SUMMARY
MicroRNA (miRNA)-deficient helper T cells exhibit abnormal IFN-γ production and decreased proliferation. However, the contributions of individual miRNAs to this phenotype remain poorly understood. We conducted a screen for miRNA function in primary T cells and identified individual miRNAs that rescue the defects associated with miRNA deficiency. Multiple members of the miR-17 and miR-92 families enhanced miRNA-deficient T cell proliferation whereas miR-29 largely corrected their aberrant interferon-γ (IFN-γ) expression. Repression of IFN-γ production by miR-29 involved direct targeting of both T-bet and Eomes, two transcription factors known to induce IFN-γ production. Although not usually expressed at functionally relevant amounts in helper T cells, Eomes was abundant in miRNA-deficient cells and was upregulated after miR-29 inhibition in wild-type cells. These results demonstrate that miR-29 regulates helper T cell differentiation by repressing multiple target genes, including at least two that are independently capable of inducing the T helper 1 (Th1) cell gene expression program.
INTRODUCTION
CD4+ helper T cells play a critical role in the coordination of effective immune responses. Upon activation, naive CD4+ T cells proliferate and differentiate into effector subsets defined primarily by distinct cytokine expression (Ansel et al., 2006; Szabo et al., 2003; Zhu et al., 2010). Because these cytokines act on many different cell types, the production and regulation of lineage-specific cytokines is fundamental to generating the appropriate immune response for different types of immune challenges. Thus, proper regulation of helper T cell proliferation and differentiation is critical for effective immune protection from pathogens. However, dysregulated T cell responses can result in immunopathology. For example, T helper 1 (Th1) cells secrete interferon-γ (IFN-γ) and mediate elimination of intracellular pathogens, but these cells can also contribute to pathologic inflammation and autoimmune disease. Examining the mechanisms of gene regulation that underlie T cell polarization has the potential to improve our understanding of cell differentiation in general and to provide insights for the development of clinically relevant immune therapies.
The differentiation fate of CD4+ T cells involves integration of antigen, costimulatory, and cytokine signals that influence the expression and duration of lineage-specific transcription factors. Enforced expression of the T-box transcription factor T-bet dominantly induces IFN-γ production, and T-bet-deficient CD4+ T cells are severely defective in Th1 cell differentiation and IFN-γ production (Szabo et al., 2000). Eomesodermin (Eomes), a closely related T-box family transcription factor, has also been shown to regulate IFN-γ production, particularly in CD8+ T cells (Pearce et al., 2003). Although it is normally expressed at relatively low amounts in CD4+ T cells, Eomes can substitute for T-bet to induce IFN-γ production and Th1 cell differentiation when its expression is enforced. Once expressed, IFN-γ initiates a positive feedback loop that reinforces its own production and T-bet expression in helper T cells.
Recent work has identified endogenously expressed micro-RNAs (miRNAs) as important contributors to the regulation of helper T cell proliferation, survival, differentiation, and cytokine production (O’Connell et al., 2010). miRNAs are ~22 nucleotide noncoding RNAs that mediate sequence-dependent posttranscriptional negative regulation of gene expression (Bartel, 2009). Primary miRNA transcripts are processed by the microprocessor complex, consisting of the RNase III enzyme Drosha and the double-stranded RNA-binding cofactor DGCR8. The resulting ~60 to 80 nucleotide hairpin precursor-miRNAs are subsequently cleaved by the RNase III enzyme Dicer to form ~22 base pair dsRNA duplexes. One strand of this duplex forms the mature miRNA, which targets mRNAs for repression by complementary base pairing, especially within the “miRNA seed” sequence at nucleotide positions 2–8. Genetic inactivation of either Dicer or Drosha results in considerable functional defects in CD4+ T cells (Chong et al., 2008; Cobb et al., 2006; Liston et al., 2008; Muljo et al., 2005; Zhou et al., 2008). Dicer-deficient cells exhibit a marked bias toward IFN-γ production as well as reduced proliferation and survival after stimulation in vitro. Similar phenotypes were observed in Drosha-deficient T cells (Chong et al., 2008). Although both Dicer and Drosha have been implicated in functions outside of miRNA biogenesis, the overlapping phenotypes of Drosha- and Dicer-deficient T cells indicate specific involvement of the miRNA pathway. These studies demonstrate the significance of miRNAs in regulating helper T cell gene expression, but the individual miRNAs responsible and their mechanisms of function remain largely undefined.
By using T cells deficient for DGCR8, we established a system for analyzing the function of individual miRNAs in otherwise miRNA-deficient cells. Side-by-side comparisons showed that Dicer- and DGCR8-deficient helper T cells exhibit identical defects in cytokine regulation and proliferation. We further demonstrated that these phenotypes result from cell-intrinsic defects and that reintroduction of DGCR8 after stimulation can partially rescue the functional defects of these cells. Building on these findings, we carried out a functional screen and identified individual miRNAs that can alleviate the defects of miRNA-deficient cells.
RESULTS
DGCR8-Deficient Helper T Cells
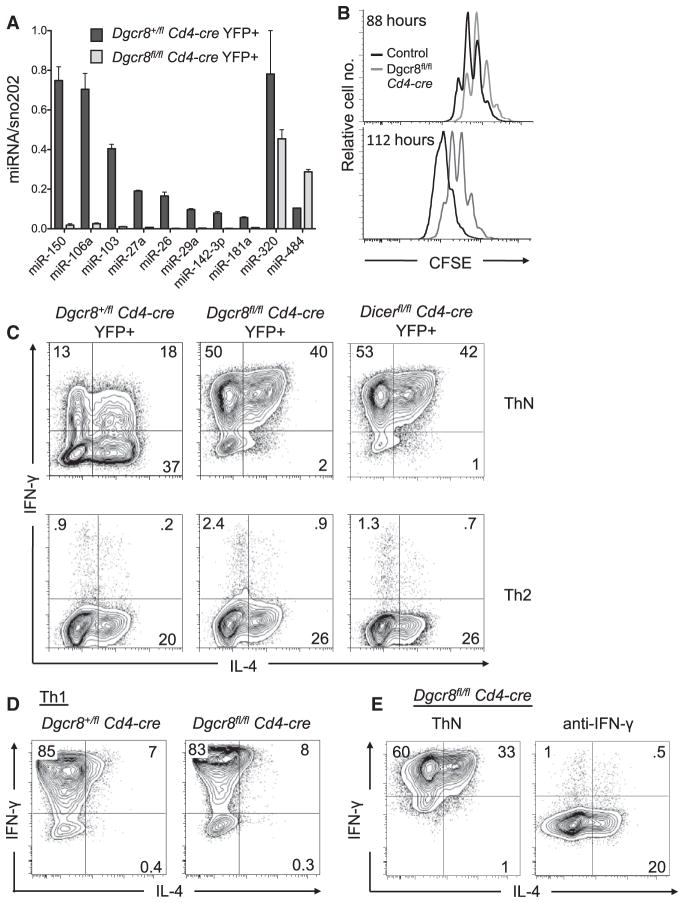
To establish a system for studying the functional effects of miRNAs in helper T cells, we intercrossed Dgcr8fl/fl mice (Wang et al., 2007) with Cd4-cre mice. Dgcr8fl/fl Cd4-cre mice were further crossed to Gt(ROSA)26Sortm1(EYFP)Cos (R26-YFP) or Gt(ROSA)26Sortm1Hjf (R26-tdRFP) mice (Luche et al., 2007) to introduce a fluorescent reporter of Cre expression. To confirm Dgcr8 inactivation, we measured miRNA expression in the YFP+ population in cultured CD4+ T cells from Dgcr8fl/fl Cd4-cre R26-YFP mice. DGCR8-dependent miRNA expression in the Dgcr8fl/fl Cd4-cre YFP+ cells was below the limit of detection, whereas control cells expressed all miRNAs analyzed (Figure 1A). miR-320 and miR-484 were still detected in Dgcr8fl/fl Cd4-cre YFP+ cells, confirming the DGCR8-independent expression of these two unusual miRNAs (Babiarz et al., 2008).
Figure 1. Decreased Proliferation and Increased IFN-γ Production by DGCR8-Deficient CD4+ T Cells.
(A) qPCR analysis of miRNA expression in flow cytometer-sorted YFP+CD4+ T cells cultured in vitro for 5 days. Bars represent miRNA expression relative to sno202; error bars represent range for replicate qPCR reactions.
