Abstract
Herpesviruses are dsDNA viruses, but their virions may additionally contain RNAs that can be transduced to recipient cells. The biological functions of herpes virion RNA species are unknown. Here we address this issue for EBV, a widespread human herpesvirus with oncogenic potential. We show that EBV-derived particles that include virions, virus-like particles, and subviral vesicles contain viral mRNAs, microRNAs, and other noncoding RNAs. Viral RNAs were transduced during infection and deployed immediate functions that enhanced EBV’s capacity to transform primary B cells. Among these transduced viral RNAs, BZLF1 transcripts transactivated viral promoters triggering the prelatent phase of EBV infection, noncoding EBV-encoded RNA transcripts induced cellular cytokine synthesis, and BNLF2a mRNA led to immune evasion that prevented T-cell responses to newly infected B cells. Hence, transduced viral RNAs govern critical processes immediately after infection of B cells with EBV and likely play important roles in herpesviral infection in general.
Herpesviruses are large enveloped viruses with a dsDNA genome. However, viral particles of several Herpesviridae members additionally contain RNA molecules, as shown for CMV, HSV1, human Kaposi sarcoma virus, and murine gammaherpesvirus 68 (1–5). Thus, it seems likely that other herpesviruses also carry RNA in their virions. How the RNA molecules are packaged into the virion is unclear, but it appears that viral RNAs are more abundant in virions than cellular RNAs, suggesting a specific sorting mechanism (3–5). In infected cells, virion mRNAs can be translated immediately and in the absence of de novo transcription. So far, no functions have been assigned to transduced virion RNAs (tvRNAs) although they were discovered more than 10 y ago.
EBV is a ubiquitous human herpesvirus that is causally associated with different types of malignant diseases (6). The virus shows a tropism for human B lymphocytes, in which it establishes latent infection. Upon receiving exogenous signals, transmitted by the B-cell receptor for example, cellular signaling pathways can activate the viral gene BZLF1, which encodes the molecular switch that induces the lytic phase (7). A cascade of viral lytic gene expression is initiated, comprising immediate-early, early, and late viral genes, resulting in the release of newly formed virions eventually.
It has recently been described that, in newly infected cells, before establishment of latency, EBV expresses a small set of viral genes that had previously been classified as immediate-early or early genes of the lytic cycle. At this prelatent stage of infection, the immediate expression of these genes activates resting B cells (8) while protecting them from immediate activation-induced apoptosis (9) and other endogenous stress response signals (10). The prelatent phase is followed by a stable latent phase, characterized by the expression of viral latent genes that support cellular proliferation and sustain the outgrowth of lymphoblastoid cells lines in vitro (11). The catalog of viral genes expressed during this prelatent phase of EBV’s life cycle is not complete, but includes classical latent genes (12) as well as certain lytic genes that, for example, comprise viral homologues of antiapoptotic Bcl-2 family members (9), a viral IL-10 homologue (13, 14), and also BZLF1 (8, 15).
The rapid, prelatent expression of a number of viral proteins in newly infected cells might increase their antigenic load and thereby the chance of immune recognition, such as elimination by virus-specific T cells. In vivo, virus-specific CD4+ and CD8+ T cells directed against epitopes of various viral proteins control EBV infection (16). In the lytic phase of infection, a number of viral factors help the virus evade the immune response. Different EBV immunoevasins interfere with different steps of the MHC class I antigen presentation pathway (17), mimic immunosuppressive cytokines such as IL-10 (18), or down-regulate Toll-like receptors (TLRs) to avoid production of proinflammatory cytokines (19, 20).
Most enveloped viruses, including EBV, exploit the exosome biogenesis pathway for their egress (21, 22). Latently EBV-infected cells also constitutively release exosomes-like particles or microvesicles carrying viral proteins or microRNA (miRNA) (23–25). We therefore hypothesized that productively EBV-infected cells release viral and subviral particles that contain viral RNAs transducing them during the process of infection. tvRNAs may improve the efficiency of infection and establishment of EBV latency. Moreover, tvRNA-encoded immunoevasins could disrupt recognition by immune cells. To test our hypothesis, we investigated the RNA content of EBV particles and potential RNA-mediated functions in newly infected B cells. We show here that EBV particles and nonviral vesicles contain viral RNAs of different classes, delivering them to target cells upon infection. tvRNA molecules were functional and became translated in newly infected cells, accounting for early viral effects and enhancing the ability to establish persistent infections. In addition, tvRNAs encoding viral immunoevasins dampened the recognition of newly EBV-infected B cells by EBV-specific CD8+ T cells. Our data suggest that tvRNAs are an integral, multifunctional part of the rich biology of EBV and probably other herpesviruses as well.
Results
Viral RNAs Are Present as Early as 2 h After B-Cell Infection.
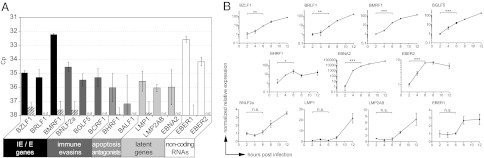
Previous studies have already described transcripts of EBV in primary B cells shortly after infection, but the origin of these RNAs was not further analyzed (8, 26). We set out to determine the time when viral transcripts appear after infection. We isolated primary B cells, infected them with EBV, and prepared total RNA at different time points starting at 2 h post infection (hpi). The samples were analyzed for the presence of selected viral transcripts. In detail, quantitative PCR (qPCR) was performed with primers that detect the cDNAs of the immediate-early and early genes BZLF1, BRLF1, and BMRF1; the immunoevasins BNLF2a, BCRF1 (vIL-10), and BGLF5; the antiapoptotic genes BHRF1 and BALF1; and the latent genes BNLF1, TP1/2, and BYRF1 encoding LMP1, LMP2A/B, and EBNA2, respectively. We also analyzed the prevalence of the noncoding EBV-encoded RNAs (EBER) 1 and 2. All transcripts were reproducibly detected in infected primary B cells with the exception of BALF1 (Fig. 1A). BMRF1 and EBER1 were the most abundant of all assessed transcripts at 2 hpi. Most transcripts increased rapidly within 2 to 4 hpi (Fig. 1B) such as those of the latent gene BYRF1 encoding EBNA2, an essential factor for B-cell transformation (27), that showed the strongest induction. In contrast, the transcripts of BNLF2a, BNLF1, LMP2A/B, and EBER1 had rather stable transcript levels up to 6 hpi, which increased subsequently (Fig. 1B, Lower).
Fig. 1.
Various EBV transcripts are present in B cells early after infection. Primary B cells were infected with B95.8 EBV and total RNA was prepared at different time points post infection. (A) qPCR with cDNA prepared from cellular RNA at 2 hpi revealed various EBV transcripts of the different indicated functional groups. When reverse transcription was omitted (striped bars), crossing-point values (Cp) were below background level or signals were not detectable (n.d.). Error bars indicate the SD of three replicates. (B) cDNA was prepared from samples at the indicated time points. qPCR was performed on different EBV transcripts as indicated and cellular glucuronidase beta (GUSB). Relative transcription levels were calculated and normalized to the value obtained 2 hpi. Student t test was performed to analyze the significances of differences (***P < 0.001,**P < 0.01, and *P < 0.05; n.s., not significant).
Although qPCR cannot discriminate between de novo transcription and potential transfer of RNAs by viral particles to B cells, these kinetic patterns suggested a biphasic process in which RNAs transferred by virions became detectable very rapidly after infection, followed by a second phase that was characterized by de novo transcription and gene expression.
EBV Particles Contain Viral RNAs.
