Abstract
In this study we demonstrated that ΔCaecm33 double mutant showed reduced biofilm formation and causes less damage to gingival mucosa tissues. This was confirmed by the reduced level of necrotic cells and Bax/Bcl2 gene expression as apoptotic markers. In contrast, parental and Caecm33 mutant strains decreased basement membrane protein production (laminin 5 and type IV collagen). We thus propose that ECM33 gene/protein represents a novel target for the prevention and treatment of infections caused by Candida.
1. Introduction
Candida species are the most frequent cause of life-threatening invasive fungal infections in the immunocompromised host [1, 2]. Under predisposing conditions, C. albicans multiplies and penetrates the host tissue to cause inflammation and tissue destruction [3]. C. albicans adherence to host cells is therefore the first step in the initiation of infection, as this enables the organism to evade the normal flushing mechanisms of body secretions [4].
The Candida cell wall is the initial point of contact and interaction with host tissues. This wall consists of a complex structure that houses a network of polysaccharides (primarily β-glucans and chitin) in which various proteins such as glycosylphosphatidylinositol (GPI) interact [5]. The key function of GPI proteins in Candida biogenesis and maintenance of the fungus cell wall involves ECM33 gene [6]. ECM33 protein is required for normal cell wall integrity and the yeast-to-hyphae transition in vitro [7]. C. albicans Caecm33Δ/Caecm33Δ mutant showed blastospores that were flocculated and were larger than those of the wild-type strain. Caecm33Δ/Caecm33Δ mutant displayed delayed hyphal formation and showed attenuated virulence in the mouse model of hematogenously disseminated candidiasis [8].
The absence of ECM33 gene may also prevent Candida from affecting the oral mucosa. Oral epithelium interacts with connective tissue via basement membrane (BM) proteins that contribute to body integrity.
Oral mucosa contains a highly complex stratified epithelia that protects the body from physical and chemical damage, infection, dehydration, and heat loss through interactions with the mesenchymal tissue via basement membrane (BM) proteins [9–11]. The BM is a thin layer of complex extracellular matrix that forms the support structure on which epithelial cells grow. This layer also provides mechanical support, divides tissue into compartments, and significantly influences cellular behavior [12]. Two major components of the BM are type IV collagen and laminin. Type IV collagen forms a network that confers the distinctive mechanical stability known to the BM [13]. Laminin binds to collagen IV to constitute a second network by interacting with nidogen. Laminin 5 is specific to the basement membrane underlying the squamous epithelium and mucosa [14]. Its primary role is to modulate stable epithelial cell attachment through interactions with integrins α 3 β 1 and α 6 β 4 [15].
The interaction between Candida and the oral epithelium is believed to be one of the most important initial events in the prevention or development of candidiasis [18]. However, following contact with C. albicans, and under certain circumstances such as reduced innate immunity, C. albicans may occasionally become pathogenic and induce lesions on the oral mucosa [19, 20]. This may occur through a deregulation of BM protein synthesis and deposition, which in turn induces a breakdown of oral homeostasis and, consequently, systemic infection [21, 22]. Tissue structure and BM protein deregulation may occur through apoptotic processes involving inducer (Bax, Bcl-xS, Bad) and inhibitor (Bcl-2, Bcl-xL) genes [23]. Indeed, host cell death following microbial infection is typically recognized as necrotic, apoptotic, or pyroptotic [24–26]. While necrosis is characterized as accidental cell death as the result of physical damage, apoptosis and pyroptosis are strictly regulated genetic and biochemical self-destruction programs that are critical during development and tissue homeostasis as well as in modulating the pathogenesis of a variety of infectious diseases [27]. A recent study reported that C. albicans stimulated the oral epithelial signaling pathways that promote early apoptotic cell death through the activation of cellular caspases, followed by late necrosis [22].
Considering the key role of oral mucosa in preventing/controlling Candida pathogenesis, and given the adhesion, which is the first stage of biofilm formation of Candida to the tissue through specific proteins such as ECM33, we sought to investigate the role of ECM33 gene on Candida biofilm formation, and its interaction with oral mucosa tissue. The central hypothesis of this study is that ECM33 gene is involved in Candida virulence leading to tissue damage and facilitating the onset of candidiasis. To test this hypothesis, we examined the ability of ECM33 isogenic strains of C. albicans to form biofilms in vitro on a catheter-associated biofilm model in order to induce tissue damage, cell necrosis, apoptosis signalling molecule (Bax and Bcl2) activation, and laminin 5 and type IV collagen modulation. To move our study closer to the clinical setting, we used an engineered human oral mucosa model, as previously described [4].
2. Materials and Methods
2.1. Candida Strains
Table 1 presents the Candida albicans strains used in this study. These strains were generously donated by Dr. C. Gil (Madrid, Spain). The parental (CAF2) C. albicans strain was genotypically identified as URA3/Δura3::imm43 [16]. Each Candida strain was cultured on Sabouraud dextrose agar plates (Becton Dickinson, Oakville, ON, Canada) at 30°C. To prepare the C. albicans suspension, one colony was used to inoculate 10 mL of Phytone-peptone medium (Becton Dickinson) supplemented with 0.1% glucose and 80 mg/L of uridine at pH 5.6. The cultures were grown under shaking conditions in a water bath for 18 h, after which time the Candida cells were collected, washed with PBS, and resuspended in the same buffer to a density of 1 × 107 cells/mL to obtain a standardized cell suspension.
Table 1.
