Abstract
The Sleeping Beauty (SB) transposon mutagenesis system is a powerful tool that facilitates the discovery of mutations that accelerate tumorigenesis. In this study, we sought to identify mutations that cooperate with MYC, one of the most commonly dysregulated genes in human malignancy. We performed a forward genetic screen with a mouse model of MYC-induced liver cancer using SB-mediated mutagenesis. We sequenced insertions in 63 liver tumor nodules and identified at least 16 genes/loci that contribute to accelerated tumor development. RNAi-mediated knockdown in a liver progenitor cell line further validate three of these genes, Ncoa2/Src-2, Zfx, and Dtnb, as tumor suppressors in liver cancer. Moreover, deletion of Ncoa2/Src-2 in mice predisposes to diethylnitrosamine-induced liver tumorigenesis. These findings reveal genes and pathways that functionally restrain MYC-mediated liver tumorigenesis and therefore may provide targets for cancer therapy.
Keywords: cancer gene identification, mouse models of cancer, Myc oncogene, hepatocellular carcinoma
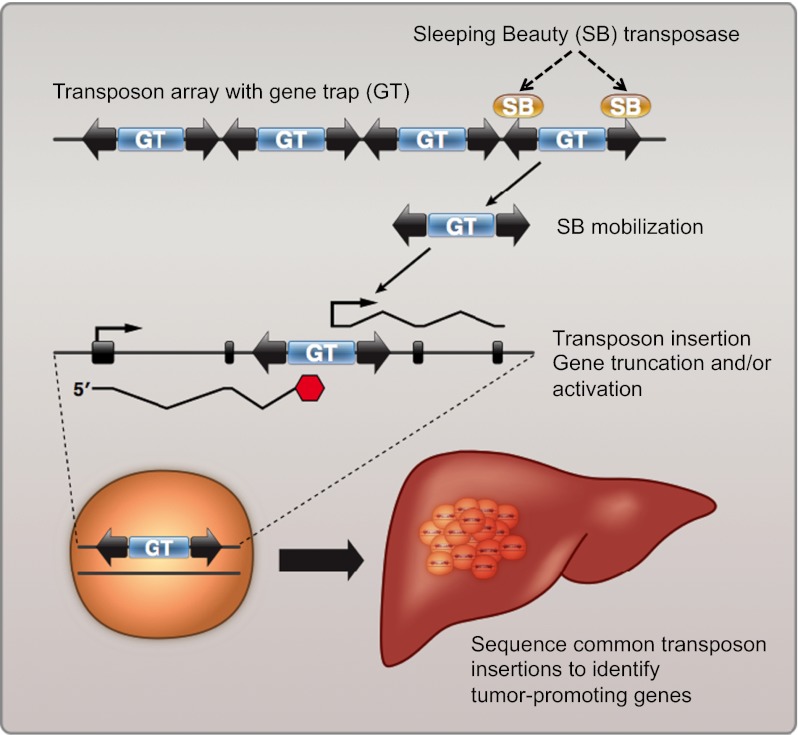
Transposable elements (TEs) are powerful genetic tools that are widely used in insertional mutagenesis screens because of the facile identification of transposon-induced mutations. The application of transposon-based approaches to cancer gene identification provides opportunities to study the consequences of specific mutations in the context of tumor cell initiation, progression, and maintenance in well-defined, genetically engineered mouse models. Sleeping Beauty (SB), a member of the Tc1/mariner superfamily of DNA transposons, is highly active in mammalian cells (1). A growing body of evidence has demonstrated that SB is an efficient tool for cancer gene discovery when used in forward genetic screens in mice (2, 3).
The SB system is based on the use of two transgenic mouse lines, one harboring a transposase and the other with a concatemerized mutagenic transposon containing sequences that can disrupt gene function through either gain-of-function or loss-of-function mechanisms. The transposase binds to the transposon ends and catalyzes its mobilization to new sites. The effect of ubiquitous transposition in mice is the development of T-cell leukemia and brain tumors (3, 4). In contrast, restricted expression of SB transposase accelerates fibrosarcoma development in p19Arf −/− mice (2).
Recently, this approach has been modified through the conditional activation of the SB transposase in specific tissues using Cre/LoxP technology (5, 6). In one of these studies, an SB screen identified 19 genes that accelerate liver cancer induced by expression of a dominant-negative Trp53 transgene. However, the SB system has not been used previously to identify genes that cooperate with oncogenes that initiate liver tumorigenesis.
Hepatocellular carcinoma (HCC) is the fifth most common solid tumor worldwide and is the third leading cause of death from cancer (7, 8). In the majority of cases, it occurs in a setting of chronic inflammation or cirrhosis. Although numerous genomic alterations have been documented in liver cancer, the key genetic alterations that drive hepatocellular transformation remain poorly understood. Nevertheless, a unifying feature of human HCC is amplification or overexpression of the MYC oncogene (8, 9). The need to identify genetic abnormalities and to understand better how they drive cellular transformation is underscored by the fact that HCC often is diagnosed at an advanced stage when no longer amenable to curative therapies. Thus, the identification of critical genes and pathways dysregulated in HCC may accelerate the development of therapeutic strategies.
We sought to identify mutations that cooperate with MYC, one of the most commonly dysregulated oncogenes in human malignancy. We bred mice containing the SB DNA transposon vector transgene and an SB transposase transgene to a well-characterized model of MYC-induced liver cancer (10–12). We hypothesized that mice harboring the active SB transposon would accumulate mutations that accelerate the rate of tumorigenesis in this sensitized liver tumor model. A cohort of quadruple-transgenic mice was generated, each containing the SB transposon harboring a gene trap element (T2/Onc), a ubiquitously expressed transposase (Rosa26-SB11), a tetracycline (tet)-repressible MYC transgene (tet-o-MYC), and a liver-specific tet-transactivator (LAPtTA) protein. Triple-transgenic mice lacking either the transposon or transposase served as a control cohort. We observed a significant acceleration of tumorigenesis in quadruple-transgenic mice compared with control animals. Sequencing of tumors from quadruple-transgenic animals identified at least 16 genes/loci associated with accelerated liver tumor development. Validation studies in mouse liver progenitor cells (13–15) confirmed that knockdown of selected genes promotes tumorigenesis in xenograft assays. Finally, we further characterized one of the genes, Nuclear receptor coactivator2 (Ncoa2/Src-2, hereafter referred to as “Ncoa2”), which facilitates transcription of Glucose 6 phosphatase (G6pc) and thus plays a critical role in mammalian glucose availability. We find that low levels of NCOA2 and G6PC expression in HCC patients are associated with poor survival. Moreover, deletion of Ncoa2 in mice promotes diethylnitrosamine (DEN)-induced liver tumorigenesis. These findings reveal genes that function to restrain MYC-induced liver tumorigenesis in multiple model systems, thereby revealing pathways that may be attractive targets for therapy.
Results
SB-Induced Mutations Accelerate Tumor Development in tet-o-MYC; LAPtTA Mice.
We used a previously described model of liver cancer in which mice harboring tet-o-MYC are crossed with mice expressing the tet-transactivator protein (tTA) driven by the liver-activator protein (LAP) promoter (10–12). Upon removal of doxycycline, double-transgenic animals express MYC specifically in the liver and subsequently develop liver tumors that resemble HCC. These mice served as a sensitized model of HCC.
Before proceeding with the transposon-mediated forward genetic screen, we performed a control experiment to determine tumor latency in the MYC liver model. The purpose of this experiment was twofold. First, the control experiment allowed us to distinguish early-developing tumors that form because of SB insertions from tumors that develop at baseline in tet-o-MYC; LAPtTA mice. Second, this experiment also provided a control for genetic background, because the SB mice are maintained on a mixed FVB/N and C57BL/6J background. We first crossed tet-o-MYC; LAPtTA mice, which are maintained on an FVB/N background, with C57BL/6J WT mice. Because of the positioning of the MYC transgene on the Y chromosome, only males are assessed in this model. MYC was induced when progeny reached 6 wk of age; mice then were dissected and scored for tumors each week from week 11 to week 22 (Fig. S1). Double-transgenic mice developed liver tumors no earlier than 13 wk on this mixed genetic background. Based on these results, we elected to analyze all mice before 16 wk of age, a time point at which liver tumor burden is low in tet-o-MYC; LAPtTA mice (Fig. S1).