(B) Proliferation of RFP+CD4+ T cells labeled with CFSE and analyzed by flow cytometry at the indicated times in culture. Cells are from Dgcr8fl/fl Cd4-cre R26-tdRFP mice (gray lines) or Dgcr8+/fl Cd4-cre R26-tdRFP mice (black lines).
(C–E) Cytokine production by restimulated YFP+ CD4+ T cells cultured in nonpolarizing (ThN) or Th2 cell-type conditions (C); CD4+ T cells cultured in Th1 cell-type conditions (D); and Dgcr8fl/fl Cd4-cre YFP+CD4+ T cells cultured ±5 μg/mL anti-IFN-γ (E). Data are representative of at least three independent experiments.
Analysis of lymph nodes and spleen revealed a decrease in the relative frequency of CD4+ and CD8+ T cells by ~50% and ~66%, respectively, in Dgcr8fl/fl Cd4-cre mice (see Figure S1A available online). This reduction is consistent with that reported for Cd4-cre-mediated deletion of Dicer or Drosha (Chong et al., 2008; Muljo et al., 2005). We observed no difference in the frequency of naive (CD62LhiCD44lo) cells among the CD4+ T cells in the lymph nodes and spleen of 4- to 7-week-old Dgcr8fl/fl Cd4-cre mice (Figure S1B). This is probably due to the relatively young age of the mice used in this study because older mice with T cell-specific deletion of miRNAs do accumulate previously activated helper T cells (Chong et al., 2008; Cobb et al., 2006; and data not shown). Moreover, counterselection against DGCR8-deficient T cells was not apparent in these mice; no difference in the frequency of YFP+ cells among CD4+ T cells was observed between Dgcr8fl/fl Cd4-cre mice and Dgcr8+/fl Cd4-cre littermate controls (Figure S1C).
Like Dicer- and Drosha-deficient helper T cells, DGCR8-deficient T cells exhibited decreased proliferation (Figure 1B) and an overwhelming bias toward IFN-γ production in vitro (Figure 1C). The cytokine expression defect of DGCR8- and Dicer-deficient T cells was essentially identical, indicating that their common products, namely miRNAs, regulate helper T cell differentiation. Also consistent with previous reports (Chong et al., 2008; Muljo et al., 2005), cytokine production by wild-type and DGCR8-deficient cells was similar in Th1 cell-type conditions and aberrant IFN-γ production was inhibited by Th2 cell-type culture conditions (Figures 1C and 1D). Further, IFN-γ blocking antibody alone was sufficient to mediate this effect without the addition of interleukin-4 (IL-4) (Figure 1E). This result demonstrates that aberrant IFN-γ production in miRNA-deficient cells is dependent on IFN-γ signaling and specifically implicates genes in this pathway as potential targets of miRNA regulation.
Cell-Intrinsic Dysregulation of Cytokine Production in miRNA-Deficient Cells
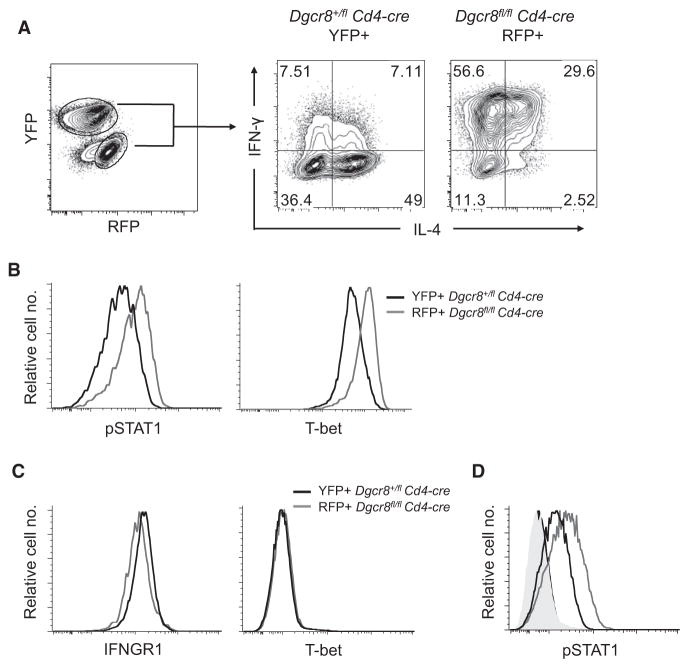
Because IFN-γ induces a positive feedback loop through STAT1 and T-bet to reinforce its own expression (Afkarian et al., 2002; Lighvani et al., 2001), we tested the possibility that an increase in early IFN-γ release could be the proximal cause of the IFN-γ production bias observed in DGCR8-deficient T cell cultures. Dgcr8fl/fl Cd4-cre R26-tdRFP cells were cocultured with Dgcr8+/fl Cd4-cre R26-YFP control cells. This system allowed us to distinguish the miRNA-deficient RFP+ cells from the control YFP+ cells at the time of analysis (Figure 2A). DGCR8-deficient T cells retained their IFN-γ production bias in coculture. However, despite exposure to the same cytokine environment as the miRNA-deficient cells, control cells did not exhibit increased IFN-γ production. To better understand this cell-intrinsic difference in cytokine production, we analyzed additional aspects of the IFN-γ feedback loop. Activated DGCR8-deficient cells exhibited higher amounts of both phosphorylated STAT1 (pSTAT1) and T-bet relative to cocultured control cells (Figure 2B). There is evidence that the gene encoding the IFN-γRα subunit (Ifngr1) can be regulated by at least one miRNA (Banerjee et al., 2010), but we did not observe increased surface expression of this receptor on unstimulated miRNA-deficient cells (Figure 2C). Additionally, naive wild-type and miRNA-deficient cells had the same basal expression of T-bet (Figure 2C) and negligible amounts of pSTAT1 (Figure 2D, shaded histograms). Notably however, after treatment with recombinant IFN-γ, naive DGCR8-deficient T cells exhibited more STAT1 phosphorylation than control cells (Figure 2D, open histograms). We conclude that cell-intrinsic defects sensitize the IFN-γ signaling pathway in miRNA-deficient T cells, thus leading to aberrant Th1 cell differentiation and IFN-γ production.
Figure 2. DGCR8-Deficient T Cells Exhibit Cell-Intrinsic Defects in Cytokine Production and IFN-γ Signaling.
(A) Cytokine production of YFP+ or RFP+CD4+ T cells from Dgcr8fl/fl Cd4-cre R26-tdRFP and Dgcr8+/fl Cd4-cre R26-YFP mice. YFP and RFP (left) were used to distinguish Dgcr8+/fl and Dgcr8fl/fl cells, respectively (right). Cells were mixed in a 4:1 ratio (Dgcr8fl/fl:Dgcr8+/fl) and co-cultured in 10 units/mL IL-4. Data are representative of three independent experiments.
(B) Intracellular protein expression of cocultured Dgcr8fl/fl Cd4-cre R26-tdRFP (Dgcr8fl/fl) and Dgcr8+/fl Cd4-cre R26-YFP (Dgcr8+/fl) cells after 68 hr of stimulation (anti-CD3 and anti-CD28).
(C) IFNGR1 and T-bet expression in freshly isolated CD4+ T cells.
(D) Phosphorylated STAT1 expression in freshly isolated CD4+ T cells ± 10 ng/mL IFN-γ for 15 min in coculture. Shaded histograms represent cells not treated with IFN-γ; Dgcr8+/fl Cd4-cre YFP+ (black) or Dgcr8fl/fl Cd4-cre RFP+ (gray).
To further examine the requirements for the miRNA pathway in the cell-intrinsic control of IFN-γ production, we used retroviral transduction to re-express DGCR8 in differentiating DGCR8-deficient T cells. Those cells that were successfully transduced with the Dgcr8-containing retrovirus exhibited improved proliferation and a decrease in the frequency of IFN-γ-producing cells (Figure S2A and data not shown). However, untransduced cells in the same culture remained biased toward IFN-γ production. Cytokine production by miRNA-deficient T cells was also not affected by a control retrovirus containing only the Thy1.1 marker gene (Figure S2A). DGCR8 expression led to the expected re-expression of mature miRNAs (Figure S2B). Taken together, these results indicate that miRNAs dynamically regulate cytokine production in helper T cells in a cell-autonomous manner and that miRNA activity has functional effects on IFN-γ production even after initial activation and polarization events have occurred.