Candida strains used in this study.
| Strain | Relevant characteristics | Reference |
|---|---|---|
| CAF2-parental strain | URA3/Δura3::imm43 | [16] |
| RML1-heterozygous ecm33 mutant | CaECM33/ΔCaecm33:: hisG-CaURA3-hisG | [17] |
| RML2-homozygous double ecm33 mutant | ΔCaecm33Δ:: hisG/Caecm33Δ:: hisG-CaURA3-hisG | [17] |
| RML3-heterozygous revertant | Caecm33Δ:: hisG/CaECM33-clz-URA3-clz-hisG | [17] |
| RML4-homozygous revertant | CaECM33-clz-hisG/CaECM33-clz-URA3-clz-hisG | [17] |
2.2. Catheter-Associated C. albicans Biofilm Formation
The first step was to prepare the substrate material (silicone elastomer) for biofilm formation. Silicone elastomer (SE) sheets were obtained from Invotec International (Jacksonville, FL, USA). Following the manufacturer's instructions, the catheter material sheet was cleaned by scrubbing it thoroughly with a clean, soft-bristled brush in a hot water/hand soap solution, rinsed with distilled water, and autoclaved. Flat circular discs 15 mm in diameter were obtained by cutting through the catheter sheets with a cork borer, as previously described [28]. The sterile SE disks were distributed into 12-well plates, precoated with fetal bovine serum (Mediatech, VA, USA) for 24 h at 37°C on a rocker table and then exposed to one of the Candida strains (1 × 107 cells/mL). To assist C. albicans adhesion, the SE discs were incubated at 37°C for 90 min, transferred to a sterile 12-well plate containing 4 mL of yeast nitrogen base medium, and subsequently cultured at 37°C for 48 h. Biofilm formation was quantified by means of a tetrazolium XTT [2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide] reduction assay [28]. Discs containing no Candida cells served as controls. To confirm biofilm formation, the Candida-seeded SE discs were transferred to microscope slides, stained with calcofluor-white (0.05% (vol/vol)) and examined under a fluorescence microscope at λ max = 432 nm.
2.3. Engineering Human Oral Mucosa Production
Gingival mucosa samples were biopsied from healthy donors following their informed consent and approval by the University Lavals ethics committee. Gingival fibroblasts and epithelial cells were subsequently extracted [29]. Following culture, the cells were used to engineer human oral mucosa (EHOM). The fibroblasts were mixed with bovine skin collagen to produce the lamina propria. Four days later, the epithelial cells were seeded on the lamina propria and grown until confluence. The EHOM was then raised to the air-liquid interface for five days for epithelium stratification and was used thereafter to study C. albicans pathogenesis.
2.4. Tissue Structure following Infection
EHOM was first grown in a serum-free, antifungal-free DMEH medium for 48 h and then placed in contact with one of the C. albicans (104 cells/cm2) strains for 24 h. Noninfected EHOM was used as the control (Ctrl). Following infection and culture, the biopsies were collected, fixed with 4% paraformaldehyde solution, embedded in paraffin, and stained with hematoxylin-eosin for analysis.
2.5. Quantitation of the Biofilms Formed on the EHOM Tissue
To quantitatively assess the differences in biofilm formation by the selected C. albicans strains, multiple specimens were collected from each infected EHOM and stained with hematoxylin-eosin. Each slide was assigned a number, and measurements were made independently by two different observers. Each blind observer measured the thickness of C. albicans biofilms at regular intervals using a calibrated image analysis system (Image-Pro Plus software, Media Cybernetics, MD, USA), as we previously reported [30]. Ten measurements were performed on the unfolded area of each slide. We then compared the thickness of the biofilms formed on the EHOM specimens infected with the mutant strains and that of the biofilms formed by the wild-type and revertant strains. A P value of <0.05 was considered statistically significant (n = 4).
2.6. Lactate Dehydrogenase Assay
The release of lactate dehydrogenase (LDH) from the EHOM into the surrounding medium was monitored as a measure of tissue damage/cell necrosis. LDH activity release into the medium of the infected tissue (IT) was measured at 24 h using a Cytox96 kit (Promega, Madison, WI, USA) according to the manufacturer's instructions. A positive control (PC) was obtained by culturing the cells for one hour in the presence of 1% Triton X-100, while a negative control (NC) was obtained by culturing cells not exposed to any Candida strain. LDH activity was calculated as the percetage of total LDH activity release = (ITabsorbance − NCabsorbance)/(PCabsorbance − NCabsorbance) × 100. Differences between groups were determined by an ANOVA Student t-test and was considered significant at a P value of <0.05 (n = 4).
2.7. Apoptotic (Bax) and Antiapoptotic (Bcl-2) Gene Expression and Protein Production
Following EHOM infection with the Candida strains, total RNA was extracted from each EHOM and used to comparatively analyze Bax and Bcl2 gene expression by qRT-PCR [31] and GAPDH gene expression. Table 2 presents the specific primer sequences used in this study. The thermocycling conditions for the Bax gene were 95°C for 5 min, followed by 40 cycles of 95°C for 15 s, 64°C for 30 s, and 72°C for 30 s, while those for the Bcl2 gene were 95°C for 3 min, followed by 40 cycles of 95°C for 10 s, 64°C for 10 s, and 72°C for 30 s. The specificity of each primer pair was verified by the presence of a single melting temperature peak. GAPDH produced uniform expression levels varying by less than 0.5 CTs between sample conditions and was therefore used as a reference gene for this study. Gene expression was supported by Bax and Bcl2 protein analyses. Candida-infected EHOM was used to prepare protein cell lysates which were then subjected to SDS polyacrylamide gel electrophoresis and electroblotting onto PVDF membranes [31]. The membranes were then blocked with 5% BSA in Tween-20/Tris-buffered saline (TTBS) for 1 h and were incubated overnight at 4°C with either primary anti-Bax or anti-Bcl2 antibodies (1 : 1000). After washing, the membranes were incubated with horseradish peroxidase-labeled secondary antibody (1 : 1000) for 1 h. For protein detection, the membranes were washed for 3 h with TBS, incubated in ECL, and analyzed on a Fujifilm Image Reader LAS-1000 Pro.
