Abstract
Targeted manipulation of complex genomes often requires the introduction of a double-strand break at defined locations by site-specific DNA endonucleases. Here, we describe a monomeric nuclease domain derived from GIY-YIG homing endonucleases for genome-editing applications. Fusion of the GIY-YIG nuclease domain to three-member zinc-finger DNA binding domains generated chimeric GIY-zinc finger endonucleases (GIY-ZFEs). Significantly, the I-TevI-derived fusions (Tev-ZFEs) function in vitro as monomers to introduce a double-strand break, and discriminate in vitro and in bacterial and yeast assays against substrates lacking a preferred 5′-CNNNG-3′ cleavage motif. The Tev-ZFEs function to induce recombination in a yeast-based assay with activity on par with a homodimeric Zif268 zinc-finger nuclease. We also fused the I-TevI nuclease domain to a catalytically inactive LADGLIDADG homing endonuclease (LHE) scaffold. The monomeric Tev-LHEs are active in vivo and similarly discriminate against substrates lacking the 5′-CNNNG-3′ motif. The monomeric Tev-ZFEs and Tev-LHEs are distinct from the FokI-derived zinc-finger nuclease and TAL effector nuclease platforms as the GIY-YIG domain alleviates the requirement to design two nuclease fusions to target a given sequence, highlighting the diversity of nuclease domains with distinctive biochemical properties suitable for genome-editing applications.
Precise genome editing often requires the introduction of a double-strand break (DSB) at defined positions (1–3), and two distinct site-specific DNA endonuclease architectures have been developed toward this goal. One of these architectures relies on reprogramming the DNA-binding specificity of naturally occurring LAGLIDADG homing endonucleases (LHEs) to target desired sequences (4, 5). The other architecture uses the reprogrammable DNA-binding specificity of zinc-finger proteins or TAL-effector domains that are fused to the nonspecific nuclease domain of the type IIS restriction enzyme FokI to create chimeric zinc-finger nucleases (ZFNs) or TAL effector nucleases (TALENs) (6–8). Regardless of the architecture, the underlying biology of the component proteins imposes design challenges and the relative merits of the LHE and the ZFN/TALEN architectures are the subject of much debate in the literature (6, 9). One notable constraint imposed by the FokI nuclease domain is the requirement to function as a dimer to efficiently cleave DNA (10, 11). For any given DNA target, this necessitates the design of two distinct ZFNs (or two TALENs), such that each pair of zinc finger or TAL effector domains is oriented for FokI dimerization and DNA cleavage (12).
Expanding the repertoire of DNA nuclease domains with distinctive properties is necessary to facilitate the development of new genome-editing reagents. Indeed, a number of recent studies have explored the potential of the PvuII restriction enzyme as an alternative site-specific nuclease domain for genome-editing applications (13, 14). The PvuII chimeras, however, share similar design constraints as ZFNs and TALENs, requiring two nuclease fusions for precise targeting. In considering alternative nuclease domains for genome editing, we were intrigued by the properties of the GIY-YIG nuclease domain that is associated with a variety of proteins of diverse cellular functions (15). The small (∼100 aa) globular GIY-YIG domain is characterized by a structurally conserved central three-stranded antiparallel β-sheet, with catalytic residues positioned to use a single metal ion to promote DNA hydrolysis (16–18). Intriguingly, the GIY-YIG homing endonucleases, typified by the isoschizomers I-TevI and I-BmoI (19), bind DNA as monomers (20), and generate a DSB with 2-nt, 3′ overhangs. It is unknown, however, if GIY-YIG homing endonucleases function as monomers in all steps of the reaction, as the oligomeric status during cleavage has yet to be studied. Notably, GIY-YIG homing endonucleases prefer a specific DNA sequence to generate a DSB (21, 22). For I-TevI, the bottom (↑) and top (↓) strand nicking sites lie within a 5′-CN↑NN↓G-3′ motif (CNNNG), with the critical G-optimally positioned ∼28 bp from where the helix-turn-helix (H-T-H) module of the I-TevI DNA-binding domain interacts with substrate (21, 22). From an engineering perspective, the modularity and sequence specificity of the GIY-YIG nuclease domain makes it an appealing candidate to create new chimeric endonucleases. Indeed, swapping of the I-BmoI and I-TevI catalytic and DNA-binding domains suggested that the GIY-YIG nuclease domain could be fused to unrelated DNA-targeting platforms (23).
To highlight the genome engineering potential of the GIY-YIG nuclease domain, we fused the domain to three-member zinc fingers to construct GIY-YIG zinc-finger endonucleases (GIY-ZFEs). The GIY-ZFEs are active in bacterial and yeast cells, and in vitro data show that they function catalytically as monomers and retain the cleavage specificity associated with the parental GIY-YIG nuclease domain. The GIY-YIG nuclease domain is also portable to the LHE platform, as we constructed monomeric GIY-LHEs that are active in vivo and possess ∼18-bp binding specificity. We selected LHEs as a DNA-targeting domain because of the greater sequence specificity compared with three-member zinc fingers, the ability to reprogram LHE DNA-binding specificity (24–26), and recent success in generating PuvII-LHE fusions (13). Collectively, our data highlight the unique biochemical properties of the GIY-YIG nuclease domain as an alternative to the FokI nuclease domain for genome-editing applications.
Results
Construction and Validation of GIY-ZFEs.
