Abstract
Rapidly declining biodiversity may be a contributing factor to another global megatrend—the rapidly increasing prevalence of allergies and other chronic inflammatory diseases among urban populations worldwide. According to the “biodiversity hypothesis,” reduced contact of people with natural environmental features and biodiversity may adversely affect the human commensal microbiota and its immunomodulatory capacity. Analyzing atopic sensitization (i.e., allergic disposition) in a random sample of adolescents living in a heterogeneous region of 100 × 150 km, we show that environmental biodiversity in the surroundings of the study subjects’ homes influenced the composition of the bacterial classes on their skin. Compared with healthy individuals, atopic individuals had lower environmental biodiversity in the surroundings of their homes and significantly lower generic diversity of gammaproteobacteria on their skin. The functional role of the Gram-negative gammaproteobacteria is supported by in vitro measurements of expression of IL-10, a key anti-inflammatory cytokine in immunologic tolerance, in peripheral blood mononuclear cells. In healthy, but not in atopic, individuals, IL-10 expression was positively correlated with the abundance of the gammaproteobacterial genus Acinetobacter on the skin. These results raise fundamental questions about the consequences of biodiversity loss for both allergic conditions and public health in general.
Keywords: biodiversity benefits, hygiene hypothesis, microbial deprivation, civilization diseases
By 2050, some predict that two-thirds of the global human population will live in urban areas with little green space and limited contact with nature and biodiversity (1). At the same time, an increasing fraction of the urban population will suffer from chronic inflammatory disorders (2, 3), of which allergic (4) and autoimmune diseases are prime examples. Building on the hygiene hypothesis (5, 6), the notion that growing up in a farming environment protects children from allergic sensitization (7, 8), and the emerging understanding of the role of microbes in the development and maintenance of epithelial cell integrity and tolerance (3, 9), the “biodiversity hypothesis” (10) proposes that reduced contact of people with natural environmental features and biodiversity, including environmental microbiota, leads to inadequate stimulation of immunoregulatory circuits. Importantly, interactions with the natural environment may influence the composition of the human commensal microbiota, the members of which are not equal in their ability to stimulate the regulatory circuits via Toll-like and other antigen-recognizing receptors to prevent or terminate inappropriate inflammatory responses (3).
To test the biodiversity hypothesis of inflammatory disorders, we studied a random sample of 118 adolescents inhabiting a small town, villages of different sizes, and isolated houses within a 100 × 150 km region in eastern Finland. The inflammatory disorder that we examined is atopic sensitization, which involves the propensity to develop IgE antibodies in response to allergen exposure (11). Here we address four questions. First, we examine whether the environmental biodiversity influences the composition of the commensal microbiota of the study subjects. Environmental biodiversity was characterized at two spatial scales, the vegetation cover of the yards and the major land use types within 3 km of the homes of the study subjects. Commensal microbiota sampling evaluated the skin bacterial flora, identified to the genus level from DNA samples obtained from the volar surface of the forearm. Second, we investigate whether atopy is related to environmental biodiversity in the surroundings of the study subjects’ homes. Third, we examine whether atopy is related to the composition of the skin microbial community. Finally, we characterize the immune function of the study subjects by in vitro measurement of IL-10 expression in peripheral blood mononuclear cells (PBMCs) and relate it to the composition of the skin microbiota. IL-10 is one of the key anti-inflammatory cytokines in immunologic tolerance.
Results
Environmental Biodiversity and Skin Microbiota.
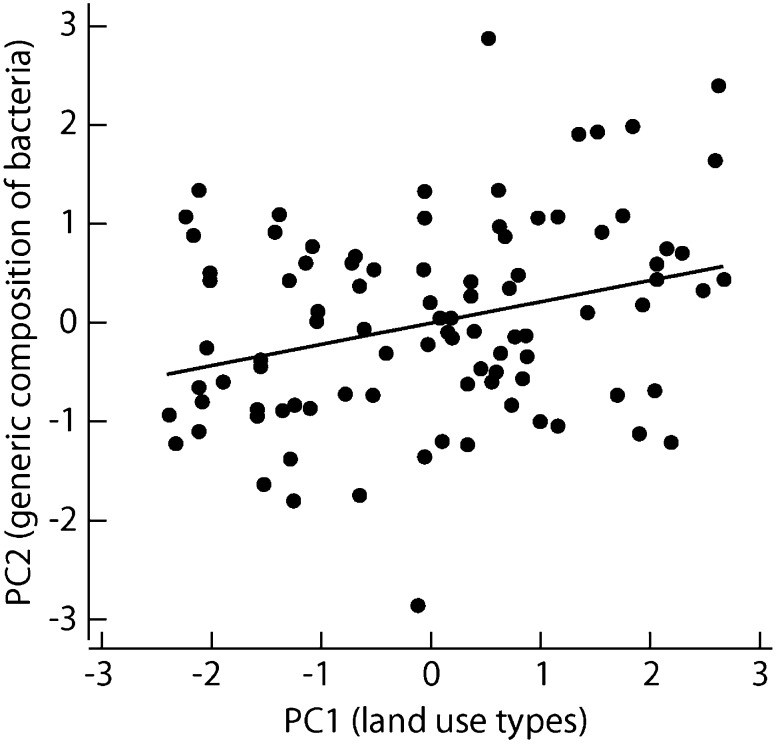