A Screen for Individual miRNA Functions in CD4+ T Cells
After observing the effect of reintroducing DGCR8, we reasoned that some subset of the miRNAs expressed in T cells must be responsible for the phenotypic rescue. In order to identify the miRNAs involved, we implemented a system to screen for individual miRNA functions in differentiating CD4+ T cells. Because of the often cooperative and redundant nature of miRNA function, there are inherent difficulties in studying individual miRNAs. To overcome some of these difficulties, we utilized Dgcr8fl/fl Cd4-cre mice to allow introduction of individual miRNAs into otherwise miRNA-deficient helper T cells, an approach that proved to be a useful and sensitive means for identifying the function of individual miRNAs in embryonic stem cells (Wang et al., 2008).
With the goal of delivering individual miRNAs to DGCR8-deficient T cells, we first optimized the transfection of primary CD4+ T cells by electroporation (Kim et al., 2008). By using siRNA to silence YFP expression in Dgcr8fl/fl Cd4-cre YFP+ cells, we were able to achieve transfection efficiency of greater than 90% with minimal effects on cell viability (data not shown). To observe changes in miRNA activity, we generated retroviral sensor constructs containing four miRNA binding sites downstream of a GFP coding region. As expected, sensor GFP expression was repressed in miRNA-expressing wild-type cells as compared to DGCR8-deficient cells (Figure S3A). We next used these sensors to validate miRNA gain of function after transfection of primary T cells with synthetic miRNA oligonucleotides. In DGCR8-deficient cells, GFP expression from a miR-29a sensor was repressed after transfection with miR-29a but not miR-150 (Figure S3B). Similarly, a sensor with miR-150 binding sites was repressed by miR-150 but not miR-29a (Figure S3B). These results indicate that synthetic miRNA oligonucleotides can be successfully introduced into primary T cells with high efficiency and exhibit sequence-specific repression of target genes, even in T cells that are otherwise deficient in their ability to generate mature miRNAs.
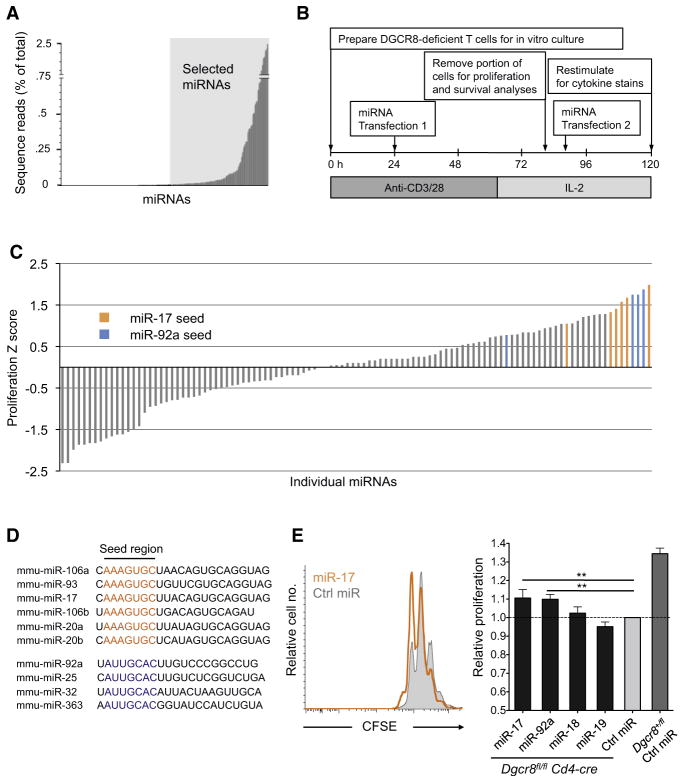
We next sought to determine the appropriate set of miRNAs for analysis in a functional screen. Small RNA libraries for deep sequencing were generated from wild-type CD4+ T cells activated in vitro for 44 hr, providing a relevant “snapshot” of the miRNAs present near the time of miRNA introduction in our screen (Figure 3A; Table S1). From these data we chose 110 miRNAs to screen for function in DGCR8-deficient helper T cells. All miRNAs for which we obtained at least 100 reads per million total reads from two independent sequencing libraries were included.
Figure 3. A Screen for miRNA Function in DGCR8-Deficient CD4+ T Cells Reveals Proliferation Rescue by miR-17 and miR-92 Family miRNAs.
(A) miRNA expression from deep sequencing analysis of small RNAs in wild-type CD4+ T cells stimulated in vitro for 44 hr. Shaded region marks miRNAs selected for screening. Unshaded region represents all miRNAs sequenced at least once. Bars are average frequency of each miRNA among all genome-matching small RNA sequences from two independent small RNA libraries.
(B) Schematic of workflow for miRNA functional screen in DGCR8-deficient CD4+ T cells (from Dgcr8fl/fl Cd4-cre R26-tdRFP mice).
(C) Proliferation index for each miRNA was determined (see Experimental Procedures) and used to calculate proliferation Z scores (Z = x-mean/SD where x represents the normalized proliferation index for each individual miRNA). All miRNAs in the miR-17 and miR-92 seed families that were screened are highlighted.
(D) miR-17 and miR-92 seed family miRNAs (ordered from highest to lowest Z score).
(E) Proliferation of CFSE-labeled, RFP+ cells 65 hr posttransfection with individual miR-17~92 cluster miRNAs or control miRNA (representative histograms in left panel). Proliferation index data were normalized to cells transfected with control miRNA. Values are means ± SD from five independent transfections in three independent experiments; **p < 0.01; ANOVA Tukey’s post-hoc test.
A schematic of the screening procedure is depicted in Figure 3B. Carboxyluorescein diacetate succinimidyl ester (CFSE)-labeled DGCR8-deficient T cells were activated in vitro, transfected with individual miRNAs after 24 hr, returned to stimulus for an additional ~40 hr, and then cultured in media containing IL-2. Proliferation was measured by CFSE dilution on the fourth day, and the remaining cells were transfected again prior to cytokine analysis to overcome miRNA turnover and dilution. The next day, cells were restimulated and stained intracellularly for analysis of cytokine production.
miR-17 and miR-92 Family miRNAs Promote T Cell Proliferation
An initial screen of our T cell miRNA library revealed several miRNAs with a positive effect on proliferation (Figure 3C; Table S2). Of the 10 miRNAs that most significantly increased proliferation, five (miR-17, miR-20a, miR-93, miR-106a, miR-106b) belong to the miR-17 seed family and an additional three (miR-25, miR-32, miR-92a) share the miR-92 seed sequence. This represents three of four miR-25 seed family miRNAs assayed and five of six miR-17 family miRNAs assayed (Figure 3D). These findings are consistent with the established role for the miR-17~92 cluster in regulating proliferation of multiple cell types (Mendell, 2008), including its ability to induce lymphoproliferative disease when overexpressed in mice (He et al., 2005; Xiao et al., 2008). The other two miRNA seed families that are represented in the miR-17~92 cluster, miR-18 and miR-19, did not promote proliferation in our screen.
To validate these results, we repeated proliferation assays with individual members of the miR-17~92 cluster (Figure 3E). These experiments verified that miR-17 and miR-92a both increased the proliferation index of DGCR8-deficient T cells approximately 10%, representing a partial but consistent rescue (p < 0.01 ANOVA, Tukey’s post hoc test). To address the mechanism of this effect, we analyzed the expression of validated miR-17~92 targets with established roles in cell proliferation and survival. Previous reports have identified Pten as a target of miR-17 and miR-19 (Olive et al., 2009; Xiao et al., 2008), Bim as a target of miR-17 and miR-92 (Koralov et al., 2008; Olive et al., 2009; Petrocca et al., 2008; Xiao et al., 2008), and p21 as a target of the miR-17 seed family (Ivanovska et al., 2008; Petrocca et al., 2008; Wang et al., 2008). However, these targets were not affected by transfection with miR-17 or miR-92a, suggesting that additional targets may be responsible for the proproliferative effects of these miRNAs in helper T cells (Figure S4A). In addition, miR-17 and miR-92a increased proliferation without observable effects on cell survival, whereas miR-19 increased cell viability relative to control transfected cells but did not augment proliferation (Figure S4B). Taken together, these results demonstrate the effectiveness of this screening approach for identifying specific functional capabilities of individual miRNAs.