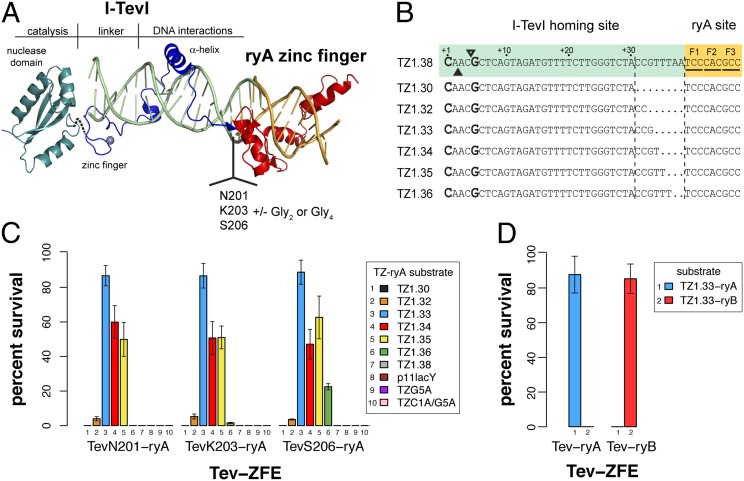
To create novel chimeric enzymes, we modeled GIY-ZFEs using existing crystal structures of the I-TevI 130C DNA binding domain and the Zif268 zinc finger (Fig. 1A) (27, 28). One notable feature of our constructs is the polarity, as the I-TevI nuclease domain is fused to the N-terminal end of the three-finger ryA zinc-finger protein to mimic its native orientation, unlike FokI constructs that are fused to the C-terminal end of zinc-finger proteins. We modeled the Zif268 zinc finger in place of the H-T-H module at the C terminus of I-TevI, providing the rationale to subsequently fuse various lengths of the I-TevI N-terminal region to the ryA zinc finger that targets a sequence in the Drosophila rosy gene to create Tev-ryA zinc-finger endonucleases (Tev-ZFEs) (Fig. 1A) (29). The Tev-zinc finger DNA substrates (TZ) consisted of 30–38 bps of the I-TevI td homing site joined to the 9-bp ryA target site. The TZ substrates differ in the distance of the CNNNG cleavage motif relative to the ryA-binding site (Fig. 1B). Each TZ substrate possesses a single zinc-finger targeting sequence, rather than two head-to-head zinc-finger sites necessary for efficient ZFN cleavage. A similar set of I-BmoI-ryA fusions (Bmo-ZFEs) and substrates (BZ) were constructed (Fig. S1).
Fig. 1.
Design and functionality of Tev-ZFEs. (A) Modeling of a Tev-zinc finger fusion with DNA substrate (light green) using structures of the I-TevI catalytic domain in green (PDB 1MK0), the I-TevI DNA-binding domain cocrystal in blue (PDB 1I3J), and the Zif268 cocrystal in red (PDB 1AAY). (B) The TZ-ryA substrate is colored according to the structural model. Shown is the top strand of the I-TevI td homing site substrate fused to the 5′ end of the ryA-binding site for all wild-type substrates tested. The substrate is numbered from the first base of the td homing site sequence (the numbering scheme is reverse of that used for the native td homing site). The substrates tested differ by insertion or deletion of td sequence at the junction of the td/ryA sites. (C) Percent survival of three representative Tev-ryA ZFEs in the bacterial two-plasmid selection. All Tev-ryA ZFEs were tested against plasmids containing various length substrates (TZ1.30–1.38), plasmids lacking a target site (p11lacY), and TZ1.33 plasmids with single or double mutations in the CNNNG motif (G5A and C1A/G5A) (Table S1). (D) Percent survival of TevN201-ryA and TevN201-ryB ZFEs on their cognate and reciprocal target sites. Data are plotted with SD for n ≥ 3.
We tested the activity of the GIY-ZFEs using a well-described two-plasmid bacterial selection system, where survival is dependent on the endonuclease cleaving a target plasmid (30, 31). Eight Tev-ZFEs were tested on seven TZ substrates cloned into the reporter plasmid (Fig. 1 B and C and Table S1). In general, the survival of all Tev-ZFEs was highest against TZ substrates where the preferred CNNNG motif was positioned between 33 and 35 bp from the ryA binding site. Low survival (∼4–6%) was observed for all Tev-ZFEs against the TZ1.32 substrate, but none survived on the TZ1.30 substrate. Similarly, there was no survival against the longer substrates, with the exception that the longest fusion (TevS206-ryA) exhibited ∼22% survival against the TZ1.36 substrate. No survival was observed when the Tev-ZFEs were tested against the target plasmid without a target site (p11lacYwtx1). Mutation of the catalytic arginine 27 of the I-TevI nuclease domain to alanine to create TevR27A-ryAs showed that survival is dependent on GIY-YIG nuclease activity as none of the Tev-R27A constructs survived (Table S1).
We also constructed and tested a fusion of the TevN201 domain to a different three-member zinc finger, the ryB zinc finger, creating TevN201-ryB. The TevN201-ryB showed survival in the bacterial selection assay against a corresponding TZ-ryB target, indicating that the I-TevI nuclease domain can function in the context of two different three-member zinc fingers, but did not survive when tested against the TZ-ryA substrate (Fig. 1D). Similarly, the TevN201-ryA fusion did not survive against the TZ-ryB substrate, indicating that the zinc finger alone directs DNA-binding. We also tested the Bmo-ZFEs in the genetic selection, but did not observe significant survival for any of the fusions, consistent with the ∼750-fold reduced activity of wild-type I-BmoI relative to I-TevI (32). However, as described below, enzymatic activity was detected in vitro using purified Bmo-ZFEs. Collectively, these data show that two different GIY-YIG nuclease domains could be fused to zinc-finger DNA-binding domains to create active site-specific chimeric nucleases.
