Abstract
We previously reported that mouse strains with lower circulating insulin-like growth factor 1 (IGF1) level at 6 mo have significantly extended longevity. Here we report that strains with lower IGF1 have significantly delayed age of female sexual maturation, measured by vaginal patency (VP). Among strains with normal lifespans (mean lifespan >600 d), delayed age of VP associated with greater longevity (P = 0.015), suggesting a genetically regulated tradeoff at least partly mediated by IGF1. Supporting this hypothesis, C57BL/6J females had 9% lower IGF1, 6% delayed age of VP, and 24% extended lifespan compared with C57BL/6J.C3H/HeJ-Igf1, which carries a C3H/HeJ allele on chromosome (Chr) 10 that increases IGF1. To identify genetic loci/genes that regulate female sexual maturation, including loci that mediate lifespan tradeoffs, we performed haplotype association mapping for age of VP and identified significant loci on Chrs 4 (Vpq1) and 16 (Vpq2 and 3). At each locus, wild-derived strains share a unique haplotype that associates with delayed VP. Substitution of Chr 16 of C57BL/6J with Chr 16 from a wild-derived strain significantly reduced IGF1 and delayed VP. Strains with a wild-derived allele at Vpq3 have significantly extended longevity compared with strains with other alleles. Bioinformatic analysis identified Nrip1 at Vpq3 as a candidate gene. Nrip1−/− females have significantly reduced IGF1 and delayed age of VP compared with Nrip1+/+ females. We conclude that IGF1 may coregulate female sexual maturation and longevity; wild-derived strains carry specific alleles that delay sexual maturation; and Nrip1 is involved in regulating sexual maturation and may affect longevity by regulating IGF1 level.
Keywords: aging, reproduction, hormone
Epidemiology studies have suggested that sexual maturation is genetically regulated (1, 2). According to evolutionary theory, natural selection plays an important role in selecting alleles that regulate female sexual maturation (3–5). The evolutionary theory of aging predicts that the timing of female sexual maturation is linked to the rate of aging by pleiotropic genes that mediate a tradeoff between sexual maturation and aging (6, 7). This theory is supported by a field population study of mammalian species ranging from mouse to elephant that identified a positive correlation between age of reproduction and lifespan (8). In rodent, caloric restriction delays female maturity and slows aging (9). Also, reduced female reproduction and extended longevity were found in most models that carry mutations in genes of the growth hormone/insulin-like growth factor 1 (IGF1) pathway (10). These studies suggest that the set of genes that regulate sexual maturation includes a subset of pleiotropic genes that mediate a life-history tradeoff between development and aging.
Previous studies have suggested that considerable genetic variance of female reproductive development exists within Mus musculus. However, before our study, the age of sexual maturation of inbred strains had not been systematically measured. In this study, we measured sexual maturation by observing the age of vaginal patency (VP) for 31 inbred strains that represent the majority of genetic diversity across inbred strains. Our results suggest that, among mouse inbred strains, sexual maturation and longevity are coregulated and that IGF1 is an important mediator. Thus, genes that regulate IGF1 may constitute a category of antagonistic pleiotropic genes that mediate an evolutionary tradeoff between development and aging. Our studies indicate that this tradeoff still operates within murine populations and that nuclear receptor-interacting protein 1 (Nrip1) may be one of the genes that regulate such a tradeoff.
Results
Updated Lifespan Data.
When we published our previous report (11) on the lifespan of the 31 inbred strains, 19.1% of the mice were still alive. We now have lifespan data for all 1,945 mice. Survival data from mice that died abnormally are treated as censored data. The current report also includes data for 32 MOLF/EiJ (MOLF) females not reported in the previous paper because they entered the study late (Table S1). Correlations between mean lifespans from previous and current studies are strong (R2 > 0.95, P < 0.001) for both females and males. Raw lifespan data are accessible from the Mouse Phenome Database (MPD; http://www.jax.org/phenome).
Age of VP Varies Among Strains, with Later VP in Wild-Derived Strains.
Results show a dramatic variation in age of VP among inbred strains (24.9 ± 0.4 to 42.9 ± 0.6 d, mean ± SEM; Table S2). Four of the five strains with the latest age of VP are wild-derived strains (PWD, WSB, CAST, and MOLF). D2 was the only domesticated strain with an age of VP within the range of the wild-derived strains. We dropped NZO/H1LtJ (NZO) because no NZO breeders were available, and measured 31 strains.
Age of VP Correlates with Lifespan, Circulating IGF1 at 6 mo, and Body Weight at 38 D.
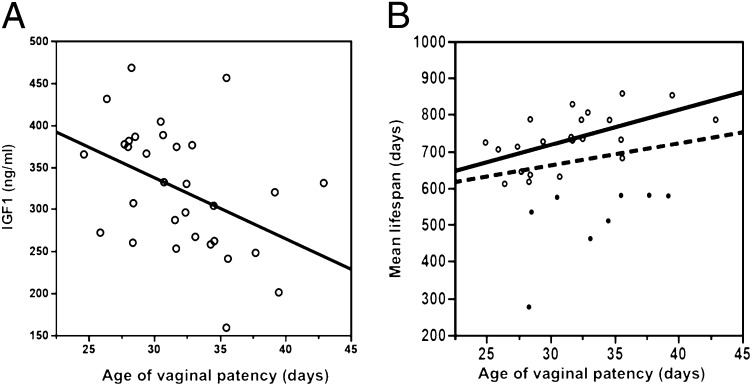
When all strains are considered, sexual maturation does not correlate with mean lifespan (R2 = 0.04, P = 0.27). Several strains have relatively short lifespans, presumably because they die from specific diseases with early onsets, not because of accelerated aging. Therefore, to study the relationship between sexual maturation and aging, we did a post hoc analysis that removed short-lived strains, using 500, 600, and 700 d as cutoffs. These cutoffs, equivalent to human ages of 52, 60, and 68 y (12, 13), removed 2, 8, and 15 strains. With these cutoffs, regressions of lifespan on age of VP are R2 = 0.03 (P = 0.38), R2 = 0.33 (P = 0.015), and R2 = 0.35 (P = 0.046), respectively (Fig. 1A). The significant relationships explaining 33–35% of the strain variance in lifespan when short-lived strains are excluded indicate that age of VP correlates with lifespan among strains with normal and long lifespans.
