnitines in Sirt3hep+/+ and Sirt3hep−/− animals in both fed (data not shown) and fasted (24 h fast) states (Fig. 2M), but again could not detect any significant difference in the acylcarnitine levels between the two genotypes.
All these findings combined suggest that the altered metabolic response of germline Sirt3-KO mice subjected to HFD14 or to fasting15,19 are not due to the absence of Sirt3 in hepatocytes, but rather stem from the indirect effect of Sirt3 ablation in some other tissue.
No metabolic phenotype in Sirt3skm−/− mice under chow or high fat diet
We next determined whether the phenotype observed in the germline Sirt3 knock-out mice could be due to another major metabolic tissue, i.e. the skeletal muscle. We therefore subjected Sirt3skm−/− and Sirt3skm+/+ mice to a similar phenotyping protocol than the one described for the liver-specific Sirt3-deficient mice (Fig. 3A). No differences in body weight (Fig. 3B) and body composition (Fig. 3C) were observed between Sirt3skm−/− and Sirt3skm+/+ mice, even under HFD conditions. Consistent with the similarities in body weight, evaluation using CLAMS indicated that food intake (Fig. 3D), mean VO2 (Fig. 3E), mean RER (Fig. 3F) and spontaneous locomotor activity (Fig. 3G) were similar between the two genotypes before and after HFD. In addition, no significant differences were observed in the weight of different tissues at sacrifice (Fig. 3H). Upon an intraperitoneal glucose tolerance test (ipGTT) the glucose excursion (Fig. 3I; left panel) and glucose-induced insulin secretion (Fig. 3I; right panel) were comparable between the two genotypes. Also, the drop and recovery of blood glucose levels upon an intraperitoneal insulin tolerance test (ipITT) were, again, not distinct between the two genotypes (Fig. 3J).
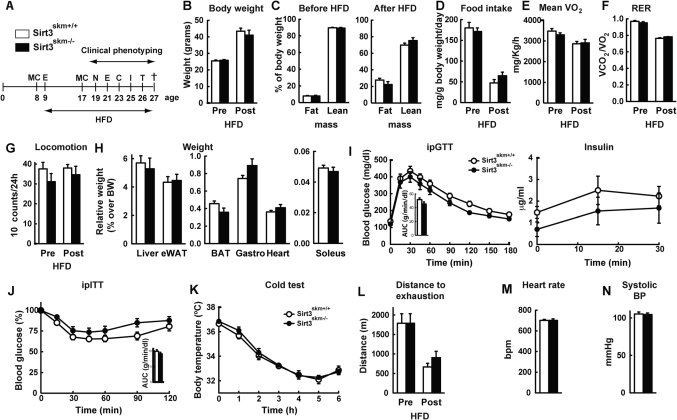
Figure 3. Metabolic phenotyping of Sirt3skm−/− mice.
(A) Experimental schedule of the clinical phenotyping protocol. For abbreviations see legend of Fig. 2A; E: endurance exercise on a treadmill; N: non-invasive blood pressure measurement (NIBP); C: cold test. (B) Body weight and (C) body composition of Sirt3skm+/+ and Sirt3skm−/− mice before and after 8 weeks of HFD. (D–G) Daily food intake (D), mean O2 consumption (VO2; E), mean respiratory exchange ratio (RER; F) and total ambulatory locomotor activity (G) as measured over a 24 h period before and after HFD. (H) Relative weight of tissues normalized by body weight in Sirt3skm+/+ and Sirt3skm−/− mice. (I) Intra-peritoneal glucose tolerance test (ipGTT, left panel) and plasma insulin during ipGTT (right panel) in mice fed a HFD for 16 weeks. (J) Intra-peritoneal insulin tolerance test (ipITT) in Sirt3skm+/+ and Sirt3skm−/− mice fed a HFD for 17 weeks. (I and J) The bar graph (inner panels) represents the average area under the curve (AUC). (K) Sirt3skm+/+ and Sirt3skm−/− mice fasted for 24h were exposed to 4°C for 6 h and rectal temperature was monitored. (L) Endurance exercise test on Sirt3skm+/+ and Sirt3skm−/− mice before and after 12 weeks of HFD. (M) Heart rate and (N) systolic blood pressure were measured in Sirt3skm+/+ and Sirt3skm−/− mice after 10 weeks on HFD. Results are presented as mean values (n = 8−10 mice for each group) ± SEM.