We estimated the areas of the following land use types within 3 km of the home of each study subject: forest, covering on average 49% of the total area around the homes; agricultural land, 12%; built areas, 16%; lakes and other water bodies, 20%; and wetlands, 3%. The first principal component (PC1env) of the land use data was positively correlated with forest and agricultural land and negatively correlated with built areas and water bodies, thus correlating with terrestrial vegetated habitats (SI Appendix, Table S1). The pooled microbiota for the 118 study subjects included 572 bacterial genera in 43 classes. Consistent with previous studies of the skin microbiota (12, 13), the dominant classes were Actinobacteria, Bacilli, Clostridia, Betaproteobacteria, Alphaproteobacteria, and Gammaproteobacteria (Table 1). We characterized the composition of the skin microbiota with a principal components analysis (PCA) of the numbers of genera in these six bacterial classes. The second principal component (PC2bac) correlated positively with the generic diversity of proteobacteria and negatively with the generic diversity of all other bacterial classes (SI Appendix, Table S2). The PC1env of the land use types was significantly (P = 0.0033) related to PC2bac (Fig. 1), indicating that the generic diversity of proteobacteria was higher on the skin of individuals living in an environment with more forest and agricultural land compared with those living in built areas and near water bodies. Repeating the PCA for the relative abundances of the bacterial classes instead of their generic diversity did not yield a significant association with PC1env, although the relative abundances of all proteobacterial classes were positively correlated, and the relative abundances of the other bacterial classes were negatively correlated, with PC1env, consistent with the pattern found for generic diversity (PC2bac).
Table 1.
Statistics for the six numerically dominant bacterial classes and their association with atopy
| Bacterial class | Relative abundance | Generic diversity | ||||
| Percentage | Sign | P | No. of genera | Sign | P | |
| Actinobacteria | 56.7 | + | 0.08 | 126 | + | 0.04 |
| Bacilli | 15.6 | + | 0.84 | 68 | + | 0.94 |
| Clostridia | 4.5 | − | 0.26 | 56 | − | 0.58 |
| Betaproteobacteria | 9.6 | − | 0.13 | 62 | + | 0.57 |
| Alphaproteobacteria | 3.8 | − | 0.51 | 76 | + | 0.76 |
| Gammaproteobacteria | 2.8 | − | 0.83 | 52 | − | 0.0003 |
| No. of study subjects | 118 | 116 | 118 | 116 | ||
The “Relative abundance” columns give the percentage of sequences in each bacterial class (total number of sequences, 1,236,839) and the significance of atopy in explaining the relative abundance of the bacterial class on the skin of the study subject (ANOVA). The “Sign” column (+ or −) indicates the direction of the effect. The “Genetic diversity” columns report the number of genera and the significance of atopy in explaining the generic diversity of the bacterial class (as in Fig. 2B, using the total number of bacterial genera as a covariate to account for variation in sample size). The total number of bacterial genera in the pooled material was 572, of which the six classes in this table accounted for 77%.
Fig. 1.
Relationship between the generic composition of skin microbiota and land use types around the home. The vertical axis shows PC2bac, which correlates positively with the generic diversity of proteobacteria and negatively with the diversity of all other bacterial classes (SI Appendix, Table S2). The horizontal axis shows PC1env, which summarizes variation in land use types within a 3-km radius of the homes of the study subjects and is positively correlated with forests and agricultural land (SI Appendix, Table S1). Regression: F = 9.12, df = 1.93, P = 0.0033.
We used 15 vegetation and other land cover types to describe the yards around the homes, with an average area of 0.17 ha. The second principal component of these data was correlated with PC2bac (P = 0.02), but because this factor was also correlated with PC1env, it did not make an independent contribution in a multiple regression model. PC2bac was not related to plant species richness in the yard or to the type, age, or condition of the house.
Determination of Atopy.
The study subjects were divided into healthy individuals and atopic individuals based on IgE antibody level in a screen with Phadiatop, a mixture of common inhalant allergens. The distribution of IgE values was bimodal (SI Appendix, Fig. S2), and in this this study we used a cutoff value of 2.5 kUA/L to identify individuals with atopic sensitization. To verify that the results were not sensitive to the exact cutoff point, we repeated the association analyses involving atopy with a lower cutoff point of 1 kUA/L (SI Appendix, Fig. S2). The same study subjects had participated in a comprehensive study of allergy 7 y earlier, including skin prick testing (SPT) against common inhalant allergens (14). Evaluation of atopic sensitization by SPT in 2003 and an IgE screen in 2010 yielded highly concordant results (χ2 = 63.2, P < 0.0001; 88% of individuals classified similarly), indicating constancy of individuals’ atopic sensitization from early to late adolescence.
Environmental Biodiversity and Atopy.