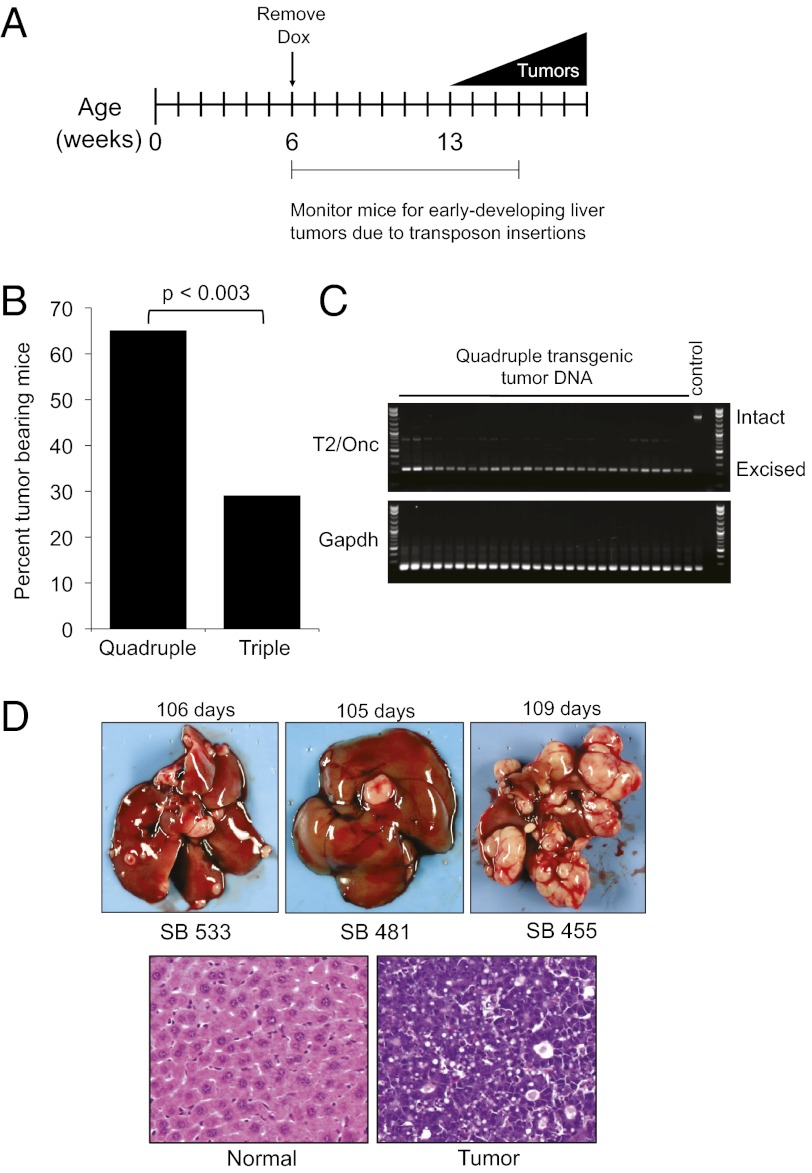
We bred mice containing the SB DNA transposon transgene to the MYC-induced liver tumor model. (The crossing scheme is depicted in Fig. S2A and the mutagenic transposon is depicted in Fig. S2B.)Transposase expression in liver had been demonstrated previously in Rosa26-SB11 mice (4). We hypothesized that mice harboring an active SB transposon would accumulate mutations that accelerate rates of liver tumorigenesis. An experimental cohort of quadruple-transgenic mice was generated, each containing the SB transposon harboring a mutagenic gene trap (T2/Onc), a ubiquitously expressed transposase (Rosa26-SB11), tet-o-MYC, and LAPtTA. We also generated a triple-transgenic control cohort lacking either the transposon or transposase. Doxycycline was withdrawn at 6 wk, and animals were monitored by palpation every 2 or 3 d for early-developing tumors. All animals were euthanized before 16 wk, by which time only 29% of triple-transgenic animals had developed liver tumors (Fig. 1 A and B and Tables S1 and S2). In contrast, 65% of the quadruple-transgenic mice developed liver tumors by this time point (P < 0.003) (Fig. 1B).
Fig. 1.
Acceleration of liver tumorigenesis in tet-o-Myc; LAPtTA mice with the SB transposon mutagenesis system. (A) Time-line of the SB mutagenesis screen. Dox, doxycycline. (B) Quantification of percentage of quadruple-transgenic (experimental) and triple-transgenic (control) animals that developed liver tumors at time of dissection. n = 37 quadruple-transgenic mice; n = 34 for triple-transgenic mice. A χ2 statistic was used for evaluating statistical significance. (C) Excision PCR assay demonstrating evidence of transposon (T2/Onc) excision in liver tumors from quadruple-transgenic animals. A T2/Onc (control) animal lacking the Rosa26-SB11 transposase is also shown. The intact donor T2/Onc amplicon is 2.2 kb, and the excised PCR amplicon is 225 bp. Gapdh serves as a control for equal loading of template gDNA. (D) (Upper) Representative images of multifocal liver tumors from quadruple-transgenic animals. (Lower) Histology of normal liver (Left) and tumor (Right) using H&E staining.
To confirm T2/Onc transgene concatemer mobilization, we performed a PCR excision assay on liver tumor genomic DNA (gDNA) isolated from quadruple-transgenic and control mice. We verified excision of the transgene in all liver tumors examined (Fig. 1C). In contrast, excision of T2/Onc was not observed in a triple-transgenic control animal lacking Rosa26-SB11 transposase (Fig. 1C). We dissected 63 liver tumors from 24 quadruple-transgenic animals (representative pictures of multifocal tumors are shown in Fig. 1D). The gross morphology as well as the age of onset of these tumors is distinct from those observed in Trp53−/− mice by Keng et al. (5). Histological analysis confirmed that these tumors resembled human HCC (Fig. 1D).
Identification of Common Insertion Sites in Liver Tumors.
To identify mutations that cooperate with MYC in the development of accelerated liver tumors, common insertion sites (CISs) were identified by sequencing tumors from quadruple-transgenic mice. We first performed ligation-mediated PCR with 63 liver tumor gDNA samples (Fig. S3) followed by 454 pyrosequencing. More than 90,000 individual sequencing reads were generated, of which 98.6% matched to barcodes that were incorporated into the ligation-mediated PCR. SB has a strong tendency to favor transposition to sites adjacent to the donor concatemer, a phenomenon termed “local hopping” (16). To control stringently for this bias, we initially excluded insertions that mapped to the same chromosome as the T2/Onc donor concatemer (chromosome 15). We further eliminated insertions that did not map to the canonical TA dinucleotide SB insertion site. We then merged all insertions that mapped to the same nucleotide from the same tumor and analyzed a total of 2,090 nonredundant insertions.
The CISs represent potential tumor-modifier genes and are defined by the presence of SB insertions at these sites significantly more frequently than would be predicted by random chance. Using Monte Carlo simulation analysis (5, 6), we defined a statistically significant CIS as a region in the genome with three or more SB insertions within a 30-kb window or four or more insertions within 155 kb. Initially, we identified 23 CISs that fit these criteria. Nine of these CISs occurred at the same nucleotide in different tumors from the same mouse and seven were eliminated from consideration as early-occurring passenger insertions less likely to be related to tumor development. Two of these CISs, Zfp189 and Bcl9, remain on the list, because they exhibited concordance with human tumor data. The remaining 14 genes/loci defined by CISs in MYC-induced liver tumors are shown in Table 1, along with two others discovered as outlined below.
Table 1.
Common insertion sites in early-developing liver tumors
| Chromosome | Gene Name | Gene Description | Range (bp) | No. of Tumors | No. of Independent insertion positions |
| 1 | Ncoa2 | Nuclear receptor coactivator 2 | 117102 | 7 | 7 |
| X | Zfx | Zinc finger protein X-linked | 34704 | 7 | 4 |
| 11 | Sfi1 | Sfi 1 homolog, spindle assembly associated | 51613 | 3 | 4 |
| 6 | Snd1 | Staphylococcal nuclease and tudor domain containing 1 | 166208 | 6 | 4 |
| 1 | AC102110.8 | Putative uncharacterized protein | 32748 | 3 | 3 |
| 1 | Rc3h1 | RING CCCH (C3H) domains 1 | 158362 | 3 | 3 |
| 10 | Ctnna3 | Catenin (cadherin associated protein), alpha 3 | 166868 | 5 | 7 |
| 3 | AC123647.9 | Uncharacterized protein | 103926 | 4 | 2 |
| 8 | AC171318.3 | Putative uncharacterized protein | 153010 | 4 | 2 |
| 8 | Atbf1/Zfhx3 | AT-motif binding factor 1 (Zinc finger homeobox 3 gene) | 161502 | 4 | 4 |
| 12 | Dtnb | Dystrobrevin, beta | 20080 | 2 | 3 |
| 2 | AL837506.3-202 | Uncharacterized protein | 4508 | 3 | 3 |
| 4 | Zfp189 | Zinc finger protein 189 | 34918 | 3 | 3 |
| 4 |
Dnajc6 |
DnaJ (Hsp40) homolog, subfamily C, member 6 |
13890 |
2 |
2 |
| 18 | Zfp608 | Zinc finger protein 608 | 1 | 3 | 1 |
| 3 | Bcl9 | B-cell CLL/lymphoma 9 | 1 | 4 | 1 |
| 15 | Nfe2 | Nuclear factor erythroid-derived 2 | 4708 | 7 | 7 |
| 15 | Ghr | Growth hormone receptor | 259059 | 7 | 7 |
Common insertion sites in liver tumors from quadruple-transgenic mice defined using Monte Carlo simulation analysis. Independent insertions refer to nonredundant insertions. The CISs below the black line represent genes with insertions at the same nucleotide in tumors from the same mouse or were located on chromosome 15. These CISs remain on the list because their expression showed concordance with HCC data.