Fig. 1.
(A) Correlation between age of vaginal patency and circulating IGF1 (R2 = 0.19, P = 0.013). (B) Correlation between age of vaginal patency and mean lifespan. Dashed line, correlation among all strains (R2 = 0.04, P = 0.27); solid line, correlation without short-lived strains (mean lifespan <600 d); solid dots, short-lived strains (R2 = 0.33, P = 0.005).
The D2 strain, widely used in aging study, has the latest age of VP among domesticated strains but a relatively shorter longevity (female mean lifespan = 580 d), suggesting that this strain carries deleterious alleles that may cause death at a relatively young age. Thus, caution must be used when considering the D2 strain for aging research. The CAST strain is another short-lived strain (female mean lifespan = 578 d). We summarized the diseases found in inbred strains at middle (12 mo) and old (20 mo) age (14); however, the major causes of early death of D2 and CAST strains have not been determined.
Strains with lower IGF1 at 6 mo (data are accessible from MPD) have significantly extended lifespans (11). Here we found that strains with lower IGF1 level have a delayed age of VP (R2 = 0.19, P = 0.013; Fig. 1B). Using mean lifespans of 500, 600, and 700 d as cutoffs, regressions of IGF1 level on age of VP are R2 = 0.25 (P = 0.015), R2 = 0.18 (P = 0.021), and R2 = 0.15 (P = 0.12), respectively.
For 27 of the 31 strains in our survey, body weights at 38 d could be obtained from the MPD. Data for NOD, MRL, P, and PWD were unavailable. Strains with lighter body weight have a delayed age of VP (R2 = 0.28, P = 0.004; Fig. S1). Residual analysis revealed that BTBR is an outlier for this correlation. Using mean lifespans of 500, 600, and 700 d as cutoffs, regressions of age of VP on body weight are R2 = 0.25 (P = 0.008), R2 = 0.45 (P = 0.0008), and R2 = 0.48 (P = 0.004), respectively.
B6 Allele Decreases IGF1, Delays Sexual Maturation, and Extends Longevity in Females.
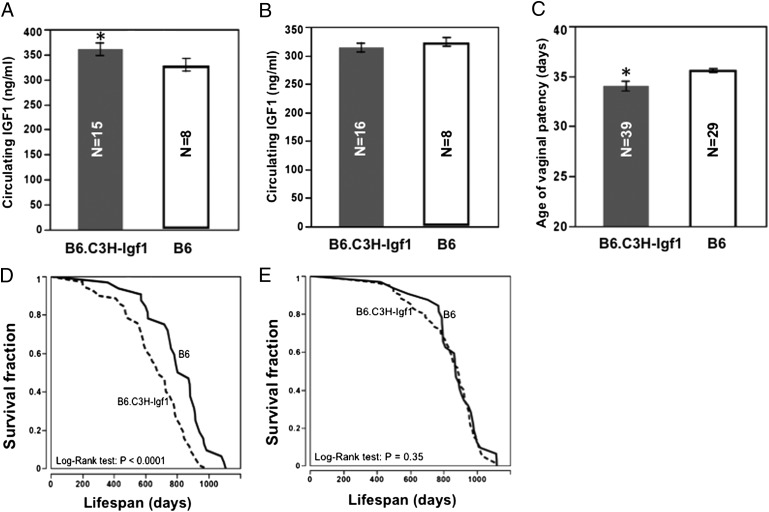
We set up B6 mice concurrently with a congenic strain, B6.C3H-Igf1, which carries a C3H allele [chromosome (Chr) 10: 65–90 Mb] on the B6 background. Previous studies identified an IGF1 quantitative trait locus (QTL) in this region, in which the B6 allele reduces IGF1 compared with the C3H allele. A lifespan QTL was also identified in this region, and the B6 allele associated with extended longevity (15). Here we confirmed that, compared with B6.C3H-Igf1 mice, B6 mice have significantly (P < 0.05) decreased IGF1 at 6 mo of age, but only in females (329.1 ± 12.6 vs. 360.9 ± 12.8 ng/mL), not in males (323.8 ± 7.0 vs. 313.8 ± 8.2 ng/mL) (Fig. 2 A and B). The age of VP of B6 is significantly delayed compared with B6.C3H-Igf1 females (36 ± 0.2 vs. 34 ± 0.5 d; P < 0.05) (Fig. 2C), and the lifespan of B6 females is greater by 24% (810 ± 32 vs. 656 ± 24 d; log-rank test, P < 0.0001). For males, however, lifespans do not differ significantly (866 ± 28 and 831 ± 24 d; log-rank test, P = 0.35) (Fig. 2 D and E and Table S3). Data are given as mean ± SEM.
Fig. 2.
Effects of the C3H allele on circulating IGF1, age of VP, and lifespan. Compared with B6, B6.C3H-Igf1 females have significantly increased circulating IGF1 (A), earlier age of VP (C), and shorter lifespan (D); there are no significant differences in IGF1 (B) and lifespan in males (E). *P < 0.05, compared with B6.
Genetic Loci Associate with Age of VP and Body Weight.
