Abstract
Oligomerization plays an important role in the function of many proteins. Thus, understanding, predicting, and, ultimately, engineering oligomerization presents a long-standing interest. From the perspective of structural biology, protein–protein interactions have mainly been analyzed in terms of the biophysical nature and evolution of protein interfaces. Here, our aim is to quantify the importance of the larger structural context of protein interfaces in protein interaction evolution. Specifically, we ask to what extent intersubunit geometry affects oligomerization state. We define a set of structural parameters describing the overall geometry and relative positions of interfaces of homomeric complexes with different oligomeric states. This allows us to quantify the contribution of direct sequence changes in interfaces versus indirect changes outside the interface that affect intersubunit geometry. We find that such indirect, or allosteric mutations affecting intersubunit geometry via indirect mechanisms are as important as interface sequence changes for evolution of oligomeric states.
Keywords: protein complex evolution, homomeric complexes, protein geometry
During the course of evolution proteins are constrained by their stability, biochemical activity, and regulation. A specific level of evolutionary constraint is added by interactions with other proteins as a greater proportion of protein structure is involved in its function (1). The basic principles of protein recognition and interface formation have been understood for many years (2), and it has been clear for a long time that considerable differences exist between two functionally distinguishable groups—obligate and transient interfaces (3–5). The evolution of protein interfaces has been related to these two groups as well as other biophysical principles. For instance, Mintseris and Weng have, by appropriately grouping types of protein complexes, shown that protein interfaces are slightly more conserved than the surface but much less than the protein core (6). This makes sense in the light of the prediction where, on average, just two substitutions are sufficient to convert a patch on a protein surface into a protein interface (7). This in turn supports work on so-called hot spot (8–10) and anchor residues (11) or conserved residue clusters (12) in protein interfaces. The shared idea in all of these publications, that there are a few key interface residues, is in agreement with the nature of protein interface packing. Residues on the interface rim, which have more conformational freedom, can accommodate sequence changes more easily than ones in the interface core. More recently, there have been contributions to the field showing how interfaces can evolve through insertions of multiple residues forming so-called enabling loops (13, 14).
Interactions put additional constraints on protein sequences (15); however, in complexes where a subunit has multiple distinct surface regions that form interfaces (16), we would not expect the increase in evolutionary constraint to be simply the sum of constraints on the individual interfaces. In addition, there may also be a constraint on their relative geometric position. Here we aim to address this gap, which exists between the two different approaches to the connection of protein interaction and sequence divergence—one focusing on protein interfaces and the other on the overall protein conservation.
From the perspective of protein complex geometry, it has been shown that on the one hand, proteins with very different structures can associate in a similar manner (17, 18). On the other hand, close homologues can have completely different binding modes (19). In this work, we use detailed geometric comparisons of close homologues with conserved binding modes to assess geometric versus straightforward interface sequence constraints on complexes with multiple interfaces. We build on protein structure geometric comparison principles, used for large-scale studies of protein complexes (20) and interdomain geometry (21), and develop a detailed set of geometric parameters necessary for detecting changes in intersubunit geometry of close homologues.
Homomeric Complexes
To compare geometric and direct interface sequence constraints on complexes with multiple interfaces, we use a system of homomeric tetramers and hexamers with dihedral symmetry, which, though simple, still exhibits the key feature of having two structurally distinct interfaces. Dihedral tetramers contain two distinct dimerization interfaces: one face-to-face and one back-to-back (Fig. 1), while hexamers contain one cyclic, face-to-back trimerization interface and one face-to-face dimerization interface. These two types of interfaces need to coexist and coevolve if they are to maintain the geometry and oligomeric state.
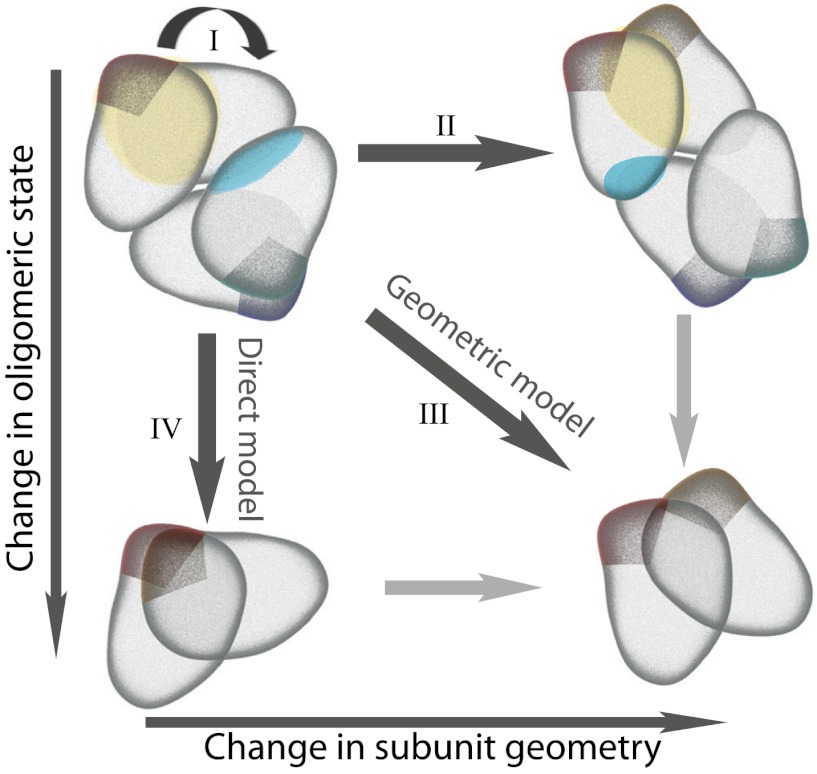
Fig. 1.