Consistent with previous studies, we find that genome-level integration of SB with respect to genes and intergenic regions is fairly random overall (Fig. S4A) (17–19). As expected, we observe the greatest number of insertions clustering around the T2/Onc transgene concatemer on chromosome 15 (Fig. S4B). Upon closer inspection of transposon integrations on this chromosome, we identified two additional genes as potential CISs that initially were excluded. These CISs correspond to the Growth Hormone Receptor (Ghr) and Nuclear Factor (erythroid-derived 2) genes (Fig. S4B and Table 1). Because these genes are located many megabases from the donor transgene, these transposon insertions cannot be explained solely by local hopping. Moreover, GHR was implicated previously in human liver cancer (20).
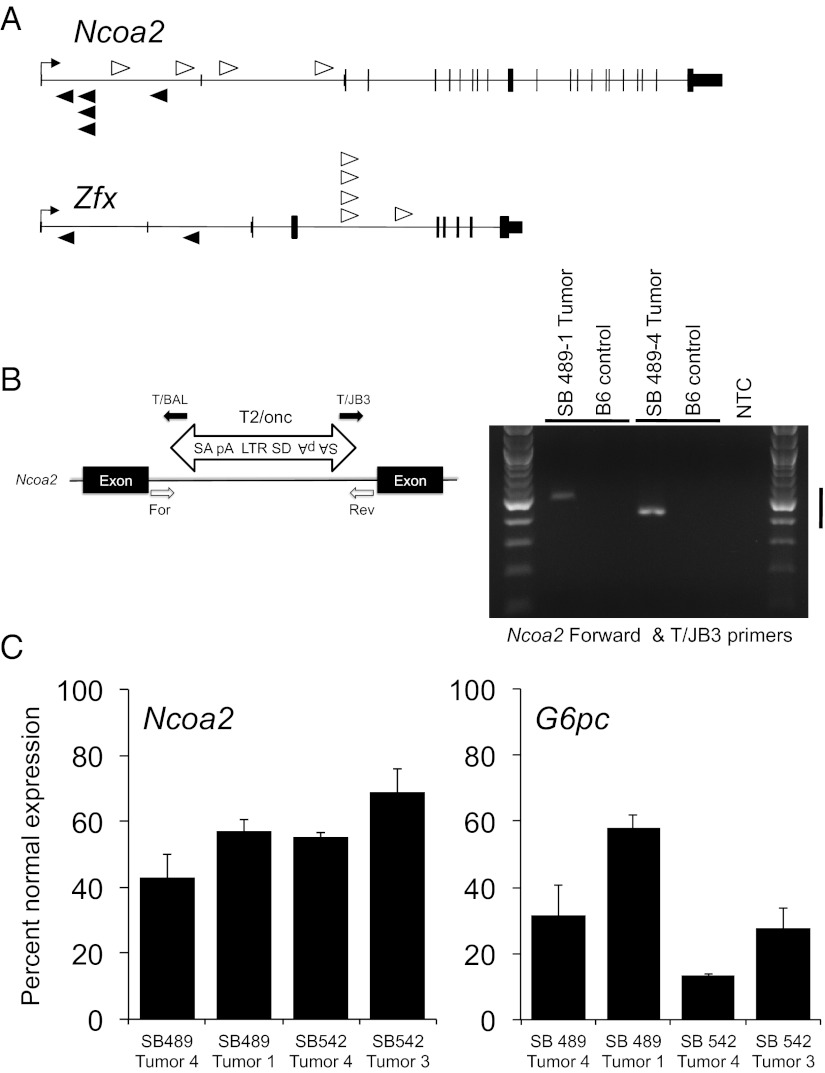
Insertions in candidate liver cancer genes identified in our screen are shown in Fig. 2A and Fig. S5A. Two examples are the Nuclear receptor coactivator2 (Ncoa2) and the Zinc finger transcription factor (Zfx). Insertions in these candidates are distributed randomly throughout the gene and occur in either a sense or antisense orientation. Thus, these insertions are likely to disrupt gene function. To confirm transposon integrations in Ncoa2, we performed PCR assays with a gene-specific primer and a T2/Onc primer. In each case, we observed the SB insertion in liver tumor gDNA but not in tail gDNA from a C57BL/6J wild-type animal (Fig. 2B).
Fig. 2.
Transposon insertions in Ncoa2 and Zfx associated with accelerated liver tumorigenesis. (A) Schematic representation of SB transposon insertions into Ncoa2 and Zfx. White arrowheads represent T2/Onc insertions in the sense orientation relative to target genes. Black arrowheads represent antisense-oriented insertions. The height of protein-coding exons is taller than the untranslated sequences for each gene. (B) Validation of SB transposon insertions in quadruple-transgenic liver tumors. PCR genotyping was performed using genomic DNA isolated from individual liver tumor nodules. For each insertion, one endogenous primer [forward (For) or reverse (Rev)] and one transposon primer (T/BAL or T/JB3) was used to validate the orientation of the transposon insertion. B6, control C57BL/6 tail gDNA; MSCV 5′LTR, long terminal repeat of the murine stem cell virus; pA, polyadenylation signal; SA, splice acceptor; SD, splice donor. (C) Real-time PCR quantitation of Ncoa2 and G6Pase mRNA expression in four liver tumors normalized to mRNA expression of the surrounding normal liver. Bar graphs represent mean Ncoa2 and G6Pase mRNA levels relative to Actin and 18S rRNA controls, respectively. Error bars represent SDs from three independent measurements.
It has been demonstrated previously that Ncoa2, a member of the p160 family of transcriptional coactivators, modulates expression of Glucose 6 phosphatase (encoding G6Pase) and other important genes by acting in concert with nuclear receptors such as RORα (21, 22). We measured mRNA expression levels of Ncoa2 and G6pc in four liver tumors with Ncoa2 insertions. Tumors with SB insertions exhibited a reduction of both Ncoa2 and its target G6pc mRNA levels (Fig. 2C). In contrast, tumors lacking SB insertions in Ncoa2 had normal or elevated expression of this gene (Fig. S6), demonstrating that Ncoa2 levels are not naturally lower in the cell of origin of HCC in this model.
SB Insertions in Lymphomas Are Distinct from Liver Tumor Insertions.
Twenty-nine percent of the quadruple-transgenic animals (11 of 37) developed lymphomas before 16 wk of age, whereas none of the triple-transgenic animals exhibited signs of hematopoietic disease. This result was not unexpected, because T2/Onc; Rosa26-SB11 mice are known to develop lymphoma and high-grade gliomas on a wild-type genetic background (4). Seven of these 11 quadruple-transgenic animals developed both liver tumors and lymphomas. To distinguish liver tumor-specific insertions from lymphoma-specific insertions in these seven mice, we performed ligation-mediated PCR with spleen gDNA and then sequenced SB insertions in these samples. Similar to our analysis with liver tumor SB insertions, we searched for CISs that occur at a greater frequency than would be predicted by random chance. We identified 11 genes defined as CISs in these lymphomas (Table S3). Despite the small number of lymphomas analyzed, two of the genes with SB insertions, Myb and Erg, had been found previously as CISs that contribute to leukemias/lymphomas in a larger cohort of 58 T2/Onc; Rosa26-SB11 animals (4). In contrast, none of the lymphoma CIS genes from the Collier et al. study (4) overlapped with liver CIS genes identified in the present study. Interestingly, the only CIS that overlapped between our lymphoma- and liver tumor-sequencing datasets was Ncoa2 (Fig. S5B). This finding underscores the potential significance of this gene in MYC-induced tumorigenesis in multiple settings. Consistent with this, we observed a robust increase in MYC protein levels in SB-induced lymphomas compared with normal spleen samples from control animals (Fig. S7). Although the source of elevated MYC in these tumors remains to be determined, it may derive from either the endogenous or transgenic loci.
Functional Validation of CIS Genes Implicates Ncoa2, Zfx, and Dtnb as Tumor Suppressors.
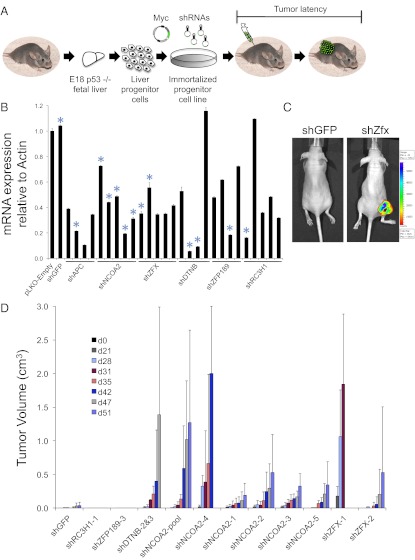
To validate functionally the tumor-suppressor activity of several of the CISs that we identified in our SB screen in an independent system, we used hepatoblasts isolated from Trp53−/− mice and immortalized by MYC. These cells do not form tumors in immunocompromised mice but provide a sensitized background in which one additional genetic alteration suffices to drive tumorigenesis (13–15). Furthermore, the Myc retrovirus used to immortalize these liver progenitor cells coexpresses GFP, allowing fluorescence imaging of subcutaneous tumors. We modified these cells through the introduction of shRNAs corresponding to candidate genes, followed by transplantation into recipient mice (Fig. 3A). Using this approach, we assess directly whether loss of function of candidate genes influences tumorigenesis and, by extension, whether these genes suppress tumor initiation. As a positive control, we knocked down the Wnt signaling pathway component Apc, a known tumor suppressor in this system (Fig. S8A). We confirmed that shRNAs targeting Apc in Trp53−/−; Myc hepatocytes gave rise to tumors within 1 mo after injection into nude mice (Fig. S8 B and C).
