Abstract
DNA primases provide oligoribonucleotides for DNA polymerase to initiate lagging strand synthesis. A deficiency in the primase of bacteriophage T7 to synthesize primers can be overcome by genetic alterations that decrease the expression of T7 gene 5.5, suggesting an alternative mechanism to prime DNA synthesis. The product of gene 5.5 (gp5.5) forms a stable complex with the Escherichia coli histone-like protein H-NS and transfer RNAs (tRNAs). The 3′-terminal sequence (5′-ACCA-3′) of tRNAs is identical to that of a functional primer synthesized by T7 primase. Mutations in T7 that suppress the inability of primase reduce the amount of gp5.5 and thus increase the pool of tRNA to serve as primers. Alterations in T7 gene 3 facilitate tRNA priming by reducing its endonuclease activity that cleaves at the tRNA–DNA junction. The tRNA bound to gp5.5 recruits H-NS. H-NS alone inhibits reactions involved in DNA replication, but the binding to gp5.5–tRNA complex abolishes this inhibition.
A 3′-hydroxyl end of an RNA or DNA strand positioned at the catalytic site of DNA polymerase functions as a primer to initiate synthesis of DNA. In most instances, primers are short oligoribonucleotides synthesized by enzymes designated DNA primases (1). DNA primases catalyze the template-directed synthesis of short oligoribonucleotides and stabilize the newly synthesized primer on the template and hand it off to DNA polymerase for initiation of DNA synthesis (2, 3).
Bacteriophage T7 has provided a model system for the study of DNA replication. Only three T7 proteins—gene 5 DNA polymerase (gp5), gene 4 primase-helicase (gp4), and gene 2.5 ssDNA-binding protein (gp2.5)—together with host Escherichia coli thioredoxin, are sufficient for leading and lagging strands synthesis in a reconstituted replication system (4). The genes encoding these proteins, together with genes 2 (E. coli RNA polymerase inhibitor), 3 (endonuclease), 3.5 (lysozyme), and 6 (exonuclease) constitute the DNA metabolism class II genes. More than a dozen additional genes within the class II group remain uncharacterized (5).
The primase activity of T7 is located in the N-terminal half of the 63-kDa gp4, whereas the C-terminal half of gene 4 encodes the DNA helicase (4). Like other prokaryotic homologs, the primase domain recognizes a specific sequence (1, 6) 5′-(G/T)(G/T)GTC-3′ in the ssDNA template to catalyze the template-directed synthesis of functional tetraribonucleotides (pppACCA, pppACCC, and pppACAC) that the lagging strand DNA polymerase can use as primers to initiate the synthesis of Okazaki fragments. Gene 4 encodes two colinear proteins, the full-length 63-kDa gp4 and a short 56-kDa form in equal amount in vivo from an internal start codon and ribosome-binding site (4). This 56-kDa gp4 has full helicase activity but lacks the N-terminal zinc-binding domain (ZBD), an indispensable motif for primer synthesis (7). Despite its inability to synthesize template-dependent tetraribonucleotides, the 56-kDa gp4 is capable of stabilizing certain preformed oligonucleotides on the cDNA template and delivering them to T7 DNA polymerase for extension (3).
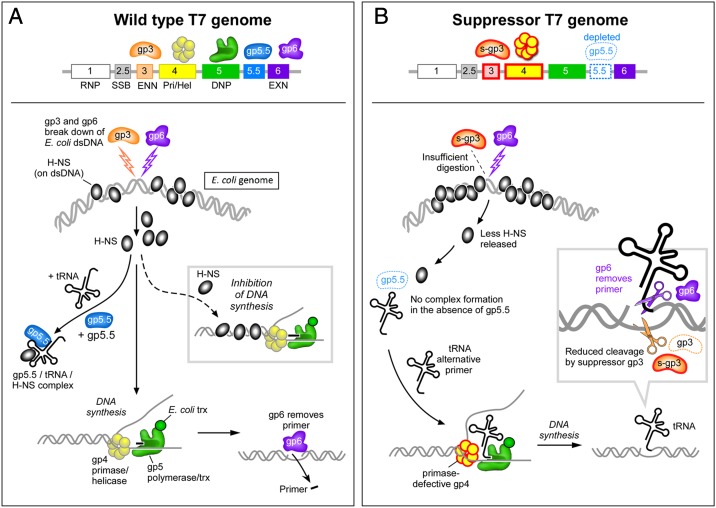
T7Δ4 phage lacking gene 4 can only grow normally on E. coli harboring the plasmid expressing full-length 63-kDa gp4 (8). On the same host expressing the primase-deficient 56-kDa gp4, T7Δ4 phage has a low efficiency of plating, but tiny phage plaques are observed. Here we show phages that grow robustly with only 56-kDa gp4 emerge after repetitive selection of single plaques, suggesting they are suppressors that overcome the primase deficiency. Indeed, many of these suppressors have mutations in gene 5.5. We show that gp5.5 forms a complex with host transfer RNAs (tRNAs) and H-NS protein. tRNAs, whose 3′ termini have the sequence ACCA identical to primers synthesized by the primase, can be delivered by the 56-kDa gp4 to prime T7 DNA synthesis. We further show that the binding of tRNA facilitates the binding of gp5.5 to E. coli H-NS.
Results
T7 Suppressors That Overcome Primase Deficiency of the 56-kDa gp4.