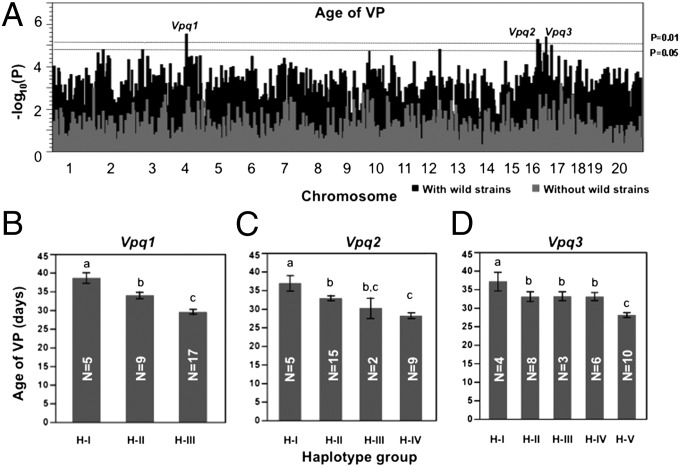
In the haplotype association mapping (HAM) study, we determined haplotypes of each single-nucleotide polymorphism (SNP) (16). When the wild-derived strains were included, we found three significant QTLs for age of VP (P < 0.01): Vpq1 (Chr 4: 62.69–62.76 Mb), Vpq2 (Chr 16: 37.68–37.7 Mb), and Vpq3 (Chr 16: 74.84 Mb) (Fig. 3A and Tables S4–S6). ANOVA analysis showed that haplotypes of Vpq1, 2, and 3 could explain 75.6% (P = 0.0003) of the variation in age of VP; even after accounting for the association with body weight, these haplotypes still explain 44.2% (P < 0.05) of the variation. No significant QTL was identified when the wild-derived strains were excluded; however, all three loci did have peaks with scores (−log10P) >2 (Fig. 3A). This suggests that these loci also may affect age of VP among the domesticated strains, but that more strains must be tested to observe significance.
Fig. 3.
Haplotype association mapping for age of vaginal patency. (A) Whole-genome scan. Dashed lines represent P = 0.01 and P = 0.05 thresholds in the permutation test (n = 1,000). Black bars represent scans including wild-derived strains; gray bars represent scans without wild-derived strains. (B–D) Haplotype groups of each QTL are defined in Tables S4–S6. Strains in haplotype group 1 (H-I) of Vpq1, 2, and 3 have significantly delayed age of VP compared with other haplotype groups. Bars are SEs. Groups that do not share a lowercase letter (at the top of the bar) differ at P ≤ 0.05. Numbers of strains for each haplotype are shown in the columns.
For body weight at 38 d, HAM analysis including the wild-derived strains detected four significant QTLs (P < 0.01): Bwq15 (Chr 4: 154.11–154.17 Mb), Bwq16 (Chr 5: 53.38–53.54 Mb), Bwq17 (Chr 10: 49.34–49.47 Mb), and Bwq18 (Chr 16: 82.27–82.58 Mb) (Fig. S2). No significant QTLs were identified when the wild-derived strains were excluded, and the QTLs for age of VP do not overlap with the QTL for body weight.
Alleles from Wild-Derived Strains Associate with Later Age of VP.
According to the haplotypes of Vpq1, 2, and 3, the 31 inbred strains could be subdivided into three, four, and five haplotype groups (Tables S4–S6). At each of the three loci, the four wild-derived strains share the same haplotype, which is different from those at the same location in domesticated inbred strains, with one exception: SM, which has the same haplotypes as the wild-derived strains at Vpq1 and Vpq2. Analysis of allele effects showed that strains with the wild-derived alleles at all three QTLs had a significantly later age of VP (P < 0.05) compared with strains with any of the other alleles at those loci. Among the haplotype groups of domesticated inbred strains, significant differences also exist (Fig. 3 B–D).
Wild-Derived Alleles on Chr 16 Reduce Circulating IGF1 and Delay Sexual Maturation.
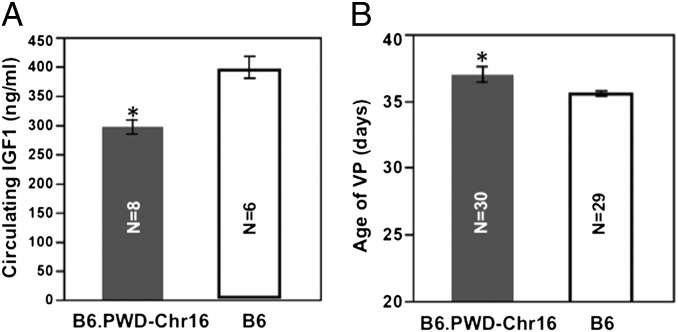
Compared with B6, B6.PWD-Chr 16, which carries Chr 16 of the wild-derived strain PWD on a B6 background, has a significantly (P < 0.05) reduced IGF1 level at 2 mo of age (296.8 ± 34.7 vs. 399.0 ± 45.4 ng/mL, n = 8 and 6) and later age of VP (40.4 ± 1.0 vs. 35.6 ± 0.2 d) (Fig. 4), although not as late as PWD (42.9 ± 0.7; P < 0.05). Data are given as mean ± SEM.
Fig. 4.
Compared with B6, B6.PWD-Chr16 females have lower IGF1 and delayed age of VP. *P < 0.05.
Bioinformatics Study Identifies Candidate Genes for Age of VP.
For Vpq1 and Vpq2, candidate genes (Table S7) have SNPs that are shared by the wild-derived strains and SM but that are different from other domesticated strains. For Vpq3, all candidate genes are those with SNPs shared only by the wild-derived strains and that are different from the SNPs of all domesticated strains.
Nrip1−/− Females Have Reduced IGF1 and Delayed Age of VP.