Fig. 3.
Functional validation of candidate genes using a mosaic mouse model. (A) Overview of validation experiment. Trp53−/− hepatoblasts were immortalized with a retrovirus that expresses Myc and GFP. Cells were infected with lentiviruses expressing shRNAs that target candidate CIS genes identified in the SB mutagenesis screen. After selection in puromycin, cells were injected subcutaneously into nude mice, and tumor volume was measured over time. (B) Real-time PCR quantitation of mRNA expression after knockdown of candidate CIS genes in Trp53−/−; Myc hepatoblasts. For every gene tested, expression of each target gene was normalized to pLKO-Empty–infected cells. Bar graphs represent mean expression levels relative to Actin. Error bars represent SDs from three independent measurements. Blue asterisks indicate shRNAs that were tested in vivo. (C) Fluorescence imaging of nude mice injected with liver progenitor cells expressing shRNAs corresponding to GFP (Left) and Zfx (Right). (D) Quantification of tumor volumes in nude mice injected with liver progenitor cells expressing candidate gene shRNAs. Bar graphs represent mean tumor volumes; error bars represent SDs from a total of four mice per shRNA tested. Independent tumorigenesis experiments yielded similar results.
We selected five genes from our SB screen to evaluate in this assay. We chose to focus on genes that had not been linked previously to liver cancer and in which the mouse shRNAs were readily available from the RNAi Consortium. We tested four or five different shRNAs per gene and chose the most potent shRNAs for our in vivo validation studies (Fig. 3B and Fig. S9 A and B). We then infected liver progenitor cells with lentiviruses expressing these shRNAs and assayed for tumor formation. Knockdown of Zfx, one of the top hits from our screen, promotes tumor formation of liver progenitor cells in nude mice (Fig. 3 C and D). We also validated Ncoa2 and β-Dystrobrevin (Dtnb) as putative tumor-suppressor genes using this approach (Fig. 3D). In contrast, the knockdown of two other genes represented by CISs, Zfp189, and Rc3h1, did not result in tumor formation in this assay.
Analysis of Candidate CISs in Human HCC.
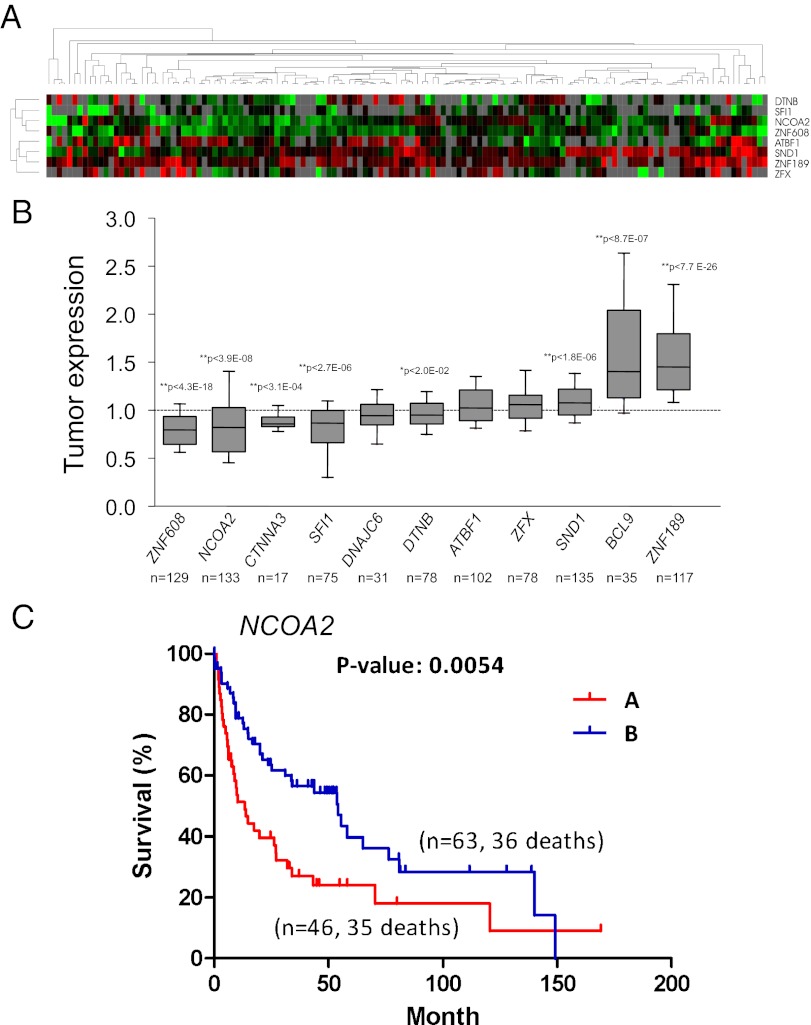
To determine the relevance of the CISs identified in our genetic screen to human liver cancer pathogenesis, we compared our SB hit list with an extensive mRNA expression profiling dataset from a large cohort of human liver tumor samples (23). We analyzed expression of 11 orthologs of the CIS genes identified in our screen in a cohort of 139 human HCC samples. Of these, five genes were expressed at statistically significantly lower levels in human tumors than in normal human liver tissue: ZNF608, NCOA2, CTNNA3, SFI1, and DTNB (Fig. 4 A and B). Three genes exhibited a statistically significant increase in expression in HCC samples: SND1, ZNF189, and Wnt pathway component BCL9 (Fig. 4 A and B).
Fig. 4.
Analysis of CIS candidates in human HCC samples. (A) Hierarchical cluster analysis of gene-expression data from 139 human HCC samples. A dendrogram (Upper) and heat map (Lower) of gene expression data were generated using eight orthologous genes from CIS candidates. Red and green cells reflect high and low expression levels, respectively; gray represents no significant information. (B) Quantification of tumor mRNA expression of CIS candidates relative to normal controls as previously described in Lee et al. (ref. 23). Each box represents the range of expression (25th to 75th percentile) observed. Error bars indicate the 10th and 90th percentiles, and the median is depicted by a horizontal line within the boxes. An independent one-sample t test was used to determine statistical significance. *P < 0.05; **P < 0.01. The P value and number of samples analyzed for each gene is shown. (C) Kaplan–Meier plots of overall survival of individuals with HCC clustered by low NCOA2 expression (cluster A) and higher NCOA2 expression (cluster B). P = 0.0054, log-rank test.
Furthermore, we observed an association between the expression of NCOA2 and its target G6PC and overall survival of HCC patients. Kaplan–Meier plot analysis and a log-rank test for overall survival in HCC based on the expression levels of these genes either independently or together revealed a statistically significant reduction in survival of HCC patients with tumors that exhibit low NCOA2 and/or G6PC expression (Fig. 4C and Fig. S10). These findings demonstrate that our transposon-mediated forward genetic screening approach identified clinically relevant genes that participate in the pathogenesis of human liver cancer.
We next assessed whether the NCOA2 gene accumulated sequence variants in a smaller panel of 14 human liver tumor samples. Paired normal liver biopsy material was available for a subset of these HCC samples. We sequenced all coding exons of NCOA2 and found one potential case of loss of heterozygosity specific to the tumor (Fig. S11A). Consistent with deletion of one allele of NCOA2 in this tumor, expression of NCOA2 was reduced by 50% in this sample relative to its paired normal liver (Fig. S11B). In contrast, NCOA2 expression was not decreased to the same extent in the other 13 liver tumor samples examined relative to matched normal controls (Fig. S11C). These results suggest that hemizygosity or other alterations affecting the NCOA2 gene may contribute to human liver tumorigenesis.
Loss of NCOA2 Enhances DEN-Induced Liver Tumorigenesis.
