Abstract
Microbial communities in the lungs of patients with cystic fibrosis (CF) and chronic obstructive pulmonary disease (COPD) have been shown to be spatially heterogeneous. Viral communities may also vary spatially, leading to localized viral populations and infections. Here, we characterized viral communities from multiple areas of the lungs of two patients with late-stage CF using metagenomics, that is, the explanted lungs from a transplant patient and lungs acquired postmortem. All regions harbored eukaryotic viruses that may infect the human host, notably herpesviruses, anelloviruses, and papillomaviruses. In the highly diseased apical lobes of explant lungs, viral diversity was extremely low, and only eukaryotic viruses were present. The absence of phage suggests that CF-associated microbial biofilms may escape top-down controls by phage predation. The phages present in other lobes of explant lungs and in all lobes of postmortem lungs comprised distinct communities, and encoded genes for clinically important microbial phenotypes, including small colony variants and antibiotic resistance. Based on the these observations, we postulate that viral communities in CF lungs are spatially distinct and contribute to CF pathology by augmenting the metabolic potential of resident microbes, as well as by directly damaging lung tissue via carcinomas and herpesviral outbreaks.
Keywords: cystic fibrosis, viruses, metagenomics, antibiotic resistance
Cystic fibrosis (CF) is the most common fatal autosomal recessive disorder in white populations. Radiographic and immunological studies demonstrated the spatial heterogeneity of disease severity and clinical manifestations in the CF lung (1, 2). The apical lobes typically suffer the most extensive deterioration and inflammatory disease; the basal lobes exhibit more air trapping (1–3). Recent studies of lung tissue from patients with CF and chronic obstructive pulmonary disease (COPD) demonstrated that microbial communities vary over the spatial landscape of the diseased lung (4, 5). However, these studies did not attempt to profile viral communities in lung tissue.
Here we report on the spatial heterogeneity of viral communities within two sets of CF lungs: one pair of explanted lungs (i.e., the discarded lung tissue after a lung transplant) and one pair of postmortem lungs. Viral communities were isolated and sequenced directly from lung tissue from spatially distinct areas. This approach yielded deep sequencing data about a limited number of samples, presented here as two case studies. The resulting viromes showed significant spatial heterogeneity, indicating that common sampling methods (e.g., induced sputum) cannot reveal the full nature of CF lung disease. In addition, phage communities in the lungs may serve as a reservoir for antibiotic resistance genes, complicating therapeutic regimens.
Materials and Methods
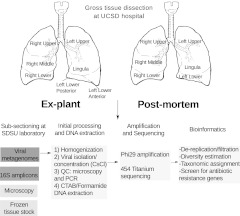
All samples and patient data were collected in accordance with the University of California Institutional Review Board (HRPP 081500) and the San Diego State University Institutional Review Board (SDSU IRB 2121). All tissues were collected, processed, and disposed of as described in the San Diego State University Biological Use Authorization (BUA 09-03-001R). Figure 1 summarizes the procedures used in this study. A more detailed discussion appears in the online supplement.
Figure 1.
Flow chart for lung dissection and subsequent sample processing. PCR, polymerase chain reaction; SDSU, San Diego State University; UCSD, University of California at San Diego.
Viral metagenomes were submitted to the National Center for Biotechnology Information (NCBI) under Genome Project Identification Number 66313. Characteristics of the sequenced metagenomes appear in Table E1 of the online supplement. Initial taxonomic assignments were made by comparison to the nonredundant database at the NCBI. Viral taxonomy was assigned based on tBLASTx similarities to a custom database of all viral genomes from the NCBI and the phage proteomic tree (http://www.phantome.org/) (E-value, <10−5). Virome sequences were compared with the Antibiotic Resistance Database, using BLASTx (e-value, <10−5 over >80% of the query length) (7).
Metagenomic reads were mapped to genomes using BLAT and coverage plots generated using Circos and the Integrated Genome Browser (8, 9). Contigs were assembled with CAP3 (10). Open reading frames in contigs were determined using Prodigal (11). Phylogenetic analyses were performed using PhyML and MrBayes version 3.1 (12, 13). The human papillomavirus (HPV), torque teno virus (TTV), and multidrug efflux pump (MexF) contigs were submitted to Genbank under accession numbers JN231328, JN231329, and JN231330.