Four models of dimer/tetramer evolution through changes in sequence and subunit geometry. (I) Selection on function and/or stability can constrain the protein complex to maintain its specific geometry. (II) Accumulated mutations can influence the orientation of subunits and thus relative positions of the two interfaces. Due to their plasticity, proteins can accommodate these changes and conserve oligomeric state. (III) Geometric model—mutations can occur that influence subunit geometry and change the oligomeric state of the complex. (IV) Direct model—oligomeric state can change without any significant changes in subunit orientation through direct interface mutations alone.
Homomeric complexes represent a significant proportion of protein complexes in the cell, as shown by analyses of known protein complex structures (22) and systematic analyses of complexes in M. pneumoniae (23). There is a large amount of structural data available for families of homomers, with ranges of sequence identities and structural conservations, which makes them good evolutionary case studies. Eleven families examined in this work, contain both dimeric and dihedral tetrameric or hexameric homologues in which the dimeric binding mode is conserved. This means the tetramers and hexamers are dimers and trimers of homologous dimers, respectively. Individual subunits of these homologues have very similar structures, as their overall sequence identities are higher than 30%. The interface conserved in both dimeric and higher oligomer homologues is referred to as the dimeric interface and is usually larger and assembles first in solution (24).
Geometric Coupling of Protein Interfaces and Evolution of Oligomeric State
Depending on functional and stability constraints, a complex experiences different selective pressure on its oligomeric state and/or geometry. Complex geometry is here defined as relative positions of subunits within a complex, and to address it we developed a set of parameters for comparisons of available crystal structures. Because all of the families contain dimers, which are structurally and evolutionary analogous to a dimeric half (or third) of their higher order oligomer homologues, comparisons of complex geometries are done as comparisons of their dimers (Fig. 2).
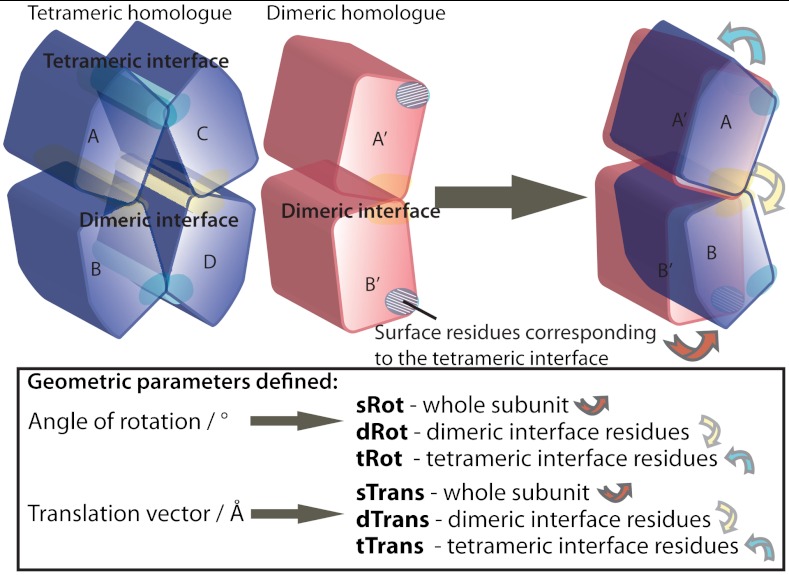
Fig. 2.
Parameters for comparison of complex geometry. Each pair of structures was superimposed in two ways, first by superimposing common dimeric interface regions and then by superimposing the evolutionary core centers of mass of subunit A’ to A. After superimposing the common dimeric residues, the dimeric interface rotation angle and translation vector (dRot and dTrans) were defined by superimposing the same residues of B’ and B subunits. After superimposing subunit evolutionary core center of mass, two types of translations were carried out: on the B subunit center of mass (to obtain sRot and sTrans) and on the A subunit tetrameric/hexameric interface residues (to obtain tRot and tTrans). dRot and dTrans show the levels of local structural differences in the common interface region. sRot and sTrans represent a global difference in complex geometry. tRot and tTrans show the extent of local differences between structural positions of tetrameric/hexameric interface residues relative to the subunit evolutionary core.
