Abstract
In sporadic clear cell renal cell carcinoma (CCRCC), the von Hippel Lindau (VHL) gene is inactivated by mutation or methylation in the majority of primary (P) tumors. Due to differing effects of wild-type (WT) and mutant (MT) VHL gene on downstream signaling pathways regulating angiogenesis, VHL gene status could impact clinical outcome. In CCRCC, comparative genomic hybridization analysis studies have reported genetic differences between paired P and metastatic (M) tumors. We thus sequenced the VHL gene in paired tumor specimens from 10 patients to determine a possible clonal relationship between the P tumor and M lesion(s) in patients with CCRCC. Using paraffin-embedded specimens, genomic DNA from microdissected samples (>80% tumor) of paired P tumor and M lesions from all 10 patients, as well as in normal tissue from 6 of these cases, was analyzed. The DNA was used for PCR-based amplification of each of the 3 exons of the VHL gene. Sequences derived from amplified samples were compared to the wild-type VHL gene sequence (GenBank Accession No. AF010238). Methylation status of the VHL gene was determined using VHL methylation-specific PCR primers after DNA bisulfite modification. In 4/10 (40%) patients the VHL gene status differed between the P tumor and the M lesion. As expected, when the VHL gene was mutated in both the P tumor and M lesion, the mutation was identical. Further, while the VHL genotype differed between the primary tumor in different kidneys or multiple metastatic lesions in the same patient, the VHL germline genotype in the normal adjacent tissue was always wild-type irrespective of the VHL gene status in the P tumor. These results demonstrate for the first time that the VHL gene status can be different between paired primary and metastatic tissue in patients with CCRCC.
Keywords: VHL genotype, renal cancer, genetic heterogeneity
Introduction
The major types of epithelial renal tumors include clear cell (75%), papillary (15%), chromophobe (5%), and oncocytoma (5%). Mutations in the von Hippel Lindau (VHL) tumor suppressor gene are associated with hereditary and sporadic forms of clear cell renal carcinoma only. The product of the VHL gene forms a heterodimeric complex with elongin C, elongin B, Cul-2, and RBX1 and targets the hypoxic inducible factors (HIF1α and HIF1β) for ubiquitin-mediated degradation. Mutation of the VHL gene in clear cell kidney cancer prevents the VHL complex from targeting HIFs for degradation, resulting in their accumulation. Increased levels of HIF result in increased transcription of downstream targets including VEGF and angiogenic pathways (Linehan et al., 2010).
Over the last few years, anti-VEGF therapies have made a major impact in the standard of care for patients with advanced clear cell renal cell carcinoma (CCRCC; Patard et al., 2011). These include bevacizumab (Yang et al., 2003), sunitinib (Motzer et al., 2006), axitinib (Rixe et al., 2007), pazopanib (Sternberg et al., 2010), and sorafenib (Motzer et al., 2007).
We had previously reported the possible impact of VHL gene mutation and promoter hypermethylation on the outcome to VEGF-targeted agents in patients with advanced CCRCC (Choueiri et al., 2008). While the overall response rate (ORR) to VEGF-targeted therapy in patients with metastatic RCC was not correlated with VHL inactivation, subset analysis suggested that loss of function VHL mutations may identify patients with increased ORR to VEGF-targeted agents.
Extensive genetic differences between matched primary and metastatic tumors in 6 out of 19 CCRCC cases following comparative genomic hybridization (CGH) analysis (Bissig et al., 1999) and intra-tumoral heterogeneity of VHL gene deletions (Moch et al., 1998) have been reported. Based on the availability of paired primary and metastatic tumors from 10 patients, the goal of this exploratory study was to determine whether the VHL genotype can indeed differ in these paired tissue samples.
Materials and Methods
DNA sequence analysis
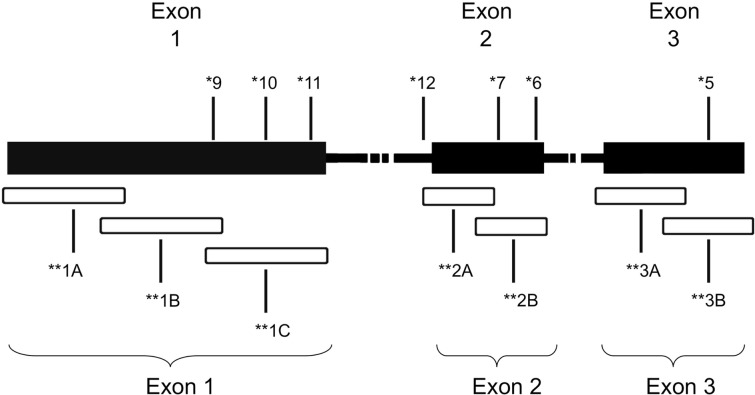
Genomic DNA was isolated from formalin fixed paraffin-embedded (FFPE) tumor biopsies as described in Choueiri et al. (2008) after manual micro dissection (Blaveri et al., 2005). Samples that were at least 80% tumor, as determined by a pathologist (HA and MZ), were included in the study. A total of seven primer sets were used to amplify overlapping fragments of the coding region of the VHL gene (Figure 1). Due to inherent problems with quality of DNA extracted from FFPE specimens, three primer sets, two primer sets, and two primer sets were used to amplify exons 1, 2, and 3, respectively. The PCR reactions consisted of 100 ng of genomic DNA, 67 mM Tris-HCL, pH 8.8, 6.7 mM magnesium chloride, 16.6 mM ammonium sulfate, 10 mM 2-mercaptoethanol, 12.5 mM dNTP’s, and 10% DMSO, 0.5 μM primers, and 0.25 units of Platinum Taq polymerase (Invitrogen, Carlsbad, CA, USA). The primer sequences used were as follows: Exon 1A–F: 5′-CGAAGACTACGGAGGTCGAC-3′; 1A–R: 5′-TCTTCAGGGCCGTACTCTTCG-3′; 1B–F: 5′-AGGCCGAGGTAGGCGCGGA-3′; 1B–R: 5′-GCAGAAGATGACCTGGGAGGGC-3′; 1C–F: 5′-CTGCGCTCGGTGAACTCG-3′; 1C–R: 5′-CTATCGTCCCTGCTGGGTCG-3′; 2A–F′: 5′-GGATTACAGGTGTGGGCCAC-3′; 2A–R′: 5′-GGCTGTCCGTCAACATTGAG-3′; 2B–F: 5′-ACGATGGGCTTCTGGTTAAC-3′; 2B–R: 5′-CCTGTACTTACCACAACAACC-3′; 3A–F: 5′-TCCTTGTACTGAGACCCTAG-3′; 3A–R: 5′-TTTGGGTGGTCTTCCAGATC-3′; 3B–F: 5′-ATTACAGGAGACTGGACATC-3′; 3B–R: 5′-CCAGTCCTGTATCTAGATC-3′.