To further evaluate possible phenotypic differences between the Sirt3skm−/− and Sirt3skm+/+ mice, we exposed the mice to a cold challenge (4°C). Mice from both genotypes similarly maintained their core body temperature for at least 6 hours after a 24 h fast (Fig. 3K), or after 14 weeks of HFD (Supl. Fig. 1A), indicating that muscle Sirt3 is not essential for cold-induced adaptive thermogenesis. Exercise increases Sirt3 expression in muscle4,5, suggesting a role for Sirt3 in exercise physiology. Sirt3skm−/− and Sirt3skm+/+ mice, however, ran a similar distance until exhaustion both before and after HFD (Fig. 3L). Also no difference in endurance running was present before and after a 24 h fast (Supp. Fig. 1B). In addition, both the heart rate (Fig. 3M) and systolic blood pressure (Fig. 3N) were similar between Sirt3skm−/− and Sirt3skm+/+ mice, indicating that the partial reduction in Sirt3 mRNA levels found in the Sirt3skm−/− hearts has no major impact on cardiac function.
Altogether, these data indicate that Sirt3skm−/− mice are metabolically indistinguishable from control mice under both chow and HFD. Thus, the absence of Sirt3 in the skeletal muscle by itself cannot explain the metabolic phenotype observed in the germline deficient Sirt3-KO animals.
Mitochondrial protein hyperacetylation but normal oxidative stress in Sirt3-deficient tissues
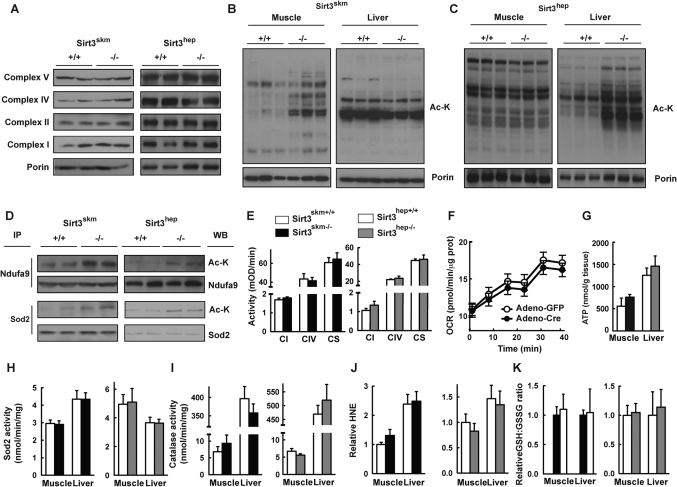
The fact that Sirt3 is a mitochondrial protein prompted us to investigate the integrity of the mitochondria in both tissue-specific Sirt3−/− mouse models after HFD treatment, a stress after which we expect the effects of Sirt3 deficiency to be accentuated. The protein levels of subunits from the mitochondrial respiratory complexes I, II, IV and V were indistinguishable in Sirt3skm−/− and Sirt3skm+/+ muscle, and in Sirt3hep−/− and Sirt3hep+/+ liver (Fig. 4A). In line with the observations in germline Sirt3−/− mice2, there was a marked increase in global mitochondrial protein acetylation in the tissues where the Sirt3 gene had been deleted (Fig. 4B and 4C). We next evaluated the acetylation levels of specific Sirt3 targets. Sirt3 interacts with and deacetylates Ndufa9, a subunit of complex I13. Consistently, we observed increased acetylation of some members of the Ndufa9-containing complex I in livers from the Sirt3hep−/− mice or quadriceps muscle from the Sirt3skm−/− mice, as compared with their respective wild type controls (Fig. 4D, top panels). However, this increased acetylation did not lead to any change in maximal complex I or complex IV activities (Fig. 4E). As an internal control, we measured the activity of citrate synthase, a mitochondrial enzyme from the Krebs cycle not belonging to the electron transport chain, whose activity was also unchanged in the absence of Sirt3 in both mouse models (Fig. 4E). To further examine mitochondrial function in the absence of Sirt3 we measured oxygen consumption in Sirt3L2/L2 primary hepatocytes-with the Sirt3 gene flanked with LoxP sites- in which we acutely deleted the Sirt3 gene by an adenoviral infection with Cre. Oxygen consumption rate (OCR) was not affected by the absence of Sirt3 (Fig. 4F and Suppl. Fig. 2A).