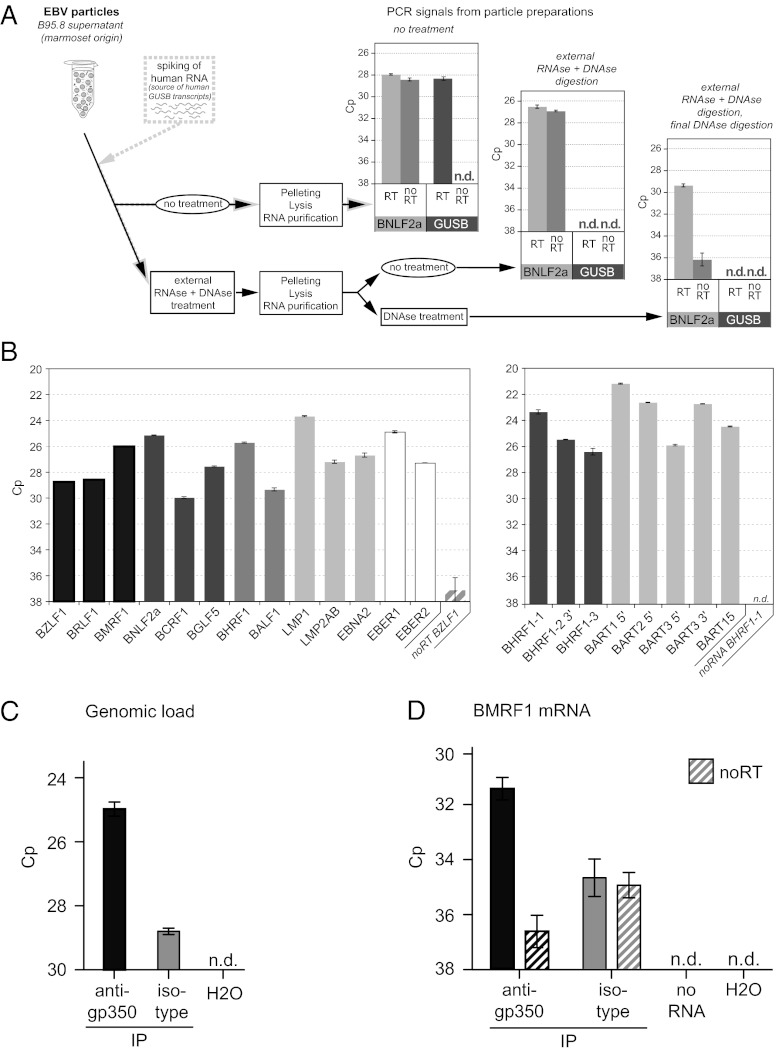
To assess whether EBV particles contain viral RNA molecules, we purified virions from supernatants of the B95.8 cell line, which spontaneously releases infectious EBV (28). The EBV transcript BNLF2a was readily detectable in particle preparations after reverse transcription and qPCR amplification (Fig. 2A). BNLF2a transcripts were still present after treatment of intact particles with DNase and RNase, and after treatment of lysed particles with DNase, confirming that EBV particles contain viral RNA molecules.
Fig. 2.
EBV particles contain viral mRNAs and microRNAs. (A) Concentrated EBV particles from B95.8 supernatants were spiked with cellular RNA derived from the EBV-negative human B-cell line BJAB. Samples were left untreated or treated with RNase and DNase to eliminate free nucleic acids. Subsequently, particles were pelleted and lysed, and RNA was extracted. After an additional DNase treatment when indicated, human GUSB and EBV BNLF2a RNAs were reverse-transcribed and the cDNAs amplified by qPCR. n.d., not detectable. (B) qRT-PCR revealed the presence of viral mRNAs assigned to different functional subgroups (Left) and microRNAs (Right) in B95.8 viral particles. Error bars indicate the SD of three replicates. (C and D) Particles from 2089 EBV stocks were precipitated with a gp350-specific antibody or an isotype control antibody and viral DNA and RNA were prepared. (C) The prevalence of viral genomes in the immunoprecipitated particles was quantified by PCR specific for the BMRF1 locus. (D) The prepared RNAs were reverse-transcribed by specific priming for the BMRF1 transcript followed by qPCR with BMRF1-specific primer pairs. Samples that were subjected to qPCR without prior reverse transcription are indicated (noRT). Reverse transcription of a sample without RNA template (noRNA) was performed to control for primer artifacts during PCR. n.d., not detected.
In the next step, we analyzed the virion RNA preparation for those transcripts we had studied in newly infected B cells (Fig. 1). Again, all transcripts were readily detectable. Transcripts of LMP1, EBER1, and BNLF2a were the most abundant RNAs (Fig. 2B, Left). EBV strain B95.8 codes for eight known pre-miRNAs giving rise to 13 mature viral miRNAs that are present in B cells early after EBV infection (29). We therefore asked whether B95.8 particles also contain viral miRNAs. To this end, we isolated small RNA species from virions and analyzed them by stem-loop PCR. As shown in Fig. 2B (Right), mature miRNAs were present in EBV particles at abundant levels.
We could also detect RNA species in virus-like particles (VLPs) released by the EBV packaging cell line TR−2/293 (Fig. S1) (30). This cell line harbors a recombinant mutant EBV genome that lacks the viral packaging signals [terminal repeats (TRs)]. Upon induction of the lytic cycle, TR–2/293 cells release infectious VLPs that are devoid of a viral DNA genome (31, 32). Taken together, B95.8 virions and TR–2/293 VLPs contained considerable amounts of viral RNAs including miRNAs.
EBV-infected cells release not only infectious virus particles but also exosomes that are less well characterized but also known to contain RNAs (24, 33). Infectious viral particles, VLPs, and exosomes have similar biophysical properties and therefore are difficult to separate properly. In case of EBV, the envelopes of virions and VLPs contain the viral glycoprotein 350 (gp350) that mediates binding of the virus to CD21 on B cells (21, 34 and refs. therein). We made use of this property. From 2089 EBV stocks (35), we selectively enriched virions and VLPs by immunoprecipitation with the gp350-specific antibody 72A1. The results of this experiment revealed that viral BMRF1 transcripts are contained within the gp350-enriched fraction (Fig. 2D) together with viral DNA (Fig. 2C), suggesting that virions, VLPs, and presumably gp350-positive exosomes that may also be present in virus stocks do indeed contain viral transcripts.
Collectively, our experiments showed that EBV particles and TR–2/293 VLPs contain considerable amounts of various viral RNAs. Because nothing was known about the role of tvRNAs in herpesvirus biology, we analyzed their immediate functions in newly infected B cells.
Viral mRNAs Are Transduced by EBV Particles and Translated Immediately Following Infection.
In a first series of experiments, we studied if packaged viral mRNAs are translated in B cells upon infection. We used an EBV-negative subclone of the Burkitt lymphoma cell line Daudi (36) that is readily infected by EBV to analyze the functional transfer and translation of virion RNAs.
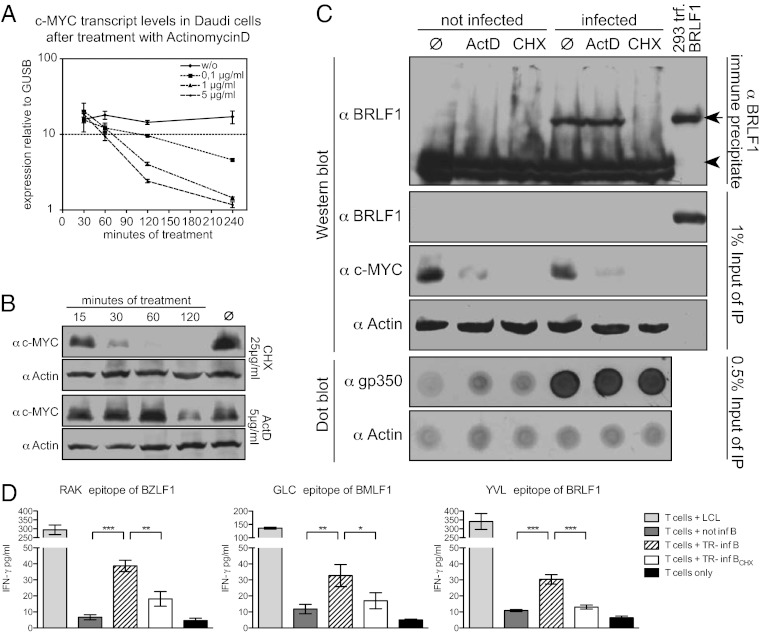
Initially, we established the required concentrations of the inhibitors actinomycin D (ActD) and cycloheximide (CHX) to fully block transcription and translation, respectively. In uninfected Daudi cells, ActD completely blocked de novo transcription at a concentration of 5 μg/mL, leading to a rapid decrease of c-myc mRNA levels (Fig. 3A) and consequently reduced c-Myc protein levels in a delayed manner (Fig. 3B). CHX at 25 μg/mL led to a reduction of c-Myc protein levels in accordance with its reported half-life of 15 min (37), indicating a complete block of translation (Fig. 3B).