Tev-ZFEs Function as Monomers to Cleave at a Specific Sequence.
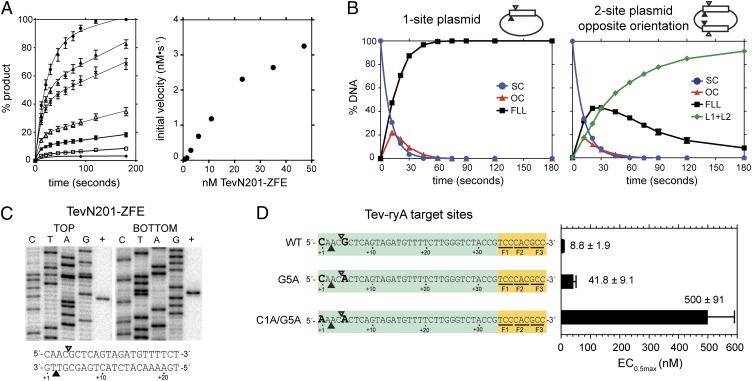
To study the GIY-ZFE biochemical characteristics in more detail, we purified TevN201-ryA for cleavage assays and in vitro mapping. We first performed cleavage assays to determine the relationship between TevN201-ryA enzyme concentration and initial reaction velocity using a plasmid substrate with a single TZ-ryA target site. The reaction progress curves indicated an initial burst of cleavage followed by a slower rate of product accumulation (Fig. 2A), consistent with product release being the rate-limiting step. The initial burst phase was used to estimate initial velocity, and plotting against protein concentration yielded a linear relationship (Fig. 2A), suggesting that DNA hydrolysis catalyzed by TevN201-ryA is first order with respect to protein concentration.
Fig. 2.
TevN201-ZFE is a monomer with a preferred cleavage site. (A) (Left) Plot of initial reaction progress for seven TevN201-ZFE concentrations expressed as percent linear product. Protein concentrations from highest to lowest are 47 nM, 32.5 nM, 23 nM, 11 nM, 6 nM, 3 nM, and 0.7 nM. (Right) Graph of initial reaction velocity (nM⋅s−1) versus TevN201-ZFE concentration (nM). (B) Graphic representation of cleavage assays with 90 nM TevN201-ZFE and 10 nM one- or two-site TZ1.33 plasmids (Left and Right, respectively). The two-site plasmid had the TZ-ryA sites in the opposite (shown) or same (Fig. S2B) orientation. FLL, full-length linear; L1+L2, linear products; OC, open-circle (nicked); SC, supercoiled. (C) Mapping of TevN201-ZFE cleavage sites on the TZ1.33 substrate, with top and bottom cleavage sites indicated below on the TZ-ryA substrate by open and closed triangles, respectively. (D) Activity of TevN201-ZFE on the wild-type TZ1.33, or the TZ1.33 G5A and TZ1.33 C1A/G5A mutant substrates. A graph of EC0.5max determinations for each substrate is shown to the right, with EC0.5max values in nanomolars. Data are plotted as averages of three independent replicates with SDs.
The model TZ-ryA substrates were designed as a single ryA zinc-finger site fused to the I-TevI target sequence. To determine if cleavage by TevN201-ryA was influenced by additional Tev-ryA target sites, we constructed two-site plasmids that differed in whether the target sites were in the same or opposite orientations relative to each other. The single- or two-site plasmids were used in time-course cleavage assays under single-turnover conditions (∼10-fold molar excess of protein to substrate) to determine reaction rates. As shown in Fig. 2B, cleavage of the one-site plasmid yielded kobs(1-site) = 0.099 ± 0.001 s−1, and cleavage of the two-site plasmids with target sites in the opposite or same (Fig. S2B) orientations generated very similar rate constants, kobs(2-site) = 0.088 ± 0.001 s−1 and 0.089 ± 0.001 s−1, respectively, to the one-site plasmid. In contrast, similar experiments with FokI showed a significant rate enhancement for two-site plasmids relative to one-site plasmids, consistent with FokI functioning as a dimer (33). We conclude that cleavage by TevN201-ryA is noncooperative and that efficient DNA hydrolysis does not require two sites, consistent with TevN201-ryA functioning catalytically as a monomer.
The I-TevI nuclease domain preferentially cleaves DNA within a 5′-CN↑NN↓G-3′ motif, with ↑ and ↓ representing the bottom- and top-strand nicking sites, respectively (22). I-TevI defaults to cleave at the wild-type distance on substrates in vitro when this motif is moved closer to, or distant from, the primary binding site, whereas mutants in the I-TevI–specific zinc finger cleave at the correct sequence rather than the wild-type distance on mutant substrates (34). To determine the cleavage preference of the TevN201-ryA construct, we mapped the bottom- and top-strand nicking sites using strand-specific end-labeled substrates to the CNNNG motif (Fig. 2C). Combined with data from the genetic assays showing no survival on substrates that displace the CNNNG motif from an optimal position, our data suggest that in the context of a ryA fusion, the TevN201 domain acts as a molecular ruler with a distance preference.