In short, geometry of a complex is defined as the relative position of two centers of mass of structurally conserved evolutionary cores of two subunits. Here the structural evolutionary core is defined as a set of residue backbone atoms, which superimpose with an rmsd of 0.5 Å or better. The global difference in geometry between two (sub)complexes is given by the vector of translation (sTrans in Å) and angle of rotation (sRot in °) needed to fit the second subunit (B’ to B in Fig. 2), after first structurally superimposing the first one (A’ to A in Fig. 2). Similarly, we can superimpose individual regions of the protein to show the extent of local structural differences. For the interface regions, we calculate the dimeric interface translation vector (dTrans) and rotation angle (dRot), and tetrameric/hexameric interface translation and rotation (tTrans and tRot).
When multiple interfaces coexist in a structure and the binding mode stays conserved, as is the case for pairs of homologues with our eleven families, interfaces need to be conserved in favorable relative positions in order to maintain the oligomeric state of the complex. A priori, we define four different models of interdependence of oligomeric state and complex geometry (Fig. 1):
Selection on function and/or stability constrains the protein complex to maintain both its specific geometry and oligomeric state.
Mutations influence the orientation of subunits and thus relative positions of the two interfaces. Protein plasticity accommodates these changes, and enables the protein to form both of the interfaces and conserve oligomeric state.
Geometric model—Mutations influence the subunit geometry and are accompanied by a switch in the oligomeric state of the complex.
Direct model—Oligomeric state changes without any significant changes in subunit orientation, through mutations, which disable the formation of the interface.
We chose whole subunit rotation (sRot) as the parameter best describing change in intersubunit geometry. Based on sRot, as well as sequence conservation of interfaces, we aimed to assign to each family a model, which best describes the evolutionary pathway of its oligomeric state change. We assigned the geometric model (III) to four families and the direct model (IV) to three families. For the remaining four families, we consider the evolution of oligomeric state to be a combination of interface residue mutation and change in subunit geometry.
Our dataset exhibits large differences in plasticity across families. Thus we also aim to elucidate the mechanisms by which intersubunit geometry change is easily accommodated in one family, while in others it results in a change in oligomeric state.
Results
Principles of Protein Interface Evolution.
We analyzed 10 SCOP (25) protein families, which, according to the 3DComplex database (22), have at least one dimer and one homologous tetramer or hexamer with the same dimeric binding mode and sequence identity higher than 40%. Phosphoribosyltransferase family has two dimeric interface binding mode subfamilies, making our final set of 11 (sub)families. We could thus define a dimeric interface as the one present throughout all members of the (sub)family and consequently define the geometry of the complex as the orientation of subunits around it. The question, we asked then, was whether and how changes in the geometry influence oligomeric state through long-range effects.
A pair of homologues with conserved binding modes has residues common to the interfaces of both structures. We define the percentage of this overlap as the ratio of overlapping residues and all of the interface residues (see SI Appendix, Fig. S1A and Table S2). The percent sequence conservation of interface residues is variable and ranges from 22% to 95%, but the binding mode is conserved, because the sequence overlap between pairs of homologues is high—from 59% to 100% (see SI Appendix, Fig. S2A).
For each family there is a shared set of interface residues at equivalent sites, which constitute around half of the family’s pool of interface residues. These represent a large proportion (60–79% of residues and 70–89% of buried surface area) of any single interface (see SI Apendix, Fig. S2D). This shows how homologous interfaces are made of a core set of structurally equivalent residues, which show different levels of conservation depending on the family (see SI Appendix, Fig. S2C). In addition, there is a small number of variable, often unique residues, which contribute to the remaining 10–30% of the buried surface. Variable residues from our dataset bury on average less surface than common residues, which implies that the variable residues form the interface rim, while the common residues form the interface core (26). In summary, the interfaces of close homologues are at roughly equivalent positions in the three-dimensional protein structures and occupy similar patches on the surfaces, confirming the familiar concepts of conserved residue clusters (12) or hot spot residues (8–10).
This might lead one to expect complexes either to have highly conserved geometry of the subunits or entirely different binding modes with unrelated interfaces. However, interfaces often comprise multiple secondary structure elements and the relative positions of separate secondary structure elements can differ between homologues. In this way, a conserved set of core interface residues can form interfaces with different geometries (see SI Appendix, Fig. S3, showing the details of dimeric interface plasticity in chemokines and triosephosphate isomerase). Also, interestingly, a recent de novo engineered interface illustrates the possible range of geometric variations for a defined set of residue contacts (27). In this work, crystal structures show that an engineered heterodimer has the predicted pairs of contacting residues but with a surprising 180° rotation relative to the intended design.