Figure 1.
Schematic of VHL gene mutations identified in the patient cohort. *Patient ID number. **Overlapping regions amplified by PCR.
The amplicons underwent Sanger sequencing using the ABI377 automated sequencer (Applied Biosystems, Foster City, CA, USA) at the Genomic’s Core Facility, Lerner Research Institute, Cleveland Clinic. Sequences derived from the amplified samples were compared to the wild-type VHL sequence (GenBank Accession No. AF010238) using LaserGene software (DNAStar, Perkin Elmer, Foster City, CA, USA) to identify mutations. A subset of samples was also sent to Transgenomics (Omaha, NE, USA) for independent analysis of the VHL genotype. Sequence variations were identified on PCR-amplified samples that were screened using the mismatch cleavage enzyme, Surveyor, and DHPLC/WAVE® analyses followed by sequencing at Transgenomic (Nickerson et al., 2008).
Methylation assay
Methylation status was determined using VHL methylation-specific PCR primers after DNA bisulfite modification. Genomic DNA was modified using the EZ DNA Methylation-Gold Kit according to the manufacturer’s protocol (Zymo Research, Orange, CA, USA). The product then underwent PCR-based amplification using methylation-specific primers described previously (Herman et al., 1996). Methylation status was determined by gel electrophoresis of the PCR products as described in Herman et al. (1996).
Results and Discussion
Paired primary and metastatic tumors were obtained from 10 patients. VHL genotype analysis in this 10 patient cohort included single biopsies from a single primary and metastatic lesion (5 patients), multiple primary and/or metastatic tumors (5 patients) and adjacent normal tissue (6 patients). Sequencing data was obtained from a total of 42 samples (18 primary tumors, 14 metastatic tumors, and 10 adjacent normal tissues). Due to the intrinsic difficulty in identifying true somatic mutations in tissues with genetic heterogeneity, a subset of 16 samples comprised of 8 primary and 8 metastatic samples were also analyzed by Transgenomic for independent assessment of VHL gene mutation status (Table 1). VHL genotype assessment of adjacent normal tissue, as well as evaluation of multiple samples where possible contributed to the validation of our data. Patient cohort included six men and four women. While the primary and metastatic tumors were obtained in most patients during a single surgery, in some patients subsequent surgery involved additional metastatic lesions or primary tumor in the contra lateral kidney (Table 1, patients 6 and 10).