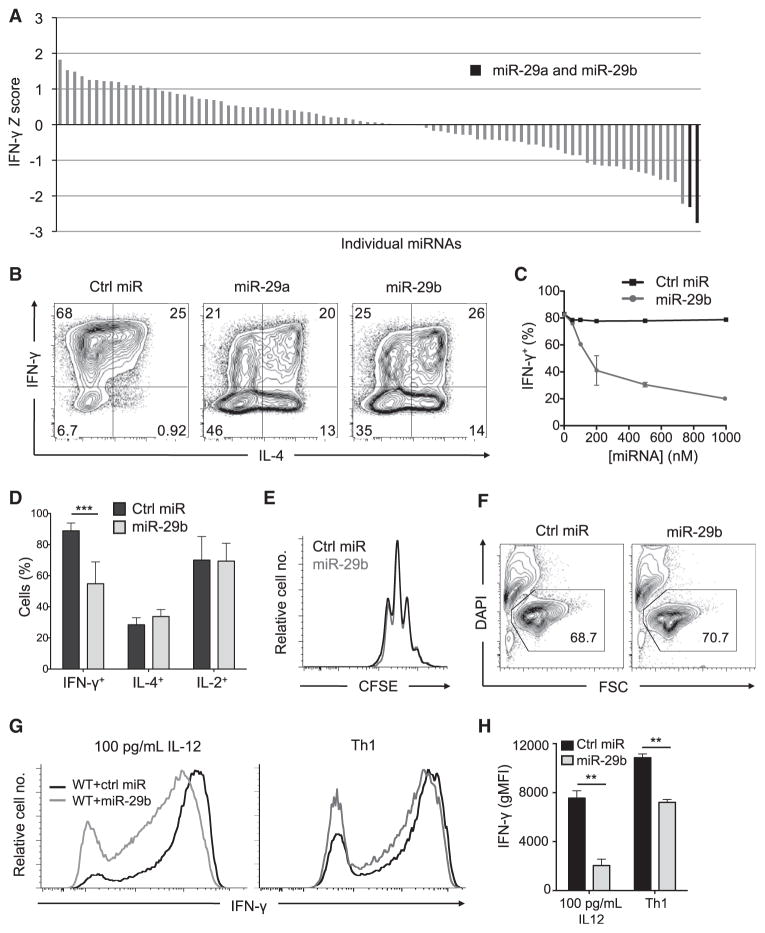
miR-29a and miR-29b Regulate IFN-γ Production
Building on our success in identifying miRNA functions that are consistent with their known biological activities, we next screened for individual miRNAs that can rescue the aberrant IFN-γ production of DGCR8-deficient T cells. In this screen, both miR-29a and miR-29b significantly decreased the frequency of IFN-γ-producing cells (Figure 4A; Table S2). The only other member of this seed family, miR-29c, was not included in our screen because of its low expression in helper T cells (Figure S5). Additional transfection experiments confirmed the ability of both miR-29a and miR-29b to reduce IFN-γ production in DGCR8-deficient T cells (Figure 4B) and that this effect was dose dependent (Figure 4C). miR-29 had no significant effect on the total frequency of IL-4- or IL-2-producing cells (Figure 4D), nor did it affect cell proliferation (Figure 4E) or viability (Figure 4F). miR-29 also repressed IFN-γ production in wild-type cells, even in conditions that strongly promote Th1 cell differentiation (Figures 4G and 4H). Taken together, these findings indicate that miR-29 mediates its effects through specific regulation of the IFN-γ production pathway rather than through general effects on T cell activation, cytokine production, or cell fitness.
Figure 4. miR-29a and miR-29b Rescue Aberrant IFN-γ Production by miRNA-Deficient CD4+ T Cells and Repress IFN-γ Production by Wild-Type Th1 Cells.
(A) Z scores for frequency of IFN-γ production among restimulated DGCR8-deficient cells transfected with individual miRNAs as described in Figure 3B. miRNAs with a proliferation score of Z < −1.5 or Z > 1.5 were not included in the IFN-γ analysis because of possible indirect effects of survival or proliferation on cytokine production.
(B) Intracellular cytokine stains for IFN-γ and IL-4 in transfected and restimulated DGCR8-deficient CD4+ T cells. Data are representative of three independent experiments.
(C) Frequency of IFN-γ production among Dgcr8fl/fl Cd4-cre CD4+ T cells transfected with increasing concentration of miR-29b. Data are representative of two independent experiments; error bars represent range from two independent transfections.
(D) Summary of cytokine production by miR-29b-transfected and restimulated Dgcr8fl/fl Cd4-cre CD4+ T cells. Values are means ± SD from four biological replicates in two independent experiments; ***p < 0.001; Student’s two-tailed t test.
(E) Proliferation analysis by CFSE dilution of transfected Dgcr8fl/fl Cd4-cre cells on day 5.
(F) Cell viability analysis of transfected Dgcr8fl/fl Cd4-cre cells on day 5. Numbers indicate percentage of total events that are viable.
(G and H) IFN-γ production by restimulated, C57BL/6 CD4+ cells cultured in 100 pg/mL IL-12 or Th1 cell-type conditions and transfected with miR-29b or control miRNA. Values in (H) represent mean ± SD from three independent transfections; data are representative of two independent experiments.
Genome-wide Analysis Identifies T-bet and Eomes as Direct Targets of miR-29
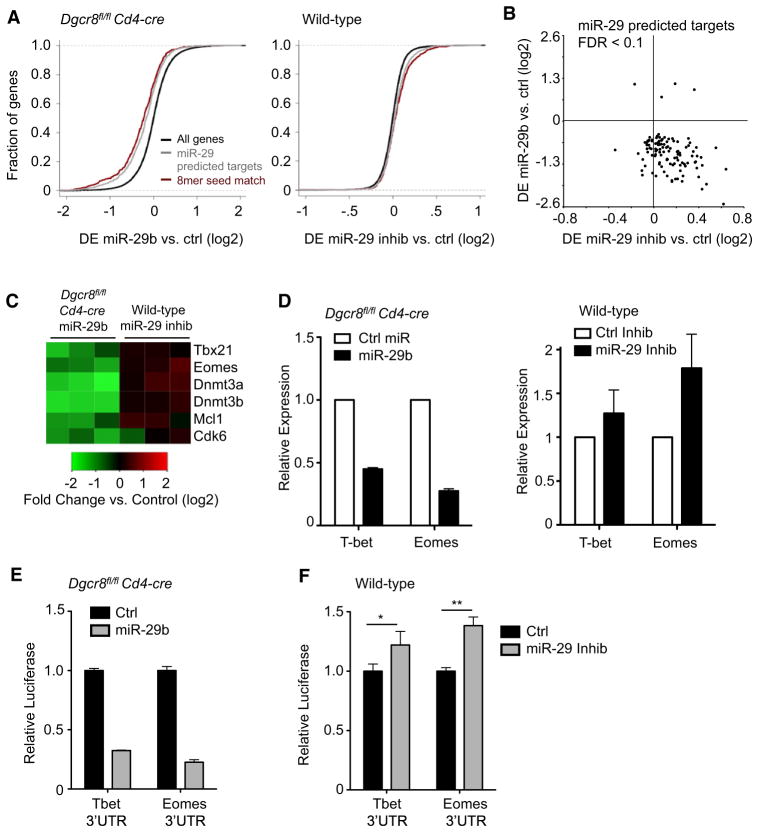
To identify miR-29 targets that contribute to IFN-γ regulation, we conducted microarray analyses of gene expression in both miR-29 gain- and loss-of-function conditions. A microarray-based approach to target identification is supported by recent evidence that miRNA-mediated changes in mRNA and protein expression are highly correlated (Guo et al., 2010) For gain-of-function experiments, we transfected DGCR8-deficient cells with synthetic miR-29b, and for loss-of-function experiments, we transfected wild-type cells with combined miR-29a and miR-29b antisense inhibitors. As a class, predicted miR-29 targets (Friedman et al., 2009) were clearly repressed subsequent to miR-29b transfection in DGCR8-deficient cells (Figure 5A, left). Of the 115 predicted miR-29 targets whose expression was markedly changed after miR-29 transfection (FDR < 0.1), 111 were downregulated and only 4 exhibited increased expression (Figure 5B; Table S3). Predicted targets were also upregulated as a class after inhibition of miR-29 in wild-type T cells (Figure 5A, right). Notably, the majority of predicted miR-29 targets that were downregulated in response to miR-29b in DGCR8-deficient cells were also upregulated in wild-type cells transfected with miR-29 inhibitors (Figure 5B). Taken together, these data indicate that many of these genes not only are capable of responding to miR-29 overexpression but are in fact subject to regulation by endogenous miR-29 in wild-type T cells. Among the genes regulated in this manner, we identified several previously validated targets of miR-29 including Dnmt3a, Dnmt3b, Cdk6, and Mcl1 (Figure 5C; Fabbri et al., 2007; Garzon et al., 2009; Mott et al., 2007). Many genes identified in this analysis probably contribute to the overall function of miR-29 in helper T cells.