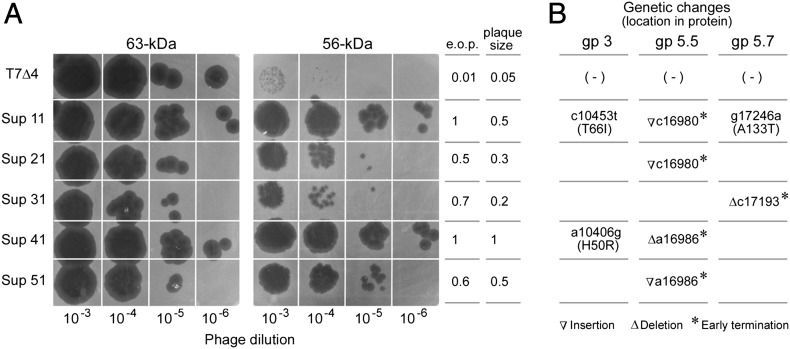
Because the 56-kDa gp4 cannot catalyze synthesis of oligoribonucleotides, its expression is not sufficient to support growth of T7Δ4 as a full-length 63-kDa gp4 does (Fig. 1A). Repetitive growth of the tiny phage plaques with 56-kDa gp4 supplied from a plasmid gave rise to suppressor phages that grow robustly on E. coli expressing only 56-kDa gp4. The plaques produced are comparable to those observed with 63-kDa gp4 (Fig. 1A). Although the initial T7Δ4 grow approximately 100 times better with 63-kDa than with 56-kDa gp4, the suppressor phages resulting from this selection (designated Sup 11–51) exhibit similar plaque size and plating efficiency with either gp4.
Fig. 1.
T7 phage suppressors that overcome primase deficiency with 56-kDa gp4. (A) Infection of wild-type and suppressor T7Δ4 phage on E. coli expressing 63-kDa or 56-kDa gp4. Numbers on the right show the relative plating efficiency and plaque size of T7Δ4 and suppressor phages in the presence of 56-kDa gp4 normalized against those in the presence of 63-kDa gp4. (B) Genetic analysis of class II genes for the presence of suppressors.
Expression of only the helicase domain of gp4 does not support normal growth of any of the suppressor phages, strongly indicating that the truncated primase domain is also necessary for the suppressors to function (Fig. S1A). The suppressor phages grew equally well with 56-kDa gp4 regardless of whether the primase catalytic site was active or mutationally inactivated (9), suggesting that primer delivery is the basis of the requirement of the 56-kDa gp4 in supporting suppressor activity (Fig. S1B).
To elucidate the alternative pathway for priming of DNA synthesis, the class II DNA metabolism genes of the suppressor phages were sequenced and compared with those of the initial T7Δ4 phage (Fig. 1B). All five phage suppressors analyzed contain mutations in gene 5.5/5.7 that result in early termination of the proteins encoded. The two suppressors showing the best growth, Sup 11 and 41, contain an additional mutation in gene 3 endonuclease.
T7 gp5.5 Forms a Stable Complex with E. coli H-NS and tRNA.
Gene 5.5 expresses a 99-aa-long protein that binds to E. coli H-NS to potentially counteract its inhibitory role to both host and phage transcription (10, 11). Gene 5.7 is located close to gene 5.5 with one overlapping nucleotide; its product has not been characterized. It has been suggested that a fusion gene 5.5/5.7 protein resulting from a frame-shift translation exists (5).
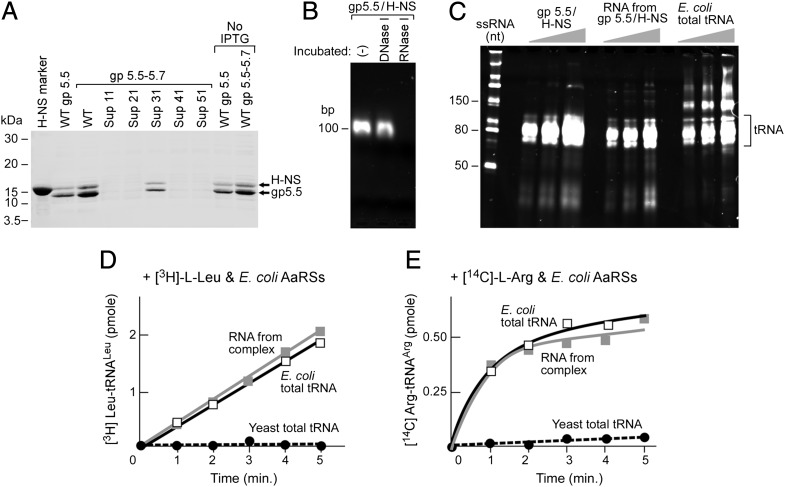
The fact that all suppressor T7Δ4 phages that allow for growth of cells expressing only the 56-kDa gp4 have mutations in or near gene 5.5 suggests that gp5.5 is involved in an alternative priming pathway. To investigate the effect of the mutations found in suppressors on the expression of gene 5.5, we purified the protein products of gene 5.5 and gene 5.5/5.7 from E. coli BL21(DE3) cells containing plasmids that express wild-type proteins as well as those expressing the proteins containing suppressor mutations. All proteins were expressed with an N-terminal His-tag. Gel analysis shows that both wild-type gene 5.5 and gene 5.5/5.7 express a His-tagged protein of the same length, presumably gp5.5 on the basis of its mobility. A gene 5.5–5.7 fusion protein is not expressed. The gp5.5 expressed from gene 5.5/5.7, however, is more abundant (Fig. 2A), suggesting that gene 5.7 stimulates the expression of gene 5.5. In both cases, host H-NS protein copurifies with gp5.5. Although all frame-shift mutations in gene 5.5 abolish full-length gp5.5 (Fig. 2A, Sup 11, 21, 41, and 51), a mutation in gene 5.7 (Sup 31) produces almost the same level of gp5.5 as that obtained without gene 5.7. Taken together, the common feature of all five suppressor phages is the decreased level of gp5.5. Interestingly, gene 5.5 expression does not require induction by isopropyl β-d-1-thiogalactopyranoside (Fig. 2A).
Fig. 2.