Table 1.
von Hippel Lindau genotype and Fuhrman grade in primary and metastatic CCRCC tumors.
| Pat. ID | Sample ID | Surgery date | Primary (P)/metastatic (M)/normal | VHL genotype | Fuhrman grade |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gr. 1/2 | Gr. 3/4 | |||||||||
| CATEGORY 1: PATIENTS WITH IDENTICAL PRIMARY AND METASTATIC VHL GENOTYPE IN TUMORS | ||||||||||
| 4 | 1 | 12/2004 | Primary, left kidney (P) | WT | Not evaluated | |||||
| 4 | 2 | 12/2004 | Metastatic, lymph node (M) | WT | Not evaluated | |||||
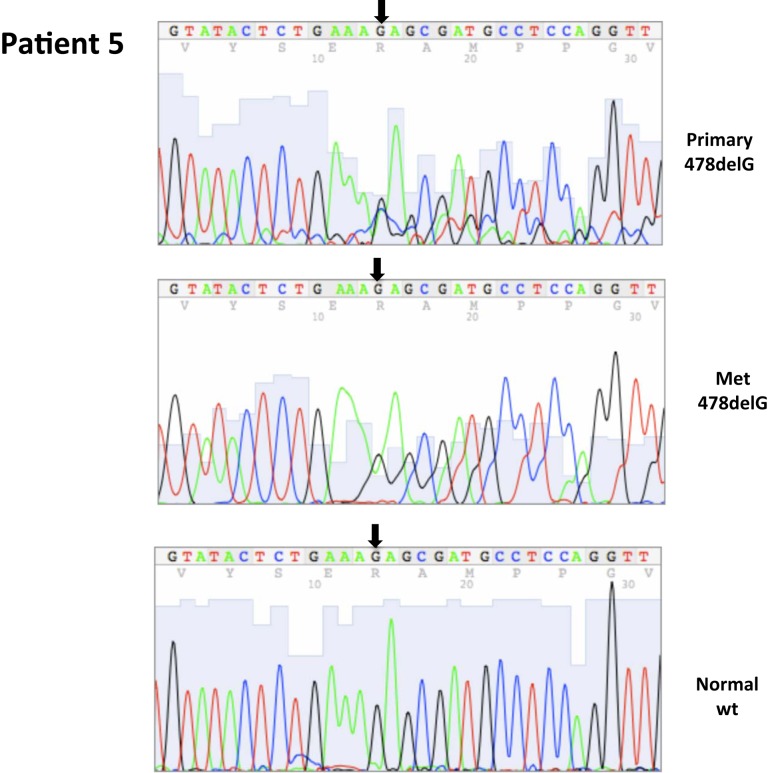
| 5 | 3 | 8/2002 | Primary, left kidney (P) | 478delG, ex3 | 0 | 100 | ||||
| 5 | 4 | 8/2002 | Metastatic, lymph node (M) | 478delG,ex3 | 0 | 100 | ||||
| 5 | 5 | 8/2002 | Adjacent normal | WT | ||||||
| 8 | 6 | 2/2005 | Primary, right kidney (P) | Methylated Promoter | 20 | 80 | ||||
| 8 | 7 | 2/2005 | Metastatic, adrenal (M) | Methylated Promoter | 0 | 100 | ||||
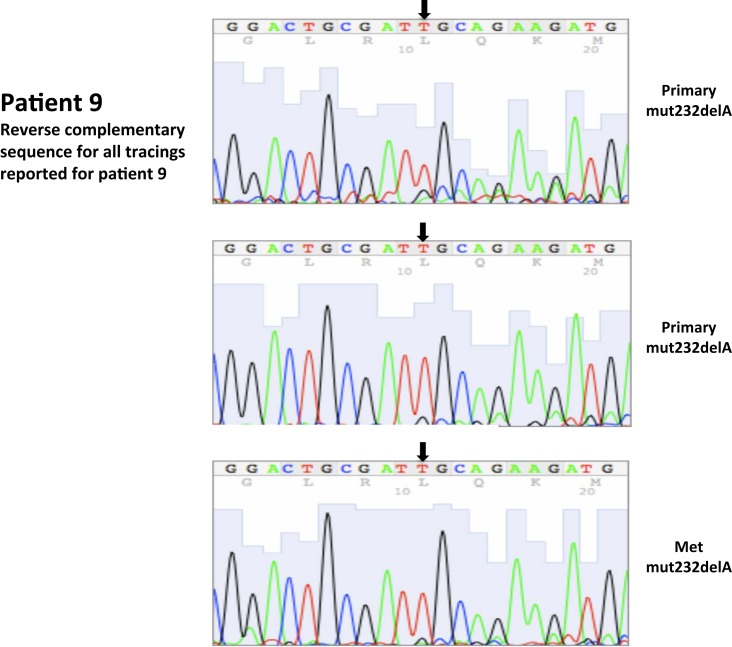
| 9* | 8 | 8/2003 | Primary, left kidney (P) | 232delA,ex1 | 10 | 90 | ||||
| Renal tumor | ||||||||||
| 9 | 9 | 8/2003 | Primary, left kidney (P) | 232delA,ex1 | 89 | 11 | ||||
| Tumor close to capsule | ||||||||||
| 9* | 10 | 8/2003 | Metastatic, lymph node (M) | 232delA, ex1 | 95 | 5 | ||||
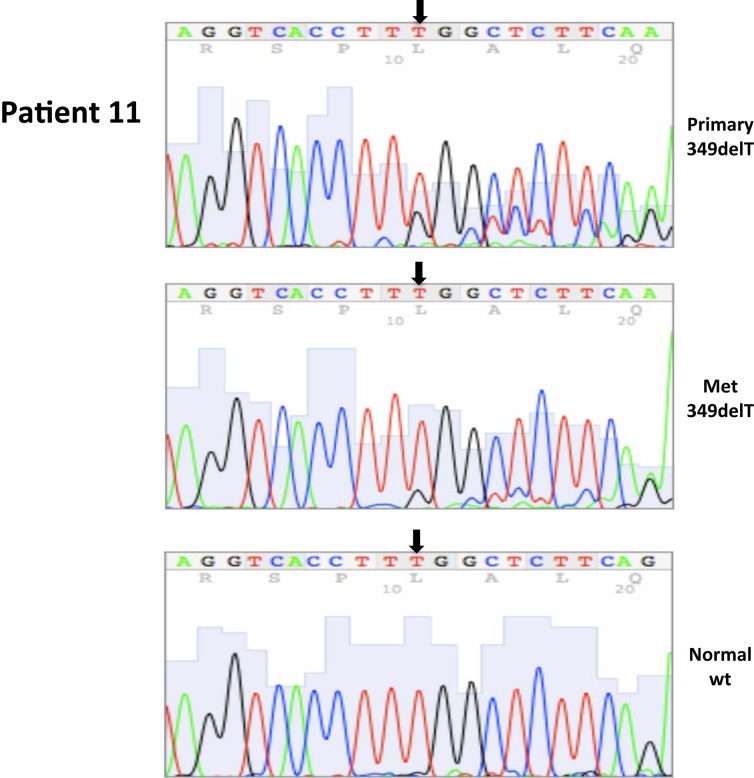
| 11 | 11 | 2/2004 | Primary, left kidney (P) | 349delT,ex1 | 10 | 90 | ||||
| 11 | 12 | 2/2004 | Metastatic, small bowel (M) | 349delT,ex1 | 50 | 50 | ||||
| 11 | 13 | 2/2004 | Adjacent normal | WT | ||||||
| 13 | 14 | 3/2004 | Primary, right kidney (P) | WT | 100 | 0 | ||||
| 13 | 15 | 3/2004 | Metastatic, left adrenal (M) | WT | 99 | 1 | ||||
| 13 | 16 | 3/2004 | Adjacent normal | WT | ||||||
| CATEGORY 2: PATIENTS WITH DIFFERENT PRIMARY AND METASTATIC VHL GENOTYPE IN TUMORS | ||||||||||
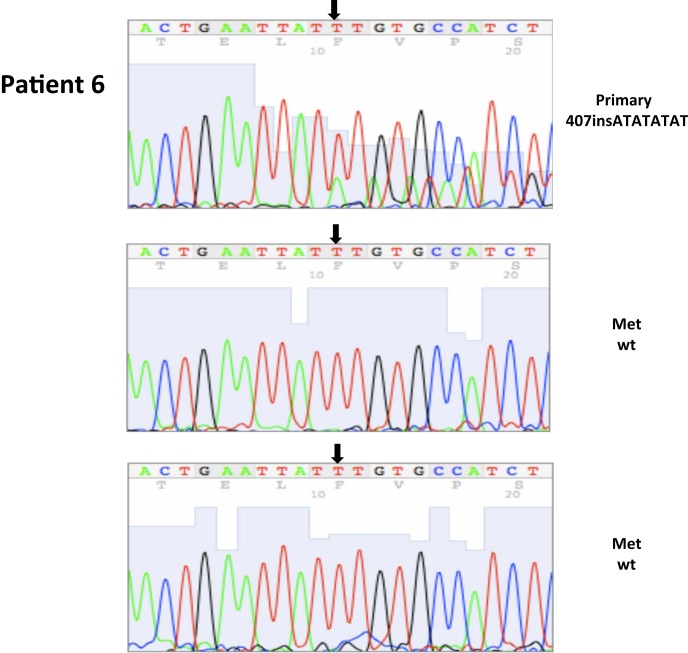
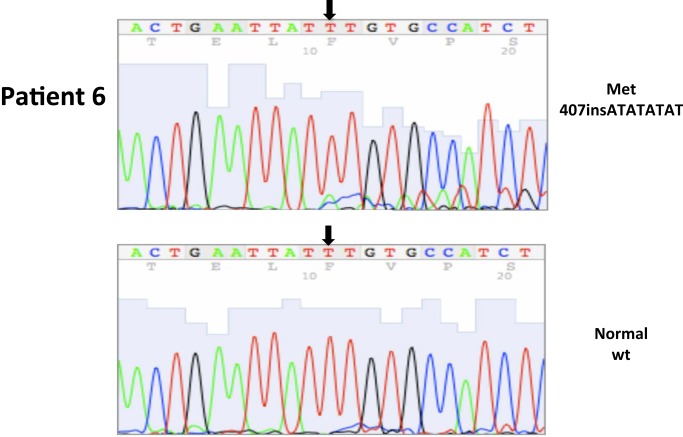
| 6* | 17 | 5/2003 | Primary, left kidney (P) | 407insATATATAT, ex2 | 100 | 0 | ||||
| 6* | 18 | 5/2004 | Metastatic, fallopian tube (M) | WT | 100 | 0 | ||||
| 6* | 19 | 5/2004 | Metastatic, fallopian tube (M) | WT | 100 | 0 | ||||
| 6* | 20 | 5/2004 | Metastatic fallopian tube (M) | 407insATATATAT, ex2 | 100 | 0 | ||||