To further demonstrate TevN201-ryA cleavage preference, we introduced mutations in the CNNNG motif that were previously shown to drastically reduce I-TevI cleavage efficiency (Fig. 2D) (21, 22). Significantly, we observed no survival under selective conditions in the two-plasmid assay on plasmids carrying either the single G5A (CNNNA) or double C1A/G5A (ANNNA) substitutions (Fig. 1C), equivalent to positions C-27 and G-23 of the I-TevI td substrate, respectively. We also performed in vitro cleavage assays on wild-type and mutant substrates with increasing concentrations of TevN201-ZFE to determine the amount of protein required for half-maximal cleavage (EC0.5max). As shown in Fig. 2D, ∼60-fold and ∼4.7-fold more protein were required to achieve half-maximal cleavage of the double- and single-mutant substrates relative to the wild-type substrate. The greater substrate discrimination observed in the genetic assay likely reflects lower in vivo protein concentrations than those used for in vitro cleavage assays. These results show that the TevN201-ryA fusion retains the cleavage specificity of the parental I-TevI enzyme and that double nucleotide substitutions significantly reduce cleavage efficiency. To determine if Bmo-ZFEs also retained substrate specificity, the bottom- and top-strand nicking sites of the BmoN221-ryA fusion were mapped to a 5′-NN↑NN↓G-3′ motif, consistent with the cleavage site preference of I-BmoI (Fig. S1D) (19).
Tev-ZFEs Function in a Yeast-Based Recombination Assay.
To extend the in vivo relevance of the Tev-ZFE fusions, we used a well-described yeast-based recombination assay to test Tev-ZFE function in a eukaryotic system (35). This assay provides a quantitative β-galactosidase readout if the nuclease cleaves its target site that is positioned between a partially duplicated lacZ gene. Furthermore, the assay allowed us to calibrate TevN201-ryA activity relative to a homodimeric FokI-Zif268 control with previously measured in vivo activity sufficient to induce recombination events for genome engineering applications (35). As shown in Fig. 3, the level of β-galactosidase activity for the TevN201-ryA fusion on its cognate TZ-ryA substrate was ∼1.4-fold higher than the Zif268 ZFN control. The TevN201-ryA or Zif268 ZFN constructs displayed no activity on each other’s substrates, and activity was dependent on a functional I-TevI nuclease domain, as the TevN201R27A catalytic mutant was unable to induce recombination. Furthermore, TevN201-ryA activity was not observed on mutant substrates where one or both of the critical residues of the CNNNG motif were mutated in the TZ1.33 substrate. Collectively, these assays show that the I-TevI nuclease domain functions in a eukaryotic system with activity on par to a characterized ZFN.
Fig. 3.
Tev-ZFEs can induce recombination in a eukaryotic system. Shown are normalized β-galactosidase units from a yeast-based recombination assay for the indicated nuclease/substrate combinations. Activity was normalized to a homodimeric FokI-Zif268 ZFN-positive control. Data are plotted with SD for n = 4.
I-TevI Nuclease Domain Is Portable to the LAGLIDADG Architecture.
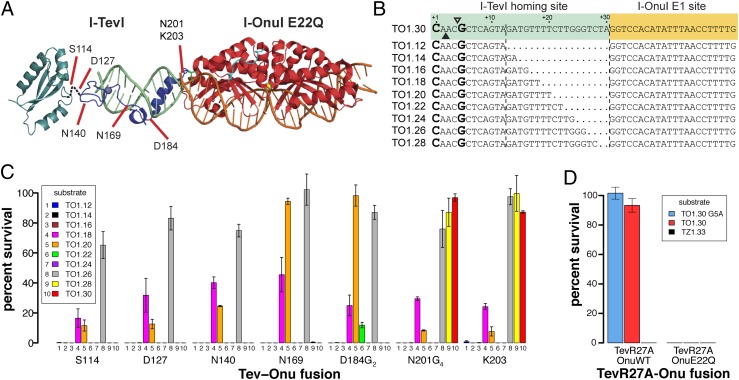
To demonstrate that the I-TevI nuclease domain functions in the context of DNA-targeting platforms with greater specificity than three-member zinc fingers, we constructed fusions of the domain to a catalytically inactive monomeric single-chain LAGLIDADG homing endonuclease (Tev-LHE). As with the Tev-ZFE constructs, we modeled a Tev-LHE chimera using the cocrystal of I-OnuI with its DNA substrate such that the I-TevI nuclease and linker domains were fused to the N terminus of I-OnuI, which is partially disordered in the structure (Fig. 4A) (25). Based on this model, we fused TevN201G4 and TevK203 fragments to a catalytically dead I-OnuI E1 E22Q mutant. A series of model DNA substrates were constructed by fusing the td target site to the I-OnuI E1 binding site in the human MAO-B gene, differing in the position of the CNNNG cleavage motif relative to the I-OnuI E1 site (TO1.12 to TO1.30) (Fig. 4B).
Fig. 4.
Design and functionality of Tev-LHEs. (A) Modeling of a Tev-Onu E1 fusion with DNA substrate (light green) using structures of the I-TevI catalytic domain in green (PDB 1MK0), the I-TevI DNA-binding domain cocrystal in blue (PDB 1I3J), and the I-OnuI cocrystal in red (PDB 3QQY). Shown are fusion points at which the I-TevI fragment has been shortened. (B) The Tev-Onu E1 (TO) substrate is colored according to the structural model. Shown is the top strand of the I-TevI td homing site substrate fused to the 5′ end of the Onu E1-binding site. The substrates are numbered from the first base of the td homing site sequence and differ by the deletion of td nucleotides at the junction of the td/Onu E1 sites. (C) Percent survival of Tev-LHEs in the bacterial two-plasmid selection with various length target sites (TO1.12–1.30). All Tev-LHEs tested were in the I-OnuI E1 E22Q background. (D) Percent survival of TevR27A(N201G4)-OnuE1 and TevR27A(N201G4)-OnuE1(E22Q) on TO1.30, TO1.30G5A, and TZ1.33. Data are plotted with SD for n = 3.
