Abstract
Endosymbiotic interactions are characterized by the formation of specialized membrane compartments, by the host in which the microbes are hosted, in an intracellular manner. Two well-studied examples, which are of major agricultural and ecological importance, are the widespread arbuscular mycorrhizal symbiosis and the Rhizobium–legume symbiosis. In both symbioses, the specialized host membrane that surrounds the microbes forms a symbiotic interface, which facilitates the exchange of, for example, nutrients in a controlled manner and, therefore, forms the heart of endosymbiosis. Despite their key importance, the molecular and cellular mechanisms underlying the formation of these membrane interfaces are largely unknown. Recent studies strongly suggest that the Rhizobium–legume symbiosis coopted a signaling pathway, including receptor, from the more ancient arbuscular mycorrhizal symbiosis to form a symbiotic interface. Here, we show that two highly homologous exocytotic vesicle-associated membrane proteins (VAMPs) are required for formation of the symbiotic membrane interface in both interactions. Silencing of these Medicago VAMP72 genes has a minor effect on nonsymbiotic plant development and nodule formation. However, it blocks symbiosome as well as arbuscule formation, whereas root colonization by the microbes is not affected. Identification of these VAMP72s as common symbiotic regulators in exocytotic vesicle trafficking suggests that the ancient exocytotic pathway forming the periarbuscular membrane compartment has also been coopted in the Rhizobium–legume symbiosis.
Keywords: infection thread, unwalled infection droplet
During the symbiosis of plants and arbuscular mycorrhizal (AM) fungi, as well as in the symbiosis between Rhizobium bacteria and legumes, the microbes are hosted intracellularly inside specialized membrane compartments of the host (1). These membrane compartments, although morphologically different, create a symbiotic interface that controls efficient exchange of nutrients and signals and therefore their formation is at the heart of endosymbiosis. Although these symbiotic interfaces have a pivotal role in endosymbiosis, the molecular and cellular mechanisms by which they are formed are still largely obscure.
In the Rhizobium–legume symbiosis, the rhizobium bacteria are hosted inside a novel organ, the root nodule. The formation of this organ, through the reprogramming of root cortical cells, is set in motion by specific lipochito-oligosaccharides called Nod factors that are secreted by rhizobia (2). At the same time, Nod factors control the formation of tubular, transcellular, cell wall-bound infection structures, called infection threads. In most of the advanced legumes infection threads originate in root hairs and guide the bacteria to nodule primordium cells that are formed from reprogrammed root cortical cells (2). There, the bacteria are released from the infection threads into the developing nodule cells (Fig. 1 A and B). In the model legume Medicago truncatula (Medicago) this process continuously occurs because of the activity of an apical nodule meristem, where invasion by infection threads and release of bacteria from these threads occur in a few cell layers just below this meristem (Fig. 1 A and B). Release of bacteria starts with the formation of a local invagination of the infection thread membrane, which is devoid of a structured cell wall, by which an unwalled infection droplet is formed (Fig. 2 C and D) (3, 4). The formation of this unwalled infection droplet is the start of the formation of a symbiotic interface; it allows the bacteria to come into close contact with the host membrane of the droplet and individual bacteria are subsequently “pinched off” by which they become surrounded by a host membrane (the symbiosome membrane) and together are called symbiosomes (1, 5). Next, symbiosomes divide and differentiate in organelle-like structures where the bacteria are able to fix atmospheric nitrogen (6). The symbiosome membrane facilitates the exchange of fixed nitrogen in return for carbohydrates from the plant (2). In more basal legume species, as well as Parasponia, the only nonlegume genus able to form a Rhizobium symbiosis, nitrogen-fixing rhizobia, are retained in highly branched intracellular thread-like structures, called fixation threads, that are continuous with the infection thread (7, 8).
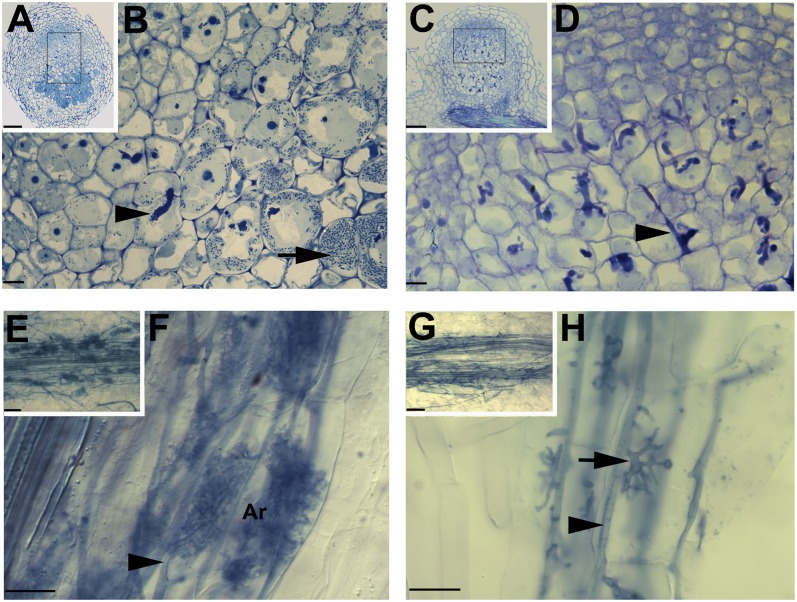
Fig. 1.
MtVAMP721d and MtVAMP721e are required for symbiosome and arbuscule formation. Light microscopic images of Medicago root nodules on control (A and B) and 35S::RNAiVAMP721d:VAMP721e transgenic roots (C and D). Longitudinal section of a control nodule 14 dpi (A) and magnification (B) of the region indicated in (A), shows cells penetrated by an infection thread (arrowhead), cells containing released and developing symbiosomes (containing S. meliloti bacteria) that are filling the host cell (arrow). Section of a 35S::RNAiVAMP721d:VAMP721e nodule (C) and the indicated magnification (D) showing cells that are penetrated by an infection thread, but symbiosomes are not formed. Light microscopic images of Medicago control (E and F) and 35S::RNAiVAMP721d:VAMP721e transgenic roots (G and H) infected by G. intraradices. Control root (E) and its magnification (F) show that the fungus forms intraradical hyphae (arrowhead) and mature fine-branched arbuscules (ar) inside inner root cortical cells. 35S::RNAiVAMP721d:VAMP721e root (G) and its magnification (H) show that AM fungal hyphae grow intraradically (arrowhead) and form trunk hyphae (arrow) that have entered inner cortical cells. A few major branches are made, but fine-branched arbuscules are not formed. (Scale bars, 100 μm in A and C, 10 μm in B and D, 50 μm in E and G, 25 μm in F and H.)