| 6 | 21 | 5/2003 | Adjacent normal | WT | ||||||
| 7* | 22 | 8/2004 | Primary, left kidney (P) | WT | 100 | 0 | ||||
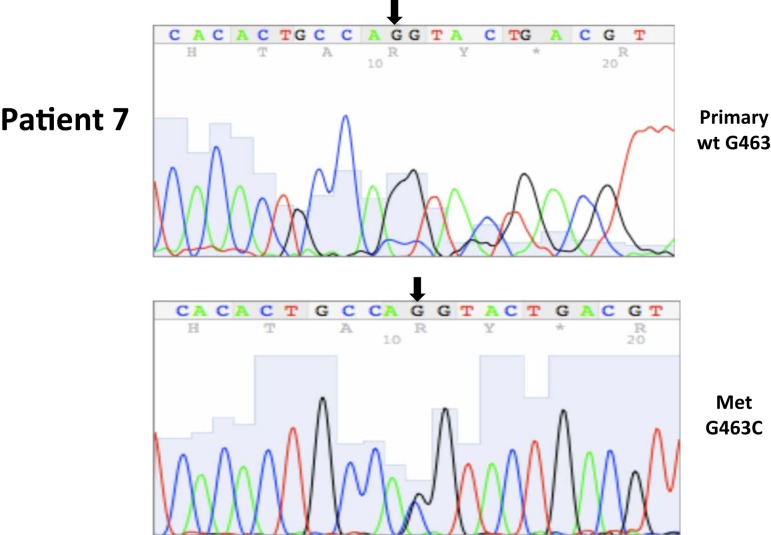
| 7* | 23 | 8/2004 | Metastatic, colon (M) | G463C, ex2 | 0 | 100 | ||||
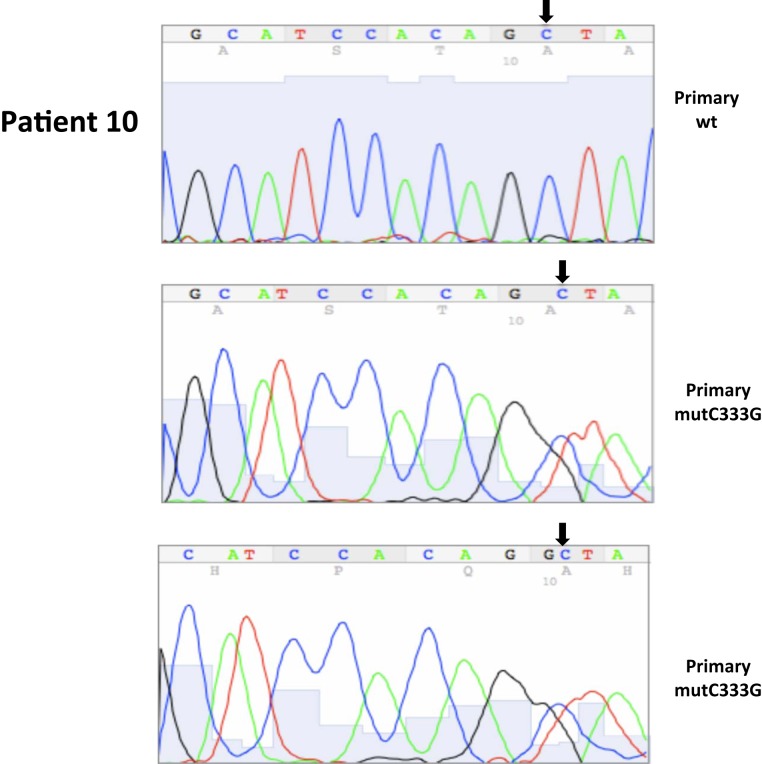
| 10* | 24 | 9/2002 | Primary, right kidney (P) | WT | 90 | 10 | ||||
| Renal vein margin with tumor | ||||||||||
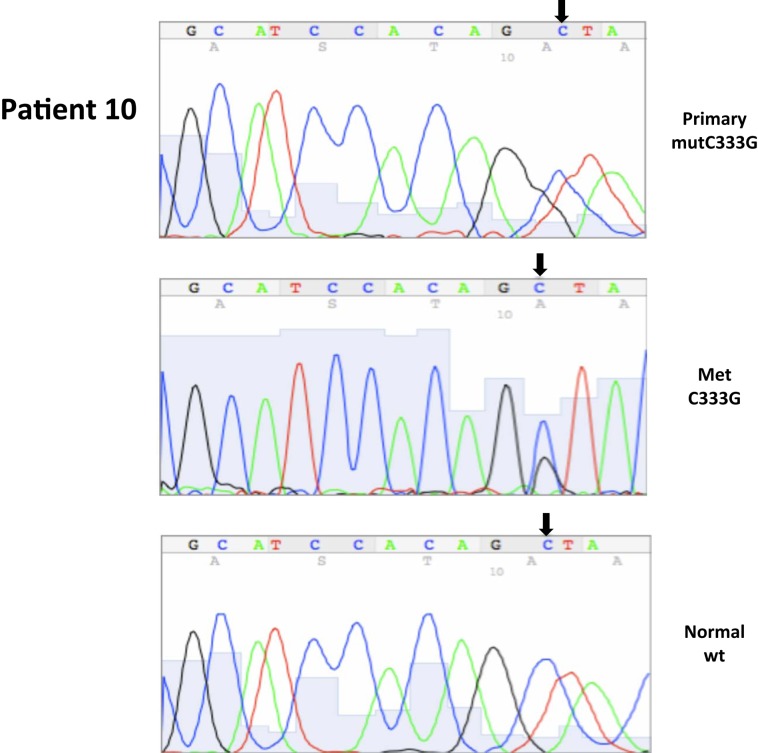
| 10* | 25 | 9/2002 | Primary, right kidney (P) | C333G, ex1 | 10 | 90 | ||||
| Spatially separated representative section of primary tumor | ||||||||||
| 10* | 26 | 12/2003 | Primary, left kidney (P) | C333G, ex1 | 45 | 55 | ||||
| Representative section of tumor in relation to capsular margin | ||||||||||
| 10* | 27 | 12/2003 | Primary, left kidney (P) | C333G, ex1 | 66 | 34 | ||||
| Representative section of tumor in relation to parenchymal margin | ||||||||||
| 10* | 28 | 12/2003 | Metastatic, lymph node (M) | C333G, ex1 | 100 | 0 | ||||
| 10 | 29 | 9/2002 | Adjacent normal | WT | ||||||
| 10 | 30 | 12/2003 | Adjacent normal | WT | ||||||
| 10 | 31 | 12/2003 | Adjacent normal | WT | ||||||
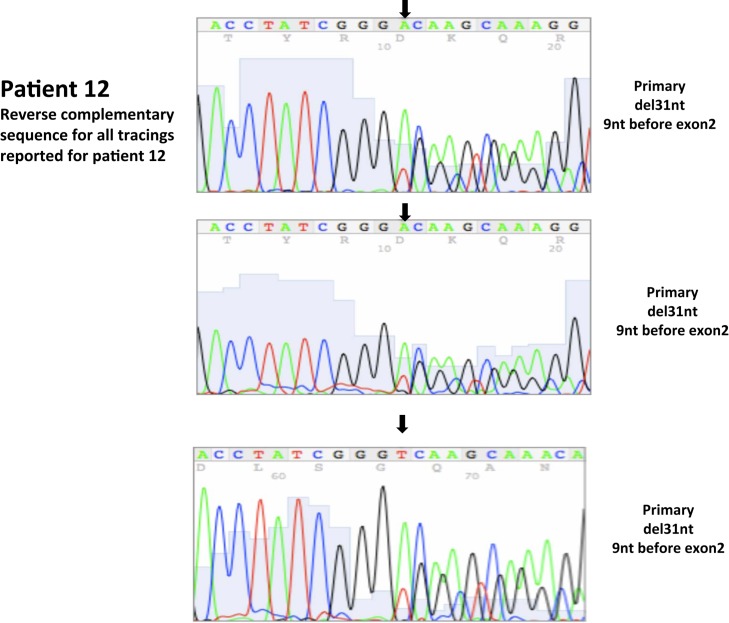
| 12 | 32 | 3/2004 | Primary, right kidney (P) Representative section of closest parenchymal margin including area with renal sinus bulge of tumor |
del31 bp, intron 1, 9nt before ex2 | 95 | 5 | ||||
| 12 | 33 | 3/2004 | Primary, right kidney (P) Representative section of closest parenchymal margin including area with renal sinus bulge of tumor |
del31 bp in intron 1, 9nt before ex2 | 95 | 5 | ||||
| 12 | 34 | 3/2004 | Primary, right kidney (P) Representative section of same primary |
del31 bp in intron 1, 9nt before ex2 | 95 | 5 | ||||
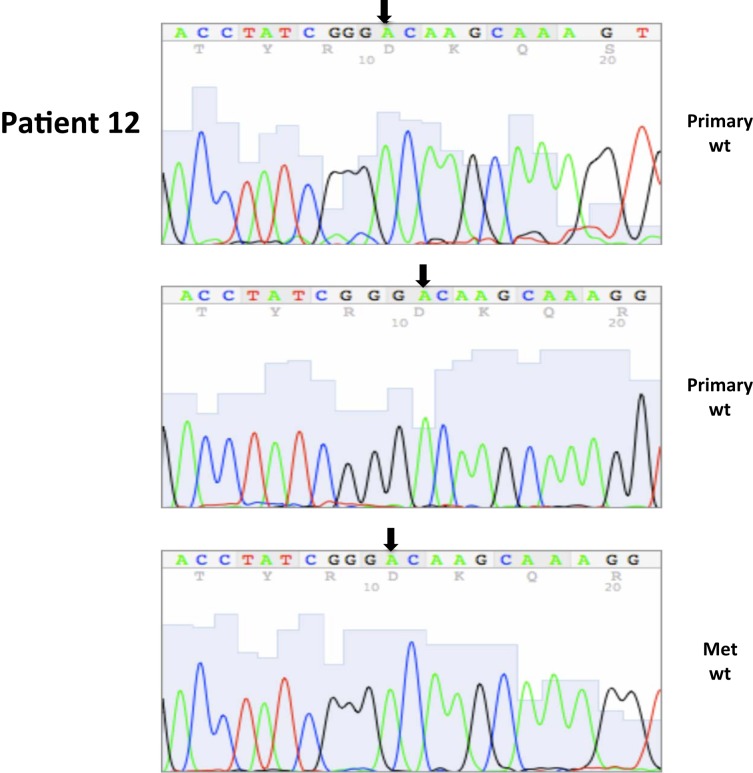
| 12* | 35 | 4/2004 | Primary, left kidney (P) | WT | 90 | 10 | ||||
| Sections of tumor with capsule | ||||||||||
| 12 | 36 | 4/2004 | Primary, left kidney (P) | WT | 50 | 50 | ||||
| Same primary | ||||||||||
| 12* | 37 | 3/2004 | Lung metastasis | WT | 10 | 90 | ||||
| Parenchymal line of resection | ||||||||||
| 12* | 38 | 3/2004 | Lung metastatsis | WT | 10 | 90 | ||||
| Pleural margin of tumor | ||||||||||
| 12 | 39 | 3/2004 | Lung metastasis | WT | 10 | 90 | ||||
| Remaining metastatic tissue | ||||||||||
| 12 | 40 | 3/2004 | Adjacent normal | WT | ||||||
| 12 | 41 | 3/2004 | Adjacent normal | WT | ||||||