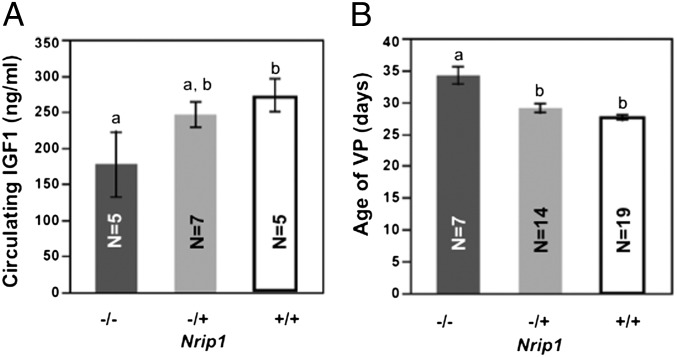
Nrip1 is a candidate gene of Vpq3. We found that Nrip1−/− females have a lower IGF1 level at 2 mo compared with Nrip1−/+ and Nrip1+/+ females (177.3 ± 44.9 vs. 245 ± 17.2 and 273.3 ± 22.6 ng/mL), although only the comparison with wild-type females reached significance (P = 0.03). Also, the age of VP of Nrip1−/− females (34.1 ± 1.3 d) is significantly (P < 0.0001) delayed compared with heterozygous females (29.0 ± 0.7 d) and wild-type-females (27.6 ± 0.4 d) (Fig. 5). Data are given as mean ± SEM.
Fig. 5.
Nrip1 knockout mice have reduced circulating IGF1 and delayed age of VP. Columns with different letters are significantly different (P < 0.05).
Haplotypes of Vpq3 Associate with Longevity Among Inbred Strains.
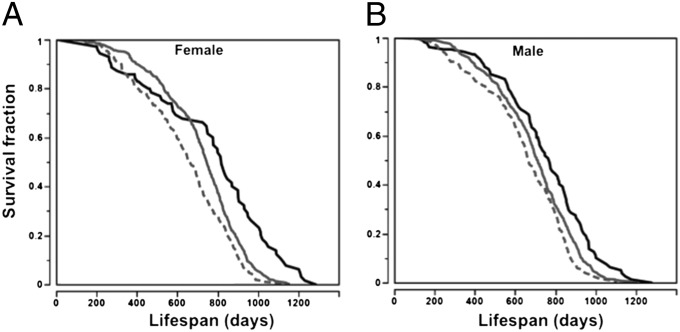
Based on age of VP (Fig. 3D) and the haplotypes of Vpq3 (Tables S4–S6), we divided the 31 inbred strains into three groups: strains with the haplotype I allele, which has the latest age of VP; strains with the haplotype V allele, which has the earliest age of VP; and strains with haplotype II, III, and IV alleles, which have ages of VP that are similar to each other but significantly different from strains with haplotype I and V alleles. Mice with the haplotype I allele have significantly extended lifespans compared with mice that carry alleles in the other haplotype groups, whereas mice with the haplotype V allele of Vpq3 have the shortest lifespans in both females and males (log-rank test, P < 0.0001; Fig. 6).
Fig. 6.
Vpq3 associates with longevity across inbred strains (Log-Rank test: P < 0.0001 for both female and male). The black curve is the H-I group, the gray solid curve is the H-II, III, and IV groups, and the gray dashed curve is the H-V group. Haplotype groups are shown in Tables S4–S6.
Discussion
Female Sexual Maturation and Body Weight Are Correlated.
Human population studies suggest that higher body mass index associates with earlier age of menarche (17). Similarly, in mice, faster maturational gain in body weight associates with earlier age of VP (18). Attaining a body weight threshold may trigger the onset of puberty (19), perhaps by altering metabolic signals to the brain (20). Bronson (21) and Miller et al. (18) separately found that female mice of domesticated stocks grew faster and reached sexual maturation earlier than females of wild-derived stocks. Our comprehensive survey of inbred strains, including both domesticated and wild-derived strains that represent M. musculus populations throughout the world, demonstrated that this inverse correlation exists among laboratory mice. This strongly suggests that either a direct causal relationship exists between body weight and female sexual maturation or that these phenotypes are genetically coregulated. We also confirmed and expanded the observation that wild-derived mice have delayed age of VP in association with lighter body weights.
IGF1 Is Involved in the Regulation of Female Sexual Maturation.
Mammalian female sexual maturation is regulated by the hypothalamus–pituitary–gonad reproductive axis (22) and its interactions with hormones that regulate growth and energy metabolism, such as leptin (23), insulin (24), and IGF1. Body fat is an important component of energy balance, and the associated leptin may be required for reproductive development (25). However, Bronson (26) found that, in a normal mouse population, neither body fat nor circulating leptin levels correlated with onset of puberty. Like leptin, insulin circulates in the periphery at levels proportionate to body fat (27, 28), and circulating levels are diminished with diet restriction, which impairs fertility. However, whether the variation of the concentration of circulating insulin affects age of female sexual maturation remains to be clarified (24, 29).
In the current report, the inverse correlation between circulating IGF1 and age of VP across inbred strains suggests that IGF1 may play an important role in regulating the timing of female sexual maturation. Supporting this hypothesis, we found that altering circulating IGF1 through various genetic means results in coordinated changes in age of VP: Age of VP is significantly accelerated in B6.C3H-Igf1 females, which have greater circulating IGF1 than B6 females, but is significantly delayed in B6.PWD-Chr16 and Nrip1−/− females, which have lower circulating IGF1 than B6 or littermate controls, respectively. The underlying mechanisms may exist at all levels of the hypothalamus–pituitary–gonad reproductive axis. For example, knocking out Igf1r in gonadotropin-releasing hormone neurons significantly delays puberty (22). In the pituitary, IGF1 stimulates the secretion of luteinizing hormone (30). In the ovary, IGF1 has also been found to be a key regulator of estrogen synthesis through regulation of aromatase expression and activity (31, 32).
IGF1 May Coregulate Female Sexual Maturation and Longevity.