Species richness was estimated using Phage Communities from Contig Spectra (PHACCs), and diversity between viromes was compared using Maxiphi (14, 15). MaxiPhi results were combined using nonmetric multidimensional scaling, with the percentage of nonshared genotypes as the distance metric. Statistically significant differences were determined empirically (please see Methods in the online supplement for further details).
Results
Clinical Background
Chest radiographic studies from the explant lung donor showed classic features of CF, with more severe disease occurring in the apical lobes (see Figure E1 in the online supplement). The CAT scan of the donor at age 44 years, 2 months before transplantation, showed extensive mucus plugging and cystic and bronchiectatic changes that were most severe in the upper lobes (Figures E1B and E1C). For purposes of comparison, the posttransplant image of the newly implanted non-CF lungs is also depicted in Figure E1D. In contrast, the postmortem lung donor's CAT scans showed extensive bronchiectasis and severe mucus plugging in all regions (Figures E1F–E1H). A limited postmortem examination of the chest revealed that the central airways, including the trachea, were almost completely obstructed with thick, purulent mucus.
Sputum cultures showed that both patients were chronically colonized with multiply antibiotic-resistant mucoid and nonmucoid strains of Pseudomonas aeruginosa (Figures E1A and E2E), as well as fungi. Both patients were previously culture-positive for Streptococcus spp., Micrococcus spp., and nonfermenting Gram-negative rods. Sputum samples from the explant donor also grew Klebsiella ozaenae, Stenotrophomonas maltophilia, and Achromobacter spp. Sputum cultures from the postmortem donor were intermittently positive for methicillin-sensitive Staphylococcus aureus and diphtheroids.
Both patients experienced intermittent pulmonary exacerbations over the course of their available history, and demonstrated a decline in FEV1% over time, typical of patients with CF (Figures E1A and E1E). The explant donor was routinely treated with ceftazidime and tobramycin (Figure E1A). Piperacillin was administered during the year before transplantation. In addition, the postmortem patient experienced periodic pancreatitis and was routinely treated with tobramycin and meropenem (Figure E1E), and intermittently with ceftazidime, ciprofloxacin, nafcillin, and piperacillin.
Characterization of Viral Communities in CF Lungs
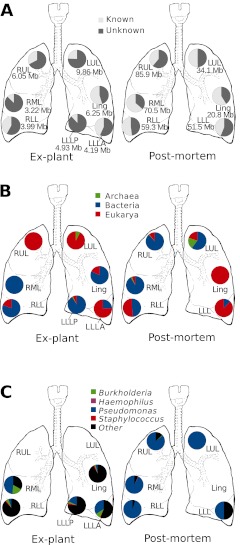
Sections were excised from both lungs to sample the right upper, middle, and lower lobes, as well as the left upper and lower lobes and lingula (Figure 1). The resultant 1.25 million reads, representing 360 megabase pairs of sequences, were distributed among the various lobes, as shown in Figure 2A. The virome sequences were compared with the nonredundant database at NCBI, using tBLASTx (e-value, <10−5). Between 36–88% of the virome sequences were unknown, an amount is comparable to the percentage of unknown sequences reported for other viral metagenomes (6, 16) (Figure 2A). In total, 138 viral taxa were identified in the explant lungs. Individual lobes each contained 6–53 taxa (Figure E2B). Similarly, 72 viral taxa were detected in the postmortem lung, with 5–45 taxa identified in each lobe (Figure E2B). The domain of the host for each of the identified viral taxa was inferred from their best BLAST similarity (Figure 2B).
Figure 2.
General characteristics of lung viromes. (A) Number of base pairs of sequenced viral DNA per lobe, and percentage of known/unknown sequences in each lobe. (B) Predicted host domains for known sequences in each lobe are shown as the percentage of annotated viral sequences attributed to a particular domain. (C) Predicted hosts of phages identified in each lobe. Ling, lingula; LLL, left lower lobe; LLLA, left lower lobe, anterior; LLLP, left lower lobe, posterior; LUL, left upper lobe; RLL, right lower lobe; RML, right middle lobe; RUL, right upper lobe.