| 12 | 42 | 3/2004 | Adjacent normal | WT | ||||||
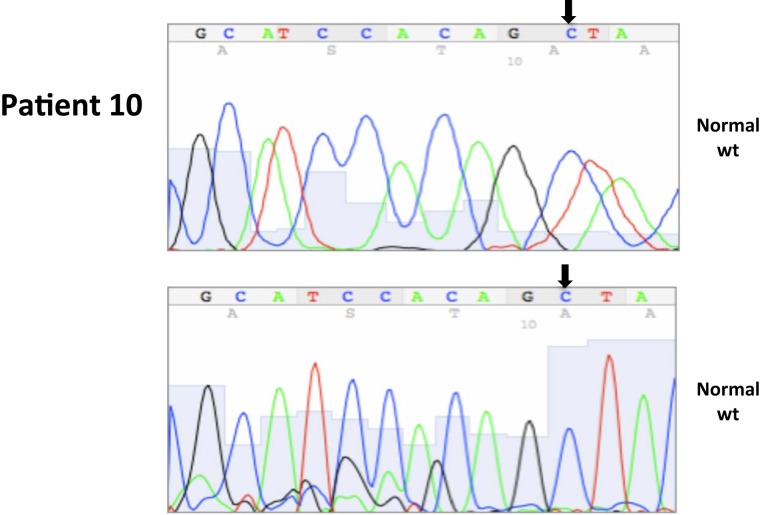
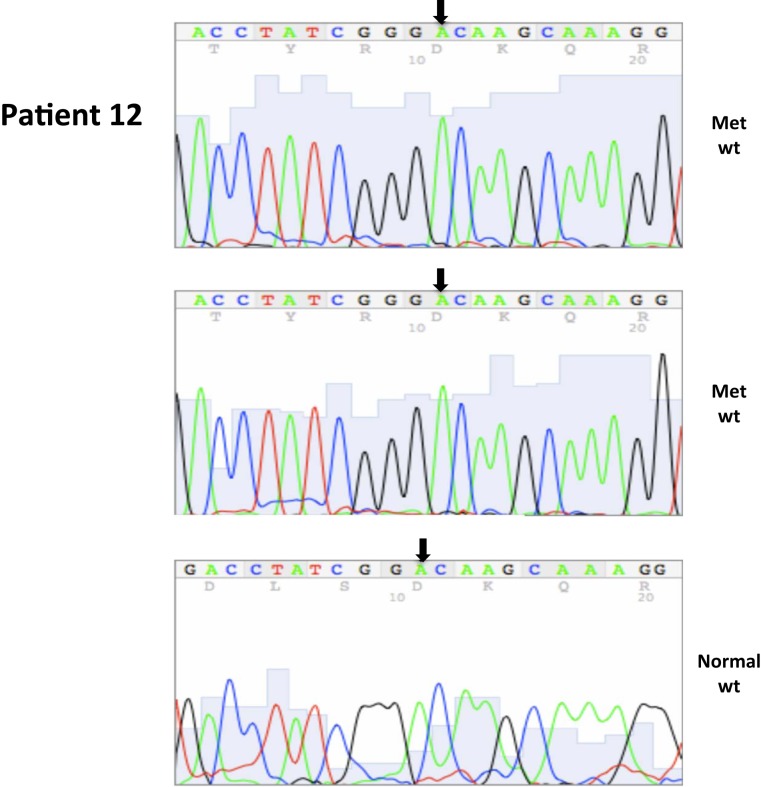
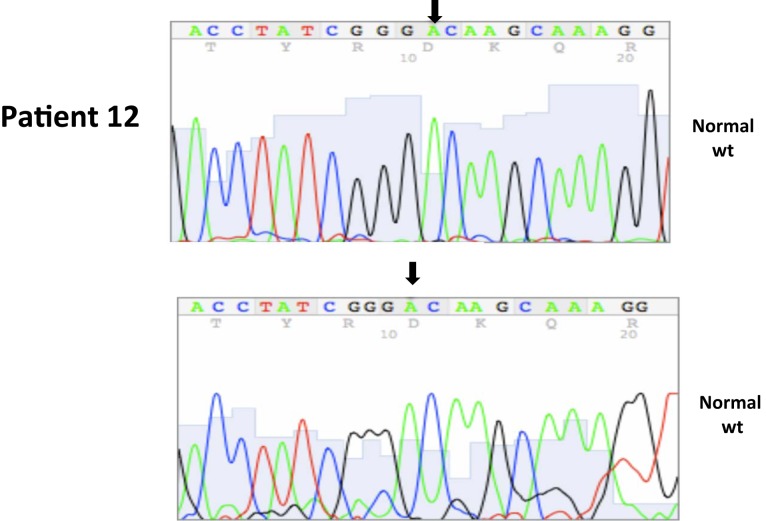
All nucleotide positions are numbered with the adenosine of the AUG start site as position number 1. This corresponds to nt position 214 in the mRNA sequence GenBank accession no. NM_000551. DNA sequencing traces are included in Appendix.
*Samples that were also sent to Transgenomic.
A total of seven patients harbored mutations in the VHL gene in the primary and/or metastatic lesions, 1 patient exhibited VHL gene methylation, and two patients had the wild-type VHL genotype (Table 1; Figure 1). Sequencing traces identifying VHL genotype for data in Table 1 are included as Figure A1 in Appendix. Mutations identified in this study were not observed in our previous larger 123 patient cohort of primary tumors (Choueiri et al., 2008) suggesting that mutations were unique to the patient in which it was first described. Further, no two patients had identical VHL mutations and methylation in this and our previous study (Choueiri et al., 2008) was observed only where the VHL genotype was wild-type. The VHL gene was methylated in 12 out 123 patients (10%) in the Choueiri study (Choueiri et al., 2008). Notably in 4 out of 10 patients the VHL genotype differed between the primary and matched metastatic lesion. Also in 2 of these patients, while the primary tumor was wild-type a mutant VHL genotype was identified in the paired metastatic lesion (patient #7 and #10, Table 1) likely due to intra-tumor heterogeneity. Inter- and intra-tumor heterogeneity in VHL genotype is exemplified in patients 10 and 12 (Table 1) between the primary tumor in different kidneys as well as metastasis removed at surgery. Additional data supporting heterogeneity in VHL genotype is based on identification of wild-type and mutant VHL in the primary tumor (patient #10 and #12) and in metastasis (patient #6) when different micro dissected tumor areas were analyzed. VHL mutations, when present, were identical between primary and metastatic sites and the VHL genotype in adjacent normal tissue in all 6/10 patients analyzed was wild-type, irrespective of VHL gene status of tumor tissue. VHL mutation status was not found to correlate with tumor grade (Table 1).