Table 2.
Description of oligonucleotide primer pairs used in PCR reactions.
| Gene | Primer sequence (5′ to 3′) | Amp size (bp) |
|---|---|---|
| Bax | Sens 5′-TGG CAG ACA TGT TTT CTG AC-3′ | 204 |
| Antisens 5′-TCA CCC AAC CAC CCT GGT CTT-3′ | ||
| Bcl2 | Sens 5′-TTT GAG TTC GGT GGG GTC AT-3′ | 274 |
| Antisens 5′-TGA CTT CAC TTG TGG CCC AG-3′ | ||
| GAPDH | Sens 5′-ATGCAACGGATTTGGTCGTAT-3′ | 221 |
| Antisens 5′-TCTCCTCCTGGAAGATGGTG-3′ |
2.8. Laminin 5 and Type IV Collagen Protein Production
Following contact for 24 h with one of the Candida strains, the EHOM protein lysates were used to assess laminin 5 and type IV collagen production by western blotting using mouse antilaminin 5 (1 : 200) or mouse antitype IV collagen (1 : 1000). Each membrane was washed in TBS-T then incubated with the appropriate secondary horseradish peroxidase-labeled antibody (1 : 1000) for 1 h at 23°C. Protein detection was performed as described above.
3. Results
3.1. Caecm33 Mutant Formed Less Biofilms
Our findings show that homozygous Caecm33 mutant tended to form less biofilms than did the isogenic wild-type strain (P = 0.052, Figure 1(a)). Fluorescence microscopy results also reveal that the biofilms formed by Caecm33 mutant were less dense and contained patchy clusters of a diffuse extracellular matrix, compared to that observed in the isogenic wild-type and revertant strains (Figures 1(b)–1(d)).
Figure 1.
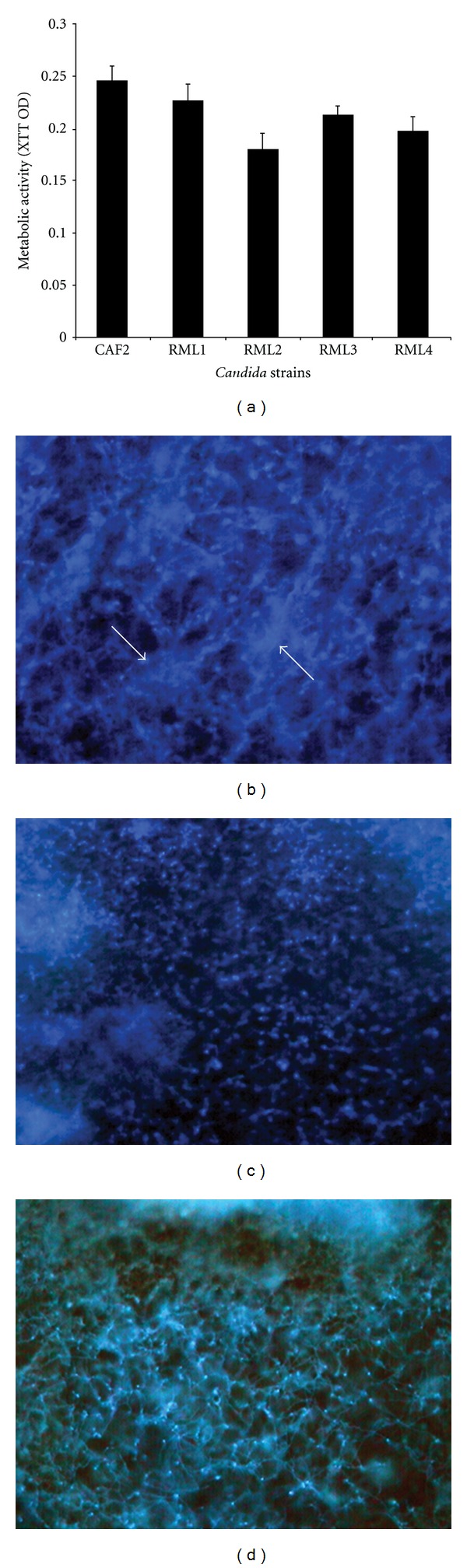
Biofilm-forming ability of isogenic wild-type, Caecm33 mutant, and revertant strains. (a) Metabolic activity of mature biofilms formed by the isogenic strains. (b–d) Fluorescence micrographs showing differences in morphology of biofilms formed by (b) wild-type, (c) ECM33 mutant, or (d) revertant strain. RML1-heterozygous Caecm33/CaECM33 mutant; RML2-homozygous Caecm33 mutant; RML3-heterozygous Caecm33/CaECM33 revertant; RML4-homozygous ECM33/ECM33 revertant.