Most of the viruses identified were classified as phages (i.e., viruses that infect bacteria) (Figure 2A). Phages of known CF pathogens were identified in all lobes of the explant lungs, except for the left upper lobe (LUL) and right upper lobe (RUL) (Figures 2C and E2A), where no phage could be detected. Phages of Pseudomonas spp. and Burkholderia spp. were found in all other lobes. Staphylococcus phages were found in the lingula and basal lobes only. In the postmortem lungs, where microbial communities were dominated by P. aeruginosa (5), most of the phages identified were related to known Pseudomonas phages (Figures 2C and E2A). The right middle lobe (RML) virome contained sequences with significant similarity to phage Pf1, a filamentous phage associated with the small colony variant phenotype of P. aeruginosa (17) (Figure E4). In addition, phages of Burkholderia spp. and Haemophilus influenzae were identified in the RUL and RML, respectively.
Viruses that infect eukaryotic cells were also found in both pairs of lungs. In the explant lungs, herpesviruses were identified in four lobes. Sequences similar to adenoviruses were found in the lower lobes and lingula (Figure E2A). In the postmortem lungs, all lobes contained herpesvirus sequences. Adenoviruses were identified in the RUL and RML (Figure E2A).
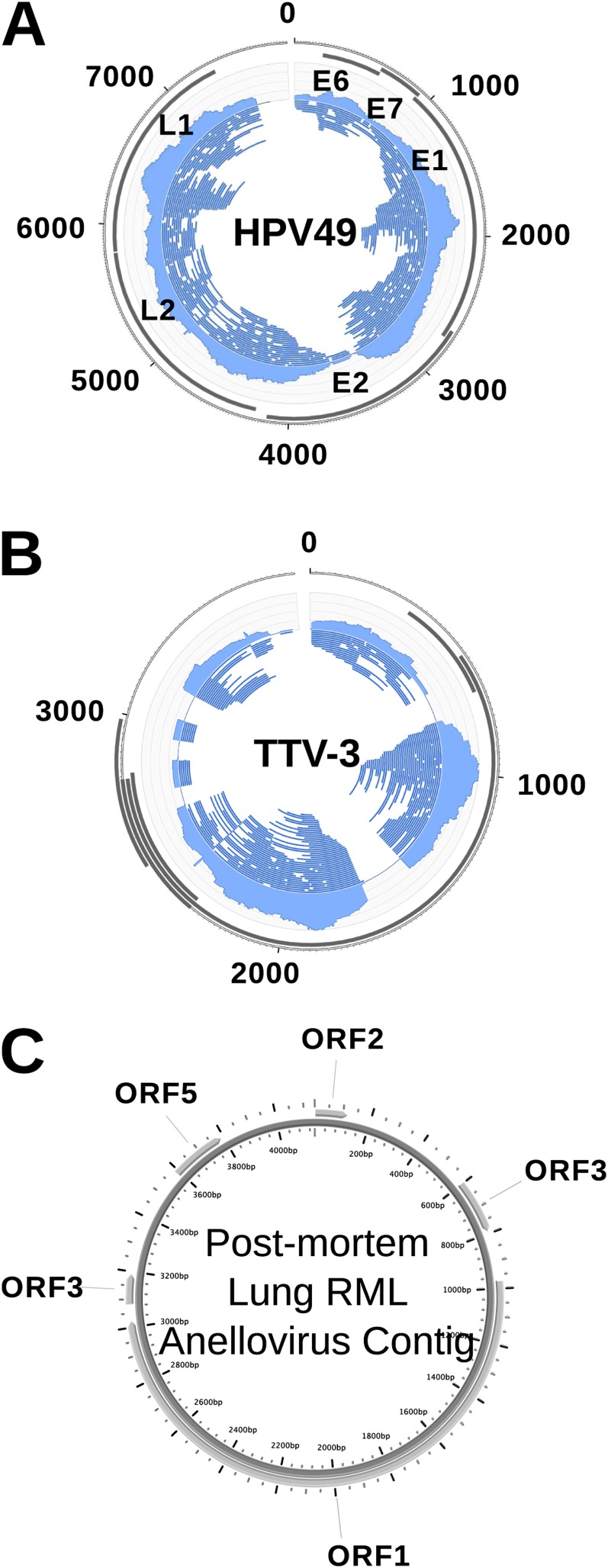
During dissection of the explant lung, cysts were apparent in the anterior portion of the left lower lobe (LLLA). The virome sequences from the LLLA yielded an approximately ×15 coverage of the HPV49 genome (Figure 3A). Assembly yielded an approximately 7.5-kb contig that was identical to the HPV49 genome over 99% of its length. Papillomaviruses were not detected in any other lobe of either set of lungs.