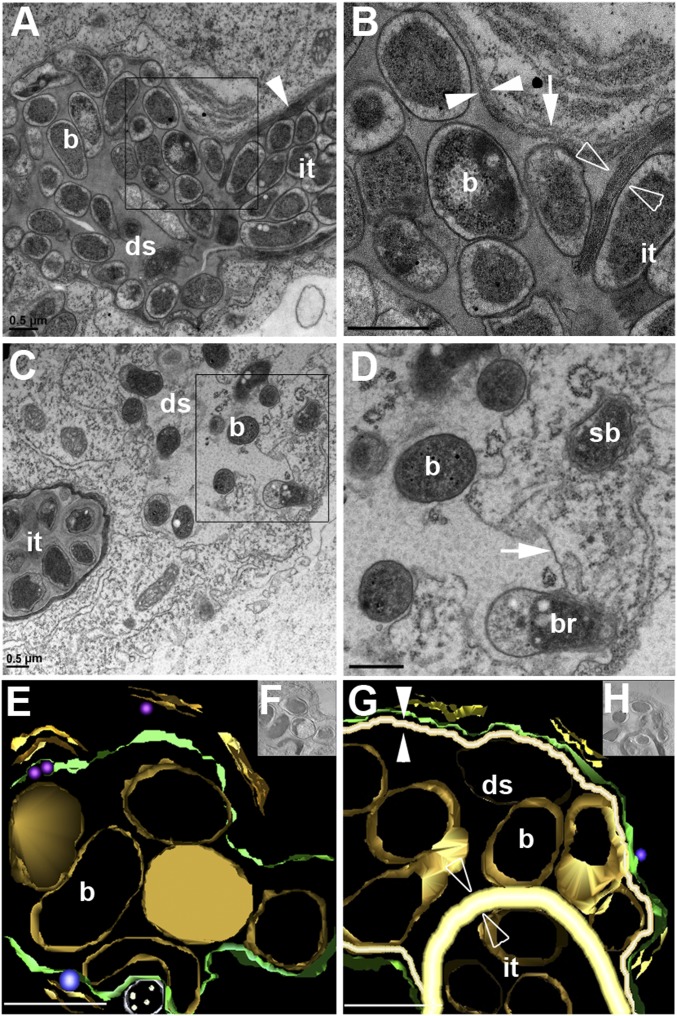
Fig. 2.
MtVAMP721d and MtVAMP721e are essential for the formation of unwalled infection droplets at infection threads. (A and B) EM images of an infection thread in a 35S::RNAiVAMP721d:VAMP721e nodule (A) and magnification of the designated area (B). The infection thread (it) is filled with bacteria and bound by a cell wall (arrowhead). At the tip of the thread a droplet-like structure (ds) filled with bacteria (b) is formed. The droplet-like structure is surrounded by a membrane (B, arrow) extending from the infection thread plasma membrane and bound by a cell wall (B, white opposed arrowheads) that is markedly thinner than the wall around the infection thread (B, open opposed arrowheads). (C and D) EM images of an infection thread in a control nodule (C) and magnification of the designated area (D). The unwalled droplet (ds) is bound by a membrane (arrow), but a cell wall is absent. Bacteria (b) are being released (br) and form symbiosomes (sb). (E–H) Electron microscopy computed tomography and 3D reconstruction of an infection thread in a control (E and F) and 35S::RNAiVAMP721d:VAMP721e (G and H) nodule. Analyses were performed on 360-nm-thick sections. In the droplet structure (ds) of RNAiVAMP721d:VAMP721e nodules (G and H) the bacteria are separated from the host membrane (green) by a cell wall (white, opposed arrowhead). This cell wall is markedly (∼3×) thinner (white opposed arrowheads) than the cell wall of the infection thread (it) at which a droplet structure is formed (open opposed arrowheads). The unwalled droplets in nodules on control roots (E and F) are bound only by the host membrane (green), which allows the bacteria (b, gold) to come into close contact with it. (Scale bars, 0.5 μm in A–G).
Like in the Rhizobium–legume symbiosis, AM fungi also enter the root (9). After traversing the epidermis and outer cortical layers, AM fungal hyphae spread mostly intercellularly in the inner cortex. Subsequently, they enter cortical cells via invagination of the plasma membrane and first “trunk” hyphae are formed that are bound by the cell wall of the host (10). In this respect they are similar to the infection threads that are formed in the Rhizobium–legume symbiosis. Subsequently, the trunk hypha branches repeatedly to develop the specialized structure known as arbuscule. This highly branched hyphal structure is enveloped by a special extension of the host plasma membrane, the periarbuscular membrane (9). Some cell-wall components typical of the primary plant cell wall (e.g., cellulose, pectins, and hemicellulose) have been immunologically detected in the space between the arbuscule and periarbuscular membranes. However, EM studies show that a structured cell wall is not present, indicating a specialized nature of this membrane compartment (10–12). The specialized nature of the periarbuscular membrane is further demonstrated by the observation that certain proteins localize specifically to the periarbuscular membrane, but not to the host-cell plasma membrane or the membrane surrounding the trunk hypha (13). Similar to the symbiosome membrane, the periarbuscular membrane is thought to facilitate the exchange of nutrients/minerals, especially phosphorus (and nitrogen) in return for photosynthates from the plant (9).
Although the molecular and cellular mechanisms of symbiotic interface formation are largely unknown, the signaling pathways that initiate these symbiotic interactions are better studied. In recent years it has become clear that several components of the signaling pathway, called the common signaling pathway, which is activated by rhizobial Nod factors in the epidermis, are also required for AM symbiosis (14). Furthermore, in nodules, this common signaling pathway is also essential for the release of the rhizobia from infection threads (symbiosome formation), but not for infection thread formation. This result has been demonstrated for the receptor-like kinase SymRK, the calcium- and calmodulin-dependent kinase, as well as its interacting partner (IPD3) (15–20). It is currently not known whether this common signaling pathway is activated by Nod-factor receptors in legume nodules. However, in the nonlegume Parasponia knock-down of a Nod-factor receptor (a single-copy gene) specifically blocked the formation of the (intracellular) rhizobial symbiotic interface (i.e., fixation threads) (21). Furthermore, knock-down of this Nod-factor receptor also blocked arbuscule formation by AM fungi, whereas intercellular colonization of the root was not affected (21). Consistent with this observation, it was recently shown that also AM fungi produce lipochito-oligosaccharides, with a structure very similar to that of Nod factors (22). Taken together, these findings strongly suggest that Rhizobium–legume symbiosis coopted the complete signaling pathway from the AM symbiosis and in both interactions this signaling pathway induces the formation of the membrane compartment, forming a symbiotic interface. Therefore, we hypothesize that this common signaling pathway activates a similar cellular process that in root cortical cells leads to the formation of fungal and in nodule cells to a rhizobial symbiotic interface. This finding would imply that in current legumes similar or even identical key regulators are required for the formation of periarbuscular and symbiosome membrane compartments, despite their—at first sight—major morphological differences.