3.2. Effect of the Caecm33 Mutants on the EHOM Structure
The noninfected EHOM showed a well-organized epithelial structure characterized by different epithelial cell layers and a fibroblast-populated lamina propria (Figure 2(a)). Following infection with the CAF2 (Figure 2(b)) and RML4-homozygous revertant strains (Figure 2(f)), the EHOM displayed a thinner, more disorganized epithelium comprised of differentiated cells (i.e., large vacuolated cells containing a large nucleus). The mutation of one Caecm33 allele partially prevented tissue damage (Figures 2(c) and 2(e)). The epithelium in the RML2-homozygous double Caecm33 mutant-infected EHOM (Figure 2(d)) continued to display a stratified appearance. Close examination of the EHOM-infected tissues revealed the formation of biofilms containing both forms of Candida (yeast and hyphae) on the outer layer of the epithelium (Figure 2). Biofilm analysis showed that the CAF2 and RML4-homozygous revertant Candida strains formed significant biofilms on the EHOM layers, resulting in disorganized tissue layers, whereas the RML1-heterozygous Caecm33 mutant, the RML2-homozygous double Caecm33 mutant, and the RML3-heterozygous revertant strains all formed less biofilms on the EHOM epithelium (Figure 2), thus causing less damage. Quantitative analysis revealed that the biofilms formed by the Caecm33 mutants were thinner than those formed by the CAF2 and RML4-homozygous revertant strains (Figure 3). Of interest is that the thinnest biofilms were obtained with the RML2-homozygous double Caecm33 mutant strain. These studies show that a disruption of the ECM33 gene resulted in a diminished ability of C. albicans to form biofilms (Figure 3) and to damage the host mucosal tissue.
Figure 2.
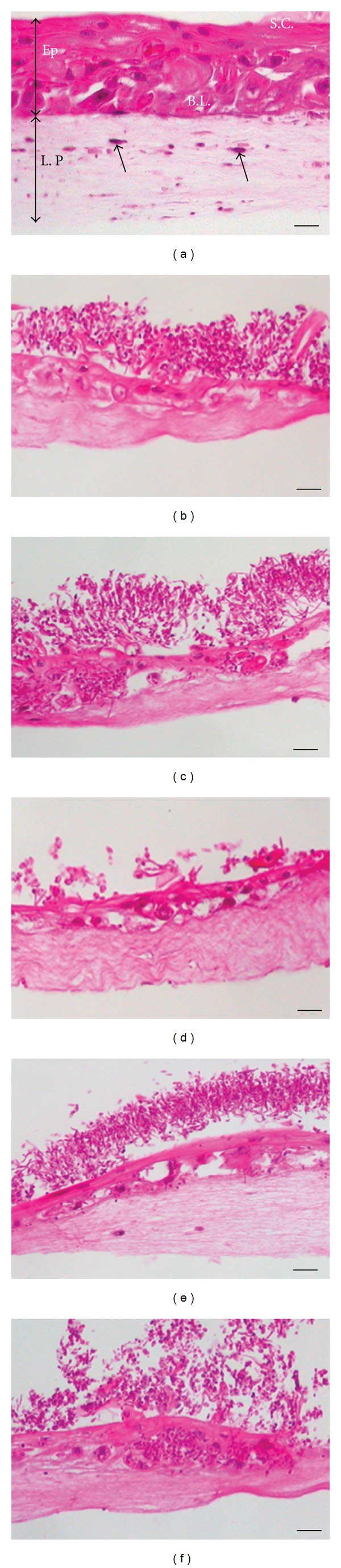
Interaction of ECM33 mutants with EHOM and biofilm formation. Histological features of the EHOM following 24 h of exposure to each C. albicans strain. (a) Noninfected EHOM tissue; (b) infected with CAF2 strain; (c) RML1; (d) RML2; (e) RML3; (f) RML4 C. albicans. The noninfected EHOM (a) displays an epithelium (Ep) containing a stratum corneum layer (SC), a basal layer (BL), and a collagen-containing lamina propria (LP) populated with fibroblasts (arrows). Scale bars, 30 μm. Representative photographs of three different experiments are shown (two EHOMs per experiment).
Figure 3.
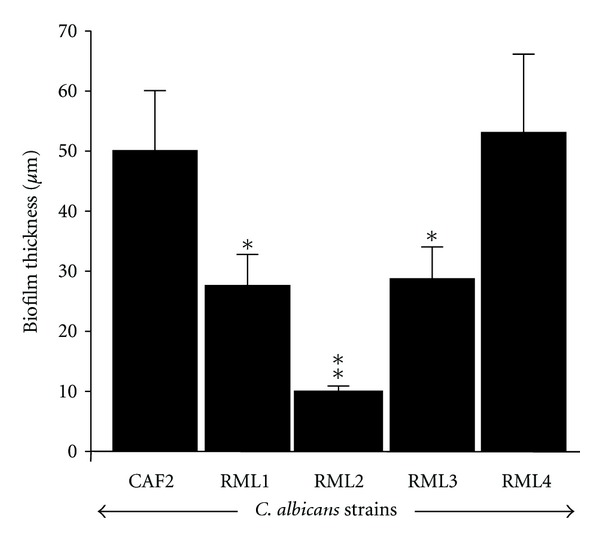
Effect of ECM33 gene on the ability of C. albicans to form biofilm on EHOM. Following infection with each Candida strain and culture for 24 h, a quantitative assessment of the thickness of the biofilm formed on the EHOM tissues was performed. Means + SD (n = 4) were plotted. Differences were obtained by comparing the biofilms obtained with the CAF2 strain and the other Candida strains. *P < 0.05; **P < 0.01.