Figure 3.
Eukaryotic viruses in the cystic fibrosis lung. (A) Human papillomavirus (HPV) genome. The left lower lobe of the explant lung provided an approximately ×15 coverage of HPV49. (B) Torque teno virus (TTV), with an approximately ×14 coverage in the virome from the right middle lobe (RML) of the postmortem lung. (C) Anellovirus. ORF, open reading frame.
Sequences with tBLASTx similarities to TTV were identified in the RML of the postmortem lung. These sequences provided an approximately ×14 coverage of a TTV-like genome (Figure 3B). Assembly yielded a 4.2-kb contig that was most similar at the nucleotide level to TTV3 (BLASTn; E-value, 0.0; 87% similarity over the entire contig length). This TTV-like contig was predicted to contain five open reading frames (ORFs), according to positive strand translation (Figure 3C). A phylogenetic analysis of the predicted ORF1 confirmed that this virus was most closely related to TTV3 (Figure E3).
Spatial Distribution of Viral Diversity
Viral community diversity was estimated using PHACCS, a computational approach that does not rely on similarities to known sequences (14). The apical lobes of the explanted lungs demonstrated an extremely low number of viral genotypes, that is, approximately three genotypes per lobe (Figure E5B). This is the lowest viral diversity observed to date in any ecosystem. In contrast, the basal lobes were estimated to contain on the order of 102 viral genotypes. This is consistent with the identification of far fewer viral taxa in the apical lobes than in the rest of the lung. In the postmortem lungs, viral diversity was more evenly distributed, with 14–105 predicted viral genotypes per lobe (Figure E5B).
The MaxiPhi program was used to estimate the interlobe similarities within both the explant and postmortem lungs (15). Multidimensional scaling, based on the percentage of viral genotypes shared, revealed that viral communities in spatially closer lobes were more similar than those in anatomically distant lobes (Figure E5 and Table E2). Overall, lobes of the postmortem lungs shared a higher proportion of viral genotypes than those in the explant lungs, and fewer lobes were significantly different from each other (Table E2).
Identification and Spatial Distribution of Antibiotic Resistance Genes
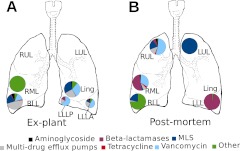
Antibiotic resistance (AR) genes were identified in the viromes by comparison to the Antibiotic Resistance Database (7). In the explant lungs, no sequences similar to AR genes were found in the RUL or LUL, the two lobes where no phage sequences were detected (Figure 4A). In the other lobes, less than 1–2% of the total virome showed a similarity to AR genes. The LLLA was enriched in sequences corresponding to aminoglycoside resistance and Class B β-lactamases (metallo–β-lactamases). In the right lower lobe (RLL), most AR sequences were related to resistance–nodulation–cell division (RND) multidrug efflux pumps, including two components of the P. aeruginosa MexE-MexF-OprN RND pump for fluoroquinolones and chloramphenicol. Metagenomic sequences from the explant RLL were assembled, and the resultant 3-kb contig contained a complete mexF gene sequence. Phylogenetic analysis indicated that this contig was most similar to the MexF transporter of Legionella pneumophila strains Corby and Philadelphia (Figure E6). The mexF gene is located in a 100-kb region of the L. pneumophila genome, termed an “efflux island” that also contains phage, transposon, and transfer RNA genes, suggesting this island is mobile and can potentially undergo phage-mediated lateral gene transfer (18).