Figure 4. Hyperacetylation of mitochondrial proteins but normal ROS markers in Sirt3hep−/− and Sirt3skm−/− mice subjected to HFD.
(A) Protein expression levels of complex I, II, IV and V in isolated mitochondria of Sirt3skm+/+ and Sirt3skm−/− muscle (left panels) and Sirt3hep+/+ and Sirt3hep−/− livers (right panels). (B, C) Acetylation status of mitochondrial proteins extracted from muscle and liver of Sirt3skm+/+ and Sirt3skm−/− mice (B) and Sirt3hep+/+ and Sirt3hep−/− mice (C). Mitochondrial protein lysates were subjected to western blotting using an anti-acetylated lysine antibody (Ac-K) and porin as a loading control. (D) Acetylation level of complex I members (top two rows of panels) and Sod2 (bottom two rows of panels) in Sirt3skm+/+ and Sirt3skm−/− muscle (left panels) or Sirt3hep+/+ and Sirt3hep−/− livers (right panels). Endogenous Ndufa9 or Sod2 were immunopurified from muscle lysates and immunoblotted using Ndufa9/Sod2 and Ac-K antibodies. (E) Complex I (CI), complex IV (CIV) and Citrate synthase (CS) activities in Sirt3skm+/+ and Sirt3skm−/− muscle (left panel) or Sirt3hep+/+ and Sirt3hep−/− (right panel). (F) Oxygen consumption rate (OCR) from primary hepatocytes obtained from Sirt3L2/L2 mice infected with the indicated viruses. (G) ATP levels were measured from the indicated tissue lysates. (H–K) Activities of mitochondrial Sod2 (H) and catalase (I) and levels of 4-hydroxy-2-nonenal (HNE; J) and GSH/GSSG ratio (K) in muscle and liver lysates of Sirt3skm+/+ and Sirt3skm−/− mice (left panels) or Sirt3hep+/+ and Sirt3hep−/− mice (right panels). Measurements in panels E–K were determined using samples from at least four mice per genotype. All the samples used for these studies come from mice subjected to the HFD treatment, sacrificed in fed state. Data are represented as mean ± SEM.
Sirt3 deficiency has also been correlated with low levels of cellular ATP13. We therefore measured ATP levels in Sirt3skm+/+ and Sirt3skm−/− gastrocnemius muscle and in Sirt3hep+/+ and Sirt3hep−/− liver, but could not detect differences between tissues from the different genotypes (Fig. 4G). Accordingly, we did not detect differences in the phosphorylation status of the AMP/ATP sensing AMP-activated protein kinase (AMPK), suggesting that Sirt3 deletion in a tissue-specific fashion does not affect intracellular energy charge (Suppl. Fig. 2B).
Sirt3 has been shown to reduce oxidative stress, mostly through deacetylating and activating the mitochondrial ROS scavenger Sod29,10,12,16. We therefore also determined Sod2 acetylation status in the absence of Sirt3; as reported, Sirt3-deficient tissues presented increased levels of acetylated Sod2 (Fig. 4D, lower panels). In contrast, Sirt3 gene ablation did not affect the activity of Sod2 (Fig. 4H), nor that of catalase, another key ROS scavenger (Fig. 4I). We next measured a few markers of oxidative stress, such as the level of 4-hydroxy-2-nonenal (HNE; a marker for lipid peroxidation) and glutathione, in liver and muscle from our mutant mouse models after HFD treatment. No difference in these parameters was observed between Sirt3-deficient tissues and their respective controls (Fig. 4J-K). These results show that tissue-specific ablation of Sirt3 results in increased acetylation of ROS scavengers, but that this alteration is not sufficient to affect their activity, which may help to explain the lack of a metabolic phenotype in these two tissue-specific Sirt3 deficient mouse models.