3.3. Toxicity of the Caecm33 Mutants on the EHOM Tissue
As Caecm33 mutants led to less tissue damage and reduced biofilm formation on the EHOM, we investigated the effect of each Candida strain on the release of LDH by the EHOM cells. It was observed (Figure 4) that infection of the EHOM with the CAF2 strain and the RML4-homozygous revertant strain resulted in high levels of LDH in the medium, which is in agreement with our observations of the damaged epithelium (Figure 2) obtained with the CAF2 strain. It should be noted that each Caecm33 mutant negatively affected LDH release by the EHOM compared to that observed with the CAF2 strain (Figure 4).
Figure 4.
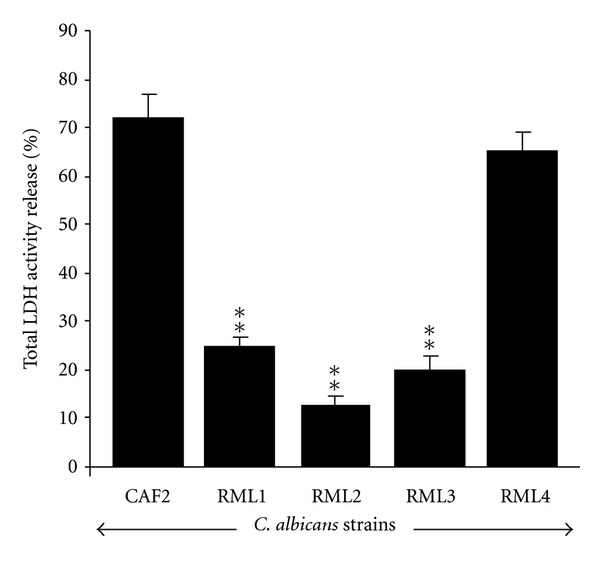
LDH release following EHOM infection with C. albicans strains. Culture media were collected from the noninfected and infected EHOMs. Supernatants were used to assess LDH release by means of specific kits. Data are means + SD, n = 5. **P < 0.01 compared to the control (CAF2-infected EHOM).
3.4. Apoptotic Gene Expression and Protein Production by the EHOM following Infection with Caecm33 Mutant C. albicans
The CAF2 and RML4-homozygous revertant strains significantly increased the Bax/Bcl2 gene expression ratio compared to that recorded by the noninfected EHOM. This was due to increased Bax gene expression which is an indicator of cell apoptosis. Following EHOM infection with the Caecm33 mutants, Bax gene expression significantly (P < 0.01) decreased compared to that observed with the CAF2-infected tissue (Figure 5(a)). Furthermore, Caecm33 double mutant recorded the lowest level of Bax gene expression. RT-PCR analyses of Bcl2 gene showed that its expression remained unchanged following tissue infection with all of the Candida strains under study (Figure 5(b)). These findings are supported by Bax and Bcl2 protein analyses using western blotting. Figure 6 shows that Bcl2 was unchanged; however, Bax protein was highly present in the EHOM infected with the CAF2 and RML4-homozygous strains compared to the noninfected and Caecm33 mutant-infected specimens. Overall data indicate that apoptotic genes and proteins' levels of expression increased when the tissues were infected with CAF2. Infection with the mutant strains nevertheless showed a reduction in apoptotic gene and protein expression, which may prevent mammalian cell death/apoptosis.
Figure 5.
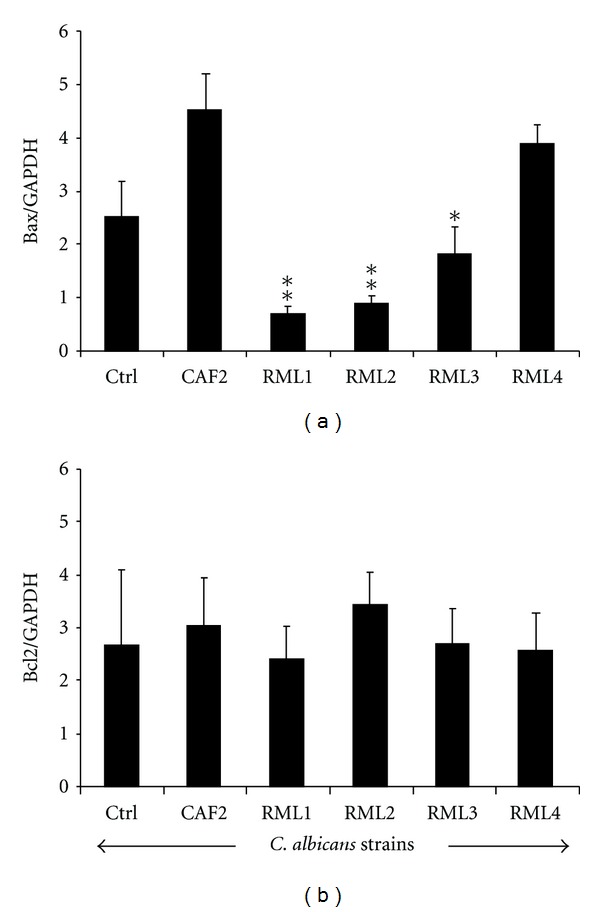
Bax and Bcl2 expression following EHOM infection with C. albicans strains. Total RNAs were extracted from the infected EHOMs and analyzed by RT-PCR for Bax and Bcl2 gene expression. Data are means + SD, n = 5. *P < 0.05; **P < 0.01 significance between the gene expression levels in the CAF2-infected and Caecm33 mutant-infected EHOMs.
Figure 6.
