Abstract
Introduction
Characterization of novel proteins in maternal serum derived from mothers carrying Down syndrome (DS) fetuses.
Material and methods
Based on last comparative proteomic analysis, five significant differences of expressed proteins in serum from four groups have been confirmed by ELISA. DAVID and GeneGo MetaCore were used to bioinformatically analyze candidate protein markers.
Results
The serum levels of ceruloplasmin (CP) and complement factor B (CFB) were significantly increased in mother carried DS fetuses (346.5 ng/ml and 466.8 ng/ml vs. 248.6 ng/ml and 293.5 ng/ml, p< 0.05). Twenty-nine proteins were mainly categorized into binding, catalytic activity and enzyme regulator activity proteins, and their biological roles were involved in biological regulation, metabolic processes, cellular processes, and response to stimuli. The immune response alternative complement pathway was the most significant GeneGo Pathway related to DS.
Conclusions
These 29 proteins have relations with the development of Down syndrome, especially CP and CFB play more important roles.
Keywords: Down syndrome, proteomic, serum, bioinformatics, biomarkers, prenatal diagnosis
Introduction
It is well known that prenatal screening and diagnosis for Down syndrome (DS) are important for prepotency. They have been widely used in pregnant women all around the world in the past 25 years. Nowadays, the main methods of prenatal screening are screening in the second trimester [1], screening in the first trimester [2] and integrated screening [3]. The biomarkers associated with DS which have been reported previously include α-fetoprotein (AFP), unconjugated estriol (uE3), free β subunit of human chorion gonadotrophin (fβhCG), pregnancy-associated plasma protein A (PAPP-A), disintegrin and metalloprotease 12 (ADAM-12), superoxide dismutase 1 (SOD1), and so on [4]. However, all of these markers still could not be sufficient, because they only achieved a detection rate of 50–85% at a 5% false-positive rate [5–8]. Not only will many pregnant women carrying DS fetuses miss diagnosis, but also some will be offered invasive and risky diagnostic procedures. Therefore, prenatal medical experts are increasingly focusing on discovery of new biomarkers which can improve the efficiency of DS screening.
In recent years, proteomic studies have proved to be a powerful platform for biomarker discovery, and the identified new biomarkers will revolutionize diagnostics for many diseases [9–13]. Nagalla et al. [14] attempted to identify potential serum biomarkers to detect DS by a comprehensive proteomic analysis firstly. However, similar studies based on maternal blood have been limited [14–17]. The protein biomarkers reported by these studies were not always reproducible at the same time. Hence, it is important to improve the research on application of proteomics in DS.
In our previous study, we used 2-DE and MS to identify the different proteins for DS in maternal serum, and successfully identified 29 protein biomarkers [18]. Moreover, we also found that TF, DES, SERPINA1, CP, APCS, CFB, C4A, CPN1, CFHR1 and PLG were noteworthy for further study, because they showed the most significant changes. Although some reports have indentified some proteins in maternal serum which were considered as new DS markers, these proteins have not been analyzed by bioinformatics methods. Thus we used bioinformatics analysis to determine whether these 29 proteins are related to the aetiology and function of DS.
This work describes the biochemical characteristics of novel proteins in maternal serum from mothers carrying DS fetuses, and explores their complicated interrelations. By ELISA, we aimed to detect the serum concentration of 5 proteins which showed the most significant differential expression, and carry out bioinformatics analysis using DAVID and MetaCore.
Material and methods
The study design and protocol were reviewed and approved by the ethics committee of Changzhou Woman and Children Health Hospital affiliated to Nanjing Medical University.
Samples collection
The maternal serum was collected from 11 pregnant women carrying a DS fetus and 10 controls with a normal fetus. They all entered the system of prenatal screening in Changzhou Women and Children Health Hospital from April 2008 to June 2010. The mothers with DS were prenatally diagnosed by the karyotype analysis of fetal cells from AF obtained by amniocentesis between gestational weeks 16 and 21. Ten women with normal fetuses were selected as the control group. The match conditions of cases and the control group were: 1) age < 1 year, 2) gestational weeks < 7 days.
Eleven DS patients (2 days – 3 months) and 10 control subjects (3 days – 1 month) underwent routine clinical and laboratory evaluation. All DS patients examined were confirmed to possess the chromosome abnormality (trisomy 21). Control subjects had no previous history of neurological deficits or any serious disorder.