An association between sexual maturation and lifespan in mouse has been reported for several mouse stocks (18, 33). The current report measures developmental traits and longevity across many inbred strains. We note here that, when lifespan is used as a phenotype that reflects underlying aging mechanisms, the lifespan of short-lived strains is unsuitable. The lifespan of a population contains information about the “aging rate” for that population because, with age, senescence mechanisms increase the likelihood of death from a variety of insults and diseases (13). Short-lived strains die of unusual vulnerability to specific causes of death, vulnerabilities not related to general mechanisms of aging. Thus, the age-specific risk of mortality contains more information about the rate of aging in strains that die at relatively later ages. For example, gerontologists now consider the AKR strain to be a very poor model for aging research because AKR mice die at middle age from early-onset thymic lymphoma (34) before senescence can influence physiological function and the risk of mortality. As a result, when using lifespan as a biomarker of aging, gerontologists often restrict the range of lifespans to “maximum” lifespan (11, 13, 35).
Unfortunately, no standard approach has been established to determine the age cutoff that would minimize uninformative survival data for aging studies. Therefore, we used a range of cutoffs, up to the mean lifespan for the strain panel, to determine whether a relationship of female reproductive maturation to lifespan could be observed for laboratory mice. The correlation is significant among strains with mean lifespans longer than 600 d, suggesting that age of female sexual maturation and longevity might be genetically coregulated.
Relationships of age of VP both with body weight (at 38 d) and with circulating IGF1 in adults (at 6 mo) are at least as strong for this edited dataset as they are for the entire dataset. Excluding the short-lived strains also increases the significance of the correlation between IGF1 and longevity (11). Thus, for inbred strains of mice with moderate and long lifespans, that is, strains with lifespans most informative with regard to senescence, relationships of other traits that were identified to be associated with longevity (11, 36) are still present. These results suggest that the genetic regulation of somatotropic signaling may account for some of the covariation of these lifespan-related phenotypes within, and possibly across, mammalian species. The significantly reduced IGF1, delayed age of VP, and extended longevity in B6 females compared with B6.C3H-Igf1 females support this hypothesis. Importantly, it also verifies the correlation between age of VP and lifespan identified by the post hoc analysis.
Although IGF1 is important in regulating development and aging in both sexes, previous studies suggested that different biological and molecular mechanisms might be involved. For example, Hamilton and Bronson found that calorie restriction, which reduces IGF1 level, delays sexual maturation only in females (37). Divall et al. showed that although both female and male mice with IGF1R knockout in the gonadotropin-releasing hormone neurons had delayed sexual maturation, the administration of IGF1 in wild-type mice accelerated puberty only in females (22). We previously reported that Igf1q8 (Chr 10: 88 Mb) associates with IGF1 level only in female mice (15). The current study confirmed that the B6 allele at this locus decreases IGF1 and extends longevity only in females. Thus, our results reinforce the hypothesis that persistent differences in somatotropic factors, such as circulating IGF1, provide a physiological mechanism that links the timing of female reproductive maturation and aging.
We note that the rationale for the hypothesis that reproductive development is genetically linked to aging is based on studies of breeding animals, whereas our tests of the hypothesis use mice that are prevented from breeding. A limited number of experiments revealed that for male mice, breeders have extended longevity compared with virgins, but that for female mice, breeders have significantly shorter lifespans than virgins (38), suggesting the possibility that different phenotypic relationships might exist for parous mice. Interestingly, our study of inbred strains and comparison of B6.C3H-Igf1 with B6 identify an association of reproductive development with lifespan consistent with evolutionary theory, indicating that parity is not required for genes that control reproductive development to also influence lifespan in a predictable way.
Wild-Derived Strains Carry Alleles That Delay Sexual Maturation.
Epidemiological studies indicate that up to 50–80% of the variance in the age of sexual maturation is genetically determined (39–41). Previous studies identified that A/J (A) alleles on Chrs 6 and 13, as well as C3H allele on Chr X (15.5 cM), associate with earlier age of VP (42, 43). Consistent with these studies, our data showed that B6 had a significantly delayed age of VP compared with A and C3H. However, the loci identified in our study do not overlap with these previously identified QTLs, probably because each of these earlier studies used only two parental genotypes, and all of the strains were domesticated. The current study identified QTLs that not only associate with the differences among domesticated inbred strains but, more importantly, also associate with the differences between wild-derived and domesticated strains.
At Vpq1, 2, and 3, wild-derived strains carry unique alleles that are different from the alleles in domesticated strains, except SM, and associated with significantly delayed age of VP. We further verified the effects of Vpq2 and 3 (both on Chr 16) by showing that B6.PWD-Chr 16 females have significantly lower IGF1 and delayed age of VP than B6 females, but not as late as PWD females. These results support the hypothesis that genes at loci on Chr 16 coregulate IGF1 and age of VP, but also demonstrate that these genes do not completely account for the differences between wild-derived and domesticated mice. Unfortunately, we could not perform a similar test for Vpq1, on Chr 4, because B6.PWD-Chr4 mice bred poorly.
The SM strain was selectively bred for small body weight during inbreeding (44), matures very slowly, and shares the same haplotypes as the wild-derived strains at Vpq1 and Vpq2. Although neither Vpq1 nor Vpq2 was identified as a significant body weight QTL in our HAM analysis, this surprising appearance of wild-derived alleles in a domesticated strain selected for small body weight indicates that genes at these loci are critical contributors to the coregulation of body weight and reproductive developmental rate. Interestingly, our study also suggests the existence of mechanisms that regulate female sexual maturation independently of body weight. First, after statistically removing the effects of body weight at 38 d, Vpq1, 2, and 3 could still explain 44.2% of the variance. Importantly, a whole-genome scan showed that the strongest QTLs for VP and for body weight do not overlap. Second, BTBR, an outlier of the correlation between body weight and age of VP, has the heaviest body weight but much delayed age of VP compared with most of the domesticated strains.
Nrip1 Is Involved in Regulating IGF1 and Female Sexual Maturation.