Polyclonality in colorectal adenomas (Thirwell et al., 2010), genetic heterogeneity in tumors with mutations in a single gene (Dalgliesh et al., 2010) and genetic diversity based on single cell sequencing (Navin et al., 2011) suggests that the focus on specific genetic lesions for personalized targeted therapy may be overly simplistic. More recently (while this manuscript was under review), intra-tumor heterogeneity in RNA expression or inactivating mutations in renal carcinoma has been reported (Gerlinger et al., 2012). The present study and previous reports (Moch et al., 1998; Bissig et al., 1999) emphasize that multiple genetically different clones are possibly present in clear cell renal carcinoma and this could contribute to the observed differences in VHL genotype between the primary and metastatic tumor in the same patient. Since the patients in this study had sporadic CCRCC, it is likely that clonal heterogeneity and mutations in the VHL gene may occur either during tumor development or subsequent tumor progression. This possibility is supported by the heterogeneity in VHL genotype within a single primary (Patient 6) or metastasis (Patient 12) when different spatially separated tumor sites was analyzed for VHL genotype. It is also becoming increasingly apparent that several targets for current cancer therapies can also display discordance in expression or mutation status between primary and metastatic sites. Previous studies in breast (Torres et al., 2007) and melanoma (Katona et al., 2007) cases have reported extensive genetic heterogeneity between primary and metastatic tumors. Detailed assessments of PIK3CA mutations between primary and matched metastatic breast tumors report not only discordance in mutations but also microheterogeneity in mutational status of the primary tumor (Dupont Jensen et al., 2011). Similarly, discordance in HER2 expression between primary and paired metastatic breast cancer tumors is reported to occur at a significant rate (Fabi et al., 2011; Houssami et al., 2011). Even in a CCRCC study of unmatched primary tumors and metastatic lesions, significant differences between primary and metastatic renal tumors in the expression levels of several proteins involved in the mTOR pathway including phos-AKT, phos-S6, 4EBP1, and c-myc were reported (Schultz et al., 2011). In summary, these studies suggest marked molecular heterogeneity between primary and metastatic solid tumors.