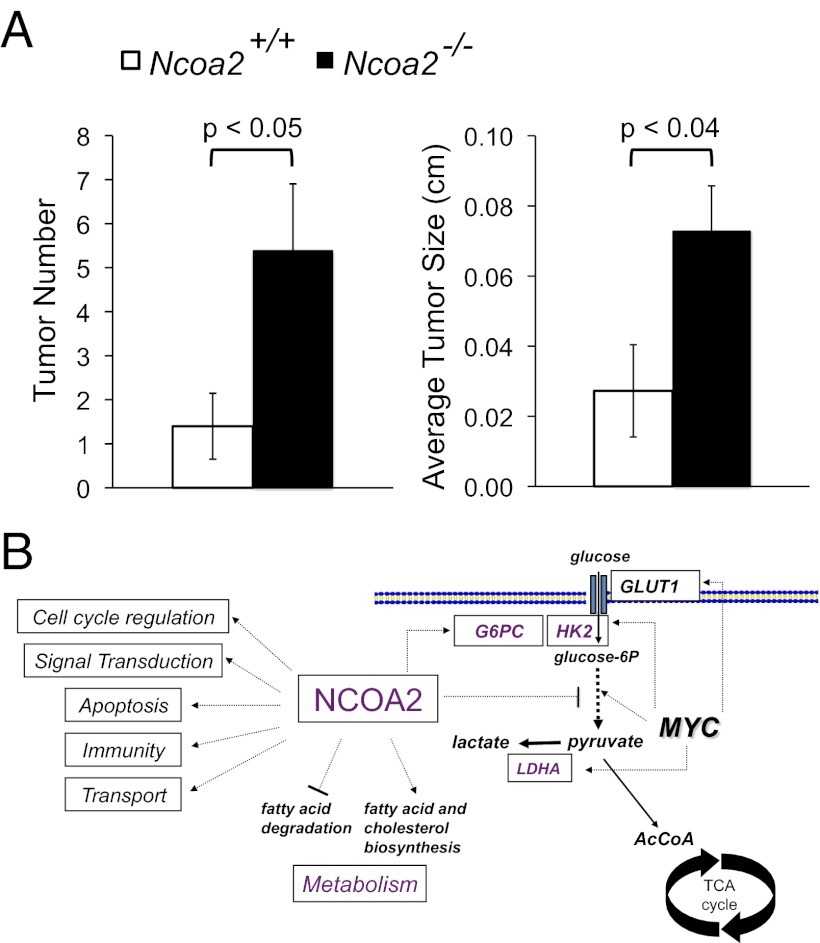
To test directly whether NCOA2 functions as a putative tumor suppressor in vivo, we treated previously described Ncoa2+/+ and Ncoa2−/− mice (24) with the chemical carcinogen DEN. Exposure to DEN causes liver damage and hepatocellular carcinogenesis in mice (25, 26). We chose to analyze males because DEN-induced tumorigenesis is known to be more effective in males than in females and because males were analyzed in the original transposon screen. Ncoa2+/+ and Ncoa2−/− littermate pups were injected with 25 mg/kg DEN on postnatal day 15 and then were euthanized at 3 or 6 mo postinjection. For each mouse, the liver was isolated, weighed, and photographed macroscopically (Fig. S12A and Table S4). DEN-treated livers were evaluated further to determine the total tumor number as well as the average and maximal tumor size. We observed that Ncoa2−/− male mice exhibited an approximately fourfold increase in tumor number and a statistically significant increase in maximal and average tumor size by 6 mo after DEN treatment (P < 0.05) (Fig. 5A and Fig. S12B). These findings demonstrate that genetic loss of NCOA2 promotes liver tumorigenesis in a mouse model that closely mimics human HCC pathogenesis.
Fig. 5.
Loss of Ncoa2 enhances DEN-induced liver tumorigenesis. (A) Quantification of liver tumor number and average liver tumor size 6 mo after DEN treatment in Ncoa2+/+ and Ncoa2−/− knockout mice. Tumor number, P = 0.04; average tumor size, P = 0.037. (B) Model for how loss of Ncoa2 may promote liver tumorigenesis in cooperation with Myc. The MYC oncogenic transcription factor reprograms metabolism in tumor cells by directly regulating genes involved in the glycolytic pathway. Loss of function of Ncoa2 may potentiate this pathway further by modulating expression of its targets, including G6pc and other genes involved in glycolysis, fatty acid degradation, and cholesterol biosynthesis. Gene-expression studies have demonstrated that NCOA2 has many additional targets, including genes involved in cell-cycle regulation, signal transduction, apoptosis, immunity, and transport, which also may contribute to tumorigenesis.
Discussion
Understanding molecular mechanisms that contribute to tumor development may provide therapeutic approaches for cancer therapy. Screens using transposons facilitate the discovery of genes and pathways contributing to tumor development in mice (5, 6, 27, 28). Thus, forward genetics provides a complementary approach to cancer genome-sequencing studies by illuminating the functional relevance of genes mutated in human tumors.
We used transgenic mouse lines that express a ubiquitous transposase and a mutagenic transposon present at low copy number to screen for insertions that accelerate MYC-induced HCC. We found a dramatic increase in the number of quadruple-transgenic animals that developed liver tumors compared with triple-transgenic controls that also expressed the tet-o-MYC and LAPtTA transgenes but lacked an active SB transposon. Sequencing of insertions from liver tumors identified at least 16 CISs/loci that contribute to accelerated tumor development.
Several of the CISs identified in our genetic screen have human orthologs dysregulated in human liver cancer. The genes ZNF608, NCOA2, CTNNA3, SFI1, and DTNB were all expressed at statistically significantly lower levels in human HCC samples. Our validation studies in p53−/−; Myc liver progenitor cells further demonstrated that inhibition of Ncoa2 and Dtnb can promote tumor formation in immunocompromised mice. DTNB, a member of the dystrobrevin protein family originally identified as components of the dystrophin–glycoprotein complex, is expressed in nonmuscle tissues including the liver, kidney, and brain (29). Dtnb may have previously unrecognized antitumorigenic activities. Interestingly, DTNB interacts with cAMP-dependent PKA and is proposed to play a scaffolding role in intracellular signal-transduction pathways (30). Atbf1, which encodes AT-motif binding factor-1 protein, also was identified in our screen. Also known as “zinc-finger homeobox 3” (ZFHX3), this protein has been shown previously to negatively regulate the expression of the liver tumor marker α-fetoprotein (AFP) in hepatocytes (31). Moreover, it was demonstrated previously that ATBF1 expression was significantly reduced in 55 of 76 HCC samples (32).
We also validated the tumor-suppressor activity of Zfx, encoding an X-linked pluripotency-associated zinc finger transcription factor that controls the self-renewal of embryonic and hematopoietic stem cells (33), as well as B-cell proliferation (34), in our functional studies. Although ZFX has not been linked previously to human liver tumors, and we did not observe altered expression of this gene in HCC samples, a potential link between the transcriptional networks coregulated by MYC and ZFX was discovered recently. A strong correlation in target gene occupancy was identified between these transcription factors (35). Transcriptional targets of these proteins are enriched for genes involved in the cell cycle, cell death, and cancer. Although future studies are necessary to elucidate the functional relationship between ZFX and MYC, previous findings and those reported here suggest that ZFX may directly oppose the activity of MYC at key transcriptional targets. Loss of ZFX may therefore remove an important negative regulator of MYC-mediated transactivation of oncogenic targets.
Genes with human orthologs that show increases in the level of mRNA expression in human liver tumor samples include SND1, ZNF189, and the Wnt pathway component BCL9. Dysregulation of the Wnt signaling pathway is a key molecular lesion in liver cancer (36–38). More than 60% of liver tumors exhibit accumulation of β-catenin, a hallmark of activated Wnt signaling. Interestingly, BCL9 was shown previously to be required for β-catenin–mediated transcriptional activity in human cancer cell lines (39). Recent findings suggest that BCL9 is an oncogene, because it has been shown to increase cell proliferation, migration, invasion, and metastatic potential of tumor cells (40). One additional gene identified in our screen, Ctnna3, also interacts with the Wnt pathway (41). Further analysis of all CIS genes using the COSMIC database analysis revealed the presence of additional mutations and/or genomic alterations that may be relevant to the pathogenesis of diverse human tumor types (Table S5). In light of the data presented here, these variants are worthy of further investigation.
An independent SB mutagenesis screen using a dominant-negative Trp53 transgene recently identified 19 genes associated with liver cancer (5). Interestingly, the only CIS gene in common between this study and the Keng study was Sfi1 (5). A meta-analysis of several SB screens indicates that this gene scores very highly in many such screens. Recent work suggests that this finding reflects an artifact caused by previously unrecognized amplification of this gene through segmental duplications and/or other mechanisms in the C57BL/6 mouse genome (42). Nevertheless, we found that SFI1 mRNA expression was down-regulated in a panel of 139 human HCC samples. Consistent with this result, Keng et al. (5) found distinct copy-number losses of SFI1 in 20 of 28 human HCC samples examined. SFI1 is a centrosomal protein with well-studied homologs present in yeast. Although SFI1 has been proposed to play a role in the formation of centrosome-associated contractile fibers (43), and the data presented here and in Keng et al. (ref. 5) are most consistent with a potential tumor-suppressive role for SFI1, additional studies are needed to determine the role of this gene in chromosome stability and to elucidate how its reduced expression may contribute to tumorigenesis.
Transposon insertions in Egfr were very common in liver tumors isolated from mice harboring a dominant-negative Trp53 transgene and a conditional SB transposase (5). In fact, SB insertions were detected most frequently in intron 24 of Egfr, giving rise to a truncated Egfr protein that has a strongly oncogenic effect. Using a PCR genotyping assay, we screened all liver tumors isolated in our genetic screen and did not find a single occurrence of SB insertions in this intron. Presumably, MYC overexpression performs a function that obviates selection for Egfr activation in this model, perhaps because EGFR signaling results in MYC up-regulation through the RAS–RAF–MEK–MAPK pathway (44, 45). Moreover, this finding underscores the importance of performing screens of this type using multiple sensitized models, because the most relevant pathways in a given tumor may depend on the initiating event that drives tumorigenesis.
Perhaps most intriguing, our findings provide evidence that the transcriptional coactivator NCOA2 acts as a tumor suppressor in liver cancer. Four lines of data support this concept. (i) Recurrent transposon insertions in SB-induced liver tumors result in decreased mRNA expression of both Ncoa2 and its previously characterized target G6pc. (ii) Inhibition of Ncoa2 using multiple independent shRNAs promotes tumor formation of liver progenitor cells in immunocompromised mice. (iii) DEN treatment of Ncoa2 −/− mice enhances liver tumorigenesis compared with Ncoa2+/+ littermates. (iv) Decreased expression of NCOA2 is observed in human liver tumor samples.