A complex of T7 gp5.5, E. coli H-NS, and tRNA. (A) Gp5.5 copurified with E. coli H-NS and its expression is eliminated by suppressor mutations. T7 gene 5.5, gene 5.5/5.7, and gene 5.5/5.7 from suppressors 11–51 were overexpressed as His-tagged proteins and purified by Ni-NTA before loading onto the SDS/PAGE gel. (B) Gp5.5 is copurified with RNA. Purified gp5.5/H-NS (1 μM) from cells expressing gp5.5 was electrophoresed through an agarose gel and then stained with ethidium bromide to detect nucleic acids. The gp5.5/H-NS complex was incubated with DNase I or RNase I (1 U/μL) for 1 h at 37 °C before loading. (C) Mobility of RNA components in gp5.5/H-NS complex is similar to E. coli tRNAs. The purified complex, RNA components extracted from the complex, and E. coli total tRNAs were compared on a 15% TBE-Urea gel. (D and E) Comparison of the RNA components in gp5.5/H-NS complex and E. coli tRNAs for their ability to be charged with leucine (D) and arginine (E) using E. coli aminoacyl-tRNA synthetases.
Because the suppressor phages need an alternative source of primers, we speculated that gp5.5 might function in the acquisition of functional primers. If RNA transcripts function as primers, one would expect enhanced transcription from higher but not decreased levels of gp5.5. On the other hand, we do find that oligonucleotides copurify with the gp5.5/H-NS complex by ethidium bromide staining of the gel containing the proteins (Fig. 2B). The gp5.5/H-NS/oligonucleotides complex is very stable throughout the purification procedure including an affinity column, a gel filtration column, and an ion-exchange column. The oligonucleotides in the complex are not sensitive to DNase I but are degraded by RNase I, suggesting that they are RNA (Fig. 2B). Further comparison of their size on a denaturing gel with E. coli tRNAs indicates that they represent host tRNAs (Fig. 2C). During the preparation of this article an independent study also described the observation of small RNAs in a gp5.5/H-NS complex, and the 3′ termini of specific tRNAs were identified (11).
Aminoacyl-tRNA synthetases (12) charge tRNAs specifically with cognate amino acids. The RNAs from the gp5.5/H-NS complex accept leucine and arginine when assayed with a preparation containing all of the aminoacyl-tRNA synthetases of E. coli. The efficiency of charging is comparable to that observed with a preparation containing the same amount of E. coli total tRNAs (Fig. 2 D and E). These results suggest that the RNA in the complex has a composition similar to that of E. coli tRNAs. We also attempted to identify the RNAs by sequencing. The efficiency of amplification of RNAs by retrotranscription is low, probably owing to the structure of tRNAs. However, among the clear sequencing results we obtained (Table S1), most of the sequences match those of E. coli tRNAs. The one exception is a 3′ fragment of an E. coli tmRNA.
tRNAs Can Be Alternative Primers for T7 DNA Replication.
The molar ratio of tRNA to the complex is 2:1. The concentration of tRNA was determined by measuring the absorbance at 260 nm in a spectrophotometer and the proteins by Bradford assay. H-NS is one of the most abundant proteins in E. coli, with a cytoplasmic concentration of more than 2 μM (13, 14). Gp5.5 is also one of the most abundant proteins in T7-infected cells and is likely to be more abundant than H-NS (5, 10). Total tRNA concentration in E. coli has been estimated at approximately 6 μM (15). Thus, maximally two-thirds of E. coli tRNAs could be sequestered in the gp5.5/H-NS complex.
These observations, together with the fact that tRNAs are used as primers by some retrovirus, prompted us to examine the possibility that tRNA can serve as a primer when the T7 primase is defective in primer synthesis. Interestingly, the 3′ termini of all tRNAs have the sequence 5′-CCA-3′, mimicking one of the major functional primers synthesized by T7 primase—5′-pppACCA-3′. Furthermore, ∼60% of the 3′ termini of all E. coli tRNAs have the sequence 5′-ACCA-3′.
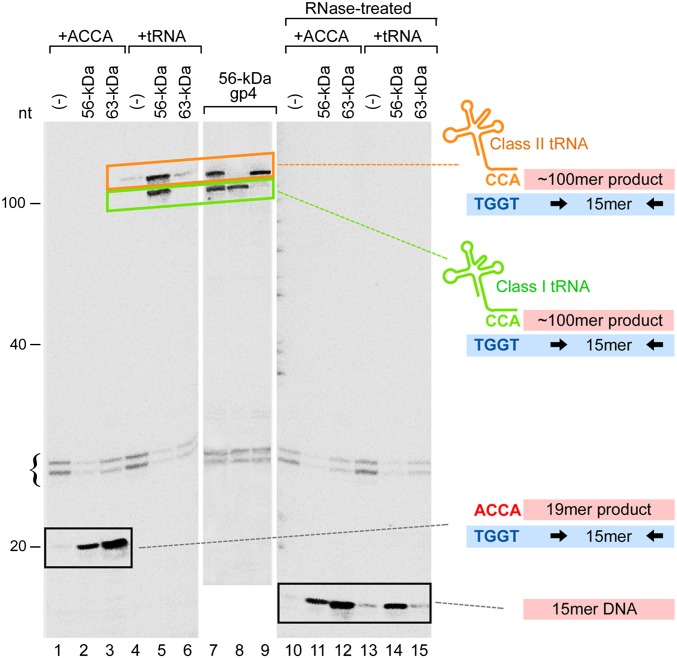
We investigated the ability of tRNA to serve as primers by measuring the extension of primers with a 25-mer ssDNA template containing the primase recognition site 5′-TGGT-3′ (Fig. 3). The conventional primer, ACCA, synthesized by T7 primase, can be delivered to T7 DNA polymerase by both the 56- and 63-kDa gp4. On the template used in this assay the primer is extended to yield a 19-mer RNA-DNA product (Fig. 3, lanes 1–3). Radioactive labeled dNMP was incorporated so that the extended RNA–DNA product can be identified on denaturing gels. The full-length 63-kDa gp4 is more efficient in delivery of ACCA than the 56-kDa gp4 because the ZBD enhances stabilization of the primer. As we expected, the E. coli tRNA can also serve as a primer, but only efficiently in the presence of the 56-kDa gp4 (Fig. 3, lanes 4–6). The two major product bands of ∼100 nt correspond to the extension of two groups of tRNA with different lengths. Consequently, the extended tRNAs are approximately 90 nt (∼75-mer class I tRNA plus 15-mer DNA) and 105 nt (∼90-mer class II tRNA plus 15-mer DNA). Use of tRNA as primer by the 63-kDa gp4 was significantly less than by the 56-kDa gp4 (Fig. 3, lane 6), consistent with previous studies suggesting that the ZBD acts as a steric hindrance to accommodate longer primers (3).