Figure 5. Genome-wide Analysis Identifies T-bet and Eomes as miR-29 Targets.
(A) Cumulative distribution of mRNA expression changes from microarray data subsequent to transfection with miR-29b in Dgcr8fl/fl Cd4-cre CD4+ T cells (left) or miR-29 inhibitor in C57BL/6 (wild-type) CD4+ T cells (right). Negative values indicate downregulation of mRNA after transfection. Black lines represent the entire microarray gene set, gray lines represent the subset of genes that are Targetscan computationally predicted miR-29 targets, and red lines represent the subset of predicted targets with 8-mer binding sites perfectly complementary to the entire miR-29 seed sequence.
(B) Scatter plot of the change in mRNA expression after miR-29 transfection in Dgcr8fl/fl Cd4-cre cells (y axis) and change in expression after miR-29 inhibitor transfection in wild-type cells (x axis). Each point represents a different gene array probe. Included are predicted miR-29 targets differentially expressed (FDR < 0.1) after miR-29 transfection.
(C) Heatmap representation of changes in gene expression after transfection for three independent biological samples of each genotype. Scale is log2 fold change in array hybridization intensity compared to control transfected cells.
(D) qPCR validation of array data for miR-29 candidate targets of interest. mRNA expression was normalized to Gapdh and is relative to expression in control transfected cells. Values are means ± SD from three biological replicates.
(E and F) Primary Dgcr8fl/fl Cd4-cre (E) or wild-type (F) CD4+ T cells were cotransfected with a dual luciferase reporter and miR-29b or control miRNA (E); miR-29 inhibitors or control inhibitor (F). Luciferase reporters contained the full-length mouse 3′UTR of Tbx21 or Eomes. Renilla luciferase activity was measured 24 hr after transfection and normalized to firefly luciferase activity. Values are relative to normalized luciferase in control transfected cells. Data are representative of three independent experiments and values are means ± SD from three independent transfections.
*p < 0.05, **p < 0.01; ANOVA Tukey’s post hoc test.
In regards to the regulation of IFN-γ specifically, we were intrigued that both Tbx21 (encoding T-bet) and Eomes appeared to be regulated by miR-29. Both T-bet and Eomes have known roles in IFN-γ production and both contain highly conserved 3′UTR miR-29 binding sites. We validated the microarray findings for these genes by quantitiative polymerase chain reaction (qPCR) and found a 50% reduction of Tbx21 mRNA after miR-29 transfection in miRNA-deficient cells and a 30% increase after miR-29 inhibition in wild-type cells (Figure 5D). Eomes mRNA was affected to an even greater extent, with expression reduced by 80% in miR-29-transfected DGCR8-deficient cells and increased by 70% after miR-29 inhibition in wild-type cells (Figure 5D).
To determine whether T-bet and Eomes are in fact direct targets of miR-29, we transfected primary T cells with dual luciferase reporters containing the full-length mouse Tbx21 or Eomes 3′UTR. In DGCR8-deficient T cells, miR-29b reduced expression of the cotransfected Tbx21 3′UTR reporter by approximately 65% and miR-29b reduced expression of the Eomes 3′UTR reporter by 80% (Figure 5E). In wild-type T cells, miR-29 inhibition significantly “derepressed” the full-length Tbx21 3′UTR and Eomes 3′UTR reporters by 20% and 40%, respectively (Figure 5F). These results demonstrate that both the Tbx21 and Eomes 3′UTRs are directly responsive to miR-29 and that physiological expression of miR-29 in wild-type helper T cells is sufficient to mediate these effects.
miR-29 Regulates T-box Transcription Factor Protein Expression In Vitro and In Vivo
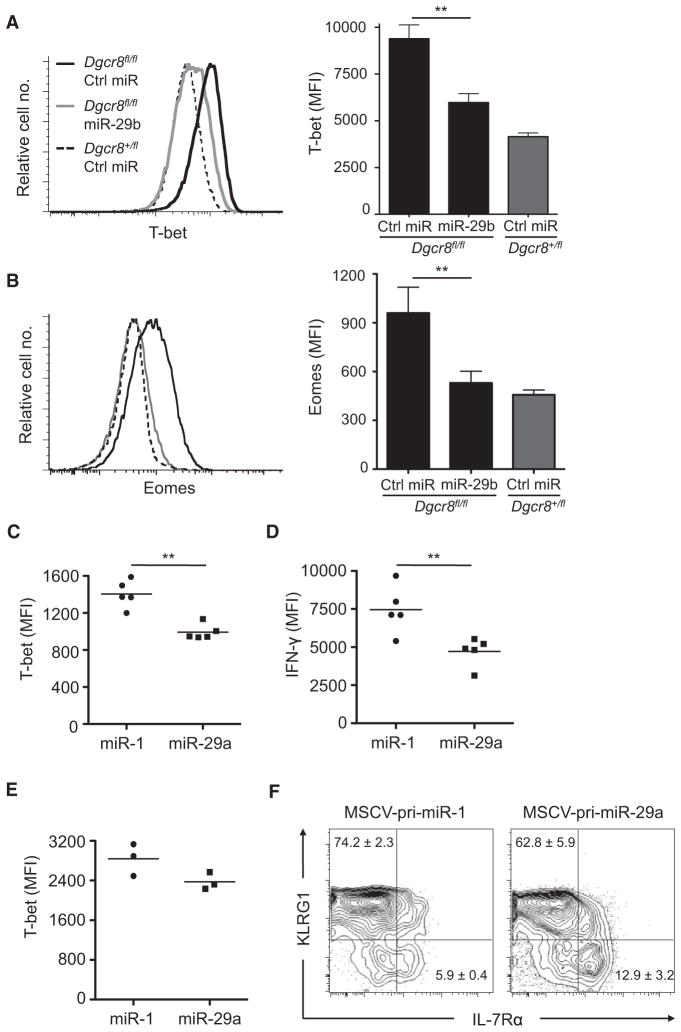
Intracellular staining confirmed that miR-29 returned the elevated T-bet protein expression of miRNA-deficient cells to an amount near that of wild-type cells (Figure 6A). Although Eomes expression is typically very low in CD4+ T cells, Eomes was also elevated in miRNA-deficient CD4+ cells and miR-29 transfection rescued this aberrant expression (Figure 6B).
Figure 6. miR-29 Regulates T-box Transcription Factor Protein Expression In Vitro and In Vivo.
(A and B) CD4+ cells from Dgcr8fl/fl Cd4-cre mice or Dgcr8+/fl Cd4-cre mice were cultured in ThN conditions and stained intracellularly for T-bet and Eomes protein 24 hr after transfection. T-bet data are on day 2 and Eomes data are on day 5. Values are means ± SD from three independent transfections; **p < 0.01; ANOVA Tukey’s post hoc test.
(C) MFI of intracellular T-bet stains for Thy1.1+ CD45.1+ cells subsequent to adoptive transfer of CD4+ SMARTA T cells and LCMV infection. Transferred and transduced cells were identified by congenic CD45.1 and retroviral Thy1.1 expression, respectively.
(D) MFI of intracellular IFN-γ for transduced and transferred CD4+ SMARTA T cells (as in C). Cells were stimulated with 1 μM gp61-80 peptide. Data represent IFN-γ MFI for TNF-α-producing cells.
(E) MFI of intracellular T-bet in CD8+ P14 T cells after LCMV infection, analyzed as in (C).
(F) Transduced CD8+ P14 T cells (as in E) were analyzed for KLRG1 and IL-7Rα expression. Values are mean frequency (%) ± SD; n = 3; p < 0.05 for both populations; Student’s two-tailed t test.
Data in (C)–(F) are representative of two independent experiments; **p < 0.01; Student’s two-tailed t test.
To analyze miR-29 function in helper T cells in vivo, we retrovirally transduced SMARTA CD4+ T cell receptor (TCR) transgenic T cells with either pri-miR-29a or control pri-miR-1. Transduced cells were adoptively transferred and recipient mice were infected with lymphocytic choriomeningitis virus (LCMV). On day 8 posttransfer, we analyzed T-bet and Eomes expression in transduced cells. As expected of wild-type CD4+ T cells, Eomes expression was negligible regardless of miR-29 overexpression (data not shown). However, T-bet expression was significantly reduced in the miR-29-transduced cells (Figure 6C). We also observed significantly decreased IFN-γ production by miR-29-overexpressing cells, as indicated by decreased mean fluorescence intensity (MFI) among CD4+ effector T cells producing both IFN-γ and tumor necrosis factor-α (TNF-α) (Figure 6D).