A major morphological difference is the fact that in advanced legumes, Rhizobium bacteria are individually internalized into symbiosome compartments, which suggests an endocytosis-like process. However, studying the localization of membrane identity markers of the various endocytic compartments did not show any association of key regulators of the default endocytosis pathway at the early steps of symbiosome formation (23). Only later in symbiosome development (as the symbiosomes differentiate) a late endosome/vacuolar marker, the small GTPase Rab7, was observed on the symbiosome membrane. In contrast, a plasma membrane t-SNARE (see below), syntaxin SYP132, has been shown to be associated with the infection thread membrane, unwalled infection droplets, and symbiosomes immediately after their release from the infection thread, and throughout symbiosome development (23, 24). This finding suggests the involvement of an exocytosis-derived process in symbiosome formation. This finding is further supported by the fact that in Parasponia and primitive legumes the membrane that surrounds the fixation threads, like the periarbuscular membrane, remains connected to the plasma membrane (3, 7, 8). Therefore, we hypothesized that the same exocytotic pathway controls the formation of the symbiotic interface in both interactions.
Exocytosis involves focalized fusion of transport vesicles (with a specific cargo) with their target (plasma) membrane. Vesicle fusion is controlled by a group of proteins named SNAREs (soluble N-ethylmaleimide sensitive factor attachment protein receptor) and it is accomplished by the formation of a stable core complex of four SNARE motifs of the interacting SNAREs (25). Generally, one SNARE protein that locates on the transport vesicle (v-SNARE), pairs with three SNARE proteins (including syntaxins), which reside on the target membrane (t-SNARE) (25). In plants, exocytotic processes are mediated by v-SNAREs belonging to the VAMP72 (vesicle-associated membrane protein) family (26, 27). Previously, it has been shown that specific VAMP72 genes have been recruited in the Arabidopsis interaction with biotrophic fungi (27). Therefore, we focused on the role of the MtVAM72 family in the formation of a symbiotic interface in the interaction of Medicago with Sinorhizobium meliloti and Glomus intraradices, respectively. Here, we show that two highly homologous Medicago VAMP72s are required for symbiosome as well as arbuscule formation.
Results and Discussion
Identification of VAMP72 Genes in Medicago.
We identified the Medicago MtVAMP72 family based on homology with Arabidopsis members (26). Mining of Medicago EST and genome sequence (28) data identified seven MtVAMP72 genes (Tables S1 and S2). Phylogenetic analysis using several genome sequences that represent major eudicot clades (29) showed that the VAMP72s can be divided into three groups, namely VAMP721, VAMP724, and VAMP727 (Fig. 3 and Table S2). The VAMP721 group can be further divided into two subgroups. One includes all Arabidopsis VAMP721 genes and Medicago VAMP721a, -b, -c, whereas the second (“symbiotic”) subgroup includes MtVAMP721d and -e, but no Arabidopsis homologs (Fig. 1). Arabidopsis has lost the ability to interact with AM fungi and this is correlated with the loss of several genes involved in the common signaling pathway (30). Furthermore, other eudicots with a sequenced genome that can interact with AM fungi do have at least one VAMP721 member that belongs to the “symbiotic” subgroup. Along this line MtVAMP721d and MtVAMP721e are the best candidates to be involved in the formation of periarbuscular and symbiosome membrane compartments. Furthermore, these two genes are expressed at relatively high levels in roots, nodules, and mycorrhized roots (Fig. S1 A–D). For these reasons we have analyzed the function of MtVAMP721d and MtVAMP721e in both interactions.
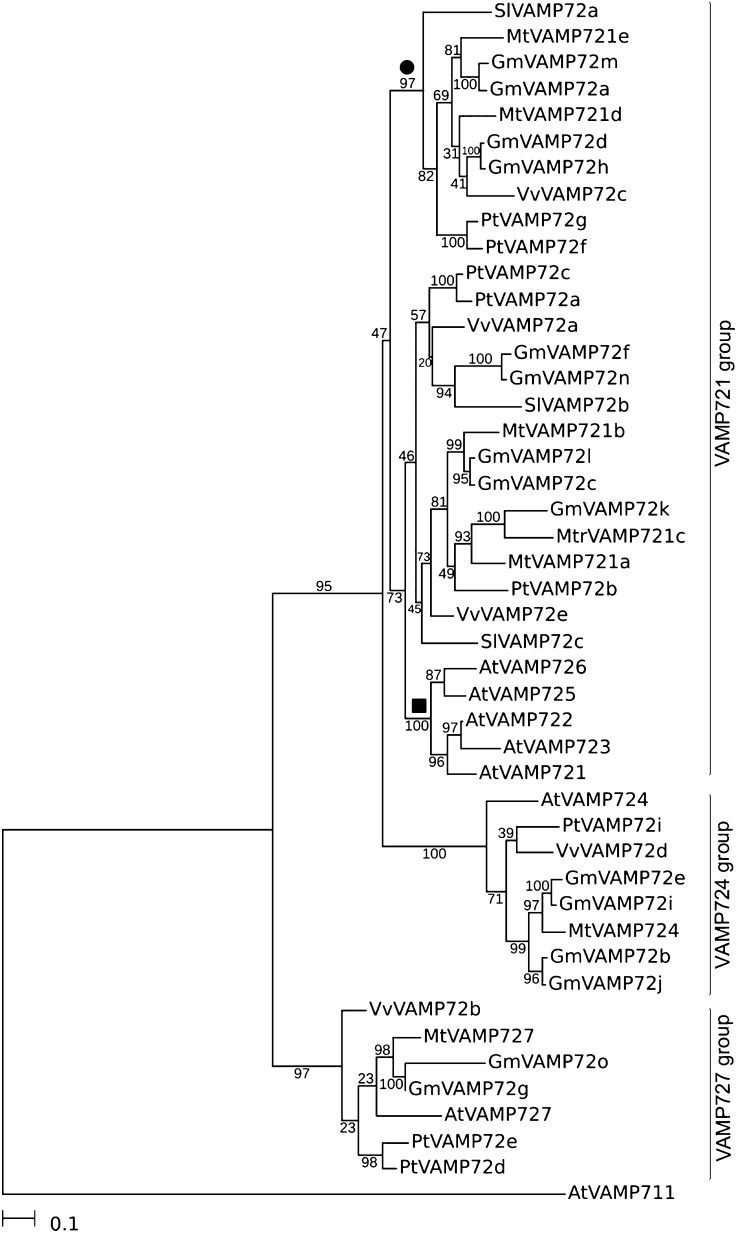
Fig. 3.