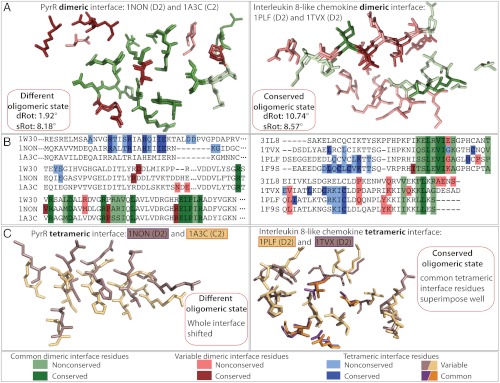
Natural interfaces also exhibit different levels of geometric variation (Fig. 3A; also see SI Appendix, Fig. S3). In the examples of PyrR and interleukin families the two pairs of structures have comparable angles of rotation of whole subunits (sRot values of 8.2° and 8.6°) as well as interface overlap (family mean values are 85% and 84%). However, in the case of the chemokine family, the interface residue conservation is much lower (approximately 40% versus approximately 80%), and common interface residue backbone atoms superimpose with an rmsd of 1.18 Å, compared to 0.31 Å in the PyrR family. Due to these differences in the interface sequence and structural conservation, dimeric interface rotation angles (dRot) for pairs of interleukine and PyrR structures are 10.7° and 1.9°, respectively. Therefore, the difference in intersubunit geometry between a pair of chemokines can be explained by the geometric difference of their dimeric interfaces. In contrast, in the case of the PyrR family, the change in relative subunit orientation between the centres of masses of two subunits in the dimeric unit must come from variations outside of the interface itself. The changes within the 3D structures of homologous PyrR subunits must also be subtle, because their backbones superimpose with an rmsd of 1.5 Å (with approximately 80% of residues superimposing with an rmsd of 0.5 Å).
Fig. 3.
Comparisons of dimeric and tetrameric interfaces in two pairs of homologues with comparable angles of rotation of subunits (sRot). All structural superpositions are of B subunit interfaces after superposition of subunit A centers of mass (see Methods and Fig. 2). Residues are colored according to the contact conservation (common (green) or variable (red) interface contacts) and shaded according to the sequence conservation (conserved or nonconserved amino acids). (A) Structural superpositions of chain B dimeric interfaces after superposition of chain A dimeric interface residues. The structural superposition of dimeric interface residues illustrates the local differences in the structure (see Methods). In the chemokine family, the dimeric interface residue conservation is lower than in the PyrR family (approximately 40% versus approximately 80% and Δrmsd of 1.18 Å versus 0.31 Å). Residues are colored according to pairwise conservation in contacts and sequence. (B) Sequence alignments show all analyzed members of the two families with residues colored according to the family conservation in contacts and sequence. Dimeric interface residues are in green (common) and red (variable) and tetrameric interface residues are in blue. (C) Structural superpositions of chain B tetrameric interface residues. An 8° rotation in the PyrR family causes the entire helix to shift uniformly. Interleukine tetrameric interface adjusted to the evolutionary 8° rotation. Its common tetrameric interface residues (in orange and purple) superimpose better than the rest of the tetrameric interface. Tetrameric interface residues are colored in purple and yellow in different structures for clarity.
Finally, our dataset comprises structures with two interfaces, meaning that a change in intersubunit geometry needs to be accompanied either by relative adjustment of the two interfaces or a change in the oligomeric state. The 8.2° intersubunit rotation in the PyrR family implies a change in oligomeric state, because the tetrameric interface helix does not adjust to the rotation but rather shifts uniformly (Figs. 3C and 4B). This is in principle an allosteric mechanism, because a structural change outside the interface itself brings about a change in oligomeric state. In the interleukin family, the 8.6° intersubunit rotation is accompanied by a structural adjustment of the common tetrameric interface residues (Fig. 3C) and the tetrameric state is conserved.
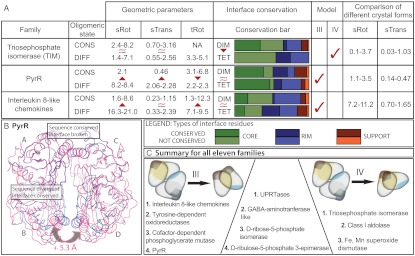
Fig. 4.
Conservation of interfaces and geometric changes. (A) Summary of geometric and sequence comparison parameters for three families. The table provides ranges of values for geometric parameters (sRot, sTrans and tRot) as well as proportions of conserved and nonconserved residues in both interfaces for all pairs with different oligomeric states within a family. Interface core, rim, and support residues are defined as in ref. 7. Higher geometric variation between homologues with different (DIFF) oligomeric state than the ones with conserved (CONS) oligomeric state indicates the geometric (III) evolutionary model. Lower sequence conservation of the tetrameric than the dimeric interface between pairs of homologues with different oligomeric state indicates the direct (IV) evolutionary model. (B) A hypothetical tetramer of B. subtilis PyrR (PDB ID code 1A3C, magenta) is constructed by superposition on to B. caldolyticus PyrR (PDB ID code 1NON, blue), showing evolutionary change in subunit orientation and the relative positions of the two interfaces. Because the residues forming the BcPyrR tetrameric interface helix are completely conserved in sequence between the two species, the 5.3-Å increase in distance between subunits B and D cannot be a matter of local change in the interface but rather due to the difference in complex geometry. (C) Summary for all 11 families. Comparison of homologues reveals high interdependence of oligomeric state and complex geometry (geometric model III) in four protein families. In three families the evolutionary change in oligomeric state is predominantly driven by sequence changes in the interface (direct model IV). The remaining four families represent an evolutionary hybrid that encompasses elements of both the geometric and the direct model.