In the bacterial two-plasmid selection, we found that the TevN201G4-Onu and TevK203-Onu fusions were active against a range of DNA substrates. Notably, the fusions displayed maximal survival on longer targets (TO1.26, TO1.28, and TO1.30), and lower survival against shorter targets (TO1.18 and TO1.20 targets). The two groups of substrate differ by approximately one helical turn of DNA, meaning that the preferred CNNNG motif would be presented on the same face of the substrate, even though the motif is closer to the I-OnuI E1 binding site on the shorter targets. Similar periodic cleavage patterns have been observed in vitro with I-TevI on substrates with a displaced CNNNG motif (36). This result also implies that the N terminus of I-OnuI possesses inherent flexibility to allow the I-TevI nuclease domain to search out the CNNNG motif, in contrast to the ruler-like behavior of the Tev-ZFE constructs, likely because the zinc-finger N terminus is inflexible. Importantly, the Tev-Onu fusions were not active against the TZ-ryA zinc finger substrates (Table S2), showing that the LHE, and not the I-TevI linker, directs DNA targeting. Survival was also dependent on an active I-TevI nuclease domain, as TevR27A fusions in the context of the I-OnuI E22Q mutant did not survive (Fig. 4D). Conversely, the targeting and activity of wild-type I-OnuI E1 was not affected by fusion of the I-TevI domain, because the TevR27A-OnuWT fusions survived against TO substrates (Fig. 4D).
The apparent flexibility of the N terminus and the greater specificity of I-OnuI prompted us to test fusions containing shorter fragments of the I-TevI nuclease domain (Fig. 4A). Based on structural and genetic data, we constructed TevS114-Onu, TevD127-Onu, TevN140-Onu, TevN169-Onu, and TevD184G2-Onu fusions, progressively removing amino acid residues of I-TevI that make specific base pair contacts to the td substrate (28) (Fig. 4A). Notably, the TevS114, TevD127, TevN140, and TevN169 removed the α-helix that binds in the minor groove, as well as residues shown by structural data to make base-specific contacts (28). The TevS114 fusion point lies at the boundary of the deletion tolerant region of the I-TevI linker, and represents a functionally minimal GIY-YIG nuclease domain (36, 37). We found that the shorter fusions were not active against the longer TO1.28 and TO1.30 substrates, yet displayed the same periodic activity on the shorter substrates (Fig. 4C and Table S2). A single exception was the TevD184G2 fusion that showed low survival against the TO1.22 substrates, against which no other fusion survived. No survival was observed on mutant substrates that contained single (CNNNA) or double (ANNNA) mutations in the CNNNG motif, recapitulating the necessity for an appropriately positioned CNNNG as seen with the Tev-ZFE fusions.
5′-CNNNG-3′ Cleavage Motif Is Not Limiting for Targeting.
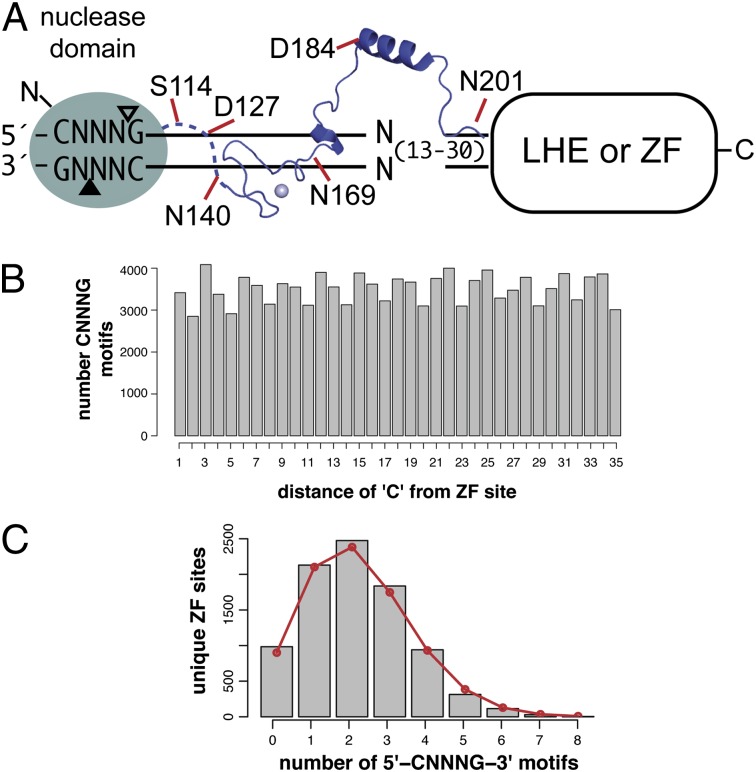
An important consideration in the design of GIY-ZFEs or GIY-LHEs for genome-editing applications is the targeting requirements, notably the need for the CNNNG di-nucleotide cleavage motif (Fig. 5A). In a complex genome of ∼3 × 109 bp, the statistically predicted occurrence of the CNNNG motif is once every 15 bp, assuming a 50% GC content. To determine if the frequency of the CNNNG motif would be limiting for targeting applications, we examined 35 bp flanking 8,829 computationally predicted ZFN sites on zebrafish chromosome 1 for the occurrence of the CNNNG motif (38). As shown in Fig. 5B, the motif is highly represented at all positions within a 35-bp window relative to the ZFN sites. Of the 8,829 sites examined, 88% (7,845) of ZFN sites possessed at least one motif within 35 bp of the predicted binding site (Fig. 5C). These requirements contrast sharply with those of the recently described PvuII-LHEs and PvuII-ZFNs that require the 6-bp 5′-CAGCTG-3′ PvuII site in addition to the LHE or ZF binding site (13, 14). Of the 8,829 ZFN sites, 97% lacked a PvuII site within the 35-bp window (Fig. S3). Thus, the requirement for a di-nucleotide cleavage motif in the context of a GIY-ZFE or GIY-LHE will not severely limit potential targeting sites.