Fig. 3.
tRNA primed DNA synthesis by T7 proteins. Lanes 1–3, extension of ACCA by T7 DNA polymerase in the absence (lane 1) or presence of 56-kDa gp4 (lane 2) or 63-kDa gp4 (lane 3). Lanes 4–6, extension of E. coli total tRNA by T7 DNA polymerase in the absence (lane 4) or presence of 56-kDa gp4 (lane 5) or 63-kDa gp4 (lane 6). Lanes 7–9, extension of E. coli total tRNA (lane 7), tRNAArgACG (lane 8), or tRNALeuCAG (lane 9) by T7 DNA polymerase in the presence of 56-kDa gp4. Lanes 10–15, samples from lanes 1–6 treated with RNase I.
Representatives of the two classes of tRNA, tRNAArgACG and tRNALeuCAG (Fig. S2A), were purified and their ability to serve as primers examined. The size of the extended products of these two tRNAs confirmed our conclusion that both classes of tRNA can be used as primer (Fig. 3, lanes 7–9). When the RNA portion of the product was removed by treatment with RNase I, both products were converted into 15-mer ssDNA (Fig. 3, lanes 1–6 vs. lanes 10–15). The bands around 25 nt in Fig. 3 arise from labeling of the 25-mer ssDNA template by sequential exonuclease and polymerase incorporation reactions of T7 DNA polymerase.
A reconstituted replisome of four proteins mediates coordinated leading and lagging strand DNA synthesis (Fig. S2B). When tRNAArg was used as primer with 56-kDa gp4, lagging-strand synthesis occurred, but the rate of lagging-strand synthesis is slower than that of the leading strand (Fig. S2B). Another consequence of tRNA-primed DNA synthesis is that the tRNA attached to the primer gives rise to strand-displacement DNA synthesis. In the experiment shown in Fig. S2C, when the preformed primer ACCA is present DNA synthesis is observed but ceases after forming double-stranded M13 DNA. Gp5/trx and gp4 cannot initiate strand-displacement synthesis unless there is a 5′-ssDNA tail (16). However, when tRNA is used as primer, the amount of incorporated dNMP exceeds the amount of M13 ssDNA template, indicating that strand-displacement DNA synthesis has occurred.
Adaption of T7 Gene 3 Endonuclease to tRNA Priming.
Two suppressor phages (Sup 11 and 41) with mutations in gene 5.5 also have acquired a mutation in gene 3 endonuclease (Fig. 1B) and grow better than do those harboring mutation only in gene 5.5. To investigate the contribution of these mutations to growth of suppressors, the two altered gp3 were purified and compared with wild-type gp3. T7 gp3 hydrolyzes ssDNA and will introduce nicks in mismatched dsDNA and at Holliday junctions (17, 18). Both altered gp3 have reduced endonuclease activity relative to wild-type gp3 as measured in a plasmid-nicking assay that measures conversion of supercoiled plasmid pUC(AT) to the nicked and linear forms (Fig. S3A).
How can reducing the activity of an endonuclease enhance the alternative priming mechanism? One possibility is that the endonuclease adapts its substrate recognition to favor the processing of the primer. Although gp3 does not hydrolyze tRNA, the flap structure (Fig. S3B) resulting from strand-displacement DNA synthesis in the tRNA-primed reaction may mimic a Holliday junction. A dsDNA containing a 5′-single-stranded tail of 16 nt was constructed with the 5′ terminus of the uninterrupted strand labeled with 32P. After incubation with wild-type gp3 a 19-nt radioactive fragment was identified; cleavage thus occurs opposite the flap and within the duplex region adjacent to the flap. The two gp3 variants have significantly reduced activity on this substrate. This reaction would essentially introduce double-strand interruptions in the lagging strand at the start of each Okazaki fragment, an event that would occur less frequently with the gp3 variants. Our recent studies show that a flap endonuclease activity of T7 gp6 can remove the flap resulted from tRNA priming.
gp5.5 Uses tRNA as Bait for H-NS Binding.
Sequestering tRNA in the gp5/H-NS complex does not seem to benefit the phage metabolism because T7 relies on the host translation system. More likely the role of gp5.5 is directly related to E. coli H-NS. H-NS interferes with phage transcription (10) and therefore is likely deleterious to several aspects of T7 metabolism, including DNA replication.