Phylogenetic analysis of MtVAMP72s. Unrooted phylogenetic neighbor-joining maximum-likelihood tree of M. truncatula (Mt) and Arabidopsis thaliana (At) Populus thrichocarpa (Pt), Glycine max (Gm), Solanum lycopersicum (Sl), and Vitis vinifera (Vv) VAMP72s. The sequences were identified using BLASTN searches in genome/EST databases and aligned at the protein level using MUSCLE v3.8.31. The phylogenetic tree was built using PHYML version 3.0 using protein-coding nucleotide sequences aligned using the protein alignment as a template. We used the general time-reversible model with γ-distributed rates of evolution with six categories. AtVAMP711 was used as outgroup, 100 bootstrap repetitions were performed to assess robustness of nodes. Note that MtVAMP721d and MtVAMP721e form a separate “symbiotic” subgroup (●, bootstrap 97%) inside VAMP721 group, whereas other MtVAMP721a, MtVAMP721b, and MtVAMP721c form other subgroup which also includes all Arabidopsis VAMP721 genes (■, bootstrap 73%). The latter are crucial in defense against fungal pathogens.
Silencing of MtVAMP721d and MtVAMP721e Blocks Symbiosome and Arbuscule Formation.
To study the function of MtVAMP721d and MtVAMP721e, we used gene-specific RNAi. Based on quantitative RT-PCR analyses of Medicago roots and nodules we also selected MtVAMP721a and MtVAMP724, because they are expressed at moderate levels in both organs (Fig. S1 A and B). As both MtVAMP721d and MtVAMP721e are highly homologous, we also created a construct (35SCaMV::RNAiVAMP721d:VAMP721e) by which both are knocked down. We obtained composite Medicago plants with transgenic roots, using Agrobacterium rhizogenes mediated transformation, that were selected through the use of a red fluorescent reporter (16, 31). The gene-specific RNAi constructs reduced the level of their corresponding target RNA to 10–40%, compared with control roots transformed with the empty vector, whereas the expression level of the other MtVAMP72 genes was not reduced (Fig. S2 A–D). The composite plants were inoculated with S. meliloti and root nodules were analyzed after 14 d. Microscopic sections of nodules (n = 20 for each construct from independently transformed roots) formed on roots expressing one of the four single gene-specific RNAi constructs showed that they all had a cytology similar to nodules formed on control roots transformed with the empty vector. Transgenic RNAiVAMP721d:VAMP721e roots had markedly reduced levels of both mRNAs (down to 10–40%) (Fig. S2E). The growth of these transgenic roots and the number of nodules that are formed were only slightly affected (Fig. S3). Light microscopic analysis of nodules collected from RNAiVAMP721d:VAMP721e transgenic roots (nroots = 15) revealed the presence of numerous infection threads in the central tissue of these nodules (Fig. 1 C and D); however, symbiosomes were absent or present at very low numbers in 18 of 37 analyzed nodules (48%). Such nodules were not observed on control roots (nnodules = 0 of 27, nroots = 15) (Fig. 1 A and B). Therefore, knock-down of both MtVAMP721d and MtVAMP721e specifically blocks symbiosome formation.
To test whether MtVAMP721d and MtVAMP721e are also required for arbuscule formation, RNAiVAMP721d, RNAiVAMP721e, RNAiVAMP721d:VAMP721e, and RNAiVAMP721a composite plants were inoculated with G. intraradices. Plants were analyzed 4 wk after inoculation. In control and all RNAi roots, fungal hyphae successfully colonized the root cortical layers and spread longitudinally along the root axis (Fig. 2 E and G, and Fig. S4). Fully developed arbuscules were efficiently formed in control roots expressing an empty vector [arbuscule presence (a) = 61%] (Fig. 2F and Fig. S4), as well as in RNAiVAMP721d, RNAiVAMP721e, and RNAiVAMP721a roots. However, silencing of both MtVAMP721d and MtVAMP721e (RNAiVAMP721d:VAMP721e) resulted in marked decrease of mature arbuscules [arbuscule presence (a) = 6%]. Small trunk-like hyphae abundantly occur in root inner cortical cells, but mature arbuscules were only rarely formed (Fig. 2H and Fig. S4). These small trunk-like hyphae are likely the result of an early arrest of arbuscule formation and not the collapse of mature arbuscules. In some plant mutants arbuscules can, at a late stage of development, be absent because of premature collapse (32). This collapse of arbuscules is associated with the formation of dense clumps (9, 32). These dense clumps of collapsed hyphae did not occur in RNAiVAMP721d:VAMP721e roots. Thus, the knock-down of both MtVAMP721d and MtVAMP721e affects the formation of arbuscules as well as symbiosomes. Arbuscule formation is even more efficiently blocked than symbiosome formation, which might imply that higher levels of MtVAMP721d and MtVAMP721e are required for arbuscule formation.
Silencing of MtVAMP721d and MtVAMP721e Affects Unwalled Droplet Formation and Bacterial Release from the Infection Thread.
The lack of symbiosomes in RNAiVAMP721d:VAMP721e nodules allowed us to predict that the formation of an unwalled droplet and subsequent release of bacteria, which marks the start of symbiosome formation, is affected. To address this finding we used EM to compare RNAiVAMP721d:VAMP721e nodules where symbiosomes were absent or present at very low numbers (Fig. 3 A and B) to nodules formed on control roots (Fig. 3 C and D). In the latter, unwalled infection droplets are formed at infection threads in the cell layer directly adjacent to the meristem (Fig. 2 A and B). The unwalled infection droplets are bound by a membrane of the host, but a cell wall is completely lacking (Fig. 3 C and D). In the RNAiVAMP721d:VAMP721e nodules infection threads contain a structure that resembles an infection droplet. However, these droplet-like structures still show the presence of a thin layer of cell wall next to the surrounding host membrane (Fig. 3 A and B). To confirm that walled infection droplets are formed, we also used EM computer tomography and made 3D reconstructions of 360-nm-thick section (Fig. 3 E–H). This finding shows that droplet-like structures have been formed and these are bound by a membrane and a cell wall with a thickness of about one-third (0.03 nm ± 0.01; ninfection threads = 10, nnodules = 3) of the infection thread wall (0.10 nm ± 0.03; ninfection threads = 10, nnodules = 3). The presence of a cell wall in the infection droplets prevents a close contact between the bacteria and the host membrane and thereby likely hampers the pinching off of symbiosomes. This finding can explain why numerous infection threads have such a walled infection droplet. The formation of droplet-like structures in the RNAiVAMP721d:VAMP721e nodules suggests that the switch to a specific MtVAMP721d/e controlled exocytosis pathway is impaired that in wild-type nodules is responsible for the formation of an unwalled infection droplet. Our explanation for walled droplet formation in RNAiVAMP721d:VAMP721e root nodules is the following: residual levels of MtVAMP721d/e vesicles (because of incomplete silencing) are present in RNAiVAMP721d:VAMP721e root nodules and these are targeted to the infection thread membrane to initiate the formation of an unwalled droplet. Their cargo [possibly cell wall-degrading enzymes (33) or altered membrane composition] leads to the formation of a cell wall-free interface. However, their number is low in comparison with other vesicles involved in growth of the plant cell. The latter may deliver cell-wall components and we postulate that they “compete” with the MtVAMP721d or -e vesicles, and therefore a thin cell wall is formed instead of a cell wall-free interface. Alternatively, the MtVAMP721d/e controlled pathway might be specifically involved in symbiosome formation. In this case, block of symbiosome formation might lead to a droplet that is an intermediate infection thread and unwalled infection droplet. Taken together, these data suggest that an exocytotic pathway involving MtVAMP721d/e controls the formation of the symbiotic interface in the Rhizobium–legume symbiosis.