Discussion
We report here the generation and phenotypic characterization of the first set of tissue-specific Sirt3 knockout mice (muscle-specific Sirt3skm−/− and liver-specific Sirt3hep−/− mice). Consistent with the role of Sirt3 as the major mitochondrial deacetylase2, mitochondrial proteins were robustly hyperacetylated in Sirt3-deficient tissues. In line with this observation, specific Sirt3 targets, such as mitochondrial complex I and the mitochondrial ROS scavenger, Sod2, were also markedly hyperacetylated in Sirt3-deficient tissues (Fig. 4D), as previously described in livers from the germline Sirt3−/− mice9,12,13. However, the activities of these enzymes (Fig. 4H–I) were not changed in Sirt3-deficient tissues, which indicates that hyperacetylation of these proteins does not necessarily result in a change in their maximal activity. Indeed, while many residues might be acetylated in a single protein, only the acetylation of a few key residues crucially determines the enzymatic activity/function of the protein, as demonstrated with LCAD6. This phenomenon might explain the lack of a metabolic phenotype in the tissue-specific Sirt3-deficient mice.
Several in vivo and in vitro studies demonstrated a role for Sirt3 in the suppression of oxidative stress3,8,9,10,11,12,16. However, different groups have reported normal ROS levels in basal state in liver, brain or cochlea9,10. In line with these last reports, we observed normal levels of surrogate markers for ROS in Sirt3-deficient tissues, as well as similar levels of activity of the mitochondrial ROS scavengers Sod2 and catalase (Fig. 4 H–I). Again, these results might explain the lack of a clear metabolic phenotype in the conditions we tested.
Despite the higher mitochondrial protein hyperacetylation, the tissue-specific Sirt3−/− mice described here were metabolically similar to control littermates in all the conditions that we tested (chow and HFD diet, 24 h fasting, exercise or cold). Our results therefore imply that the previously described phenotypes of germline Sirt3−/− animals under a short fasting or HFD6,9,10,11,13,14,15,19 are not due to the absence of Sirt3 specifically in liver or muscle. Rather, our data indicate that either Sirt3 deficiency in another tissue or the coordinated defect of Sirt3 in multiple tissues accounts for these effects in the germline Sirt3 knock-out mice. In this regard, our results do also not rule out the possibility that latent defects in muscle or liver function in our models can be fully compensated by another tissue or sirtuin function. In this sense, further tissue-specific Sirt3 deficient mouse models exploring the brain, white or brown adipose, or macrophage function of Sirt3 will be key to fully evaluate the etiology of the metabolic alterations in the germline Sirt3 knock-out mice. In this context it is interesting to note that no change in Sirt3 mRNA or protein expression in non-targeted tissues (Fig. 1B, 1C, 1D and 1E), nor any change in the mRNA levels of the other mitochondrial sirtuins (Sirt4 and Sirt5) in the Sirt3-targeted tissues (Fig. 1F and 1G) was present, which suggest that there is no compensation by other mitochondrial sirtuins. These observations, however, do not rule out a change in the activity, rather than in mRNA or protein levels, of these sirtuins as the possible mechanism of compensation.
Of note is also that Sirt3skm−/− mice, when subjected to a 24 h fast and, subsequently, to a cold test, do not suffer hypothermia, as previously reported for the whole-body Sirt3-KO mice6. Since the two main tissues responsible for temperature homeostasis are muscle and brown fat, and Sirt3 is highly expressed in this last tissue, it is tempting to speculate that Sirt3 in brown fat, but not in skeletal muscle, plays an important role in temperature homeostasis, most probably via regulation of mitochondrial uncoupling. Brown fat is also an important tissue in fat and glucose clearance, and increased brown fat amounts or activity can significantly improve global metabolism20,21. Thus, one possibility is that the brown fat tissue in Sirt3hep−/− and Sirt3skm−/− mice could compensate for the tissue-specific ablation of Sirt3 in liver or muscle.