Figure 4.
Spatial distribution of antibiotic resistance genes in the viromes from the explant (A) and postmortem (B) lungs.
In the postmortem lungs, AR sequences were found in all lobes (except the lingula), and represented 1–27% of total viromes (Figure 4B). Nearly all of the putative AR genes in the LUL were similar to ribosomal RNA [adenine-N-6-]-methyltransferase (ermC), which confers resistance to macrolide-lincosamide-streptogramin antibiotics. No sequences similar to β-lactamases were identified in the LUL, whereas β-lactamases accounted for most of the AR sequences in the RML and left lower lobe. This patient had been treated with a variety of β-lactam antibiotics that could have driven the acquisition of a larger repertoire of resistance genes via lateral gene transfer (19).
Discussion
Viral Diversity and CF Lung Disease
Previous studies found that regional differences in the severity of CF lung disease are more pronounced in stable patients, and that the apical regions are the most affected (2). Consistent with this finding, the apical lobes in the explant lungs suffered the most severe disease (Figures 2B and 2C). These lobes also demonstrated the lowest viral diversity, and harbored no detectable phages. The polysaccharide matrix of biofilms has been hypothesized to present a barrier that protects susceptible bacteria from top-down control by phage predation (20). Our data support this hypothesis, and suggest that the apical lobes of patients with stable CF may house a unique viral community characterized by extremely low richness and the absence of phage.
The less diseased basal lobes and lingula of the explant lungs demonstrated higher viral diversity, including phages of known CF pathogens, thus suggesting that some phage predation occurs in these more functional regions. The transitory binding of environmental viruses probably accounts for the greater diversity observed and the presence of more viral genotypes not shared with other lobes. The binding may be attributable to increased air trapping in the basal lobes, as previously observed (1). The severity of disease in the postmortem lungs was more similar throughout, accompanied by pervasive viral infection and a more uniform distribution of viral taxa. A recent study using sputum samples demonstrated that microbial species richness is correlated with lung function, and that CF-associated microbial communities consist of core (i.e., resident) and satellite (i.e., transient) taxa (21). Our results indicate that the same model could apply to viral communities in CF lung tissue.
Viral Infections of Host Tissue
Both sets of lungs exhibited disseminated herpesvirus infection as well as other highly localized eukaryotic viral infections. Herpesviruses were detected in CF and non-CF lung tissue, and were previously identified as core constituents of the respiratory virome (15, 22). Although herpesviruses have been linked to complications in lung transplant patients, the roles they play in the progression of CF lung disease, if any, are largely unknown (23).
Infection with HPV49 was observed in the LLLA of the explant lung in conjunction with evident cysts. HPV was not found in any other sample. Papillomaviruses are ubiquitous human commensals, but also cause diseases of the skin, genital tract, and respiratory tract (24). HPV infections in the airways can lead to recurrent respiratory papillomatosis (RRP), which in rare cases leads to cysts in the lung itself, especially in the lower lobes (25). Papillomaviruses were detected in lung carcinomas in several studies (26), but causal associations have not been determined. HPV49 was not previously linked to lung carcinomas or RRP (27). However, the unique immunological state of the CF lung might allow for infection, persistence, and cyst formation by nonrespiratory papillomaviruses, and the explant patient may have had an undiagnosed papilloma or carcinoma.
The RML of the postmortem lungs contained sequences that were most closely related to TTV3. Although TTV and other anelloviruses were found to be associated with respiratory disease, bronchiectasis, and loss of lung function, whether TTV plays a specific role in CF remains unknown (28).