Fig. 3.
Transduced virion RNAs are translated in newly infected cells. (A) Daudi cells were treated with different concentrations of ActD and mRNA levels of short-lived c-MYC and GUSB were measured by qRT-PCR. Shown are the levels of c-MYC mRNA in relation to GUSB at the indicated time points. (B) Inhibition of translation by CHX was assessed in Daudi cells. Cells were treated with ActD or CHX at final concentrations of 5 μg/mL or 25 μg/mL, respectively. c-MYC protein levels were measured by immunoblotting at the indicated time points. Actin served as a loading control. (C) Daudi cells were treated with ActD or CHX for 15 min or left untreated (Ø) and infected with B95.8 EBV or left uninfected in the continued presence of the respective inhibitor. Cells were lysed 2 hpi, BRLF1 protein was immunoprecipitated, and precipitates were analyzed by Western blot. Total cell lysates of 293 cells, transiently transfected with a BRLF1 expression plasmid, were included as positive control. Signals for BRLF1 (arrow) and the Ig heavy chain (arrowhead) are indicated. One percent of the IP-input lysate was used for Western blot analysis on BRLF1, c-MYC, and actin to control for BRLF1 enrichment during immunoprecipitation, successful inhibition of transcription or translation, and equal loading. A dot blot of input lysate against gp350 illustrates comparable infection of the samples; actin signals served as loading control. (D) CD8+ T-cell clones specific for lytic EBV epitopes recognize VLP-infected B cells. Primary B cells were infected overnight with TR–2/293 VLPs and, in parallel, treated with CHX or left untreated. Cells were washed and incubated with HLA-matched CD8+ T-cell clones specific for the RAK, GLC, or YVL epitopes derived from BZLF1, BMLF1, or BRLF1, respectively, at an effector-to-target ratio of 1:2 for 20 h. IFN-γ levels in the supernatants were assessed by ELISA.
Having established these conditions, we infected Daudi cells with B95.8 virus stocks in the presence or absence of ActD or CHX. After 15 min of pretreatment with inhibitors, cells were incubated with virus for 2 h in the continued presence of inhibitors. Thereafter, we immunoprecipitated BRLF1 protein from cell lysates and performed Western blot analyses. Similar amounts of BRLF1 protein were detected in lysates of untreated and ActD-treated Daudi cells (Fig. 3C). Treatment with CHX completely eliminated the BRLF1 signal, indicating that it originated from tvRNAs translated in the infected Daudi cells. The viral gp350, a structural component of EBV virions, served as a control for infection of Daudi cells (Fig. 3C).
We wished to functionally assess de novo translation of tvRNAs in an additional experimental model. Cells that synthesize proteins can present peptide epitopes derived from these proteins to CD8+ T cells. Antigen-specific CD8+ T-cell clones sensitively detect these peptides in conjunction with HLA class I molecules on the surface of protein-synthesizing cells.
We used CD8+ T-cell clones specific for the RAKFKQLL (RAK) peptide epitope of the BZLF1 protein, the GLCTLVAML (GLC) epitope of the BMLF1 protein, and the YVLDHLIVV (YVL) epitope of the BRLF1 protein (38). These EBV specific CD8+ T-cell clones were coincubated with primary, HLA-matched B cells that had been infected with purified TR–2/293 VLPs in the presence or absence of CHX. After 20 h of coincubation, we quantified the levels of IFN-γ in the supernatant of the cultures as an indicator of T-cell activation. All EBV-specific CD8+ T-cell clones recognized VLP-infected B cells on day 1 post infection (pi), documenting early antigen expression and epitope presentation. Treatment with CHX during infection completely abolished recognition (Fig. 3D). Thus, the three EBV proteins BRLF1, BZLF1, and BMLF1 are translated in newly infected cells from tvRNAs delivered by VLPs.
Transduced Viral RNAs Support Latent Infection of Primary Human B Lymphocytes.
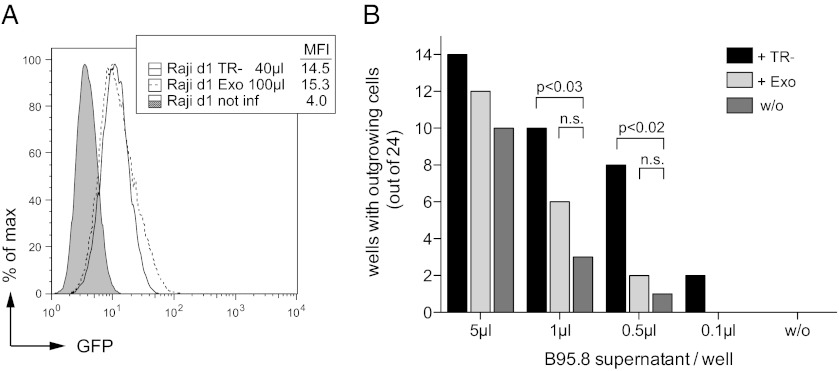
The transfer of virion RNAs could be beneficial to the success of EBV by contributing to critical rate-limiting steps during the initial phase of infection. The frequency of clonal outgrowth of human B cells infected with EBV in vitro at limiting dilution (39) is a quantitative measure of the potency of EBV to establish latent infection (9). We used limiting dilution assays to elucidate if virion RNAs supported latent infections of B cells with B95.8 EBV. We quantitatively compared the transformation capacity of B95.8 alone or in combination with VLPs derived from TR–2/293 cells as a source of tvRNAs. However, there was the possibility that, independent of tvRNAs, VLPs would contribute to B-cell activation and transformation by binding to the cell surface triggering the cellular receptor CD21 (40). To control for such effects, we used exosomes, i.e., subcellular vesicles that are spontaneously released by many cells. To produce exosomes with B-cell tropism that could be phenotypically traced, we transiently cotransfected HEK293 cells with two expression plasmids encoding gfp and the viral glycoprotein gp350 (41) and harvested and purified the exosomes from the cell supernatants.
First, we assessed and indirectly compared the concentration of gp350+/GFP+ exosomes and VLP preparations by flow cytometry (Fig. 4A). A pilot experiment shown in Fig. S2 revealed that GFP is a reliable surrogate marker because gp350+/GFP+ exosomes and VLPs derived from HEK293 cells and TR–2/293 cells, respectively, delivered comparable amounts of GFP and gp350 to Raji cells [TR–2/293 cells are a derivative of HEK293 cells (42)]. Next, we infected primary B cells in 96-well plates with serial dilutions of B95.8 EBV. We added adjusted, equal amounts of VLPs or gp350+/GFP+ exosomes to selected samples as indicated in Fig. 4B. The results documented that VLPs, but not gp350+/GFP+ exosomes, significantly supported the outgrowth of EBV-infected B cells in vitro (Fig. 4B and Fig. S3). The contribution of VLPs was especially pronounced when infectious EBV was limiting. This outcome suggested that tvRNAs directly improve EBV’s capacity to transform primary B cells and establish latent infections.
Fig. 4.
VLPs enhance the transformation potential of B95.8 virus stocks. (A) Raji cells were infected with different amounts of TR–2/293 VLPs (TR−) or gp350+/GFP+ exosomes (Exo) and analyzed for GFP and gp350 by flow cytometry (Fig. S2). (B) After appropriate adjustment of the amounts of gp350+/GFP+ exosomes and VLPs primary B cells were seeded into 96-well microwell plates and infected with serial dilutions of B95.8 supernatant. To investigate the effect of tvRNAs, TR−2/293 VLPs or gp350+/GFP+ exosomes were added to selected samples. After 6 wk of culture, numbers of wells with proliferating cells were determined by a viability assay. Significances of differences (P values) were calculated by Fisher exact test. n.s., not significant (P > 0.05). A single representative experiment is shown. Three independent experiments were performed with adenoid B cells from three different donors, which all showed the supportive effect of VLPs but not of gfp350+/GFP+ exosomes in a statistically significant manner in this experimental setting (Fig. S3).
Transduced Viral mRNAs Control a BZLF1-Responsive Gene Immediately After Infection.