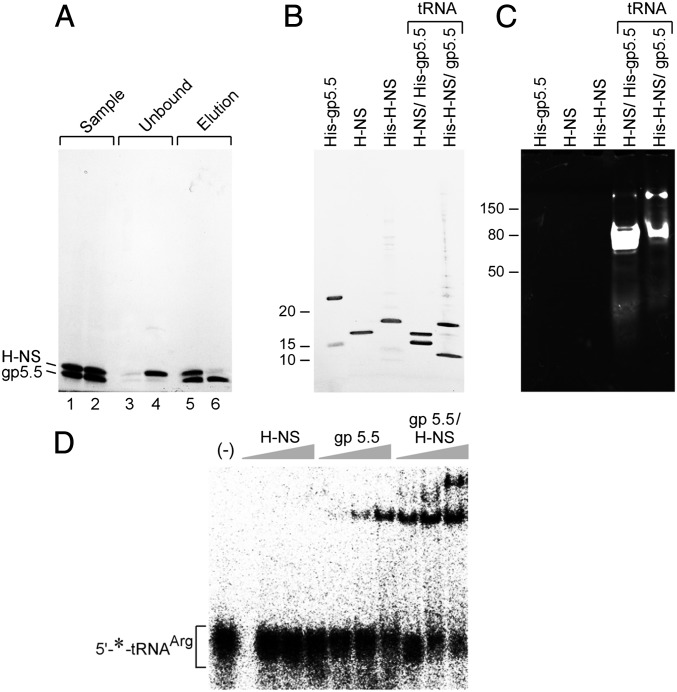
We find that the tRNA component is necessary for gp5.5/H-NS complex formation. Because the gp5.5 used in these experiments is His-tagged, the gp5.5/H-NS complex should also bind to Ni-NTA agarose. In the experiment shown in Fig. 4A the complex is fully bound to Ni-NTA agarose after mixing and centrifugation. In the supernatant there is no significant amount of protein, as shown by gel analysis of the fraction (Fig. 4A, lane 3). The complex is eluted from the resin by a high concentration of imidazole (Fig. 4A, lane 5). However, if the tRNA component is removed by RNase I treatment during the binding to Ni-NTA resin, H-NS is released into the supernatant (Fig. 4A, lane 4), whereas His-tagged gp5.5 remains bound to the Ni-NTA resin and can be eluted from the resin by imidazole (Fig. 4A, lane 6).
Fig. 4.
tRNA is indispensible for formation of the gp5.5/H-NS complex. (A) Disruption of gp5.5/H-NS/tRNA complex by removing tRNA. The same amount of purified gp5.5/H-NS complex (30 μg in 200 μL) was mixed with 50 μL Ni-NTA resin in two tubes and incubated at room temperature for 30 min. In one of the tubes RNase I (1 U/μL) was mixed together with the complex and resin. After centrifugation the supernatants containing unbound protein were collected. The resin was then mixed with 150 μL 500 mM imidazole and centrifuged. The eluted proteins in the supernatants were collected. Collected fractions were then analyzed by SDS/PAGE gel electrophoresis. Lanes 1 and 2, two batches of gp5.5/H-NS complex before mixing with Ni-NTA resin; lane 3, supernatant containing unbound protein from the tube without RNase I; lane 4, supernatant containing unbound protein from the tube with RNase I; lane 5, supernatant containing eluted protein from the tube without RNase I; lane 6, supernatant containing eluted protein from the tube with RNase I. (B) SDS/PAGE analysis of purified proteins. (C) Agarose gel analysis of RNA component from purified proteins. The samples in B were analyzed on TBE-Urea gel followed by ethidium bromide staining. (D) Complex formation of tRNA with purified proteins. Binding of His-tagged gp5.5 (110 nM, 330 nM, 1 μM) or/and E. coli H-NS (110 nM, 330 nM, 1 μM) to [5′-32P] tRNAArgACG (20 nM) was determined by mobility shift assay on a 10% TBE gel.
In an attempt to reconstitute the gp5.5/H-NS/tRNA complex from purified components, we first purified H-NS protein and two individual tRNAs (Fig. S2A). However, when tRNA and H-NS are removed from the complex by RNase or urea, gp5.5 irreversibly precipitates. Soluble refolded gp5.5 was obtained by keeping the protein concentration under 10 μM. Interestingly, the majority of gp5.5 in this preparation forms a stable dimer that is not separated by an SDS-denaturing gel (Fig. 4B, lane 1). The dimer is not likely a disulfide bond because it remains intact at a high concentration of DTT. H-NS, His-tagged H-NS, and gp5.5/H-NS with His-tag on gp5.5 or H-NS, all purified under nondenaturing conditions, are shown in the gel in Fig. 4B. All samples in Fig. 4B are also analyzed on an agarose gel stained with ethidium bromide (Fig. 4C). Only the protein complexes contain tRNA, the His-tag can reside on either gp5.5 or H-NS. Because the gp5.5 purified under denaturing/renaturing condition still contains a small amount of soluble monomer (Fig. 4B, lane 1), we used it to reconstitute a gp5.5/H-NS complex with tRNA. Complexes were detected by a mobility shift assay. As shown in Fig. 4D, H-NS alone did not bind any tRNAArg, whereas the gp5.5 mixture of monomer and dimer bound tRNAArg weakly. However, the maximal binding of tRNA occurs with a mixture of gp5.5 and H-NS, and a larger complex formed, suggesting that H-NS is incorporated into the complex.
Inhibition of T7 DNA Helicase and DNA Replication by H-NS.
We constructed a T7 phage in which gene 5.5 is deleted (T7Δ5.5). We also used an E. coli strain in which H-NS is inactivated (Keio collection) to examine the effect of H-NS on phage growth (Fig. S4). Even in the presence of gp5.5, a higher amount of H-NS expressed from a plasmid can reduce the efficiency of plating of T7 by twofold. Although still in the presence of gp5.5, phage infection increases twofold upon depletion of H-NS. These results suggest that H-NS does have an inhibitory effect on phage growth. When gp5.5 is depleted, the phage shows defects in both efficiency of plating and plaque size. These defects become more apparent when the amount of H-NS is increased, indicating that gp5.5 plays an antagonistic role against H-NS.
We find that H-NS binds to a preformed Y-shaped dsDNA (Fig. S5) with a higher affinity (Kd = 620 nM) than it does to ssDNA (Kd >2 μM), dsDNA with a single overhang (Kd = 1.2 μM), or curved dsDNA (Kd = 1.1 μM) (19). In the experiment presented in Fig. S6 we have examined the effect of H-NS on the unwinding of dsDNA by the T7 gene 4 helicase. With increasing concentrations of helicase a radioactively labeled 75mer is displaced from its complementary 95mer. The addition of 200 nM H-NS to the reaction impedes the unwinding. H-NS in complex with gp5.5 and tRNA does not exhibit such an inhibitory effect (Fig. S6A). H-NS does not interfere with DNA synthesis catalyzed by T7 DNA polymerase on circular ssDNA bearing a primer (Fig. S6B). However, when the polymerase and helicase together mediate leading-strand DNA synthesis on circular dsDNA bearing a replication fork, H-NS reduces synthesis to approximately fourfold (Fig. S6B). H-NS in complex with gp5.5 and tRNA does not exhibit such an inhibitory effect, even at the higher concentration.