Previous studies of Ncoa2 illuminate our mechanistic understanding of its tumor-suppressor activity. Mice lacking Ncoa2 develop glycogen storage disease type 1 (Von Gierke’s disease) and exhibit decreased expression and enzymatic activity of G6pase (21). Similarly, a genetic deficiency of G6PC leads to Von Gierke’s disease both in humans and in mice (46, 47). Several observations demonstrate that loss of function of G6Pase also promotes liver tumorigenesis. For example, enzymatic activity of G6Pase is reduced in human and rodent liver tumors (48, 49), and G6pc−/− mice develop hepatic tumors (47). Furthermore, a significant fraction of patients with glycogen storage disease type 1 develop hepatic adenomas and have an additional risk of undergoing malignant transformation (50). Finally, inhibition of hexokinase, which has the equivalent functional effect as increasing G6Pase activity, is antitumorigenic (51, 52).
Based on these findings, loss of Ncoa2 may promote liver tumorigenesis at least in part through a subsequent reduction in G6Pase activity (Fig. 5B). This reduction, in turn, may increase the intracellular pool of glucose-6-phosphate that can enter the glycolytic pathway. Our data indicate that loss of Ncoa2 is particularly protumorigenic in settings of MYC hyperactivity, which is consistent with the known ability of MYC to stimulate glycolytic metabolism potently through the direct induction of expression of numerous genes in the glycolytic pathway (53–55). Despite the potential relevance of this mechanism to tumorigenesis, we were unable to detect expression of G6Pase in the hepatoblast cells, suggesting that NCOA2 tumor-suppressor activity is independent of G6Pase in the hepatoblast system. Although this finding does not rule out the potential significance of G6Pase as a relevant target of NCOA2 in vivo, we acknowledge that mechanisms of NCOA2-mediated tumor suppression are complex and likely involve multiple targets and pathways. Indeed, a previously reported microarray analysis of gene expression in livers of Ncoa2−/− mice revealed a signature of increased energy expenditure including the stimulation of genes involved in fatty acid degradation and suppression of genes involved in cholesterol and fatty acid biosynthesis (22). Thus, Ncoa2 deficiency likely contributes to a broader protumorigenic reprogramming of metabolism beyond the glycolytic pathway. Moreover, loss of function of Ncoa2 influences gene-expression programs relevant to other important aspects of tumor biology including cell-cycle regulation, signal transduction, and apoptosis. Therefore future studies are warranted to further dissect the mechanism(s) by which dysregulated expression of NCOA2 contributes to tumorigenesis.
Methods
Transgenic Animals.
LAPtTA and tet-o-MYC mice were obtained from Dean Felsher (Stanford School of Medicine, Stanford, CA). T2/Onc transgenic mice with the donor concatemer on chromosome 15 were bred with LAPtTA mice to obtain T2/Onc; LAPtTA animals. Simultaneously, Rosa26-SB11 females were bred to tet-o-MYC males to obtain Rosa26-SB11; tet-o-MYC mice. The two double-transgenic lines were first intercrossed with littermates to produce homozygous transgenes when possible. In the final cross, LAPtTA; T2/Onc females were bred with Rosa26-SB11; tet-o-MYC males to obtain quadruple (experimental) or triple (control) transgenic cohorts. The MYC transgene is on chromosome Y, precluding analysis of females. The Johns Hopkins Animal Care and Use Committee approved all procedures described in this work.
PCR Genotyping.
Genomic DNA was isolated from tail clippings using the DNeasy Blood and Tissue Kit (Qiagen) according to the manufacturer’s instructions. PCR genotyping was performed using 10–100 ng of genomic DNA as template. Primers used were tet-o-MYC, forward 5′-TAGTGAACCGTCAGATCGCCTG-3′ and reverse 5′-TTTGATGAAGGTCTCGTCGTCC-3′ (amplicon 500 bp); LAPtTA, forward 5′-GCTGCTTAATGAGGTCGG-3′ and reverse 5′-CTCTGCACCTTGGTGATC-3′ (amplicon 500 bp); T2/Onc, forward 5′-CCTTGCAAAATGGCGTTACT-3′ and reverse 5′-GCCGCACTGGTTGTTAGCAA-3′ (amplicon 200 bp); Rosa26-SB11 WT, forward 5′-CTGTTTTGGAGGCAGGAA-3′, reverse 5′-CCCCAGATGACTACCTATCCTCCC-3′ (amplicon 420 bp); Rosa26-SB11 SB reverse 5′-CTAAAAGGCCTATCACAAAC-3′ (SB knock in amplicon 317 bp ); Hprt, forward 5′-CTTTCTTGTCACTCCCACTTTTCC-3′ and reverse 5′-CACATCCTTCATTCAGGTGTCACT-3′ (amplicon 340 bp). PCR conditions for Ex Taq (Takara) were used according to the manufacturer’s instructions. PCR products were separated on a 1.2% agarose gel.
Liver Tumor Analysis.
The whole liver was dissected from the euthanized animal, washed, and placed in ice-cold PBS. The number of liver tumor nodules was counted for each animal. Tumor nodules then were dissected away from surrounding normal liver tissue. When large enough, tumor nodules were split into three pieces for gDNA isolation, RNA isolation, and histological analysis. DNA isolation was performed using the All-in-One Purification Kit (Norgen) or by phenol-chloroform extraction. Total RNA was isolated from tumor and normal tissue using TRIzol (Invitrogen) followed by additional cleanup and DNase digestion using the RNeasy kit (Qiagen) according to the manufacturers’ protocols. For histological analysis, tissues were fixed in 10% formalin, embedded in paraffin, and sectioned. Tissue section slides then were stained with H&E using standard protocols.
SB Excision Assay.
To confirm excision of the T2/Onc transgene from the chromosome 15 concatemer, we used the following forward 5′-TGTGCTGCAAGGCGATTA-3′ and reverse 5′-ACCATGATTACGCCAAGC-3′ primers (amplicon 233 bp); Gapdh, forward 5′-GGAGCCAAACGGGTCATCATCTC-3′ and reverse 5′-GAGGGGCCATCCACAGTCTTCT-3′. PCR conditions for Ex Taq (Takara) were used according to the manufacturer’s instructions. PCR products were separated on a 1.2% agarose gel.
Ncoa2 Insertion Validation Assay.
For each insertion that was validated, 1 kb of sequence surrounding each insertion position was downloaded (500 bp on each side of the insertion), and an ∼800-bp amplicon was designed. To confirm the orientation of the SB insertion, different combinations of Ncoa2 and T2/Onc primers were used in the PCR. Primers used to confirm SB insertion at position 13272606 were forward 5′-CACACACACACACAGCAGTAGTGAAGCA-3′ and reverse 5′-CGCCAGAAAACTGATTTTGAAGTTGCAG-3′; to confirm the SB insertion at position 13300371, the primers used were forward 5′-GAGCGATGGTTTCTACTCCCACAGACCT-3′ and reverse 5′- CTGTACCGTGGTGAGCATTGGGAGTAAT-3′.
Sequence Processing and Annotation.
The 47,343 initial 454 sequencing reads were processed and analyzed using a custom semiautomated processing pipeline first described in ref. 6. Briefly, Fasta-formatted sequences from 454 runs were assigned to libraries allowing one mismatch in the 10-bp barcode. All raw sequences were scanned for inverted repeat/direct repeat (IRDR) and linker recognition sequences using EMBOSS Vectorstrip (56) with successively less stringent parameters (10%, 15%, and finally 20% mismatches allowed) until the maximum number of constructs (i.e., IRDR: GTGTATGTAAACTTCCGACTTCAACTG and linker sequence: GTCCCTTAAGCGGAGCCCT) were recognized and trimmed off. Only 44,294 sequences that had a matched IRDR with an insert sequence of at least 16 bp were carried further for mapping to the mouse genome. These trimmed insert sequences were mapped to the mouse genome (National Center for Biotechnology Information build 37) using BLASTN (DeCypher's TeraBLASTN; Active Motif), requiring query sequences to align within 1 bp of the end of the LTR sequence that was trimmed. Additionally, the query sequence was required to match with at least 95% identity. Ambiguous sequences that mapped to multiple genomic loci were removed, leaving 2,090 uniquely mappable nonredundant insertions. Unambiguously mapped nonredundant insertions were assigned to clusters of CISs if the local density of insertions in a given window size exceeded the density that would be expected by chance, as determined by Monte Carlo simulation. A CIS is defined as statistically significant if a region in the genome has three or more SB insertions with a 30-kb window or four or more insertions within 155 kb. CISs that occurred at the same nucleotide in different tumors from the same mouse were removed with the exceptions of Zfp608 and Bcl9. Nonredundant insertion positions were annotated using the EnsEMBL API (57) with the name of the gene whose start site was closest to the position. CIS positions were annotated similarly using the median insertion position within the CIS as a reference point.
Bioinformatic Analysis of SB Insertions.
To analyze mouse genomic features that harbor SB insertions, additional computational analysis was performed using the Python programming language (www.python.org) and the R statistical environment (http://www.R-project.org/).
Cell Culture.
HEK293T and Trp53 −/−; Myc cells were cultured in high-glucose (4.5 g/L) DMEM supplemented with 10% FBS, penicillin, and streptomycin.
Lentiviral Infection of PHM Cells.