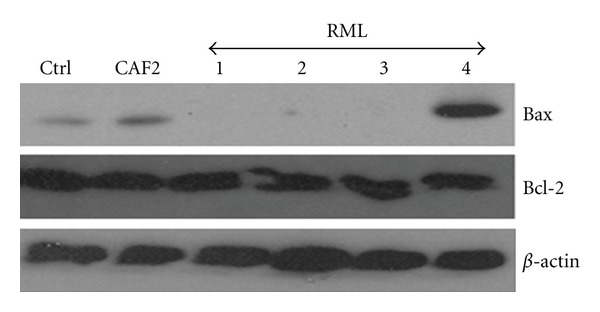
Bax and Bcl2 protein analyses following EHOM infection with C. albicans strains. Total proteins were extracted from the infected EHOMs and analyzed by western blotting for Bax and Bcl2 production. Gels are representative (n = 5).
3.5. The C. albicans Strains Modulated Laminin 5 and Type IV Collagen Synthesis
Western blot results show elevated basal levels of laminin 5 and type IV collagen proteins in the noninfected EHOM (Figure 7(a)), whereas both proteins were significantly (P < 0.05) down regulated in the infected tissues. It is important to note that all of the tested Candida strains significantly decreased laminin 5 and type IV collagen levels in the infected EHOM (Figure 7(b)) compared to the noninfected specimens. Thus ECM33 gene mutation did not prevent the laminin 5 and type IV collagen decreases.
Figure 7.
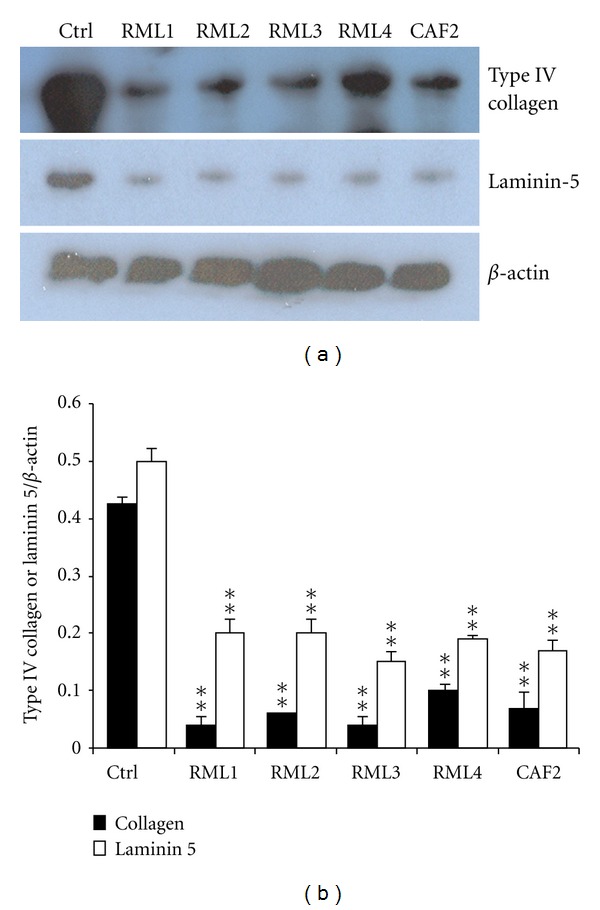
C. albicans downregulated basement membrane protein production. Total proteins were extracted from the infected EHOMs and analyzed by western blotting for the presence of laminin 5 and type IV collagen. (a) Representative gel of three separate experiments. Unstimulated tissue was used as a control (Ctrl). Protein production modulation was determined by band scanning using public domain NIH Image software. (b) Histograms are the means + SD (n = 4). Statistical differences were found by comparing the values obtained from the infected and noninfected tissues. **P < 0.01.
4. Discussion
C. albicans adhesion to various surfaces, including mucosal tissue and catheter disks, is the first step in biofilm formation and host infection [32, 33]. Our study demonstrates that the Candida adhesion to and biofilm formation on catheter discs were modulated by ECM33 gene, and that the disruption of this gene led to decreased biofilm formation, which supports previously reported data with medical devices [3, 34]. Thus adhesion and biofilm formation on a catheter that is in contact with host tissue may contribute to tissue disorganization and facilitate yeast penetration into the deep tissue and subsequent systemic invasion [35].
C. albicans adhesion to mammalian cells and tissue is under the control of various genes, such as ECM33. Using an EHOM model, we demonstrated that Caecm33 mutant was unable to damage tissue structure, compared to the CAF2 strains. This inability to cause tissue damage may be linked to a defect in the ability of Caecm33 mutant to form hyphae. Indeed, it was reported that Caecm33 mutants displayed filamentation defects in both liquid and solid media [17]. The inability of ECM33 mutant to damage the EHOM tissue may also be due to cell wall architectural changes that hamper interactions with host cell/tissue. Caecm33 mutants have in fact been shown to exhibit significant cell changes, including an abnormally electron-dense outer mannoprotein layer [17]. This architectural modification likely contributed to the reduced adherence level, decreased biofilm formation, and less extensive tissue damage observed in this study.
Caecm33 mutants were also less capable of promoting cell death/apoptosis by reducing LDH release and Bax gene expression. These findings support the reduced tissue disorganization when placed in contact with the Caecm33 mutants. Recent studies suggest that the decreased host cell damage caused by Caecm33 mutants is likely due in part to the reduced endocytosis of these strains [8]. Caecm33 mutant strains may also secrete less lytic enzymes, such as secreted aspartyl proteases and phospholipases [8], thus contributing to the host tissue cell damage defect of these mutants. Further studies are necessary to determine the mechanisms underlying the decreased virulence of the Caecm33 mutant strains.