The VHL gene is unique in that somatic mutations are observed only with kidney tumors and the aberrant signaling due to VHL mutations has a direct effect on the angiogenic pathway. Although VHL is classified as a tumor suppressor, nearly 20% of sporadic CCRCC harbor the wild-type VHL genotype. While the observed differences in VHL genotype between the primary and metastatic tumor could be ascribed to technical issues, results from the present study were independently validated in 13/16 samples by Transgenomic (Table A1 in Appendix) using previously reported methodology (Nickerson et al., 2008) and further, the adjacent normal tissue in all cases independent of the tumor VHL genotype was wild-type VHL since the patients had sporadic CCRCC. In summary, in CCRCC, since primary tumor nephrectomy can precede a subsequent primary in the contralateral kidney and/or metastatic disease, the present results on intra- and inter-tumor heterogeneity in wild-type or mutant VHL gene suggest that reliance on VHL genotype of the primary tumor for treatment strategy may not be completely informative.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Appendix
Table A1.
Comparison of VHL genotype calls – Cleveland Clinic and Transgenomic (n = 16 samples).
| Patient ID | Sample ID | DNA sequenced at Cleveland Clinic | DNA sequenced and reported by Transgenomic |
|---|---|---|---|
| 6Pa | 407insATATATAT, ex2 | 412inATATATAT, ex2 | |
| 6Mb | 18 | wt | wt |
| 6M | 19 | wtc | 412insATATATAT, ex2 |
| 6M | 20 | 407insATATATAT, ex2 | 412insATATATAT, ex2 |
| 7P | wtd | G463C | |
| 7M | G463C | G463C | |
| 9P | 232delA | 232 delA | |
| 9M | 232delAe | P25S, 10% RSIf | |
| 10P | 24 | wt | wt |
| 10P | 25 | C333G | C333G |
| 10P | 26 | C333G | C333G |
| 10P | 27 | C333G | C333G |
| 10M | C333G | C333G | |
| 12P | wt | wt | |
| 12M | 37 | wt | wt |
| 12M | 38 | wt | wt |
aPrimary.
bMetastatic.
c,d,eCalls based on sequence traces presented in Figure A1.
fRelative signal intensity (RSI).
Figure A1.
Somatic VHL mutation sequencing in normal and tumor tissue of representative patients from Table 1.
References
- Bissig H., Richter J., Desper R., Meier V., Schraml P., Schaffer A. A., Sauter G., Mihatsch M. J., Moch H. (1999). Evaluation of the clonal relationship between primary and metastatic renal cell carcinoma by comparative genomic hybridization. Am. J. Pathol. 155, 267–274 10.1016/S0002-9440(10)65120-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaveri E., Simko J. P., Korkola J. E., Brewer J. L., Baehner F., Mehta K., Devries S., Koppie T., Pejavar S., Carroll P., Waldman F. M. (2005). Bladder cancer outcome and subtype classification by gene expression. Clin. Cancer Res. 11, 4044–4055 10.1158/1078-0432.CCR-05-0177 [DOI] [PubMed] [Google Scholar]
- Choueiri T. K., Vaziri S. A., Jaeger E., Elson P., Wood L., Bhalla I. P., Small E. J., Weinberg V., Sein N., Simko J., Golshayan A. R., Sercia L., Zhou M., Waldman F. M., Rini B. I., Bukowski R. M., Ganapathi R. (2008). von Hippel-Lindau gene status and response to vascular endothelial growth factor targeted therapy for metastatic clear cell renal cell carcinoma. J. Urol. 180, 860–865; discussion 865–866. 10.1016/j.juro.2008.05.015 [DOI] [PubMed] [Google Scholar]
- Dalgliesh G. L., Furge K., Greenman C., Chen L., Bignell G., Butler A., Davies H., Edkins S., Hardy C., Latimer C., Teague J., Andrews J., Barthorpe S., Beare D., Buck G., Campbell P. J., Forbes S., Jia M., Jones D., Knott H., Kok C. Y., Lau K. W., Leroy C., Lin M. L., McBride D. J., Maddison M., Maguire S., McLay K., Menzies A., Mironenko T., Mulderrig L., Mudie L., O’Meara S., Pleasance E., Rajasingham A., Shepherd R., Smith R., Stebbings L., Stephens P., Tang G., Tarpey P. S., Turrell K., Dykema K. J., Khoo S. K., Petillo D., Wondergem B., Anema J., Kahnoski R. J., Teh B. T., Stratton M. R., Futreal P. A. (2010). Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 463, 360–363 10.1038/nature08672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont Jensen J., Laenkholm A. V., Knoop A., Ewertz M., Bandaru R., Liu W., Hackl W., Barrett J. C., Gardner H. (2011). PIK3CA mutations may be discordant between primary and corresponding metastatic disease in breast cancer. Clin. Cancer Res. 17, 667–677 10.1158/1078-0432.CCR-10-1133 [DOI] [PubMed] [Google Scholar]
- Fabi A., Di Benedetto A., Metro G., Perracchio L., Nistico C., Di Filippo F., Ercolani C., Ferretti G., Melucci E., Buglioni S., Sperduti I., Papaldo P., Cognetti F., Mottolese M. (2011). HER2 protein and gene variation between primary and metastatic breast cancer: significance and impact on patient care. Clin. Cancer Res. 17, 2055–2064 10.1158/1078-0432.CCR-10-1920 [DOI] [PubMed] [Google Scholar]
- Gerlinger M., Rowan A. J., Horswell S., Larkin J., Endesfelder D., Gronroos E., Martinez P., Matthews N., Stewart A., Tarpey P., Varela I., Phillimore B., Begum S., McDonald N. Q., Butler A., Jones D., Raine K., Latimer C., Santos C. R., Nohadani M., Eklund A. C., Pencer-Dene B., Clark G., Pickering L., Stamp G., Gore M., Szallasi Z., Downward J., Futreal P. A., Swanton C. (2012). Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892 10.1056/NEJMoa1113205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman J. G., Graff J. R., Myohanen S., Nelkin B. D., Baylin S. B. (1996). Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. U.S.A. 93, 9821–9826 10.1073/pnas.93.18.9821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houssami N., Macaskill P., Balleine R. L., Bilous M., Pegram M. D. (2011). HER2 discordance between primary breast cancer and its paired metastasis: tumor biology or test artefact? Insights through meta-analysis. Breast Cancer Res. Treat. 129, 659–674 10.1007/s10549-011-1632-x [DOI] [PubMed] [Google Scholar]