Spatial Heterogeneity of Phage-Encoded Antibiotic Resistance
Phages were previously implicated in the rampant transfer of antibiotic resistance genes in vivo, and the transduction of antibiotic resistance by phages has also been demonstrated in vitro (29, 30). The varied distribution of phage-encoded antibiotic resistance genes observed here demonstrates that CF lungs are spatially heterogenous with respect to the presence of clinically relevant factors. Previous studies found that treatment based on the common practice of randomly picking bacterial colonies cultured from sputum and assessing their antibiotic resistance is often unsuccessful (31). In addition, the dispersal of aerosolized antibiotics such as tobramycin was shown to be uneven in the lungs, which may create isolated antibiotic-resistant niches (32). Phages likely maintain a reservoir of antibiotic-resistance genes that can be mobilized and disseminated. Notably, the administration of antibiotics has been shown to induce phage and phage-mediated lateral gene transfer (33). Thus, even if infection by a particular host is cleared from a lobe, phage may reintroduce antibiotic-resistant genes from other regions or hosts.
Supplementary Material
Acknowledgments
The authors are grateful to Derek Vosten for providing the artwork in Figures 2 and 4.
Footnotes
This work was supported by grant #09-002 from Cystic Fibrosis Research Incorporated and NIH R01 GM095384-01.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2011-0253OC on October 6, 2011
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Brody AS, Klein JS, Molina PL, Quan J, Bean JA, Wilmott RW. High-resolution computed tomography in young patients with cystic fibrosis: distribution of abnormalities and correlation with pulmonary function tests. J Pediatr 2004;145:32–38 [DOI] [PubMed] [Google Scholar]
- 2.Meyer K, Sharma A. Regional variability of lung inflammation in cystic fibrosis. Am J Respir Crit Care Med 1997;156:1536–1540 [DOI] [PubMed] [Google Scholar]
- 3.Gurney JW, Habbe TG, Hicklin J. Distribution of disease in cystic fibrosis. Chest 1997;112:357–362 [DOI] [PubMed] [Google Scholar]
- 4.Erb-Downward JR, Thompson DL, Han MK, Freeman CM, McCloskey L, Schmidt LA, Young VB, Toews GB, Curtis JL, Sundaram B, et al. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS ONE 2011;6:e16384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Willner D, Haynes M, Furlan M, Schmieder R, Lim YW, Rainey P, Rohwer F, Conrad D. Spatial distribution of microbial communities in the cystic fibrosis lung. ISME J (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu B, Pop M. ARDB: Antibiotic Resistance Genes Database. Nucleic Acids Res 2009;37:D443–D447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krzywinski M, Schein J, Birol İ, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res 2009;19:1639–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nicol JW, Helt GA, Blanchard SG, Raja A, Loraine AE. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics 2009;25:2730–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang X, Madan A. CAP3: a DNA sequence assembly program. Genome Res 1999;9:868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hyatt D, Chen G-L, LoCascio P, Land M, Larimer F, Hauser L. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 2010;11:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 2003;52:696–704 [DOI] [PubMed] [Google Scholar]
- 12.Huelsenbeck JP, Ronquist F. MRBAYES: bayesian inference of phylogenetic trees. Bioinformatics 2001;17:754–755 [DOI] [PubMed] [Google Scholar]
- 13.Angly F, Rodriguez-Brito B, Bangor D, McNairnie P, Breitbart M, Salamon P, Felts B, Nulton J, Mahaffy J, Rohwer F. PHACCS, an online tool for estimating the structure and diversity of uncultured viral communities using metagenomic information. BMC Bioinformatics 2005;6:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, Gordon JI. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 2010;466:334–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willner D, Furlan M, Haynes M, Schmieder R, Angly FE, Silva J, Tammadoni S, Nosrat B, Conrad D, Rohwer F. Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PLoS ONE 2009;4:e7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dinsdale EA, Edwards RA, Hall D, Angly F, Breitbart M, Brulc JM, Furlan M, Desnues C, Haynes M, Li L, et al. Functional metagenomic profiling of nine biomes. Nature 2008;452:629–632 [DOI] [PubMed] [Google Scholar]