EHOM disorganization following contact with the ECM33-positive gene may also modulate some physiological mechanisms such as BM protein production. Indeed, following contact with the CAF2 and Caecm33 mutant strains, both laminin 5 and type IV collagen levels decreased significantly. By decreasing BM protein production, Candida may thus deregulate epithelial and connective tissue structural interactions, thereby reducing their capacity to prevent its invasion [36]. It has been shown that Candida invasion requires adhesion to BM proteins as well as their degradation [21]. Although Candida is capable of degrading these proteins, this does not necessarily mean that Caecm33 mutant will, in turn, downregulate laminin 5 and type IV collagen protein production. The deletion of single or double Caecm33 alleles led to the same effect as that produced by the CAF2 strains on BM protein expression. This suggests that ECM33 mutation does not reduce the ability of Candida to decrease BM protein synthesis by gingival cells. Further research will be conducted to shed light on this possible mechanism.
In conclusion, we used an engineered human oral mucosa model to demonstrate that Caecm33 mutant was not able to damage tissue structure and promote cell apoptosis. We also demonstrated that biofilm formation was reduced by Caecm33 mutants compared to parental strains. Finally, we showed that parental and Caecm33 mutant strains downregulated laminin 5 and type IV collagen production. Overall data thus suggest that ECM33 gene plays an active role in Candida-host interactions and is responsible for tissue damage that ultimately leads to candidiasis.
Acknowledgments
This paper was supported by grants from the NIH/R01-DE017486-01A1, BRS-ACURE Q0600136 (OHARA), NIH/NIAID, R21-AI74077-01, and the NSERCs Discovery Program. The authors declare no potential conflicts of interests with respect to the authorship and/or publication of this paper.
References
- 1.Shorr AF, Gupta V, Sun X, Johannes RS, Spalding J, Tabak YP. Burden of early-onset candidemia: analysis of culture-positive bloodstream infections from a large U.S. database. Critical Care Medicine. 2009;37(9):2519–2526. doi: 10.1097/CCM.0b013e3181a0f95d. [DOI] [PubMed] [Google Scholar]
- 2.Cannon RD, Chaffin WL. Oral colonization by Candida albicans . Critical Reviews in Oral Biology and Medicine. 1999;10(3):359–383. doi: 10.1177/10454411990100030701. [DOI] [PubMed] [Google Scholar]
- 3.Naglik JR, Moyes D. Epithelial cell innate response to Candida albicans . Advances in dental research. 2011;23(1):50–55. doi: 10.1177/0022034511399285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rouabhia M, Mukherjee PK, Lattif AA, Curt S, Chandra J, Ghannoum MA. Disruption of sphingolipid biosynthetic gene IPT1 reduces Candida albicans adhesion and prevents activation of human gingival epithelial cell innate immune defense. Medical Mycology. 2011;49(5):458–466. doi: 10.3109/13693786.2010.535031. [DOI] [PubMed] [Google Scholar]
- 5.Chaffin WL, López-Ribot JL, Casanova M, Gozalbo D, Martínez JP. Cell wall and secreted proteins of Candida albicans: identification, function, and expression. Microbiology and Molecular Biology Reviews. 1998;62(1):130–180. doi: 10.1128/mmbr.62.1.130-180.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Groot PW, De Boer AD, Cunningham J, et al. Proteomic analysis of Candida albicans cell walls reveals covalently bound carbohydrate-active enzymes and adhesins. Eukaryotic Cell. 2004;3(4):955–965. doi: 10.1128/EC.3.4.955-965.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis RE. Overview of the changing epidemiology of candidemia. Current Medical Research and Opinion. 2009;25(7):1732–1740. doi: 10.1185/03007990902990817. [DOI] [PubMed] [Google Scholar]
- 8.Martinez-Lopez R, Park H, Myers CL, Gil C, Filler SG. Candida albicans Ecm33p is important for normal cell wall architecture and interactions with host cells. Eukaryotic Cell. 2006;5(1):140–147. doi: 10.1128/EC.5.1.140-147.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cherepanova OA, Kalmykova N, Petrov YP, Blinova M, Pinaev G. Contribution of α2β1, α3β1, α6β4 integrins and 67 kDa laminin receptor to the interaction of epidermoid carcinoma A-431 cells with laminin-2/4. Cell Biology International. 2006;30(10):784–792. doi: 10.1016/j.cellbi.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 10.Merker HJ. Morphology of the basement membrane. Microscopy Research and Technique. 1994;28(2):95–124. doi: 10.1002/jemt.1070280203. [DOI] [PubMed] [Google Scholar]
- 11.Yurchenco PD. Basement membranes: cell scaffoldings and signaling platforms. Cold Spring Harbor Perspectives in Biology. 2011;3(2) doi: 10.1101/cshperspect.a004911. Article ID a004911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sasaki T, Fässler R, Hohenester E. Laminin: the crux of basement membrane assembly. Journal of Cell Biology. 2004;164(7):959–963. doi: 10.1083/jcb.200401058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sacca B, Sinner EK, Kaiser J, Lubken C, Eble JA, Moroder L. Binding and docking of synthetic heterotrimeric collagen type IV peptides with alpha1beta1 integrin. ChemBioChem. 2002;3(9):904–907. doi: 10.1002/1439-7633(20020902)3:9<904::AID-CBIC904>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 14.Rousselle P, Golbik R, Van der Rest M, Aumailley M. Structural requirement for cell adhesion to kalinin (laminin-5) Journal of Biological Chemistry. 1995;270(23):13766–13770. doi: 10.1074/jbc.270.23.13766. [DOI] [PubMed] [Google Scholar]