Three millimetres blood samples of all the cases were collected by simple needle aspiration. The samples were centrifuged at 3000 rpm for 5 min to remove cells. The serum was stored at –86°C until further analysis.
Methods
2DE-MS and protein identification
Firstly, we depleted high-abundance proteins in serum, including albumin, IgG, and so on. Then, 2-DE and MS were used to identify the proteins for DS in maternal serum. Twenty-nine proteins were identified successfully in the end. The methods of 2-DE, MS and protein identification were performed as in our report [18].
ELISA
Among the 29 protein biomarkers, we selected 5 proteins to detect their serum concentration. They were Ceruloplasmin (CP, P00450), Complement factor H-related protein 1 (CFHR1, Q03591), Complement factor B (CFB, P00751), Desmin (DES, P17661) and Plasminogen (PLG, P00747).
Enzyme-linked immunosorbent assays for these 5 proteins were based on the non-competitive sandwich ELISA method using commercially available kits purchased from R&D, USA. The main protocol was performed as described previously [19].
Bioinformatics analysis
DAVID (http://david.abcc.ncifcrf.gov/, version: 6.7) [20, 21] was used to process the bioinformatics analysis of these candidate protein markers, including protein classification (based on Biological Process Ontology and Molecular Function Ontology, respectively), enrichment analysis for significant gene ontology categories, KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway mapping and significant pathway computing, and so on. GeneGo MetaCore (http://www.genego.com, version: 6.5) [22] was used to analyze the pathways of these proteins and related genes. At the same time, we generated biological association networks with Meta-Core.
Statistical analysis
All data were collected and statistically analyzed using SPSS 13.0 software. Results of parameters were expressed as mean and SD. We compared differences in serum level of proteins by means of the t statistic. A value of p less than 0.05 was considered to be statistically significant.
Results
Differential expression of five proteins
A total of 29 proteins were identified successfully in maternal serum coming from DS cases compared with the control group, including 14 proteins that were up-regulated, while 15 proteins were decreased (Table I). More results of the 29 proteins were descried in our last study [18]. In the present study, we selected 5 proteins for further analysis, including CP, CFHR1, CFB, DES and PLG. Their entry name, protein name, molecular weight (MW), PI, score, coverage, expect and the fold change of expression density are shown in Table II.
Table I.
Twenty-nine proteins differentially expressed in serum of mothers with DS fetuses
| Gene names | Entry name | Accession | Protein names |
|---|---|---|---|
| A1BG | A1BG_HUMAN | P04217 | α1B-glycoprotein |
| AFM | AFAM_HUMAN | P43652 | Afamin |
| AMBP | AMBP_HUMAN | P02760 | Protein AMBP |
| APCS | SAMP_HUMAN | P02743 | Serum amyloid P-component |
| APOA1 | APOA1_HUMAN | P02647 | Apolipoprotein A-I |
| APOH | APOH_HUMAN | P02749 | β2-glycoprotein 1 |
| C4A | CO4A_HUMAN | P0C0L4 | Complement C4-A |
| CFB | CFAB_HUMAN | P00751 | Complement factor B |
| CFH | CFAH_HUMAN | P08603 | Complement factor H |
| CFHR1 | FHR1_HUMAN | Q03591 | Complement factor H-related protein 1 |
| CFHR2 | FHR2_HUMAN | P36980 | Complement factor H-related protein 2 |
| CFI | Q8WW88_HUMAN | Q8WW88 | CFI protein |
| CLU | CLUS_HUMAN | P10909 | Clusterin |
| CP | CERU_HUMAN | P00450 | Ceruloplasmin |
| CPN1 | CBPN_HUMAN | P15169 | Carboxypeptidase N catalytic chain |
| DES | DESM_HUMAN | P17661 | Desmin |
| GC | VTDB_HUMAN | P02774 | Vitamin D-binding protein |
| GRIN1 | GRIN1_HUMAN | Q7Z2K8 | G protein-regulated inducer of neurite outgrowth 1 |
| HPX | HEMO_HUMAN | P02790 | Hemopexin |
| KLC2 | KLC2_HUMAN | Q9H0B6 | Kinesin light chain 2 |
| LRG1 | A2GL_HUMAN | P02750 | Leucine-rich α2-glycoprotein |
| MASP2 | MASP2_HUMAN | O00187 | Mannan-binding lectin serine protease 2 |
| PEPD | PEPD_HUMAN | P12955 | Xaa-Pro dipeptidase |
| PLG | PLMN_HUMAN | P00747 | Plasminogen |
| RIMS3 | RIMS3_HUMAN | Q9UJD0 | Regulating synaptic membrane exocytosis protein 3 |
| SERPINA1 | A1AT_HUMAN | P01009 | α1-antitrypsin |
| TF | TRFE_HUMAN | P02787 | Serotransferrin |
| VTN | VTNC_HUMAN | P04004 | Vitronectin |
| ZNF485 | ZN485_HUMAN | Q8NCK3 | Zinc finger protein 485 |
Table II.