Geometric Comparisons Reveal Different Models of Oligomeric State Evolution.
We compared complex geometries of all homologous protein pairs (see SI Appendix, Table S3) using simple parameters we have developed (Fig. 2 and SI Appendix, Fig. S1B). The analysis was done on crystal structures, and we first wanted to address the limitations of this type of data. Crystal structures represent a snapshot of the native protein structure, which is in its nature dynamic, and may not represent its most stable or most populated state. However, for some proteins, multiple crystal structures are available—in different crystallographic (space group) and/or biological (ligands) contexts—and can be used to explore the dynamics of the biological structure (28, 29). Thus, throughout this work, we calculate geometric variation between homologues and compare it to the variation between multiple crystal structures of the same protein wherever possible. This allows us to distinguish geometric variation that corresponds to functional allosteric changes or simply flexibility of a protein, from genuine variation in evolution across homologues (see SI Appendix).
Based on the obtained geometric and sequence parameters, we assigned to each family a model, which best describes the evolutionary pathway of its oligomeric state change—meaning either the geometric (III) or the direct model (IV) (Fig. 1).
The whole subunit rotation (sRot) parameter describes the difference between geometry of homologues on a general and robust level. This parameter is also a good predictor of change in oligomeric state (see SI Appendix, Fig. S5). Larger sRot values between pairs of homologues with changed oligomeric state, than between ones with conserved oligomeric state imply that different oligomeric states evolve with a concomitant change in geometry—as described by the geometric model III. On the other hand, we compared the conservation of the interface core residues of the dimeric interface with the conservation between tetrameric/hexameric interface core residues (in tetramer/hexamer) versus surface residues in the homologous dimer. Lower conservation of the tetrameric/hexameric interface core implies that a change in interface sequence is the driving force for change in oligomeric state—as described by the direct model IV. We assign the geometric model (III) to four families and the direct model (IV) to three families (see SI Appendix, Fig. S4). It is interesting to note that in these families, the models are mutually exclusive. In the remaining four families, oligomeric state evolved through a combination of interface residue mutation and change in subunit geometry. The relative contributions of the two mechanisms in these families are difficult to quantify, because dimers are less evolutionarily constrained in their surface regions and lack the geometric coupling of interfaces seen in tetramers/hexamers. Better phylogenetic coverage of homologous structures might pinpoint specific pathways in these families, because there may have been sequential mutations conforming to a combination of the different pathways (pale arrows in Fig. 1).
Three Protein Families Illustrate the Wide Range in Geometric Conservation with Oligomeric State Change.
Triosephosphate isomerase (TIM) family.
TIM family members are obligate dimers, and our dataset contains one tetrameric orthologue from a thermophilic Thermotoga maritima (30). No cooperativity is observed between the two catalytic subunits in the dimers, but dimerization is essential for enzymatic activity, because known inactivating mutations impair oligomerization (31). Some results support the idea of the activity being facilitated by rotational flexibility around the dimeric interface, which transmits to the dynamics of a loop covering the active site (32).
TIM homologues exhibit rotations around the dimeric interface, sRot, ranging from 1.4° to 8.2° (Fig. 4) and the magnitude of sRot is very similar across homologues with conserved or variable oligomeric states. There are comparable levels of geometric variability among different crystal forms of individual proteins (see SI Appendix, Table S4), where sRot ranges from 0.1° to 3.7°. On the other hand, the sequence conservation between the dimeric homologues and the tetrameric Thermotoga maritima TIM is greater at the dimeric interface than at the tetrameric interface/surface patch. Taken together, these two observations—small geometric difference and low conservation of the variable (tetrameric) interface—point toward sequence changes in the tetrameric interface/surface patch as causal for the change in quaternary structure. Thus, we classify the thermophilic TIM as having evolved by the direct model IV, in which changes in the tetrameric interface lead to a change in oligomeric state.
PyrR family.
PyrR protein is a mRNA-binding operon expression attenuator, homologous to the pyrimidine synthesis enzymes it regulates (33). Our dataset includes two tetrameric and one dimeric orthologue.