Fig. 5.
Cleavage requirements do not limit GIY-ZFE and GIY-LHE applicability. (A) A diverse set of monomeric and sequence specific reagents can be generated by fusing distinct GIY-YIG domain linker lengths to engineered DNA-binding platforms, including zinc-finger arrays and inactive LAGLIDADGs. (B) Shown is the distribution of the CNNNG motif in a 35-bp window flanking 8,829 predicted ZFN sites on zebrafish chromosome 1. The number of occurrences of the “C” of the motif at each distance is indicated. (C) Unique ZFN sites were grouped according to the number of occurrences of the CNNNG motif in the 35-bp window. The red line is the expected number of ZFN sites for each group based on a binomial distribution.
Discussion
Here, we provide evidence that the GIY-YIG nuclease domain is a potential alternative to the currently used FokI nuclease domain for genome-editing applications. We show that the I-TevI GIY-YIG nuclease domain is portable to two reprogrammable DNA-binding scaffolds, the three-member zinc fingers, and LAGLIDADG homing endonucleases. The Tev-ZFE and Tev-LHE fusions are active in vitro and in vivo, with the activity of the Tev-ZFE in a yeast-based recombination assay on par with that of a characterized ZFN. We foresee the monomeric nature of the Tev-LHEs and Tev-ZFEs as a key advantage over existing ZFNs and TALENs, because a single fusion protein need be designed to target a given sequence, rather than two ZFNs or TALENs required to promote dimerization of the FokI nuclease domain (12). Moreover, the fact that the I-TevI nuclease domain possesses a preferred cleavage motif adds another layer of specificity to targeting requirements, potentially limiting DSBs at off-target sites that do not posses the cleavage motif.
One targeting consideration for chimeric GIY-YIG endonucleases is the DNA sequence requirement of the I-TevI linker. The I-TevI linker is a complex structure, consisting of defined structural elements with distinct roles in I-TevI function (28, 34, 36). The primary role of the linker is to position the nuclease domain on substrate for cleavage at the CNNNG motif, which is found at a defined distance from the binding site on naturally occurring I-TevI substrates. However, the linker can direct the nuclease domain in vitro to search out displaced CNNNG motifs on both native and nonnative substrates with insertions or deletions, albeit with reduced cleavage efficiency. Our Tev-LHE fusions recapitulate this distance versus sequence behavior in vivo, as the fusions can cleave displaced CNNNG motifs with a periodicity that parallels the helical nature of DNA. We partially attribute this ability of the Tev-Onu fusions to the flexible N terminus of I-OnuI. The substrate flexibility of different length Tev-Onu fusions is an important consideration for targeting, as CNNNG motifs at various positions relative to the LHE binding site would be accessible by the choice of the appropriate Tev-LHE fusion. In contrast, the apparently inflexible N terminus of the three-member zinc fingers constrains cleavage to a distance of 33–36 bp from the ryA-binding site, mimicking the spacing of the CNNNG motif on native td substrate. Our longest Tev-ZFE and Tev-LHE fusions encompass all of the known elements of the I-TevI linker that make multiple base-specific and nonspecific contacts to DNA (28). However, biochemical studies revealed that I-TevI retains significant cleavage activity on substrates with multiple substitutions in the central region of its cognate DNA substrate that is contacted by the linker, equivalent to positions 6–33 of our longest chimeric substrates (39). The shortest Tev-LHE fusions do not contain any linker elements that are known to make base-specific DNA contacts, and cleave only at the preferred CNNNG motif. This observation implies that the I-TevI linker may contact substrate nucleotides adjacent to the CNNNG motif. Potential contacts may play a role in the positioning of the nuclease domain, rather than being necessary for cleavage, and any preference may be related to regulating the position of the nuclease domain on substrate or the maintenance of DNA-structure (36).
Future work on Tev-ZFEs and Tev-LHEs will require a detailed dissection of binding affinity and specificity, and characterization of cellular toxicity that results from cleavage at off-target sites. In their current form, the targeting specificity of the Tev-ZFEs is a function of the three-zinc finger domain, which could be further enhanced by addition of zinc fingers to generate a four-, five-, or six-zinc finger fusion to increase specificity, as has been done with a variety of ZFNs (40). In contrast, the ∼18-bp specificity of LHEs is sufficient to direct targeting and cleavage at endogenous loci in human cells. LHEs, however, are tolerant of nucleotide substitutions within their recognition sequence, and I-OnuI E1 cleaves off-target sites that differ by one or two nucleotide substitutions (25). In the context of Tev-LHEs, decoupling of DNA-cleavage and DNA-binding activity by using a catalytically dead LHE scaffold, combined with the requirement for a preferred I-TevI CNNNG cleavage motif, would significantly reduce cleavage at off-target sites (Fig. S4). Another advantage of the decoupled activities of Tev-LHEs is that they would not require reoptimization of catalytic activity that is often necessary in LHEs that have been reprogrammed to bind nonnative target sites (25, 41). Similar to the exploration of alternative DNA-binding platforms (2), it is imperative to incorporate nuclease domains with distinct biochemical properties into the genome-engineering pipeline to create highly precise tools. With further optimization, the I-TevI nuclease domain may become an alternative to the FokI-derived ZFNs and TALENs.