- 15.Colognato H, Yurchenco PD. Form and function: the laminin family of heterotrimers. Developmental Dynamics. 2000;218(2):213–234. doi: 10.1002/(SICI)1097-0177(200006)218:2<213::AID-DVDY1>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 16.Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans . Genetics. 1993;134(3):717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez-Lopez R, Monteoliva L, Diez-Orejas R, Nombela C, Gil C. The GPI-anchored protein CaEcm33p is required for cell wall integrity, morphogenesis and virulence in Candida albicans . Microbiology. 2004;150(10):3341–3354. doi: 10.1099/mic.0.27320-0. [DOI] [PubMed] [Google Scholar]
- 18.Calderone R, Diamond R, Senet JM, Warmington J, Filler S, Edwards JE. Host cell-fungal cell interactions. Journal of Medical and Veterinary Mycology, Supplement. 1994;32(1):151–168. doi: 10.1080/02681219480000801. [DOI] [PubMed] [Google Scholar]
- 19.Nomanbhoy F, Steele C, Yano J, Fidel PL., Jr. Vaginal and oral epithelial cell anti-Candida activity. Infection and Immunity. 2002;70(12):7081–7088. doi: 10.1128/IAI.70.12.7081-7088.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rouabhia M, Ross G, Pagé N, Chakir J. Interleukin-18 and gamma interferon production by oral epithelial cells in response to exposure to Candida albicans or lipopolysaccharide stimulation. Infection and Immunity. 2002;70(12):7073–7080. doi: 10.1128/IAI.70.12.7073-7080.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Claveau I, Mostefaoui Y, Rouabhia M. Basement membrane protein and matrix metalloproteinase deregulation in engineered human oral mucosa following infection with Candida albicans . Matrix Biology. 2004;23(7):477–486. doi: 10.1016/j.matbio.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 22.Villar CC, Zhao XR. Candida albicans induces early apoptosis followed by secondary necrosis in oral epithelial cells. Molecular Oral Microbiology. 2010;25(3):215–225. doi: 10.1111/j.2041-1014.2010.00577.x. [DOI] [PubMed] [Google Scholar]
- 23.Boise LH, Gonzalez-Garcia M, Postema CE, et al. Bcl-x, A bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell. 1993;74(4):597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- 24.Kanduc D, Mittelman A, Serpico R, et al. Cell death: apoptosis versus necrosis. International Journal of Oncology. 2002;21(1):165–170. [PubMed] [Google Scholar]
- 25.Lian LH, Milora KA, Manupipatpong KK, Jensen LE. The double-stranded RNA analogue polyinosinic-polycytidylic acid induces keratinocyte pyroptosis and release of IL-36γ . Journal of Investigative Dermatology. 2012;132(5):1346–1353. doi: 10.1038/jid.2011.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsujimoto Y. Multiple ways to die: non-apoptotic forms of cell death. Acta Oncologica. 2012;51(3):293–300. doi: 10.3109/0284186X.2011.648340. [DOI] [PubMed] [Google Scholar]
- 27.Thompson CB. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267(5203):1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- 28.Chandra J, Mukherjee PK, Ghannoum MA. In vitro growth and analysis of Candida biofilms. Nature protocols. 2008;3(12):1909–1924. doi: 10.1038/nprot.2008.192. [DOI] [PubMed] [Google Scholar]
- 29.Rouabhia M, Deslauriers N. Production and characterization of an in vitro engineered human oral mucosa. Biochemistry and Cell Biology. 2002;80(2):189–195. doi: 10.1139/o01-237. [DOI] [PubMed] [Google Scholar]
- 30.Mukherjee PK, Mohamed S, Chandra J, et al. Alcohol dehydrogenase restricts the ability of the pathogen Candida albicans to form a biofilm on catheter surfaces through an ethanol-based mechanism. Infection and Immunity. 2006;74(7):3804–3816. doi: 10.1128/IAI.00161-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Semlali A, Jacques E, Rouabhia M, Milot J, Laviolette M, Chakir J. Regulation of epithelial cell proliferation by bronchial fibroblasts obtained from mild asthmatic subjects. Allergy. 2010;65(11):1438–1445. doi: 10.1111/j.1398-9995.2010.02376.x. [DOI] [PubMed] [Google Scholar]
- 32.Chandra J, Mukherjee PK, Leidich SD, et al. Antifungal resistance of Candidal biofilms formed on denture acrylic in vitro . Journal of Dental Research. 2001;80(3):903–908. doi: 10.1177/00220345010800031101. [DOI] [PubMed] [Google Scholar]
- 33.Hawser SP, Douglas LJ. Biofilm formation by Candida species on the surface of catheter materials in vitro . Infection and Immunity. 1994;62(3):915–921. doi: 10.1128/iai.62.3.915-921.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nett JE, Lepak AJ, Marchillo K, Andes DR. Time course global gene expression analysis of an in VivoCandida biofilm. Journal of Infectious Diseases. 2009;200(2):307–313. doi: 10.1086/599838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mostefaoui Y, Claveau I, Rouabhia M. In vitro analyses of tissue structure and interleukin-1β expression and production by human oral mucosa in response to Candida albicans infections. Cytokine. 2004;25(4):162–171. doi: 10.1016/j.cyto.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 36.Pärnänen P, Meurman JH, Virtanen I. Laminin-511 and fibronectin degradation with Candida yeast. Journal of Oral Pathology and Medicine. 2009;38(10):768–772. doi: 10.1111/j.1600-0714.2009.00785.x. [DOI] [PubMed] [Google Scholar]