Whole subunit rotations around the dimeric interface (sRot) are in the range of 2.1° to 8.4°, with a clear distinction between comparisons within tetramers, or between a tetramer and a dimer. To illustrate the impact of the intersubunit rotation on the tetramer formation, we show the structural superposition of the dimer and one of the tetramers. In the case of dimeric PyrR (PDB ID code 1A3C), an 8.2° rotation around the dimeric interface, when compared to the tetrameric PyrR (PDB ID code 1NON) pulls two helices, that would otherwise form the tetrameric interface, more than 5 Å apart (Figs. 3C and 4B). When comparing sequences of the two interfaces, there is a surprising situation: The tetrameric interface/surface patch is more conserved in sequence than the dimeric interface, which is conserved in its structure and function. Thus, the PyrR family scenario conforms to the geometric model III, where the change in the oligomeric state is accompanied by a change in geometry.
In the case of the other tetrameric homolog (Mycobacterium tuberculosis PyrR, PDB ID code 1W30), a smaller 2.1° subunit rotation around the dimeric interface (sRot) is accompanied by a 6.8° rotation of the tetrameric interface (tRot). The small change in geometry between the two tetramers can be described by model II from Fig. 1. This suggests that oligomeric state can be maintained up to a certain degree of rotation between subunits, but that rotations beyond this result in a change of oligomeric state.
Interleukin 8-like chemokine family.
The interleukin 8-like chemokine superfamily covers a range of paralogues with functions in immunophysiology (34). Chemokines are small proteins, which activate different G protein-coupled receptors and cause migration of cells. They all belong to the same superfamily (35) and some of them exhibit high sequence and structural similarity. Our dataset consists of three tetramers (of which two are orthologues—human and bovine PF4), and one dimer.
Whole subunit rotations around the dimeric interface (sRot) range from 1.6° to 8.6° and 21.9° to 31.6° for conserved and changed oligomeric state, respectively. Our control set consists of three different conformations of IP-10, a chemokine that exists as both dimer and tetramer and which has a different tetrameric interface than the other tetrameric chemokines. IP-10 has considerable flexibility around the dimeric interface (see SI Appendix, Table S4) with values of whole subunit rotations ranging from 7.2 to 11.2°.
Greater rotation compared to the PyrR family is not surprising, because the different functions of paralogues in the chemokine family also imply that functional constraints are put on different parts of the structure. The greater structural variability is evident in superimposed dimeric interfaces in Fig. 3. There are relatively large geometric changes in the dimeric interface (dRot) across homologues, but the interface overlap remains substantial, as a consequence of common ancestry. Likewise, the sequence conservations of the dimeric and tetrameric interfaces in the interleukin family are comparable (Fig. 4). The geometric changes are largest for family members with different oligomeric states, and thus geometric model III also best describes the quaternary structure changes for this family.
Conclusions
Based on our analysis of a high confidence set of quaternary structures from 11 protein families we reveal the importance of a geometric component in the evolution of oligomeric state, through coupling between the two interfaces of a protein complex. We illustrate how changes in intersubunit geometry change the relative positions of interfaces, which can consequently impact oligomerization (Figs. 3C and 4B). The mutations, which bring about these geometric changes, can be outside the tetrameric interface itself and their effect is thus allosteric. We also show how the evolutionary dynamics of complex geometry is dependent on the particular structural and functional features of a protein family. Some families can accommodate large geometric changes, while in others much smaller values imply a change in oligomeric state. Sometimes this change in geometry can be brought about by changes in one of the interfaces—when sequence changes in the interface do not significantly change the interface stability but rather the orientation of subunits. In other cases, the changes in geometry are caused by changes outside of both of the interfaces—in these cases both interfaces superimpose well, but their relative positions change.
Overall, we can summarize the evolutionary pathways of oligomeric state by four models of interdependence between geometry and oligomeric state (Fig. 1). While some families conserve their intersubunit geometry (models I and IV), others exhibit much more structural plasticity. This plasticity enables some families to accommodate large geometric changes and maintain the same oligomeric state (model II). In other families the plasticity presents a base for evolutionary geometric dynamics, which leads to the change in the oligomeric state (model III).
From our analysis, two main principles emerge. In one, the evolutionary change in oligomeric state is predominantly driven by sequence changes in the interface (direct model IV), and three out of 11 families conform to this principle. In the second major pathway, comparison of homologues reveals high interdependence of oligomeric state and complex geometry (geometric model III). Four out of 11 families follow this pathway. The remaining four families represent an evolutionary hybrid that encompasses elements of both the geometric model III and the direct model IV.
Sequence changes in interfaces have been widely and frequently used in engineering of protein interactions (36, 37). The analysis here highlights the role of distant mutations that influence intersubunit geometry and thus have an allosteric effect on oligomeric state. These allosteric changes need not necessarily be caused by a large number of gradual mutations. There are many cases where ligand binding is known to allosterically affect protein interactions (e.g., G-protein coupled receptors) and allosteric effects of ligands (e.g., small molecule inhibitors) have been probed by mutations, which introduce structural transitions similar to those induced by the ligand (38).