Genetic data suggested that the early expression of BZLF1 has a critical role in driving the proliferation of resting B cells (8). However, tvRNAs were previously not considered as a source of BZLF1 protein immediately after infection and before de novo transcription. The transfer and concomitant translation of BZLF1 tvRNAs could also explain the immediate expression of lytic genes in newly infected B cells.
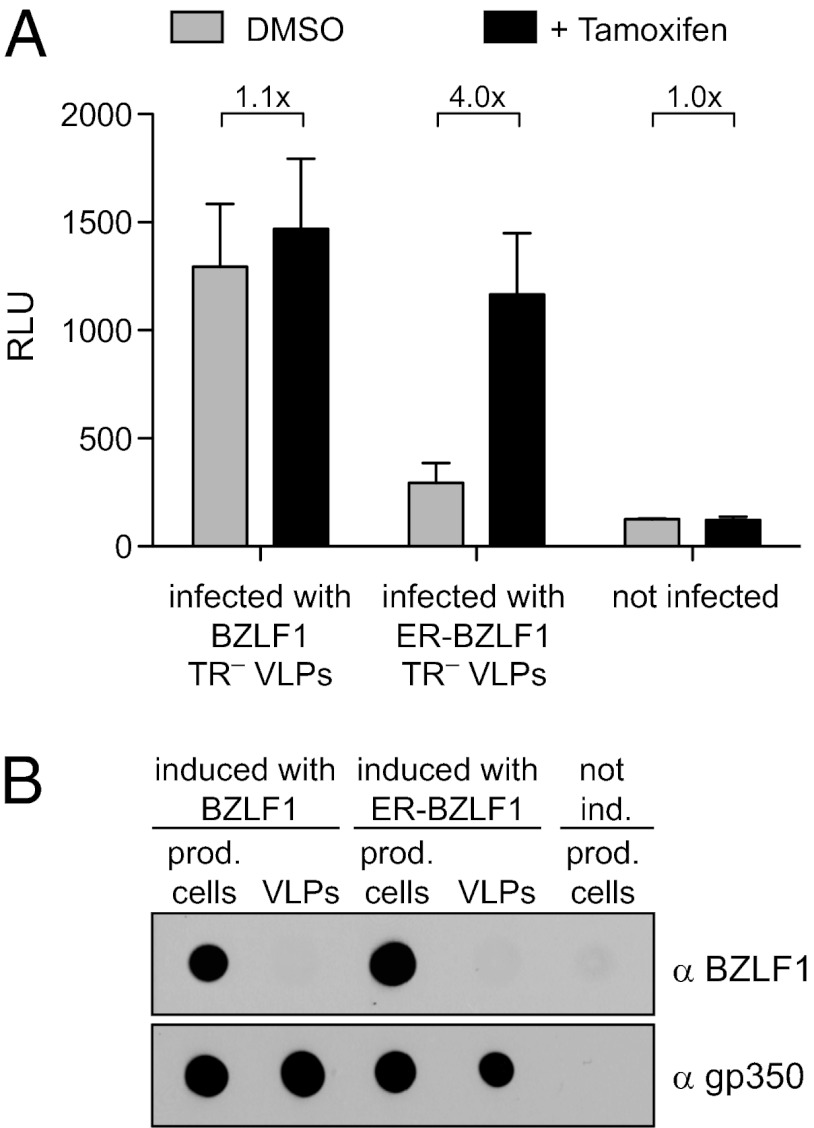
We wanted to investigate if VLP-derived BZLF1 transcripts regulate downstream genes and chose the BMRF1 gene as a representative example. This gene is prominently up-regulated early after infection (8) presumably because its promoter contains several BZLF1-responsive elements (43). EBV-negative Daudi cells with a BMRF1 promoter-driven luciferase gene served as reporter for the early expression of BZLF1. We prepared VLPs by transient transfection of TR–2/293 cells with an expression plasmid encoding WT BZLF1 or a fusion of the coding domains of BZLF1 and the hormone-binding domain of the modified estrogen receptor (BZLF1-ER). The function of the BZLF1-ER fusion protein depends on the addition of tamoxifen (44). We purified and concentrated VLPs and infected the Daudi reporter cells in the presence or absence of tamoxifen for 24 h (Fig. 5A). VLPs generated with WT BZLF1 clearly induced the activity of the BMRF1 promoter, but VLPs that were generated with BZLF1-ER and presumably transduced high levels of BZLF1-ER mRNA activated the BMRF1 promoter in the presence of tamoxifen only. The VLP preparations were free of BZLF1 protein as determined by dot blot analysis (Fig. 5B).
Fig. 5.
Transduced BZLF1 mRNA transactivates the early lytic BMRF1 promoter. TR–2/293 VLPs were generated by the transient transfection of expression plasmids encoding WT BZLF1 or BZLF1 fused to the modified estrogen receptor, BZLF1-ER, in the presence of tamoxifen and VLPs were isolated 3 d later. EBV-negative Daudi cells were stably transfected with a reporter plasmid, in which the luciferase gene is under the control of the BMRF1 promoter (Daudi/BMRF1/LUC). Daudi-reporter cells were cultured in the presence or absence of tamoxifen and infected with TR–2/293 VLPs generated with BZLF1 or BZLF1-ER in the presence or absence of tamoxifen. (A) Relative light units (RLU) were determined 24 h after infection and indicated reporter activity. Error bars indicate the SD of three experiments. (B) Dot blots with lysates of VLPs and producing cells were performed with BZLF1- and gp350-specific antibodies to exclude the transfer of BZLF1 protein to Daudi/BMRF1/LUC cells and as loading controls for lytically induced producer cells or VLP preparations.
We concluded from this experiment that VLP-transduced BZLF1 mRNAs (and, by implication, those contained in viral particles) suffice to transactivate the BMRF1 early lytic promoter. Our results in Fig. 3C show that another viral transcription factor, BRLF1, is also translated from tvRNAs, suggesting that several tvRNA-encoded viral transactivators likely trigger downstream EBV gene transcription, inducing the prelatent phase in newly EBV-infected B cells.
Packaged EBERs Induce IFN-α Production in Newly Infected Cells.
EBER1 and -2 are abundantly expressed in all cells latently infected with EBV (11). EBER RNA molecules fold into secondary structures with prominent stretches of dsRNA. Hence, EBERs are recognized as pathogen-associated molecular pattern by RIG-I in the cytoplasm (45) or TLR3 in endosomes (46) and activate downstream signaling pathways (47), leading to production of IFN-α (48). EBER1 and -2 are abundant in EBV virions (Fig. 2), suggesting that virion-carried EBERs might trigger their cellular receptors upon infection.
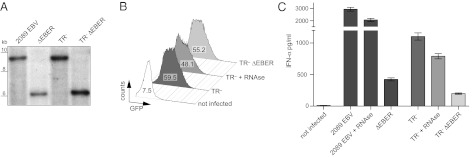
To address the effects of EBERs in B cells, we generated two EBV mutants in which both EBER genes were deleted. The EBV mutant ΔEBER is based on the recombinant wild-type 2089 EBV (35), the mutant ΔEBER TR–2/293 on the TR–2/293 EBV genome (30). Both mutant EBV genomes were cloned, stably introduced into HEK293 cells, and their genotypes were confirmed by Southern blot hybridizations (Fig. 6A). ΔEBER-carrying cells were used to generate virions with the mutant EBV genome, and ΔEBER TR–2/293-carrying cells produced VLPs devoid of EBV genomes. Both species, ΔEBER virions and ΔEBER TR–2/293 VLPs, contained viral RNAs except EBER1 and -2.
Fig. 6.
Transduced EBERs induce IFN-α production in infected B cells. (A) Genomic DNA of 2089 EBV, ΔEBER, TR–2/293, and TR–2/293 ΔEBER producer cells was cut with BamHI and BglII and analyzed by Southern blot hybridizations probing for oriP adjacent to the EBER genes, which confirmed their deletion in the respective samples. (B) VLPs were generated in TR–2/293 cells and TR–2/293 ΔEBER cells as described. Raji cells were infected with TR–2/293 VLPs, TR–2/293 VLPs treated with RNase, or TR–2/293 ΔEBER VLPs. The VLP-mediated GFP fluorescence revealed comparable amounts of VLPs in the supernatants. Numbers within the histograms indicate mean fluorescence intensities of GFP. (C) Primary B cells were infected at a multiplicity of infection of 0.1 with 2089 EBV or EBER-deficient recombinant EBV (ΔEBER), or infected with equal amounts of TR–2/293 VLPs or EBER-deficient TR–2/293 VLP (TR– ΔEBER). An RNase treated sample controlled for effects of free EBERs in the supernatant. IFN-α levels in the supernatant were determined by ELISA 3 d later. Error bars indicate the SD of three replicates.