To further assess the in vivo role of T-bet regulation by miR-29, we conducted additional experiments with P14 LCMV-specific TCR transgenic CD8+ T cells. Again we observed a decrease in T-bet in miR-29-overexpressing cells (Figure 6E). Furthermore, we analyzed surface expression of Killer cell lectin-like G1 (KLRG1) and the IL-7 receptor alpha-chain (IL-7Rα) as markers of CD8+ effector cell subsets. In miR-29-overexpressing cells, we observed fewer KLRG1+IL-7Rα− cells and an increased frequency of KLRG1−IL-7Rα+ cells (Figure 6F). These results are consistent with reports that KLRG1+IL-7Rα− short-lived effector cells (SLECs) are particularly sensitive to graded expression of T-bet (Joshi et al., 2007). Taken together, these results demonstrate the ability of miR-29 to functionally contribute to T-bet regulation in both CD4+ and CD8+ T cells in vivo.
Regulation of IFN-γ by miR-29 Involves Both T-bet and Eomes
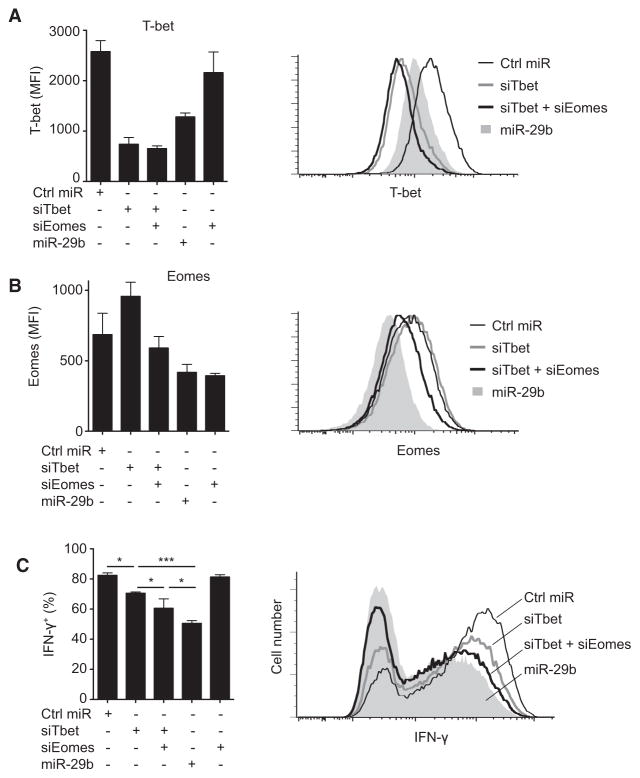
To measure the functional relevance of T-bet and Eomes as miR-29 targets, we determined whether direct T-bet and Eomes silencing with siRNA would phenocopy miR-29 gain of function in DGCR8-deficient T cells. T-bet siRNA reduced T-bet expression to an even greater extent than miR-29 (Figure 7A) and did lead to a significant reduction in IFN-γ production (Figure 7C). Surprisingly, however, T-bet reduction alone was not sufficient to reduce IFN-γ production to the same extent achieved by miR-29b (Figure 7C). Direct silencing of Eomes alone had no effect on IFN-γ production (Figures 7B and 7C), probably because of a dominant effect of T-bet on IFN-γ production in these cells. However, combined silencing of both T-bet and Eomes reduced IFN-γ production to a greater extent than T-bet siRNA alone (Figure 7C). Thus, T-bet expression does influence IFN-γ production and miR-29 does repress T-bet, but T-bet repression alone cannot fully account for the ability of miR-29 to regulate IFN-γ. Although we must acknowledge that additional miR-29 targets may also contribute, we conclude that miR-29 regulates IFN-γ production by simultaneously targeting T-bet and Eomes, two proteins with overlapping transcriptional activity.
Figure 7. miR-29-Mediated Repression of IFN-γ Involves Regulation of Both T-bet and Eomes.
Dgcr8fl/fl Cd4-cre CD4+ T cells were transfected with control miRNA, miR-29b, siRNA targeting T-bet (siTbet), and/or siRNA targeting Eomes (siEomes). Cells were cultured in ThN conditions and transfected at 24 hr and 96 hr. T-bet (A), Eomes (B), and IFN-γ (C) protein expression was analyzed subsequent to restimulation at 120 hr. Values are means ± SD from three independent transfections; ***p < 0.001, *p < 0.05; ANOVA Tukey’s post hoc test.
DISCUSSION
We and others have previously shown that miRNA-deficient helper T cells exhibit defects in proliferation and the regulation of cytokine production. However, defining the contributions of specific miRNAs to these processes has been challenging because of cooperation and redundancy among miRNAs. In this study, we utilized T cells from Dgcr8fl/fl Cd4-cre mice as a source of miRNA-deficient cells into which individual miRNAs could be reintroduced and assayed for their ability to rescue phenotypes associated with miRNA deficiency. Optimization of primary T cell transfection allowed an efficient functional screen of more than 100 miRNAs in primary T cells. Starting with this screen, we identified individual miRNAs that rescue the defects of miRNA-deficient T cells and further determined the molecular mechanism of this activity. miR-17 and miR-92 family members supported proliferation of DGCR8-deficient T cells, whereas miR-29 family members potently and specifically inhibited IFN-γ production. miR-29 directly targeted T-bet and its close relative Eomes. Both endogenous and overexpressed miR-29 inhibited the expression of these key transcriptional regulators of IFN-γ expression. This coordinated repression of functional homologs by a single miRNA represents an intriguing mode of cell fate regulation and may apply broadly to miRNA function in other settings.
Introduction of miRNAs into otherwise miRNA-deficient cells proved to be an especially sensitive system for detecting bona fide functional effects. However, we also considered the possibility that a screen of all known miRNAs could reveal “artificial” functions for miRNAs not normally expressed at meaningful concentrations. For this reason, we limited our screen to miRNAs expressed in activated CD4+ T cells. Further analysis of the regulation of miRNA expression during T cell differentiation may reveal optimal conditions for modulating miRNA activity and analyzing functional effects and target genes in wild-type T cells.
Multiple members of the miR-17 and miR-92 miRNA families were able to promote proliferation of DGCR8-deficient helper T cells. All three clusters containing these miRNAs are expressed in helper T cells, and two of them (miR-17~92 and miR-106a~363) are strongly induced upon T cell activation (Barski et al., 2009; Kuchen et al., 2010). Overexpression of the entire miR-17~92 cluster promotes lymphomagenesis in B cells and can substitute for costimulation to promote helper T cell proliferation (He et al., 2005; Xiao et al., 2008). Our screening system allowed us to dissect the functional contribution of individual miRNAs within these clusters. miR-17 and miR-92 family members were key players in regulating the rapid proliferation of activated T cells. Of note, miRNAs with the miR-17 seed sequence also promote cell cycle progression in embryonic stem cells and human cell lines (Ivanovska et al., 2008; Petrocca et al., 2008; Wang et al., 2008). Clarifying the regulatory networks that enable miR-17~92 miRNAs to cooperatively regulate cell proliferation and survival will be important to fully understand their role in immunity and immune dysfunction.
miRNA effects on cell proliferation and survival appear to be nearly universal in diverse somatic cell types. In contrast, the increased propensity of both Dicer- and DGCR8-deficient helper T cells to produce IFN-γ reveals a noteworthy case of a cell type-specific immune function that is regulated by the miRNA pathway. Our findings demonstrate that miR-29 is an important regulator of the IFN-γ pathway in helper T cells and that this regulation is mediated in part through the Th1 cell transcription factor T-bet. T-bet heterozygous mice, which exhibit only a 50% reduction in T-bet protein, spontaneously develop Th2 cell-mediated airway inflammation (Finotto et al., 2002). Thus, even moderate repression of T-bet by miR-29 may have important physiological implications. Consistent with the well-established role of T-bet in promoting IFN-γ production, siRNA-mediated reduction of T-bet did repress IFN-γ in our experiments. Surprisingly, however, miR-29 had a far more potent effect than T-bet reduction alone. These findings indicate that the effects of miR-29 on IFN-γ are mediated through both T-bet and additional targets and support the notion that miRNAs rarely function through repression of a single target gene.