Finally, when comparing studies from different laboratories, it is important to bear in mind that the genetic background and/or the exact composition of the diets are key determinants of phenotypic outcome, which is the result of gene-environment interactions22,23. In this regard, our mice were on an obesity-prone background (C57BL/6)23,24,25 and fed with a standard HFD, with 60% calories coming from fat, both conditions being different from those used in other reports14. Furthermore, differences in the developmental onset of the Sirt3 gene deletions need to be considered, as germline Sirt3-KO mouse model lacks Sirt3 from the beginning of embryonic development, while the tissue-specific promoters used in our work are activated only from a certain developmental stage.
In summary, our work confirms the critical impact of Sirt3 on global mitochondrial acetylation. It is unclear why these alterations do not have a major physiological impact when occurring in a tissue-specific manner in muscle or liver, as in our study. Further investigations to understand this conundrum are therefore warranted.
Methods
Generation and clinical phenotyping of the Sirt3hep−/− and Sirt3skm−/− mice
Sirt3 floxed (Sirt3L2/L2) mice were generated using standard gene targeting procedures26. These animals were crossed with liver-specific Cre-expressing mice (Alb-Cre)18 to obtain the Sirt3hep−/− mouse line; or with the skeletal muscle-specific Cre-expressing line (HSA-Cre)17 to obtain the Sirt3skm−/− line. These two mouse lines were backcrossed onto a pure C57BL/6J background. All animal work in the manuscript has been performed according to the validated standard operating procedures (SOPs)23, as defined and validated by the Eumorphia program (see: http://empress.har.mrc.ac.uk/). Briefly, we subjected our mice to non-invasive monitoring of body fat and lean mass by EchoMRI; energy expenditure analysis by indirect calorimetry (CLAMS system); intraperitoneal glucose tolerance test (ipGTT); insulin tolerance test (ipITT); blood sampling before and after a 24 fast; endurance exercise on a treadmill; non-invasive blood pressure measurement; and cold test. A summary of these methods is given in the supplementary materials and methods section. Animal experiments were approved by the ethic veterinary committee of the canton of Vaud - Switzerland (Permit IDs 2307 and 2307-1).
Quantitative RT-PCR
qRT-PCR was performed as already described27,28. Briefly, RNA was extracted from tissues using TRIzol reagent (Invitrogen). Complementary DNA was generated using QuantiTect Rev (Qiagen). The real-time PCR measurement of individual cDNAs was performed using SYBR green dye to measure duplex DNA formation with the LightCycler System (Roche Diagnostics). Primer sequences are listed in the supplementary materials and methods section.
Complex I, complex IV and citrate synthase activity assays
Complex I, complex IV and citrate synthase enzymatic activities were measured in homogenates of gastrocnemius muscle of Sirt3skm+/+ and Sirt3skm−/− or of livers of Sirt3hep+/+ and Sirt3hep−/− mice using the respective enzyme activity assay kits (Mitosciences), as reported elsewhere29.
Mitochondrial isolation, immunoprecipitation and western blot analysis
Mitochondria were isolated from tissues following a previously described protocol30. Isolated mitochondria were lysed in RIPA lysis buffer containing protease inhibitors and supplemented with 5 mM NAM and 1 mM sodium butyrate. Immunoprecipitations were performed as described previously28. Protein extracts were separated by SDS-PAGE. α-Sirt3 (home-made rabbit polyclonal against the peptides CDLMQRERGKLDGQDR and STPSGIPDFRSPGSGL), α-Total OxPhos (MitoSciences, MS604), α-Ndufa9 (Abcam, ab14713), α-AcK (Cell Signaling, 9441), α-Sod2 (Santa Cruz, sc-133254 for IP, sc-3008 for WB), and α-porin (Calbiochem, 529536) antibodies were used to detect the respective proteins.
Analysis of blood metabolites
For β-hydroxybutyrate (BHBA) analysis, plasma samples were precipitated with 1 M perchloric acid on ice. After centrifugation the supernatant was neutralized using 2M KOH, 0.5 M 2-(N- morpholino)ethanesulfonic acid (MES). BHBA concentrations were measured using LC-MS/MS in the neutralized supernatant after removal of KClO4. Amino acids and acylcarnitines were analyzed in plasma according to established LC-MS/MS methods31,32.