MtVAMP721d and MtVAMP721e Localize at the Site of Bacterial Release on Symbiosome and Periarbuscular Membranes.
To find further support for a role of MtVAMP721d/e vesicles in symbiotic interface formation, their subcellular localization was determined in transgenic roots expressing translational fusions GFP-MtVAMP721d or GFP-MtVAMP721e under the control of their native promoters. Promoter–β-glucuronidase (GUS) analyses showed that these promoters are active in the nodule region where symbiosomes are formed (Fig. 4 C and D). Confocal microscopy of GFP-MtVAMP721e-expressing nodules (nnodules = 5) revealed accumulation of GFP-fluorescence on dot-like structures (Fig. 5A). These dot-like structures accumulated at local regions near infection threads where unwalled droplets are formed and symbiosomes start to develop. The subcellular localization of GFP-VAMP721d is very similar to that of GFP-VAMP721e (Fig. S5A). The nature of the labeled dot-like structures was investigated using EM immunodetection with an antibody against GFP or an antibody that recognize both MtVAMP721d and MtVAMP721e (Figs. S5 C and D and S6 A and B). This finding showed that MtVAMP721d and MtVAMP721e were present on small (∼50 nm) vesicles in close association with unwalled infection droplets (Fig. S5D) and on or near symbiosome membranes of young symbiosomes (Fig. S5C). When we used both antibodies simultaneously we could distinguish the signals using secondary antibodies linked to gold particles with a different size. This result showed that the signals coincided (Fig. S5C) and indicates the specificity of the anti- MtVAMP721d/e antibodies. Thus, these data are consistent with focal MtVAMP721d/e-mediated delivery of exocytotic vesicles to unwalled droplets and symbiosomes.
Fig. 4.
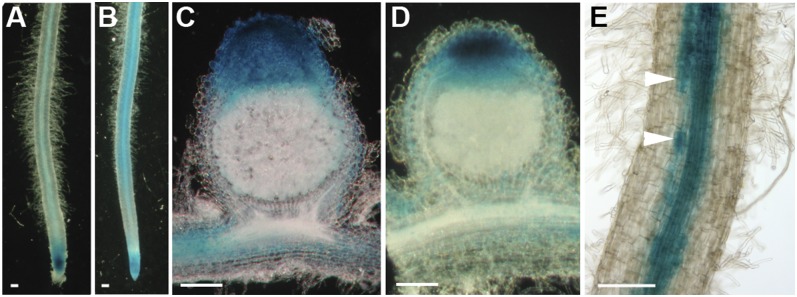
Activity of MtVAMP721d and MtVAMP721e promoters. The 2.5-kb region upstream of the translational start of MtVAMP721d (A and C) and of MtVAMP721e (B, D, and E), respectively, were fused to GUS and transgenic roots expressing these genetic constructs were inoculated by S. meliloti 2011 (C and D) or G. intraradices (E). Nodules were harvested 14 dpi and were analyzed for GUS activity. Nodules were hand-sectioned. Promoters of MtVAMP721d and MtVAMP721e are active in the meristem and the infection zone of the nodule where release of bacteria from the infection threads and symbiosome development occurs. Mycorrhized roots were harvested and analyzed for GUS activity 28 dpi. Promoter MtVAMP721e shows activity in vascular tissue and arbuscule-containing cells (E, arrowheads). (Scale bars, 100 μm in A–E).
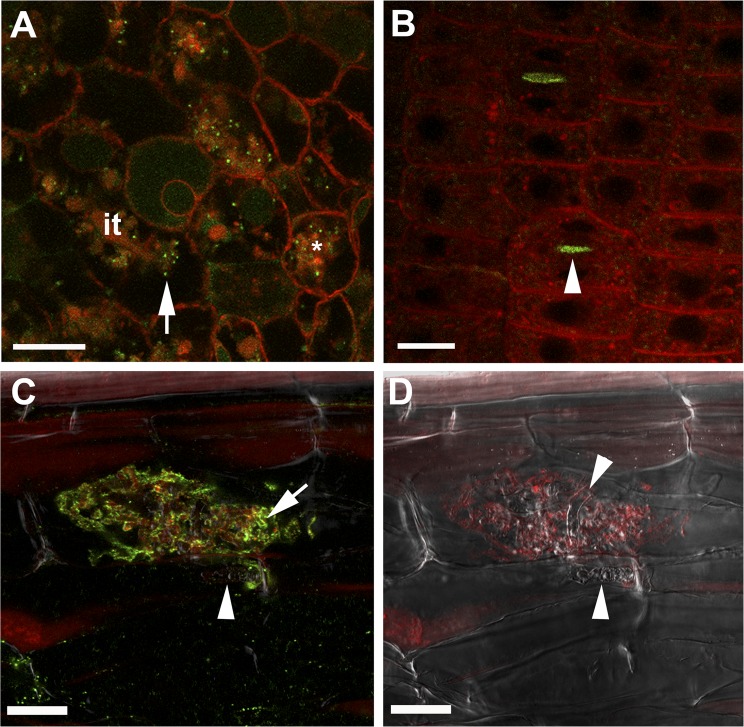
Fig. 5.