Microarray analysis after miR-29 gain of function and loss of function demonstrated that many genes with canonical miR-29 binding sites are markedly repressed by both overexpressed and endogenous miR-29. These data confirm previously validated targets of miR-29 and identify many additional candidate targets. At least one previously validated target, Dnmt3a, has been implicated in the regulation of cytokine production (Gamper et al., 2009) and may also be involved in the observed function of miR-29 reported here. Of particular interest to this study was Eomes, which encodes a T-box transcription factor closely related to T-bet. Although Eomes is known to be important for IFN-γ production in CD8+ T cells, it is normally expressed at very low amounts in CD4+ effector cells. However, ectopic expression of Eomes can promote IFN-γ production in CD4+ cells and mechanisms for the transcriptional repression of Eomes in CD4+ cells have been described (Pearce et al., 2003; Suto et al., 2006; Yagi et al., 2010). These findings underscore the importance of maintaining Eomes silencing for proper control of helper T cell differentiation and cytokine production. Conceivably, even transient or minimal expression of Eomes could initiate the IFN-γ and T-bet positive feedback loop that drives Th1 cell differentiation. We found that miRNA-deficient CD4+ cells did not appropriately restrict T-bet or Eomes, and RNAi experiments showed that both contributed to aberrant IFN-γ production. The direct targeting of both T-bet and Eomes by miR-29 implicates a central role for this miRNA in the phenotype of miRNA-deficient CD4+ T cells. However, additional miRNAs probably contribute as well, perhaps through effects on transcription factors such as GATA3 and Runx3 that can also regulate Eomes and IFN-γ expression (Yagi et al., 2010). Ifng itself has also been described as a computationally predicted target of miR-29 (Betel et al., 2008). However, we could not confirm any effect of miR-29 on a full-length Ifng 3′UTR reporter in primary T cells, perhaps because of the dominant miRNA-independent regulation of the Ifng 3′UTR in helper T cells (Villarino et al., 2011).
Taken together, our data suggest that miR-29 may have evolved as an important regulator of CD4+ effector T cell cytokine production, both by modulating expression of the lineage-specific transcription factor T-bet and by restricting leaky transcription of the functionally overlapping T-box transcription factor Eomes. CD4+ T cells are distinguished by their ability to undergo functional polarization into diverse effector subsets, some of which depend on silencing of Ifng for proper immune function. Therefore, CD4+ T cells must have mechanisms available for robust regulation of both T-bet and Eomes. In contrast, miRNA-deficient NK cells and CD8+ T cells do not exhibit increased IFN-γ production as CD4+ T cells do (Bezman et al., 2010; Zhang and Bevan, 2010). However, because activated NK and CD8+ T cells express T-bet and Eomes, miR-29 probably regulates their expression in these cells as well. Indeed, we observed decreased T-bet in CD8+ T cells overexpressing miR-29, and this corresponded with a significant decrease in the T-bet-dependent SLEC population during an in vivo immune response to LCMV.
Current models of helper T cell differentiation emphasize the role of cytokine and transcription factor positive feedback loops that polarize gene expression patterns. This feature makes T cell fate decisions very sensitive to small changes in the expression of key genes in the regulatory circuit and therefore especially amenable to regulation by miRNAs. Indeed, multiple molecular players in the Th1 cell pathway have now been characterized as targets of miRNAs. In addition to the current findings regarding miR-29, miR-155 and miR-146 have been shown to target Ifngr1 and Stat1, respectively, and miR-155-deficient T cells display dysregulated Th2 cell differentiation (Banerjee et al., 2010; Lu et al., 2010; Rodriguez et al., 2007; Thai et al., 2007). We did not observe increased IFNGR1 expression in DGCR8-deficient T cells, and miR-155 was excluded from our primary cytokine screen because it also compromised T cell proliferation. STAT1 was increased in DGCR8-deficient cells, and miR-146 transfection 24 hr after T cell activation consistently but modestly repressed IFN-γ production. It is possible that even earlier introduction of miR-146 would have had a larger effect on IFN-γ production. To this extent, naive DGCR8-deficient T cells exhibited increased phosphorylated STAT1 in response to IFN-γ as compared to wild-type cells. This indicates a role for miRNA regulation of IFN-γ signaling prior to the time when miRNAs were reintroduced into T cells in this study. Thus, our screen may underestimate the role of miR-146 and other miRNAs whose targets may be most critical for very early events in T cell activation and differentiation.
In conclusion, we have identified individual miRNAs that have significant effects on proliferation and cytokine production by helper T cells. These processes are central to appropriate immune system function and their dysregulation can have important pathological consequences. As such, miR-29 regulation of IFN-γ production and helper T cell differentiation has important implications for human diseases that are associated with T cell-mediated immunity such as asthma, type 1 diabetes, and multiple sclerosis. Understanding the complete set of functions for miR-29 and other miRNAs in helper T cells will probably provide useful insights for the development of novel immune therapies. Such therapies may focus on the miRNAs themselves or may modulate pathways implicated through characterization of miRNA target genes. Our results broaden our understanding of genome regulation by miR-29 and may have implications for other biological and disease processes that have also been linked to miR-29 function including fibrosis, HIV latency, and leukemia (Croce, 2009; Nathans et al., 2009; van Rooij et al., 2008).
EXPERIMENTAL PROCEDURES
Mice
Dgcr8fl/fl and Gt(ROSA)26Sortm1Hjf (R26-tdRFP) have been described previously (Luche et al., 2007; Wang et al., 2007). C57BL/6 (NCI), Cd4-cre (Taconic), Gt(ROSA)26Sortm1(EYFP)Cos (R26-YFP), SMARTA, and P14 mice (Jax) were purchased. All mice were housed and bred in specific-pathogen-free conditions in the Animal Barrier Facility at the University of California, San Francisco. All animal experiments were approved by the Institutional Animal Care and Use Committee of the University of California, San Francisco.
T Cell Stimulation and Transfection
CD4+ T cells from spleen and lymph nodes of young mice (4–7 weeks old) were isolated by magnetic bead selection (Dynal), stimulated with anti-CD3 and anti-CD28, and cultured in polarizing conditions as indicated. Cells were transfected by the Neon electroporation transfection system (Invitrogen) with an optimized version of the manufacturers’ recommended protocol. miRIDIAN miRNA mimics, inhibitors, controls, and T-bet siRNA were from Dharmacon. Eomes siRNA was from QIAGEN. See Supplemental Information for details.
miRNA Screening and Analysis
Cells were stimulated at 1 × 106 cells/mL for 24 hr in 6-well plates. Cells were pooled and 0.2 × 106 cells were transfected with an individual miRNA. After transfection, cells were placed in goat anti-hamster IgG-coated wells of a 24-well plate in 0.5 ml media with anti-CD3 and anti-CD28. Availability of mice and cells allowed 16–24 miRNAs to be screened at one time such that the complete screen of all miRNAs consisted of five batches. CFSE analysis on day 4 was used to calculate a proliferation index. Proliferation index is the average number of cell divisions among cells that underwent at least one division (Flowjo analysis software). 0.2 × 106 cells of the remaining cells from each well were transfected again on day 4 and restimulated for intracellular staining on day 5. For each batch, the proliferation index and the frequency of IFN-γ-producing cells was normalized to the batch median to give a proliferation score and IFN-γ score, respectively. These values were then used to generate Z scores for the entire set of miRNAs screened (Z = x-mean/SD, where x is the proliferation score or IFN-γ score for each individual miRNA).
Adoptive Transfer and LCMV Infection
CD4+ T cells from SMARTA mice (CD45.1) were isolated by magenetic bead selection, activated in vitro as described, and transduced with pri-miR retroviral vectors (pri-miR-29a or pri-miR-1) at 40 hr. At 45 hr cells were washed and resuspended in PBS, and 5 × 104 cells were injected intravenously into recipient CD45.2 C57BL/6 mice (NCI). One day later, mice were infected intraperitoneally with 2 × 105 plaque forming units of LCMV. On day 8 posttransfer, total splenocytes were harvested and immediately fixed for analysis. CD8+ T cells were activated in vivo, transduced, and transferred as described (Hu et al., 2011) and splenocytes were analyzed on day 7 posttransfer.
Microarray Procedures and Analysis
Sample preparation, labeling, array hybridizations, and false discovery rate calculations were performed according to standard protocols of the UCSF Shared Microarray Core Facilities (http://www.arrays.ucsf.edu) and Agilent (http://www.agilent.com) as previously described (Price et al., 2010).
Luciferase Reporter Assays
The full-length 3′UTR of Tbx21 and Eomes were cloned into the psiCHECK-2 luciferase reporter construct (Promega). CD4+ T cells were cotransfected on day 4 of culture with reporter constructs and miRNA mimics or inhibitors. Luciferase activity was measured 24 hr after transfection with the Dual Luciferase Reporter Assay System (Promega) and a FLUOstar Optima plate-reader (BMG Labtech).