There is a wide range of mechanisms through which distant mutations can impact oligomerization via conformational changes, which can in turn influence interface coupling. Acknowledging and understanding them can directly aid the engineering of protein interactions, as well as contribute to insights into the diversity of evolutionary pathways that generate protein interactions.
Methods
When comparing the crystal structures within a family, four regions of the protein were defined:
the dimeric interface—the interface conserved in all of the homologues within the family, both dimers and tetramers/hexamers;
the tetrameric (or hexameric) interface—the interface which exists only in homologues with higher oligomeric state;
the region on the surface of the dimeric homolog that corresponds to the residues involved in the interface in homologues of higher oligomeric state (tetrameric or hexameric);
evolutionary conserved core of a protein subunit.
The subunit evolutionary core was defined using a sieve fit method developed by Lesk (39). In this method, all atoms (or in this case all residue backbone atoms) that superimpose with an rmsd lower than some threshold (here an empirical value of 0.5 Å) are referred to as the subunit core and only those are used for the structural fit. A schematic illustration of all the structural fits is provided in Fig. 2 (also see SI Appendix). First A’ to A superposition was done using only common dimeric residues, which correspond to green residues in Fig. 3. The dimeric interface rotation angle and translation vector were defined by superimposing the same residues of the B subunits—these parameters illustrate the contribution of local differences within the conserved (dimeric) interface to the overall structure geometry.
After superimposing the centers of mass of subunit evolutionary cores (A and A’), two types of translations were done. First, we translated and rotated subunit B to fit its evolutionary core—these geometric parameters show the difference in relative orientations of subunits around the dimeric interface. Secondly, we translated and rotated tetrameric (or hexameric) interface residues of subunit A/A’—these parameters yield the differences in position of the interface residues relative to the subunit evolutionary core center of mass.
Supplementary Material
Acknowledgments.
The authors wish to thank Emmanuel D. Levy and Joseph A. Marsh for constructive and helpful comments on the manuscript. This work was supported by the Medical Research Council (MRC file reference number U105161047).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1120028109/-/DCSupplemental.
References
- 1.Fraser HB, Hirsh AE, Steinmetz LM, Scharfe C, Feldman MW. Evolutionary rate in the protein interaction network. Science. 2002;296:750–752. doi: 10.1126/science.1068696. [DOI] [PubMed] [Google Scholar]
- 2.Chothia C, Janin J. Principles of protein-protein recognition. Nature. 1975;256:705–708. doi: 10.1038/256705a0. [DOI] [PubMed] [Google Scholar]
- 3.Janin J, Bahadur RP, Chakrabarti P. Protein-protein interaction and quaternary structure. Q Rev Biophys. 2008;41:133–180. doi: 10.1017/S0033583508004708. [DOI] [PubMed] [Google Scholar]
- 4.Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J Mol Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 5.Nooren IMA, Thornton JM. Diversity of protein-protein interactions. EMBO J. 2003;22:3486–3492. doi: 10.1093/emboj/cdg359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mintseris J, Weng Z. Structure, function, and evolution of transient and obligate protein-protein interactions. Proc Natl Acad Sci USA. 2005;102:10930–10935. doi: 10.1073/pnas.0502667102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levy ED. A simple definition of structural regions in proteins and its use in analyzing interface evolution. J Mol Biol. 2010;403:660–670. doi: 10.1016/j.jmb.2010.09.028. [DOI] [PubMed] [Google Scholar]
- 8.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 9.Bogan A, Thorn K. Anatomy of hot spots in protein interfaces. J Mol Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 10.Ma B, Elkayam T, Wolfson H, Nussinov R. Protein-protein interactions: Structurally conserved residues distinguish between binding sites and exposed protein surfaces. Proc Natl Acad Sci USA. 2003;100:5772–5777. doi: 10.1073/pnas.1030237100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajamani D, Thiel S, Vajda S, Camacho CJ. Anchor residues in protein-protein interactions. Proc Natl Acad Sci USA. 2004;101:11287–11292. doi: 10.1073/pnas.0401942101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guharoy M, Chakrabarti P. Conserved residue clusters at protein-protein interfaces and their use in binding site identification. BMC Bioinformatics. 2010;11:286. doi: 10.1186/1471-2105-11-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akiva E, Itzhaki Z, Margalit H. Built-in loops allow versatility in domain-domain interactions: Lessons from self-interacting domains. Proc Natl Acad Sci USA. 2008;105:13292–13297. doi: 10.1073/pnas.0801207105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hashimoto K, Panchenko AR. Mechanisms of protein oligomerization, the critical role of insertions and deletions in maintaining different oligomeric states. Proc Natl Acad Sci USA. 2010;107:20352–20357. doi: 10.1073/pnas.1012999107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teichmann SA. The constraints protein-protein interactions place on sequence divergence. J Mol Biol. 2002;324:399–407. doi: 10.1016/s0022-2836(02)01144-0. [DOI] [PubMed] [Google Scholar]