We isolated primary B cells from adenoids and infected them with 2089 EBV or ΔEBER mutant EBV. Before infection, fractions of the virion preparations were treated with RNase to eliminate free EBER RNAs (46). Similarly, VLPs were prepared and purified from induced TR–2/293 and ΔEBER TR–2/293 cells and quantified by flow cytometry of Raji cells, which turn dim GFP-positive upon infection (21). The concentration of the VLP preparations were adjusted and reevaluated by flow cytometry as shown in Fig. 6B.
Three days after infection, we measured the IFN-α production of the infected cells. B cells infected with 2089 EBV released high concentrations of IFN-α, in contrast to cells infected with ΔEBER mutant EBV (Fig. 6C). Similarly, cells incubated with TR–2/293 derived VLPs produced high concentrations of IFN-α in contrast to cells incubated with ΔEBER TR–2/293 VLPs (Fig. 6C). Pretreatment of the supernatants with RNase slightly reduced IFN-α production, indicating the presence of free EBER molecules in the virus stocks and VLP preparations that had a minor effect on IFN-α release (Fig. 6C). Together, the results showed that the transfer of virion-contained EBER RNAs induced the production of IFN-α in newly infected B cells in the absence of de novo transcription of viral genes.
Packaged BNLF2a Transcripts Inhibit Recognition of Infected Cells by CD8+ T Cells.
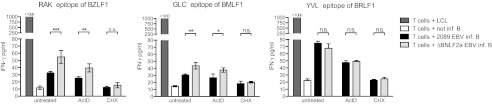
BNLF2a is an immunoevasin of EBV that inhibits loading of antigenic epitopes on HLA class I molecules (49). Thus, early expression of BNLF2a could dampen T-cell recognition of newly EBV-infected B cells. We detected BNLF2a mRNA in particle preparations and B cells immediately after infection (Fig. 1), supporting this hypothesis.
We generated a BNLF2a-negative EBV mutant and assessed the recognition of mutant and 2089 EBV-infected B cells by three different EBV epitope-specific CD8+ T-cell clones. B cells infected with the ΔBNLF2a mutant virus activated BZLF1-specific RAK CD8+ T cells significantly better than cells infected with 2089 EBV (Fig. 7, Left). Importantly, the improved recognition of the ΔBNLF2a mutant was maintained in the presence of ActD preventing de novo transcription but not translation of tvRNAs. Thus, transduced BNLF2a mRNA contributed to immunoevasion in WT EBV-infected cells. As expected, recognition was completely abrogated after CHX treatment. Thus, virion-mediated transfer of BNLF2a RNA interfered with the activation of RAK-specific CD8+ T cells, whereas a putative transfer of BNLF2a protein did not occur or was not sufficient for this effect.
Fig. 7.
Virally transduced BNLF2a mRNA inhibits the recognition of newly infected cells by EBV-specific CD8+ T-cell clones. Primary B cells were infected with 2089 EBV or ΔBNLF2a mutant EBV or were left uninfected. Selected samples were treated with ActD or CHX or left untreated and cocultivated with HLA-matched CD8+ T-cell clones specific for the RAK, GLC, or YVL epitopes for 20 h. IFN-γ levels in the supernatant were determined by ELISA. Error bars indicate the SD of three replicates. Significance was analyzed by Student t test (*P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001; n.s., not significant).
We obtained comparable results with the BMLF1-specific CD8+ T-cell clone GLC (Fig. 7, Middle). The BRLF1-specific CD8+ T-cell clone YVL is an interesting exception because this peptide is loaded onto HLA class I molecules independent of TAP (50). Accordingly, no difference was observed between 2089 EBV- and ΔBNLF2a EBV-infected B cells (Fig. 7, Right).
Our experiments demonstrate the immunological functions of tvRNAs: translation of virion-delivered BNLF2a mRNA contributes to immune evasion of newly infected B cells by interfering with the activation of antigen-specific CD8+ T cells.
Discussion
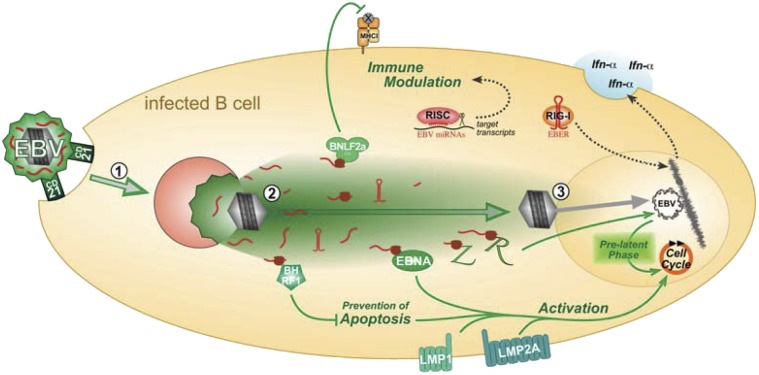
EBV establishes latent infection in its preferred target cells, human B lymphocytes, characterized by the expression of few viral genes only. Recent findings revealed that a stable latent infection is preceded by a prelatent phase, during which the virus expresses a subset of immediate-early and early genes together with latent genes (8, 9, 15, 51). The relevance of this transient prelatent phase is not completely understood, but findings indicate that it efficiently supports latent infection of quiescent B cells (8, 9).
Our data suggest that the prelatent phase is initiated by tvRNAs, which control critical processes upon infection before and independent of de novo transcription. We show that packaged viral mRNAs are instantaneously translated, and their products, as well as noncoding tvRNAs, induce viral and cellular genes and modify pathways related to innate and adaptive immune responses. Thus, tvRNAs can have an unexpectedly rich spectrum of different functions. Our study provides a basis for more extended explorations of this promising field. For instance, it seems plausible that translation of delivered BZLF1 transcripts activates resting cells and induces their cell-cycle entry (8, 52), translated BHRF1 and BALF1 transcripts might protect the infected cells from activation-induced cell death (9), and noncoding packaged RNAs like miRNAs might control detrimental antiviral responses of the infected cell. tvRNAs, which encode viral IL-10, could also shape the cellular microenvironment (53) and thereby protect the EBV-infected cell from antiviral responses of the innate and adaptive immune system. The coordinated action of tvRNA-encoded immunoevasins (viral IL-10, BGLF5, BNLF2a) could protect the newly infected cell from antigen-specific T-cell responses that might otherwise eliminate the newly infected cell before latency can be established. Hence, tvRNAs likely constitute a precisely timed stealth strategy to secure the initial success of EBV infection by blunting immunogenicity.
Several herpesviruses have been reported to deliver virion RNAs to infected cells (1, 2, 4, 5), but how the virion RNAs reach the cytosol and become translated (in the case of mRNAs), whether they interfere with translation and abrogate mRNA functions (in the case of miRNAs), or whether they activate cellular pattern recognition signaling (in the case of noncoding RNAs) awaits further investigations. Surprisingly little is known about the cellular entry of EBV. Adsorption of virions mediated by viral envelope glycoproteins and the cellular surface receptor CD21 (54) triggers viral endocytosis into clathrin-coated pits, where membrane fusion occurs at low pH (34, 55). Alternatively, viral envelopes could directly fuse with the plasma membrane in a pH-independent manner (56). Ultimately, the entry process delivers the uncoated virion, i.e., the capsid with its tegument, into the cytosol of the infected cell. The mode of entry protects virion RNAs, as the viral particles stay intact when they pass or bypass the endosomal–lysosomal pathway.