Atopic individuals resided throughout the study area (SI Appendix, Fig. S3), with no spatial autocorrelation in their occurrence in the study population (Moran’s I, Z = −0.75, P = 0.45). Atopy was somewhat more frequent in the small town of Joensuu than in the rest of the study area (0.51 vs. 0.38), but the difference was not statistically significant (χ2 = 1.97, P = 0.16). However, atopy was significantly explained by environmental biodiversity around the homes of the study subjects. Atopy decreased with PC1env of the land use types, and thus with the amount of forested and agricultural land within 3 km of the home (Table 2). The types of vegetation in the yard had no effect, but species richness of one group of plants—uncommon native flowering plants—was significantly negatively correlated with atopy (Table 2). This finding is illustrated in Fig. 2A, showing that the number of uncommon native flowering plant species was ∼25% higher in the yards of healthy individuals compared with the yards of atopic individuals. In this analysis, we used the total number of all plant species identified in the yard as a covariate to account for differences in size and type of yard. The effect of uncommon native flowering plant species was an exception, given that species richness in other plant categories was unrelated to atopy (SI Appendix, Table S3).
Table 2.
Logistic regression models of atopy
| Variable | Stepwise model | Regression model | |||
| Deviance | Difference | P | Coefficient | P | |
| Constant | 158.0 | −0.58 | 0.023 | ||
| Land use types, PC1env | 117.6 | 40.4 | <0.0001 | −0.52 | 0.0059 |
| Flowering plants | 107.4 | 10.1 | 0.0014 | −0.10 | 0.0016 |
| Gammaproteobacteria | 100.9 | 6.5 | 0.011 | −0.31 | 0.015 |
| P value of the model | 0.20 | ||||
| Positive cases/N | 38/94 | ||||
The three explanatory variables are the first principal component of land use types (PC1env; SI Appendix, Table S1), the number of uncommon native flowering plant species in the yard (residual, accounting for the effects of field assistants and the total number of vascular plant species in the yard; Fig. 2A), and the generic diversity of gammaproteobacteria (residual, accounting for the effect of the total number of bacterial genera; Fig. 2B).
Fig. 2.
Relationships among environmental biodiversity, skin microbiota, and atopy in the study subjects. (A) The number of uncommon native flowering plant species plotted against the total number of plant species in the yard of atopic individuals (solid symbols) and healthy individuals (open symbols). The number of plant species on the vertical axis is the residual accounting for variation in the results of five pairs of field assistants. The effect of atopy is significant (t = −3.14, P = 0.0022, n = 94; without correcting for the effect of field assistants, t = −2.83, P = 0.0058). (B) The number of genera of gammaproteobacteria plotted against the total number of bacterial genera in the skin microbiota of atopic individuals (solid symbols) and healthy individuals (open symbols). The effect of atopy is highly significant (t = −3.72, P = 0.0003, n = 112).
Using the data collected in 2003 (14), we examined a number of additional factors that might have affected atopy in the study population (e.g., family member smoking, living on a farm, frequent contact with pets). None of these factors was significantly related to atopy (SI Appendix, Table S4).
Skin Microbiota and Atopy.
Atopy was not related to PC2bac, which we used to characterize the skin microbiota, but a strikingly strong, more specific association was detected at the level of bacterial class. Atopic individuals had highly significantly (P = 0.0003) lower generic diversity of gammaproteobacteria on the skin compared with healthy individuals (Fig. 2B and Tables 1 and 2). In this analysis, we used the total number of bacterial genera in the sample to account for variation in sample size (i.e., number of sequences), which is more effective here than using rarefaction (SI Appendix). Note that the correlation was detected specifically with generic diversity, rather than with relative abundance of gammaproteobacteria (Table 1). For the other bacterial classes, there were no correlations that even approached significance with either generic diversity or relative abundance (Table 1). IgE tests for specific common inhalant allergens (cat, dog, horse, birch, timothy grass, and mugwort) were all negatively correlated with the generic diversity of gammaproteobacteria, but none of these correlations was much stronger than the others, with P values ranging from 0.01 to 0.10 (SI Appendix, Table S5). These results imply that the strong and highly significant negative correlation between the generic diversity of gammaproteobacteria and atopy (Fig. 2B) is not due solely to any particular allergen.
We repeated the analysis of the correlation of atopy with environmental biodiversity and the richness of gammaproteobacteria using two other criteria for atopy: a different cutoff point for IgE values (SI Appendix, Fig. S2) and the SPT done in 2003. The results remained essentially the same (SI Appendix, Table S6).
Gammaproteobacteria and Immune Function.