Five proteins differentially expressed in serum of mothers with DS fetuses
| Entry name | MW | PI | Score | Coverage | Fold change | Expect |
|---|---|---|---|---|---|---|
| CFAB_HUMAN | 86847 | 6.67 | 264 | 43% | 17.5/0 | 2.70E-22 |
| FHR1_HUMAN | 38766 | 7.38 | 82 | 31% | 1.0/0 | 0.00045 |
| CERU_HUMAN | 122983 | 5.44 | 119 | 21% | 5.43 | 8.60E-08 |
| DESM_HUMAN | 32382 | 4.97 | 78 | 39% | 7.0 | 0.001 |
| PLMN_HUMAN | 15776 | 8.73 | 92 | 44% | 0.7/0 | 3.90E-05 |
Serum concentration
In order to verify the results of 5 proteins as identified by MALDI-TOF-TOF/MS, we detected the serum concentrations of them by ELISA. Table III shows the ELISA results of 5 proteins in the four groups.
Table III.
Serum concentrations of proteins by ELISA
| n | CP [ng/ml] | DES [nmol/l] | PLG [ng/ml] | CFB [ng/ml] | CFHR1 [ng/ml] | |
|---|---|---|---|---|---|---|
| Mothers with DS fetuses | 11 | 346.5 ±111.81 | 21.1 ±13.5 | 2.1 ±1.3 | 466.8 ±216.41 | 109.0 ±35.5 |
| Mothers with normal fetuses | 10 | 248.6 ±78.3 | 20.1 ±11.7 | 1.7 ±1.2 | 293.5 ±75.0 | 88.6 ±51.9 |
| t | 2.301 | 0.191 | 0.786 | 2.400 | 1.058 | |
| P | 0.033 | 0.851 | 0.442 | 0.027 | 0.303 | |
| DS patients | 11 | 166.1 ±55.02 | 6.5 ±2.31 | 0.9 ±0.4 | 174.3 ±55.02 | 44.6 ±17.41 |
| Normal babies | 10 | 244.0 ±36.0 | 12.3 ±7.8 | 0.9 ±0.4 | 311.8 ±102.4 | 67.4 ±15.2 |
| t | –3.873 | –2.273 | –0.608 | –3.776 | –3.218 | |
| P | 0.001 | 0.035 | 0.550 | 0.001 | 0.005 |
compared with control group p < 0.05
compared with control group p < 0.001
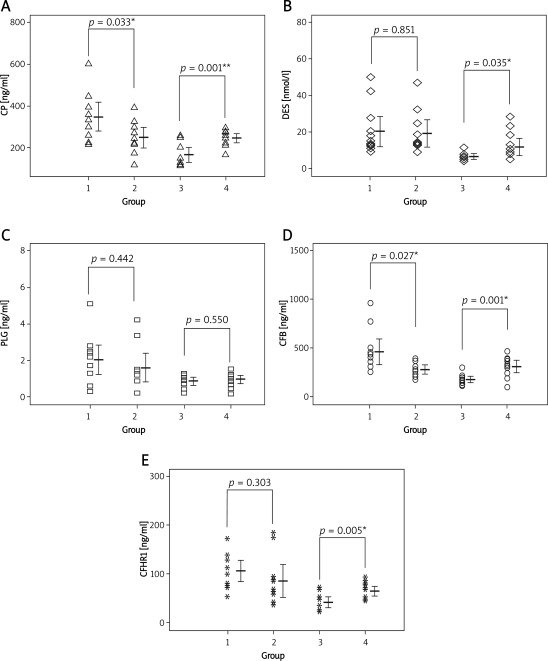
Compared with women with normal fetuses, the serum levels of CP and CFB were significantly increased in mothers carrying DS fetuses (p < 0.05). The mean concentrations were 346.5 ng/ml and 466.8 ng/ml respectively, vs. 248.6 ng/ml and 293.5 ng/ml in the control group, respectively (Figures 1 A, D). There were no significant differences in the amount of CFHR1, DES and PLG between the two groups (p > 0.05) (Figures 1 B, C, E). However, the levels of CP, CFB, DES and CFHR1 were decreased in DS patients. There were significant difference between DS patients and normal babies (p < 0.05). Especially, CP and CFB were significantly reduced (p < 0.001). The level of PLG still had no significant changes (p > 0.05).