Entry of EBV does not go unnoticed by immune and pattern recognition receptors (PRR). Our experiments suggested that the noncoding EBERs constitute a considerable fraction of virion RNAs of EBV (Fig. 2), which contribute to the induction of the antiviral innate type I IFN, IFN-α (Fig. 6). Release of IFN-α triggered by EBERs has been described previously but in the context of free or protein-complexed RNA molecules, which, perhaps by endocytic uptake, activate TLR3 in EBV-transformed B cells and peripheral mononuclear cells (46). However, TLR3 appears not to be expressed in resting human B cells (57). As EBV’s mode of entry into B cells likely excludes the activation of endosomal PRRs, virion-transduced EBERs are presumably recognized by the cytoplasmic PRR RIG-I upon their delivery to the cytosol (45). Type I interferons are classically considered to be antiviral cytokines and modulate a range of immune responses, but, counterintuitively, they have been regarded important for a sustained coexistence of certain persistent viruses and their hosts (58). Along this line, EBERs curtail autocrine IFN-α signaling in EBV-infected cells. IFN-α induces the autophosphorylation of RNA-dependent protein kinase, which, in turn, phosphorylates the eukaryotic initiation factor 2α abrogating protein synthesis. By binding to RNA-dependent protein kinase and preventing its autophosphorylation, EBERs act as inhibitors of intracellular IFN-α signaling (59). EBERs are universally expressed in EBV’s different programs of cellular infection, suggesting a central function in the viral life cycle. It is therefore conceivable that EBV uses EBERs as a means to trigger selected PRR pathways to favor B-cell activation, proliferation, and viral latency (60, 61).
Latently EBV-infected cells spontaneously release exosomes that are gp350-positive and contain viral proteins or miRNAs (23–25). Like many enveloped viruses (22), EBV uses the exosome biogenesis pathway for egress during de novo virus synthesis, and virions, VLPs, and exosomes all contain characteristic polyproteins of this pathway (21). We thus propose that productively EBV-infected cells not only release complete virions but also a heterogeneous mixture of subviral particles including VLPs, exosomes, or other microvesicles that may also contain viral RNAs. The exact composition of particles released from productively EBV-infected cells (or cells that had been engineered to release VLPs) is currently unclear because virions, subviral particles, and nonviral exosomes have similar biophysical properties and many biochemical components in common. We postulate that these cells release virions, heterogeneous subviral particles like VLPs and gp350+ exosomes that all share the intrinsic potential to transduce viral RNAs to EBV’s target cells and together enhance the viral success of infection.
Taken together, our data provide strong arguments that packaged virion RNAs are of importance during the prelatent phase of EBV infection. We show that this virus selectively transduces viral RNAs to prime the cell for persistent infection and to prevent its elimination by the host’s immune system. Bridging the time gap from virus entry to de novo transcription of the viral DNA genome tvRNAs are a critical part of the biology of EBV and, presumably, of other herpesviruses as well. The exact in vivo role of tvRNAs remains to be elucidated.
Materials and Methods
Cell Culture.
All cells were maintained in RPMI medium with 10% FBS (PAA Laboratories) at 37 °C in a 5% (vol/vol) CO2 incubator.
Preparation of Virus, VLP, and Exosome Stocks.
The cell line B95.8 (28) was used to generate WT virus stocks. Recombinant 2089 EBV and TR–2/293 VLPs were produced as described (30, 35) by transient transfection of expression plasmids encoding BZLF1 (62) and BALF4 (63). Supernatants were harvested 3 d after transfection, cleared of cellular debris, and passed through a 0.8-μm nylon filter. Titers were calculated as “green Raji units” as described previously (9). Exosomes were harvested from supernatants of HEK293 cells transiently transfected with expression plasmids for gp350 and GFP on day 3 after transfection and treated like the recombinant virus stocks as described earlier. To determine particle concentrations in preparations of VLPs and gp350+/GFP+ exosomes, Raji cells were incubated with supernatants and analyzed by flow cytometry for GFP fluorescence after 1 d. When required, supernatants were concentrated by centrifugation at 100,000 × g for 2 h and resuspended in PBS solution.
Construction of Recombinant EBV.
The construction and genetic confirmation of EBER-1– and EBER-2–deficient EBV (ΔEBER) and VLPs (TR–2/293 ΔEBER) and of the BNLF2a-deficient EBV (ΔBNLF2a) is described in detail in SI Materials and Methods.
Isolation and Infection of Human Primary B Cells.
Surgically removed adenoids from anonymous donors were obtained from the department of otorhinolaryngology. The use of this material was approved by the local ethics committee. Single cell suspensions were prepared by dissecting the tissues and passing them through nylon mesh. T cells were rosetted with sheep erythrocytes (Life Technologies) and Ficoll gradient centrifugation enriched for mononuclear cells containing more than 95% B cells. B cells from peripheral blood were purified by using the MACS B-cell isolation kit II (Miltenyi Biotec). Infections were carried out overnight at 37 °C. For inhibition of transcription and translation, cells were treated with indicated concentrations of ActD (Merck) and CHX (Sigma), respectively, 15 min before infection, and treatment was maintained during infection.
Preparation of RNA, Reverse Transcription, and qPCR.
Total RNA and microRNA were extracted by standard procedures using the RNeasy Kit (Qiagen) and the mirVana Kit (Ambion), respectively. Details on RNA isolation from EBV particles and qPCR and primers are described in SI Materials and Methods (Tables S1 and S2).
Immunoprecipitation and Immunodetection.
Whole cell lysates from 2 × 108 Daudi cells were prepared in RIPA buffer with 0.2% SDS, protease inhibitors (Complete Mini; Roche), and DNase I. BRLF1 protein was immunoprecipitated with 15 μL Sepharose G beads (GE Healthcare) coupled to the BRLF1-specific antibody 5A9. Beads were washed in RIPA buffer, and bound protein was eluted in Laemmli buffer. Proteins were separated by SDS/PAGE and blotted to nitrocellulose (Millipore). One percent of the input lysate was used for control blots. Immunodetection was performed with antibodies against actin (I-19; Santa Cruz Biotechnology), BRLF1 (5A9), c-Myc (9E10), and gp350 (72A1). gp350+ particles were immunoprecipitated by incubating 2089 EBV stocks with biotinylated gp350-antibody 72A1 and bound to streptavidin-coupled Dynabeads (Invitrogen). Biotinylated isotype antibody (6A1) served as negative control. Beads were magnetically immobilized, extensively washed with PBS solution, and finally resuspended in TRIzol (Invitrogen). RNA and DNA were extracted according to the manufacturer’s recommendations.
T-Cell Recognition Assays.
EBV-specific CD8+ T-cell clones recognizing the epitopes HLA-B*0801/RAKFKQLL (RAK) from BZLF1 (64), HLA-A*0201/GLCTLVAML (GLC) from BMLF1 (38), and HLA-A*0201/YVLDHLIVV (YVL) from BRLF1 (65) were generated from peripheral blood of healthy, HLA-typed EBV carriers. PBMCs were stimulated overnight with antigenic peptides, and specifically reactive T cells were isolated on the following day by IFN-γ capture and immunomagnetic separation (Miltenyi Biotech) (66). Specific CD8+ T-cell clones were obtained from this enriched cell population by limiting dilution (67). For T-cell recognition assays, 1 × 104 clonal T cells were coincubated with 2 × 104 HLA-matched EBV-infected B cells in a total volume of 200 μL at 37 °C in V-shaped 96-well plates overnight. The assay was carried out in triplicates and supernatants of each well were analyzed for IFN-γ content by ELISA (Mabtech). Noninfected B cells and established HLA-matched lymphoblastoid cells lines were included as negative and positive control, respectively.
Limiting Dilution Assays.
B cells from adenoids were infected with serial dilutions of B95.8 supernatant and plated at a density of 105 cells per well in 24 replicates in 96-well microtiter plates. After incubation for 6 wk and weekly change of medium, wells with proliferating cells were quantified by a viability assay using 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (68). When indicated, selected samples were additionally treated with VLPs or equal amounts of gp350+/GFP+ exosomes that had been adjusted on Raji cells for equal GFP fluorescence and gp350 surface levels (Fig. S2). Controls included uninfected cells treated with VLPs or gp350+/GFP+ exosomes only.