Supplementary Material
Acknowledgments
The authors thank S. Abdul-Wajid and M. Panduro-Sicheva for technical assistance; H.J. Fehling (Institute for Immunology, University Clinics Ulm) for providing R26-tdRFP reporter mice; R. Barbeau, J. Pollack, A. Barczak, and D. Erle for advice and expert assistance with microarray experiments; O. Solberg and P. Woodruff for help with bioinformatic analysis; E. Fink for advice on the use of statistics; A. Weiss and L. Jeker for critical reading of the manuscript; and members of the UCSF miRNA in lymphocytes interest group for helpful discussions. This work was supported by the Dana Foundation, the Burroughs Wellcome Fund (CABS 1006173), the UCSF Program for Breakthrough Biomedical Research, the UCSF/NIH Medical Scientist Training Program (D.F.S. and M.F.T.), and the Susan G. Komen Foundation.
Footnotes
ACCESSION NUMBERS
The microarray data are available in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/gds) under the accession number GSE30525.
Supplemental Information includes Supplemental Experimental Procedures, five figures, and three tables and can be found with this article online at doi:10.1016/j.immuni.2011.07.009.
References
- Afkarian M, Sedy JR, Yang J, Jacobson NG, Cereb N, Yang SY, Murphy TL, Murphy KM. T-bet is a STAT1-induced regulator of IL-12R expression in naïve CD4+ T cells. Nat Immunol. 2002;3:549–557. doi: 10.1038/ni794. [DOI] [PubMed] [Google Scholar]
- Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A, Schambach F, DeJong CS, Hammond SM, Reiner SL. Micro-RNA-155 inhibits IFN-gamma signaling in CD4+ T cells. Eur J Immunol. 2010;40:225–231. doi: 10.1002/eji.200939381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Jothi R, Cuddapah S, Cui K, Roh TY, Schones DE, Zhao K. Chromatin poises miRNA- and protein-coding genes for expression. Genome Res. 2009;19:1742–1751. doi: 10.1101/gr.090951.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36(Database issue):D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezman NA, Cedars E, Steiner DF, Blelloch R, Hesslein DG, Lanier LL. Distinct requirements of microRNAs in NK cell activation, survival, and function. J Immunol. 2010;185:3835–3846. doi: 10.4049/jimmunol.1000980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong MM, Rasmussen JP, Rudensky AY, Littman DR. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. J Exp Med. 2008;205:2005–2017. doi: 10.1084/jem.20081219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb BS, Hertweck A, Smith J, O’Connor E, Graf D, Cook T, Smale ST, Sakaguchi S, Livesey FJ, Fisher AG, Merkenschlager M. A role for Dicer in immune regulation. J Exp Med. 2006;203:2519–2527. doi: 10.1084/jem.20061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finotto S, Neurath MF, Glickman JN, Qin S, Lehr HA, Green FH, Ackerman K, Haley K, Galle PR, Szabo SJ, et al. Development of spontaneous airway changes consistent with human asthma in mice lacking T-bet. Science. 2002;295:336–338. doi: 10.1126/science.1065544. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamper CJ, Agoston AT, Nelson WG, Powell JD. Identification of DNA methyltransferase 3a as a T cell receptor-induced regulator of Th1 and Th2 differentiation. J Immunol. 2009;183:2267–2276. doi: 10.4049/jimmunol.0802960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Liu S, Fabbri M, Liu Z, Heaphy CE, Callegari E, Schwind S, Pang J, Yu J, Muthusamy N, et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JK, Kagari T, Clingan JM, Matloubian M. Expression of chemokine receptor CXCR3 on T cells affects the balance between effector and memory CD8 T-cell generation. Proc Natl Acad Sci USA. 2011;108:E118–E127. doi: 10.1073/pnas.1101881108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovska I, Ball AS, Diaz RL, Magnus JF, Kibukawa M, Schelter JM, Kobayashi SV, Lim L, Burchard J, Jackson AL, et al. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol Cell Biol. 2008;28:2167–2174. doi: 10.1128/MCB.01977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, Hagman J, Gapin L, Kaech SM. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27:281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JA, Cho K, Shin MS, Lee WG, Jung N, Chung C, Chang JK. A novel electroporation method using a capillary and wire-type electrode. Biosens Bioelectron. 2008;23:1353–1360. doi: 10.1016/j.bios.2007.12.009. [DOI] [PubMed] [Google Scholar]
- Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, Jensen K, Cobb BS, Merkenschlager M, Rajewsky N, Rajewsky K. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132:860–874. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- Kuchen S, Resch W, Yamane A, Kuo N, Li Z, Chakraborty T, Wei L, Laurence A, Yasuda T, Peng S, et al. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity. 2010;32:828–839. doi: 10.1016/j.immuni.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lighvani AA, Frucht DM, Jankovic D, Yamane H, Aliberti J, Hissong BD, Nguyen BV, Gadina M, Sher A, Paul WE, O’Shea JJ. T-bet is rapidly induced by interferon-gamma in lymphoid and myeloid cells. Proc Natl Acad Sci USA. 2001;98:15137–15142. doi: 10.1073/pnas.261570598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston A, Lu LF, O’Carroll D, Tarakhovsky A, Rudensky AY. Dicer-dependent microRNA pathway safeguards regulatory T cell function. J Exp Med. 2008;205:1993–2004. doi: 10.1084/jem.20081062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu LF, Boldin MP, Chaudhry A, Lin LL, Taganov KD, Hanada T, Yoshimura A, Baltimore D, Rudensky AY. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 2010;142:914–929. doi: 10.1016/j.cell.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luche H, Weber O, Nageswara Rao T, Blum C, Fehling HJ. Faithful activation of an extra-bright red fluorescent protein in “knock-in” Cre-reporter mice ideally suited for lineage tracing studies. Eur J Immunol. 2007;37:43–53. doi: 10.1002/eji.200636745. [DOI] [PubMed] [Google Scholar]
- Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott JL, Kobayashi S, Bronk SF, Gores GJ. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26:6133–6140. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathans R, Chu CY, Serquina AK, Lu CC, Cao H, Rana TM. Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol Cell. 2009;34:696–709. doi: 10.1016/j.molcel.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, Cordon-Cardo C, Li QJ, Lowe SW, Hannon GJ, He L. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009;23:2839–2849. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, Zediak VP, Banica M, DiCioccio CB, Gross DA, Mao CA, et al. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 2003;302:1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini M, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- Price AE, Liang HE, Sullivan BM, Reinhardt RL, Eisley CJ, Erle DJ, Locksley RM. Systemically dispersed innate IL-13-expressing cells in type 2 immunity. Proc Natl Acad Sci USA. 2010;107:11489–11494. doi: 10.1073/pnas.1003988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suto A, Wurster AL, Reiner SL, Grusby MJ. IL-21 inhibits IFN-gamma production in developing Th1 cells through the repression of Eomesodermin expression. J Immunol. 2006;177:3721–3727. doi: 10.4049/jimmunol.177.6.3721. [DOI] [PubMed] [Google Scholar]
- Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- Szabo SJ, Sullivan BM, Peng SL, Glimcher LH. Molecular mechanisms regulating Th1 immune responses. Annu Rev Immunol. 2003;21:713–758. doi: 10.1146/annurev.immunol.21.120601.140942. [DOI] [PubMed] [Google Scholar]
- Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci USA. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarino AV, Katzman SD, Gallo E, Miller O, Jiang S, McManus MT, Abbas AK. Posttranscriptional silencing of effector cytokine mRNA underlies the anergic phenotype of self-reactive T cells. Immunity. 2011;34:50–60. doi: 10.1016/j.immuni.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet. 2008;40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi R, Junttila IS, Wei G, Urban JF, Jr, Zhao K, Paul WE, Zhu J. The transcription factor GATA3 actively represses RUNX3 protein-regulated production of interferon-gamma. Immunity. 2010;32:507–517. doi: 10.1016/j.immuni.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Bevan MJ. Dicer controls CD8+ T-cell activation, migration, and survival. Proc Natl Acad Sci USA. 2010;107:21629–21634. doi: 10.1073/pnas.1016299107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Jeker LT, Fife BT, Zhu S, Anderson MS, McManus MT, Bluestone JA. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J Exp Med. 2008;205:1983–1991. doi: 10.1084/jem.20080707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations. Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.