Evaluation of oxidative stress
Sod2 and catalase enzyme activity; HNE; and the GSH/GSSG ratio were measured in liver or gastrocnemius muscle homogenates using a Sod2 and catalase enzyme activity assays (Cayman Chemicals); the OxiSelect HNE-His Adduct ELISA Kit (Cell Biolabs Inc.); or the Bioxytech GSH/GSSG-412 kit (OxisResearch TM), respectively, following the manufacturer's protocols.
ATP measurements
ATP was measured in samples from liver or skeletal muscle using the ATP determination kit (Molecular Probes, Invitrogen), following previous reports33 and the manufacturer's instructions. Briefly, tissues were weighed and lysed in 600 μl of 6% (v/v) perchloric acid, and centrifuged at 4000xg for 10 minutes at 4°C. 500 μl of the supernatant were neutralized with 200 μl of 2.5 M KOH, and precipitate was removed by centrifugation at 4000 xg for 5 minutes at 4°C. 10 μl of a 1:10 dilution of the samples in 0.1 M Tris-HCl – 2 mM EDTA buffer (TE) were placed in an appropriate bioluminescence plate, and luciferase activity was measured after addition of 100μl of the reaction solution, with a lag time of 1 second and an integration time of 3 seconds using a Glomax luminometer (Promega).
Preparation and culture of primary mouse hepatocytes and determination of oxygen consumption rates
Liver cells were prepared by the two-step collagenase method34 from post-absorptive male mice (25–30 g) after anaesthesia with ketamin/xylazin (1 mg/100 g body weight). Cell viability (>80%) was checked by trypan blue exclusion. Primary culture of hepatocytes was seeded at 1.5×104 per well on rat-ail collagen type 1 (BDpharmingen) either on coated XF-96 cell culture plates (Seahorse) or on 12-well plates in serum-free DMEM (4.5 g/l glucose) medium. Four hours after seeding, the medium was replaced by a glucose and serum free DMEM medium, supplemented with lactate 10 mM and pyruvate 1 mM and the cells were infected with either an adenovirus containing a GFP construct (Adeno-GFP) or an adenovirus containing a recombinase Cre (Adeno-Cre) construct at a multiplicity of infection (MOI) of 15. Cellular oxygen consumption rate (OCR) was measured by Seahorse XF96 forty-eight hours after infection of cells as previously reported35.
Statistical analyses
Statistical analyses were performed with non-parametric Student's t-test. Data are expressed as mean ± SEM and p values smaller than 0.05 were considered as statistically significant. Statistical significance is displayed as * (p < 0.05) or ** (p < 0.01).
Author Contributions
PJF-M and EJ (+) contributed equally to this work. PJF-M and EJ directed and performed all the experiments. CC participated in the immunoprecipitation experiments. PA participated in the mitochondrial complex activities assays. TH and NM performed the primary hepatocyte experiments. CC, EP and HY participated in the metabolic phenotyping of the mouse models. VB and SH performed the blood metabolite analysis. KS and JA directed all the work. PJF-M, CC, KS and JA wrote the manuscript. All authors reviewed the manuscript.
Supplementary Material
Supplementary Figures and Materials&Methods
Acknowledgments
This study was supported by grants of the Ecole Polytechnique Fédérale de Lausanne, the Swiss National Science Foundation (31003A-124713), and the European Research Council Ideas program (Sirtuins; ERC-2008-AdG23118). JA is the Nestle Chair in Energy Metabolism (NCEM). PJFM benefited from a FEBS fellowship. The authors thank all the members of the Auwerx lab for inspiring discussions, and Dr. Manuel Serrano (CNIO, Spain) for his help and support.
Footnotes
None of the authors have competing financial interests.