Supplementary Material
Acknowledgments
We thank Dagmar Pich and Judith Dünzkofer for technical help, Elisabeth Kremmer for monoclonal antibodies, and Bill Sugden for the BZLF1-ER expression plasmid. This work was supported by institutional intramural grants; Deutsche Krebshilfe Grants 107277 and 107793; Deutsche Forschungsgemeinschaft Grants SPP1230, SFB455, SFBTR5, SFBTR36, and Ze 419/10-1; José-Carreras Leukämie-Stiftung Grant DJCLS R 09/17; and National Institutes of Health Grant CA70723.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. P.M.L. is a guest editor invited by the Editorial Board.
See Author Summary on page 7970 (volume 109, number 21).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1115906109/-/DCSupplemental.
References
- 1.Bechtel J, Grundhoff A, Ganem D. RNAs in the virion of Kaposi’s sarcoma-associated herpesvirus. J Virol. 2005;79:10138–10146. doi: 10.1128/JVI.79.16.10138-10146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bresnahan WA, Shenk T. A subset of viral transcripts packaged within human cytomegalovirus particles. Science. 2000;288:2373–2376. doi: 10.1126/science.288.5475.2373. [DOI] [PubMed] [Google Scholar]
- 3.Cliffe AR, Nash AA, Dutia BM. Selective uptake of small RNA molecules in the virion of murine gammaherpesvirus 68. J Virol. 2009;83:2321–2326. doi: 10.1128/JVI.02303-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greijer AE, Dekkers CA, Middeldorp JM. Human cytomegalovirus virions differentially incorporate viral and host cell RNA during the assembly process. J Virol. 2000;74:9078–9082. doi: 10.1128/jvi.74.19.9078-9082.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sciortino MT, Suzuki M, Taddeo B, Roizman B. RNAs extracted from herpes simplex virus 1 virions: apparent selectivity of viral but not cellular RNAs packaged in virions. J Virol. 2001;75:8105–8116. doi: 10.1128/JVI.75.17.8105-8116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 7.Countryman J, Miller G. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc Natl Acad Sci USA. 1985;82:4085–4089. doi: 10.1073/pnas.82.12.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalla M, Schmeinck A, Bergbauer M, Pich D, Hammerschmidt W. AP-1 homolog BZLF1 of Epstein-Barr virus has two essential functions dependent on the epigenetic state of the viral genome. Proc Natl Acad Sci USA. 2010;107:850–855. doi: 10.1073/pnas.0911948107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altmann M, Hammerschmidt W. Epstein-Barr virus provides a new paradigm: A requirement for the immediate inhibition of apoptosis. PLoS Biol. 2005;3:e404. doi: 10.1371/journal.pbio.0030404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nikitin PA, et al. An ATM/Chk2-mediated DNA damage-responsive signaling pathway suppresses Epstein-Barr virus transformation of primary human B cells. Cell Host Microbe. 2010;8:510–522. doi: 10.1016/j.chom.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rickinson A, Kieff E. Epstein–Barr virus. In: Knipe DM, Howley P, editors. Fields Virology. 5th Ed. Philadelphia: Lippincott Williams and Wilkins; 2007. pp. 2655–2700. [Google Scholar]
- 12.Allday MJ, Crawford DH, Griffin BE. Epstein-Barr virus latent gene expression during the initiation of B cell immortalization. J Gen Virol. 1989;70:1755–1764. doi: 10.1099/0022-1317-70-7-1755. [DOI] [PubMed] [Google Scholar]
- 13.Miyazaki I, Cheung RK, Dosch HM. Viral interleukin 10 is critical for the induction of B cell growth transformation by Epstein-Barr virus. J Exp Med. 1993;178:439–447. doi: 10.1084/jem.178.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeidler R, et al. Downregulation of TAP1 in B lymphocytes by cellular and Epstein-Barr virus-encoded interleukin-10. Blood. 1997;90:2390–2397. [PubMed] [Google Scholar]
- 15.Wen W, et al. Epstein-Barr virus BZLF1 gene, a switch from latency to lytic infection, is expressed as an immediate-early gene after primary infection of B lymphocytes. J Virol. 2007;81:1037–1042. doi: 10.1128/JVI.01416-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hislop AD, Taylor GS, Sauce D, Rickinson AB. Cellular responses to viral infection in humans: Lessons from Epstein-Barr virus. Annu Rev Immunol. 2007;25:587–617. doi: 10.1146/annurev.immunol.25.022106.141553. [DOI] [PubMed] [Google Scholar]
- 17.Rowe M, Zuo J. Immune responses to Epstein-Barr virus: Molecular interactions in the virus evasion of CD8+ T cell immunity. Microbes Infect. 2010;12:173–181. doi: 10.1016/j.micinf.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salek-Ardakani S, Arrand JR, Mackett M. Epstein-Barr virus encoded interleukin-10 inhibits HLA-class I, ICAM-1, and B7 expression on human monocytes: Implications for immune evasion by EBV. Virology. 2002;304:342–351. doi: 10.1006/viro.2002.1716. [DOI] [PubMed] [Google Scholar]
- 19.Fathallah I, et al. EBV latent membrane protein 1 is a negative regulator of TLR9. J Immunol. 2010;185:6439–6447. doi: 10.4049/jimmunol.0903459. [DOI] [PubMed] [Google Scholar]
- 20.van Gent M, et al. EBV lytic-phase protein BGLF5 contributes to TLR9 downregulation during productive infection. J Immunol. 2011;186:1694–1702. doi: 10.4049/jimmunol.0903120. [DOI] [PubMed] [Google Scholar]
- 21.Ruiss R, et al. A virus-like particle-based Epstein-Barr virus vaccine. J Virol. 2011;85:13105–13113. doi: 10.1128/JVI.05598-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Welsch S, Müller B, Kräusslich HG. More than one door - budding of enveloped viruses through cellular membranes. FEBS Lett. 2007;581:2089–2097. doi: 10.1016/j.febslet.2007.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meckes DG, Jr, et al. Human tumor virus utilizes exosomes for intercellular communication. Proc Natl Acad Sci USA. 2010;107:20370–20375. doi: 10.1073/pnas.1014194107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pegtel DM, et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci USA. 2010;107:6328–6333. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vallhov H, et al. Exosomes containing glycoprotein 350 released by EBV-transformed B cells selectively target B cells through CD21 and block EBV infection in vitro. J Immunol. 2011;186:73–82. doi: 10.4049/jimmunol.1001145. [DOI] [PubMed] [Google Scholar]
- 26.Cheung RK, Miyazaki I, Dosch HM. Unexpected patterns of Epstein-Barr virus gene expression during early stages of B cell transformation. Int Immunol. 1993;5:707–716. doi: 10.1093/intimm/5.7.707. [DOI] [PubMed] [Google Scholar]
- 27.Kieff E, Rickinson A. Epstein-Barr virus and its replication. In: Knipe DM, Howley P, editors. Fields Virology. 5th Ed. Philadelphia: Lippincott Williams and Wilkins; 2007. pp. 2604–2654. [Google Scholar]
- 28.Miller G, Shope T, Lisco H, Stitt D, Lipman M. Epstein-Barr virus: Transformation, cytopathic changes, and viral antigens in squirrel monkey and marmoset leukocytes. Proc Natl Acad Sci USA. 1972;69:383–387. doi: 10.1073/pnas.69.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seto E, et al. Micro RNAs of Epstein-Barr virus promote cell cycle progression and prevent apoptosis of primary human B cells. PLoS Pathog. 2010;6:e1001063. doi: 10.1371/journal.ppat.1001063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delecluse HJ, Pich D, Hilsendegen T, Baum C, Hammerschmidt W. A first-generation packaging cell line for Epstein-Barr virus-derived vectors. Proc Natl Acad Sci USA. 1999;96:5188–5193. doi: 10.1073/pnas.96.9.5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feederle R, Shannon-Lowe C, Baldwin G, Delecluse HJ. Defective infectious particles and rare packaged genomes produced by cells carrying terminal-repeat-negative Epstein-Barr virus. J Virol. 2005;79:7641–7647. doi: 10.1128/JVI.79.12.7641-7647.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hettich E, et al. Genetic design of an optimized packaging cell line for gene vectors transducing human B cells. Gene Ther. 2006;13:844–856. doi: 10.1038/sj.gt.3302714. [DOI] [PubMed] [Google Scholar]