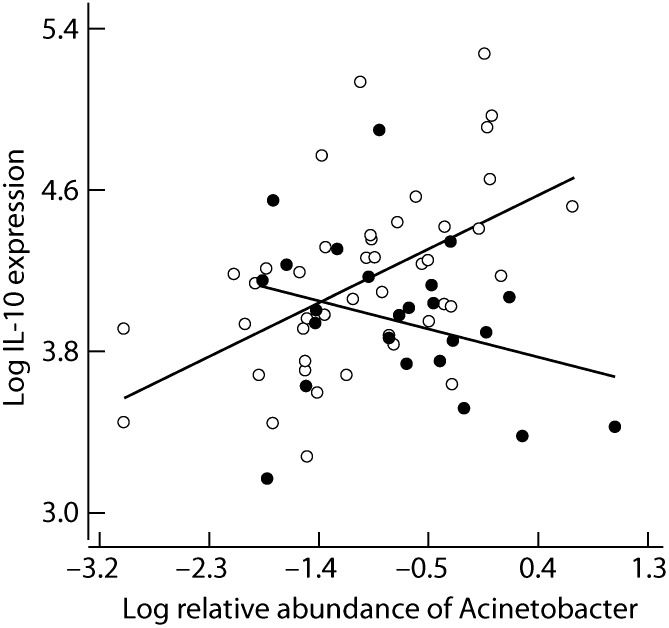
The foregoing observational results (Fig. 2B and Tables 1 and 2) imply a strong allergy-protective effect for gammaproteobacteria. This conclusion is supported by in vitro measurements of baseline levels of IL-10 mRNA in PBMCs in a large subset of the study subjects (n = 69). We analyzed correlations between IL-10 expression and the relative abundance and generic diversity of the different bacterial classes in the skin microbiota separately for healthy individuals and atopic individuals. We found one significant correlation, between the relative abundance of gammaproteobacteria and IL-10 expression in healthy individuals (P = 0.015) (SI Appendix, Table S7). Examining this association at the generic level revealed one highly significant correlation, between the relative abundance of Acinetobacter and IL-10 expression (P = 0.0004) (SI Appendix, Table S8), which was reversed in atopic individuals (Fig. 3). At present, we do not know which cells are mainly responsible for IL-10 secretion in this material. In addition to allergen-specific T regulatory cells (15), T effector cells, B cells, and innate cells (monocytes/macrophages) (16) secrete substantial amounts of IL-10 and thus potentially influence the development of immunologic tolerance.
Fig. 3.
Cytokine IL-10 expression against the relative abundance of Acinetobacter in the skin community of healthy (open symbols) and atopic individuals (filled dots). The interaction term is highly significant (P = 0.0009 in a linear regression model, with adjusted R2 = 0.23) (SI Appendix, Table S8).
Discussion
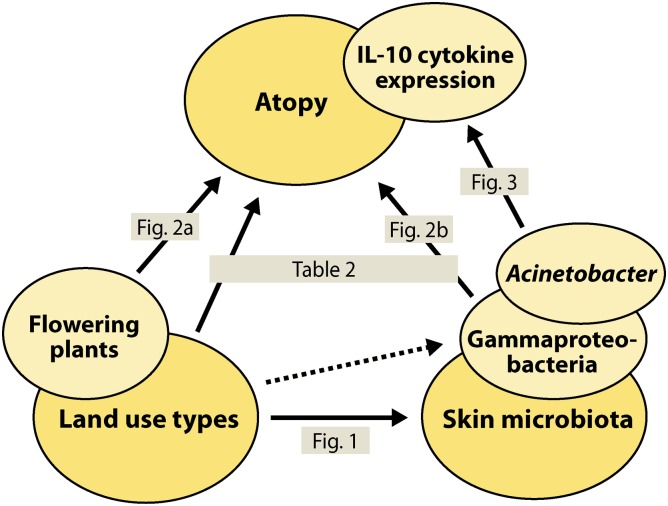
Environmental biodiversity, the human commensal microbiota, and the human immune system are complex systems with numerous components (i.e., species and molecules) that interact with each other. One might assume that interactions between these three systems might lead to intractable dynamics and defy any attempts to identify general patterns, a necessary starting point for developing a mechanistic understanding of these dynamics. Nonetheless, the present study involving a modest number of study subjects revealed several significant correlations (Fig. 4). We conjecture that this study had sufficient power to detect these associations, because the study subjects had been randomly selected among the school children inhabiting a large region with widely varying environmental conditions, and because the study subjects had lived in the same dwellings during their entire childhood and thus were exposed to the same surrounding environmental conditions for a long period. We realize that the skin microbiota might be affected by the study subjects’ washing habits and the kinds of soaps and detergents they used, but a correlation between these factors and the strong relationship between the skin microbiota and atopy is not plausible.
Fig. 4.
Summary graph of the associations among environmental biodiversity, skin microbiota, and atopy. The solid arrows refer to the results in Figs. 1–3 and Table 2. The dashed-line arrow indicates a less significant effect of PC1env on the generic diversity of gammaproteobacteria (t = 1.91, P = 0.059, n = 95, with total number of bacterial genera as a covariate as in Fig. 2B).
We hypothesize that the associations between environmental biodiversity and atopy reflect immunologic responses developed by individuals with long-term exposure to particular assortments of environmental microbiota and natural allergens. The structure of the commensal microbiota showed much variation among the study subjects and was influenced by the environment, consistent with previous studies demonstrating significant differences between persons in commensal microbiota (12, 13, 15). Our results do not reveal the mechanism of the environmental influence on atopy, but it is possible that this effect is due to the influence of environmental microbiota on commensal microbiota. Microbes are readily transmitted via pollen grains, dust, and ambient air (17), and the microbes may act as both adjuvants (18, 19) and triggers of the regulatory circuits (3, 9). Other environmental features, such as the amount and diversity of pollen, may play a role, although a simple relationship between pollen exposure and allergic diseases is unlikely (20).