Discussion
Most organisms use oligoribonucleotides synthesized by DNA primases as primers for DNA polymerase to initiate DNA synthesis on the lagging strand. However, longer RNA transcripts are also used as in the replication of mitochondria DNA and RNA viruses. The reverse-transcriptase of HIV uses host tRNALys as the specific primer for the initiation of cDNA synthesis (20). Our current studies show that tRNA can also serve as primers for T7 DNA polymerase. The extension of tRNA priming to bacterial systems suggests an ancient common priming mechanism using tRNA. The observation that the C termini of all tRNAs are 5′-CCA-3′, with 60% having the sequence 5′-ACCA-3′, is particularly intriguing because this is precisely the sequence of one of the tetraribonucleotides synthesized by T7 primase.
tRNAs are one of the most abundant small RNAs in cells, and they have unique sequences with stable structures, thus making them good candidates as primers. In the case of HIV, annealing of the tRNA primer to the RNA genome involves a large portion of the tRNALys upstream of the CCA 3′ terminus (20). In T7 the ACCA terminus of the tRNA that matches the primase recognition site 5′-TGGT-3′ is sufficient for delivery to T7 DNA polymerase by the primase. tRNAArg and tRNALeu with a different sequence both served as primer with comparable efficiency. Although priming of DNA synthesis by tRNA would seem to be an economic solution, it has disadvantages in the case of T7 DNA replication. First, the synthesis of oligoribonucleotides by the T7 primase is important in the coordination of leading- and lagging-strand synthesis (21). Second, the tetranucleotide synthesized by T7 DNA primase leaves only a nick between Okazaki fragments, at which point T7 DNA polymerase dissociates from the DNA. The 5′-attached tRNA may require strand-displacement synthesis, thus interfering with the coordination mechanism (22). Despite those shortcomings, tRNA may serve as a backup source of primers and might even be used for priming at the chromosomal origin of replication.
We show that upon T7 infection of E. coli gp5.5 binds to tRNA, and the gp5.5/tRNA then binds tightly to E. coli H-NS to form a gp5.5/tRNA/H-NS complex. At least one of the purposes of such a complex is to inhibit H-NS. H-NS functions as a transcription inhibitor to down-regulate the expression of hundreds of genes. It is also thought to be a sentinel against invading DNA in bacterial cells owing to its preference for A/T-rich dsDNA often found in phage genomes (13, 14). The location of gp5.5 on the T7 genome is interesting because the expression of T7 genes is a sequential event (Fig. 5). Gene 5.5 precedes gene 6, whose product is an exonuclease important in the degradation of host genome and in removal of primers (23). Before the appearance of gp6 most of the H-NS molecules are likely bound to the E. coli genome (Fig. 5). After degradation of the host chromosome the T7 genome represents the only target of H-NS; the ratio could be as high as one H-NS molecule for every two nucleotides in T7 DNA. Recent evidence suggests that H-NS is a potential inhibitor for DNA replication and recombination (24). We have shown that these high concentrations of H-NS can indeed inhibit the unwinding of DNA by the T7 helicase and impede movement of the replication fork.
Fig. 5.
Schematic representation of the interplay of T7 gp5.5, gp3, E. coli H-NS, and tRNA in cells infected with phage T7. (A) After infection of E. coli by T7 the gene 3 endonuclease and gene 6 exonuclease degrade the E. coli genome to provide dNMPs for T7 DNA synthesis. T7 gp5 binds tightly to the abundant host thioredoxin to acquire processivity. The gp5/trx complex along with the gp4 helicase-primase and the gene 2.5 ssDNA binding protein form a replisome that mediates coordinated DNA synthesis. The degradation of the host genome releases the histone-like H-NS, which in turn can bind to the phage DNA, inhibiting DNA replication. However, the abundant gp5.5 binds to another abundant component in E. coli, tRNA, and acquires a high affinity for H-NS, resulting in a gp5.5/tRNA/H-NS complex. Thus, the excess H-NS is sequestered by gp5.5, eliminating its binding to T7 DNA. The short primers, synthesized by T7 primase, at the 5′ termini of Okazaki fragments are removed by gp6, and the Okazaki fragments are ligated. (B) In the absence of a functioning DNA primase an alternative pathway for priming of the lagging strand DNA polymerase is required. Suppressor mutations arise within gene 5.5 that deplete the pool of gp5.5. As a consequence, the pool of tRNA is not reduced. The similarity of the 3′ terminus of many tRNAs to the tetraribonucleotides synthesized by the T7 primase allows them to function as primers. The tRNA primed DNA structure, however, is a target for gp3 endonuclease. Some suppressors therefore also contain mutations in gene 3 that reduce the activity of gp3.
The strategy of T7 to counteract H-NS is interesting in that gp5.5 uses tRNA, another abundant molecule in the cell, as bait to bind H-NS (Fig. 5A). H-NS is known to bind RNA (19), but the affinity is much lower than that to dsDNA. The affinity of H-NS to various nucleic acids is in the order of dsDNA > ssDNA > tRNA > rRNA (25). However, in the presence of gp5.5, the affinity of H-NS to tRNA is stimulated, presumably owing to a relaxing of the compact tertiary structure of tRNA by gp5.5 to expose the branching secondary structure. The formation of gp5.5/H-NS/tRNA tertiary complex blocks the function of both tRNA and H-NS; no aminoacylation or DNA-binding activity can be detected. H-NS in the tertiary complex no longer inhibits DNA unwinding by gp4 helicase or strand-displacement DNA synthesis mediated by T7 helicase and polymerase. Nonetheless, gp5.5 seems to have functions in addition to the silencing of H-NS. In an hns− E. coli strain, lacking H-NS, T7 phage lacking gp5.5 still have a growth defect (Fig. S4). Although tRNAs are the dominant RNA species in the complex, smaller fragments of RNA are also observed on gel. Our limited sequencing results (Table S1) also revealed a 3′-fragment of a tmRNA (26). The existence of tmRNA in the complex suggests that the proteins recognize structural features of tRNA-like molecules.