- Katona T. M., Jones T. D., Wang M., Eble J. N., Billings S. D., Cheng L. (2007). Genetically heterogeneous and clonally unrelated metastases may arise in patients with cutaneous melanoma. Am. J. Surg. Pathol. 31, 1029–1037 10.1097/PAS.0b013e31802b3488 [DOI] [PubMed] [Google Scholar]
- Linehan W. M., Srinivasan R., Schmidt L. S. (2010). The genetic basis of kidney cancer: a metabolic disease. Nat. Rev. Urol. 7, 277–285 10.1038/nrurol.2010.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moch H., Schraml P., Bubendorf L., Richter J., Gasser T. C., Mihatsch M. J., Sauter G. (1998). Intratumoral heterogeneity of von Hippel-Lindau gene deletions in renal cell carcinoma detected by fluorescence in situ hybridization. Cancer Res. 58, 2304–2309 [PubMed] [Google Scholar]
- Motzer R. J., Hoosen S., Bello C. L., Christensen J. G. (2006). Sunitinib malate for the treatment of solid tumours: a review of current clinical data. Expert Opin. Investig. Drugs 15, 553–561 10.1517/13543784.15.5.553 [DOI] [PubMed] [Google Scholar]
- Motzer R. J., Michaelson M. D., Rosenberg J., Bukowski R. M., Curti B. D., George D. J., Hudes G. R., Redman B. G., Margolin K. A., Wilding G. (2007). Sunitinib efficacy against advanced renal cell carcinoma. J. Urol. 178, 1883–1887 10.1016/j.juro.2007.07.030 [DOI] [PubMed] [Google Scholar]
- Navin N., Kendall J., Troge J., Andrews P., Rodgers L., McIndoo J., Cook K., Stepansky A., Levy D., Esposito D., Muthuswamy L., Krasnitz A., McCombie W. R., Hicks J., Wigler M. (2011). Tumour evolution inferred by single-cell sequencing. Nature 472, 90–94 10.1038/nature09807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson M. L., Jaeger E., Shi Y., Durocher J. A., Mahurkar S., Zaridze D., Matveev V., Janout V., Kollarova H., Bencko V., Navratilova M., Szeszenia-Dabrowska N., Mates D., Mukeria A., Holcatova I., Schmidt L. S., Toro J. R., Karami S., Hung R., Gerard G. F., Linehan W. M., Merino M., Zbar B., Boffetta P., Brennan P., Rothman N., Chow W. H., Waldman F. M., Moore L. E. (2008). Improved identification of von Hippel-Lindau gene alterations in clear cell renal tumors. Clin. Cancer Res. 14, 4726–4734 10.1158/1078-0432.CCR-07-4921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patard J. J., Pignot G., Escudier B., Eisen T., Bex A., Sternberg C., Rini B., Roigas J., Choueiri T., Bukowski R., Motzer R., Kirkali Z., Mulders P., Bellmunt J. (2011). ICUD-EAU International Consultation on Kidney Cancer 2010: treatment of metastatic disease. Eur. Urol. 60, 684–690 10.1016/j.eururo.2011.06.017 [DOI] [PubMed] [Google Scholar]
- Rixe O., Bukowski R. M., Michaelson M. D., Wilding G., Hudes G. R., Bolte O., Motzer R. J., Bycott P., Liau K. F., Freddo J., Trask P. C., Kim S., Rini B. I. (2007). Axitinib treatment in patients with cytokine-refractory metastatic renal-cell cancer: a phase II study. Lancet Oncol. 8, 975–984 10.1016/S1470-2045(07)70285-1 [DOI] [PubMed] [Google Scholar]
- Schultz L., Chaux A., Albadine R., Hicks J., Kim J. J., De Marzo A. M., Allaf M. E., Carducci M. A., Rodriguez R., Hammers H. J., Argani P., Reuter V. E., Netto G. J. (2011). Immunoexpression status and prognostic value of mTOR and hypoxia-induced pathway members in primary and metastatic clear cell renal cell carcinomas. Am. J. Surg. Pathol. 35, 1549–1556 10.1097/PAS.0b013e31822895e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg C. N., Davis I. D., Mardiak J., Szczylik C., Lee E., Wagstaff J., Barrios C. H., Salman P., Gladkov O. A., Kavina A., Zarba J. J., Chen M., McCann L., Pandite L., Roychowdhury D. F., Hawkins R. E. (2010). Pazopanib in locally advanced or metastatic renal cell carcinoma: results of a randomized phase III trial. J. Clin. Oncol. 28, 1061–1068 10.1200/JCO.2009.23.9764 [DOI] [PubMed] [Google Scholar]
- Thirwell C., Will O. C., Domingo E., Graham T. A., McDonald S. A., Oukrif D., Jeffrey R., Gorman M., Rodriguez-Justo M., Chin-Aleong J., Clark S. K., Novelli M. R., Jankowski J. A., Wright N. A., Tomlinson I. P., Leedham S. J. (2010). Clonality assessment and clonal ordering of individual neoplastic crypts shows polyclonality of colorectal adenomas. Gastroenterology 138, 1441–1454 10.1053/j.gastro.2010.01.033 [DOI] [PubMed] [Google Scholar]
- Torres L., Ribeiro F. R., Pandis N., Andersen J. A., Heim S., Teixeira M. R. (2007). Intratumor genomic heterogeneity in breast cancer with clonal divergence between primary carcinomas and lymph node metastases. Breast Cancer Res. Treat. 102, 143–155 10.1007/s10549-006-9317-6 [DOI] [PubMed] [Google Scholar]
- Yang J. C., Haworth L., Sherry R. M., Hwu P., Schwartzentruber D. J., Topalian S. L., Steinberg S. M., Chen H. X., Rosenberg S. A. (2003). A randomized trial of bevacizumab, an anti-vascular endothelial growth factor antibody, for metastatic renal cancer. N. Engl. J. Med. 349, 427–434 10.1056/NEJMoa021491 [DOI] [PMC free article] [PubMed] [Google Scholar]