Turning to the significant association between gammaproteobacteria and atopy, it is conceivable that atopy affects the composition of the skin microbiota. Changes in gut microbiota have been reported in patients with allergic diseases (21, 22), although it is also possible that these changes precede the development of allergic manifestations and thus are a cause rather than (or as well as) a consequence (23, 24). An affect of allergic disease on skin microbiota is particularly plausible in atopic eczema, which is commonly treated topically with corticosteroid ointments that include antibacterial agents. However, atopic eczema is unlikely to explain the association between atopy and gammaproteobacteria in the present study, given the lack of significant association between atopy and self-reported atopic eczema in our study population (χ2 = 3.16, P = 0.08). Previous studies have found increased abundance of Staphylococcus on the skin of individuals with atopic eczema (25, 26). We found the same, with staphylococci accounting for 5.7% of all sequences in individuals with atopic eczema versus 4.4% in healthy individuals, a non-statistically significant difference (F = 1.53, P = 0.22).
Although we cannot definitely exclude the possibility that atopy can somehow encourage a more diverse assemblage of gammaproteobacteria in the skin microbiota, the reverse causality is much more likely, for several reasons. First, several previous studies found that high diversity of the commensal microbiota is associated with low risk for allergic diseases (27–29), and a recent study, in line with ours, suggested a role for proteobacteria in this context (30). Second, endotoxin derived from Gram-negative bacteria, such as gammaproteobacteria, is known to have immunomodulatory and allergy-protective potential (31). Third, the positive association between the abundance of the gammaproteobacterial genus Acinetobacter and IL-10 expression in PBMCs in healthy individuals, but not in atopic individuals, is consistent with IL-10’s central role in maintaining immunologic tolerance to harmless substances (16). Hessle et al. (32) reported that seven Gram-negative bacteria (including four gammaproteobacteria) significantly stimulated IL-10 secretion in PBMCs obtained from healthy blood donors (see also refs. 33 and 34), and Zhang et al. (35) found significant IL-10 expression in Acinetobacter lwoffii-pulsed dendritic cells. The lack of association between Acinetobacter and IL-10 expression in atopic individuals in the present study might reflect a breakdown of the regulatory mechanisms and an imbalance in the ratio of T regulatory cells to T effector memory cells in atopic individuals (36, 37). Finally, a series of experimental studies using the mouse model have demonstrated strong allergy-protective properties for A. lwoffii (38–40). These latter studies leave no doubt about causality.
Thus, our results imply that gammaproteobacteria, although representing only 3% of the sequences in the skin community (Table 1), may play a special role in the development and maintenance of the skin homeostasis and healthy barrier function, similar to that of certain gut bacteria (41, 42). Gammaproteobacteria are common in the soil but are particularly dominant in above-ground vegetation, such as flowering plants (43), and they have been detected on, for example, grass pollen grains (17). Why atopy is associated with the gammaproteobacteria’s generic diversity rather than with their relative abundance remains an open question warranting further study. It is possible that microbial diversity as such is important (29, 30, 44), but it also is possible that our results reflect the history of atopic sensitization in the study subjects. The microbial sample was obtained in 2010, long after the development of atopy in these individuals. Therefore, any feature of the commensal microbiota that we have sampled must exhibit long-term constancy to be a serious candidate as the causal agent. It is possible that the generic diversity of a bacterial class is a more constant feature than its relative abundance.
In conclusion, the present results demonstrate that biodiversity can be surprisingly strongly associated with atopy, a common immune dysfunction of modern era. This association is observed both at the scale of the macrobiota, here in the form of species richness of native flowering plants and land use types in the wider environment, and at the scale of the microbiota, in the form of the diversity of one class of bacteria in the skin microbial community. The mechanisms underlying these associations remain to be clarified, but their implications are profound. Interactions with natural environmental features not only may increase general human well being in urban areas (45), but also may enrich the commensal microbiota and enhance its interaction with the immune system, with far-reaching consequences for public health.
Materials and Methods
Study Subjects.
The study subjects were a random sample of 14- to 18-y-old school children living within a 100 × 150 km area in eastern Finland (SI Appendix, Fig. S3). The subjects were originally recruited to a comprehensive study of allergy in 2003 (14), and their families continued to live, with a few exceptions, in the same dwellings in 2010–2011, when data and samples for the present study were collected. The subjects’ homes included apartments, row homes, and individual houses in the town of Joensuu (73,000 inhabitants) and villages of varying populations, as well as isolated houses in the sparsely populated countryside (SI Appendix, Fig. S3).
Allergy Tests.
Blood samples (n = 116) were collected in September 2010 and screened with Phadiatop (a mixture of common inhalant allergens) and ImmunoCAP (Phadia). Samples with an IgE antibody level ≥0.35 kUA/L were also analyzed for allergen-specific IgE antibodies. In the previous study of the same subjects in 2003, SPT was performed against a standard set of eight inhalant allergens (ALK-Abello) with negative (solvent) and positive controls (histamine dihydrochloride 10 mg/mL). Individuals positive (i.e., wheal diameter ≥3 mm) for at least one of the eight inhalant allergens tested were classified as atopic. Background and clinical data were obtained using questionnaires (14). The characteristics of the study subjects with respect to atopy as defined by SPT are presented in SI Appendix, Table S4.