MtVAMP721e marked vesicles accumulate at the site of symbiosome, periarbuscular membrane formation and at cell plates. Confocal microscopy images showing localization of GFP-MtVAMP721e expressed under the control of its native promoter. (A) Accumulation of GFP-MtVAMP721e labeled dot-like structures (green) in infected Medicago nodule cells at local regions of infection threads (it) where bacteria are released (unwalled droplet, arrow) and around newly formed symbiosomes (*). (B) Accumulation of GFP-MtVAMP721e at the cell plate (arrowhead) in dividing meristematic cells of a Medicago root. (C and D) Confocal microscopy picture of immunolocalization of GFP-MtVAMP721e (C) and corresponding Bright-field image (D) in a root cortical cell containing an arbuscule. GFP was visualized by anti-GFP antibodies (green fluorescence). The signal is present near fine arbuscule branches (arrow) and absent on intraradical hypha (arrowhead). Samples were contrasted by FM4-64 (red) to visualize membranes. (Scale bars, 20 μm in A, C, and D; 10 μm in B).
Medicago roots expressing MtVAMP721d::GUS or MtVAMP721e::GUS showed that these promoters are active in the inner cortex, where arbuscules can be formed (Fig. 4 A and B). In MtVAMP721e::GUS transgenic roots colonized by G. intraradices, GUS activity was slightly higher in cells with arbuscules than in noninfected inner cortical cells (Fig. 4E). Roots expressing GFP-VAMP721e under the control of its native promoter (nroots = 3) were infected by G. intraradices and the fusion protein was detected using anti-GFP or anti-MtVAMP721d/e antibodies. This finding showed that GFP-VAMP721e was abundantly present in root cells containing arbuscules, whereas the level of MtVAMP721e in noninfected inner cortical cells was rather low. In cells containing arbuscules, the signal was accumulating over the periarbuscular membrane, especially at the fine branches (Fig. 5 C and D and Fig. S5B).
Therefore, the knockdown and localization studies show that arbuscule and symbiosome formation are specifically controlled by the MtVAMP721d/e-regulated exocytotic pathway. This finding suggests that symbiosomes, like the periarbuscular endomembrane compartments, represent an apoplastic compartment despite their intracellular nature. At a later stage of development the symbiosome membrane obtains the identity marker Rab7, which also occur at the late endosome/vacuole, whereas vacuolar SNAREs accumulate on symbiosome membrane when senescence is initiated (23). Therefore, we conclude that functional symbiosomes do not seem to have a true vacuolar nature but remain as an apoplastic-like compartment. The switch to the MtVAMP721d/e controlled exocytotic pathway allows the targeting of vesicles with a different cargo, and this facilitates the formation of a symbiotic interface with specific protein composition (34).
In root and nodule meristems we observed MtVAMP721d and MtVAMP721e accumulation at the site of cell plate formation (Fig. 5B). The cell plate is a transient membrane compartment created by membrane vesicle fusion in which a cellulose-based cell wall is not yet formed (35). Therefore, it is tempting to speculate that part of the machinery that is involved in the formation of a cell plate is coopted to build the symbiotic interface. MtVAMP721d and MtVAMP721e might not be essential for cell plate formation, because RNAiVAMP721d:VAMP721e has only a slight effect on root growth and nodule formation. However, we cannot exclude that the residual levels of MtVAMP721d and MtVAMP721e in the RNAi roots are sufficient for rather normal root growth. Furthermore, the expression level of MtVAMP721a is slightly higher in MtVAMP721d and MtVAMP721e knockdown lines (Fig. S2E) and this might have a compensating effect. This finding is well in line with studies of Kwon et al., who showed that in Arabidopsis two VAMP721 genes from the “nonsymbiotic” subgroup (AtVAMP721 and AtVAMP722) have been recruited in defense against powdery mildew (Fig. S1E) and marked reduction of their expression blocked the defense response, but had no effect on plant growth (27). However, a complete loss of function of both AtVAMP721 and AtVAMP722 causes severe dwarfism of the plant. Recently, it has been shown that these AtVAMP72s play a crucial role in cell plate formation (36).
The interaction of plants with AM fungi is about 450 million years old (37), whereas the Rhizobium symbiosis evolved about 60 million years ago (38). Recent studies show that AM fungi produce Nod factor-like lipochito-oligosaccharides (22), and that the same Parasponia Nod-factor receptor is required in both symbiotic interactions (21). This finding strongly suggests that during evolution, rhizobia acquired the ability to make a factor (Nod factor) with a structure similar to that of more ancient mycorrhizal factors. This finding is consistent with the idea that rhizobia obtained the genes to make Nod factors by horizontal gene transfer (39). Here we show that the exocytotic pathway that is required for arbuscule formation is also essential for Medicago symbiosome formation. Therefore, it is probable that during evolution the acquisition to produce Nod factors provided rhizobia with the ability to use the ancient AM machinery to establish an intracellular interface. In this study we focused on the Medicago symbiosis. Whether the VAMP721d/VAMP721e controlled exocytosis pathway is also used in other legumes to establish symbiosome formation, or fixation threads, remains to be demonstrated. Our studies on Medicago strongly suggest that most plants have genes encoding the core of a mechanism that can support a Rhizobium infection process. Nevertheless, although the AM fungal symbiosis is widespread across the plant kingdom, it is perplexing that the Rhizobium–legume symbiosis is restricted to legumes and Parasponia.
Materials and Methods
Plant Transformation and Inoculation.
A. rhizogenes MSU440-mediated hairy root transformation was used to obtain transgenic roots (31). M. truncatula accession Jemalong A17 was grown in perlite saturated with Färhaeus medium without nitrate in a growth chamber at 21 °C and 16/8-h light/darkness. These plants were inoculated with S. meliloti 2011 (OD600 0.1, 2 mL per plant). Root nodules were collected for analysis 10–14 d postinoculation (dpi). Medicago plants that were inoculated with G. intraradices were cocultivated with Allium schoenoprasum nurse plants in sand/hydrobeads mixture saturated by Hoagland medium. To quantify the infection by the fungus, roots were stained with 0.2% Trypan blue and roots were analyzed by light microscopy. In total, 75 cm of each transgenic root system was analyzed and the level of infection was quantified as previously described (39).
Cloning.
DNA fragments were generated via PCR on Medicago genomic DNA or cDNA made from root nodules as a template using Phusion High-Fidelity DNA Polymerase (Finnzymes) and gene-specific primers (Table S3). The Gateway technology (Invitrogen) was used to create genetic constructs for GFP-fusion, promoter-GUS and RNA interference (40). TOPO cloning (Invitrogen) was used to create entry clones. To create RNAi constructs, entry clones were recombined into the modified Gateway pK7GWIWG2(II)-Q10::DsRED binary vector (16, 31). To generate GFP-MtVAMP721d and GFP-MtVAMP721e translational fusions, GFP was fused to the N-terminal end of MtVAMP721d or MtVAMP721e. Expression of these fusions were driven by 2.5-kb 5′ regulatory sequence (VAMP721d::GFP-VAMP721d and VAMP721e::GFP-VAMP721e).