- 16.Kim WK, Henschel A, Winter C, Schroeder M. The many faces of protein-protein interactions: A compendium of interface geometry. PLoS Comput Biol. 2006;2:e124. doi: 10.1371/journal.pcbi.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keskin O, Nussinov R. Favorable scaffolds: proteins with different sequence, structure and function may associate in similar ways. Protein Eng Des Sel. 2005;18:11–24. doi: 10.1093/protein/gzh095. [DOI] [PubMed] [Google Scholar]
- 18.Zhang QC, Petrey D, Norel R, Honig BH. Protein interface conservation across structure space. Proc Natl Acad Sci USA. 2010;107:10896–10901. doi: 10.1073/pnas.1005894107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dayhoff JE, Shoemaker BA, Bryant SH, Panchenko AR. Evolution of protein binding modes in homooligomers. J Mol Biol. 2010;395:860–870. doi: 10.1016/j.jmb.2009.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aloy P, Ceulemans H, Stark A, Russell RB. The relationship between sequence and interaction divergence in proteins. J Mol Biol. 2003;332:989–998. doi: 10.1016/j.jmb.2003.07.006. [DOI] [PubMed] [Google Scholar]
- 21.Han JH, Kerrison N, Chothia C, Teichmann SA. Divergence of interdomain geometry in two-domain proteins. Structure. 2006;14:935–945. doi: 10.1016/j.str.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 22.Levy ED, Pereira-Leal JB, Chothia C, Teichmann SA. 3D complex: a structural classification of protein complexes. PLoS Comput Biol. 2006;2:e155. doi: 10.1371/journal.pcbi.0020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kühner S, et al. Proteome organization in a genome-reduced bacterium. Science. 2009;326:1235–1240. doi: 10.1126/science.1176343. [DOI] [PubMed] [Google Scholar]
- 24.Levy ED, Boeri Erba E, Robinson CV, Teichmann SA. Assembly reflects evolution of protein complexes. Nature. 2008;453:1262–1265. doi: 10.1038/nature06942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: A structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 26.Chakrabarti P, Janin J. Dissecting protein-protein recognition sites. Proteins. 2002;47:334–343. doi: 10.1002/prot.10085. [DOI] [PubMed] [Google Scholar]
- 27.Karanicolas J, et al. A de novo protein binding pair by computational design and directed evolution. Mol Cell. 2011;42:250–260. doi: 10.1016/j.molcel.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Best RB, Lindorff-Larsen K, DePristo MA, Vendruscolo M. Relation between native ensembles and experimental structures of proteins. Proc Natl Acad Sci USA. 2006;103:10901–10906. doi: 10.1073/pnas.0511156103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perica T, Chothia C. Ubiquitin—molecular mechanisms for recognition of different structures. Curr Opin Struct Biol. 2010;20:367–376. doi: 10.1016/j.sbi.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 30.Maes D, et al. The crystal structure of triosephosphate isomerase (TIM) from Thermotoga maritima: A comparative thermostability structural analysis of ten different TIM structures. Proteins. 1999;37:441–453. [PubMed] [Google Scholar]
- 31.Mainfroid V, et al. Three hTIM mutants that provide new insights on why TIM is a dimer. J Mol Biol. 1996;257:441–456. doi: 10.1006/jmbi.1996.0174. [DOI] [PubMed] [Google Scholar]
- 32.Cansu S, Doruker P. Dimerization affects collective dynamics of triosephosphate isomerase. Biochemistry. 2008;47:1358–1368. doi: 10.1021/bi701916b. [DOI] [PubMed] [Google Scholar]
- 33.Turnbough CL, Jr, Switzer RL. Regulation of pyrimidine biosynthetic gene expression in bacteria: repression without repressors. Microbiol Mol Biol Rev. 2008;72:266–300. doi: 10.1128/MMBR.00001-08. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fernandez EJ, Lolis E. Structure, function, and inhibition of chemokines. Annu Rev Pharmacol Toxicol. 2002;42:469–499. doi: 10.1146/annurev.pharmtox.42.091901.115838. [DOI] [PubMed] [Google Scholar]
- 35.de Lima Morais DA, et al. SUPERFAMILY 1.75 including a domain-centric gene ontology method. Nucleic Acids Res. 2011;39:D427–434. doi: 10.1093/nar/gkq1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fleishman SJ, et al. Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science. 2011;332:816–821. doi: 10.1126/science.1202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grueninger D, et al. Designed protein-protein association. Science. 2008;319:206–209. doi: 10.1126/science.1150421. [DOI] [PubMed] [Google Scholar]
- 38.Hardy JA, Wells JA. Dissecting an allosteric switch in caspase-7 using chemical and mutational probes. J Biol Chem. 2009;284:26063–26069. doi: 10.1074/jbc.M109.001826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lesk AM, editor. Biosequences: Perspectives and User Services in Europe. 1986. Integrated access to sequence and structural data; pp. 23–28. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.