Materials and Methods
See detailed SI Materials and Methods for further discussion, Fig. S5 for amino acid sequences of GIY-ZFEs and Tev-LHEs, Table S3 for strains and plasmids used in this study, and Table S4 for oligonucleotides used in this study. Tev-ZFE and Tev-LHE fusions and hybrid target sites were modeled in PyMOL using the I-TevI 130C (PDB 1I3J), Zif268 (PDB 1AAY), and I-OnuI (PDB 3QQY) cocrystal structures (25, 27, 28). The in vivo activity of fusions was determined using a two-plasmid bacterial selection (31), and a yeast-based reporter assay was used to calibrate activity of the Tev-ZFE against a characterized ZFN (35). TevN201-ryA was purified using nickel affinity chromatography to determine the in vitro biochemical properties of Tev-ZFEs. Cleavage assays were performed as previously described (42). A custom Perl script (Dataset S1) was created to determine CNNNG site occurrences relative to 8,829 predicted ZFN sites on zebrafish chromosome 1 (38).
Supplementary Material
Acknowledgments
We thank Dr. Adam Bogdanove for providing Saccharomyces cerevisiae strains and reporter assay plasmids; Dr. Chris Brandl for yeast culture and assay reagents; and Dr. Greg Gloor, Dr. Megan Davey, Clarice Schmidt, Julie Genereaux, and Russell Dickson for invaluable advice and assistance. This work was supported by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (to D.R.E.), and an Alexander Graham Bell Canada Graduate Scholarship from the Natural Sciences and Engineering Research Council of Canada (to B.P.K.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1117984109/-/DCSupplemental.
References
- 1.Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300:764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- 2.Bogdanove AJ, Voytas DF. TAL effectors: Customizable proteins for DNA targeting. Science. 2011;333:1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- 3.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 4.Marcaida MJ, Muñoz IG, Blanco FJ, Prieto J, Montoya G. Homing endonucleases: From basics to therapeutic applications. Cell Mol Life Sci. 2010;67:727–748. doi: 10.1007/s00018-009-0188-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stoddard BL. Homing endonucleases: From microbial genetic invaders to reagents for targeted DNA modification. Structure. 2011;19:7–15. doi: 10.1016/j.str.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cathomen T, Joung JK. Zinc-finger nucleases: The next generation emerges. Mol Ther. 2008;16:1200–1207. doi: 10.1038/mt.2008.114. [DOI] [PubMed] [Google Scholar]
- 7.Christian M, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halford SE, Catto LE, Pernstich C, Rusling DA, Sanders KL. The reaction mechanism of FokI excludes the possibility of targeting zinc finger nucleases to unique DNA sites. Biochem Soc Trans. 2011;39:584–588. doi: 10.1042/BST0390584. [DOI] [PubMed] [Google Scholar]
- 10.Bitinaite J, Wah DA, Aggarwal AK, Schildkraut I. FokI dimerization is required for DNA cleavage. Proc Natl Acad Sci USA. 1998;95:10570–10575. doi: 10.1073/pnas.95.18.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vanamee ES, Santagata S, Aggarwal AK. FokI requires two specific DNA sites for cleavage. J Mol Biol. 2001;309:69–78. doi: 10.1006/jmbi.2001.4635. [DOI] [PubMed] [Google Scholar]
- 12.Smith J, et al. Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic Acids Res. 2000;28:3361–3369. doi: 10.1093/nar/28.17.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fonfara I, Curth U, Pingoud A, Wende W. Creating highly specific nucleases by fusion of active restriction endonucleases and catalytically inactive homing endonucleases. Nucleic Acids Res. 2012;40:847–860. doi: 10.1093/nar/gkr788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schierling B, et al. A novel zinc-finger nuclease platform with a sequence-specific cleavage module. Nucleic Acids Res. 2012;40:2623–2638. doi: 10.1093/nar/gkr1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunin-Horkawicz S, Feder M, Bujnicki JM. Phylogenomic analysis of the GIY-YIG nuclease superfamily. BMC Genomics. 2006;7:98. doi: 10.1186/1471-2164-7-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mak AN, Lambert AR, Stoddard BL. Folding, DNA recognition, and function of GIY-YIG endonucleases: Crystal structures of R.Eco29kI. Structure. 2010;18:1321–1331. doi: 10.1016/j.str.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sokolowska M, Czapinska H, Bochtler M. Hpy188I-DNA pre- and post-cleavage complexes—Snapshots of the GIY-YIG nuclease mediated catalysis. Nucleic Acids Res. 2011;39:1554–1564. doi: 10.1093/nar/gkq821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Roey P, Meehan L, Kowalski JC, Belfort M, Derbyshire V. Catalytic domain structure and hypothesis for function of GIY-YIG intron endonuclease I-TevI. Nat Struct Biol. 2002;9:806–811. doi: 10.1038/nsb853. [DOI] [PubMed] [Google Scholar]