Gene Expression.
Total RNA was extracted from roots, root nodules, and mycorrhized roots using E.Z.N.A. Plant RNA Mini Kit (Omega Bio-Tek). Equal amounts of total RNA was used to analyze gene expression of MtVAMP72s by quantitative real-time PCR using iQ SYBR Green Supermix (Bio-Rad) and gene-specific primers (Table S3). Detection of fluorescent signal was performed on a My iQ Real-Time Detection System (Bio-Rad). Gene-expression profiles were normalized against the transcription level of reference gene MtUBQ10. Data were compared with M. truncatula Gene Expression Atlas data (41) (http://mtgea.noble.org/v2/).
Antibodies Against MtVAMP721d and MtVAMP721e.
Affinity-purified polyclonal rabbit anti-MtVAMP721d antibodies were generated by GenScript against the peptides QKLPSTNNKFTYNC. Protein extraction and immunoprecipitation from transgenic roots expressing GFP-VAMP721a, GFP-VAMP721d, or GFP-VAMP721e was performed using GFP Trap_A Kit (Chromotek) according to the manufacturer’s instructions. To detect GFP fusion protein, proteins were separated on SDS-polyacrylamid gel and probed with anti-GFP antibody (Molecular Probes) in dilution 1:2,000 or anti-VAMP721d in dilution 1:100/1:500 and secondary anti-rabbit antibodies conjugated with alkaline phosphatase (Promega).
Confocal Laser-Scanning Microscopy.
GFP-fused proteins were visualized on transgenic roots and hand sectioned nodules. Imaging was done on a Zeiss LSM 5 Pascal confocal laser-scanning microscope (Carl Zeiss). Immunodetection was performed as described previously (23). Goat serum or 3% (vol/wt) BSA was used as blocking agent. Polyclonal rabbit anti-GFP antibodies (Molecular Probes) in dilution 1:200 or anti-VAMP721d/VAMP721e in dilution 1:50–100 and secondary anti-rabbit Alexa 488 antibodies (Molecular Probes) in dilution 1:200 were used. Sections were counterstained by FM4-64 (30 μg/mL).
Sample Preparation for Light and Transmission Electron Microscopy (TEM) and TEM Immunodetection.
Tissue preparation was performed as described previously (23). Semithin (0.6 μm) for light and thin sections (60 nm) for EM of the same nodule were cut using a Leica Ultracut microtome (Leica). Nickel grids with the sections were blocked in normal goat serum with 1% milk or 2% BSA in PBS and incubated with the primary antibody according to dilutions given above. Goat anti-rabbit coupled with 15-nm gold (BioCell) (1:50 dilution) were used as secondary antibody. Sections were examined using a JEOL JEM 2100 transmission electron microscope equipped with a Gatan US4000 4K×4K camera.
TEM Computer Tomography and 3D Reconstruction.
The method of electron tomographic analysis adapted for plant tissue was used (42). Sections of 300–360 nm thickness were mounted on 100-nm mesh copper grids coated by Formvar. Tilted images were collected from −55 to 55° with a tilt-rotate specimen holder using Serial EM software. Obtained image series were used for 3D reconstruction using IMOD (Cygwin, Linux) software package. The membrane boundaries were traced manually and 3D models were computed. Rendering of the obtained images was performed automatically by IMOD mesh command. Four tomograms were reconstructed.
Supplementary Material
Acknowledgments
We thank Dr. R. Geurts and Prof. Dr. A. M. C. Emons for critical discussion; Dr. J. van Lent for technical support with electron microscopic computer tomography. Electron microscopy imaging and sample preparation were performed at the Wageningen Electron Microscopy Centre (Wageningen University). This study was supported by The Netherlands Organization for Scientific Research/Russian Federation for Basic Research grant for Centre of Excellence 047.018.001.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1200407109/-/DCSupplemental.
References
- 1.Parniske M. Intracellular accommodation of microbes by plants: A common developmental program for symbiosis and disease? Curr Opin Plant Biol. 2000;3:320–328. doi: 10.1016/s1369-5266(00)00088-1. [DOI] [PubMed] [Google Scholar]
- 2.Oldroyd GE, Murray JD, Poole PS, Downie JA. The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet. 2011;45:119–144. doi: 10.1146/annurev-genet-110410-132549. [DOI] [PubMed] [Google Scholar]
- 3.Brewin NJ. Plant cell wall remodelling in the Rhizobium-legume symbiosis. Crit Rev Plant Sci. 2004;23:293–316. [Google Scholar]
- 4.Rae AL, Bonfante-Fasolo P, Brewin NJ. Structure and growth of infection threads in the legume symbiosis with Rhizobium leguminosarum. Plant J. 1992;2:385–395. [Google Scholar]
- 5.Roth LE, Stacey G. Bacterium release into host cells of nitrogen-fixing soybean nodules: The symbiosome membrane comes from three sources. Eur J Cell Biol. 1989;49:13–23. [PubMed] [Google Scholar]
- 6.Vasse J, de Billy F, Camut S, Truchet G. Correlation between ultrastructural differentiation of bacteroids and nitrogen fixation in alfalfa nodules. J Bacteriol. 1990;172:4295–4306. doi: 10.1128/jb.172.8.4295-4306.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sprent JI, James EK. Legume evolution: Where do nodules and mycorrhizas fit in? Plant Physiol. 2007;144:575–581. doi: 10.1104/pp.107.096156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Faria SM, McInroy SG, Sprent JI. The occurrence of infected cells, with persistent infection threads, in legume root nodules. Can J Bot. 1987;65:553–558. [Google Scholar]
- 9.Smith SE, Read D. Mycorrhizal Symbiosis. San Diego: Academic Press; 2008. [Google Scholar]
- 10.Bonfante-Fasolo P, Vian B, Perotto S, Faccio A, Knox LP. Cellulose and pectin localization in root of mycorrhizal Allium porrum: Labeling continuity between host cell wall and interfacial material. Planta. 1990;180:537–547. doi: 10.1007/BF02411452. [DOI] [PubMed] [Google Scholar]
- 11.Balestrini R, Bonfante P. The interface compartment in arbuscular mycorrhizae: A special type of plant cell wall? Plant Biosyst. 2005;139:8–15. [Google Scholar]
- 12.Balestrini R, Cosgrove DJ, Bonfante P. Differential location of alpha-expansin proteins during the accommodation of root cells to an arbuscular mycorrhizal fungus. Planta. 2005;220:889–899. doi: 10.1007/s00425-004-1431-2. [DOI] [PubMed] [Google Scholar]