Another unexpected finding has been the involvement of gene 3 endonuclease in the alternative priming pathway. The two altered residues in gene 3 endonuclease, Tyr66 and His50, are involved in the coordination of the metal ion at the active site and in substrate recognition, respectively (18). Studies with the altered endonucleases suggest that they are less destructive than wild-type protein to the tRNA-primed DNA structure, a structure resembling a Holliday junction (Fig. 5B). Interestingly, similar genetically altered gp3 were found to suppress the phenotype arising from an altered T7 DNA polymerase (27). Thus, any abnormal replication structures seem to be targets for gene 3 endonuclease, perhaps providing a check-point for faithful replication.
Materials and Methods
Proteins and RNAs.
DNA encoding genes from wild-type or suppressor T7 phages and E. coli H-NS were inserted into pET24a, pET17b, or pET28b plasmids. Ni-NTA affinity column (Qiagen) was used for the purification of His-tagged gp5.5 (complexed with H-NS and tRNAs), the His-tagged H-NS, and His-tagged H-NS/gp5.5/tRNAs complex. The proteins were further purified by S-200 gel filtration and Mono Q ion-exchange column chromatography. Purification of His-tagged gp5.5 alone was according to a denaturing/refolding procedure (Qiagen). The refolded protein was immediately diluted to 10 μM to prevent protein precipitation. H-NS (24) and gp3 (27) were purified as described. T7 gp4, gp5/trx were overproduced and purified as described previously (28, 29). E. coli aminoacyl-tRNA synthetases were purchased from Sigma-Aldrich. RNAs complexed with gp5.5 and H-NS were extracted from the purified complex with phenol/chloroform. E. coli total tRNA was purchased from Roche Applied Science. E. coli tRNALeuCAG and tRNAArgACG were prepared as previously described (30).
Assays.
Phage growth complementation assay was performed as previously described (27). tRNA aminoacylation assay was performed as previously described (31), except that 20 μM RNA from the complex or E. coli total tRNA, 20 μM [3H] leucine or [3H] arginine (15 Ci/mmol), and 2 μM E. coli aminoacyl-tRNA synthetases were used. RNA-primed DNA polymerase reactions were carried out in a reaction mixture containing 40 mM Tris·HCl (pH 7.5), 10 mM DTT, 10 mM MgCl2, 50 mM potassium glutamate, 300 μM dNTP containing [α-32P] dGTP, 30 μM 25-mer template ssDNA (5′-CGT AAT CTG CAG GCA TGGT GAA TTT-3′), 20 μM RNA primer (ACCA, total tRNA, or single tRNA species), and 1 μM T7 DNA polymerase (exonuclease deficient) in the absence or presence of 2 μM T7 56-kDa or 63-kDa gp4. Reactions were incubated at room temperature for 30 min. RNase I (Ambion) 1 U/μL was then added into some reactions, and an additional incubation at 37 °C for 20 min was performed. Reaction products were separated on a 15% (wt/vol) Tris-borate-EDTA (TBE) gel containing 7 M urea. For gel mobility shift assay, the reaction mixtures contained 40 mM Tris·HCl (pH 7.5), 10 mM DTT, 10 mM MgCl2, 50 mM potassium glutamate, 20 nM 5′-[32P] radiolabeled DNA or tRNA, and the indicated concentration of E. coli H-NS and/or gp5.5. After incubation at room temperature for 20 min, samples were electrophoresed on a 10% polyacrylamide gel in TBE buffer at 80 V and 4 °C for 3–5 h. The gel was dried and radioactivity measured. For 5′-[32P] labeled tRNAArgACG, the tRNAArgACG was first treated with Antarctic phosphatase (New England Biolabs) and then radioactively labeled using T4 polynucleotide kinase. Materials and methods for supporting data are listed in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Steven Moskowitz (Advanced Medical Graphics) for illustrations and Udi Qimron for advice. This work was supported by National Institutes of Health Grant GM54397 (to C.C.R.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205990109/-/DCSupplemental.