- 19.Edgell DR, Shub DA. Related homing endonucleases I-BmoI and I-TevI use different strategies to cleave homologous recognition sites. Proc Natl Acad Sci USA. 2001;98:7898–7903. doi: 10.1073/pnas.141222498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mueller JE, Smith D, Bryk M, Belfort M. Intron-encoded endonuclease I-TevI binds as a monomer to effect sequential cleavage via conformational changes in the td homing site. EMBO J. 1995;14:5724–5735. doi: 10.1002/j.1460-2075.1995.tb00259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bryk M, Belisle M, Mueller JE, Belfort M. Selection of a remote cleavage site by I-tevI, the td intron-encoded endonuclease. J Mol Biol. 1995;247:197–210. doi: 10.1006/jmbi.1994.0133. [DOI] [PubMed] [Google Scholar]
- 22.Edgell DR, Stanger MJ, Belfort M. Coincidence of cleavage sites of intron endonuclease I-TevI and critical sequences of the host thymidylate synthase gene. J Mol Biol. 2004;343:1231–1241. doi: 10.1016/j.jmb.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 23.Liu Q, Derbyshire V, Belfort M, Edgell DR. Distance determination by GIY-YIG intron endonucleases: Discrimination between repression and cleavage functions. Nucleic Acids Res. 2006;34:1755–1764. doi: 10.1093/nar/gkl079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosen LE, et al. Homing endonuclease I-CreI derivatives with novel DNA target specificities. Nucleic Acids Res. 2006;34:4791–4800. doi: 10.1093/nar/gkl645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeuchi R, et al. Tapping natural reservoirs of homing endonucleases for targeted gene modification. Proc Natl Acad Sci USA. 2011;108:13077–13082. doi: 10.1073/pnas.1107719108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacoby K, et al. Expanding LAGLIDADG endonuclease scaffold diversity by rapidly surveying evolutionary sequence space. Nucleic Acids Res. 2012 doi: 10.1093/nar/gkr1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elrod-Erickson M, Rould MA, Nekludova L, Pabo CO. Zif268 protein-DNA complex refined at 1.6 A: A model system for understanding zinc finger-DNA interactions. Structure. 1996;4:1171–1180. doi: 10.1016/s0969-2126(96)00125-6. [DOI] [PubMed] [Google Scholar]
- 28.Van Roey P, Waddling CA, Fox KM, Belfort M, Derbyshire V. Intertwined structure of the DNA-binding domain of intron endonuclease I-TevI with its substrate. EMBO J. 2001;20:3631–3637. doi: 10.1093/emboj/20.14.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161:1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Z, Zhao H. A highly sensitive selection method for directed evolution of homing endonucleases. Nucleic Acids Res. 2005;33:e154. doi: 10.1093/nar/gni148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kleinstiver BP, Fernandes A, Gloor GB, Edgell DR. A unified genetic, computational, and experimental framework identifies noon-conserved residues as critical for function of the homing endonuclease I-BmoI. Nucleic Acids Res. 2010;38:2411–2427. doi: 10.1093/nar/gkp1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edgell DR, Stanger MJ, Belfort M. Importance of a single base pair for discrimination between intron-containing and intronless alleles by endonuclease I-BmoI. Curr Biol. 2003;13:973–978. doi: 10.1016/s0960-9822(03)00340-3. [DOI] [PubMed] [Google Scholar]
- 33.Catto LE, Ganguly S, Milsom SE, Welsh AJ, Halford SE. Protein assembly and DNA looping by the FokI restriction endonuclease. Nucleic Acids Res. 2006;34:1711–1720. doi: 10.1093/nar/gkl076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dean AB, et al. Zinc finger as distance determinant in the flexible linker of intron endonuclease I-TevI. Proc Natl Acad Sci USA. 2002;99:8554–8561. doi: 10.1073/pnas.082253699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cermak T, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Q, et al. Role of the interdomain linker in distance determination for remote cleavage by homing endonuclease I-TevI. J Mol Biol. 2008;379:1094–1106. doi: 10.1016/j.jmb.2008.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kowalski JC, et al. Configuration of the catalytic GIY-YIG domain of intron endonuclease I-TevI: Coincidence of computational and molecular findings. Nucleic Acids Res. 1999;27:2115–2125. doi: 10.1093/nar/27.10.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Foley JE, et al. Rapid mutation of endogenous zebrafish genes using zinc finger nucleases made by Oligomerized Pool ENgineering (OPEN) PLoS ONE. 2009;4:e4348. doi: 10.1371/journal.pone.0004348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bryk M, et al. The td intron endonuclease I-TevI makes extensive sequence-tolerant contacts across the minor groove of its DNA target. EMBO J. 1993;12:2141–2149. doi: 10.1002/j.1460-2075.1993.tb05862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shimizu Y, et al. Adding fingers to an engineered zinc finger nuclease can reduce activity. Biochemistry. 2011;50:5033–5041. doi: 10.1021/bi200393g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takeuchi R, Certo M, Caprara MG, Scharenberg AM, Stoddard BL. Optimization of in vivo activity of a bifunctional homing endonuclease and maturase reverses evolutionary degradation. Nucleic Acids Res. 2009;37:877–890. doi: 10.1093/nar/gkn1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kleinstiver BP, Bérubé-Janzen W, Fernandes AD, Edgell DR. Divalent metal ion differentially regulates the sequential nicking reactions of the GIY-YIG homing endonuclease I-BmoI. PLoS ONE. 2011;6:e23804. doi: 10.1371/journal.pone.0023804. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.