Nrip1, a regulator of nuclear receptors that have broad functions in regulating cell growth, cell death, and metabolism, is expressed at a high level in metabolic tissue such as liver, fat, and muscle. Microarray analysis found that the mRNA level of Nrip1 varies dramatically among inbred strains, demonstrating the existence of a genetic factor in its regulation (45). The expression of Nrip1 in ovary is essential for successful reproduction, and knocking out Nrip1 results in female infertility due to ovulation failure (46). Nrip1 null (−/−) mice are smaller and leaner, have resistance to high-fat diet–induced obesity, and show increased glucose tolerance and insulin sensitivity (46, 47). Most of these characteristics of Nrip1−/− mice are also found in mice that are calorie-restricted. Maintaining mitochondrial biogenesis during diet restriction through the activation of peroxisome proliferator-activated receptor gamma coactivator 1α (PGC-1α) has been suggested as one of the mechanisms related to the lifespan extension (48). Nrip1, as a transcriptional repressor of PGC-1α and β, could promote senescence by suppressing mitochondrial biogenesis (49).
Comparing Nrip−/− mice with their littermates, our results verified the hypothesis, generated by genetic and bioinformatic analyses, that knocking out Nrip1 could significantly decrease IGF1 level and delay age of VP. Although the lifespan of Nrip1−/− mice is still under observation, haplotype analysis suggests that at the Vpq3 locus, the haplotypes of alleles also associate with longevity. Strains with wild-derived alleles have significantly delayed age of VP and extended longevity, whereas strains with the haplotype V allele have earlier age of VP and shorter longevity. Our results suggest that Nrip1 is a strong candidate for inclusion in the category of pleiotropic genes—first proposed by Williams more than 50 y ago (7)—that influence resource allocation to mediate evolutionary tradeoffs between development and aging.
Materials and Methods
Mice.
Mice of 32 inbred strains, B6.C3H-Igf1, and B6.PWD-Chr 16 were obtained from The Jackson Laboratory. Mouse husbandry details have been fully described previously (11). Pups were weaned at 21 d. To avoid influence of male pheromonal and tactile cues on female sexual maturation, females and males were separated and housed in pressurized individually ventilated polycarbonate cages (four or five mice per pen) with high-efficiency particulate air–filtered air (Thoren Caging Systems). Mice were inspected at least once daily. If a mouse was moribund—severely ill and judged likely not to survive another 48 h—it was euthanized (11). Animal use adhered to National Institutes of Health Laboratory Animal Care Guidelines and was approved by the Animal Care and Use Committee (ACUC) of The Jackson Laboratory.
Age of Vaginal Patency.
Beginning at 18 d of age, mice were examined daily until vaginal patency was observed. Determination of age of VP of Nrip1−/−, Nrip1−/+, and Nrip1+/+ mice was conducted by M.G.P.’s group.
IGF1.
Mice were fasted for 4 h (8:00 AM to 12:00 PM) before blood was collected. IGF1 levels were measured by an RIA as previously described (11).
Statistical Analysis.
Survival curves were drawn using the Kaplan–Meier method. Differences were analyzed between curves by the log-rank method. One-way ANOVA was used to compare age of VP among strains; to correct for multiple testing, significance was adjusted by the Tukey HSD (Honestly Significant Difference) method. The Student’s t test was used to compare age of VP and IGF1 between groups. All statistical analyses were performed using JMP 7.0 (SAS Institute).
Haplotype Association Mapping.
The genotype data of the inbred strains were downloaded from http://cgd.jax.org/datasets/popgen/70KSNP.shtml, and the procedures have been fully described previously (11). Briefly, mean values of age of VP for each strain were input as vectors, and genotype data across multiple inbred mouse strains were input as a matrix. A hidden Markov model was applied to impute missing genotypes and identify haplotypes (16). Regression-based test statistics were computed to measure the strength of association between genotype and age of VP. The type I error rate for multiple testing, which results from a genome-wide search, was controlled for using family-wise error rate control. Strain labels in the phenotype data were shuffled, and the genotype data were kept intact. The minimum P value was recorded on each permutation. A total of 1,000 permutations was performed, and the distribution of these P values provided approximate multiple test-adjusted thresholds.
Bioinformatics Analysis.
Currently, there is no widely accepted method for determining the confidence interval of QTLs identified by HAM analysis. Our strategy was to extend the QTL by 2 Mb on both sides. At Vpq1 and 2, the wild-derived strains share the same haplotypes at the loci that associate with the age of VP; these haplotypes are different from those of domesticated inbred strains—except for SM. Because of this, our assumption was that, at Vpq1 and 2, wild-derived strains and SM share the same genetic polymorphisms, which are different from those of other inbred strains. At Vpq3, all of the wild-derived strains share the same SNP, which differs from the SNP shared by all domesticated inbred strains, including SM, leading to the assumption that the wild-derived strains share a genetic polymorphism at Vpq3 that is different from that of all domesticated inbred strains, including SM. Under these assumptions, a bioinformatics study compared high-density SNPs from the MPD among haplotype groups of each allele.
Supplementary Material
Acknowledgments
We thank J. Currer for editing the paper and D. Schultz, M. Stanwood, E. Malay, and D. Godfrey for outstanding technical help. We appreciate Drs. Rich Miller and Elissa Chesler for reviewing the manuscript and providing valuable suggestions. This work was funded by Grants AG025707 and AG038070 (to The Jackson Aging Center), AG038560 (to D.E.H.), and AG034349 (to R.Y.) from the National Institute of Aging, National Institutes of Health, as well as by grants from The Ellison Medical Foundation (to B.P., D.E.H., R. Woychik, and S.-W.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. A.B. is a guest editor invited by the Editorial Board.
Data deposition: The lifespan and age of sexual maturation data reported in this paper have been deposited in the Mouse Phenome Database, www.jax.org/phenome.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1121113109/-/DCSupplemental.