- 17.Webb JS, Lau M, Kjelleberg S. Bacteriophage and phenotypic variation in Pseudomonas aeruginosa biofilm development. J Bacteriol 2004;186:8066–8073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chien M, Morozova I, Shi S, Sheng H, Chen J, Gomez SM, Asamani G, Hill K, Nuara J, Feder M, et al. The genomic sequence of the accidental pathogen Legionella pneumophila. Science 2004;305:1966–1968 [DOI] [PubMed] [Google Scholar]
- 19.Payne DJ, Du W, Bateson JH. Beta-lactamase epidemiology and the utility of established and novel beta-lactamase inhibitors. Expert Opin Investig Drugs 2000;9:247–261 [DOI] [PubMed] [Google Scholar]
- 20.Doolittle MM, Cooney JJ, Caldwell DE. Tracing the interaction of bacteriophage with bacterial biofilms using fluorescent and chromogenic probes. J Ind Microbiol 1996;16:331–341 [DOI] [PubMed] [Google Scholar]
- 21.van der Gast CJ, Walker AW, Stressmann FA, Rogers GB, Scott P, Daniels TW, Carroll MP, Parkhill J, Bruce KD. Partitioning core and satellite taxa from within cystic fibrosis lung bacterial communities. ISME J 2011;5:780–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costa C, Delsedime L, Solidoro P, Curtoni A, Bergallo M, Libertucci D, Baldi S, Rinaldi M, Cavallo R. Herpesviruses detection by quantitative real-time polymerase chain reaction in bronchoalveolar lavage and transbronchial biopsy in lung transplant: viral infections and histopathological correlation. Transplant Proc 2010;42:1270–1274 [DOI] [PubMed] [Google Scholar]
- 23.Puchhammer-Stöckl E. Herpesviruses and the transplanted lung: looking at the air side. J Clin Virol 2008;43:415–418 [DOI] [PubMed] [Google Scholar]
- 24.Antonsson A, Forslund O, Ekberg H, Sterner G, Hansson BG. The ubiquity and impressive genomic diversity of human skin papillomaviruses suggest a commensalic nature of these viruses. J Virol 2000;74:11636–11641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruan S-Y, Chen K-Y, Yang P-C. Recurrent respiratory papillomatosis with pulmonary involvement: a case report and review of the literature. Respirology 2009;14:137–140 [DOI] [PubMed] [Google Scholar]
- 26.Klein F, Amin Kotb WFM, Petersen I. Incidence of human papilloma virus in lung cancer. Lung Cancer 2009;65:13–18 [DOI] [PubMed] [Google Scholar]
- 27.Berkhout R, Tieben L, Smits H, Bavinck J, Vermeer B, ter Schegget J. Nested PCR approach for detection and typing of epidermodysplasia verruciformis–associated human papillomavirus types in cutaneous cancers from renal transplant recipients. J Clin Microbiol 1995;33:690–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pifferi M, Maggi F, Caramella D, De Marco E, Andreoli E, Meschi S, Macchia P, Bendinelli M, Boner AL. High torquetenovirus loads are correlated with bronchiectasis and peripheral airflow limitation in children. Pediatr Infect Dis J 2006;25:804–808 [DOI] [PubMed] [Google Scholar]
- 29.Blahová J, Králiková K, Krcméry V, Schäfer V. Bacteriophages transducing antibiotic resistance from a cluster of lysogenic strains of Pseudomonas aeruginosa isolated from patients. J Chemother 2001;13:331–333 [DOI] [PubMed] [Google Scholar]
- 30.Colomer-Lluch M, Jofre J, Muniesa M. Antibiotic resistance genes in the bacteriophage DNA fraction of environmental samples. PLoS ONE 2011;6:e17549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Foweraker JE, Laughton CR, Brown DF, Bilton D. Comparison of methods to test antibiotic combinations against heterogeneous populations of multi-resistant Pseudomonas aeruginosa from patients with acute infective exacerbations in cystic fibrosis. Antimicrob Agents Chemother 2009;53:4809–4815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mukhopadhyay S, Staddon GE, Eastman C, Palmer M, Davies ER, Carswell F. The quantitative distribution of nebulized antibiotic in the lung in cystic fibrosis. Respir Med 1994;88:203–211 [DOI] [PubMed] [Google Scholar]
- 33.Goerke C, Köller J, Wolz C. Ciprofloxacin and trimethoprim cause phage induction and virulence modulation in Staphylococcus aureus. Antimicrob Agents Chemother 2006;50:171–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.