HEK293T (1 × 106) cells were cotransfected with pLKO shRNA constructs, Gag/Pol, VSV-G, and Rev helper plasmids using the FuGene6 reagent (Roche). Following transfection, the lentiviral supernatant was collected, filtered, and supplemented with 8μg/mL hexadimethrene bromide (Sigma). Trp53−/−; Myc cells (1 × 105) were infected overnight twice and, 24 h after the second infection, were plated into medium containing 1.5 μg/mL puromycin and selected for at least 7 d. Cells then were injected into nude mice and harvested for RNA analysis. The following TRC clone IDs were used for knockdown experiments: Apc: (B4) TRCN0000042533, (B5) TRCN0000042534, (B6) TRCN0000042535, (B7) TRCN0000042536; Ncoa2: (i) TRCN0000096169, (ii) TRCN0000096170, (iii) TRCN0000096171, (iv) TRCN0000096172, (v) TRCN0000096173; Zfx: (i) TRCN0000095620, (ii) TRCN0000095619, (iii) TRCN0000095621, (iv) TRCN0000095622, (v) TRCN0000095623; Dtnb: (i) TRCN0000108760, (ii) TRCN0000108761, (iii) TRCN0000108762, (iv), TRCN0000108764; Zpf189: (i) TRCN00000082103, (ii) TRCN0000082105, (iii) TRCN0000082106, (iv) TRCN0000082107; Rc3h1: (i) TRCN0000200305; (ii) TRCN0000177880, (iii) TRCN0000198362, (iv) TRCN0000200175, (v) TRCN0000177150.
RT-PCR Analysis of CIS Genes.
All quantitative real-time PCR analysis was performed using the Step One Plus Real Time PCR system (Applied Biosystems). Analysis of Ncoa2 in SB liver tumors was performed using a predeveloped TaqMan assay with One-Step RT-PCR Master Mix Reagents (Applied Biosystems) or by SYBR green analysis of cDNA samples using primers that cross an exon–exon junction. The Ncoa2 TaqMan assay was performed with Mm00500749_m1 (Applied Biosystems). Primers used for SYBR green were forward 5′-TCACTGCATTGGCTCTTCTG-3′ and reverse 5′-TGTTTTCTCCCATCCCACTC-3′. Analysis of G6Pase mRNA in SB liver tumors was performed with a TaqMan assay using the Universal Probe #26 (Roche) and forward 5′-GAGGAAAGAAAAAGCCAACGTA-3′ and reverse 5′-GGGACAGACAGACGTTCAGC-3′ primers. Expression was normalized to 18SrRNA with the Ribosomal RNA control assay (Applied Biosystems) or Actin. All PCR assays were performed in triplicate.
To assess the efficiency of shRNAs in Trp53−/−; Myc cells, RNA was isolated from cell pellets using the RNeasy Kit (Qiagen), and 2 μg total RNA was reverse transcribed using SuperScript III First Strand (Invitrogen) according to the manufacturers’ protocols. SYBR green analysis was performed with cDNA samples using primers that cross exon–exon junctions for each gene. Primers used for Apc were forward 5′-AAGACGGTATGCTGGAATGG-3′ and reverse 5′-AGTGCTCTCATGCAGCCTTT-3′; Ncoa2 forward 5′-TGGACATGATCAAGCAGGAG-3′ and reverse 5′-CCCAGTGGGTTGAAGAAAGA-3′; Zfx forward 5′-CGGCAGACTGGCTAAACAA-3′ and reverse 5′-CCACAAATCATGCAAGGGTA-3′; Dtnb forward 5′-CATGTGTGGTGGAAAAATGC-3′ and reverse 5′-CCTTCAGGAACTGGTCAAGC-3′; Zfp189 forward 5′-GGCTGCTATTCCTGAACGAG-3′ and reverse 5′-AGGCTTCTCGGAGCTGGAT-3′; Rc3h1 forward 5′-GCAGCTGAACCATCAGATCA-3′ and reverse 5′-GGGTGTTCACCTCCCTCTGT-3′; Actin forward 5′-CGGTTCCGATGCCCTGAGGCTCTT-3′ and reverse 5′-CGTCACACTTCATGATGGAATTGA-3′. All SYBR green PCR assays were performed in triplicate.
Xenograft Assay.
Trp53−/−; Myc cells (2 × 106) expressing shRNA lentiviruses were injected subcutaneously into the right flank of 6- to 8-wk-old female nude mice (National Cancer Institute-Frederick). Tumor volume was measured with calipers every 3–5 d until the average tumor mass reached 2 cm3. Tumor volume was calculated with the formula (length × width2)/2. A total of four mice were injected per shRNA tested. At least two independent tumorigenesis experiments were performed for each gene.
GFP Imaging.
Whole-body GFP imaging was performed using the IVIS Spectrum Optical Imaging system (Xenogen) according to the manufacturer’s instructions.
Microarray Analysis of Human HCC Samples.
The mRNA expression data of 12 genes in human HCC were obtained from Gene Expression Omnibus with accession nos. GSE4024 and GSE1898 (23). Hierarchical cluster analysis was conducted using Cluster 3.0, and the image analysis was performed with TreeView 1.60 software (Michael Eisen Laboratory, Lawrence Berkeley National Laboratory and University of California, Berkeley, CA; http://rana.lbl.gov/eisen/). We applied Kaplan–Meier plot analysis and a log rank test for overall survival in HCC by using Prism v. 5.03 (GraphPad Software, Inc.).
DEN Tumorigenesis Experiment.
Fifteen-day-old male Ncoa2+/+ and Ncoa2−/− littermate pups were injected intraperitoneally with a 25 mg/kg dose of DEN (Sigma) in 0.9% sterile saline as previously described (58). DEN-treated mice were fed normal chow (Teklad 2920×) and euthanized at 3 or 6 mo postinjection. Mice were weighed, and liver was isolated, weighed, and photographed macroscopically. Livers were evaluated further to determine the total number of tumors as well as average and maximal tumor size using a handheld micrometer. Samples of liver tissue were prepared for histological analysis as previously described (59).
Supplementary Material
Acknowledgments
We thank Dean Felsher for tet-o-MYC and LAPtTA mice; Scott Lowe for Trp53 −/−; Myc liver progenitor cells; Adam Dupuy, Nancy Jenkins, and Neal Copeland for Rosa26-SB11 mice; Eric Cooper for Gag/Pol, Rev, and VSV-G helper plasmids; and J. Mendell and C. Dang for critical reading of the manuscript. The Minnesota Supercomputing Institute provided system administration support and the computational research infrastructure used to perform the analysis. This work was supported by National Institutes of Health Grant CA16519 (to J.D.B.) and National Cancer Institute Grant R01CA113535 (to D.A.L.). K.A.O. was a Damon Runyon fellow supported by the Damon Runyon Cancer Research Foundation Grant DRG-1918-06 and currently is a Cancer Prevention Research Institute of Texas Scholar in Cancer Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences from this study have been submitted to the National Center for Biotechnology Information (NCBI) Short Read Archive, http://www.ncbi.nlm.nih.gov/sra (accession no. SRA051795.1).
See Author Summary on page 7966 (volume 109, number 21).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1115433109/-/DCSupplemental.