References
- 1.Meyer JM, Eaves LJ, Heath AC, Martin NG. Estimating genetic influences on the age-at-menarche: A survival analysis approach. Am J Med Genet. 1991;39(2):148–154. doi: 10.1002/ajmg.1320390207. [DOI] [PubMed] [Google Scholar]
- 2.Sharma K. Genetic basis of human female pelvic morphology: A twin study. Am J Phys Anthropol. 2002;117:327–333. doi: 10.1002/ajpa.10055. [DOI] [PubMed] [Google Scholar]
- 3.Reznick D, Bryant M, Holmes D. The evolution of senescence and post-reproductive lifespan in guppies (Poecilia reticulata) PLoS Biol. 2006;4:e7. doi: 10.1371/journal.pbio.0040007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stearns SC, Ackermann M, Doebeli M, Kaiser M. Experimental evolution of aging, growth, and reproduction in fruitflies. Proc Natl Acad Sci USA. 2000;97:3309–3313. doi: 10.1073/pnas.060289597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Austad S. Retarded senescence in an insular population of Virginia opossums (Didelphis virginiana) J Zool. 1993;229:695–708. [Google Scholar]
- 6.Hamilton WD. The moulding of senescence by natural selection. J Theor Biol. 1966;12(1):12–45. doi: 10.1016/0022-5193(66)90184-6. [DOI] [PubMed] [Google Scholar]
- 7.Williams GC. Pleiotropy, natural selection, and the evolution of senescence. Evolution. 1957;11:398–411. [Google Scholar]
- 8.Harvey PH, Zammuto RM. Patterns of mortality and age at first reproduction in natural populations of mammals. Nature. 1985;315:319–320. doi: 10.1038/315319a0. [DOI] [PubMed] [Google Scholar]
- 9.Merry BJ, Holehan AM. Onset of puberty and duration of fertility in rats fed a restricted diet. J Reprod Fertil. 1979;57:253–259. doi: 10.1530/jrf.0.0570253. [DOI] [PubMed] [Google Scholar]
- 10.Brown-Borg HM. Hormonal control of aging in rodents: The somatotropic axis. Mol Cell Endocrinol. 2009;299(1):64–71. doi: 10.1016/j.mce.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan R, et al. Aging in inbred strains of mice: Study design and interim report on median lifespans and circulating IGF1 levels. Aging Cell. 2009;8:277–287. doi: 10.1111/j.1474-9726.2009.00478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finch CE. Longevity, Senescence, and the Genome. Chicago: Univ of Chicago Press; 1990. [Google Scholar]
- 13.Flurkey K, Currer JM, Harrison DE. The mouse in aging research. In: Fox JG, editor. The Mouse in Biomedical Research (Academic Press, New York) 2nd Ed. 2007. pp. 637–672. [Google Scholar]
- 14.Sundberg JP, et al. The mouse as a model for understanding chronic diseases of aging: The histopathologic basis of aging in inbred mice. Pathobiol Aging Age Relat Dis. 2011;1:7179. doi: 10.3402/pba.v1i0.7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leduc MS, et al. Identification of genetic determinants of IGF-1 levels and longevity among mouse inbred strains. Aging Cell. 2010;9:823–836. doi: 10.1111/j.1474-9726.2010.00612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szatkiewicz JP, et al. An imputed genotype resource for the laboratory mouse. Mamm Genome. 2008;19(3):199–208. doi: 10.1007/s00335-008-9098-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perry JR, et al. Meta-analysis of genome-wide association data identifies two loci influencing age at menarche. Nat Genet. 2009;41:648–650. doi: 10.1038/ng.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller RA, Harper JM, Dysko RC, Durkee SJ, Austad SN. Longer life spans and delayed maturation in wild-derived mice. Exp Biol Med (Maywood) 2002;227:500–508. doi: 10.1177/153537020222700715. [DOI] [PubMed] [Google Scholar]
- 19.Frisch RE, Revelle R. Height and weight at menarche and a hypothesis of critical body weights and adolescent events. Science. 1970;169:397–399. doi: 10.1126/science.169.3943.397. [DOI] [PubMed] [Google Scholar]
- 20.Cameron JL, Koerker DJ, Steiner RA. Metabolic changes during maturation of male monkeys: Possible signals for onset of puberty. Am J Physiol. 1985;249:E385–E391. doi: 10.1152/ajpendo.1985.249.4.E385. [DOI] [PubMed] [Google Scholar]
- 21.Bronson FH. Energy allocation and reproductive development in wild and domestic house mice. Biol Reprod. 1984;31(1):83–88. doi: 10.1095/biolreprod31.1.83. [DOI] [PubMed] [Google Scholar]
- 22.Divall SA, et al. Divergent roles of growth factors in the GnRH regulation of puberty in mice. J Clin Invest. 2010;120:2900–2909. doi: 10.1172/JCI41069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cunningham MJ, Clifton DK, Steiner RA. Leptin’s actions on the reproductive axis: Perspectives and mechanisms. Biol Reprod. 1999;60:216–222. doi: 10.1095/biolreprod60.2.216. [DOI] [PubMed] [Google Scholar]
- 24.Smith CP, et al. Relationship between insulin, insulin-like growth factor I, and dehydroepiandrosterone sulfate concentrations during childhood, puberty, and adult life. J Clin Endocrinol Metab. 1989;68:932–937. doi: 10.1210/jcem-68-5-932. [DOI] [PubMed] [Google Scholar]
- 25.Chehab FF, Lim ME, Lu R. Correction of the sterility defect in homozygous obese female mice by treatment with the human recombinant leptin. Nat Genet. 1996;12:318–320. doi: 10.1038/ng0396-318. [DOI] [PubMed] [Google Scholar]
- 26.Bronson FH. Puberty in female mice is not associated with increases in either body fat or leptin. Endocrinology. 2001;142:4758–4761. doi: 10.1210/endo.142.11.8495. [DOI] [PubMed] [Google Scholar]