- 13.Pumplin N, Harrison MJ. Live-cell imaging reveals periarbuscular membrane domains and organelle location in Medicago truncatula roots during arbuscular mycorrhizal symbiosis. Plant Physiol. 2009;151:809–819. doi: 10.1104/pp.109.141879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kouchi H, et al. How many peas in a pod? Legume genes responsible for mutualistic symbioses underground. Plant Cell Physiol. 2010;51:1381–1397. doi: 10.1093/pcp/pcq107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Capoen W, Goormachtig S, De Rycke R, Schroeyers K, Holsters M. SrSymRK, a plant receptor essential for symbiosome formation. Proc Natl Acad Sci USA. 2005;102:10369–10374. doi: 10.1073/pnas.0504250102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Limpens E, et al. Formation of organelle-like N2-fixing symbiosomes in legume root nodules is controlled by DMI2. Proc Natl Acad Sci USA. 2005;102:10375–10380. doi: 10.1073/pnas.0504284102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Godfroy O, Debellé F, Timmers T, Rosenberg C. A rice calcium- and calmodulin-dependent protein kinase restores nodulation to a legume mutant. Mol Plant Microbe Interact. 2006;19:495–501. doi: 10.1094/MPMI-19-0495. [DOI] [PubMed] [Google Scholar]
- 18.Yano K, et al. CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc Natl Acad Sci USA. 2008;105:20540–20545. doi: 10.1073/pnas.0806858105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horváth B, et al. Medicago truncatula IPD3 is a member of the common symbiotic signaling pathway required for rhizobial and mycorrhizal symbioses. Mol Plant Microbe Interact. 2011;24:1345–1358. doi: 10.1094/MPMI-01-11-0015. [DOI] [PubMed] [Google Scholar]
- 20.Ovchinnikova E, et al. IPD3 controls the formation of nitrogen-fixing symbiosomes in pea and Medicago Spp. Mol Plant Microbe Interact. 2011;24:1333–1344. doi: 10.1094/MPMI-01-11-0013. [DOI] [PubMed] [Google Scholar]
- 21.Op den Camp R, et al. LysM-type mycorrhizal receptor recruited for rhizobium symbiosis in nonlegume Parasponia. Science. 2011;331:909–912. doi: 10.1126/science.1198181. [DOI] [PubMed] [Google Scholar]
- 22.Maillet F, et al. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature. 2011;469:58–63. doi: 10.1038/nature09622. [DOI] [PubMed] [Google Scholar]
- 23.Limpens E, et al. Medicago N2-fixing symbiosomes acquire the endocytic identity marker Rab7 but delay the acquisition of vacuolar identity. Plant Cell. 2009;21:2811–2828. doi: 10.1105/tpc.108.064410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Catalano CM, Czymmek KJ, Gann JG, Sherrier DJ. Medicago truncatula syntaxin SYP132 defines the symbiosome membrane and infection droplet membrane in root nodules. Planta. 2007;225:541–550. doi: 10.1007/s00425-006-0369-y. [DOI] [PubMed] [Google Scholar]
- 25.Lang T, Jahn R. Core proteins of the secretory machinery. Handb Exp Pharmacol. 2008;184:107–127. doi: 10.1007/978-3-540-74805-2_5. [DOI] [PubMed] [Google Scholar]
- 26.Sanderfoot A. Increases in the number of SNARE genes parallels the rise of multicellularity among the green plants. Plant Physiol. 2007;144:6–17. doi: 10.1104/pp.106.092973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwon C, et al. Co-option of a default secretory pathway for plant immune responses. Nature. 2008;451:835–840. doi: 10.1038/nature06545. [DOI] [PubMed] [Google Scholar]
- 28.Young ND, et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature. 2011;480:520–524. doi: 10.1038/nature10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soltis PS, Soltis DE, Chase MW. Angiosperm phylogeny inferred from multiple genes as a tool for comparative biology. Nature. 1999;402:402–404. doi: 10.1038/46528. [DOI] [PubMed] [Google Scholar]
- 30.Zhu H, Riely BK, Burns NJ, Ané JM. Tracing nonlegume orthologs of legume genes required for nodulation and arbuscular mycorrhizal symbioses. Genetics. 2006;172:2491–2499. doi: 10.1534/genetics.105.051185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Limpens E, et al. RNA interference in Agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. J Exp Bot. 2004;55:983–992. doi: 10.1093/jxb/erh122. [DOI] [PubMed] [Google Scholar]
- 32.Javot H, Penmetsa RV, Terzaghi N, Cook DR, Harrison MJ. A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA. 2007;104:1720–1725. doi: 10.1073/pnas.0608136104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie F, et al. Legume pectate lyase required for root infection by rhizobia. Proc Natl Acad Sci USA. 2012;109:633–638. doi: 10.1073/pnas.1113992109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whitehead LF, Day DA. The peribacteroid membrane. Physiol Plant. 1997;100:30–44. [Google Scholar]
- 35.Jürgens G. Plant cytokinesis: Fission by fusion. Trends Cell Biol. 2005;15:277–283. doi: 10.1016/j.tcb.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 36.Zhang L, et al. Arabidopsis R-SNARE proteins VAMP721 and VAMP722 are required for cell plate formation. PLoS ONE. 2011;6:e26129. doi: 10.1371/journal.pone.0026129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bonfante P, Genre A. Plants and arbuscular mycorrhizal fungi: An evolutionary-developmental perspective. Trends Plant Sci. 2008;13:492–498. doi: 10.1016/j.tplants.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 38.Sprent JI. 60Ma of legume nodulation. What’s new? What’s changing? J Exp Bot. 2008;59:1081–1084. doi: 10.1093/jxb/erm286. [DOI] [PubMed] [Google Scholar]
- 39.Sullivan JT, Ronson CW. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc Natl Acad Sci USA. 1998;95:5145–5149. doi: 10.1073/pnas.95.9.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trouvelot A, Kough J, Gianinazzi-Pearson V. Research for estimation methods having a functional significance. In: Gianinazzi-Pearson V, Gianinazzi S, editors. Physiological and Genetical Aspects of Mycorrhizae. Paris: INRA; 1986. pp. 217–221. [Google Scholar]
- 41.Karimi M, Inzé D, Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- 42.Benedito VA, et al. A gene expression atlas of the model legume Medicago truncatula. Plant J. 2008;55:504–513. doi: 10.1111/j.1365-313X.2008.03519.x. [DOI] [PubMed] [Google Scholar]
- 43.Seguí-Simarro JM, Austin JR, 2nd, White EA, Staehelin LA. Electron tomographic analysis of somatic cell plate formation in meristematic cells of Arabidopsis preserved by high-pressure freezing. Plant Cell. 2004;16:836–856. doi: 10.1105/tpc.017749. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.