Real-Time Quantitative PCR Analysis.
Blood samples (n = 69) were collected in September 2011. PBMCs were separated from whole blood in BD Vacutainer CPT cell preparation tubes with sodium heparin, frozen, and shipped to the analysis site. This approach provides unique opportunities to stimulate cells in vitro by different compounds, as well as to analyze cytokine and surface molecule expression using state-of-the-art biomedical methods. The thawed PBMCs were cultured in 24-well plates at 1 × 106/mL in complete RPMI-1640 medium with 10% heat-inactivated FBS at 37 °C and 5% CO2. After 24 h of culture, total RNA was extracted from the PBMCs using Trisure (Bioline) according to the manufacturer’s instructions. The RNA was reverse-transcribed into cDNA, and the mRNA expression level of IL-10 was measured by real-time quantitative PCR using the Applied Biosystems 7500 Fast Real-Time PCR System. PCR amplification of the endogenous 18S rRNA was performed for each sample to control sample loading and allow normalization between the samples. The results were expressed as relative units, calculated by the comparative CT method according to the manufacturer’s instructions.
Skin Microbiota.
We sampled 118 individuals in September 2010 by lightly pressing a sterile nylon swab (Copan Diagnostics) dipped in sterile 0.15 M NaCl + 0.1% Tween 20 solution against the skin on a 5 × 5 cm area of the volar surface of the forearm of the subject’s writing hand, midway between the wrist and elbow. Total DNA was extracted using FastDNA Spin Kit for Soil (MP Biomedicals), according to the manufacturer’s instructions. PCR amplification was carried out in a PTC-225 thermal cycler (MJ Research). The V1–V3 regions of the 16S rRNA gene were amplified with modified universal bacterial primers pA (AGAGTTTGATCMTGGCTCAG; ref. 46) and pD′ (GTATTACCGCGGCTGCTG; ref. 47). Phusion polymerase (Thermo Fisher Scientific/Finnzymes) with the HF buffer and 2.5% DMSO were used. Cycling conditions consisted of an initial denaturation at 98 °C for 30 s, followed by 30 cycles of 98 °C for 10 s, 65 °C for 30 s, and 72 °C for 10 s, and then a final extension for 5 min. Between 10 and 15 ng of template was used for each reaction. PCR products were processed as described previously (48) and sequenced using the 454-GS FLX Titanium protocol, with an average read length of ∼400 bp (Roche Diagnostics). The sequence data were analyzed using mothur (49). Tag and primer sequences, as well as low-quality sequences (i.e., ambiguous nucleotides, homopolymers longer than eight nucleotides, average quality score <25) were removed. Sequences were aligned and clustered, and OTUs were defined. The sample-specific sequences were uploaded into the RDP Classifier (50) to identify the bacterial classes and genera, with 80% as the threshold value. The sequences have been deposited in the Sequence Read Archive at the European Bioinformatics Institute (accession no. ERP001059).
Environmental Biodiversity.
Data on environmental biodiversity were collected in June and July 2010. For each home, the type (apartment building, row house, single/double house, or farmhouse), age, and condition (good, moderate, bad) were recorded. The area of the yard surrounding the house was estimated (average, 0.17 ha). Ten field assistants working in pairs recorded the species of vascular plants, which were classified into five functional groups and into “common” and “uncommon” species (SI Appendix, Table S3). The percentages of 15 land cover types in the yards were estimated. Land use types in the broader environment surrounding the home within a radius of 3 km were characterized using the CORINE2000 land cover database (51).
Statistical Analyses.
Because of logistical problems, complete data including blood samples, skin swabs, and the environmental features could be obtained for only 95 individuals. The data were normally distributed after logarithmic transformation of the relative abundances of the six main bacterial classes. Data were analyzed by parametric ANOVA, linear and logistic regression, and PCA.
Supplementary Material
Acknowledgments
We thank T. Vlasoff and S. Lipponen for obtaining DNA and blood samples; E.-M. Turkki, K. Lipponen, and H. Kangas for technical assistance in processing the bacterial samples; and J. Corander, C. Godfray, H. Jacobs, E. von Mutius, and G. Rook for comments on the manuscript. This work was supported by European Research Council Advanced Grant 232826 (to I.H.), European Commission Seventh Framework Programme Grant Agreement 261357, the Finnish CoE Programme 2006–2011, Academy of Finland Grants 131155 and 138932, Helsinki University Hospital Research Grant 8361, the Juselius Foundation, and the Liv och Hälsa Foundation.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the Sequence Read Archive at the European Bioinformatics Institute (accession no. ERP001059).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205624109/-/DCSupplemental.