References
- 1.Frick DN, Richardson CC. DNA primases. Annu Rev Biochem. 2001;70:39–80. doi: 10.1146/annurev.biochem.70.1.39. [DOI] [PubMed] [Google Scholar]
- 2.Yuzhakov A, Kelman Z, O’Donnell M. Trading places on DNA—a three-point switch underlies primer handoff from primase to the replicative DNA polymerase. Cell. 1999;96:153–163. doi: 10.1016/s0092-8674(00)80968-x. [DOI] [PubMed] [Google Scholar]
- 3.Zhu B, Lee SJ, Richardson CC. Direct role for the RNA polymerase domain of T7 primase in primer delivery. Proc Natl Acad Sci USA. 2010;107:9099–9104. doi: 10.1073/pnas.1004220107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamdan SM, Richardson CC. Motors, switches, and contacts in the replisome. Annu Rev Biochem. 2009;78:205–243. doi: 10.1146/annurev.biochem.78.072407.103248. [DOI] [PubMed] [Google Scholar]
- 5.Dunn JJ, Studier FW. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983;166:477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- 6.Lee SJ, Zhu B, Hamdan SM, Richardson CC. Mechanism of sequence-specific template binding by the DNA primase of bacteriophage T7. Nucleic Acids Res. 2010;38:4372–4383. doi: 10.1093/nar/gkq205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akabayov B, et al. DNA recognition by the DNA primase of bacteriophage T7: A structure-function study of the zinc-binding domain. Biochemistry. 2009;48:1763–1773. doi: 10.1021/bi802123t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mendelman LV, Notarnicola SM, Richardson CC. Roles of bacteriophage T7 gene 4 proteins in providing primase and helicase functions in vivo. Proc Natl Acad Sci USA. 1992;89:10638–10642. doi: 10.1073/pnas.89.22.10638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee SJ, Richardson CC. Acidic residues in the nucleotide-binding site of the bacteriophage T7 DNA primase. J Biol Chem. 2005;280:26984–26991. doi: 10.1074/jbc.M504817200. [DOI] [PubMed] [Google Scholar]
- 10.Liu Q, Richardson CC. Gene 5.5 protein of bacteriophage T7 inhibits the nucleoid protein H-NS of Escherichia coli. Proc Natl Acad Sci USA. 1993;90:1761–1765. doi: 10.1073/pnas.90.5.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ali SS, Beckett E, Bae SJ, Navarre WW. The 5.5 protein of phage T7 inhibits H-NS through interactions with the central oligomerization domain. J Bacteriol. 2011;193:4881–4892. doi: 10.1128/JB.05198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ibba M, Söll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 13.Dorman CJ. H-NS, the genome sentinel. Nat Rev Microbiol. 2007;5:157–161. doi: 10.1038/nrmicro1598. [DOI] [PubMed] [Google Scholar]
- 14.Navarre WW, McClelland M, Libby SJ, Fang FC. Silencing of xenogeneic DNA by H-NS-facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes Dev. 2007;21:1456–1471. doi: 10.1101/gad.1543107. [DOI] [PubMed] [Google Scholar]
- 15.Dong H, Nilsson L, Kurland CG. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J Mol Biol. 1996;260:649–663. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- 16.Zhu B, Lee SJ, Richardson CC. Bypass of a nick by the replisome of bacteriophage T7. J Biol Chem. 2011;286:28488–28497. doi: 10.1074/jbc.M111.252023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Center MS, Studier FW, Richardson CC. The structural gene for a T7 endonuclease essential for phage DNA synthesis. Proc Natl Acad Sci USA. 1970;65:242–248. doi: 10.1073/pnas.65.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hadden JM, Déclais AC, Carr SB, Lilley DM, Phillips SE. The structural basis of Holliday junction resolution by T7 endonuclease I. Nature. 2007;449:621–624. doi: 10.1038/nature06158. [DOI] [PubMed] [Google Scholar]
- 19.Brescia CC, Kaw MK, Sledjeski DD. The DNA binding protein H-NS binds to and alters the stability of RNA in vitro and in vivo. J Mol Biol. 2004;339:505–514. doi: 10.1016/j.jmb.2004.03.067. [DOI] [PubMed] [Google Scholar]
- 20.Tisné C. Structural bases of the annealing of primer tRNA(3Lys) to the HIV-1 viral RNA. Curr HIV Res. 2005;3:147–156. doi: 10.2174/1570162053506919. [DOI] [PubMed] [Google Scholar]
- 21.Hamdan SM, Loparo JJ, Takahashi M, Richardson CC, van Oijen AM. Dynamics of DNA replication loops reveal temporal control of lagging-strand synthesis. Nature. 2009;457:336–339. doi: 10.1038/nature07512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.López de Saro FJ, Georgescu RE, O’Donnell M. A peptide switch regulates DNA polymerase processivity. Proc Natl Acad Sci USA. 2003;100:14689–14694. doi: 10.1073/pnas.2435454100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Serwer P, Watson RH, Son M. Role of gene 6 exonuclease in the replication and packaging of bacteriophage T7 DNA. J Mol Biol. 1990;215:287–299. doi: 10.1016/S0022-2836(05)80347-X. [DOI] [PubMed] [Google Scholar]
- 24.Sharadamma N, Harshavardhana Y, Singh P, Muniyappa K. Mycobacterium tuberculosis nucleoid-associated DNA-binding protein H-NS binds with high-affinity to the Holliday junction and inhibits strand exchange promoted by RecA protein. Nucleic Acids Res. 2010;38:3555–3569. doi: 10.1093/nar/gkq064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedrich K, Gualerzi CO, Lammi M, Losso MA, Pon CL. Proteins from the prokaryotic nucleoid. Interaction of nucleic acids with the 15 kDa Escherichia coli histone-like protein H-NS. FEBS Lett. 1988;229:197–202. doi: 10.1016/0014-5793(88)80826-3. [DOI] [PubMed] [Google Scholar]
- 26.Moore SD, Sauer RT. The tmRNA system for translational surveillance and ribosome rescue. Annu Rev Biochem. 2007;76:101–124. doi: 10.1146/annurev.biochem.75.103004.142733. [DOI] [PubMed] [Google Scholar]
- 27.Lee SJ, Chowdhury K, Tabor S, Richardson CC. Rescue of bacteriophage T7 DNA polymerase of low processivity by suppressor mutations affecting gene 3 endonuclease. J Virol. 2009;83:8418–8427. doi: 10.1128/JVI.00855-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bernstein JA, Richardson CC. Purification of the 56-kDa component of the bacteriophage T7 primase/helicase and characterization of its nucleoside 5′-triphosphatase activity. J Biol Chem. 1988;263:14891–14899. [PubMed] [Google Scholar]
- 29.Tabor S, Huber HE, Richardson CC. Escherichia coli thioredoxin confers processivity on the DNA polymerase activity of the gene 5 protein of bacteriophage T7. J Biol Chem. 1987;262:16212–16223. [PubMed] [Google Scholar]
- 30.Yong L, Enduo W, Yinglai W. Overproduction and purification of Escherichia coli tRNA(Leu) Sci China C Life Sci. 1998;41:225–231. doi: 10.1007/BF02895095. [DOI] [PubMed] [Google Scholar]
- 31.Tan M, et al. tRNA-dependent pre-transfer editing by prokaryotic leucyl-tRNA synthetase. J Biol Chem. 2010;285:3235–3244. doi: 10.1074/jbc.M109.060616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.