Figure 1 A-E.
The levels of five proteins in four groups *p < 0.05, **p < 0.01, group 1 as women with DS fetus, group 2 as women with normal fetus, group 3 as DS patients, group 4 as normal babies
Gene ontology analysis
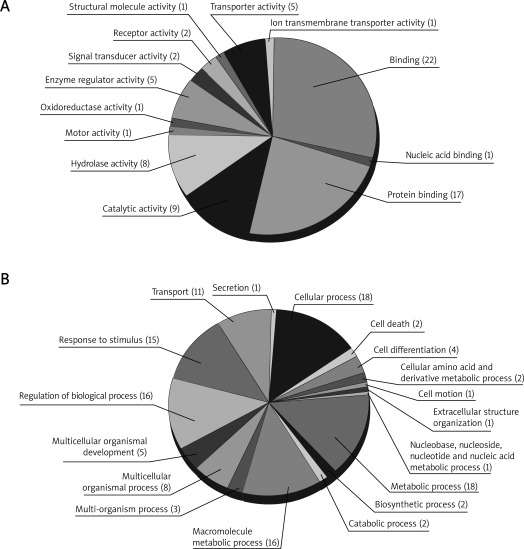
By DAVID, 29 proteins were mapped to at least one annotation term within the GO molecular function category, including 40 (53.3%) binding proteins, 9 proteins (12.9%) with catalytic activity, 8 proteins (10.7%) with hydrolase activity, 5 proteins (6.7%) with enzyme regulator activity, 5 proteins (6.7%) with transporter activity, and 9 others (12.0%), including proteins with signal transducer activity, receptor activity, motor activity, oxidoreductase activity, structural molecule activity, and ion transmembrane transporter activity, as shown in Figure 2 A. On the other hand, they were also mapped within the GO biological process category: biological regulation (18, 13.5%), metabolic process (18, 13.5%), cellular process (18, 13.5%), response to stimulus (15, 11.3%), establishment of localization (11, 8.3%), localization (11, 8.3%), immune system process (8, 6.0%), multicellular organismal process (8, 6.0%), developmental process (7, 5.3%), cellular component organization (6, 4.5%) and other proteins (13, 9.8%) related to the categories of multi-organism process, biological adhesion, death, cellular component biogenesis, locomotion, reproductive process, reproduction, and growth, as shown in Figure 2 B.
Figure 2.
Classification of identified proteins based on GOA: A– Molecular Function Ontology, B– Biological Process Ontology
KEGG pathway qnalysis
Relating the gene symbol of these proteins to the KEGG GENE ID, a total of 12 proteins corresponded to 15 pathways of KEGG, of which the pathway of Complement and coagulation cascades was the most significantly enriched. There were 7 proteins participating in the pathway: C4A, CFB, CFAH, CFI, MASP2, PLG and SERPINA1.
Enrichment analysis
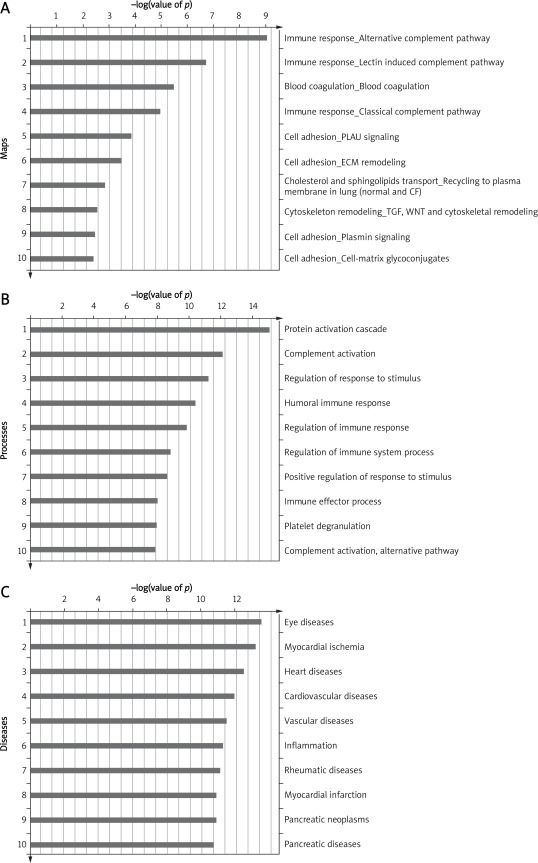
Enrichment analysis consists of matching gene IDs of proteins in functional ontologies by Meta-Core. The probability of a random intersection between a set of IDs the size of the target list with ontology entities was estimated with the p value of the hypergeometric intersection. A lower p value means higher relevance of the entity to the dataset, which shows in higher rating for the entity. All maps were drawn by GeneGo. The height of the histogram corresponded to the relative expression value for a particular gene/protein.