References
- 1.United Nations . World Urbanization Prospects: The 2007 Revision. New York: United Nations; 2008. [Google Scholar]
- 2.Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med. 2002;347:911–920. doi: 10.1056/NEJMra020100. [DOI] [PubMed] [Google Scholar]
- 3.Rook GAW. 99th Dahlem conference on infection, inflammation and chronic inflammatory disorders: Darwinian medicine and the “hygiene” or “old friends” hypothesis. Clin Exp Immunol. 2010;160:70–79. doi: 10.1111/j.1365-2249.2010.04133.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graham-Rowe D. When allergies go west. Nature. 2011;479:S2–S4. [Google Scholar]
- 5.Wills-Karp M, Santeliz J, Karp CL. The germless theory of allergic disease: Revisiting the hygiene hypothesis. Nat Rev Immunol. 2001;1:69–75. doi: 10.1038/35095579. [DOI] [PubMed] [Google Scholar]
- 6.Strachan DP. Hay fever, hygiene, and household size. BMJ. 1989;299:1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ege MJ, et al. GABRIELA Transregio 22 Study Group Exposure to environmental microorganisms and childhood asthma. N Engl J Med. 2011;364:701–709. doi: 10.1056/NEJMoa1007302. [DOI] [PubMed] [Google Scholar]
- 8.von Mutius E, Vercelli D. Farm living: Effects on childhood asthma and allergy. Nat Rev Immunol. 2010;10:861–868. doi: 10.1038/nri2871. [DOI] [PubMed] [Google Scholar]
- 9.Rook GAW. Review series on helminths, immune modulation and the hygiene hypothesis: The broader implications of the hygiene hypothesis. Immunology. 2009;126:3–11. doi: 10.1111/j.1365-2567.2008.03007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.von Hertzen L, Hanski I, Haahtela T. Natural immunity: Biodiversity loss and inflammatory diseases are two global megatrends that might be related. EMBO Rep. 2011;12:1089–1093. doi: 10.1038/embor.2011.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arshad SH, Tariq SM, Matthews S, Hakim E. Sensitization to common allergens and its association with allergic disorders at age 4 years: A whole population birth cohort study. Pediatrics. 2001;108:E33. doi: 10.1542/peds.108.2.e33. [DOI] [PubMed] [Google Scholar]
- 12.Costello EK, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grice EA, et al. NISC Comparative Sequencing Program Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.von Hertzen L, et al. Growing disparities in atopy between the Finns and the Russians: A comparison of 2 generations. J Allergy Clin Immunol. 2006;117:151–157. doi: 10.1016/j.jaci.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 15.Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011;9:244–253. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lloyd CM, Hawrylowicz CM. Regulatory T cells in asthma. Immunity. 2009;31:438–449. doi: 10.1016/j.immuni.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heydenreich B, et al. Gram-positive bacteria on grass pollen exhibit adjuvant activity inducing inflammatory T cell responses. Clin Exp Allergy. 2012;42:76–84. doi: 10.1111/j.1365-2222.2011.03888.x. [DOI] [PubMed] [Google Scholar]
- 18.Herrick CK, Bottomly K. To respond or not to respond: T cells in allergic asthma. Nat Rev Immunol. 2003;3:405–412. doi: 10.1038/nri1084. [DOI] [PubMed] [Google Scholar]
- 19.Napolitani G, Rinaldi A, Bertoni F, Sallusto F, Lanzavecchia A. Selected Toll-like receptor agonist combinations synergistically trigger a T helper type 1-polarizing program in dendritic cells. Nat Immunol. 2005;6:769–776. doi: 10.1038/ni1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ariano R, Canonica GW, Passalacqua G. Possible role of climate changes in variations in pollen seasons and allergic sensitizations during 27 years. Ann Allergy Asthma Immunol. 2010;104:215–222. doi: 10.1016/j.anai.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 21.Penders J, et al. Gut microbiota composition and development of atopic manifestations in infancy: The KOALA Birth Cohort Study. Gut. 2007;56:661–667. doi: 10.1136/gut.2006.100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gore C, et al. Bifidobacterium pseudocatenulatum is associated with atopic eczema: A nested case-control study investigating the fecal microbiota of infants. J Allergy Clin Immunol. 2008;121:135–140. doi: 10.1016/j.jaci.2007.07.061. [DOI] [PubMed] [Google Scholar]
- 23.Vael C, Vanheirstraeten L, Desager KN, Goossens H. Denaturing gradient gel electrophoresis of neonatal intestinal microbiota in relation to the development of asthma. BMC Microbiol. 2011;11:68. doi: 10.1186/1471-2180-11-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakayama J, et al. Aberrant structures of fecal bacterial community in allergic infants profiled by 16S rRNA gene pyrosequencing. FEMS Immunol Med Microbiol. 2011;63:397–406. doi: 10.1111/j.1574-695X.2011.00872.x. [DOI] [PubMed] [Google Scholar]
- 25.Maintz L, Novak N. Modifications of the innate immune system in atopic dermatitis. J Innate Immun. 2011;3:131–141. doi: 10.1159/000323963. [DOI] [PubMed] [Google Scholar]
- 26.Cho SH, Strickland I, Boguniewicz M, Leung DY. Fibronectin and fibrinogen contribute to the enhanced binding of Staphylococcus aureus to atopic skin. J Allergy Clin Immunol. 2001;108:269–274. doi: 10.1067/mai.2001.117455. [DOI] [PubMed] [Google Scholar]