- 27.Baura GD, et al. Saturable transport of insulin from plasma into the central nervous system of dogs in vivo. A mechanism for regulated insulin delivery to the brain. J Clin Invest. 1993;92:1824–1830. doi: 10.1172/JCI116773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwartz MW, Peskind E, Raskind M, Boyko EJ, Porte D., Jr Cerebrospinal fluid leptin levels: Relationship to plasma levels and to adiposity in humans. Nat Med. 1996;2:589–593. doi: 10.1038/nm0596-589. [DOI] [PubMed] [Google Scholar]
- 29.Caprio S, et al. Increased insulin secretion in puberty: A compensatory response to reductions in insulin sensitivity. J Pediatr. 1989;114:963–967. doi: 10.1016/s0022-3476(89)80438-x. [DOI] [PubMed] [Google Scholar]
- 30.Waterston RH, et al. Mouse Genome Sequencing Consortium Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 31.Erickson GF, Garzo VG, Magoffin DA. Insulin-like growth factor-I regulates aromatase activity in human granulosa and granulosa luteal cells. J Clin Endocrinol Metab. 1989;69:716–724. doi: 10.1210/jcem-69-4-716. [DOI] [PubMed] [Google Scholar]
- 32.Hiney JK, Srivastava V, Dearth RK, Dees WL. Influence of estradiol on insulin-like growth factor-1-induced luteinizing hormone secretion. Brain Res. 2004;1013(1):91–97. doi: 10.1016/j.brainres.2004.03.054. [DOI] [PubMed] [Google Scholar]
- 33.Harper JM, Durkee SJ, Dysko RC, Austad SN, Miller RA. Genetic modulation of hormone levels and life span in hybrids between laboratory and wild-derived mice. J Gerontol A Biol Sci Med Sci. 2006;61:1019–1029. doi: 10.1093/gerona/61.10.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karpova GV, et al. Hemopoietic and lymphoid organs in AKR/JY mice with thymic lymphoma. Bull Exp Biol Med. 2002;134(1):69–72. doi: 10.1023/a:1020669007938. [DOI] [PubMed] [Google Scholar]
- 35.Flurkey K, Papaconstantinou J, Miller RA, Harrison DE. Lifespan extension and delayed immune and collagen aging in mutant mice with defects in growth hormone production. Proc Natl Acad Sci USA. 2001;98:6736–6741. doi: 10.1073/pnas.111158898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller RA, Harper JM, Galecki A, Burke DT. Big mice die young: Early life body weight predicts longevity in genetically heterogeneous mice. Aging Cell. 2002;1(1):22–29. doi: 10.1046/j.1474-9728.2002.00006.x. [DOI] [PubMed] [Google Scholar]
- 37.Hamilton GD, Bronson FH. Food restriction and reproductive development: Male and female mice and male rats. Am J Physiol. 1986;250:R370–R376. doi: 10.1152/ajpregu.1986.250.3.R370. [DOI] [PubMed] [Google Scholar]
- 38.Klebanov S, et al. Heritability of life span in mice and its implication for direct and indirect selection for longevity. Genetica. 2000;110:209–218. doi: 10.1023/a:1012790600571. [DOI] [PubMed] [Google Scholar]
- 39.Palmert MR, Hirschhorn JN. Genetic approaches to stature, pubertal timing, and other complex traits. Mol Genet Metab. 2003;80(1-2):1–10. doi: 10.1016/s1096-7192(03)00107-0. [DOI] [PubMed] [Google Scholar]
- 40.Parent AS, et al. The timing of normal puberty and the age limits of sexual precocity: Variations around the world, secular trends, and changes after migration. Endocr Rev. 2003;24:668–693. doi: 10.1210/er.2002-0019. [DOI] [PubMed] [Google Scholar]
- 41.Plant TM, Barker-Gibb ML. Neurobiological mechanisms of puberty in higher primates. Hum Reprod Update. 2004;10(1):67–77. doi: 10.1093/humupd/dmh001. [DOI] [PubMed] [Google Scholar]
- 42.Krewson TD, et al. Chromosomes 6 and 13 harbor genes that regulate pubertal timing in mouse chromosome substitution strains. Endocrinology. 2004;145:4447–4451. doi: 10.1210/en.2004-0543. [DOI] [PubMed] [Google Scholar]
- 43.Zhu W, et al. A dominant X-linked QTL regulating pubertal timing in mice found by whole genome scanning and modified interval-specific congenic strain analysis. PLoS One. 2008;3:e3021. doi: 10.1371/journal.pone.0003021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.MacArthur JW. Genetics of body size and related characters. Am Nat. 1944;78(775):142–157. [Google Scholar]
- 45.Shockley KR, Witmer D, Burgess-Herbert SL, Paigen B, Churchill GA. Effects of atherogenic diet on hepatic gene expression across mouse strains. Physiol Genomics. 2009;39(3):172–182. doi: 10.1152/physiolgenomics.90350.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.White R, et al. The nuclear receptor co-repressor nrip1 (RIP140) is essential for female fertility. Nat Med. 2000;6:1368–1374. doi: 10.1038/82183. [DOI] [PubMed] [Google Scholar]
- 47.Christian M, White R, Parker MG. Metabolic regulation by the nuclear receptor corepressor RIP140. Trends Endocrinol Metab. 2006;17:243–250. doi: 10.1016/j.tem.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 48.López-Lluch G, et al. Calorie restriction induces mitochondrial biogenesis and bioenergetic efficiency. Proc Natl Acad Sci USA. 2006;103:1768–1773. doi: 10.1073/pnas.0510452103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Powelka AM, et al. Suppression of oxidative metabolism and mitochondrial biogenesis by the transcriptional corepressor RIP140 in mouse adipocytes. J Clin Invest. 2006;116(1):125–136. doi: 10.1172/JCI26040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.