The top three most significant GeneGo Pathway Maps were: 1) Immune response_Alternative complement pathway, 2) Immune response_Lectin induced complement pathway, and 3) Blood coagulation_Blood coagulation (Figure 3 A). Meanwhile, protein activation cascade, complement activation and regulation of response to stimulus were the most significant enriched GO processes of the proteins (Figure 3 B). With the Disease folders, representing over 500 human diseases annotated by GeneGo, these 29 proteins were mainly related to eye diseases and some kinds of heart diseases (Figure 3 C).
Figure 3.
Enrichment analysis of the proteins by GeneGo MetaCore: A– GeneGo Pathway Maps, B– GO Processes, C– GeneGo Diseases (by Biomarkers)
Network connectivity analysis
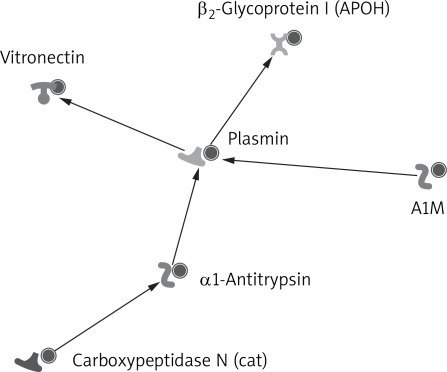
GeneGo MetaCore was used to generate biological association networks. A total of 15 relevant networks were constructed. The one with the highest score is shown in Figure 4, which was constituted by 6 proteins with direct interaction. The 6 proteins were PLG, APOH, Vitronectin, Carboxypeptidase N (cat), α1-antitrypsin and A1M. The PLG was the center of the network.
Figure 4.
A small network constituted by six proteins with direct interaction
Discussion
Two-dimensional (2-D) gel electrophoresis and tandem mass spectrometry (MS-MS) have been used to search for new biomarkers, including in DS screening and diagnosis [12, 14–17, 23–27]. But limited studies have been focused on this screening in maternal blood [14–17]. In a past study of this lab, 29 protein biomarkers for DS in maternal serum were successfully identified by the two methods [18]. The top 5 increased proteins were TF, A1BG, DES, SERPINA1 and CP, while APCS was the most down-regulated one. In the present study, we selected 5 proteins (CP, CFHR1, CFB, DES and PLG) for further analysis (bioinformatics analysis and ELISA). The reasons why we selected them were: 1) the degree of differential expression, 2) biological function of protein, 3) the relationship between protein and disease, 4) learning from published literature. Based on maternal serum, we found that only the serum levels of CP and CFB were significantly increased, while there were no significant differences in the amount of CFHR1, DES and PLG. Some studies have also reported different results between proteomics and ELISA [28, 29]. Perhaps the reasons were: 1) there are differences in the detection sensitivity of the two methods between MS and ELISA, 2) related to the small number of samples of ELISA tests, 3) some technological limitations of 2-DE and MS. Meanwhile, we found that the levels of CP, CFB, DES and CFHR1 were significantly decreased in DS patients, while PLG still had no changes. Combined with the two ELISA tests, we suggest that CP and CFB play more important roles in DS. They might be used to improve the efficacy of the prenatal screening and diagnosis of DS, and they might relate to the disease phenotype and molecular mechanism of DS. However, the expressions of protein were inconsistent in the serum from mothers and newborn babies. This might be caused by the biosynthesis and metabolism of the proteins, and it is worthy of further research.