- 27.Bisgaard H, et al. Reduced diversity of the intestinal microbiota during infancy is associated with increased risk of allergic disease at school age. J Allergy Clin Immunol. 2011;128:646–652, e1–e5. doi: 10.1016/j.jaci.2011.04.060. [DOI] [PubMed] [Google Scholar]
- 28.Sjögren YM, Jenmalm MC, Böttcher MF, Björkstén B, Sverremark-Ekström E. Altered early infant gut microbiota in children developing allergy up to 5 years of age. Clin Exp Allergy. 2009;39:518–526. doi: 10.1111/j.1365-2222.2008.03156.x. [DOI] [PubMed] [Google Scholar]
- 29.Wang M, et al. Reduced diversity in the early fecal microbiota of infants with atopic eczema. J Allergy Clin Immunol. 2008;121:129–134. doi: 10.1016/j.jaci.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 30.Abrahamsson TR, et al. Low diversity of the gut microbiota in infants with atopic eczema. J Allergy Clin Immunol. 2012;129:434–440, 440, e1–e2. doi: 10.1016/j.jaci.2011.10.025. [DOI] [PubMed] [Google Scholar]
- 31.Doreswamy V, Peden DB. Modulation of asthma by endotoxin. Clin Exp Allergy. 2011;41:9–19. doi: 10.1111/j.1365-2222.2010.03628.x. [DOI] [PubMed] [Google Scholar]
- 32.Hessle C, Andersson B, Wold AE. Gram-positive bacteria are potent inducers of monocytic interleukin-12 (IL-12) while gram-negative bacteria preferentially stimulate IL-10 production. Infect Immun. 2000;68:3581–3586. doi: 10.1128/iai.68.6.3581-3586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arvå E, Andersson B. Induction of phagocyte-stimulating cytokines by in vitro stimulation of human peripheral blood mononuclear cells with Haemophilus influenzae. Scand J Immunol. 1999;49:411–416. doi: 10.1046/j.1365-3083.1999.00480.x. [DOI] [PubMed] [Google Scholar]
- 34.Arvå E, Andersson B. Induction of phagocyte-stimulating and Th1-promoting cytokines by in vitro stimulation of human peripheral blood mononuclear cells with Streptococcus pneumoniae. Scand J Immunol. 1999;49:417–423. doi: 10.1046/j.1365-3083.1999.00481.x. [DOI] [PubMed] [Google Scholar]
- 35.Zhang M, Liu M, Luther J, Kao JY. Helicobacter pylori directs tolerogenic programming of dendritic cells. Gut Microbes. 2010;1:325–329. doi: 10.4161/gmic.1.5.13052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akdis M. Immune tolerance in allergy. Curr Opin Immunol. 2009;21:700–707. doi: 10.1016/j.coi.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 37.Akdis M, et al. Immune responses in healthy and allergic individuals are characterized by a fine balance between allergen-specific T regulatory 1 and T helper 2 cells. J Exp Med. 2004;199:1567–1575. doi: 10.1084/jem.20032058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brand S, et al. Epigenetic regulation in murine offspring as a novel mechanism for transmaternal asthma protection induced by microbes. J Allergy Clin Immunol. 2011;128:618–625, e1–e7. doi: 10.1016/j.jaci.2011.04.035. [DOI] [PubMed] [Google Scholar]
- 39.Conrad ML, et al. Maternal TLR signaling is required for prenatal asthma protection by the nonpathogenic microbe Acinetobacter lwoffii F78. J Exp Med. 2009;206:2869–2877. doi: 10.1084/jem.20090845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Debarry J, et al. Acinetobacter lwoffii and Lactococcus lactis strains isolated from farm cowsheds possess strong allergy-protective properties. J Allergy Clin Immunol. 2007;119:1514–1521. doi: 10.1016/j.jaci.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 41.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. 2010;330:1768–1773. doi: 10.1126/science.1195568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Junker RR, et al. Composition of epiphytic bacterial communities differs on petals and leaves. Plant Biol (Stuttg) 2011;13:918–924. doi: 10.1111/j.1438-8677.2011.00454.x. [DOI] [PubMed] [Google Scholar]
- 44.von Mutius E. A fascinating look at the world with a new microscope. J Allergy Clin Immunol. 2012 doi: 10.1016/j.jaci.2011.12.994. [DOI] [PubMed] [Google Scholar]
- 45.Matsuoka RH, Kaplan R. People needs in the urban landscape: Analysis of landscape and urban planning contributions. Landsc Urban Plan. 2008;84:7–19. [Google Scholar]
- 46.Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic Acid Techniques in Bacterial Systematics. New York: Wiley; 1991. pp. 115–175. [Google Scholar]
- 47.Edwards U, Rogall T, Blöcker H, Emde M, Böttger EC. Isolation and direct complete nucleotide determination of entire genes: Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koskinen K, et al. Spatially differing bacterial communities in water columns of the northern Baltic Sea FEMS. Microb Ecol. 2011;75:99–110. doi: 10.1111/j.1574-6941.2010.00987.x. [DOI] [PubMed] [Google Scholar]
- 49.Schloss PD, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haakana M, et al. Finnish Corine 2006 Project: Determining changes in land cover in Finland between 2000 and 2006. In: Michel U, Civco DL, Ehlers M, Kaufmann HJ, editors. Remote Sensing for Environmental Monitoring, GIS Applications and Geology VIII. Bellingham, WA: SPIE; 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.