References
- Schwer B., North B. J., Frye R. A., Ott M. & Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol 158, 647–657 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard D. B. et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol 27, 8807–8814 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi T., Wang F., Stieren E. & Tong Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J Biol Chem 280, 13560–13567 (2005). [DOI] [PubMed] [Google Scholar]
- Hokari F. et al. Muscle contractile activity regulates Sirt3 protein expression in rat skeletal muscles. J Appl Physiol 109, 332–340 (2010). [DOI] [PubMed] [Google Scholar]
- Palacios O. M. et al. Diet and exercise signals regulate SIRT3 and activate AMPK and PGC-1alpha in skeletal muscle. Aging (Albany NY) 1, 771–783 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschey M. D. et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 464, 121–125 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing E. et al. Sirtuin-3 (Sirt3) regulates skeletal muscle metabolism and insulin signaling via altered mitochondrial oxidation and reactive oxygen species production. Proceedings of the National Academy of Sciences of the United States of America 108, 14608–14613 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. S. et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell 17, 41–52 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X., Brown K., Hirschey M. D., Verdin E. & Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab 12, 662–667 (2010). [DOI] [PubMed] [Google Scholar]
- Someya S. et al. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell 143, 802–812 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan N. R. et al. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J Clin Invest 119, 2758–2771 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao R. et al. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol Cell 40, 893–904 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn B. H. et al. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad Sci U S A 105, 14447–14452 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschey M. D. et al. SIRT3 Deficiency and Mitochondrial Protein Hyperacetylation Accelerate the Development of the Metabolic Syndrome. Mol Cell (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallows W. C. et al. Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol Cell 41, 139–149 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X. et al. Sirtuin 3, a new target of PGC-1alpha, plays an important role in the suppression of ROS and mitochondrial biogenesis. PLoS One 5, e11707 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miniou P. et al. Gene targeting restricted to mouse striated muscle lineage. Nucleic Acids Res 27, e27 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postic C. et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. The Journal of biological chemistry 274, 305–315 (1999). [DOI] [PubMed] [Google Scholar]
- Shimazu T. et al. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab 12, 654–661 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P. et al. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. The Journal of clinical investigation 121, 96–105 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Molina A. et al. Pten positively regulates brown adipose function, energy expenditure, and longevity. Cell metabolism 15, 382–394 (2012). [DOI] [PubMed] [Google Scholar]
- Champy M. F. et al. Mouse functional genomics requires standardization of mouse handling and housing conditions. Mammalian genome : official journal of the International Mammalian Genome Society 15, 768–783 (2004). [DOI] [PubMed] [Google Scholar]
- Champy M. F. et al. Genetic background determines metabolic phenotypes in the mouse. Mamm Genome 19, 318–331 (2008). [DOI] [PubMed] [Google Scholar]
- Donovan M. J., Paulino G. & Raybould H. E. Activation of hindbrain neurons in response to gastrointestinal lipid is attenuated by high fat, high energy diets in mice prone to diet-induced obesity. Brain Res 1248, 136–140 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- West D. B., Boozer C. N., Moody D. L. & Atkinson R. L. Dietary obesity in nine inbred mouse strains. Am J Physiol 262, R1025–1032 (1992). [DOI] [PubMed] [Google Scholar]
- Argmann C. A., Chambon P. & Auwerx J. Mouse phenogenomics: the fast track to "systems metabolism". Cell metabolism 2, 349–360 (2005). [DOI] [PubMed] [Google Scholar]
- Yamamoto H. et al. NCoR1 is a conserved physiological modulator of muscle mass and oxidative function. Cell 147, 827–839 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C. et al. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458, 1056–1060 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper R. H. et al. The metabolic footprint of aging in mice. Sci Rep 1, 134 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frezza C., Cipolat S. & Scorrano L. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat Protoc 2, 287–295 (2007). [DOI] [PubMed] [Google Scholar]
- Koutnikova H. et al. Identification of the UBP1 locus as a critical blood pressure determinant using a combination of mouse and human genetics. PLoS Genet 5, e1000591 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piraud M. et al. ESI-MS/MS analysis of underivatised amino acids: a new tool for the diagnosis of inherited disorders of amino acid metabolism. Fragmentation study of 79 molecules of biological interest in positive and negative ionisation mode. Rapid Commun Mass Spectrom 17, 1297–1311 (2003). [DOI] [PubMed] [Google Scholar]
- Khan H. A. Bioluminometric assay of ATP in mouse brain: Determinant factors for enhanced test sensitivity. J Biosci 28, 379–382 (2003). [DOI] [PubMed] [Google Scholar]
- Berry M. N. & Friend D. S. High-yield preparation of isolated rat liver parenchymal cells: a biochemical and fine structural study. The Journal of cell biology 43, 506–520 (1969). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M. et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature 439, 484–489 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures and Materials&Methods