The CP is an important iron transport protein for physiological iron homeostasis in the brain and neuronal survival [30], and it is reported to be associated with neurodegenerative disease [31]. Recently, Perluigi et al. [32] found that oxidative damage is an early event in DS pathogenesis and might contribute to the development of deleterious DS phenotypes. Moreover, its expression is adjusted by SOD1 [33], which is located on chromosome 21. Complement factor B (CFB, P00751) plays an important role in the alternative pathway for complement activation. Strohmeyer et al. [34] reported that CFB was present in the frontal cortex of AD patients, and was significantly increased, indicating alternative pathway activation. It suggested that conditions conducive to chronic alternative pathway activation may exist in the AD brain. Furthermore, as a temporal model for studying the development of AD, the brain in DS often had similar changes as in AD brain [35]. According to our study, both the 2 proteins seemed associated with the brain damage of DS, but the exact mechanisms need further research.
Pathway and network analyses in this study related the immune response alternative complement pathway to DS. Some previous studies have also reported that there were relationships between DS and the complement system [36, 37]. For example, C1q was positive in DS cases and was associated with activated microglia. It provides evidence for antibody-mediated inflammatory factors contributing to the rapid accumulation of neuropathology in DS brain [36]. It was very interesting that we first found CFB also to be associated with DS. Surprisingly, the enrichment analysis of differentially expressed proteins in DS using the MetaCore database showed the most strongly related disease to be eye diseases and some kinds of heart diseases. About 3% of DS newborns are associated with cataract, and the rate of congenital heart disease in DS patients is up to 40%. Thus, these proteins may play important roles in the development of the disease. Fifteen relevant networks were constructed by MetaCore. The highest scoring network was constituted by 6 proteins with direct interaction. The PLMN was located in the center of the network. However, the serum concentration from both mothers and patients showed no significant change. But the reason for this needs to be more deeply explored in future studies.
Regarding the 29 proteins we found in this study, we think that it needs clinical verification whether these proteins could be used for prenatal screening. Based on the study, we analyzed the characterization of differential proteins by bioinformatics analysis, and found something very interesting. These proteins may play important roles in the development of DS, for example mental retardation. They may help to explain the new mechanism of the disease.
In conclusion, based on comparative proteomic analysis, five significant differences of expressed proteins in maternal serum between normal and DS cases have been confirmed by ELISA. DAVID and GeneGo MetaCore were used for bioinformatics analysis of these candidate protein markers. The combined use of ELISA and bioinformatics analyses proved the different expression of the proteins, and revealed their biological processes and functional network in DS. These 29 proteins have relations with the development of Down syndrome, especially CP and CFB play more important roles.
Acknowledgments
We thank all the project participants for their contributions. At the same time, we are grateful for professional help from the Institutes of Biomedical Sciences, Fudan University and Biorefer Pte Ltd. This study was supported by grants from Changzhou Health Bureau (ZD200904, QN200801) and Graduate Innovation Project of Jiangsu Province (CXLX11-0607).
References
- 1.Wald NJ, Hackshaw AK. Combining ultrasound and biochemistry in first-trimester screening for Down's syndrome. Prenat Diagn. 1997;17:821–9. [PubMed] [Google Scholar]
- 2.Cuckle HS, Wald NJ, Thompson SG. Estimating a woman's risk of having a pregnancy associated with Down's syndrome using her age and serum alphafetoprotein level. BJOG. 1987;94:387–402. doi: 10.1111/j.1471-0528.1987.tb03115.x. [DOI] [PubMed] [Google Scholar]