References
- 1.Ivics Z, Hackett PB, Plasterk RH, Izsvák Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 2.Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436:272–276. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
- 3.Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–226. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- 4.Collier LS, et al. Whole-body sleeping beauty mutagenesis can cause penetrant leukemia/lymphoma and rare high-grade glioma without associated embryonic lethality. Cancer Res. 2009;69:8429–8437. doi: 10.1158/0008-5472.CAN-09-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keng VW, et al. A conditional transposon-based insertional mutagenesis screen for genes associated with mouse hepatocellular carcinoma. Nat Biotechnol. 2009;27:264–274. doi: 10.1038/nbt.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Starr TK, et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science. 2009;323:1747–1750. doi: 10.1126/science.1163040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts LR. Sorafenib in liver cancer—just the beginning. N Engl J Med. 2008;359:420–422. doi: 10.1056/NEJMe0802241. [DOI] [PubMed] [Google Scholar]
- 8.Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339–346. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- 9.Buendia MA. Genetics of hepatocellular carcinoma. Semin Cancer Biol. 2000;10:185–200. doi: 10.1006/scbi.2000.0319. [DOI] [PubMed] [Google Scholar]
- 10.Beer S, et al. Developmental context determines latency of MYC-induced tumorigenesis. PLoS Biol. 2004;2:e332. doi: 10.1371/journal.pbio.0020332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell. 1999;4:199–207. doi: 10.1016/s1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- 12.Shachaf CM, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- 13.Zender L, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zender L, et al. Generation and analysis of genetically defined liver carcinomas derived from bipotential liver progenitors. Cold Spring Harb Symp Quant Biol. 2005;70:251–261. doi: 10.1101/sqb.2005.70.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zender L, et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. 2008;135:852–864. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keng VW, et al. Region-specific saturation germline mutagenesis in mice using the Sleeping Beauty transposon system. Nat Methods. 2005;2:763–769. doi: 10.1038/nmeth795. [DOI] [PubMed] [Google Scholar]
- 17.Carlson CM, et al. Transposon mutagenesis of the mouse germline. Genetics. 2003;165:243–256. doi: 10.1093/genetics/165.1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horie K, et al. Characterization of Sleeping Beauty transposition and its application to genetic screening in mice. Mol Cell Biol. 2003;23:9189–9207. doi: 10.1128/MCB.23.24.9189-9207.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vigdal TJ, Kaufman CD, Izsvák Z, Voytas DF, Ivics Z. Common physical properties of DNA affecting target site selection of sleeping beauty and other Tc1/mariner transposable elements. J Mol Biol. 2002;323:441–452. doi: 10.1016/s0022-2836(02)00991-9. [DOI] [PubMed] [Google Scholar]
- 20.García-Caballero T, et al. Increased expression of growth hormone and prolactin receptors in hepatocellular carcinomas. Endocrine. 2000;12:265–271. doi: 10.1385/ENDO:12:3:265. [DOI] [PubMed] [Google Scholar]
- 21.Chopra AR, et al. Absence of the SRC-2 coactivator results in a glycogenopathy resembling Von Gierke’s disease. Science. 2008;322:1395–1399. doi: 10.1126/science.1164847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeong JW, et al. The genomic analysis of the impact of steroid receptor coactivators ablation on hepatic metabolism. Mol Endocrinol. 2006;20:1138–1152. doi: 10.1210/me.2005-0407. [DOI] [PubMed] [Google Scholar]
- 23.Lee JS, et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med. 2006;12:410–416. doi: 10.1038/nm1377. [DOI] [PubMed] [Google Scholar]
- 24.Gehin M, et al. The function of TIF2/GRIP1 in mouse reproduction is distinct from those of SRC-1 and p/CIP. Mol Cell Biol. 2002;22:5923–5937. doi: 10.1128/MCB.22.16.5923-5937.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fausto N, Campbell JS. Mouse models of hepatocellular carcinoma. Semin Liver Dis. 2010;30:87–98. doi: 10.1055/s-0030-1247135. [DOI] [PubMed] [Google Scholar]
- 26.Maeda S, Kamata H, Luo JL, Leffert H, Karin M. IKKbeta couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell. 2005;121:977–990. doi: 10.1016/j.cell.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 27.Rad R, et al. PiggyBac transposon mutagenesis: A tool for cancer gene discovery in mice. Science. 2010;330:1104–1107. doi: 10.1126/science.1193004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Starr TK, et al. A Sleeping Beauty transposon-mediated screen identifies murine susceptibility genes for adenomatous polyposis coli (Apc)-dependent intestinal tumorigenesis. Proc Natl Acad Sci USA. 2011;108:5765–5770. doi: 10.1073/pnas.1018012108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loh NY, Nebenius-Oosthuizen D, Blake DJ, Smith AJ, Davies KE. Role of beta-dystrobrevin in nonmuscle dystrophin-associated protein complex-like complexes in kidney and liver. Mol Cell Biol. 2001;21:7442–7448. doi: 10.1128/MCB.21.21.7442-7448.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ceccarini M, et al. Association of dystrobrevin and regulatory subunit of protein kinase A: A new role for dystrobrevin as a scaffold for signaling proteins. J Mol Biol. 2007;371:1174–1187. doi: 10.1016/j.jmb.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 31.Yasuda H, Mizuno A, Tamaoki T, Morinaga T. ATBF1, a multiple-homeodomain zinc finger protein, selectively down-regulates AT-rich elements of the human alpha-fetoprotein gene. Mol Cell Biol. 1994;14:1395–1401. doi: 10.1128/mcb.14.2.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim CJ, et al. Down-regulation of ATBF1 is a major inactivating mechanism in hepatocellular carcinoma. Histopathology. 2008;52:552–559. doi: 10.1111/j.1365-2559.2008.02980.x. [DOI] [PubMed] [Google Scholar]
- 33.Galan-Caridad JM, et al. Zfx controls the self-renewal of embryonic and hematopoietic stem cells. Cell. 2007;129:345–357. doi: 10.1016/j.cell.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arenzana TL, Smith-Raska MR, Reizis B. Transcription factor Zfx controls BCR-induced proliferation and survival of B lymphocytes. Blood. 2009;113:5857–5867. doi: 10.1182/blood-2008-11-188888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu G, et al. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009;23:837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee HC, Kim M, Wands JR. Wnt/Frizzled signaling in hepatocellular carcinoma. Front Biosci. 2006;11:1901–1915. doi: 10.2741/1933. [DOI] [PubMed] [Google Scholar]
- 37.Merle P, et al. Oncogenic role of the frizzled-7/beta-catenin pathway in hepatocellular carcinoma. J Hepatol. 2005;43:854–862. doi: 10.1016/j.jhep.2005.05.018. [DOI] [PubMed] [Google Scholar]
- 38.Satoh S, et al. AXIN1 mutations in hepatocellular carcinomas, and growth suppression in cancer cells by virus-mediated transfer of AXIN1. Nat Genet. 2000;24:245–250. doi: 10.1038/73448. [DOI] [PubMed] [Google Scholar]
- 39.de la Roche M, Worm J, Bienz M. The function of BCL9 in Wnt/beta-catenin signaling and colorectal cancer cells. BMC Cancer. 2008;8:199. doi: 10.1186/1471-2407-8-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mani M, et al. BCL9 promotes tumor progression by conferring enhanced proliferative, metastatic, and angiogenic properties to cancer cells. Cancer Res. 2009;69:7577–7586. doi: 10.1158/0008-5472.CAN-09-0773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Busby V, et al. Alpha-T-catenin is expressed in human brain and interacts with the Wnt signaling pathway but is not responsible for linkage to chromosome 10 in Alzheimer’s disease. Neuromolecular Med. 2004;5:133–146. doi: 10.1385/NMM:5:2:133. [DOI] [PubMed] [Google Scholar]
- 42.Quinlan AR, et al. Genome-wide mapping and assembly of structural variant breakpoints in the mouse genome. Genome Res. 2010;20:623–635. doi: 10.1101/gr.102970.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinez-Sanz J, et al. Binding of human centrin 2 to the centrosomal protein hSfi1. FEBS J. 2006;273:4504–4515. doi: 10.1111/j.1742-4658.2006.05456.x. [DOI] [PubMed] [Google Scholar]
- 44.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 45.Soucek L, et al. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chou JY, Matern D, Mansfield BC, Chen YT. Type I glycogen storage diseases: Disorders of the glucose-6-phosphatase complex. Curr Mol Med. 2002;2:121–143. doi: 10.2174/1566524024605798. [DOI] [PubMed] [Google Scholar]
- 47.Mutel E, et al. Targeted deletion of liver glucose-6 phosphatase mimics glycogen storage disease type 1a including development of multiple adenomas. J Hepatol. 2011;54:529–537. doi: 10.1016/j.jhep.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 48.Tsubouchi H, Kamibeppu A, Fujisaki K, Nagahama J, Hashimoto S. Hepatic gluconeogenic key enzymes in patients with hepatic cancer. Gastroenterol Jpn. 1980;15:564–569. doi: 10.1007/BF02773759. [DOI] [PubMed] [Google Scholar]
- 49.Weber G, Cantero A. Glucose-6-phosphatase activity in normal, pre-cancerous, and neoplastic tissues. Cancer Res. 1955;15:105–108. [PubMed] [Google Scholar]
- 50.Lee PJ. Glycogen storage disease type I: Pathophysiology of liver adenomas. Eur J Pediatr. 2002;161(Suppl 1):S46–S49. doi: 10.1007/s00431-002-1002-0. [DOI] [PubMed] [Google Scholar]
- 51.Kim W, et al. Apoptosis-inducing antitumor efficacy of hexokinase II inhibitor in hepatocellular carcinoma. Mol Cancer Ther. 2007;6:2554–2562. doi: 10.1158/1535-7163.MCT-07-0115. [DOI] [PubMed] [Google Scholar]
- 52.Pelicano H, Martin DS, Xu RH, Huang P. Glycolysis inhibition for anticancer treatment. Oncogene. 2006;25:4633–4646. doi: 10.1038/sj.onc.1209597. [DOI] [PubMed] [Google Scholar]
- 53.Kim JW, et al. Evaluation of myc E-box phylogenetic footprints in glycolytic genes by chromatin immunoprecipitation assays. Mol Cell Biol. 2004;24:5923–5936. doi: 10.1128/MCB.24.13.5923-5936.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shim H, et al. c-Myc transactivation of LDH-A: Implications for tumor metabolism and growth. Proc Natl Acad Sci USA. 1997;94:6658–6663. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Osthus RC, et al. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem. 2000;275:21797–21800. doi: 10.1074/jbc.C000023200. [DOI] [PubMed] [Google Scholar]
- 56.Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 57.Stabenau A, et al. The Ensembl core software libraries. Genome Res. 2004;14:929–933. doi: 10.1101/gr.1857204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sakurai T, Maeda S, Chang L, Karin M. Loss of hepatic NF-kappa B activity enhances chemical hepatocarcinogenesis through sustained c-Jun N-terminal kinase 1 activation. Proc Natl Acad Sci USA. 2006;103:10544–10551. doi: 10.1073/pnas.0603499103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.York B, et al. Reprogramming the posttranslational code of SRC-3 confers a switch in mammalian systems biology. Proc Natl Acad Sci USA. 2010;107:11122–11127. doi: 10.1073/pnas.1005262107. [DOI] [PMC free article] [PubMed] [Google Scholar]