- 33.Meckes DGJ, Jr, Raab-Traub N. Microvesicles and viral infection. J Virol. 2011;85:12844–12854. doi: 10.1128/JVI.05853-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanner J, Weis J, Fearon D, Whang Y, Kieff E. Epstein-Barr virus gp350/220 binding to the B lymphocyte C3d receptor mediates adsorption, capping, and endocytosis. Cell. 1987;50:203–213. doi: 10.1016/0092-8674(87)90216-9. [DOI] [PubMed] [Google Scholar]
- 35.Delecluse HJ, Hilsendegen T, Pich D, Zeidler R, Hammerschmidt W. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc Natl Acad Sci USA. 1998;95:8245–8250. doi: 10.1073/pnas.95.14.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nanbo A, Inoue K, Adachi-Takasawa K, Takada K. Epstein-Barr virus RNA confers resistance to interferon-alpha-induced apoptosis in Burkitt’s lymphoma. EMBO J. 2002;21:954–965. doi: 10.1093/emboj/21.5.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eick D, et al. Aberrant c-myc RNAs of Burkitt’s lymphoma cells have longer half-lives. EMBO J. 1985;4(13B):3717–3725. doi: 10.1002/j.1460-2075.1985.tb04140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steven NM, et al. Immediate early and early lytic cycle proteins are frequent targets of the Epstein-Barr virus-induced cytotoxic T cell response. J Exp Med. 1997;185:1605–1617. doi: 10.1084/jem.185.9.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frisan T, Levitsky V, Masucci M. Limiting dilution assay. Methods Mol Biol. 2001;174:213–216. doi: 10.1385/1-59259-227-9:213. [DOI] [PubMed] [Google Scholar]
- 40.Sinclair AJ, Farrell PJ. Host cell requirements for efficient infection of quiescent primary B lymphocytes by Epstein-Barr virus. J Virol. 1995;69:5461–5468. doi: 10.1128/jvi.69.9.5461-5468.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruiss R, Jochum S, Mocikat R, Hammerschmidt W, Zeidler R. EBV-gp350 confers B-cell tropism to tailored exosomes and is a neo-antigen in normal and malignant B cells—a new option for the treatment of B-CLL. PLoS ONE. 2011;6:e25294. doi: 10.1371/journal.pone.0025294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Graham FL, Smiley J, Russell WC, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 43.Bergbauer M, et al. CpG-methylation regulates a class of Epstein-Barr virus promoters. PLoS Pathog. 2010;6:e1001114. doi: 10.1371/journal.ppat.1001114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feil R, Wagner J, Metzger D, Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun. 1997;237:752–757. doi: 10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- 45.Samanta M, Iwakiri D, Kanda T, Imaizumi T, Takada K. EB virus-encoded RNAs are recognized by RIG-I and activate signaling to induce type I IFN. EMBO J. 2006;25:4207–4214. doi: 10.1038/sj.emboj.7601314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iwakiri D, et al. Epstein-Barr virus (EBV)-encoded small RNA is released from EBV-infected cells and activates signaling from Toll-like receptor 3. J Exp Med. 2009;206:2091–2099. doi: 10.1084/jem.20081761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kawai T, Akira S. Toll-like receptor and RIG-I-like receptor signaling. Ann N Y Acad Sci. 2008;1143:1–20. doi: 10.1196/annals.1443.020. [DOI] [PubMed] [Google Scholar]
- 48.Samanta M, Takada K. Modulation of innate immunity system by Epstein-Barr virus-encoded non-coding RNA and oncogenesis. Cancer Sci. 2010;101:29–35. doi: 10.1111/j.1349-7006.2009.01377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hislop AD, et al. A CD8+ T cell immune evasion protein specific to Epstein-Barr virus and its close relatives in Old World primates. J Exp Med. 2007;204:1863–1873. doi: 10.1084/jem.20070256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lautscham G, et al. Identification of a TAP-independent, immunoproteasome-dependent CD8+ T-cell epitope in Epstein-Barr virus latent membrane protein 2. J Virol. 2003;77:2757–2761. doi: 10.1128/JVI.77.4.2757-2761.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shannon-Lowe C, et al. Features distinguishing Epstein-Barr virus infections of epithelial cells and B cells: Viral genome expression, genome maintenance, and genome amplification. J Virol. 2009;83:7749–7760. doi: 10.1128/JVI.00108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo Q, et al. Transactivators Zta and Rta of Epstein-Barr virus promote G0/G1 to S transition in Raji cells: A novel relationship between lytic virus and cell cycle. Mol Immunol. 2010;47:1783–1792. doi: 10.1016/j.molimm.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 53.Suzuki T, et al. Viral interleukin 10 (IL-10), the human herpes virus 4 cellular IL-10 homologue, induces local anergy to allogeneic and syngeneic tumors. J Exp Med. 1995;182:477–486. doi: 10.1084/jem.182.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chandran B, Hutt-Fletcher L. Gammaherpesviruses entry and early events during infection. In: Arvin A, et al., editors. Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis. Cambridge, UK: Cambridge Univ Press; 2007. chap 23. [PubMed] [Google Scholar]
- 55.Nemerow GR, Cooper NR. Early events in the infection of human B lymphocytes by Epstein-Barr virus: The internalization process. Virology. 1984;132:186–198. doi: 10.1016/0042-6822(84)90102-8. [DOI] [PubMed] [Google Scholar]
- 56.Miller N, Hutt-Fletcher LM. Epstein-Barr virus enters B cells and epithelial cells by different routes. J Virol. 1992;66:3409–3414. doi: 10.1128/jvi.66.6.3409-3414.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Muzio M, et al. Differential expression and regulation of toll-like receptors (TLR) in human leukocytes: Selective expression of TLR3 in dendritic cells. J Immunol. 2000;164:5998–6004. doi: 10.4049/jimmunol.164.11.5998. [DOI] [PubMed] [Google Scholar]
- 58.García-Sastre A, Biron CA. Type 1 interferons and the virus-host relationship: A lesson in détente. Science. 2006;312:879–882. doi: 10.1126/science.1125676. [DOI] [PubMed] [Google Scholar]
- 59.McKenna SA, Lindhout DA, Shimoike T, Aitken CE, Puglisi JD. Viral dsRNA inhibitors prevent self-association and autophosphorylation of PKR. J Mol Biol. 2007;372:103–113. doi: 10.1016/j.jmb.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Iskra S, Kalla M, Delecluse HJ, Hammerschmidt W, Moosmann A. Toll-like receptor agonists synergistically increase proliferation and activation of B cells by Epstein-Barr virus. J Virol. 2010;84:3612–3623. doi: 10.1128/JVI.01400-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Iwakiri D, Takada K. Role of EBERs in the pathogenesis of EBV infection. Adv Cancer Res. 2010;107:119–136. doi: 10.1016/S0065-230X(10)07004-1. [DOI] [PubMed] [Google Scholar]
- 62.Hammerschmidt W, Sugden B. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell. 1988;55:427–433. doi: 10.1016/0092-8674(88)90028-1. [DOI] [PubMed] [Google Scholar]
- 63.Neuhierl B, Feederle R, Hammerschmidt W, Delecluse HJ. Glycoprotein gp110 of Epstein-Barr virus determines viral tropism and efficiency of infection. Proc Natl Acad Sci USA. 2002;99:15036–15041. doi: 10.1073/pnas.232381299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bogedain C, Wolf H, Modrow S, Stuber G, Jilg W. Specific cytotoxic T lymphocytes recognize the immediate-early transactivator Zta of Epstein-Barr virus. J Virol. 1995;69:4872–4879. doi: 10.1128/jvi.69.8.4872-4879.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saulquin X, et al. A global appraisal of immunodominant CD8 T cell responses to Epstein-Barr virus and cytomegalovirus by bulk screening. Eur J Immunol. 2000;30:2531–2539. doi: 10.1002/1521-4141(200009)30:9<2531::AID-IMMU2531>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 66.Moosmann A, et al. Effective and long-term control of EBV PTLD after transfer of peptide-selected T cells. Blood. 2010;115:2960–2970. doi: 10.1182/blood-2009-08-236356. [DOI] [PubMed] [Google Scholar]
- 67.Wiesner M, et al. Conditional immortalization of human B cells by CD40 ligation. PLoS ONE. 2008;3:e1464. doi: 10.1371/journal.pone.0001464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mosmann T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]