- 3.Weisz B, Pandya P, Chitty L, Jones P, Huttly W, Rodeck C. Practical issues drawn from the implementation of the integrated test for Down syndrome screening into routine clinical practice. BJOG. 2007;114:493–7. doi: 10.1111/j.1471-0528.2007.01238.x. [DOI] [PubMed] [Google Scholar]
- 4.Pennings JL, Koster MP, Rodenburg W, Schielen PC, de Vries A. Discovery of novel serum biomarkers for prenatal Down syndrome screening by integrative data mining. PLoS One. 2009;4:e8010. doi: 10.1371/journal.pone.0008010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spencer K. Second trimester prenatal screening for Down's syndrome using alpha-fetoprotein and free beta hCG: a seven year review. BJOG. 1999;106:1287–93. doi: 10.1111/j.1471-0528.1999.tb08183.x. [DOI] [PubMed] [Google Scholar]
- 6.Wang YY, Luo J, Zhu MW. Second-trimester double or triple screening for Down syndrome: a comparison of Chinese and Caucasian populations. Int J Gynaecol Obstet. 2006;94:67–72. doi: 10.1016/j.ijgo.2006.04.030. [DOI] [PubMed] [Google Scholar]
- 7.Liu QL, Hu YL, Xu ZF, et al. A study on population-based prenatal screening and diagnosis of Down's syndrome in Jiangsu province. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2010;27:340–2. doi: 10.3760/cma.j.issn.1003-9406.2010.0.023. [DOI] [PubMed] [Google Scholar]
- 8.Hu YL. Serum screening of fetal chromosome abnormality during second pregnancy trimester: results of 26,803 pregnant women in Jiangsu Province. Zhonghua Yi Xue Za Zhi. 2007;87:2476–80. [PubMed] [Google Scholar]
- 9.Choolani M, Narasimhan K, Kolla V, Hahn S. Proteomic technologies for prenatal diagnostics: advances and challenges ahead. Expert Rev Proteomics. 2009;6:87–101. doi: 10.1586/14789450.6.1.87. [DOI] [PubMed] [Google Scholar]
- 10.Buhimschi IA, Zhao G, Rosenberg VA, Abdel-Razeq S, Thung S, Buhimschi CS. Multidimensional proteomics analysis of amniotic fluid to provide insight into the mechanisms of idiopathic preterm birth. PLoS One. 2008;3:e2049. doi: 10.1371/journal.pone.0002049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park JS, Oh KJ, Norwitz ER, Han JS, Choi HJ, Seong HS. Identification of proteomic biomarkers of preeclampsia in amniotic fluid using SELDI-TOF mass spectrometry. Reproduct Sci. 2008;15:457–68. doi: 10.1177/1933719108316909. [DOI] [PubMed] [Google Scholar]
- 12.Oh JE, Fountoulakis M, Juranville JF, Rosner M, Hengstschläger M, Lubec G. Proteomic determination of metabolic enzymes of the amnion cell: basis for a possible diagnostic tool? Proteomics. 2004;4:1145–58. doi: 10.1002/pmic.200300654. [DOI] [PubMed] [Google Scholar]
- 13.Gravett MG, Novy MJ, Rosenfeld RG, Reddy AP, Jacob T, Turner M. Diagnosis of intra-amniotic infection by proteomic profiling and identification of novel biomarkers. JAMA. 2004;292:462–9. doi: 10.1001/jama.292.4.462. [DOI] [PubMed] [Google Scholar]
- 14.Nagalla SR, Canick JA, Jacob T, Schneider KA, Reddy AP, Thomas A. Proteomic analysis of maternal serum in down syndrome: identification of novel protein biomarkers. J Proteome Res. 2007;6:1245–57. doi: 10.1021/pr060539h. [DOI] [PubMed] [Google Scholar]
- 15.Kolla V, Jenö P, Moes S, Tercanli S, Lapaire O, Choolani M. Quantitative proteomics analysis of maternal plasma in Down syndrome pregnancies using isobaric tagging reagent (iTRAQ) J Biomed Biotechnol. 2010;95:2047. doi: 10.1155/2010/952047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolialexi A, Tsangaris GT, Papantoniou N, Anagnostopoulos AK, Vougas K, Bagiokos V. Application of proteomics for the identification of differentially expressed protein markers for Down syndrome in maternal plasma. Prenat Diagn. 2008;28:691–8. doi: 10.1002/pd.2040. [DOI] [PubMed] [Google Scholar]
- 17.Lopez MF, Kuppusamy R, Sarracino DA, Prakash A, Athanas M, Krastins B. Mass spectrometric discovery and selective reaction monitoring (SRM) of putative protein biomarker candidates in first trimester trisomy 21 maternal serum. J Proteomes Res. 2011;10:133–42. doi: 10.1021/pr100153j. [DOI] [PubMed] [Google Scholar]
- 18.Bin Y, Bin Z, Jing W, Qiu-wei W, Rui-ping H, Shi-he S. Proteomic-based identification of novel protein for Down syndrome in maternal serum. Exp Biol Med. 2012 doi: 10.1258/ebm.2012.011312. pending publication. [DOI] [PubMed] [Google Scholar]
- 19.Liao Q, Zhao L, Chen X, Deng Y, Ding Y. Serum proteome analysis for profiling protein markers associated with carcinogenesis and lymph node metastasis in nasopharyngeal carcinoma. Clin Exp Metastasis. 2008;25:465–76. doi: 10.1007/s10585-008-9152-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 21.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zoltán Dezso, Yuri Nikolsky. A comprehensive functional analysis of tissue specificity of human gene expression. BMC Biology. 2008;6:49. doi: 10.1186/1741-7007-6-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsangaris GT, Karamessinis P, Kolialexi A, Garbis SD, Antsaklis A, Mavrou A. Proteomic analysis of amniotic fluid in pregnancies with Down syndrome. Proteomics. 2006;6:4410–9. doi: 10.1002/pmic.200600085. [DOI] [PubMed] [Google Scholar]
- 24.Mange A, Desmetz C, Bellet V, Molinari N, Maudelonde T, Solassol J. Proteomic profile determination of autosomal aneuploidies by mass spectrometry on amniotic fluids. Proteome Sci. 2008;11:1. doi: 10.1186/1477-5956-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho CK, Smith CR, Diamandis EP. Amniotic fluid proteome analysis from Down syndrome pregnancies for biomarker discovery. J Proteomes Res. 2010;9:3574–82. doi: 10.1021/pr100088k. [DOI] [PubMed] [Google Scholar]
- 26.Wang TH, Chao AS, Chen JK, Chao A, Chang YL, Cheng PJ. Network analyses of differentially expressed proteins in amniotic fluid supernatant associated with abnormal human karyotypes. Fertil Steril. 2009;92:96–107. doi: 10.1016/j.fertnstert.2008.05.038. [DOI] [PubMed] [Google Scholar]
- 27.Wang TH, Chang YL, Peng HH, Wang ST, Lu HW, Teng SH. Rapid detection of fetal aneuploidy using proteomics approaches on amniotic fluid supernatant. Prenat Diagn. 2005;25:559–66. doi: 10.1002/pd.1186. [DOI] [PubMed] [Google Scholar]
- 28.Cheng PJ, Wang TH, Huang SY, et al. Differential proteomics analysis of amniotic fluid in pregnancies of increased nuchal translucency with normal caryotype. Prenat Diagn. 2011;31:274–81. doi: 10.1002/pd.2719. [DOI] [PubMed] [Google Scholar]
- 29.Cho EH, Kim MR, Kim HJ, et al. The discovery of bio-markers for type 2 diabetic nephropathy by serum proteome analysis. Proteomics Clin Appl. 2007;1:352–61. doi: 10.1002/prca.200600608. [DOI] [PubMed] [Google Scholar]
- 30.Ke Y, Qian ZM. Brain iron metabolism: neurobiology and neurochemistry. Prog Neurobiol. 2007;83:149–73. doi: 10.1016/j.pneurobio.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 31.Vassiliev V, Harris ZL, Zatta P. Ceruloplasmin in neurode-generative diseases. Brain Res Brain Res Rev. 2005;49:633–40. doi: 10.1016/j.brainresrev.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 32.Perluigi M, di Domenico F, Fiorini A, et al. Oxidative stress occurs early in Down syndrome pregnancy: a redox proteomics analysis of amniotic fluid. Proteomics Clin Appl. 2011;5:167–78. doi: 10.1002/prca.201000121. [DOI] [PubMed] [Google Scholar]
- 33.Torsdottir G, Kristinsson J, Snaedal J, et al. Case-control studies on ceruloplasmin and superoxide dismutase (SOD1) in neurodegenerative diseases: a short review. J Neurol Sci. 2010;299:51–4. doi: 10.1016/j.jns.2010.08.047. [DOI] [PubMed] [Google Scholar]
- 34.Strohmeyer R, Shen Y, Rogers J. Detection of complement alternative pathway mRNA and proteins in the Alzheimer's disease brain. Brain Res Mol Brain Res. 2000;81:7–18. doi: 10.1016/s0169-328x(00)00149-2. [DOI] [PubMed] [Google Scholar]
- 35.Hyman BT. Down syndrome and Alzheimer disease. Prog Clin Biol Res. 1992;379:123–42. [PubMed] [Google Scholar]
- 36.Head E, Azizeh BY, Lott IT, Tenner AJ, Cotman CW, Cribbs DH. Complement association with neurons and beta-amyloid deposition in the brains of aged individuals with Down Syndrome. Neurobiol Dis. 2001;8:252–65. doi: 10.1006/nbdi.2000.0380. [DOI] [PubMed] [Google Scholar]
- 37.Stoltzner SE, Grenfell TJ, Mori C, et al. Temporal accrual of complement proteins in amyloid plaques in Down's syndrome with Alzheimer's disease. Am J Pathol. 2000;156:489–99. doi: 10.1016/S0002-9440(10)64753-0. [DOI] [PMC free article] [PubMed] [Google Scholar]